Highlights:
-
•
The comparison used 207 images from both research and clinical datasets.
-
•
When retrained, NicMSlesion, a convolutional network, was the most accurate.
-
•
Performance of this deep learning method severely dropped on clinical routine data.
-
•
On clinical routine data, regression and clustering methods were the top-ranked methods.
-
•
SLS was the most robust to artifacted images, and BIANCA to scanners variability.
Abbreviations: WMH, White Mater Hyperintensities; WM, White Matter; ADNI, Alzheimer's Disease Neuroimaging Initiative; DSC, Dice similarity coefficient
Keywords: White matter hyperintensity, Dementia, Artificial intelligence, Segmentation, Microvascular
Abstract
Background
Manual segmentation is currently the gold standard to assess white matter hyperintensities (WMH), but it is time consuming and subject to intra and inter-operator variability.
Purpose
To compare automatic methods to segment white matter hyperintensities (WMH) in the elderly in order to assist radiologist and researchers in selecting the most relevant method for application on clinical or research data.
Material and Methods
We studied a research dataset composed of 147 patients, including 97 patients from the Alzheimer's Disease Neuroimaging Initiative (ADNI) 2 database and 50 patients from ADNI 3 and a clinical routine dataset comprising 60 patients referred for cognitive impairment at the Pitié-Salpêtrière hospital (imaged using four different MRI machines). We used manual segmentation as the gold standard reference. Both manual and automatic segmentations were performed using FLAIR MRI. We compared seven freely available methods that produce segmentation mask and are usable by a radiologist without a strong knowledge of computer programming: LGA (Schmidt et al., 2012), LPA (Schmidt, 2017), BIANCA (Griffanti et al., 2016), UBO detector (Jiang et al., 2018), W2MHS (Ithapu et al., 2014), nicMSlesion (with and without retraining) (Valverde et al., 2019, Valverde et al., 2017). The primary outcome for assessing segmentation accuracy was the Dice similarity coefficient (DSC) between the manual and the automatic segmentation software. Secondary outcomes included five other metrics.
Results
A deep learning approach, NicMSlesion, retrained on data from the research dataset ADNI, performed best on this research dataset (DSC: 0.595) and its DSC was significantly higher than that of all others. However, it ranked fifth on the clinical routine dataset and its performance severely dropped on data with artifacts. On the clinical routine dataset, the three top-ranked methods were LPA, SLS and BIANCA. Their performance did not differ significantly but was significantly higher than that of other methods.
Conclusion
This work provides an objective comparison of methods for WMH segmentation. Results can be used by radiologists to select a tool.
1. Introduction
White matter hyperintensities (WMH) are signal abnormalities of white matter (WM) on T2-weighted (T2w) magnetic resonance imaging (MRI) sequences. They are commonly seen in the brain of elderly people. In such populations, the majority of these abnormalities are presumed to be of vascular origin. The STandards for Reporting Vascular changes on nEuroimaging (STRIVE) have provided recommendations to standardize their interpretations (Wardlaw et al., 2013). In clinical practice, visual rating scales are used to evaluate WMH linked to microvascular pathology, the most common being the Fazekas scale (Fazekas et al., 1987). However, it does not give a precise information about the spatial localization and volume of WMH. Manual segmentation is currently the gold standard to evaluate the volume of WMH, but it is time consuming and subject to intra and inter-operator variability (Commowick et al., 2018, Grimaud et al., 1996).
Automated segmentation of WMH is thus potentially very useful, as it would allow large scales analyses which could progress our understanding of the relationship between pathologies and localized WMH. In the clinics, automated segmentation can represent a gain of the radiologist time and may speed up the evaluation of the patient state. Many approaches (see (Caligiuri et al., 2015) for a review) have been proposed for automatic segmentation of WMH, mostly in the context of vascular abnormalities of the elderly and multiple sclerosis. Several of them are implemented in freely available software. However, currently, none of these approaches is recognized as a reference standard. Therefore, radiologists willing to use such tools have little information on performance or dos and don’ts. Caligiuri and colleagues reviewed the methods behind automatic WMH segmentation but did not compare their performance (Caligiuri et al., 2015). The MICCAI 2017 WMH Segmentation Challenge (https://wmh.isi.uu.nl/) (Kuijf et al., 2019) has evaluated 20 methods on a dataset of 170 images (60 for training and 110 for testing) from memory cohorts of three different institutes (UMC Utrecht, NUHS Singapore, VU Amsterdam). However, most of these techniques require preprocessing that is very specific to the dataset at hand, which is difficult to adapt to another dataset. R. Heinen and colleagues (Heinen et al., 2019) performed a comparison of five methods including LPA and LGA on a dataset of 60 patients, but did not evaluate some of the most recent tools (NicMSlesion, UBO, BIANCA) . None of the previous publications evaluated the performance on routine data, with artifacted images.
In this paper, we aimed to determine which are the best freely and user-friendly available software tools for segmenting WMH in the elderly. To that purpose, we benchmarked the performances of seven tools on a large subset of 137 images from the ADNI research dataset. In addition, we evaluated the performances of the tools in a clinical routine context on sixty patients, using off-the-shelf algorithms optimized on ADNI. We further evaluated the robustness of the algorithms in presence of artifacts or for data collected across multiple scanners.
2. Material and methods
2.1. Participants
We used two different datasets: a research dataset from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu) (Mueller et al., 2005) and a clinical routine dataset. For the research dataset, we randomly selected 97 participants from ADNI 2 and 50 participants from ADNI 3. We extracted a training subset by randomly selecting 20 patients from ADNI 2 and 20 patients from ADNI 3. More details about the ADNI are presented in Supplementary Text 1.
The clinical routine dataset was composed of 60 patients from the Pitié-Salpêtrière hospital. Specifically, we included the last 15 consecutive patients (at the date of May 15, 2019) who were referred for assessment of cognitive impairment on each of the four MRI machines currently in use in the Department of Neuroradiology. We excluded patients with stroke, tumor, hematoma, inflammatory and infectious pathology. For the clinical routine dataset, all clinical and biological data were generated during a routine clinical workup and were retrospectively extracted for the purpose of this study. Therefore, according to French legislation, explicit consent was waived.
The main characteristics of these three populations are summarized in Table 1 and detailed information is provided in Supplementary Tables 1 and 2.
Table 1.
Demographic information for the research dataset (from ADNI) and the clinical routine dataset. Continuous values are displayed as average with the min–max range within parentheses. For ADNI, we also display the characteristics of the training and testing datasets separately.
| ADNI |
ROUTINE |
|||
|---|---|---|---|---|
| All | Training | Testing | All | |
| N | 147 | 40 | 107 | 60 |
| Age (range) |
74 (58–90) |
74.7 (58–90) |
73.7 (59–90) |
78.2 (52–101) |
| Sex | 85 F/51 M |
19 F / 21 M |
66 F / 41 M |
30 F / 30 M |
2.2. MRI acquisition
In the research dataset (ADNI), all patients had a 3D T1-weighted (T1w) and a FLAIR sequence acquired at 3 T. FLAIR sequences were 2D in ADNI2 and 3D in ADNI 3. Acquisition protocols have been previously described (Jack et al., 2015, Jack et al., 2008) (http://adni.loni.usc.edu/methods/documents/mri-protocols/).
In the clinical routine dataset, all patients had a 3D T1-weighted sequence and a 3D FLAIR sequence (except for two patients who had a 2D FLAIR). The acquisitions were performed on four different MRI machines (GE Signa HDxt 3 T, Siemens Skyra 3 T, GE Optima MR450w 1.5 T, GE Signa PET/MR 3 T) and parameters were heterogeneous (see Supplementary Tables 3 and 4), thereby reflecting the reality of clinical routine.
We assessed visually the presence of artifacts on T1 and FLAIR images. Specifically, a participant was assigned to the “artifact group” if either the T1 or the FLAIR image had artifacts that could limit the interpretation. This assessment was made independently by two radiologists (QV, EX) and both readers agreed on all cases. Ten participants were assigned to the “artifact group”. Three participants had artifacts on the T1w image, two had artifacts on the FLAIR image while for the remaining five participants, both images were artifacted. One example of an artifacted image is shown in Supplementary Fig. 1 and the characteristics of those participants are reported in Supplementary Table 2.
Table 2.
Intra- and inter-rater reproducibility assessed on the training dataset from ADNI (comprising 40 patients).
| DSC | Volume similarity | Intraclass correlation | Volume error rate | False positive rate | False negative rate | |
|---|---|---|---|---|---|---|
| Intra-operator reproducibility | 0.744 (0.723–0.766) |
0.899 (0.875–0.922) |
0.987 (0.971–0.994) |
0.185 (0.145–0.226) |
0.196 (0.164–0.228) |
0.292 (0.259–0.325) |
| First segmentation first operator vs second operator | 0.723 (0.699–0.747) |
0.884 (0.856–0.914) |
0.984 (0.962–0.992) |
0.277 (0.199–0.355) |
0.324 (0.286–0.362) |
0.199 (0.168–0.231) |
| Second segmentation first operator vs second operator | 0.701 (0.674–0.729) |
0.844 (0.815–0.871) |
0.974 (0.951–0.986) |
0.310 (0.256–0.364) |
0.262 (0.216–0.307) |
0.290 (0.238–0.341) |
DSC: Dice similarity coefficient. For each metric, the table displays the average and the 95% confidence interval within parentheses.
2.3. Automatic segmentation tools
We selected segmentation methods by reviewing the literature from 2012 to November 2018, from which we identified 33 different methods. A summary of our literature review can be found in Supplementary Table 5. For inclusion in the comparison, methods needed to be freely available and to produce the segmentation mask as output (and not only the volume), which reduced the list to fourteen methods. Moreover, we included only user-friendly methods, which had to be usable by a radiologist without a strong knowledge of computer programming. Thus, we removed methods for which the user needed to perform specific image preprocessing. This left seven methods that we considered in our study.
Table 5.
Summary of evaluation, and some selected information to choose a method.
| Ranking on research data | Ranking on routine data | Robustness artifacts | Robustness different scanner | Sequences needed | Need training data | Limitations/ Requirements | Proc. time | |
|---|---|---|---|---|---|---|---|---|
| LPA | 2 | 1* | – | – | FLAIR | No | Matlab | 1 min |
| LGA | 4 | 5 | – | FLAIR/ T1w | No | Matlab | 6 min | |
| BIANCA | 4 | 1 | – | FLAIR/ T1w | Yes | Need mask of WM | 17 min1 | |
| SLS | 2 | 1 | – | FLAIR/ T1w | No | Matlab | 8 min | |
| W2MHS | 8 | 8 | – | FLAIR/ T1w | No | Matlab | 5 min | |
| nicMSlesion (original) | 4 | 7 | – | FLAIR/ T1w | No | GPU | 10 min2,3 | |
| nicMSlesion (retrained) | 1* | 5 | – | – | FLAIR/ T1w | Yes | GPU | 10 min2,3 (23.5 h2,4) |
| UBO | 4 | 4 | – | – | FLAIR/ T1w | No | Matlab | 9 min |
Ranking performed using t-test comparison on the primary criterion (DSC) (see Supplementary Tables 7 and 9 for details). We started by looking at the method with the best DSC. Then all methods not significantly different from it were given the same rank classified, and so on.
Processing time were evaluated on MacBook Pro laptop with a 2.2 GHz Intel Core i7 2018 CPU, without a graphic processing unit (GPU), with 16 Go RAM except for the nicMSlesion for which we used a GPU-equipped computer, namely a Linux workstation with an Intel Xeon E5-2699 @ 2.30 GHz CPU, with NVIDIA Quadro M4000 GPU, 256 Go RAM.
– indicates that the DSC is sensitive to artifacts or scanner type at p < 0.05 uncorrected for multiple comparisons, on routine dataset.
-- indicates that the DSC is sensitive to artifacts or scanner type at after correction for multiple testing, on routine dataset.
Best DSC in our evaluation (though not necessarily significantly better which explains equal first).
2 min for segmentation and 15 min for generation of the exclusion mask.
With graphic processing unit (GPU, NVIDIA Quadro M4000).
3.5 min for segmentation and 6.5 min for preprocessing
Retraining time.
We included :1) the lesion growth algorithm (LGA) (Schmidt et al., 2012) from the lesion segmentation toolbox (LST) (www.statisticalmodelling.de/lst.html), included in SPM12 and based on probabilistic modeling and a region growing algorithm; 2) the lesion prediction algorithm (LPA) (Schmidt, 2017) (Schmidt, 2017, Chapter 6.1), also from the SPM LST toolbox and based on logistic regression; 3) the Brain Intensity AbNormality Classification Algorithm (BIANCA) (Griffanti et al., 2016) included in FSL, based on the K nearest neighbors (K-nn) algorithm; 4) the UBO detector (Jiang et al., 2018), also based on K-nn; 5) the Wisconsin White Matter Hyperintensities Segmentation Toolbox (W2MHS) (Ithapu et al., 2014) a method based on the random forest algorithm; 6) the multiple Sclerosis Lesion Segmentation toolbox (Roura et al., 2015) (SLS) based on thresholding of a WM segmentation map; 7) the nicMSlesion toolbox based on a cascade of two 3D patch-wise convolutional neural networks (Valverde et al., 2019, Valverde et al., 2017). For nicMSlesion, we used two different models: the original model (trained by the authors of the original publication on a multiple sclerosis dataset and directly available), a retrained model for which we retrained the last three fully connected layers using our training dataset. To note, BIANCA specifically requires a training set, to create a set of feature vectors for lesion and non-lesion classes, for which we also used the ADNI training subset. Computation times for the different methods are reported in Supplementary Text 2.
2.4. Determination of hyper-parameters
Some methods have hyper-parameters that can be adjusted. We determined the optimal value of the parameters which maximized the DSC on the ADNI training subset of 40 patients. We performed a “grid-search”, i.e. testing several possible combination of parameters using fixed intervals within the range of possible hyperparameters. We determined the optimal parameters for all methods, except for SLS for which there is no adjustable parameter. For all other methods, which return a continuous prediction, we estimated the optimal probability threshold used to define WMH. Other hyper parameters included the number of K-nn neighbours (UBO), a threshold on the result of the registration of the segmentation mask to FLAIR space (nicMSlesion, LGA), a threshold of the WM mask registered to FLAIR space (BIANCA), a “cleaning threshold” which removes hyperintensities that are closer than a given distance from the grey matter (W2MHS). More details regarding the parameters and their optimization are provided in Supplementary Text 3.
2.5. Manual segmentation
The reference standard was built using manual segmentation of WMH by a radiology resident trained rater (QV). WMH masks were manually segmented from the FLAIR sequence using the ITK SNAP editing tool (Yushkevich et al., 2006). A segmentation protocol was designed from the advice of two experienced neuroradiologists (SS and DD). It included the following rules: 1) exclusion of hyperintense lines adjacent to the ventricles that are one voxel thick; 2) exclusion of WMH in the septum pellucidum, at the junction of the genu of the corpus callosum and the septum pellucidum and at the junction of the splenium of the corpus callosum and the ventricles.
We evaluated inter and intra-rater reproducibility on the ADNI training subset of 40 patients. For inter-rater agreement, images were segmented by two raters (QV and EX). For intra-rater, QV segmented the images twice, with a minimum interval of 4 weeks between the two evaluations.
2.6. Statistical analysis
The primary outcome for assessing segmentation accuracy was the Dice similarity coefficient (DSC) calculated as :
We used paired t-tests to compare DSC between methods, or between clinical images with and without artifact. In post-hoc analyses, we further adjusted for WMH volume and site/scanner, we also stratified the analysis by high/low WMH volumes (cut off at 10,000 mm3, which correspond to Fazekas score<3 (Hernández et al., 2013) commonly used in clinical practice). On the clinical routine dataset, we estimated the proportion of DICE variance attributable to scanner variability (partial eta-square effect sizes) and tested its significance using ANOVAs. The significance level was corrected for multiple comparisons using Bonferroni correction.
Secondary outcomes included the following metrics:
-
-
Volume Similarity (Taha and Hanbury, 2015):
-
-
Absolute volume error rate :
-
-
Voxel-level false positive ratio :
-
-
Voxel-level false negative ratio : .
-
-
Intra-class correlation coefficient between volumes using a two-way model with absolute agreement definition and single rater (Koo and Li, 2016)(Shrout and Fleiss, 1979)
3. Results
3.1. Reproducibility of manual segmentation
Table 2 displays the intra and inter-observer agreement on 40 patients from the training ADNI dataset. In all cases, the DSC indicated a substantial, nonetheless imperfect agreement of WMH maps, Though the intraclass correlation between volumes indicated an excellent agreement of WMH volumes (Koo and Li, 2016).
3.2. WMH volume distribution
The average WMH volume was 9.6 ml (SD 14.3, median 3.8) on the ADNI training dataset and 8.0 ml (SD 11.9, median 4.2) on ADNI testing dataset. WMH distributions were highly skewed to the right and with many outliers (Supplementary Fig. 2) which led us test to use the Mann-Whitney-Wilcoxon test to test for difference between groups. WMH volumes were not significantly different between the ADNI training and testing dataset (p = 0.5, Mann-Whitney-Wilcoxon test). The average WMH volume was 16.2 ml (SD 24.8, median 8.1) on the clinical routine dataset, which was significantly higher than in the ADNI training dataset (p = 0.001, Mann-Whitney-Wilcoxon test).
3.3. Determination of hyper-parameters
Initially, we aimed to determine optimal parameters separately for 2D (from ADNI2) and 3D data (from ADNI3). However, we found that the optimal parameter were very similar for 2D and 3D data (Supplementary Fig. 3) and yielded comparable DSC. Thus, we used a single optimal value based on the merged dataset of 2D and 3D training dataset and did not separate 2D and 3D scans in the rest of the analysis. We report the distribution of absolute volume error rate and intraclass correlation according to the different parameter values on Supplementary Fig. 4 (LPA), 5 (DSC), 6 (Volume error rate) and 7 (Intraclass correlation).
We report the best DSC results and the optimal parameters for each method in Supplementary Table 6. We used these parameters for our evaluations in both research and clinical routine datasets.
3.4. Performance on the research dataset (ADNI)
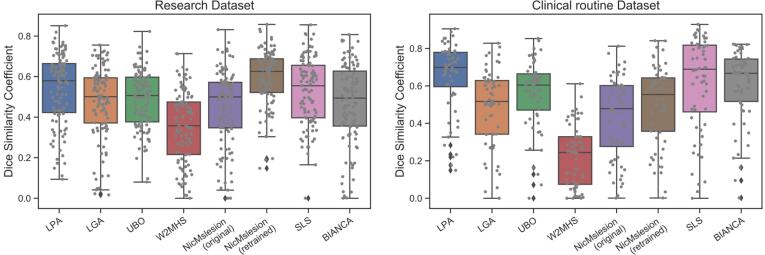
The performance of retrained nicMSlesion was significantly better than that of algorithm LPA that came second (DSC of 0.595 vs. 0.535 Fig. 1, Table 3; p < 0.001 Supplementary Table 7). nicMSlesion also achieved the best performance according to secondary outcomes except for the false negative ratio, where it was superseded by LPA, UBO and SLS (Supplementary Figure 8, Table 3). The DSC difference between LPA and SLS (that came third) did not reach significance, however they performed better than all other algorithms (p < 0.001). Adjusting for site and WMH volume load did change the ranking, though the superiority of retrained nicMSlesion over LPA (1st vs. 2nd) and SLS (3rd) over LGA, nicMSlesion (original) and UBO could not be deemed significant anymore (Supplementary Table 7).
Fig. 1.
DSC performance of the different automatic segmentation methods. Left : ADNI research dataset Right : clinical routine dataset. The boxplots show the median and the 25% and 75% percentiles of the metrics distribution. Values outside the whiskers indicate outliers. Gray dots show the value for individual participants. .
Table 3.
Performance of the different automatic segmentation methods on the research dataset ADNI.
| ADNI | DSC | Volume similarity | Volume error rate | Intraclass correlation | False positive rate | False negative rate |
|---|---|---|---|---|---|---|
| LPA | 0.539 (0.505–0.573) |
0.734 (0.691–0.775) |
0.850 (0.570–1.131) |
0.812 (0.709–0.876) |
0.438 (0.399–0.477) |
0.366 (0.321–0.410) |
| LGA | 0.474 (0.441–0.509) |
0.759 (0.719–0.798) |
0.426 (0.361–0.490) |
0.680 (0.561–0.770) |
0.444 (0.408–0.480) |
0.535 (0.494–0.574) |
| BIANCA | 0.469 (0.430–0.506) |
0.638 (0.588–0.686) |
0.760 (0.609–0.912) |
0.417 (0.249–0.560) |
0.393 (0.349–0.436) |
0.481 (0.428–0.533) |
| SLS | 0.527 (0.495–0.559) |
0.732 (0.696–0.766) |
0.903 (0.729–1.078) |
0.890 (0.507–0.957) |
0.564 (0.531–0.596) |
0.277 (0.239–0.314) |
| W2MHS | 0.351 (0.318–0.385) |
0.603 (0.551–0.654) |
2.219 (1.139–3.299) |
0.292 (0.108–0.456) |
0.539 (0.482–0.594) |
0.569 (0.529–0.608) |
| nicMSlesion(original) | 0.454 (0.419–0.490) |
0.787 (0.746–0.826) |
0.694 (0.382–1.007) |
0.948 (0.924 –0.964) |
0.517 (0.476–0.557) |
0.503 (0.463–0.543) |
| nicMSlesion(retrained) |
0.595 (0.357–0.921) |
0.889 (0.867–0.910) |
0.270 (0.159–0.381) |
0.979 (0.968–0.986) |
0.384 (0.351–0.416) |
0.402 (0.376–0.427) |
| UBO | 0.486 (0.459–0.514) |
0.762 (0.730–0.793) |
0.907 (0.575–1.239) |
0.881 (0.652–0.945) |
0.587 (0.559–0.615) |
0.360 (0.328–0.392) |
For each metric, we present the average and the 95% confidence interval within parentheses. DSC: Dice similarity coefficient. Results in bold indicates the best score for each metric
To complement our analysis controlling for WMH volume load, we studied the performance separately for patients with low (<10,000 mm3, which correspond to Fazekas score<3 (Hernández et al., 2013)) and high WMH volume load. Again, the retrained nicMSlesion performed best for both low and high volume load groups followed by SLS and LPA. (Supplementary Table 8).
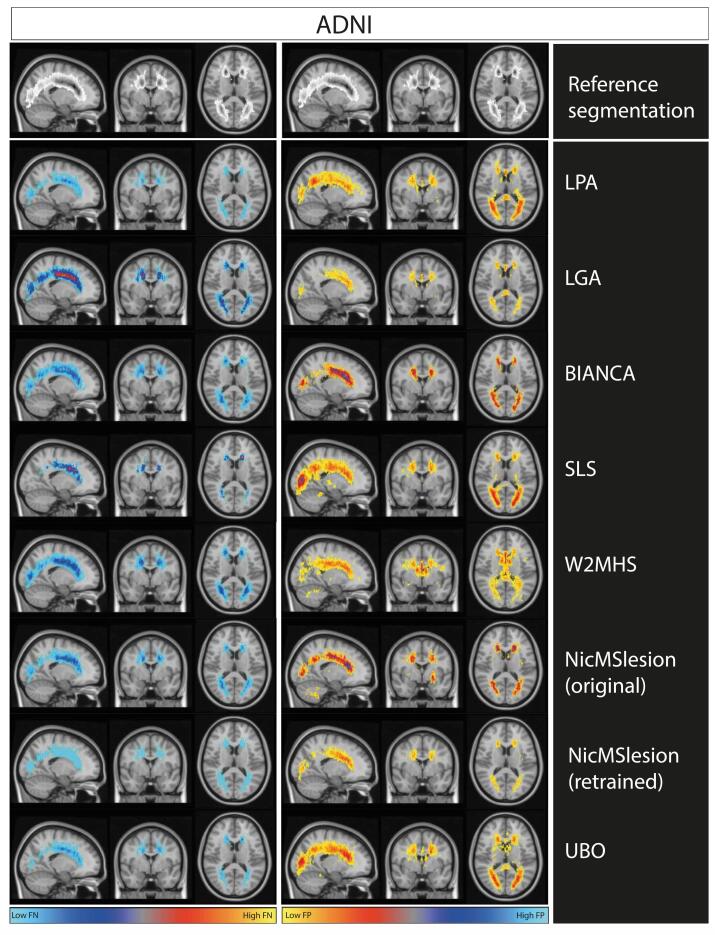
In order to appreciate the spatial distribution of errors, we constructed maps of false positive and false negative rate for each algorithm (Fig. 2). A comparison of manual and automatic segmentation for a single individual is shown in Supplementary figure 9. Overall, we note that the errors remain localized around the true WMH location. We observe that retrained nicMSlesion had a low level of false negative and that all errors remained around true WMH location. We note that retraining nicMSlesion reduced massively the frequency of errors and avoided large errors in unusual locations (e.g. cerebellum). LPA and SLS good DSC performance came from a relatively low false negative rate (Fig. 2, Table 3). However, LPA and SLS resulted in false positives located mainly in posterior regions.
Fig. 2.
Maps of False negative and False positive rate from each method on the ADNI research dataset. We represent masks of segmentation on MNI template. The first row of the plot represents an overlay of manual segmentation in the ADNI testing set. The greyscale ranges from 0%(white) to 33% (black) of WMH at any particular voxel. The left column of the plot represents the false negative rate map for each method in ADNI testing set. The right column shows the false positive rate map for each method on ADNI dataset. Scale ranges from 0 to 33% of errors at each voxel, which corresponds to the maximal error rates observed.
3.5. Performance on the clinical routine dataset
On the clinical routine dataset, LPA ranked first on the primary outcome (DSC of 0.652), followed by SLS (0.613) and BIANCA (0.607; Table 4 and Fig. 1), though the differences were not statistically significant (Supplementary Table 9). However, LPA significantly superseded all other methods (p < 0.001) (Supplementary Table 9). Of note, the size of the clinical sample was smaller than the ADNI sample, leading to reduced power in detecting significant differences. As for secondary outcomes, LPA performed best on average for absolute volume error rate and false negative ratio, while SLS minimized the false positive ratio, retrained nicMSlesion maximized the intraclass correlation between WMH volumes, and UBO showed the maximal volume similarity (Table 4, Supplementary Figure 10).
Table 4.
Performance of the different automatic segmentation methods on the clinical routine dataset.
| Routine | DSC | Volume similarity | Volume error rate | Intraclass correlation | False positive rate | False negative rate |
|---|---|---|---|---|---|---|
| LPA |
0.652 (0.604–0.701) |
0.790 (0.733–0.846) |
1.011 (0.469–1.552) |
0.727 (0.546–0.836) |
0.402 (0.346–0.459) |
0.189 (0.149–0.229) |
| LGA | 0.490 (0.437–0.543) |
0.729 (0.664–0.794) |
2.533 (0.615–4.451) |
0.287 (0.050–0.497) |
0.560 (0.502–0.618) |
0.354 (0.305–0.403) |
| BIANCA | 0.607v(0.556–0.657) | 0.788 (0.733–0.843) |
0.709 (0.431–0.987) |
0.859 (0.774–0.913) |
0.404 (0.346–0.463) |
0.296 (0.247–0.344) |
| SLS | 0.613 (0.546–0.679) |
0.738 (0.676–0.801) |
0.515 (0.367–0.662) |
0.815 (0.708–0.885) |
0.289 (0.231–0.346) |
0.368 (0.288–0.448) |
| W2MHS | 0.223 (0.181–0.266) |
0.448 (0.382–0.515) |
0.682 (0.621–0.743) |
0.510 (0.157–0.719) |
0.461 (0.377–0.546) |
0.844 (0.812–0.877) |
| nicMSlesion(original) | 0.433 (0.377–0.489) |
0.647 (0.571–0.723) |
4.498 (0.667–8.33) |
0.396 (0.109–0.61) |
0.616 (0.558–0.674) |
0.351 (0.295–0.407) |
| nicMSlesion(retrained) | 0.500 (0.446–0.555) |
0.781 (0.722–0.841) |
1.349 (0.221–2.477) |
0.922 (0.868–0.954) |
0.505 (0.439–0.571) |
0.433 (0.39–0.476) |
| UBO | 0.560 (0.512–0.608) |
0.836 (0.789–0.882) |
0.569 (0.211–0.926) |
0.734 (0.584–0.834) |
0.471 (0.422–0.52) |
0.353 (0.301–0.405) |
For each metric, the table displays the average and the 95 % confidence interval within parentheses. DSC: Dice similarity coefficient. Results in bold indicates the best score for each metric.
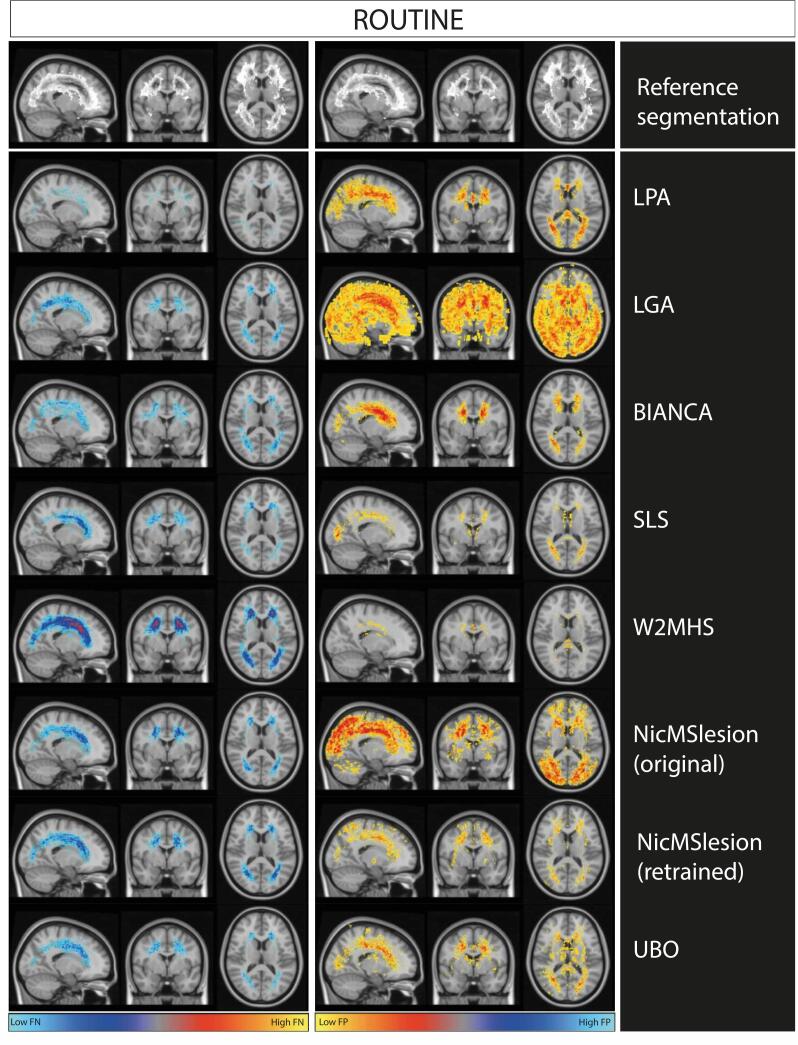
Fig. 3 shows that LPA resulted in few false negatives but many false positives (consistent with Table 4), similar to what we observed in the ADNI research dataset (Fig. 2). It was the opposite for SLS (limited false positives, Fig. 3), which contrasted with results of the research dataset. Original model of NicMSlesion and LGA had a lot of false positive segmentations in particularly in the parietal and occipital cortex. Those were not entirely removed by retraining NicMSlesion (with ADNI data). Widespread false positives could explain the drop of performance observed on the clinical routine dataset. W2MHS showed extreme false negative rate, thus misses a lot of WMH (consistent with Table 4)
Fig. 3.
Maps of False negative and False positive rate from each method on the clinical routine dataset. We represent masks of segmentation on MNI template. The first row of the plot represents an overlay of manual segmentation in the ADNI testing set. The greyscale ranges from 0%(white) to 33% (black) of WMH at any particular voxel. The left column of the plot represents the false negative rate map for each method in ADNI testing set. The right column shows the false positive rate map for each method on ADNI dataset. Scale ranges from 0 to 33% of errors at each voxel, which corresponds to the maximal error rates observed.
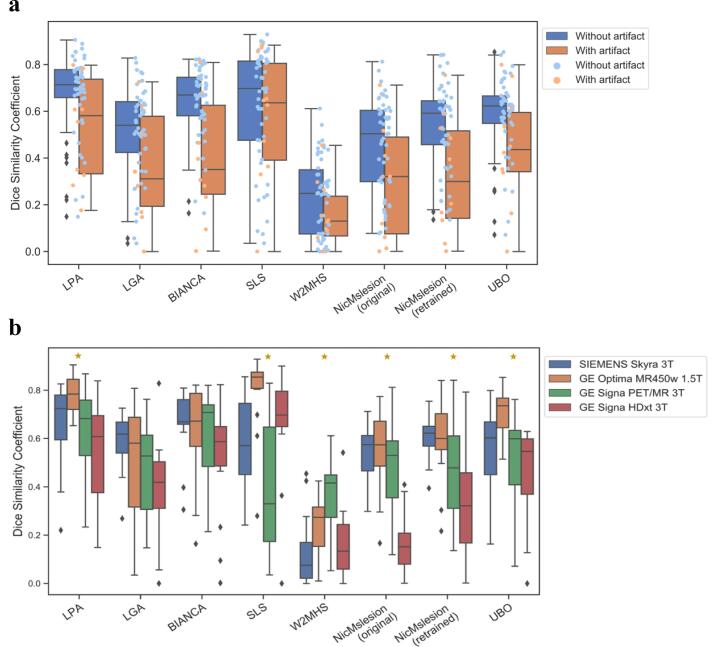
Breaking down the sample into high and low volume load did not affect the conclusions (Supplementary Table 10). However, when comparing images with and without artifacts, we found that the DSC performance of BIANCA and nicMSlesion significantly dropped in the artifact group (Supplementary Table 11). On images with artifacts, SLS performed best for the primary criterion (Fig. 4.a, Supplementary Table 12). Due to low number of the images with artifacts (10), it was not possible to test whether this superior performance was statistically significant.
Fig. 4.
Boxplots of DSC performance across Artifact and Scanner subgroups. a. DSC distributions with and without artifact. The box shows the median and the 25% and 75% percentiles. The whiskers indicate the distribution in function of the inter-quartile range. Orange boxplot and dots show data without strong artifact. Blue boxplot and dots show results with artifact. N artifact image = 10 and N without artifact = 50. b. DSC distributions for the different MRI scanners. The box shows the median and the 25% and 75% percentiles. The whiskers indicate the distribution in function of the inter-quartile range. Outliers are presented as black rhombus. Yellow Stars indicates a significant effect of scanner type on DSC variance. N = 15 per scanner. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In addition, we found a significant effect of scanner type on the performance of most methods (Supplementary Table 13, Fig. 4.b). In particular, NicMSlesion (original) was the most sensitive to having different scanners, with about 50% of the DICE variance being attributable to scanner types. The performances of LPA, SLS, W2MHS, NicMSlesion retrained, and UPO were also significantly associated with scanner types (Supplementary Table 13, Fig. 4.b), though this only explained 21–32% of the variability in performance. In contrast, LGA and BIANCA seemed more robust to the different scanners used to collect brain MRIs (Partial eta-square effect sizes of 8 and 11%, non-significantly different from 0).
4. Discussion
We compared seven tools for automatic WMH segmentation to determine which is the most efficient. All tools are freely available and usable by a radiologist without advanced knowledge in computer programming. Our evaluation used both a research dataset (ADNI) and a routine practice dataset of patients with cognitive impairment.
On the research dataset, nicMSlesion, a cascade of convolutional networks (with a specific re-training on a subset of the sample) achieved the highest performance on the primary criterion (DSC). However, its performance did not generalize well on clinical routine images and in particular on data with strong artifacts (Table 3, Fig. 1, Fig. 3.a). One important lesson from our study is that complex models (such as neural network) may be the most accurate when trained on data similar to the data used for testing but they do not generalize well. Valverde and colleagues (Valverde et al., 2019), already demonstrated this for NicMSlesion, on two different multiple sclerosis datasets, that one obtains lower performances when using a model trained on a dataset that is too different from the test set.
On the clinical routine dataset, LPA, SLS and BIANCA exhibited the highest DSC and their performances were not significantly different (respectively 0.65, 0.61,0.61). To note, LPA and SLS ranked second in term of DSC performance on the ADNI sample (0.54, 0.53), which suggests they generalize well to clinical samples even after we optimized their hyper-parameters on a subset of the ADNI sample. However, LPA performance drop on images with artifacts and results dependence of WMH and scanner type was not statistically significant. SLS appears very robust to artifacts (achieving the highest DSC on the artifacted dataset) but not to the heterogeneity of scanner (Fig. 3, Supplementary Table 13. To BIANCA, Additional to his top result on clinical routine dataset, BIANCA was the most robust to the scanner heterogeneity, however his performance significantly drop on artifacted images and it was not a top ranked method in the research dataset. To note, we demonstrate that BIANCA had no performance drop on routine dataset even though training data came from the research dataset.
Overall, our results demonstrate that several tools achieved acceptable performances on both research and clinical datasets. A reasonable first choice of WMH segmentation tool can be either LPA or SLS, even though one drawback is that they require a Matlab license (Table 5). Based on our results when the image dataset could contain patients with artifacts, SLS may be the method of choice. (Fig. 3 a, Table 5). On the other hand, in a dataset with many different scanners, BIANCA may be preferred because of its robustness (Fig. 3 b, Table 5). As for the neural network nicMSlesion, it seems to be performant only when retrained on data that is similar to the data to be segmented (Fig. 1, Table 5).
Overall, we reported lower performances than in the majority of previously published papers (Griffanti et al., 2016, Jiang et al., 2018, Kuijf et al., 2019). However, Rachmadi and colleagues obtained results similar to ours when evaluating LST LGA on the ADNI data (Rachmadi et al., 2018). In the same way, Heinen and coworkers had also similar result on their evaluation of LST LGA and LPA (Heinen et al., 2019). One of the reasons could be the lower volume of WMH in our two datasets (and especially in ADNI). However, Jiang and colleagues (Jiang et al., 2018), in the original publication describing the UBO software, also reported higher performance for low WMH volume. Another possible explanation is that the methods and performance reported may not generalize that well and may be somewhat optimistic or simply not comparable between publications or with our results that used different training and test samples. We also benchmarked prediction accuracy on the same research and clinical samples. Finally, we cannot rule out that the lower performances we report are attributable to differences in manual segmentation protocols between our study and previous ones.
One should note that the performances we report of most tools is moderate (DSC between 0.4 and 0.6 in most cases, Table 3, Table 4, Figure 11) and always below intra and inter-rater reproducibility (Table 2). This suggests more work is needed to improve performance of automated algorithms for WMH parcellation, which may include considering other models (Supplementary Table 5) or larger training samples. We provided maps of false positive and false negative rates for each of the methods (Fig. 2, Fig. 3), which may represent a useful feedback for method developers. In short, we note that on the ADNI dataset, errors were really close to the WMH identified by the radiographers which suggests some of the errors are on the boundaries of WMH regions. To improve performance of their methods, apart from the fact that the medical definition of WMH could be better homogenized, they could use a mask to eliminate some regions where WMH is impossible or presumed to be of vascular origin (e.g. septum pellucidum, cortex or cerebellum white matter). Secondly, they should find ways to better standardize white matter intensity, either using extensive training datasets or using the intensity of the cortex for instance. In clinical use, it might be important to progress preprocessing to reduce artifact rate.
Importantly, we should not necessarily discard those methods because of low DSC, as DSC may be overly sensitive to limited WMH parcellation errors. The boundaries of WMH regions are always being debated. Thus, we reported several metrics throughout the manuscript that may advise on which method is best for different applications. For example, several algorithms achieved good performance on WMH volume evaluation [Table 3, Table 4], which is the main criterion used in clinical assessment. For example, one could choose to use UBO, given that it obtained good volumetric results and that it directly provides additional features, such as segmenting WMH by vascular territories or anatomical regions. In addition, one could select the method that minimizes the false negative rate (SLS or LPA) as a way of reducing the search space for manual segmentation.
Many teams develop automatic WMH segmentation methods, mainly for multiple sclerosis or microvascular pathology associated with aging and cognitive impairment. However, few have compared the performance of these different methods. Many challenges are comparing automatic WMH segmentation methods, both for multiple sclerosis (MS segmentation challenge MICCAI 2008 (http://www.ia.unc.edu/MSseg/), ISBI 2015 longitudinal multiple sclerosis lesion segmentation challenge (Carass et al., 2017) (http://iacl.ece.jhu.edu/index.php/MSChallenge), MSSEG MICCAI Challenge 2016 (Commowick et al., 2018) (https://portal.fli-iam.irisa.fr/msseg-challenge)) and age-related WMH (MICCAI 2017 WMH Challenge (Kuijf et al., 2019)). Such challenges are very useful to assess which are the most efficient methodological approaches. But most of the participants to these challenges do not provide easily usable codes implementing their tools. Thereby, while these challenges are very useful to the methodological community of researchers developing new algorithms, they are of less use to a radiologist who would like to choose an easy-to-use tool.
To our knowledge, this is the first study to compare software while including data with artifacts, which reflects the reality of clinical routine. Indeed, artifacts are common, in particular during MRI acquisitions of patients with cognitive impairment. For all methods, there was a performance drop on data with artifacts (Fig. 3a). Such reduction in performance was significant for the deep neural network and BIANCA, losing over 0.2 point of DSC. On the other hand, SLS performs best on these data and the performance drop between data without and with artifacts was only 0.05 points of DSC.
Beyond sheer performances of the algorithms, we also evaluated how robust their performances were when using several MRI scanners, which is one of the principal factor of heterogeneity in MRI intensity. We found that the performance of most algorithms was sensitive to scanner types, though, LGA and BIANCA appeared the most robust. Our results align and extent those of a recent publication which suggested a possible scanner effect on algorithms performance on a sample of 42 participants from 7 different scanners (Heinen et al., 2019). Robustness may be an important criterion for algorithm selection in multi-centric studies, and in particular when the proportion of cases and controls varies between sites/scanners, thus when site/scanner may confound WMH association analyses because. More work is needed to further study if training on each/several scanner type could improve the performance of algorithms sensitive to scanner type. We evaluated heterogeneity related to the type of scanner, although it is a major factor of heterogeneity, it is not the only one. Thus, many factors, such as TE, TR, matrix size, etc., can influence image quality and contrast, and warrant further investigation.
Our study has the following limitations. First, the imperfect reproducibility of manual parcellation (at a vertex level, Table 2) calls for a more precise definition of the WMH and their boundaries, even though it was similar to that reported in the literature (Commowick et al., 2018, Coupé et al., 2018, Kuijf et al., 2019). When we visually inspected our different manual segmentation, we found that most of the differences to be at the limit of the WMH regions with an unclear intensity gradient. There is no precise standardized recommendation about the periventricular hyperintensities, whether they should be considered microvascular pathology or not. To overcome this limitation, we designed a protocol with experienced neuroradiologists. We discussed the pathogenesis of some hyperintensities, such as one-pixel hyperintensities close to the ventricles or in touch with the genu of the corpus callosum. Whether these very thin WMH represent microvascular pathology remains debated in the field. Kim et al. demonstrated that non-ischemic WMH are often located in juxtaventricular areas because they likely result from cerebrospinal fluid leakage (Kim et al., 2008) while Hernandez et al. came to more mixed conclusions (Hernández et al., 2014). Overall, the segmentation accuracy remains substantially lower than what is reported for other medical image segmentation tasks, such as subcortical grey matter structures (Pagnozzi et al., 2019) or brain tumors (Wadhwa et al., 2019). This illustrates that WMH segmentation remains a challenging task. Secondly, having only one reader for the test set may also be seen as a limitation. Nevertheless, considering the limited inter- and intra-observer reproducibility that we report it may have led to an overly conservative definition of WMH. Lastly, another limitation is to have limited ourselves to a set of methods, which are directly usable and allowing access to the segmentation mask. For example, this led us to exclude the tool which ranked first in the MICCAI 2017 WMH Segmentation Challenge (Li et al., 2018) because it was not directly usable. It uses a U-net convolutional neural network (Ronneberger et al., 2015) architecture. Notably, Duong and colleagues (Duong et al., 2019) and four of the top ten challengers of this challenge used this architecture, but they have not released a user-friendly version. We also did not include the promising LesionBRAIN (Coupé et al., 2018) (Supplementary Table 4) because it does not provide the required segmentation mask. More work is needed to make new software available and user-friendly to the community. In addition, future work could focus on the integration in the clinical workflow, allowing for example, to use these tools directly with DICOM. We have released all hyper-parameters used in our analysis, which should facilitate replication of our results and reuse of the algorithms we considered here.
In conclusion, we compared seven automatic WMH segmentation algorithms on both research and clinical routine data. These results can provide useful information to researchers and radiologists looking to choose an automatic WMH segmentation method.
Source of funding
The research leading to these results has received funding from the program “Investissements d’avenir” reference ANR-19-P3IA-0001 (PRAIRIE 3IA Institute) and reference ANR-10-IAIHU-06 (Agence Nationale de la Recherche-10-IA Institut Hospitalo-Universitaire-6), from the European Union H2020 program (project EuroPOND, grant number 666992, from the joint NSF/NIH/ANR program “Collaborative Research in Computational Neuroscience” (project HIPLAY7, grant number ANR-16-NEUC-0001-01), from Agence Nationale de la Recherche (project PREVDEMALS, grant number ANR-14-CE15-0016-07), from Fondation pour la Recherche sur Alzheimer (project HistoMRI), from the Abeona Foundation (project Brain@Scale), from the Fondation Vaincre Alzheimer (grant number FR-18006CB), and from the “Contrat d’Interface Local” program (to Dr Colliot) from Assistance Publique-Hôpitaux de Paris (AP-HP). Q. Vanderbecq is supported by a “Bourse de Recherche Alain Rahmouni” from the Société Française de Radiologie-Collège des Enseignants de Radiologie de France (SFR-CERF). BCD is supported by the NHMRC (CJ Martin Fellowship, APP1161356).
CRediT authorship contribution statement
Quentin Vanderbecq: Conceptualization, Methodology, Software, Investigation, Formal analysis, Data curation, Writing - original draft, Writing - review & editing, Visualization, Funding acquisition. Eric Xu: Conceptualization, Methodology, Software, Investigation, Formal analysis, Data curation, Writing - original draft, Writing - review & editing, Visualization, Funding acquisition. Sebastian Ströer: Methodology, Validation, Investigation, Resources, Writing - review & editing, Supervision. Baptiste Couvy-Duchesne: Formal analysis, Writing - original draft, Writing - review & editing, Visualization, Supervision, Formal analysis, Writing - original draft, Writing - review & editing, Visualization, Supervision. Mauricio Diaz Melo: Software, Resources, Writing - review & editing. Didier Dormont: Methodology, Resources, Validation, Writing - review & editing, Supervision. Olivier Colliot: Conceptualization, Methodology, Software, Formal analysis, Investigation, Resources, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Acknowledgments
Acknowledgements
Ours thoughts go to Anne Bertrand, MD, PhD, a brilliant radiologist, professor and researcher, without whom this project would never have taken shape.
We thank Simona Bottani, Johann Faouzi, Alexis Guyot, Arnaud Marcoux, Benoit Martin, Alexandre Routier and Adam Wild for the IT support and assistance in programming during this study. We also thank Ninon Burgos for her helpful advice.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2020.102357.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Caligiuri M.E., Perrotta P., Augimeri A., Rocca F., Quattrone A., Cherubini A. Automatic Detection of White Matter Hyperintensities in Healthy Aging and Pathology Using Magnetic Resonance Imaging: A Review. Neuroinformatics. 2015;13:261–276. doi: 10.1007/s12021-015-9260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carass A., Roy S., Jog A., Cuzzocreo J.L., Magrath E., Gherman A., Button J., Nguyen J., Prados F., Sudre C.H., Jorge Cardoso M., Cawley N., Ciccarelli O., Wheeler-Kingshott C.A.M., Ourselin S., Catanese L., Deshpande H., Maurel P., Commowick O., Barillot C., Tomas-Fernandez X., Warfield S.K., Vaidya S., Chunduru A., Muthuganapathy R., Krishnamurthi G., Jesson A., Arbel T., Maier O., Handels H., Iheme L.O., Unay D., Jain S., Sima D.M., Smeets D., Ghafoorian M., Platel B., Birenbaum A., Greenspan H., Bazin P.-L., Calabresi P.A., Crainiceanu C.M., Ellingsen L.M., Reich D.S., Prince J.L., Pham D.L. Longitudinal multiple sclerosis lesion segmentation: Resource and challenge. NeuroImage. 2017;148:77–102. doi: 10.1016/j.neuroimage.2016.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commowick O., Istace A., Kain M., Laurent B., Leray F., Simon M., Pop S.C., Girard P., Améli R., Ferré J.-C., Kerbrat A., Tourdias T., Cervenansky F., Glatard T., Beaumont J., Doyle S., Forbes F., Knight J., Khademi A., Mahbod A., Wang C., McKinley R., Wagner F., Muschelli J., Sweeney E., Roura E., Lladó X., Santos M.M., Santos W.P., Silva-Filho A.G., Tomas-Fernandez X., Urien H., Bloch I., Valverde S., Cabezas M., Vera-Olmos F.J., Malpica N., Guttmann C., Vukusic S., Edan G., Dojat M., Styner M., Warfield S.K., Cotton F., Barillot C. Objective Evaluation of Multiple Sclerosis Lesion Segmentation using a Data Management and Processing Infrastructure. Sci. Rep. 2018;8:13650. doi: 10.1038/s41598-018-31911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupé, P., Tourdias, T., Linck, P., Romero, J.E., Manjón, J.V., 2018. LesionBrain: An Online Tool for White Matter Lesion Segmentation, in: Bai, W., Sanroma, G., Wu, G., Munsell, B.C., Zhan, Y., Coupé, P. (Eds.), Patch-Based Techniques in Medical Imaging, Lecture Notes in Computer Science. Springer International Publishing, pp. 95–103.
- Duong M.T., Rudie J.D., Wang J., Xie L., Mohan S., Gee J.C., Rauschecker A.M. Convolutional Neural Network for Automated FLAIR Lesion Segmentation on Clinical Brain MR Imaging. Am. J. Neuroradiol. 2019 doi: 10.3174/ajnr.A6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas F., Chawluk J.B., Alavi A., Hurtig H.I., Zimmerman R.A. MR signal abnormalities at 1.5 T in Alzheimer’s dementia and normal aging. AJR Am. J. Roentgenol. 1987;149:351–356. doi: 10.2214/ajr.149.2.351. [DOI] [PubMed] [Google Scholar]
- Griffanti L., Zamboni G., Khan A., Li L., Bonifacio G., Sundaresan V., Schulz U.G., Kuker W., Battaglini M., Rothwell P.M., Jenkinson M. BIANCA (Brain Intensity AbNormality Classification Algorithm): A new tool for automated segmentation of white matter hyperintensities. Neuroimage. 2016;141:191–205. doi: 10.1016/j.neuroimage.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud J., Lai M., Thorpe J., Adeleine P., Wang L., Barker G.J., Plummer D.L., Tofts P.S., McDonald W.I., Miller D.H. Quantification of MRI lesion load in multiple sclerosis: a comparison of three computer-assisted techniques. Magn. Reson. Imaging. 1996;14:495–505. doi: 10.1016/0730-725x(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Heinen R., Steenwijk M.D., Barkhof F., Biesbroek J.M., van der Flier W.M., Kuijf H.J., Prins N.D., Vrenken H., Biessels G.J., de Bresser J. Performance of five automated white matter hyperintensity segmentation methods in a multicenter dataset. Sci Rep. 2019;9:1–12. doi: 10.1038/s41598-019-52966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández, M. del C.V., Morris, Z., Dickie, D.A., Royle, N.A., Maniega, S.M., Aribisala, B.S., Bastin, M.E., Deary, I.J., Wardlaw, J.M., 2013. Close Correlation between Quantitative and Qualitative Assessments of White Matter Lesions. Neuroepidemiology 40, 13–22. https://doi.org/10.1159/000341859. [DOI] [PubMed]
- Hernández M.C.V., Piper R.J., Bastin M.E., Royle N.A., Maniega S.M., Aribisala B.S., Murray C., Deary I.J., Wardlaw J.M. Morphologic, Distributional, Volumetric, and Intensity Characterization of Periventricular Hyperintensities. Am. J. Neuroradiol. 2014;35:55–62. doi: 10.3174/ajnr.A3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithapu V., Singh V., Lindner C., Austin B.P., Hinrichs C., Carlsson C.M., Bendlin B.B., Johnson S.C. Extracting and summarizing white matter hyperintensities using supervised segmentation methods in Alzheimer’s disease risk and aging studies. Hum. Brain Mapp. 2014;35:4219–4235. doi: 10.1002/hbm.22472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Barnes J., Bernstein M.A., Borowski B.J., Brewer J., Clegg S., Dale A.M., Carmichael O., Ching C., DeCarli C., Desikan R.S., Fennema-Notestine C., Fjell A.M., Fletcher E., Fox N.C., Gunter J., Gutman B.A., Holland D., Hua X., Insel P., Kantarci K., Killiany R.J., Krueger G., Leung K.K., Mackin S., Maillard P., Malone I.B., Mattsson N., McEvoy L., Modat M., Mueller S., Nosheny R., Ourselin S., Schuff N., Senjem M.L., Simonson A., Thompson P.M., Rettmann D., Vemuri P., Walhovd K., Zhao Y., Zuk S., Weiner M. Magnetic resonance imaging in Alzheimer’s Disease Neuroimaging Initiative 2. Alzheimers Dement. J. Alzheimers Assoc. 2015;11:740–756. doi: 10.1016/j.jalz.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, C.R., Bernstein, M.A., Fox, N.C., Thompson, P., Alexander, G., Harvey, D., Borowski, B., Britson, P.J., L Whitwell, J., Ward, C., Dale, A.M., Felmlee, J.P., Gunter, J.L., Hill, D.L.G., Killiany, R., Schuff, N., Fox-Bosetti, S., Lin, C., Studholme, C., DeCarli, C.S., Krueger, G., Ward, H.A., Metzger, G.J., Scott, K.T., Mallozzi, R., Blezek, D., Levy, J., Debbins, J.P., Fleisher, A.S., Albert, M., Green, R., Bartzokis, G., Glover, G., Mugler, J., Weiner, M.W., 2008. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J. Magn. Reson. Imaging JMRI 27, 685–691. https://doi.org/10.1002/jmri.21049. [DOI] [PMC free article] [PubMed]
- Jiang J., Liu T., Zhu W., Koncz R., Liu H., Lee T., Sachdev P.S., Wen W. UBO Detector – A cluster-based, fully automated pipeline for extracting white matter hyperintensities. NeuroImage. 2018;174:539–549. doi: 10.1016/j.neuroimage.2018.03.050. [DOI] [PubMed] [Google Scholar]
- Kim, MacFall, Payne Classification of white matter lesions on magnetic resonance imaging in elderly persons. Biolog. Psychiatry. 2008;64:273–280. doi: 10.1016/j.biopsych.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo T.K., Li M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016;15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijf, H.J., Biesbroek, J.M., de Bresser, J., Heinen, R., Andermatt, S., Bento, M., Berseth, M., Belyaev, M., Cardoso, M.J., Casamitjana, A., Collins, D.L., Dadar, M., Georgiou, A., Ghafoorian, M., Jin, D., Khademi, A., Knight, J., Li, H., Llado, X., Luna, M., Mahmood, Q., McKinley, R., Mehrtash, A., Ourselin, S., Park, B.-Y., Park, H., Park, S.H., Pezold, S., Puybareau, E., Rittner, L., Sudre, C.H., Valverde, S., Vilaplana, V., Wiest, R., Xu, Y., Xu, Z., Zeng, G., Zhang, J., Zheng, G., Chen, C., van der Flier, W., Barkhof, F., Viergever, M.A., Biessels, G.J., 2019. Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities; Results of the WMH Segmentation Challenge. IEEE Trans. Med. Imaging. https://doi.org/10.1109/TMI.2019.2905770.
- Li H., Jiang G., Zhang J., Wang R., Wang Z., Zheng W.-S., Menze B. Fully convolutional network ensembles for white matter hyperintensities segmentation in MR images. NeuroImage. 2018;183:650–665. doi: 10.1016/j.neuroimage.2018.07.005. [DOI] [PubMed] [Google Scholar]
- Mueller S.G., Weiner M.W., Thal L.J., Petersen R.C., Jack C., Jagust W., Trojanowski J.Q., Toga A.W., Beckett L. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin. N. Am. 2005;15(869–877):xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnozzi A.M., Fripp J., Rose S.E. Quantifying deep grey matter atrophy using automated segmentation approaches: A systematic review of structural MRI studies. NeuroImage. 2019;201 doi: 10.1016/j.neuroimage.2019.116018. [DOI] [PubMed] [Google Scholar]
- Rachmadi M.F., Valdés-Hernández M.D.C., Agan M.L.F., Di Perri C., Komura T. Alzheimer’s Disease Neuroimaging Initiative, Segmentation of white matter hyperintensities using convolutional neural networks with global spatial information in routine clinical brain MRI with none or mild vascular pathology. Comput. Med. Imaging Graph Off. J. Comput. Med. Imaging Soc. 2018;66:28–43. doi: 10.1016/j.compmedimag.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Ronneberger, O., Fischer, P., Brox, T., 2015. U-Net: Convolutional Networks for Biomedical Image Segmentation. ArXiv150504597 Cs.
- Roura E., Oliver A., Cabezas M., Valverde S., Pareto D., Vilanova J.C., Ramió-Torrentà L., Rovira À., Lladó X. A toolbox for multiple sclerosis lesion segmentation. Neuroradiology. 2015;57:1031–1043. doi: 10.1007/s00234-015-1552-2. [DOI] [PubMed] [Google Scholar]
- Schmidt, P., 2017. Bayesian inference for structured additive regression models for large-scale problems with applications to medical imaging (Text.PhDThesis). Ludwig-Maximilians-Universität München.
- Schmidt P., Gaser C., Arsic M., Buck D., Förschler A., Berthele A., Hoshi M., Ilg R., Schmid V.J., Zimmer C., Hemmer B., Mühlau M. An automated tool for detection of FLAIR-hyperintense white-matter lesions in Multiple Sclerosis. NeuroImage. 2012;59:3774–3783. doi: 10.1016/j.neuroimage.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Shrout P.E., Fleiss J.L. Intraclass correlations: uses in assessing rater reliability. Psychol. Bull. 1979;86:420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Taha A.A., Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med. Imaging. 2015;15 doi: 10.1186/s12880-015-0068-x. 29 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde S., Cabezas M., Roura E., González-Villà S., Pareto D., Vilanova J.C., Ramió-Torrentà L., Rovira À., Oliver A., Lladó X. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage. 2017;155:159–168. doi: 10.1016/j.neuroimage.2017.04.034. [DOI] [PubMed] [Google Scholar]
- Valverde S., Salem M., Cabezas M., Pareto D., Vilanova J.C., Ramió-Torrentà L., Rovira À., Salvi J., Oliver A., Lladó X. One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks. NeuroImage Clin. 2019;21 doi: 10.1016/j.nicl.2018.101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa A., Bhardwaj A., Singh Verma V. A review on brain tumor segmentation of MRI images. Magn. Reson. Imaging. 2019;61:247–259. doi: 10.1016/j.mri.2019.05.043. [DOI] [PubMed] [Google Scholar]
- Wardlaw J.M., Smith E.E., Biessels G.J., Cordonnier C., Fazekas F., Frayne R., Lindley R.I., O’Brien J.T., Barkhof F., Benavente O.R., Black S.E., Brayne C., Breteler M., Chabriat H., Decarli C., de Leeuw F.-E., Doubal F., Duering M., Fox N.C., Greenberg S., Hachinski V., Kilimann I., Mok V., van Oostenbrugge R., Pantoni L., Speck O., Stephan B.C.M., Teipel S., Viswanathan A., Werring D., Chen C., Smith C., van Buchem M., Norrving B., Gorelick P.B., Dichgans M., STandards for ReportIng Vascular changes on nEuroimaging (STRIVE v1), Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 2013;12:822–838. doi: 10.1016/S1474-4422(13)70124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P.A., Piven J., Hazlett H.C., Smith R.G., Ho S., Gee J.C., Gerig G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.