Abstract
The nematode Caenorhabditis elegans has been instrumental in the identification of evolutionarily conserved mechanisms of aging. C. elegans also has recently been found to have evolutionarily conserved extracellular vesicle (EV) signaling pathways. We have been developing tools that allow for the detailed study of EV biology in C. elegans. Here we apply our recently published method for high specificity purification of EVs from C. elegans to carry out target-independent proteomic and RNA analysis of nematode EVs. We identify diverse coding and non-coding RNA and protein cargo types commonly found in human EVs. The EV cargo spectrum is distinct from whole worms, suggesting that protein and RNA cargos are actively recruited to EVs. Gene ontology analysis revealed C. elegans EVs are enriched for extracellular-associated and signaling proteins, and network analysis indicates enrichment for metabolic, immune, and basement membrane associated proteins. Tissue enrichment and gene expression analysis suggests the secreted EV proteins are likely to be derived from intestine, muscle, and excretory tissue. An unbiased comparison of the EV proteins with a large database of C. elegans genome-wide microarray data showed significant overlap with gene sets that are associated with aging and immunity. Taken together our data suggest C. elegans could be a promising in vivo model for studying the genetics and physiology of EVs in a variety of contexts including aging, metabolism, and immune response.
Electronic supplementary material
The online version of this article (10.1007/s11357-020-00204-1) contains supplementary material, which is available to authorized users.
Keywords: Aging, LC-MS-MS analysis, Small RNAseq, Gene enrichment analysis, Metabolism, Immune response, Basement membrane glycoproteins, Membrane raft proteins
Introduction
The release of extracellular vesicles (EVs) into the external environment appears to be a universal feature of all living cells. In eukaryotes, this can occur directly from the plasma membrane (microvesicles) or through the endosomal pathway (exosomes) (Jeppesen et al. 2019; Kowal et al. 2016). Common EV cargos include small non-coding RNAs and proteins such as translation elongation factors, heat shock proteins, metabolic enzymes, membrane raft proteins, and miRNAs (Brown and London 2000; Kim et al. 2015; Mathivanan et al. 2012).
EV biology has gained significant attention in recent years due to growing evidence that EVs play important roles in, and may also serve as useful predictive biomarkers for, many human diseases (Shah et al. 2018). This is particularly true for age-related diseases, such as cancers (Kinoshita et al. 2017; Xu et al. 2018), and Alzheimer’s disease and related dementias (D'Anca et al. 2019; DeLeo and Ikezu 2018; Li et al. 2019). There is also growing evidence that EVs may be directly involved in the biological aging process itself by mediating inflammatory processes and other cell non-autonomous signaling and thereby contributing to a degradation in intercellular communication (Go et al. 2020; Robbins 2017; Takasugi 2018), one of the hallmarks of aging (Lopez-Otin et al. 2013).
Recently, C. elegans has emerged as a potentially useful genetic model for identifying EV signaling pathways in vivo (Wang and Barr 2018). For instance, a flippase was shown to induce EV biogenesis in C. elegans embryos, which led to its subsequent discovery as a mechanism that influences EV release in human cells (Naik et al. 2019; Wehman et al. 2011). Likewise, EV-dependent Hedgehog signaling was first identified in C. elegans and subsequently shown to also impact development in zebrafish, mice, and humans (Liegeois et al. 2006; Qi et al. 2017; Sigg et al. 2017; Simon et al. 2016). These studies demonstrate that C. elegans shares key EV signaling mechanisms with humans. However, outside of a small number of reports, the composition and function of C. elegans EVs remain almost completely uncharacterized.
Given the long track record of C. elegans as a premier model organism for the study of aging biology (Kenyon 1996; Tissenbaum 2015; Yanos et al. 2012), we believe that C. elegans could become a powerful system to understand the role of EVs in longevity and healthspan of animals. Toward this goal, we have developed a scalable method for purifying high-quality secreted EVs from nematodes using filtration and size exclusion chromatography (Russell et al. 2020a). This procedure is considered the most effective means for bulk EV isolation, providing more pure preparations and less prone to artifacts than other commonly used approaches like ultracentrifugation and PEG-assisted precipitation (Lee et al. 2019). Here we provide the first large-scale report of RNA and protein cargos contained in young adult C. elegans EVs. We identify many different RNA and protein cargo types also observed in human EVs and show that these cargos are enriched for proteins involved in aging, metabolism, and immunity.
Methods
Strains and EV preparation
For this study we utilized wild type (N2) C. elegans. Worms were cultured and maintained using standard methods (Brenner 1974). EVs were isolated from young adult N2 animals using our recently developed method for large-scale EV purification from C. elegans (Russell et al. 2020b). Three independent biological replicates were analyzed for RNA and protein composition. For each replicate 500,000 worms were incubated at 20 °C on high growth plates seeded with live NA22 Escherichia coli until they reached young adulthood. Animals were then washed free of bacteria and incubated in 500 mL of sterile S basal media. The media was placed in 2 L cross-bottom baffled flasks, rotated at 100 rpm in a 20 °C incubator. After 24 h the worms were separated, and the conditioned media was concentrated 1000-fold over a 10-kDa filter. The concentrate was then passed over a 10-mL Sepharose CL-2B column. The elution volume between 2 and 6 mL, which contains the majority of the EVs (Russell et al. 2020a), was then concentrated 10-fold over 10-kDa filters. Nanoparticle tracking analysis, transmission electron microscopy, and flow cytometry analysis demonstrated that this protocol separates EVs from freely soluble proteins and lipoprotein aggregates (Russell et al. 2020b). To prepare for LC-MS-MS the EV samples were run into an SDS-PAGE gel, digested with trypsin, and analyzed on a Velos Pro spectrometer.
Proteomics
Total protein in the EV samples was quantified by QuBit analysis, and samples were normalized for protein concentration. These were then extracted in RIPA buffer with vortexing for 30 min before adding NuPage 4X sample buffer and heating to 70 °C for 10 min. Samples were then spun at 18 kG for 15 min to pellet any precipitates. About 50 μg of protein was loaded onto a large volume capacity SDS-PAGE gel and run at 80 mV for 10 min and then 100 mV until the dye front was ~ 1 cm below the start of the resolving gel. The gels were then washed twice with 250 mL dH2O for 60 min to remove any detergent. The samples were then cut out of the gel with a new razor blade and kept in a low-protein binding Eppi-tube (ThermoFisher, San Jose, CA USA, Cat no. 90410).
In-gel digestion
Gel bands were washed in 100 mM ammonium bicarbonate, reduced with DTT, and alkylated with IAA. Gel bands were then shrunk with acetonitrile and speed-vacuumed to dry the gel bands. The gel bands were then digested with 1 μg of trypsin overnight at 37 °C with shaking. The next day proteins were extracted from gel bands with 60% acetonitrile, 0.1% trifluoroacetic acid, and then reconstituted in 0.1% formic acid.
Liquid chromatography and mass spectrometry
Fused silica microcapillary columns of 75 μm inner diameter (Polymicro Technologies, Phoenix, AZ) were packed in-house by pressure loading 30 cm of Repro sil-pur C18 material (Dr. Maisch, Gmbh, Germany). Kasil (PQ Corporation, Malvern, PA) frit microcapillary column traps of 150 μm inner diameter with a 2-mm Kasil frit were packed with 4 cm of Repro sil-pur C18 material. A retention time calibration mixture (Pierce, Rockford, IL) was used to assess quality of the column before and during analysis. Three of these quality control runs are analyzed prior to any sample analysis, and then after every six sample runs another quality control run is analyzed. Samples were loaded onto the trap and column by the NanoACQUITY UPLC (Waters Corporation, Milford, MA) system. Buffer solutions used were 0.1% formic acid in water (Buffer A) and 0.1% formic acid in acetonitrile (Buffer B). The 60-min gradient of the quality control consisted of 30 min of 98% buffer A and 2% buffer B, 5 min of 65% buffer A and 35% buffer B, 6 min of 40% buffer A and 60% buffer B, 5 min of 95% buffer A and 5% buffer B, and 18 min of 98% buffer A and 2% buffer B at a flow rate of 0.3 μL /min. The 180-min gradient for the sample digest consisted of 130 min of 98% buffer A and 2% buffer B, 10 min of 60% buffer A and 40% buffer B, 1 min of 40% buffer A and 60% buffer B, 6 min of 5% buffer A and 95% buffer B, and 33 min of 98% buffer A and 2% buffer B at a flow rate of 0.3 μL/min. Peptides are eluted from the column and electrosprayed directly into a Velos Pro mass spectrometer (ThermoFisher, San Jose, CA) with the application of a distal 3-kV spray voltage. For the quality control analysis, a full-scan mass spectrum (400–1600 m/z) is measured followed by a 17 SRM spectra all at 35% normalized collision energy and a 2 m/z isolation window. For the sample digests, a full-scan mass spectrum (400–1600 m/z) followed by 17 data-dependent MS/MS spectra on the top 16 most intense precursor ions at 35% normalized collision energy with a 2 m/z isolation window. Application of the mass spectrometer and UPLC solvent gradients was controlled by the ThermoFisher XCalibur data system.
Data analysis
The quality control data were analyzed using Skyline (MacLean et al. 2010). The DDA MS/MS data were searched using COMET with dynamic modification searches of 15.994915 methionine and a static modification of 57.021464 Cysteine against a FASTA database containing all the protein sequences from the WS250 freeze of C. elegans from WormBase plus contaminant proteins (Eng et al. 2013). Peptide spectrum match false discovery rates were determined using Percolator (Kall et al. 2007) at a threshold of 0.01, and peptides were assembled into protein identifications using an in-house implementation of IDPicker (Zhang et al. 2007). The results of these mass spectrometry runs are available at chorusproject (https:/chorusproject.org/anonymous/download/experiment/d41c70cbecfe42a0a00050488903a163).
The comparison of proteins between EV and whole worm was conducted by filtering the 224 proteins identified in all replicates against a protein data set of young adult whole worm lysate obtained from the same Velos Pro spectrometer as the EV proteomics was conducted (data in supplemental spreadsheet). The data set contained abundance values for 118 of the 224 EV proteins. A scatterplot of the EV and whole worm abundance values of these 118 proteins was generated and the linear trendline and R2 were calculated. The EV and whole worm proteomic data and a table of the analyzed proteins with their abundance values are given in the supplemental spreadsheet.
To assess the ontology associations of EV protein cargos and characterize their protein interaction network structure we used WormBase enrichment analysis, STRING V11, and WormExp WS235 (Angeles-Albores et al. 2016; Croft et al. 2011; Mi et al. 2013; Szklarczyk et al. 2019; Yang et al. 2016). Human homologs of C. elegans proteins were identified through WORMHOLE (wormhole.jax.org) (Sutphin et al. 2016).
RNA sequencing
To quantify EV-associated RNA samples were processed with total exosome protein and RNA isolation kit (Invitrogen, Carlsbad CA, USA, Cat no. 4478545). We then characterized the abundance and size of the eluates on an Agilent 2200 TapeStation (Agilent, Santa Clara CA, USA; Cat no. G2991AA) using high-sensitivity screen tape (Agilent, Santa Clara USA Cat no. 5067-5579) along with a small RNA calibration ladder (Agilent, Santa Clara CA, USA; Cat no. 5067-1550). The vesicle samples displayed substantial small RNAs as well as 16 s and 28 s rRNA species at the expected sizes. To determine what RNA species are packaged into EVs we conducted RNAseq. We used a Qiagen small RNA Sample Preparation kit to prepare a cDNA library from our EV-associated RNA (Qiagen Germantown MD, USA Cat no. 331502), while an Illumina MiSeq v2 kit (300 cycles) was used to prepare the sequencing library with 5′ adapter sequences (San Diego USA; Cat no. MS-102-2002). The sample was then submitted to single-end sequencing on an Illumina MiSeq desktop sequencer.
Small RNAseq data analysis
Adapter trimming and sequence alignment were conducted in the sRNAnalyzer (Wu et al. 2017), which provides comprehensive small RNA profiling. The adapter trimming resulted in RNA with a peak sequence length of 19 nucleotides (nt) dropping off to zero reads around 55 nt. For miRNA profiling, sequences were mapped against miRBase version 21 (Kozomara and Griffiths-Jones 2014; Zerbino et al. 2018). For other RNA profiling, FASTA files for C. elegans transcriptome and genome sequences were retrieved from NCBI genome database (assembly WBcel235) (Kozomara and Griffiths-Jones 2014; Zerbino et al. 2018). The FASTA files were transformed with bowtie-index files. All the alignments to microRNAs, RNAs, and DNAs were performed under 0 to 2 mismatch allowance. About 8M reads were mapped to the C. elegans genome. The complete list of RNA cargos is given publicly available at Gene Expression Ominibus (Accession ID: GSE146973). A spreadsheet with all reads is provided in supplemental file.
To compare the abundances of miRNA between EV and whole worms we chose three miRBase data sets obtained on young adult worms that were not subjected to an experimental treatment (accession nos. dER0000000402, ER0000000403, ER0000000408) (Stoeckius et al. 2009). The miRNAs contained in these three sets were then filtered those that were in all three replicates and had a confidence checkmark. This list was then filtered miRNAs that were present in the EVs resulting in 87 miRNAs with EV and three replicates of whole worm readings. The average reads/million was calculated for the three whole worm readings. EV miRNA abundances were expressed as reads per million by multiplying the number of raw reads a factor of 1 million divided by the total no. (22,042) of miRNA reads. A scatterplot of the EV and whole worm abundances of these 87 miRNAs was then generated, and the linear trendline and R2 value were determined. A list of the abundances used to do the calculation is in the supplemental spreadsheet.
Results
Proteomic analysis of C. elegans EVs
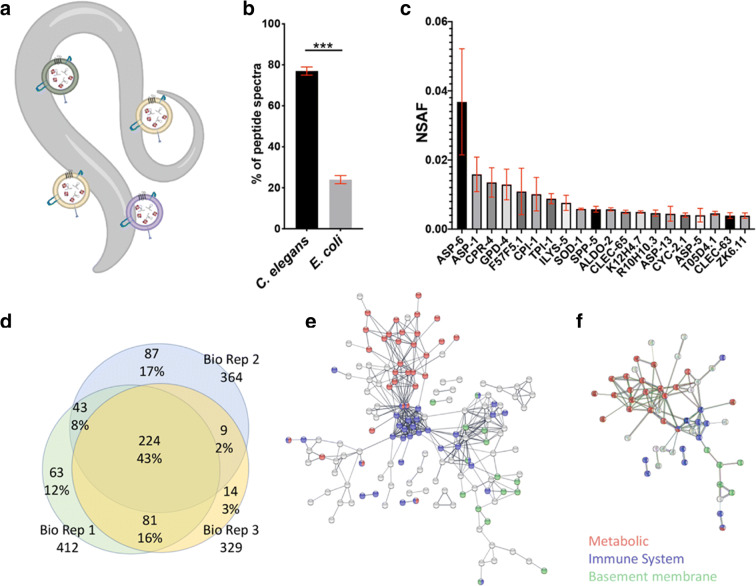
We conducted proteomic analysis of triplicate biological replicates for EVs purified from wild type young adult C. elegans. Secreted EVs were purified from synchronized cultures of ~ 500,000 worms at day 1 of adulthood (Fig. 1a). Greater than 80% of the peptide spectra corresponded to C. elegans proteins with the remainder mapping to E. coli proteins, presumably from the residual food source (Fig. 1b). We ranked the identified C. elegans proteins according to their average Normalized Spectral Abundance Factor (NSAF) (McIlwain et al. 2012) and found that the 20 most abundant proteins displayed relatively consistent abundance levels across biological replicates (Fig. 1c). Each biological replicate identified hundreds of EV protein cargos via unique peptide hits (Supplemental File 1).
Fig. 1.
Proteomic analysis of C. elegans EVs. a Caenorhabditis elegans secrete EVs outside their bodies allowing for their isolation without disrupting tissues. b Comparison of detected EV peptides associated with C. elegans and E. coli. c Absolute NSAF scores of the 20 most-abundant proteins detected across all three EV replicates. Error bars are standard error of the mean. d Venn diagram of the intersection of proteins identified in biological replicates. About 43% of protein hits were found in all biological replicates (n = 3). e Protein interaction network of 224 C. elegans EV proteins identified in every biological replicate. f Protein interaction network for the 44 EV proteins with clear human homologs as determined by Ortholist 2. The 223 EV proteins that were identified in all biological replicates were filtered for those appearing in all of the six C. elegans orthology databases queried by Ortholist 2. For both analyses the STRING association stringency was set to 0.7 (Strong), and the thickness of the bar indicates the strength of interaction. Unconnected proteins have been removed. The colored circles correspond to metabolic (red), immune (blue), and basement membrane-associated (green) proteins
To assess the ontology associations of EV protein cargos and characterize their protein interaction network structure we used WormBase enrichment analysis, STRING V11, and WormExp WS235 (Angeles-Albores et al. 2016; Croft et al. 2011; Mi et al. 2013; Szklarczyk et al. 2019; Yang et al. 2016). We limited our analyses to the set of 224 proteins (43%) that were identified in each of the three biological replicates (Fig. 1d). The most enriched ontology terms in WormBase and STRING were “extracellular region” (FDR 2.23E-12) and “signal” (FDR 7.35E-51), respectively. Tissue enrichment analysis showed that “intestine” (q-value 2.4E-14) and “muscular system” (q value 2.9E-8) were the most associated tissues. The top ontology associations are presented in Supplemental Table 1. Protein interaction network analysis identified three clusters consisting of metabolic, immune, and basement membrane associated proteins (Fig. 1e). Eighty-eight of these proteins are considered strong orthologs because based on reciprocal best hits with human genes using WORMHOLE (Sutphin et al. 2016). The sets of 224 EV proteins as well as the 88 with clear human orthologs consist mainly of metabolic, immune response, and basement membrane protein interaction networks (Fig. 1f; Supplemental Table 2).
C. elegans EVs are enriched for typical human EV cargos
To determine whether C. elegans EVs contain similar cargos as human EVs, we compared our proteomic results with a list of the 100 most-frequently identified human EV proteins in Exocarta.org (Mathivanan et al. 2012). A reciprocal BLAST best hit approach was used to identify worm orthologs corresponding to the 100 human EV proteins, which yielded 46 C. elegans proteins. Although the reciprocal BLAST best hit approach is a stringent means for determining orthology, this approach can overlook protein families with strongly conserved sequences, because none of them emerge as the best single hit (e.g., actin). Therefore, we conducted BLAST analysis on the 54 human EV proteins and filtered for E-values smaller than E < − 30. The reciprocal BLAST best hits and E < − 30 hits were combined into a final set in which 93 human EV proteins were associated with 70 C. elegans proteins (Supplemental Table 3). These 70 C. elegans proteins were then queried against EV proteins identified in any of the replicates and as well as proteins identified all three replicates. This matched 9 proteins that were identified in every replicate (12%) and 21 proteins identified in any replicate (42%). In both cases, the overlap between known human EV cargos and C. elegans EV cargos observed in this study was significantly more than expected by chance (p < 1 E-9, Fisher’s exact tests) (Supplemental Fig. 1).
Sequence analysis of small RNAs in C. elegans EVs
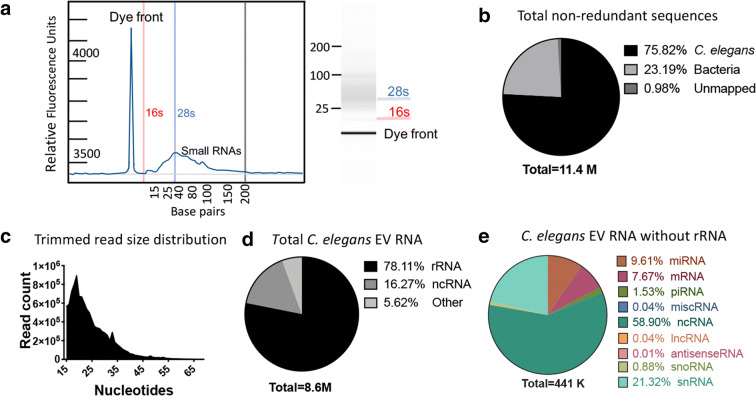
While EVs from other species are known to contain small RNA cargos it has not yet been determined whether C. elegans EVs contain similar RNA species. We therefore conducted an RNA isolation protocol on purified C. elegans EVs and then analyzed the product by TapeStation. We observed 16 s and 28 s rRNA peaks as well as a broad peak below 30 bp (Fig. 2a). Given the presence of abundant RNA in nematode EVs, we proceeded to generate a cDNA library and conduct single-end next-generation sequencing. This resulted in ~ 14 million total reads mapping to ~ 8.5 million C. elegans and ~ 2.5 million E. coli sequences (Fig. 2b). The majority of the 8.5 million C. elegans trimmed reads were < 35 bp (Fig. 2c) with rRNA species comprising about two thirds of the reads (Fig. 2d). We filtered our results to exclude rRNA and any sequences with mismatches. The remaining 440,392 reads corresponded to a wide variety of RNA types including mRNA, miRNA, piRNA, snRNA, and snoRNA among others (Fig. 2e).
Fig. 2.
RNAseq analysis of C. elegans EVs reveals different classes of RNA: a Bioanalyzer reading of RNA extracted from C. elegans EVs. The RFU profile is on the left, while the generated agarose gel illustration is on the right. b Distribution of three major categories of total non-redundant reads with up to 2 nucleotide mismatches. c Trimmed read size distribution of high-quality RNA reads. d Distribution of total non-redundant C. elegans reads with no mismatches. e Distribution of RNA species distribution excluding rRNA
EV composition differs from whole animal composition
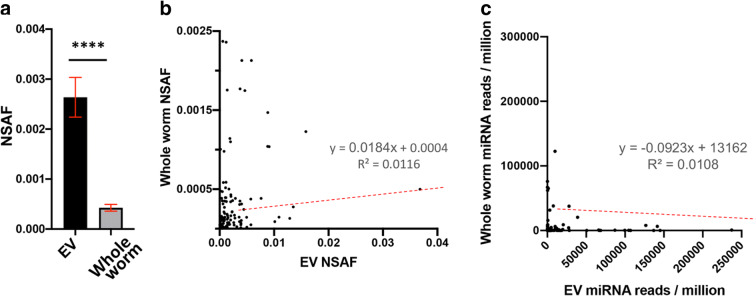
We next compared the composition of EV cargos to global levels within the worm in order to gain insight into whether cargos are actively selected or result from passive uptake from the cellular milieu. To compare protein abundance between EVs and whole worms, we filtered the proteomic data from whole animals for the 224 EV proteins that were identified in every replicate (Fig. 1d). Of these, 106 were not identified in whole worm extract, indicating they are likely of low abundance in whole worms. The remaining 118 proteins that were identified in both EVs and whole worm extract were compared for relative NSAF (Supplemental Table 4A). On average, NSAF values were 27-fold higher in EVs than in whole worms (Fig. 3a), and there was no significant correlation between protein abundance in EVs versus whole worms (Fig. 3b; r2 = 0.01). Consistent with this analysis, EVs did not contain any of the numerous ribosome-associated proteins that are among the most-abundant proteins identified in whole worms (Wang et al. 2015).
Fig. 3.
Comparison of relative levels of EV protein and RNA cargos with reported levels in whole worm lysate. a Normalized abundance spectrum (NSAF) values of 118 EV proteins compared to NSAF values obtained from age-matched whole worm LC-MS-MS data set. b Scatterplot of the EV proteins identified in all three replicates against their global levels in age-matched whole worms. c Scatterplot comparing the EV (X-axis) vs whole worm (Y-axis) miRNA expression levels
To evaluate relative abundance of specific miRNA species in EV compared to whole animals, we compared the EV miRNA levels to three miRNA data sets from miRBase (Kozomara and Griffiths-Jones 2014) corresponding to day one adult C. elegans (accession nos. ER0000000402, ER0000000403, ER0000000408) (Stoeckius et al. 2009). The whole worm miRNA data sets were filtered for miRNAs that were in all replicates, had high confidence reads, and were among the EV miRNAs detected in our analysis. This resulted in 87 miRNAs that could be compared between EVs and whole worms (Supplemental Table 4B). No significant correlation was found between EV miRNA and whole worm miRNA abundance (Fig. 3c; r2 = 0.01).
Discussion
To the best of our knowledge, this is the first large-scale characterization of EV protein and RNA cargos in an invertebrate model organism. The ability to obtain C. elegans EVs of sufficient quality and quantity for downstream-omic analyses will allow for detailed studies of EV biology in a variety of different genetic and environmental contexts. We have been intentionally conservative in this analysis by only considering the RNA sequences with no mismatches and proteins that had unique peptide hits in every biological replicate (n = 3). Even this conservative approach yielded 224 proteins and greater than 2000 different non-coding RNA transcripts including piRNA, miRNA, ncRNA (non-coding RNA), snoRNA, and others (see Supplemental Data File 1).
Although additional studies will be needed to determine the physiological relevance of the EV cargos identified here, several categories of proteins appear to be present in EVs that we speculate may be functionally important. For example, the EV proteome contained an abundance of enzymes involved in metabolic pathways including gluconeogenesis, glycolysis, and amino-acid metabolism. About half (108 of 224) of the C. elegans EV proteins identified in every replicate were associated with carbon metabolism. This is in line with prior reports that metabolic proteins constitute ~ 15% of Escherichia coli EV proteins and about half of Saccharomyces cerevisiae or human EV protein cargos (Lee et al. 2007; Oliveira et al. 2010; Thompson et al. 2018). C. elegans has been shown to secrete metabolites into the external environment, referred to as the exo-metabolome (Butler et al. 2010), and these exo-metabolites have been implicated in longevity determination (Mishur et al. 2016). Based on our data, it seems plausible that a significant portion of the nematode exo-metabolome is derived from EVs, suggesting one mechanism by which EVs could impact lifespan. In the future, it will be of interest to compare the metabolites in the EV and non-EV secretate fractions among wild-type and long-lived mutants.
Another category that stood out from our analysis of EV cargos is proteins involved in immunity. The top Reactome and many top GO “Biological function” pathways were associated with immune response (Supplemental Table 1C). Our analysis indicates that C. elegans EVs are especially enriched for C-type lectin protein cargos, a family of proteins known for recognizing diverse infectious ligands and thus contributing to an antimicrobial immune response (Mallo et al. 2002). We also found that EV proteins are enriched for genes that change expression after exposure to pathogenic bacteria (Supplemental Table 1K) (Sinha et al. 2012; Yang et al. 2015). These connections suggest that C. elegans, like humans, may also enact innate immune responses though EVs (Chen et al. 2019).
Protein interaction analysis identified a small node of four laminins (LAM-1 LAM-2 UNC-52, and EPI-1) and two collagen alpha-1(IV) chain proteins (LET-2 and EMB-9) that localize to the basement membrane (Fig. 1f). Five of these have reciprocal best hits with human proteins as determined by WORMHOLE (Sutphin et al. 2016). Each of these proteins have also been identified in human saliva and urinary EVs, as well as from various types of cancer (Demory Beckler et al. 2013; Gonzales et al. 2009; Gonzalez-Begne et al. 2009). Laminins are known to drive human EV trafficking through interacting with fibrous protein–polysaccharides like fibronectin and functioning with the integrin signaling network (French et al. 2017). These observations suggest that, as in other animals (Antonyak et al. 2011; Desrochers et al. 2016; Hoshino et al. 2015; Sabatier et al. 2002), extracellular matrix–integrin interactions likely influence EV binding and uptake by cells in C. elegans.
Our analysis revealed that membrane raft proteins are enriched in C. elegans EVs. Membrane rafts form when sphingolipids and cholesterol aggregate into discrete punctate nanodomains within the lipid bilayer membrane. In vertebrate EVs these nanodomains facilitate colocalization of proteins that function together and are critical for both EV biogenesis and for targeting recipient cells (Head et al. 2014). Twentyfive out of forty-three previously identified nematode membrane raft proteins (Rao et al. 2011) were identified in EVs (Supplemental Table 5). The enrichment of membrane raft proteins suggests that, like humans, C. elegans EVs also contain these lipid nanodomains, and once again points to substantial similarities in EV structure and function between worms and people.
While not unexpected, the presence of abundant RNA species in C. elegans EVs indicates that this major EV signaling modality is likely conserved in C. elegans. One of the advantages of RNAseq analysis is that different small RNA families can be studied simultaneously in a comprehensive manner. In addition to abundant rRNA, mammalian EV cargos can include mRNAs, ncRNAs including miRNAs, long non-coding RNA (lncRNA), single-stranded DNA, double-stranded DNA, mitochondrial DNA, and oncogene amplifications (i.e., c-myc) (Guescini et al. 2010; Janas et al. 2015; Thakur et al. 2014; Yáñez-Mó et al. 2015). Our RNAseq analysis yielded more than 440,000 no-mismatch reads identifying other RNA species, including ncRNA, mRNA, miRNA, piRNA, and snRNA among others in roughly similar percentages of RNA subtypes as previously reported from human EVs. Almost a third of the ncRNA reads were for three uncharacterized species, M02F4.12, C44B7.15, and B0244.13. snRNA followed by miRNA were the next most-abundant families of RNA comprising 25% and ~ 10% of the non-rRNA reads, respectively (Fig. 2d). Small ncRNA and rRNA have been shown to also be enriched in human exosomes (Elsemüller et al. 2019). The piRNAs identified were notable because of their great variety, with more than 1200 different transcripts from 6776 no-mismatch reads. In contrast, we only identified 139 miRNA species from 42,444 no-mismatch miRNA reads. The miRNA reads mapped to 139 miRNA species all encoded from the plus strand. The miRNA families most over-represented in EVs were miR-51 and miR-58 (Supplemental Table 4B). Studies have shown that the miR-51 family functions in developmental timing and tissue adhesion, suggesting the possibility that EV transport of these miRNAs may contribute to their cell non-autonomous functions (Brenner et al. 2012; Shaw et al. 2010). The miR-58 family has been shown to interact with the TGF-beta pathway and suggested to play housekeeping roles to influence tissue-specific gene expression (de Lucas et al. 2015; Pagano et al. 2015). Intriguingly, one of the most abundant miRNAs was let-7, which has been recently been identified in human cancer cells and breast milk EVs (Ohshima et al. 2010; van Herwijnen et al. 2018). Each of the human orthologs of the miRNAs contained in C. elegans EVs (Supplemental Table 4B) have been previously identified in EVs based on datasets uploaded to microvesicles.org and EVmiRNA (Kalra et al. 2012; Liu et al. 2019). Our EV RNA sequencing results also contained over 5000 mRNA no mismatch hits. In general, the most abundant mRNAs correspond to the most-abundant proteins identified in C. elegans whole worm lysate in the pax.db database (Wang et al. 2015), including numerous ribosomal subunits as well as collagens, histones, and actin/tubulin genes (Supplemental Data File 1). Although little is known about ncRNA functions this enrichment in EVs suggests they may play a role in cell-to-cell signaling and comparison with previous human EV RNA datasets suggests that C. elegans is likely to share many aspects of canonical RNA-mediated EV signaling with humans (Kim et al. 2017).
To date, the only C. elegans ciliated neurons have been shown to secrete EVs outside the body (Wang et al. 2014), but our data suggests that multiple other tissues also likely contribute to this EV subset. The miRNA with the highest EV expression from our dataset is mir-82 (90,651 reads/million), which is known to be expressed specifically in the amphid neurons, excretory gland cell, and a subset of neurons in the tail, while a miR-81expression is neuron-specific (Isik et al. 2010; Martinez et al. 2008). The protein with the highest abundance score ASP-6 is localized to the extracellular pseudocoelomic space and intestine. Tissue enrichment analysis of EV protein cargo identified the intestine as the most represented tissue, followed by muscle and pharynx (Supplemental Table 1A). Together, this suggests that the intestine and excretory duct are likely the major producers of secreted EVs, although neurons, muscle, and other tissues contribute as well.
One particularly intriguing feature of the proteomic analysis is an association with genes regulated in the context of aging. We used WormExp to determine the correlation of the EV protein set with published genome-wide gene expression data sets in C. elegans. Although aging-related microarray experiments comprised just 4% of the data sets (97 of 2297), four of the ten most-correlated studies were age-related (Supplemental Table 6). Two of these studies identified genes whose mRNA levels decrease significantly with age (Budovskaya et al. 2008; Pu et al. 2015), and two studies identified genes that are downstream targets of transcription factors that have been shown to influence aging in C. elegans (Habacher et al. 2016; Mann et al. 2016; Thyagarajan et al. 2010). The most abundant miRNA cargos were also enriched for miRNA families that have been shown to function in longevity pathways. For instance, miR-58 has been shown to increase longevity in daf-2 mutants through daf-16 ((FOXO6)) signaling (Zhang et al. 2018). Three members of the miR-58 miRNA family (miR-58, miR-80, miR-82) were all among the top ten most-abundant miRNAs in EVs, and miR-71, the fourth-most abundant EV miRNA, is known to influence longevity in a cell non-autonomous manner via the insulin/IGF pathway (Boulias and Horvitz 2012).
We envision at least two general ways that EVs could be important for aging in C. elegans and other animals. First, there is abundant evidence that many longevity pathways act through cell non-autonomous signals to regulate lifespan. In worms, for example, long-lived mutants involved in mitochondrial function (Durieux et al. 2011), insulin signaling (Apfeld and Kenyon 1998), mTOR signaling (Zhang et al. 2019), and hypoxic signaling (Leiser et al. 2015) all involve signals arising in neurons and mediating downstream molecular changes in the intestine. EVs are a likely candidate for transducing these signals. For instance, our studies showed that miR-83 was enriched 30-fold in EVs compared to whole worm lysate (Supplemental Table 4B) and intestinal miR-83 was recently shown to control the age-related decrease in macroautophagy in other tissues, potentially through EV signaling (Zhou et al. 2019). Second, EVs may serve as a mechanism for removing toxic misfolded or aggregated proteins that accumulate with age. Such a cellular self-defense mechanism would allow cells to reduce the intracellular proteotoxic burden but could also inadvertently contribute to the spreading of pathogenic proteinaceous species if taken up by other cells. There is evidence that such EV-mediated spreading may be important during Alzheimer’s disease (Pluta et al. 2018; Sardar Sinha et al. 2018), and large EV “exophers” are produced by aged nematodes (Melentijevic et al. 2017). Exploring these and other potential roles for C. elegans EVs may shed light on fundamental mechanisms of aging in nematodes as well as human age-related disorders.
The methods we have developed and implemented here result in abundant, relatively pure EVs sufficient for conducting multiple types of analyses. We note that in order to separate the worms from bacterial contaminants, the animals were incubated in buffer free of bacteria for 24 h prior to collection of EVs. We determined that greater than 99% of the animals were healthy and active following the incubation period, suggesting that little to no death occurred and isolated EVs came from live animals. Indeed, we have previously shown that complete removal of bacterial food during adulthood increases lifespan in C. elegans and related species (Smith et al. 2008; Sutphin and Kaeberlein 2008). However, we recognize that the absence of food may also impact the composition of the EVs and different cargo compositions are likely in fully fed adult worms. Future efforts will be directed toward purification of secreted EVs from fed C. elegans, either through affinity-based approaches or utilizing axenic media.
Conclusion
Human EV cargos are increasingly being investigated as biomarkers of, and contributing factors for, health and disease. Until now, however, the EV field has been lacking a tractable invertebrate genetic model. In this study, we performed the first large-scale identification of EV proteins and RNA cargo from C. elegans. Our experiments uncovered a diverse spectrum of small non-coding RNAs and proteins, the composition of which indicates significant evolutionary conservation with humans. The genetic tractability of C. elegans, coupled with the ability to isolate EVs for proteomic, metabolomic, and RNAseq analysis, suggests that C. elegans could be a useful model system for dissecting genetic pathways and physiological processes that impact EV signaling in vivo. In particular, the use of C. elegans to understand the role of EVs in aging and age-related disease appears to be a fruitful line of investigation.
Electronic supplementary material
(PDF 1344 kb)
(XLSX 2372 kb)
Acknowledgments
We gratefully acknowledge Nick Terzopoulos for worm plates and reagents, the Caenorhabditis Genetics Center (CGC) for the N2 nematode line, Safyie Celik for help with statistical analysis, and Albert Tai and Matthew Fierman of the Tufts University Bioseq facility for RNA sequencing. Sequencing was sponsored by the National Institutes of Health Science Education Partnership Award and the Cummings Foundation.
Author contributions
JCR conceived the study, carried out the experiments in the study unless otherwise indicated, and wrote the manuscript. AG carried out purification of EVs. GEM, JER, and MJM conducted the mass-spectrometry experiments and analyzed the raw proteomics data to determine the statistically significant protein hits, TK, KW, AN, and JCR analyzed the RNAseq data. MK supervised the study and wrote the manuscript.
Funding information
This work was supported by NIH grants P30AG013280 and P50AG005136 to MK and NIH grant F32AG054098 to JCR.
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Joshua C. Russell, Email: jcr32@uw.edu
Matt Kaeberlein, Email: kaeber@uw.edu.
References
- Angeles-Albores D, Lee RYN, Chan J, Sternberg PW. Tissue enrichment analysis for C. elegans genomics. BMC Bioinformatics. 2016;17:366. doi: 10.1186/s12859-016-1229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonyak MA, et al. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc Natl Acad Sci U S A. 2011;108:4852–4857. doi: 10.1073/pnas.1017667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/S0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- Boulias K, Horvitz HR. The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 2012;15:439–450. doi: 10.1016/j.cmet.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner JL, Kemp BJ, Abbott AL. The mir-51 family of microRNAs functions in diverse regulatory pathways in Caenorhabditis elegans. PLoS One. 2012;7:e37185. doi: 10.1371/journal.pone.0037185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–4. 10.1074/jbcR000005200. [DOI] [PubMed]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, et al. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed]
- Butler JA, Ventura N, Johnson TE, Rea SL. Long-lived mitochondrial (Mit) mutants of Caenorhabditis elegans utilize a novel metabolism. FASEB J. 2010;24:4977–4988. doi: 10.1096/fj.10-162941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Larregina AT, Morelli AE. Impact of extracellular vesicles on innate immunity. Curr Opin Organ Transplant. 2019;24:670–678. doi: 10.1097/MOT.0000000000000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Anca M, Fenoglio C, Serpente M, Arosio B, Cesari M, Scarpini EA, et al. Exosome determinants of physiological aging and age-related neurodegenerative diseases. Front Aging Neurosci. 2019;11:232. 10.3389/fnagi.2019.00232. [DOI] [PMC free article] [PubMed]
- de Lucas MP, Saez AG, Lozano E. miR-58 family and TGF-beta pathways regulate each other in Caenorhabditis elegans. Nucleic Acids Res. 2015;43:9978–9993. doi: 10.1093/nar/gkv923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo AM, Ikezu T. Extracellular vesicle biology in Alzheimer’s disease and related tauopathy. J NeuroImmune Pharmacol. 2018;13:292–308. doi: 10.1007/s11481-017-9768-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demory Beckler M, et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343–55. 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed]
- Desrochers LM, Bordeleau F, Reinhart-King CA, Cerione RA, Antonyak MA. Microvesicles provide a mechanism for intercellular communication by embryonic stem cells during embryo implantation. Nat Commun. 2016;7:11958. doi: 10.1038/ncomms11958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsemüller A-K, et al. Characterization of mast cell–derived rRNA-containing microvesicles and their inflammatory impact on endothelial cells. FASEB J. 2019;33:5457–5467. doi: 10.1096/fj.201801853RR. [DOI] [PubMed] [Google Scholar]
- Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- French KC, Antonyak MA, Cerione RA. Extracellular vesicle docking at the cellular port: Extracellular vesicle binding and uptake. Semin Cell Dev Biol. 2017;67:48–55. doi: 10.1016/j.semcdb.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go V, et al. Extracellular vesicles from mesenchymal stem cells reduce microglial-mediated neuroinflammation after cortical injury in aged Rhesus monkeys. Geroscience. 2020;42:1–17. doi: 10.1007/s11357-019-00115-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–79. 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed]
- Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE, et al. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J Proteome Res. 2009;8:1304–14. 10.1021/pr800658c. [DOI] [PMC free article] [PubMed]
- Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm. 2010;117:1. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- Habacher C, Guo Y, Venz R, Kumari P, Neagu A, Gaidatzis D, et al. Ribonuclease-mediated control of body fat. Dev Cell. 2016;39:359–69. 10.1016/j.devcel.2016.09.018. [DOI] [PubMed]
- Head BP, Patel HH, Insel PA. Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim Biophys Acta. 2014;1838:532–545. doi: 10.1016/j.bbamem.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isik M, Korswagen HC, Berezikov E. Expression patterns of intronic microRNAs in Caenorhabditis elegans. Silence. 2010;1:5. doi: 10.1186/1758-907X-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas T, Janas MM, Sapoń K, Janas T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015;589:1391–1398. doi: 10.1016/j.febslet.2015.04.036. [DOI] [PubMed] [Google Scholar]
- Jeppesen DK, et al. Reassessment of exosome composition. Cell. 2019;177:428–445. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JC, Noori A, Merrihew GE, Robbins JE, Golubeva A, Wang K, MacCoss MJ, Kaeberlein M Purification and analysis of Caenorhabditis elegans extracellular vesicles. J Vis Exp (2020a) [DOI] [PMC free article] [PubMed]
- Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Kalra H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. Ponce d_elegans—genetic quest for the fountain of youth. Cell. 1996;23:501–504. doi: 10.1016/S0092-8674(00)81024-7. [DOI] [PubMed] [Google Scholar]
- Kim DK, et al. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics. 2015;31:933–939. doi: 10.1093/bioinformatics/btu741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, Abdelmohsen K, Mustapic M, Kapogiannis D, Gorospe M. RNA in extracellular vesicles. Wiley Interdiscip Rev: RNA. 2017;8:e1413. doi: 10.1002/wrna.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Yip KW, Spence T, Liu FF. MicroRNAs in extracellular vesicles: potential cancer biomarkers. J Hum Genet. 2017;62:67–74. doi: 10.1038/jhg.2016.87. [DOI] [PubMed] [Google Scholar]
- Kowal J, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EY, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7:3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- Lee YXF, Johansson H, Wood MJA, El Andaloussi S. Considerations and implications in the purification of extracellular vesicles—a cautionary tale. Front Neurosci. 2019;13:1067. doi: 10.3389/fnins.2019.01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser SF, et al. Cell nonautonomous activation of flavin-containing monooxygenase promotes longevity and health span. Science. 2015;350:1375–1378. doi: 10.1126/science.aac9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T-R, Wang X-N, Sheng C, Li Y-X, Li FZ-T, Sun Y, et al. Extracellular vesicles as an emerging tool for the early detection of Alzheimer’s disease. Mech Ageing Dev. 2019;184:111175. [DOI] [PubMed]
- Liegeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J Cell Biol. 2006;173:949–961. doi: 10.1083/jcb.200511072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang Q, Zhang J, Li C, Miao YR, Lei Q, et al. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019;47:D89–93. [DOI] [PMC free article] [PubMed]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–14. [DOI] [PubMed]
- Mann FG, Van Nostrand EL, Friedland AE, Liu X, Kim SK. Deactivation of the GATA transcription factor ELT-2 is a major driver of normal aging in C. elegan. PLoS Genet. 2016;12:e1005956. doi: 10.1371/journal.pgen.1005956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, Walhout AJ. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008;18:2005–2015. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain S, Mathews M, Bereman MS, Rubel EW, MacCoss MJ, Noble WS. Estimating relative abundances of proteins from shotgun proteomics data. BMC Bioinformatics. 2012;13:308. doi: 10.1186/1471-2105-13-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melentijevic I, et al. C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature. 2017;542:367–371. doi: 10.1038/nature21362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishur RJ, et al. Mitochondrial metabolites extend lifespan. Aging Cell. 2016;15:336–348. doi: 10.1111/acel.12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik J, et al. The P4-ATPase ATP9A is a novel determinant of exosome release. PLoS One. 2019;14:e0213069. doi: 10.1371/journal.pone.0213069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DL, et al. Characterization of yeast extracellular vesicles: evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS One. 2010;5:e11113. doi: 10.1371/journal.pone.0011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano DJ, Kingston ER, Kim DH. Tissue expression pattern of PMK-2 p38 MAPK is established by the miR-58 family in C. elegans. PLoS Genet. 2015;11:e1004997. doi: 10.1371/journal.pgen.1004997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta R, Ułamek-Kozioł M, Januszewski S, Czuczwar SJ. Exosomes as possible spread factor and potential biomarkers in Alzheimer's disease: current concepts. Biomark Med. 2018;12:1025–1033. doi: 10.2217/bmm-2018-0034. [DOI] [Google Scholar]
- Pu M, et al. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span genes. Genes Dev. 2015;29:718–731. doi: 10.1101/gad.254144.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, et al. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth through hedgehog signaling pathway cell. Physiol Biochem. 2017;42:2242–2254. doi: 10.1159/000479998. [DOI] [PubMed] [Google Scholar]
- Rao W, Isaac RE, Keen JN. An analysis of the Caenorhabditis elegans lipid raft proteome using geLC-MS/MS. J Proteome. 2011;74:242–253. doi: 10.1016/j.jprot.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Robbins PD. Extracellular vesicles and aging. Stem Cell Investig. 2017;4:98. doi: 10.21037/sci.2017.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JC, Postupna N, Golubeva A, Keene CD, Kaeberlein M. Purification and analysis of Caenorhabditis elegans extracellular vesicles. J Visual Exp : JoVE. 2020b. 10.3791/60596. [DOI] [PMC free article] [PubMed]
- Sabatier F, Roux V, Anfosso F, Camoin L, Sampol J, Dignat-George F. Interaction of endothelial microparticles with monocytic cells in vitro induces tissue factor–dependent procoagulant activity blood. J Am Soc Hematol. 2002;99:3962–3970. doi: 10.1182/blood.v99.11.3962. [DOI] [PubMed] [Google Scholar]
- Sardar Sinha M, et al. Alzheimer's disease pathology propagation by exosomes containing toxic amyloid-beta oligomers. Acta Neuropathol. 2018;136:41–56. doi: 10.1007/s00401-018-1868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R, Patel T, Freedman JE. Circulating extracellular vesicles in human disease. N Engl J Med. 2018;379:958–966. doi: 10.1056/NEJMra1704286. [DOI] [PubMed] [Google Scholar]
- Shaw WR, Armisen J, Lehrbach NJ, Miska EA. The conserved miR-51 microRNA family is redundantly required for embryonic development and pharynx attachment in Caenorhabditis elegans. Genetics. 2010;185:897–905. doi: 10.1534/genetics.110.117515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigg MA, et al. Evolutionary proteomics uncovers ancient associations of cilia with signaling pathways. Dev Cell. 2017;43:744–762. doi: 10.1016/j.devcel.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon E, Aguirre-Tamaral A, Aguilar G, Guerrero I. Perspectives on intra- and intercellular trafficking of hedgehog for tissue patterning. J Dev Biol. 2016;4:34. doi: 10.3390/jdb4040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A, Rae R, Iatsenko I, Sommer RJ. System wide analysis of the evolution of innate immunity in the nematode model species Caenorhabditis elegans and Pristionchus pacificus. PLoS One. 2012;7:e44255. doi: 10.1371/journal.pone.0044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ED, Kaeberlein TL, Lydum BT, Sager J, Welton KL, Kennedy BK, et al. Age- and calorie-independent life span extension from dietary restriction by bacterial deprivation in Caenorhabditis elegans. BMC Dev Biol. 2008;8:49. 10.1186/1471-213X-8-49. [DOI] [PMC free article] [PubMed]
- Stoeckius M, Maaskola J, Colombo T, Rahn HP, Friedländer MR, Li N, et al. Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nat Methods. 2009;6:745–51. [DOI] [PMC free article] [PubMed]
- Sutphin GL, Kaeberlein M. Dietary restriction by bacterial deprivation increases life span in wild-derived nematodes. Exp Gerontol. 2008;43:130–135. doi: 10.1016/j.exger.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphin GL, Mahoney JM, Sheppard K, Walton DO, Korstanje R. WORMHOLE: novel least diverged ortholog prediction through machine learning. PLoS Comput Biol. 2016;12:e1005182. doi: 10.1371/journal.pcbi.1005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasugi M. Emerging roles of extracellular vesicles in cellular senescence and aging. Aging Cell. 2018;17:e12734. doi: 10.1111/acel.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur BK, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AG, et al. UFLC-derived CSF extracellular vesicle origin and proteome. Proteomics. 2018;18:1800257. doi: 10.1002/pmic.201800257. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Blaszczak AG, Chandler KJ, Watts JL, Johnson WE, Graves BJ. ETS-4 is a transcriptional regulator of life span in Caenorhabditis elegans. PLoS Genet. 2010;6:e1001125. doi: 10.1371/journal.pgen.1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA. Using C. elegans for aging research. Invertebr Reprod Dev. 2015;59:59–63. doi: 10.1080/07924259.2014.940470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Herwijnen MJC, et al. Abundantly present miRNAs in milk-derived extracellular vesicles are conserved between mammals. Front Nutr. 2018;5:81. doi: 10.3389/fnut.2018.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Barr MM. Cell-cell communication via ciliary extracellular vesicles: clues from model systems. Essays Biochem. 2018;62:205–213. doi: 10.1042/EBC20170085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Silva M, Haas LA, Morsci NS, Nguyen KCQ, Hall DH, et al. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal. Commun Curr Biol. 2014;24:519–25. [DOI] [PMC free article] [PubMed]
- Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr Biol. 2011;21:1951–1959. doi: 10.1016/j.cub.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WS, Tu BW, Chen TT, Hou SW, Tseng JT. CSmiRTar: condition-specific microRNA targets database. PLoS One. 2017;12:e0181231. doi: 10.1371/journal.pone.0181231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer—implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15:617. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- Yáñez-Mó M, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Dierking K, Esser D, Tholey A, Leippe M, Rosenstiel P, et al. Overlapping and unique signatures in the proteomic and transcriptomic responses of the nematode Caenorhabditis elegans toward pathogenic Bacillus thuringiensis. Dev Comp Immunol. 2015;51:1–9. 10.1016/j.dci.2015.02.010. [DOI] [PubMed]
- Yang W, Dierking K, Schulenburg H. WormExp: a web-based application for a Caenorhabditis elegans-specific gene expression enrichment analysis. Bioinformatics. 2016;32:943–945. doi: 10.1093/bioinformatics/btv667. [DOI] [PubMed] [Google Scholar]
- Yanos ME, Bennett CF, Kaeberlein M. Genome-wide RNAi longevity screens in Caenorhabditis elegans. Curr Genomics. 2012;13:508–518. doi: 10.2174/138920212803251391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Neuronal TORC1 modulates longevity via AMPK and cell nonautonomous regulation of mitochondrial dynamics in C. elegans. eLife. 2019;8:e49158. doi: 10.7554/eLife.49158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang W, Dong M. The miR-58 microRNA family is regulated by insulin signaling and contributes to lifespan regulation in Caenorhabditis elegans. Sci China Life Sci. 2018;61:1060–1070. doi: 10.1007/s11427-018-9308-8. [DOI] [PubMed] [Google Scholar]
- Zhou Y, et al. A secreted microRNA disrupts autophagy in distinct tissues of Caenorhabditis elegans upon ageing. Nat Commun. 2019;10:1–14. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1344 kb)
(XLSX 2372 kb)