Abstract
Chrysanthemum (Chrysanthemum morifolium) is an ideal model species for studying petal morphogenesis because of the diversity in the flower form across varieties; however, the molecular mechanisms underlying petal development are poorly understood. Here, we show that the brassinosteroid transcription factor BRI1-EMS-SUPPRESSOR 1 (CmBES1) in chrysanthemum (C. morifolium cv. Jinba) is important for organ boundary formation because it represses organ boundary identity genes. Chrysanthemum plants overexpressing CmBES1 displayed increased fusion of the outermost ray florets due to the loss of differentiation of the two dorsal petals, which developed simultaneously with the ventral petals. RNA-seq analysis of the overexpression lines revealed potential genes and pathways involved in petal development, such as CUP-SHAPED COTYLEDON (CUC2), CYCLOIDEA 4 (CYC4), genes encoding MADS-box transcription factors and homeodomain-leucine zippers (HD-Zips) and auxin pathway-related genes. This study characterizes the role of CmBES1 in ray floret development by its modulation of flower development and boundary identity genes in chrysanthemum.
Subject terms: Plant regeneration, Plant breeding
Introduction
Organogenesis is the process of tissue production by multipotent progenitor cells and is common to all multicellular organisms1. Lateral organs such as leaves and flowers are formed by lateral organ primordia, where cells are recruited from the periphery of the meristem2. In the central meristem, cell division maintains the stem cell population, while the growth of the surrounding cellular organ primordia is restricted and enters a quiescent state to form an organ boundary that separates itself from the central meristem and adjacent organs3,4. The boundary is important for organ shape because it allows for different growth patterns, and defects in the formation of organ boundaries can lead to organ-fusion phenotypes5.
Organ boundaries are regulated by complex networks comprising transcription factors and miRNAs and the spatial distribution of growth-promoting hormones such as auxin and brassinosteroids (BRs)1,6. The key genes involved in boundary regulation include CUP-SHAPED COTYLEDON 1/2/3 (CUC), LATERAL ORGAN FUSION (LOF), GROWTH REGULATING FACTOR (GRF), and LATERAL ORGAN BOUNDARY (LOB), and the corresponding mutants of these genes have organ fusion phenotypes that contribute to our understanding of the importance of their roles in boundary regulation7–10. CUC genes encode NAC family transcription factors (NAM, ATAF1/2, and CUC2), which are regulated by miR164, and can control the expression of other boundary genes7, such as KNAT6 (KNOTTED-like gene from Arabidopsis thaliana), which contributes to shoot apical meristem (SAM) maintenance and boundary establishment11. Organ boundaries are characterized by low cell expansion rates, and overexpression of Arabidopsis thaliana homeobox 12 (ATHB12) leads to increased leaf cell expansion rates, indicating that ATHB12 is a positive regulator of cell expansion12. Auxin concentration is higher in the meristem and developing primordia compared to that in the organ boundaries, where it limits the growth rate of boundary cells13. Spatial regulation of the brassinosteroid (BR) pathway is necessary for normal development of organ boundaries6. The BRASSINAZOLE RESISTANT 1 (BZR1) and BRI1-EMS SUPPRESSOR 1 (BES1) genes are components of the core regulators of BR signaling14. BR inhibits the expression of CUC at the organ boundary through transcriptional inhibition of the core transcription factor BZR1. A low level of BZR1 in boundary cells ensures the appropriate expression of CUC and the normal formation of organ boundary shapes5. The development of lateral organs, such as flowers, is regulated by various transcription factors15–18. Specifically, the MADS-box family of transcription factors plays a major role in the control of flower architecture and induction15. Ectopic expression of SUPPRESSOR OF OVEREXPRESSION OF CO1 (GhSOC1), which is an AtSOC1-like1 MADS-box gene, in Gerbera hybrida led to a partial loss of floral organ identity16. lfy mutants, which have a mutation in the floral meristem identity gene LEAFY (LFY), show partial flower-to-shoot conversion17, and AINTEGUMENTA-LIKE 6 (AIL6) is critical for cell differentiation in flowers18.
Chrysanthemum flowers are typical representatives of capitula, with many small individual flowers clustered at the top of the inflorescence axis. The shape resembles a large single flower but is actually composed of many flowers19. The capitulum contains two types of florets: outer ray florets and inner disc florets20. The ray florets are bilaterally symmetrical, with two dorsal petals that are degenerated and three fused petals that form an elongated ventral ligule, while the disc florets are radially symmetrical with five equal petals21. The shape of the ray floret is important for the chrysanthemum flower type and can be classified as flat, spoon, or tubular according to the degree of corolla tube merging (CTMD), which ranges from 0 to 0.2, 0.2 to 0.6, and 0.6 to 1.0 22.
Many species in the Asteraceae family are ideal for the study of flower development. Mutants for inflorescence morphology, of which the tubular-rayed (tub) mutant of sunflower is a typical example23, have helped us to understand the floral traits of the capitulum. The tub mutant is caused by the loss of function of the HaCYC2c (CYCLOIDEA-like) gene, which leads to the generation of tubular ray florets23. Overexpression of SvRAY2 (CYC-like genes) in Senecio vulgaris produced tubular ray florets24. In G. hybrida, GhCYC2, GhCYC3, and GhCYC4 have redundant functions in regulating ray flower identity and promoting petal development in ray flowers25. In addition, CmCYC2c controls ray floret identity in chrysanthemum20.
Although the function of BES1 in Arabidopsis has been well characterized, it is poorly understood in chrysanthemum. Here, we present evidence for the importance of CmBES1 in the petal development of chrysanthemum ray florets. We show that when CmBES1 is overexpressed in transgenic chrysanthemum plants, the two dorsal petals do not degenerate but instead develop simultaneously with the ventral petals and increase the degree of fusion of the outermost ray florets. RNA-seq analysis revealed that several genes associated with flower development were differentially expressed, including CUC2, CYC4, genes encoding MADS-box transcription factors and HD-ZIP proteins, and auxin pathway-related genes. Overall, the present study links CmBES1 to ray floret development through the regulation of flower development and organ boundary identity gene expression in chrysanthemum.
Results
CmBES1 sequence characteristics
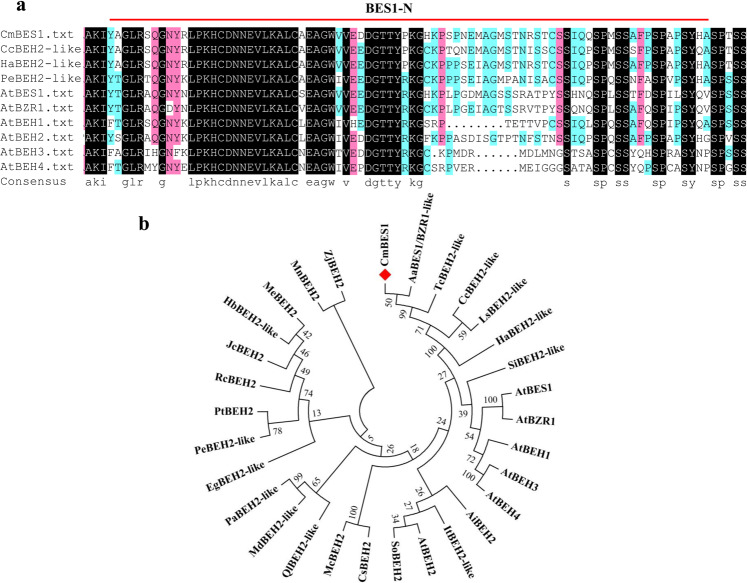
To better understand the effects of BES1 family genes on chrysanthemum, a homolog of the BES1 gene was identified. The genomic sequence of CmBES1 was cloned from leaves of C. morifolium cv. Jinba; the gene had a predicted open reading frame (ORF) of 921 bp and encoded a 306-amino acid protein. A BLAST nucleotide sequence search of CmBES1 in The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/) revealed that the BES1 family of genes in Arabidopsis most closely related to CmBES1 was the AtBES1 family. The deduced polypeptide of CmBES1 was similar in various plant species and had a highly conserved BES1-N domain in the N terminus (Fig. 1a, Supplementary Fig. S1). Phylogenetic analysis (Fig. 1b) showed that CmBES1 and AaBES1/BZR1-like were closely related, with an amino acid sequence similarity of 71.12%. The levels of peptide identity between CmBES1 and other proteins were 70.48% (TcBEH2-like), 66.83% (HaBEH2-like), 65.16% (CcBEH2-like), and 45.11% (AtBES1).
Fig. 1. Deduced amino acid sequence comparison and phylogenetic analysis of CmBES1.
a Amino acid sequence alignment of CmBES1 with BES1 sequences from various plant species. The single red lines indicate the conserved BES1-N region in BES1-like proteins. The black color indicates 100% identity; red, 75% identity; and blue, 50% identity. The sequences compared to CmBES1 were Arabidopsis thaliana AtBES1 (AT1G19350), AtBZR1 (AT1G75080), AtBEH1 (AT3G50750.1), AtBEH2 (AT4G36780), AtBEH3 (AT4G18890), and AtBEH4 (AT1G78700); Helianthus annuus BES1/BZR1 homolog protein 2-like (HaBEH2-like, LOC110908961); Cynara cardunculus BES1/BZR1 homolog protein 2-like (CcBEH2-like, LOC112512677); and Populus euphratica BES1/BZR1 homolog protein 2-like (PeBEH2-like, LOC105107647). b Phylogenetic tree comprising the following CmBES1 and BES1 family proteins: Arabidopsis thaliana AtBES1 (AT1G19350), AtBZR1 (AT1G75080), AtBEH1 (AT3G50750.1), AtBEH2 (AT4G36780), AtBEH3 (AT4G18890), and AtBEH4 (AT1G78700); Arachis ipaensis AiBEH2 (XP_016201823.1); Artemisia annua AaBES1/BZR1-like (PWA37330.1); Cucumis sativus CsBEH2 (XP_004143497.1); Cynara cardunculus CcBEH2-like (LOC112512677); Eucalyptus grandis EgBEH2-like (NP_001306904.1); Helianthus annuus HaBEH2-like (LOC110908961); Hevea brasiliensis HbBEH2-like (XP_021646974.1); Ipomoea triloba ItBEH2-like (XP_031095547.1); Jatropha curcas JcBEH2 (XP_012079081.1); Lactuca sativa LsBEH2-like (XP_023759843.1); Malus domestica MdBEH2-like (XP_008338783.2); Manihot esculenta MeBEH2 (XP_021632984.1); Momordica charantia McBEH2 (XP_022132946.1); Morus notabilis MnBEH2 (XP_024020611.1); Populus euphratica PeBEH2-like (LOC105107647); Populus trichocarpa PtBEH2 (XP_002310201.1); Prunus avium PaBEH2-like (XP_021809451.1); Quercus lobata QlBEH2-like (XP_030949816.1); Ricinus communis RcBEH2 (XP_002525100.1); Sesamum indicum SiBEH2-like (XP_011097209.1); Spinacia oleracea SoBEH2 (XP_021835401.1); Tanacetum cinerariifolium TcBEH2-like (GEY43666.1); and Ziziphus jujuba ZjBEH2 (XP_015890203.1). The phylogenetic tree was constructed using the neighbor-joining method and bootstrap test with 1000 replicates. The divergence of each branch is indicated by the bootstrap values
Subcellular localization, expression patterns, and transcriptional activity analysis of CmBES1
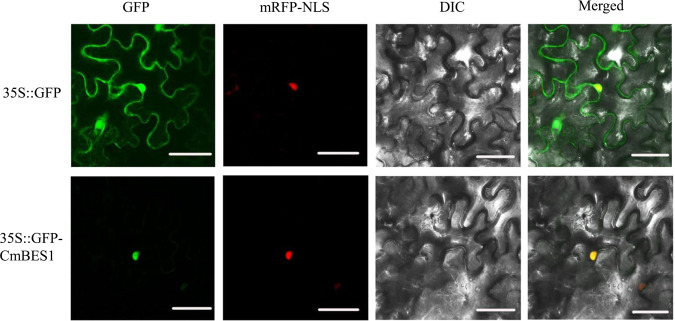
To examine the subcellular localization of CmBES1, a 35S::GFP-CmBES1 construct was developed and introduced into Nicotiana benthamiana epidermal cells, along with a 35S::GFP construct as a control. The 35S::GFP construct alone was detected in both the cytoplasm and the nucleus of the tobacco cells (Fig. 2), while the CmBES1-GFP fusion protein was detected only in the nucleus. These data suggest that CmBES1 is localized in the nucleus.
Fig. 2. Subcellular localization of CmBES1 in N. benthamiana epidermal cells.
The images correspond to the GFP (left), mRFP-NLS, DIC (middle), and merged (right). 35S::D53-RFP was used as a nuclear marker (mRFP-NLS). Bars: 50 μm
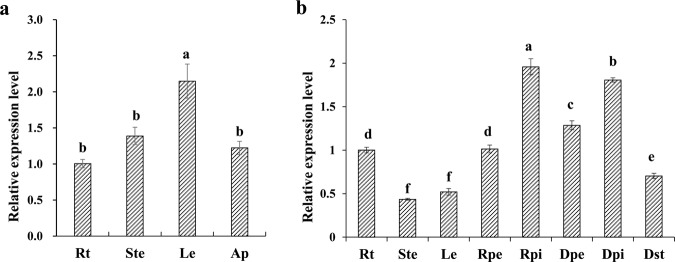
We further examined changes in CmBES1 gene expression by quantitative real-time polymerase chain reaction (qRT-PCR) during vegetative and reproductive periods of Jinba chrysanthemum plant organs to predict their possible functions (which have not been predicted) (Fig. 3). CmBES1 transcripts were detected in all organs at all developmental stages. During the vegetative period, the transcript level of CmBES1 was abundant in the leaves and showed similar expression in the roots, stems, and shoot apex (Fig. 3a). During reproductive growth, the transcript level of CmBES1 was high in the ray floret pistils and disc floret pistils; moderate in the ray floret petals, disc floret petals, and roots; and low in the stems, leaves, and disc floret stamens (Fig. 3b).
Fig. 3. Expression pattern analysis of CmBES1.
a Expression patterns of CmBES1 during vegetative growth in the roots (Rt), stems (Ste), leaves (Le) and shoot apex (Ap). bCmBES1 expression patterns during the reproductive stage in the roots (Rt), stems (Ste), leaves (Le), ray floret petals (Rpe), ray floret pistil (Rpi), disc floret petals (Dpe), disc floret pistil (Dpi), and disc floret stamens (Dst). EF-1α expression in chrysanthemum was used as an internal control. The error bars indicate SEs (n = 3). The differences were analyzed by Duncan’s multiple range test. The different lowercase letters indicate significant differences (P < 0.05)
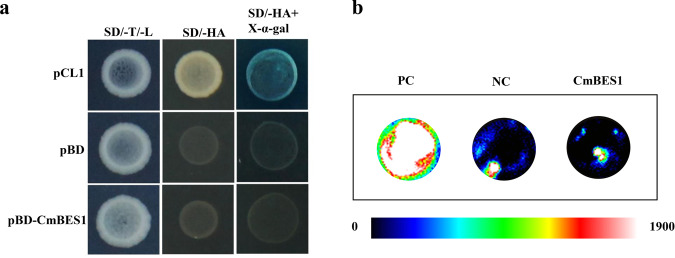
To test the transactivation activity of the CmBES1 protein, a transactivation assay was performed in yeast. Yeast expressing the pCL1 plasmid grew well on SD/-His-Ade media and turned blue on SD/-His-Ade media supplemented with X-a-gal (Fig. 4a); however, the negative control pGBKT7 and pGBKT7-CmBES1 constructs did not turn blue on this selective medium. To further confirm these results, we transfected CmBES1 as an effector plasmid into Arabidopsis protoplasts, and the results showed that the relative luciferase (LUC) activity of 35S::GAL4DB-AtARF5 was significantly higher than that of 35S::GAL4DB-CmBES1 (Fig. 4b, Supplementary Fig. S2, P < 0.01). The relative LUC activity of 35S::GAL4DB-CmBES1 was lower than that of 35S::GAL4DB but was not significantly different (Fig. 4b, Supplementary Fig. S2), indicating that CmBES1 acts as a repressor of transcription.
Fig. 4. Analysis of CmBES1 transactivation.
a Transcriptional activity of CmBES1 measured in a yeast assay system. Y2H cells expressing pCL1 grew on SD/-His-Ade media and served as positive controls. Y2H cells expressing pGBKT7 could not grow on this media and served as negative controls. SD/-T/-L: SD/-Trp media (pGBKT7 and genes) or SD/-Leu media (pCL1), SD/-HA: SD/-His/-Ade media, SD/-HA + X-α-gal: SD/-His/-Ade media+X-α-gal. b Relative luciferase activities in Arabidopsis mesophyll protoplasts after transfection with 35 S::GAL4DB-CmBES1; the 35 S::GAL4DB-AtARF5 construct served as a positive control, and 35 S::GAL4DB served as a negative control
CmBES1 is involved in chrysanthemum petal development
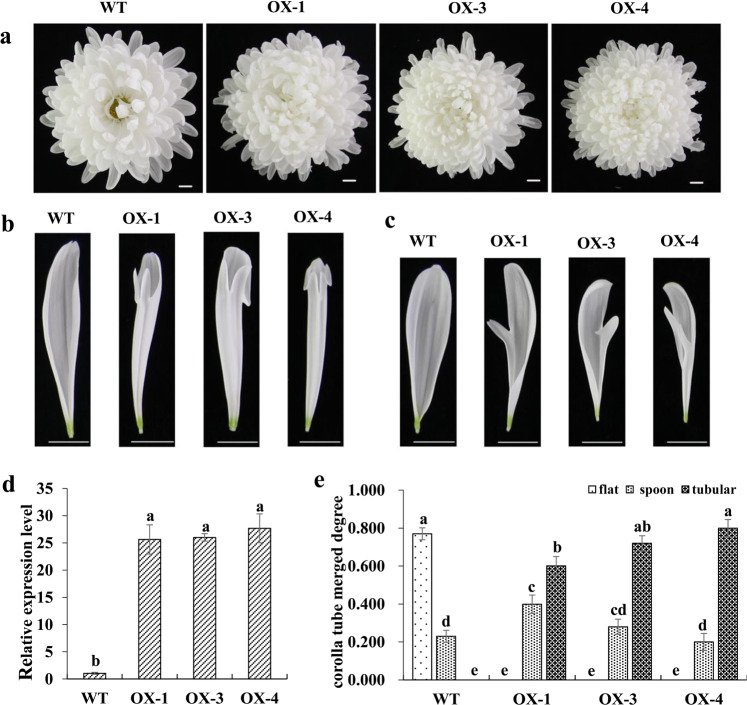
To further investigate the function of CmBES1 in chrysanthemum growth, a population of seven CmBES1-overexpressing lines was validated by confirming the increase in CmBES1 expression in transgenic lines compared with wild-type (WT) plants by qRT-PCR analysis (Supplementary Fig. S3), and three representative CmBES1-overexpressing lines (OX-1, OX-3, and OX-4) were selected for functional analysis (Supplementary Fig. S3, Fig. 5d). We evaluated the inflorescences of OX-CmBES1 transgenic plants and found that the ray floret shape was different from that of WT plants (Fig. 5a–c). The CTMD of the outermost ray florets increased, while the degree of the middle ray floret fusion did not change; however, the shape of the petals was altered at the tips of ray florets, and the petal tips exhibited petalized protrusions (Fig. 5a–c). To further study the changes in the degree of merging of the outermost ray florets, 15 of the outermost ray florets from each inflorescence of 15 individual plants were randomly selected from each transgenic line to measure the degree of ray floret fusion. The chrysanthemum ray floret shape can be flat, spoon shaped, or tubular, with the CTMD ranging from 0 to 0.2 (flat), 0.2 to 0.6 (spoon shaped), and 0.6 to 1.0 (tubular)22. Among the WT chrysanthemum plants, 77.0% of the total number of the outermost ray florets were flat, and 23.0% were spoon shaped; there were no tubular outermost ray florets (Fig. 5e). In transgenic overexpression lines, the outermost ray florets were mainly tubular. The proportion of spoon ray florets in the transgenic lines ranged from 20.0 to 39.9%, and the proportion of tubular ray florets ranged from 60.1 to 80.0%.
Fig. 5. Ectopic expression of CmBES1 affected the corolla tube merged degree.
a Top view of wild type (WT) and transgenic overexpression (OX) lines 1, 3, and 4. The positive transgenic lines had a relatively high degree of corolla tube merging (corolla tube length/ray floret length). Bar: 1 cm. b Top view of the outermost ray florets of WT and OX lines. Bar: 1 cm. c Top view of the middle ray florets of WT and OX transgenic plants. Bar: 1 cm. d Expression level of CmBES1 in WT and OX transgenic plants. The error bars indicate SEs (n = 3). The differences were analyzed by Duncan’s multiple range test. The different lowercase letters indicate significant differences (P < 0.05). e Corolla tube merged degree between the outermost ray florets. The error bars indicate SEs (n > 100). The differences were analyzed by Duncan’s multiple range test. The different lowercase letters indicate significant differences (P < 0.01)
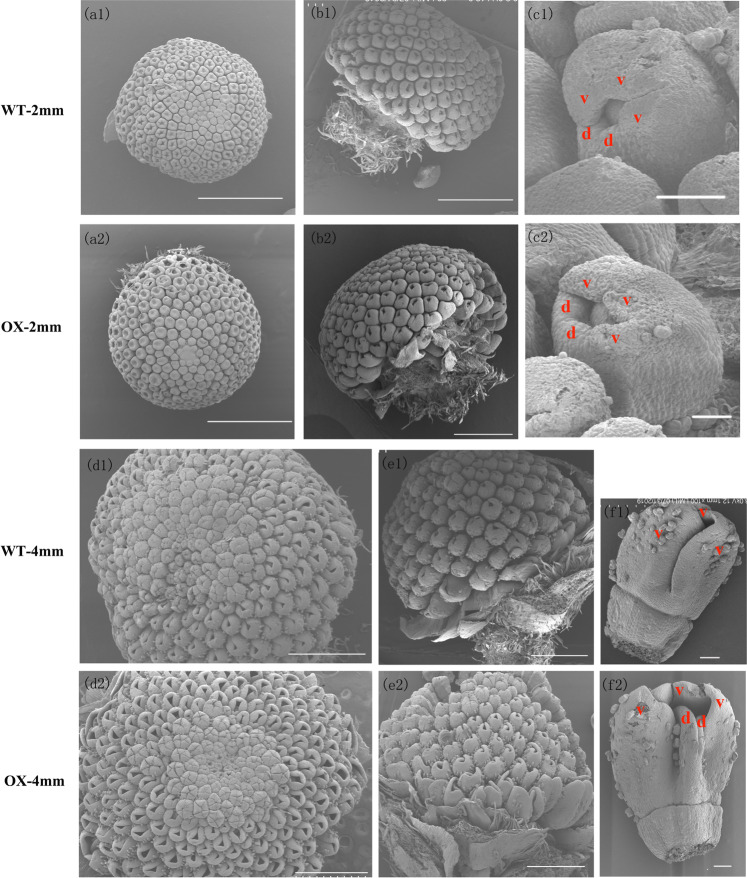
To detect the onset of this different phenotype, images of the OX-4 transgenic line were taken with a scanning electron microscope (SU8010 device, Hitachi, Japan). A whole inflorescence (Fig. 6a1, a2, b1, b2) or only the outermost ray florets were sampled when inflorescences were ~2 and ~4 mm long. When inflorescences whose diameter was ~2 mm were sampled, the OX-4 transgenic outermost ray florets were not significantly different from those of the WT (Fig. 6c1, c2). Petals were sampled when their diameter was ~4 mm (Fig. 6f1, f2). In the WT line, the peripherally located ray florets were bilaterally symmetrical to two rudimentary dorsal petals and an elongated ventral ligule was formed by three fused petals. In the transgenic line, the two dorsal petals did not degenerate and developed simultaneously with the ventral ligule, which increased the degree of fusion of the outermost ray florets. We also imaged the fresh samples at the ~4, ~6, and ~8 mm stages as well as the relevant petals using light microscopy (Supplementary Fig. S4) to evaluate their development, and characterize the fusion phenotype of the transgenic chrysanthemum plants. The degree of fusion of the outermost ray florets was increased in transgenic lines compared with the WT. These results suggested that overexpression of CmBES1 affects the degree to which the ray florets fused to form the corolla tube and alters the pattern of the outermost ray florets.
Fig. 6. Morphological analysis of the inflorescence of the wild type and CmBES1-overexpressing transgenic lines at the 2 and 4 mm stages.
a1, a2 Top view of the inflorescence at the 2 mm stages. Bar: 1 mm. b1, b2 Side view of the inflorescence at the 2 mm stage. Bar: 1 mm. c1, c2 SEM images of ray florets at the 2 mm stage. Bar: 100 μm. d and v in red letters represent dorsal and ventral petals, respectively. d1, d2 Top view of the inflorescence at the 4 mm stage. Bar: 1 mm. e1, e2 Side view of the inflorescence at the 4 mm stage. Bar: 1 mm. f1, f2 SEM images of ray florets at the 4 mm stage. Bar: 100 μm. d and v in red letters represent dorsal and ventral petals, respectively. Ectopic expression of CmBES1 increased the corolla tube merged degree
Global expression of downstream CmBES1-associated transcripts in chrysanthemum outermost ray florets
To better understand the mechanisms by which CmBES1 mediates the regulation of floral development, a large-scale screen of genes differentially expressed in WT and OX-4 transgenic lines was performed using RNA sequencing (RNA-seq). The RNA from the outermost ray florets of WT and OX-4 plants was sampled from inflorescences with a diameter of ~4 mm. A summary of the sequence reads is given in Supplementary Table S1, with the contents of each library ranging from 62.74 to 65.52 million clean reads. After assembly, the outcome involved a set of 98,155 unigene sequences with a mean length of 987 nucleotides (nt); the N50 (genome splicing quality) was 1407 nt. The total number of unigenes obtained from the six libraries (WT-1, WT-2, WT-3, OX4-1, OX4-2, and OX4-3) was 65,984, 59,431, 62,636, 67,405, 64,908, and 65,476, respectively. Differentially expressed genes (DEGs) were identified from pairwise comparisons between WT and OX-4, with a false discovery rate (FDR) less than 0.05. A total of 9722 DEGs were obtained from the transcriptomic data, of which 4986 were upregulated and 4736 were downregulated (Supplementary Fig. S5a). In addition, NR species distribution, KEGG pathway classification, and GO classification analyses were performed (Supplementary Fig. S5).
Based on the CmBES1 regulation of the CTMD of the ray florets, we focused our analysis on the DEGs related to organ boundary growth and flower development (Supplementary Table S2), which revealed genes related to the auxin pathway, such as the homologs of AUXIN RESPONSE FACTOR (ARF) and INDOLEACETIC ACID-INDUCED PROTEIN (IAA). The expression levels of some boundary genes varied between the WT and overexpression lines; the central boundary gene is a homolog of the Helianthus annuus protein CUP-SHAPED COTYLEDON 2-like (CL12058.Contig3_All), which was downregulated in OX-4 compared to WT. Additionally, other boundary-related genes, such as CmKNAT2 (CL3389.Contig2_All) and the WUSCHEL-related homeobox homologs WOX3 (CL9508.Contig2_All) and WOX4 (Unigene1685_All), were also downregulated in OX-4. CmCYC2 controls ray floret identity in chrysanthemum20, and we found the CmCYC4 homolog (CL4616.Contig3_All) also had relatively low expression, suggesting that it also plays a role in regulating the degree of chrysanthemum ray floret fusion. The MADS-box transcription factor plays an important role in flower development because it regulates the characteristics of floral organs and influences their subsequent development26. Homologs of the Agamous-like MADS-box protein and MADS-box transcription factor were differentially expressed, with CmBES1 being overexpressed. Homeodomain-leucine zipper (HD-Zip) genes that contain homeodomains with highly conserved leucine zipper motifs are unique to plants27. Arabidopsis thaliana homeobox 12 (ATHB12) is an HD-Zip I gene that is mainly expressed in leaves and stems and represses the expression of gibberellin 20 oxidase 1 (GA20ox1), thereby inhibiting stem elongation during the early development of inflorescence stems12. Five chrysanthemum homeobox (HB) genes were found in our transcriptomic data, as well as other genes related to flower development that were also evaluated.
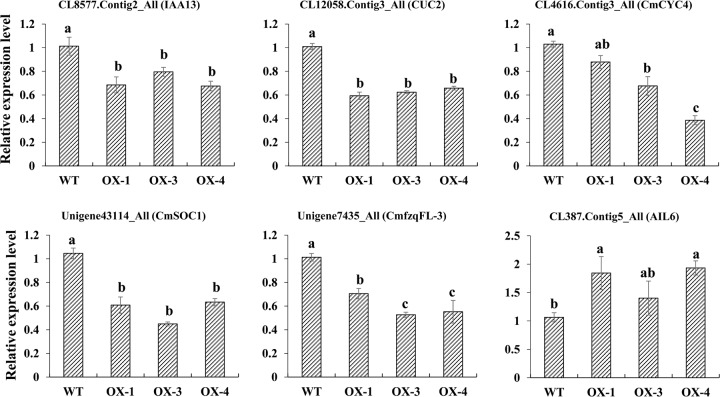
The above results led us to examine the expression of genes that can serve as markers for ray floret petal development. The expression of several identified genes, such as CUC2, CYC4, SOC1-like, CmfzqFL-3, AIL6, and IAA13, was confirmed by qRT-PCR (Fig. 7). RNA was prepared from the outermost ray florets of WT, OX-1, OX-3, and OX-4 samples collected from florets whose diameter was ~4 mm. Although the expression fold changes indicated by RNA-seq and qRT-PCR differed in detail, the overall sequencing data were reliable (Fig. 7).
Fig. 7. Validation of the RNA-seq classification of differential transcription via qRT-PCR.
The error bars indicate SEs (n = 3). The differences were analyzed by Duncan’s multiple range test. The different lowercase letters indicate significant differences (P < 0.05)
Discussion
Boundary formation is critical to the function of mature organs because it allows for correct patterning and separation of different activities. These boundaries are regulated by complex networks that include transcription factors and the spatial distribution of growth-promoting hormones such as auxin and BRs1,6. In Arabidopsis, the core gene involved in BR signaling and the node of multiple signal cascades is BZR1, which is closely related to organ fusion. Indeed, the bzr1-1D mutant has general defects in various organ boundaries, such as stems that bend toward the axillary branches and cauline leaves, bending of siliques at the silique-pedicel junction, and fused stamens; however, its petals develop normally5. Little is known about the genetic and molecular mechanisms underlying the key transcription factor regulating BR signaling during organ development in chrysanthemum. We showed that the chrysanthemum BES1 gene is involved in regulating the fusion of the ray floret corolla but does not affect the development of other organs, which is significantly different from the fusion regulated by BZR1 in Arabidopsis thaliana5. Overexpression of CmBES1 in transgenic chrysanthemum plants promoted an increased degree of fusion of the outermost ray florets because the two dorsal petals developed simultaneously with the ventral ligule rather than differentiating. In the WT cut flower chrysanthemum Jinba, more than 70% of the outermost ray florets are flat, and only part of the higher degree of fusion presents a spoon-like phenotype. The transgenic overexpression line does not produce flat outermost ray florets; instead, they are mainly tubular. The pattern of CmBES1 accumulation in the ray floret pistil is consistent with its inhibitory role in boundary formation and its activation role during cell growth promotion.
To gain a better understanding of how CmBES1 affects organ development, we conducted RNA-seq-based transcript profiling to identify potential genes and pathways involved in organ fusion. In the present study, RNA-seq-based transcript profiling showed that the expression levels of a number of gene homologs to known components of flower development and boundary regulation were differentially expressed in transgenic chrysanthemum overexpressing CmBES1 compared with those of WT plants. In Arabidopsis, mutants with increased BR signaling showed severe organ fusions across a wide range of boundary tissues. These phenotypes are caused by BZR1 suppression of the organ boundary identity genes CUC and LOF. BZR1 is mainly expressed in the SAM but is expressed at low levels at the boundary, which ensures the proper expression of boundary identity genes during normal boundary formation5. BES1 in chrysanthemum is a typical transcription factor with the highest homology with AtBES1, compared with other BES1 family genes (AtBEH1-AtBEH4, AtBZR1) in Arabidopsis. Its subcellular localization is in the nucleus, with no transcriptional activation activity, and mainly in the ray floret pistil during the reproductive growth period. The activity pattern of CmBES1 is similar to that of Arabidopsis BZR1; CmBES1 inhibits CUC2 transcription, causing the downregulation of CUC2 at the boundary of ray florets and the appearance of the petal fusion phenotype and architectural alteration. As the dorsal petals of the ray florets elongated dramatically, the expression of several genes related to flower development also changed significantly in the transgenic chrysanthemum plants. MADS-box genes play a major role in the control of flower architecture15; for example, GhSOC1 in G. hybrida controls floral organ identity16, and the APETALA 1 (AP1) gene in Arabidopsis, which has an A-class homeotic function, is required for sepal and petal development28. Homologs of MADS-box genes (SOC1-like, AP1-like) showed significant changes in expression in transgenic chrysanthemum. We also identified several HD-ZIP genes; given the role of ATHB12 in cell expansion12, it is tempting to speculate that the identified chrysanthemum HD-ZIP protein may function in the regulation of petal development (Supplementary Table S2). Of primary importance for flower primordium initiation is the ARF MONOPTEROS protein (MP/ARF5); MP induces the expression of LFY, which specifies floral fate, and of two ANT/AIL6 transcription factors, which are key regulators of floral growth29. RNA-seq-based transcript profiling suggests that the roles of the LFY-like (CmfzqFL-3), AIL6, and ARF-like genes are significantly altered in transgenic chrysanthemum and could be related to the fusion of the ray floret corolla. These findings suggest that CmBES1 expression influences ray floret development by modulating flower development and the expression of genes related to boundary regulation.
In addition to the changes in ray floret petal development, we also found that the number of ray florets in transgenic CmBES1-OX lines is different significantly from those of the WT (Supplementary Fig. S6). There was no significant difference in the total number of florets in the capitulum between WT and OX lines; however, in the transgenic lines, the number of disc florets was significantly decreased compared to those in the WT, but the number of ray florets significantly increased, indicating that the BES1 expression level affects ray floret identity. Previous studies have shown that variation in the capitulum type is dependent on the replacement of ray florets by disc florets or vice versa30. The phenotype of the fusion of the ray floret corolla in transgenic chrysanthemum plants overexpressing CmBES1 is similar to the phenotype of the tubular-rayed (tub) mutant of sunflower, while the phenotype of disc florets that turn into ray florets is similar to that of the double-flowered (dbl) sunflower mutants23. Research has shown that misexpression of the HaCYC2c gene causes a dbl phenotype, whereas loss of gene function causes a tub phenotype23. In fact, the CYC gene has been widely studied in the Asteraceae family, and overexpression of GhCYC2 in G. hybrida causes disc florets to present a morphology similar to that of ray florets25. However, in S. vulgaris, overexpression of SvRAY1 (a CYC-like gene) repressed ray floret development and overexpression of SvRAY2 produced tubular ray florets24. Transgenic analysis showed that overexpression of CmCYC2c in chrysanthemum led to a significant increase in the number of ray florets per inflorescence compared to the number in WT20. Further molecular analysis suggests that CmCYC2-like transcription factors may interact with each other or bind to the promoter to regulate floral symmetry development in Chrysanthemum morifolium31. Differences in transgenic phenotypes in the Asteraceae family suggest that the CYC gene may play a role in promoting or inhibiting the development of ray florets. The results indicate that CYC plays a critical role in controlling ray flower identity and floral organ development in the Asteraceae family. RNA-seq analysis showed that the CYC homolog CYC4 was downregulated in the transgenic CmBES1-OX lines, suggesting that it might be a new candidate gene that regulates ray flower identity and petal development. The relationship between CmBES1 and CYC4 and their regulation of inflorescence development will need further investigation.
Some genes involved in the BR biosynthetic pathway are suppressed by the final product of the BR signaling pathway through a feedback loop, such as CYP90B1 (DWF4), Constitutive photomorphogenesis and dwarfism (CPD), CYP85A1 (BR6ox1), and Rotundifolia 3 (ROT3), which are downregulated by BZR1 when it directly binds to the promoter region of these genes32. Through further mining of the transcriptomic data, we found that the expression level of CPD (CL5486.Contig2_All) was upregulated, while the expression levels of BR6ox1 (CL13153.Contig1_All) homologs and ROT3 (Unigene11129_All) were slightly upregulated but did not significantly differ (Supplementary Table S3, Supplementary Fig. S7). Based on the above data, we found that CmBES1 feedback regulates the expression of BR synthesis genes in chrysanthemum, suggesting that this gene is similar to the homologous gene of Arabidopsis but that there are some differences in its regulation of downstream effectors that may be species specific.
Our findings provide insight into the mechanism of ray flower identity and petal development through CmBES1, which regulates flower development and boundary identity genes in chrysanthemum. This study provides important evidence for the molecular mechanism underlying inflorescence development and is potentially relevant for molecular breeding strategies.
Materials and methods
Plant materials and growth conditions
Samples of the C. morifolium cv. Jinba, which is a popular variety as a cut flower on the market, was obtained from the Chrysanthemum Germplasm Resource Conservation Centre (Nanjing Agricultural University, China). The plants were cultured in a greenhouse under a 16-h light/8-h dark photoperiod (80–100 μmol/m2/s irradiation) with 70% relative humidity and a day/night temperature of 26/15 °C. Petals were sampled from the outermost whorl of the inflorescence and used for both RNA extraction and morphological analysis.
Cloning and phylogenetic analysis of CmBES1
Total RNA was extracted from leaves and used for cDNA synthesis as previously described33. A CmBES1-specific primer pair (BES1-F and -R, Supplementary Table S4) was designed with Primer 5.0 software (www.bbioo.com/Soft/2005/114.htm) according to the Unigene19979 sequence in the Jinba chrysanthemum transcriptome to amplify the ORF. The resulting PCR product was purified and cloned into pMD19-T (TaKaRa, Tokyo, Japan) for sequencing. Homologs of BES1 in other species were obtained using BLAST searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and the BES1 transcription factor family members in Arabidopsis were obtained from TAIR (http://www.arabidopsis.org/). Phylogenetic trees were constructed using MEGA 5.0 software with the neighbor-joining method and bootstrap test using 1000 replicates34. The BES1 family genes comprised the following: Arabidopsis thaliana AtBES1 (AT1G19350), AtBZR1 (AT1G75080), AtBEH1 (AT3G50750.1), AtBEH2 (AT4G36780), AtBEH3 (AT4G18890), and AtBEH4 (AT1G78700); Arachis ipaensis AiBEH2 (XP_016201823.1); Artemisia annua AaBES1/BZR1-like (PWA37330.1); Cucumis sativus CsBEH2 (XP_004143497.1); Cynara cardunculus CcBEH2-like (LOC112512677); Eucalyptus grandis EgBEH2-like (NP_001306904.1); Helianthus annuus HaBEH2-like (LOC110908961); Hevea brasiliensis HbBEH2-like (XP_021646974.1); Ipomoea triloba ItBEH2-like (XP_031095547.1); Jatropha curcas JcBEH2 (XP_012079081.1); Lactuca sativa LsBEH2-like (XP_023759843.1); Malus domestica MdBEH2-like (XP_008338783.2); Manihot esculenta MeBEH2 (XP_021632984.1); Momordica charantia McBEH2 (XP_022132946.1); Morus notabilis MnBEH2 (XP_024020611.1); Populus euphratica PeBEH2-like (LOC105107647); Populus trichocarpa PtBEH2 (XP_002310201.1); Prunus avium PaBEH2-like (XP_021809451.1); Quercus lobata QlBEH2-like (XP_030949816.1); Ricinus communis RcBEH2 (XP_002525100.1); Sesamum indicum SiBEH2-like (XP_011097209.1); Spinacia oleracea SoBEH2 (XP_021835401.1); Tanacetum cinerariifolium TcBEH2-like (GEY43666.1); and Ziziphus jujuba ZjBEH2 (XP_015890203.1). The conserved domain of the CmBES1 sequence was inferred by querying the Conserved Domains Database (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and DNAMAN software was used for multiple sequence alignment.
Subcellular localization, expression patterns, and transcriptional activity analysis of CmBES1
The amplicon of CmBES1 and the pENTR 1A vector (Invitrogen, Carlsbad, CA, USA) were digested with SalI and EcoRI endonucleases (the primers used are listed in Supplementary Table S4) and then ligated with T4 DNA ligase (TaKaRa, Tokyo, Japan) before sequencing. The plasmid pENTR1A-CmBES1 was recombined with the binary vector pMDC43 (35S::GFP) using an LR reaction (Invitrogen, Carlsbad, CA, USA) to obtain a 35S::GFP-CmBES1 green fluorescent protein fusion construct35. The 35S::GFP-CmBES1 and free GFP (35S::GFP) plasmids were transiently coexpressed together with a nuclear marker (35S::D53-RFP construct)36 into Nicotiana benthamiana leaves. The leaf epidermal cells were monitored with a Zeiss LSM 780 confocal microscope (Zeiss, Jena, Germany) after 3 days.
Temporal and spatial expression characteristics of CmBES1 during the vegetative and reproductive periods were analyzed. Samples from the vegetative period comprised roots (Rt), stems (Ste), leaves (Le), and the shoot apex (Ap), and were collected from rooted chrysanthemum seedlings at the 9–10 leaf stage grown under LDs (16 h light/8 h dark). Samples from the reproductive period comprised Rt, Ste, Le, ray floret petals (Rpe), ray floret pistil (Rpi), disc floret petals (Dpe), disc floret pistil (Dpi), and disc floret stamens (Dst) and were collected from WT chrysanthemum plants at the early opening stage. The EF-1α (GenBank: AB548817.1) gene in chrysanthemum was used as the internal expression control.
The transcriptional activity of CmBES1 was examined using a yeast assay system and a luminescence assay. The ORFs (1 to 918 bp nucleotides) of the CmBES1 gene were amplified using primers that added EcoR I and Sal I sites. The primers used are listed in Supplementary Table S4. The PCR products and pGBKT7 vector were digested with EcoR I and Sal I and then ligated with T4 DNA ligase (TaKaRa, Tokyo, Japan) to produce a pGBKT7-CmBES1 expression vector. The pGBKT7-CmBES1, pCL1 (positive control), and pGBKT7 (negative control) constructs were subsequently transformed into a Y2H Gold Saccharomyces cerevisiae strain (Clontech, Tokyo, Japan). The yeast transformants expressing pGBKT7-CmBES1 or pGBKT7 were incubated on SD/-Trp media at 30 °C for 3 days, and those expressing pCL1 were incubated on SD/-Leu media. The transformed yeast cells were then transferred to SD/-His-Ade media for 8 h in the presence of X-α-gal to screen for positive transformants.
For the luminescence assay, a 35S::GAL4DB-CmBES1 plasmid was constructed with the pENTR1A-CmBES1 plasmid (the primers used are listed in Supplementary Table S4) by the LR reaction (Invitrogen). The transient expression assay was performed in Arabidopsis protoplasts as previously described37. For each transformation, 5 μg of 35S::GAL4DB-AtARF5 (positive control), 35S::GAL4DB (negative control) or 35S::GAL4DB-CmBES1 and 5 μg of 5× GAL4-LUC plasmid (luminescence reporter) were used. The luciferase activity was measured as previously reported38. Three independent experiments were performed.
Genetic transformation of chrysanthemum
The pMDC43-CmBES1 overexpression plasmid was introduced into WT chrysanthemum Jinba via Agrobacterium-mediated genetic transformation39. After screening by PCR with the appropriate vector primers (Supplementary Table S4), positive lines were further analyzed with qRT-PCR using BES1-RT-F/R primers to determine the relative expression levels. Each sample was tested for three biological and three technical replicates. Overexpression and WT chrysanthemum lines during the same growth period were grown in a growth chamber for 1 week under 16 h light/8 h dark day/night conditions at 25 °C/18 °C (day/night) before they were transferred to a greenhouse and subjected to standard management practices.
Morphological and histological analyses
The outermost ray florets at the open-flower stage of WT and CmBES1 transgenic chrysanthemum plants were used to measure the CTMD, which was defined as the corolla tube length/ray floret length and measured as described previously22. The main characteristics compared between the ray florets and disc florets are listed in Supplementary Table S5. The whole inflorescence (either 2 or 4 mm in diameter) or only the outermost ray florets was sampled and fixed with 2.5% (v/v) glutaraldehyde after removing the bracts. The samples were dried to the critical point and then coated with gold before they were observed by scanning electron microscopy (SU8010 device, Hitachi, Japan). Fresh inflorescence and ray florets were also imaged with an S8AP0 optical microscope (Leica Camera AG, Germany).
RNA extraction and RNA-seq analysis
The outermost ray florets of WT and CmBES1 transgenic chrysanthemum were sampled at 4 mm and because of their small size, each sample consisted of nine inflorescences. Total RNA was extracted using an RNA Isolation Kit (Waryong, Beijing, China) and subjected to an Illumina HiSeq™2000 instrument located at the Beijing Genomics Institute (Shenzhen, China; http://www.genomics.cn/index) for sequencing according to the manufacturer’s instructions. Adaptor sequences and low-quality reads were removed from the raw sequence data, and the transcriptomic analysis was conducted with the Trinity program40. For unigene annotations, homology searches employed the NCBI nonredundant nucleotide (NT), NCBI nonredundant protein (NR), Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), COG (Cluster of Orthologous Groups), and Pfam databases. In the present analysis, an FDR below 0.05 was identified as the criterion for DEGs41.
Quantitative RT-PCR analysis
To verify the expression characteristics of CmBES1 and the expression differences of key DEGs in the three transgenic lines and WT chrysanthemum, qRT-PCR analysis was performed. The primers used for qRT-PCR were designed by Primer 5 software (Supplementary Table S4). The reference gene EF-1α (GenBank: KF305681) was used as an expression control. Each sample was represented by three biological and three technical replicates. The specific experimental qRT-PCR methods used have been described previously42. The relative transcript levels of DEGs were calculated using the 2−ΔΔCT method43.
Statistical analysis
Analysis of variance by Duncan’s multiple range test was used to identify significant differences between genotypes and/or treatments. The statistical analyses were performed by SPSS v17.0 software (SPSS Inc., Chicago, IL).
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31930100), the National Natural Science Foundation of China (31701959), the Natural Science Fund of Jiangsu Province (BK20170717), and the Fundamental Research Funds for the Central Universities (KJQN201815).
Author contributions
P.C., J.J., and L.D. designed the experiments, and P.C. performed most of the experiments and wrote the manuscript; F.C. and S.C. conceived the study, participated in its design, and contributed to revisions of the manuscript. Other authors participated in the preparation of the experimental materials and helped with the RNA extractions. All the authors have read and approved the final version of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-00351-8).
References
- 1.Richardson A, Hake S. Drawing a line: grasses and boundaries. Plants. 2019;8:4. doi: 10.3390/plants8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brand U, Hobe M, Simon R. Functional domains in plant shoot meristems. Bioessays. 2001;23:134–141. doi: 10.1002/1521-1878(200102)23:2<134::AID-BIES1020>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Aida M, Tasaka M. Morphogenesis and patterning at the organ boundaries in the higher plant shoot apex. Plant Mol. Biol. 2006;60:915–928. doi: 10.1007/s11103-005-2760-7. [DOI] [PubMed] [Google Scholar]
- 4.Hamant O, et al. Developmental patterning by mechanical signals in Arabidopsis. Science. 2008;322:1650–1655. doi: 10.1126/science.1165594. [DOI] [PubMed] [Google Scholar]
- 5.Gendron JM, et al. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl Acad. Sci. USA. 2012;109:21152–21157. doi: 10.1073/pnas.1210799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hibara KI, et al. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell. 2006;18:2946–2957. doi: 10.1105/tpc.106.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee DK, Geisler M, Springer PS. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development. 2009;136:2423–2432. doi: 10.1242/dev.031971. [DOI] [PubMed] [Google Scholar]
- 10.Lee BH, Jeon JO, Lee MM, Kim JH. Genetic interaction between GROWTH-REGULATING FACTOR and CUP-SHAPED COTYLEDON in organ separation. Plant Signal. Behav. 2015;10:e988071. doi: 10.4161/15592324.2014.988071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belles-Boix E, et al. KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell. 2006;18:1900–1907. doi: 10.1105/tpc.106.041988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Son O, et al. ATHB12, an ABA-inducible homeodomain-leucine zipper (HD-Zip) protein of Arabidopsis, negatively regulates the growth of the inflorescence stem by decreasing the expression of a gibberellin 20-oxidase gene. Plant Cell Physiol. 2010;51:1537–1547. doi: 10.1093/pcp/pcq108. [DOI] [PubMed] [Google Scholar]
- 13.Heisler MG, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 14.Li QF, et al. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim. Biophys. Acta Gene Regul. Mech. 2018;1861:561–571. doi: 10.1016/j.bbagrm.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Ng M, Yanofsky MF. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001;2:186–195. doi: 10.1038/35056041. [DOI] [PubMed] [Google Scholar]
- 16.Ruokolainen S, Ng YP, Albert VA, Elomaa P, Teeri TH. Over-expression of the Gerbera hybrida At-SOC1-like1 gene Gh-SOC1 leads to floral organ identity deterioration. Ann. Bot. 2011;107:1491–1499. doi: 10.1093/aob/mcr112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-N. [DOI] [PubMed] [Google Scholar]
- 18.Krizek BA, Eadd M. AINTEGUMENTA-LIKE6 regulates cellular differentiation in flowers. Plant Mol. Biol. 2012;78:199–209. doi: 10.1007/s11103-011-9844-3. [DOI] [PubMed] [Google Scholar]
- 19.Funk, V. A., Susanna, A., Stuessy, T. & Bayer, R. Systematics, Evolution and Biogeography of Compositae (International Association for Plant Taxonomy, 2009).
- 20.Huang D, et al. Identification and characterization of CYC-like genes in regulation of ray floret development in Chrysanthemum morifolium. Front. Plant Sci. 2016;7:1633. doi: 10.3389/fpls.2016.01633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bremer, K. & Anderberg, A. A. Asteraceae: Cladistics & Classification (Timber Press, Inc., 1994).
- 22.Song X, et al. Quantitative classification of the morphological traits of ray florets in large-flowered Chrysanthemum. HortScience. 2018;53:1258–1265. doi: 10.21273/HORTSCI13069-18. [DOI] [Google Scholar]
- 23.Chapman MA, et al. Genetic analysis of floral symmetry in Van Gogh’s sunflowers reveals independent recruitment of CYCLOIDEA genes in the Asteraceae. PLoS Genet. 2012;8:3. doi: 10.1371/journal.pgen.1002628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim M, et al. Regulatory genes control a key morphological and ecological trait transferred between species. Science. 2008;322:1116–1119. doi: 10.1126/science.1164371. [DOI] [PubMed] [Google Scholar]
- 25.Juntheikki-Palovaara I, et al. Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae) Plant J. 2014;79:783–796. doi: 10.1111/tpj.12583. [DOI] [PubMed] [Google Scholar]
- 26.Ding L, et al. Comprehensive characterization of a floral mutant reveals the mechanism of hooked petal morphogenesis in Chrysanthemum morifolium. Plant Biotechnol. J. 2019;17:2325–2340. doi: 10.1111/pbi.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schena M, Davis RW. Structure of homeobox-leucine zipper genes suggests a model for the evolution of gene families. Proc. Natl Acad. Sci. USA. 1994;91:8393–8397. doi: 10.1073/pnas.91.18.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi N, et al. A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell. 2013;24:271–282. doi: 10.1016/j.devcel.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 30.Gillies, A. C., Cubas, P., Coen, E. S. & Abbott, R. J. in Developmental Genetics and Plant Evolution (eds. Cronk, Q. C. B. et al.) Ch. 6 (Taylor & Francis, 2002).
- 31.Yuan, C. et al. CmCYC2-like transcription factors may interact with each other or bind to the promoter to regulate floral symmetry development in Chrysanthemum morifolium. Plant Mol. Biol.103, 159–171 (2020). [DOI] [PubMed]
- 32.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren L, et al. A transcriptomic analysis of Chrysanthemum nankingense provides insights into the basis of low temperature tolerance. BMC Genomics. 2014;15:844. doi: 10.1186/1471-2164-15-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhou F, et al. D14–SCF D3-dependent degradation of D53 regulates strigolactone signalling. Nature. 2013;504:406–410. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 38.Song A, et al. An isoform of eukaryotic initiation factor 4E from Chrysanthemum morifolium interacts with chrysanthemum virus B coat protein. PLoS ONE. 2013;8:e57229. doi: 10.1371/journal.pone.0057229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, et al. The CmTCP20 gene regulates petal elongation growth in Chrysanthemum morifolium. Plant Sci. 2019;280:248–257. doi: 10.1016/j.plantsci.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren L, et al. Transcriptomic analysis of differentially expressed genes in the floral transition of the summer flowering chrysanthemum. BMC Genomics. 2016;17:673. doi: 10.1186/s12864-016-3024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.