Abstract
The global health emergency generated by coronavirus disease 2019 (COVID-19) has prompted the search for preventive and therapeutic treatments for its pathogen, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). There are many potential targets for drug discovery and development to tackle this disease. One of these targets is the main protease, Mpro or 3CLpro, which is highly conserved among coronaviruses. 3CLpro is an essential player in the viral replication cycle, processing the large viral polyproteins and rendering the individual proteins functional. We report a biophysical characterization of the structural stability and the catalytic activity of 3CLpro from SARS-CoV-2, from which a suitable experimental in vitro molecular screening procedure has been designed. By screening of a small chemical library consisting of about 150 compounds, the natural product quercetin was identified as reasonably potent inhibitor of SARS-CoV-2 3CLpro (Ki ~ 7 μM). Quercetin could be shown to interact with 3CLpro using biophysical techniques and bind to the active site in molecular simulations. Quercetin, with well-known pharmacokinetic and ADMET properties, can be considered as a good candidate for further optimization and development, or repositioned for COVID-19 therapeutic treatment.
Keywords: SARS-CoV-2 3CL protease, Structural stability, Drug discovery
Highlights
-
•
SASR-CoV-2 3CLpro shows cooperative unfolding in which dimers or tetramers predominate depending on the pH.
-
•
SARS-CoV-2 3CLpro shows a pH-dependent hydrolytic activity that correlates with its structural stability.
-
•
SARS-CoV-2 3CLpro is an appropriate target for drug discovery given its high sequence identity among different coronaviruses.
-
•
Quercetin has been identified as a SARS-CoV-2 3CLpro inhibitor by an activity-based experimental screening.
-
•
Experimental and computational target engagement for quercetin was gathered by ITC, spectroscopy, TSA, and molecular docking.
1. Introduction
December 2019 was the starting point for a global pandemic, caused by a newly identified coronavirus, SARS-CoV-2, rising to dramatic numbers of infections and deaths worldwide [1,2]. Much effort and many resources have been devoted to combat this health emergency on many fronts: 1) better diagnosis and patient management [3]; 2) improving protective and palliative care [4]; 3) reducing damage in the different organs affected by the virus and minimizing collateral harmful effects of the immune response [5]; and 4) developing preventive vaccines and therapeutic treatments [6], to mention the more relevant. In addition, very strict and severe social and economic measures, with the potential danger of causing a profound global crisis, have been taken [7]. In this context, even if an effective vaccine is promisingly close to becoming available, drugs are needed for infected people [8]. From experience with other pathogens, we have learnt that the best therapeutic strategy consists in administrating a combination of several drugs acting through different mechanisms, in order to minimize the probability of drug resistance appearance [9]. At the same time, cost-efficient drugs (if possible, developed by repositioning or repurposing known drugs) are urgently needed for reaching all the population [10].
The SARS-CoV-2 genome is 82% identical to that of SARS-CoV, the causative virus of the 2002 coronavirus outbreak [11]. The genomic differences are reflected in different infectivity and mortality rates for SARS-CoV-2 [12]. Among all the potential protein targets within coronaviruses, the main protease (Mpro, or 3C-like protease, 3CLpro) stands out as a highly conserved gene (96% sequence identity between the SARS-CoV-2 and SARS-CoV), making 3CLpro a good target for developing effective drugs against SARS-CoV-2 (and other future coronavirus variants) [13,14]. Together with PLpro, 3CLpro is responsible for the processing of the viral polyproteins synthesized from the viral RNA after infection, rendering the individual viral proteins active and functional [15]. Remarkably, 3CLpro must process at least 11 cleavage sites on polyprotein 1ab (replicase 1ab), most of them sharing a common cleavage sequence LQ↓(S/A/G), quite unusual for human proteases [16]. Therefore, molecules able to inhibit SARS-CoV-2 3CLpro in a specific manner would hinder viral replication and represent appropriate candidates to develop low-toxicity drugs against this devastating pathogen [17].
Searching for drugs against a protein target always benefits from a comprehensive structural and functional characterization [18]. Experimental and computational knowledge gathered along this process is instrumental for identifying weak points and key interaction sites in the target, and developing suitable screening procedures for finding molecules interfering with or modulating its function [19]. Here we have characterized some structural and functional features of SARS-CoV-2 3CLpro by employing biophysical techniques (spectroscopy and calorimetry), and we have implemented a fast in vitro screening procedure based on 3CLpro hydrolytic activity using a Förster resonance energy transfer (FRET) substrate, which allows the ready identification of small molecules blocking the enzymatic activity of 3CLpro. Starting from a small chemical library consisting of about 150 compounds, collected from successful screening programs previously carried out in our laboratory, a natural compound was identified as an inhibitor of SARS-CoV-2 3CLpro with enough potency (inhibition constant Ki ~ 7 μM) to be considered a good candidate for further optimization and development. Quercetin, with known pharmacokinetic and ADMET properties, exhibits anti-oxidant, anti-allergic, anti-inflammatory, and anti-proliferative indications, and can be directed for drug repositioning.
2. Materials & methods
2.1. Protein expression and purification
A pET22b plasmid containing the SARS-CoV-2 His-tagged 3CLPRO sequence was transformed into BL21 (DE3) Gold E. coli strain. Bacterial cultures were grown in 250 mL of LB/ampicillin (100 μg/mL) media at 37 °C overnight with gentle shaking. Then, 4 L of LB/ampicillin (100 μg/mL) were inoculated and incubated under the same conditions until reaching OD = 0.6 at a wavelength of 600 nm. Protein expression was induced with 1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG) at 18 °C for 5 h. Cells were harvested by centrifugation at 4 °C for 10 min at 10,000 rpm (using a Beckman Coulter Avanti J-26 XP Centrifuge) and then resuspended in lysis buffer (sodium phosphate 50 mM, pH 8, sodium chloride 500 mM). Cell rupture was achieved by sonication (using a Sonics Vibra-Cell Ultrasonic Liquid Processor) in ice, and 20 U/mL of benzonase (Merck-Millipore) were added to remove nucleic acids. To remove cellular debris, the extract was centrifuged at 4 °C for 30 min at 20,000 rpm, and filtered through a 0.45 μm-pore membrane. After increasing imidazole concentration up to 10 mM, the protein was purified using immobilized metal ion affinity chromatography (IMAC) in a ÄKTA FPLC System (GE Healthcare Life Sciences) using a cobalt HiTrap TALON column (GE-Healthcare Life Sciences), eluting in an imidazole 10–250 mM gradient. Purity was checked by SDS-PAGE, and pure protein fractions were dialyzed to remove imidazole and reach the protein storage condition (sodium phosphate 50 mM, pH 8, sodium chloride 150 mM). The identity of the protein was assessed by mass spectrometry (LC-ESI-MS/MS). Potential DNA contamination was assessed by determining the UV absorption 260 nm/280 nm ratio. An extinction coefficient of 32,890 M−1 cm−1 at 280 nm was employed for quantification. When needed, buffer exchange was performed using a PD-10 pre-packed Sephadex G-25 M (GE Healthcare Life Sciences).
2.2. Circular dichroism and fluorescence spectroscopy
Circular dichroism (CD) spectra were recorded in a Chirascan spectropolarimeter (Applied Photophysics) at 25 °C. Far-UV spectrum was recorded at wavelengths between 190 and 260 nm in a 0.1-cm path-length cuvette. Near-UV spectrum was recorded at wavelengths between 250 and 310 nm in a 1-cm path-length cuvette. Protein concentration was 10 μM in all cases. Fluorescence measurements were performed in a Cary Eclipse fluorescence spectrophotometer (Agilent Technologies), monitoring the intrinsic tryptophan fluorescence of the protein solution at 2 μM concentration. An excitation wavelength of 290 nm was used, with excitation and emission bandwidths of 5 nm, and recording fluorescence emission between 300 and 400 nm. All spectroscopic measurements were made in sodium phosphate 50 mM, pH 8.
Thermal denaturations were monitored by CD and fluorescence, employing a protein concentration of 10 μM and 2 μM, respectively, and performing thermal scans with a scanning rate of 60 °C/h.
2.3. Dynamic light scattering
Hydrodynamic radius of SARS-CoV-2 3CLpro was measured at different pH values for estimating the size of the oligomeric native state in a DynaPro NanoStar equipment (Wyatt Technology), employing a protein concentration of 3 μM at pH 5, 7 and 8.
2.4. Size-exclusion chromatography
Self-association state of SARS-CoV-2 3CLpro was assessed employing an ÄKTA FPLC (GE Healthcare) using a Superdex Increase 75 10/30 chromatographic column, at pH 8 and 150 mM NaCl.
2.5. Differential scanning calorimetry
Thermal stability of SARS-CoV-2 3CLpro was assessed by temperature unfolding transitions monitored by high-precision differential scanning calorimetry (DSC). The partial molar heat capacity of the protein in solution was measured as a function of temperature in an Auto-PEAQ-DSC (MicroCal, Malvern-Panalytical). Experiments were performed with a protein solution at a concentration of 14 μM in buffer, and scanning from 15 to 95 °C at a rate of 60 °C/h. Different buffers were employed to get protein stability information under different conditions: acetate 50 mM at pH 5, and phosphate at pH 7 and pH 8.
2.6. Protein unfolding
2.6.1. Model-free calorimetric analysis
The first approach in DSC experimental analysis consists of applying a model-free data analysis for discriminating between different possibilities: two-state unfolding, non-two-state unfolding, and oligomer unfolding of the protein. From the thermogram (excess molar heat capacity of the protein as a function of the temperature, <ΔCP(T)>), the calorimetric unfolding enthalpy, ΔHcal, the unfolding temperature, Tm, and the maximal unfolding heat capacity, CP,max, can be readily evaluated, from which it is possible to calculate the van't Hoff enthalpy, ΔHvH:
| (1) |
From the ratio ΔHvH/ΔHcal different possibilities may arise: 1) if ΔHvH/ΔHcal = 1, the thermogram would reflect a single transition and the protein unfolds according to a two-state model (i.e., the protein contains a single energetic domain); 2) if ΔHvH/ΔHcal < 1, the thermogram would reflect at least two (partially) overlapping transitions and the protein unfolds according to a non-two-state model (i.e., the protein contains at least two domains which unfold in an independent manner); and 3) if ΔHvH/ΔHcal > 1, the thermogram would reflect an unfolding transition coupled to subunit dissociation (i.e., the protein is oligomeric and unfolds into monomers).
In the case of SARS-CoV-2 3CLpro, the thermograms under the different conditions assayed showed that ΔHvH/ΔHcal > 1 and, therefore, the protein is not monomeric and dissociates upon thermal unfolding. In fact, according to the crystallographic structure of the unliganded species, the protein is dimeric in its native state, although it has been reported that SARS-CoV-1 3CLpro can populate tetramers and octamers.
2.6.2. Cooperative homo-oligomeric protein unfolding into isolated unfolded monomers
The conformational equilibrium is governed by the equilibrium constant, KU:
| (2) |
where Nn is the oligomeric native state constituted of n monomeric subunits, and U is the unfolded state of each identical monomeric subunit. The total protein concentration can be expressed (per monomer) as:
| (3) |
from which the molar fractions of the different species can be calculated:
| (4) |
Given the equilibrium constant and the total protein concentration, Eq. (3) can be solved numerically for the unknown [U], and the molar fraction of both protein species can be calculated at any temperature, from which the excess average molar unfolding enthalpy, <ΔH>(T) can be determined:
| (5) |
where ΔHU is the enthalpy of the unfolded state, considering that of the native state as a reference. The temperature derivative of the excess molar unfolding enthalpy is the excess unfolding heat capacity at constant pressure, <ΔCP>(T):
| (6) |
taking into account the temperature dependence of the different magnitudes:
| (7) |
where ΔSU, ΔGU and ΔCP,U are the entropy, the Gibbs energy and the heat capacity of the unfolded state (taking the native state as a reference), respectively, and T0 is an appropriate reference temperature. In this case, T0, is the temperature at which the unfolding Gibbs energy is zero, and does not coincide with the temperature for maximal heat capacity, Tmax, nor with the temperature for half denaturation, T1/2 (i.e., the temperature at which FU = 0.5, or median temperature in the thermogram). This model particularized to n = 1 corresponds to the two-state model, for which T0 is the apparent unfolding temperature, Tm, which is usually very close to both Tmax and T1/2.
For a spectroscopically monitored unfolding of a homo-oligomeric protein, the procedure is similar, but the final observable quantity is the spectroscopic signal, S(T), which can be calculated as:
| (8) |
where SN(T) and SU(T) are the intrinsic spectroscopic signal values for the native and unfolded protein states, and can be considered linear functions of the temperature:
| (9) |
2.7. Proteolytic activity assay
In vitro catalytic activity of 3CLpro was determined using a fluorescence resonance energy transfer (FRET) cleavage continuous assay with the peptide substrate (Dabcyl)KTSAVLQSGFRKME(Edans)-NH2 (Biosyntan GmbH) [[20], [21], [22]]. Briefly, enzyme was at 0.2–2 μM final concentration in sodium phosphate 50 mM, NaCl 150 mM, at pH 8, and the enzymatic reaction was initiated by adding the substrate at 20 μM final concentration, in a final volume of 100 μL. Fluorescence was continuously measured in a FluoDia T70 microplate reader (Photon Technology International) for 20 min (excitation wavelength, 380 nm; emission wavelength, 500 nm). Enzyme activity was quantified as the initial slope of the time evolution curve of the fluorescence signal. The slope ratio between the activity in the presence and absence of a given compound provides the percentage of inhibition. For comparison, the hydrolytic activity was also measured at different pH values. The Michaelis-Menten constant, Km, and the catalytic rate constant or turnover number, kcat, were estimated by measuring the initial enzymatic rate at different substrate concentrations at fixed 0.2 enzyme concentration and applying non-linear regression data analysis.
2.8. Activity-based screening
Screening of a small chemical library was performed based on the catalytic activity continuous FRET assay. In a 96-well microplate, enzyme at 1 μM final concentration was incubated in the presence of chemical compounds (at 125 μM final concentration) solubilized previously at 5 mM concentration in DMSO. The final concentration of DMSO was 2.5% v/v. The same amount of DMSO was added to control wells (no compounds added). Several hits were identified as those compounds reducing the enzymatic activity below a threshold (average enzymatic activity of controls minus twice their standard deviation). In order to determine the inhibition constants of the selected compounds, 2-fold serial dilutions of each compound were tested (ranging from 0 to 125 μM) under the same conditions described above.
2.9. Inhibition assay
To assess the in vitro inhibition potency of the selected compounds, the inhibition constants were estimated from the experimental curves. Inhibition curves were obtained by measuring the enzyme activity as a function of compound concentration: monitoring the substrate fluorescence emission as a function of time, fixing the enzyme concentration at 2 μM, the substrate concentration at 20 μM, and varying the compound concentration from 0 to 125 μM, while maintaining constant the percentage of DMSO. The enzymatic activity was quantitated as the initial slope of the substrate fluorescence emission time curve, and was plotted as a function of compound concentration. Non-linear regression analysis employing a simple inhibition model allowed us to estimate the apparent inhibition constant for both compounds, according to this equation:
| (10) |
where v is the initial slope of the enzymatic activity trace at a (free) compound concentration [I], Km is the Michaelis-Menten constant for the enzyme-substrate interaction, [S] is the substrate concentration, and Ki app is the apparent inhibition constant for the compound. If the inhibitor acts through a purely competitive mechanism, the previous equation can be substituted by the following one:
| (11) |
where Ki is the intrinsic (i.e., substrate concentration-independent) inhibition constant.
2.10. Thermal shift assay
For the thermal shift assay (TSA), SYPRO Orange (Thermo Fisher Scientific) was employed as an extrinsic fluorescent probe for reporting protein unfolding. A reaction mixture containing 3CLpro at 1 μM of final concentration in sodium phosphate 50 mM, pH 8, and SYPRO Orange at a final concentration of 5×, was freshly prepared and dispensed into 96-well microplates (96-well PCR plate, non-skirted, from 4titude) at a final volume of 100 μL. Compound from a 5 mM stock solution in pure DMSO were added to the mixture of protein and fluorescence probe to a final concentration of 250 μM each. Controls were considered by adding the same volume of 100% DMSO instead of any compound. Unfolding curves were registered from 25 to 99 °C at a 1 °C/min scan rate in a Stratagene Mx3005P qPCR real-time thermal cycler (Agilent Technologies). Thermal denaturation was followed by fluorescence emission of the fluorescent probe, using 492 and 610 nm as excitation and emission wavelength filters, respectively. The apparent midpoint unfolding temperature, Tm, was calculated in each well as the temperature of maximum slope (i.e., the inflection point), and compared to the controls. To assess the concentration dependence of the stability change induced by the selected compound, 2-fold serial dilutions (ranging from 0 to 125 μM) for each compound were assayed by following the same protocol described above.
2.11. Isothermal titration calorimetry
The interaction between the compound and 3CLpro was further assessed by isothermal titration calorimetry. Calorimetric titrations were performed using a high precision Auto-iTC200 calorimeter (MicroCal, Malvern-Panalytical). Protein at 15 μM and compound at 150 μM were dissolved in sodium phosphate 50 mM, pH 8, with and without NaCl 150 mM. Compound was titrated into protein solution by performing 19 injections of 2 μL each, and mixed using a stirring speed of 750 rpm, maintaining a spacing between injections of 150 s, and applying a reference power of 10 μcal/s. The association constant, Ka, the binding enthalpy, ΔH, and the binding stoichiometry, N, were estimated through non-linear least squares regression analysis of the data, by using a model considering a single ligand binding site, implemented in Origin 7.0 (OriginLab). The dissociation constant, Kd, the binding Gibbs energy, ΔG, and the binding entropy, −TΔS, were obtained from basic thermodynamic relationships.
2.12. Molecular docking
Molecular docking was performed by using the simulation software AutoDock Vina [23] and the supporting suite AutoDock Tools [24]. The structure of 3CLpro was extracted from the entries 6Y2E and 6Y2F [22] of the Protein Data Bank (PDB), which contain the crystallographic conformation of the protein in unliganded form and with an α-ketoamide inhibitor bound in the active site, respectively. The protein structures were considered in all cases free from any ligand and water molecules. A few missing residues in the entry 6Y2F were reconstructed in silico, and the structure of quercetin was also built by using the molecular editor Avogadro [25]. A blind search was carried out on the whole protein surface, by considering a volume of size 50 Å × 60 Å × 60 Å. Extensive sampling was performed in all cases, using an exhaustiveness 16 times larger than the value normally recommended [26].
3. Results
3.1. SARS-CoV-2 3CLpro is properly folded
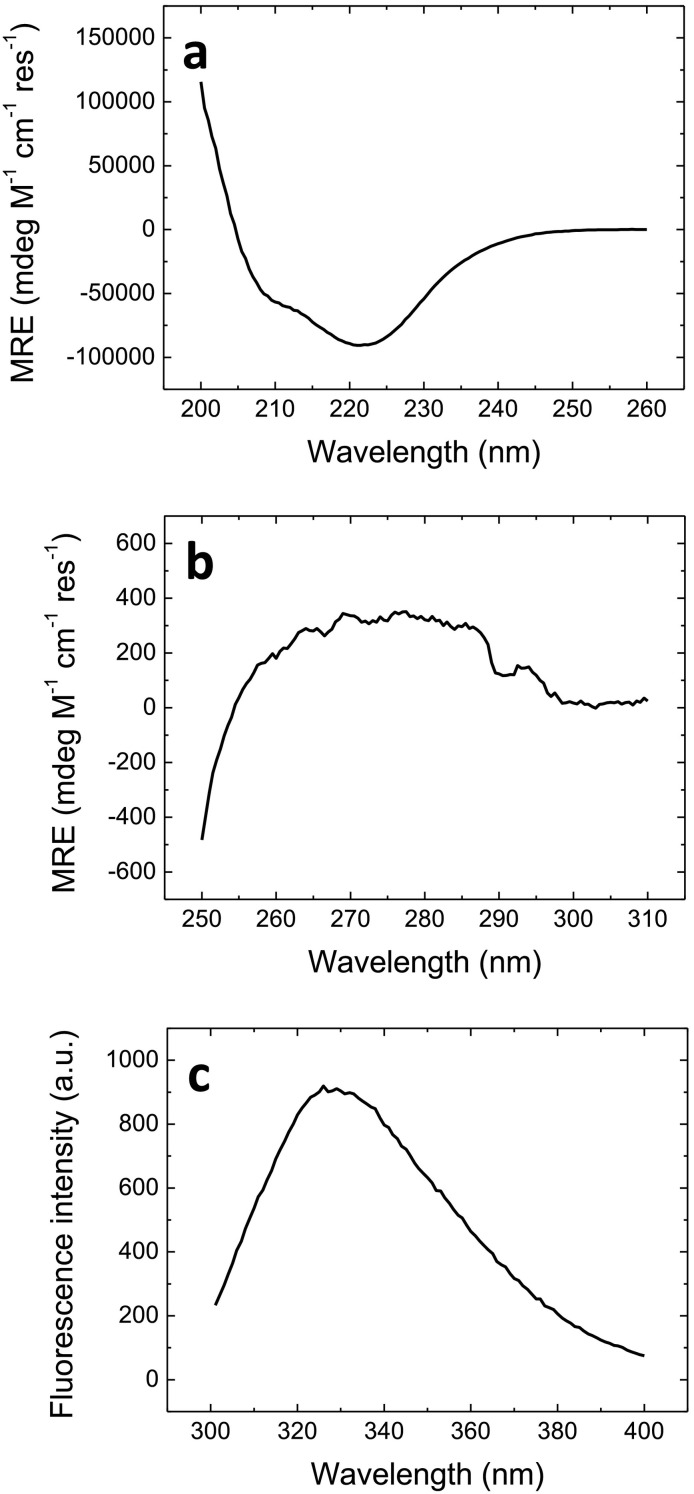
Recombinant 3CLpro was expressed in E. coli at sufficiently high yield for initiating a biophysical characterization and an activity-based molecular screening (Fig. S1). The far-UV circular dichroism (CD) spectrum showed negative bands around 208 and 222 nm, typical of proteins with α-helical and β-sheet content (Fig. 1a), in agreement with the crystallographic structure [22]. The near-UV CD spectrum indicated aromatic residues, especially tryptophans, are enclosed within an asymmetric environment (Fig. 1b). The fluorescence spectrum monitoring the intrinsic emission of the tryptophans (there are three tryptophan residues, two located in the α-helical domain and one in the β-sheet domain) showed a maximum around 330 nm, confirming that some of them are partially exposed to the solvent (Fig. 1c). These results confirmed 3CLpro adopted a folded conformation with well-defined secondary and tertiary structures.
Fig. 1.
Spectroscopic properties of SARS-CoV-2 3CLpro. (a) Far-UV CD at 10 μM protein concentration. (b) Near-UV CD at 10 μM protein concentration. (c) Fluorescence at 2 μM protein concentration. All the spectra were recorded at pH 8.
3.2. SARS-CoV-2 3CLpro self-associates
3CLpro from other coronaviruses (SARS-CoV and MERS) dimerizes, and such dimerization is known to be essential for the enzymatic activity; even higher-level oligomerization structures (tetramers and octamers) have been reported [27]. Among other experimental techniques providing information about protein oligomerization (such as analytical ultracentrifugation and gel filtration chromatography), thermal denaturation monitored by spectroscopy or calorimetry can be used to detect the self-association of proteins, since the thermal unfolding of monomeric and oligomeric proteins show markedly different features [28].
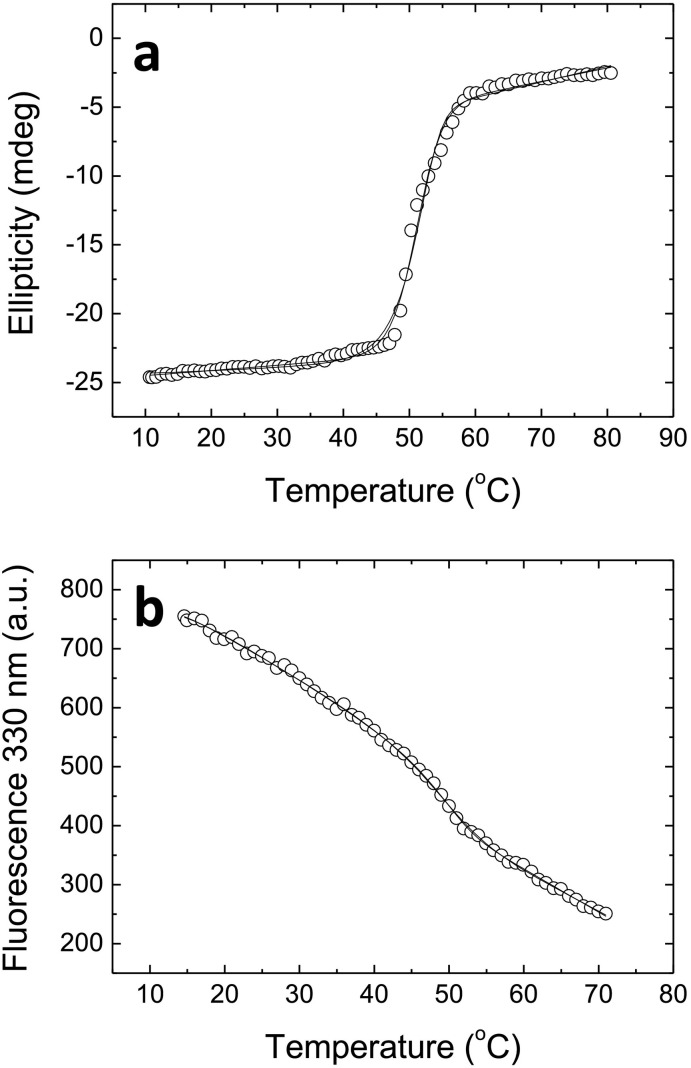
Thermal unfolding of 3CLpro followed by far-UV CD showed a single transition with an apparent unfolding temperature close to 51 °C (Fig. 2a). The same process followed by fluorescence also showed a single transition with an apparent unfolding temperature close to 48.5 °C (Fig. 2b). The small difference in the unfolding temperature could be justified by the use of a different protein concentration in CD and fluorescence assays and considering the oligomer-monomer equilibrium coupled to the unfolding process. Non-linear regression analysis of the unfolding traces according to different two-state models for a monomeric (n = 1, see Eq. (1)), a dimeric (n = 2), a tetrameric protein (n = 4), and an octameric protein (n = 8) were performed (see Materials & methods section). These experiments confirmed 3CLpro adopts a well-folded conformation under native conditions and becomes unstructured upon temperature stress. However, it was not possible to discriminate which model reproduces better the experimental data, because the fitting curves from the different models overlapped. The fitting was marginally better (according to the residual sum of squares; see Table S1) for the two-state monomer model in the case of CD unfolding, and for the two-state tetramer model in the fluorescence unfolding. Therefore, spectroscopic unfolding traces were not sensitive enough for discriminating the self-association nature of 3CLpro native state.
Fig. 2.
Spectroscopic thermal unfolding of SARS-CoV-2 3CLpro. (a) Far-UV CD thermal unfolding monitoring the (raw) ellipticity at 222 nm, at 10 μM protein concentration. (b) Fluorescence thermal unfolding monitoring the intrinsic tryptophan fluorescence emission at 330 nm, at 2 μM protein concentration. Both experiments were recorded at pH 8. The experimental data (circles) and the non-linear regression analysis fitting curves (continuous lines) according to two-state monomeric, dimeric and tetrameric protein are shown (indistinguishable in the plot).
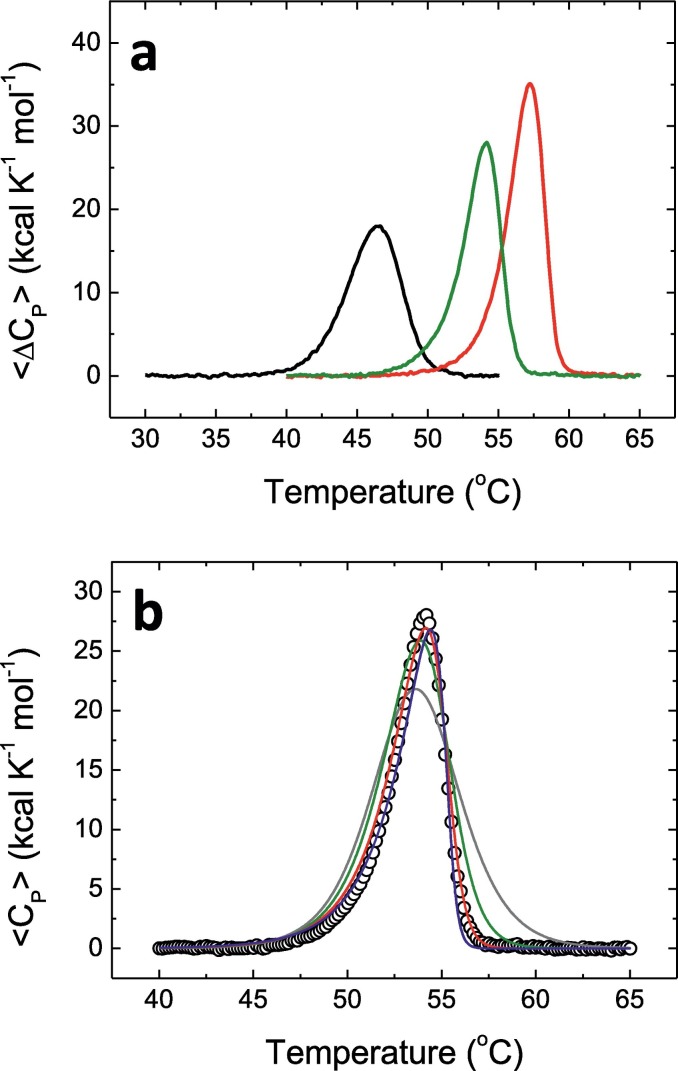
Thermal unfolding monitored by differential scanning calorimetry (DSC) also showed a single apparent unfolding transition with a marked dependence on the pH (Fig. 3a). The lower thermal stability observed at pH 5 and 8, compared to that at pH 7, may reflect the protonation/deprotonation equilibrium of ionizable sidechains titratable around pH 6–7 (e.g., histidines). Very importantly, the transitions are quite asymmetric, a clear indication of the oligomeric nature of the native state of 3CLpro. A model-free analysis (see Materials & methods section) allowed estimating an apparent unfolding enthalpy of 106 kcal/mol and an unfolding temperature of 54.2 °C, at pH 8. Again, the difference in the unfolding temperature compared to those obtained by using spectroscopic techniques could be justified by the use of a different protein concentration in the DSC assays and considering the oligomer-monomer equilibrium coupled to the unfolding process. From this model-free analysis, the ratio between the van't Hoff enthalpy and the calorimetric enthalpy was estimated to be >1, which is another clear indication of the oligomeric nature of the native state of the protein. A small influence of the ionic strength of the solution on the thermal stability of the protein was observed (Fig. S2A). The increase in NaCl concentration to 150 mM caused a reduction in the apparent unfolding temperature of 0.6 °C, which suggested the uptake of salt ions upon protein unfolding.
Fig. 3.
Calorimetric thermal unfolding of SARS-CoV-2 3CLpro. (a) Molar excess heat capacity as a function of temperature at pH 5 (black), pH 7 (red), and pH 8 (green), at 14 μM protein concentration. (b) Molar excess heat capacity as a function of temperature at pH 8. The experimental data (circles) and the non-linear regression analysis fitting curves (continuous lines) according to two-state monomeric (gray), dimeric (green), tetrameric (red), and octameric (blue) protein unfolding models are shown.
The scan rate along the DSC assay may distort the stability parameters (i.e., the DSC profiles may show a scan rate dependence) if the unfolding kinetic equilibration for the transitions between the conformational states is sufficiently slow. In this case, irreversible kinetic models should be applied instead of equilibrium models. In our experiments on SARS-CoV-2 3CLpro, a small scan rate dependence could be observed (Fig. S2B). The data analysis according to the Lumry-Eyring model for protein thermal denaturation (Fig. S2B, inset) allowed estimating a high kinetic activation energy barrier for the irreversible denaturation process, close to 200 kcal/mol [29]. This energy barrier corresponds to a half-life for irreversible denaturation extremely long (close to 30 years at 37 °C, and much larger at 25 °C). These results suggested the analysis employing equilibrium models may be appropriate.
The two-state tetramer (n = 4) unfolding model reproduced significantly better the experimental data at pH 7 and 8 (Fig. 3b), while the two-state dimer (n = 2) unfolding model reproduced better the experimental data at pH 5 (according to the residual sum of squares; see Table S1). Application of a more complex model considering the coexistence of tetramers and dimers (T → 2D → 4U) did not improve the results (Table S1). In addition, other models considering a two-step unfolding process where the dimeric or tetrameric states undergo a first transition with no dissociation (Nn → Nn*) followed by a second transition with coupled unfolding-dissociation (Nn* → nU) did not improve the results either. This indicates that any unfolding model considering more than one step lacks the sufficient cooperativity to reproduce the experimental results. Therefore, calorimetric unfolding traces were sensitive enough for discriminating the self-association nature of 3CLpro native state, and very likely there is a pH dependence of the quaternary rearrangement in 3CLpro.
3.3. SARS-CoV-2 3CLpro activity profile correlates with the thermal stability profile
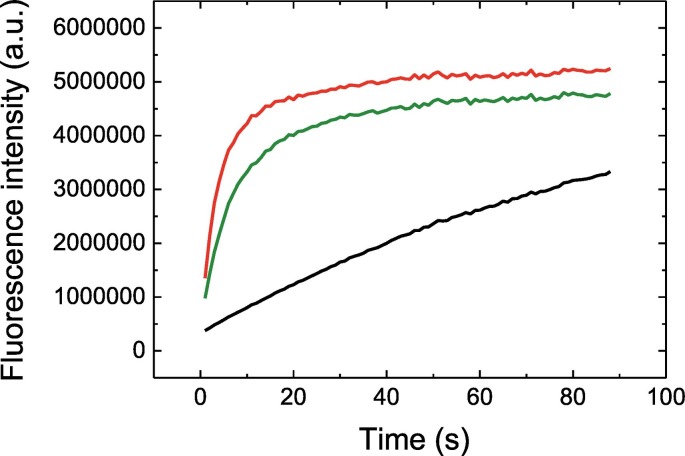
The enzymatic activity of 3CLpro was measured using a FRET substrate containing Edans/Dabcyl as the donor/acceptor FRET pair. Continuous assays allowed us monitoring the time evolution of the substrate fluorescence emission as a function of time at different pH values. The enzymatic activity was higher at pH 7, compared to pH 5 and 8 (Fig. 4 ), which correlates with the pH dependence of the structural stability observed by DSC assays. Among other factors, the pH dependence of the enzyme activity may reflect structural changes (either secondary, tertiary or quaternary, or a combination of them) and, very importantly, the ionization state of titratable sidechains from the catalytic dyad residues (Cys-145 and His-41). Very likely, the catalytic histidine is responsible for this pH effect.
Fig. 4.
Catalytic activity of SARS-CoV-2 3CLpro. Time evolution of the substrate fluorescence emission as a function of time at pH 5 (black), pH 7 (red), and pH 8 (green), at protein concentration of 1 μM and substrate concentration of 20 μM. The increase in fluorescence emission reflects the decrease in Dabcyl quenching on Edans emission (i.e., reduction of FRET phenomenon) due to the increase in the spatial separation of the donor-acceptor pair as a result of the catalytic cleavage of the substrate molecule.
The enzymatic parameters were estimated from the initial enzymatic rate dependence on the substrate concentration at pH 8 (Fig. S3): Km = 11 μM and kcat = 0.040 s−1, resulting in a catalytic efficiency, kcat/Km of 3640 M−1 s−1, comparable to reported values considering the differences in the experimental conditions [21,22].
3.4. Screening of a small chemical library
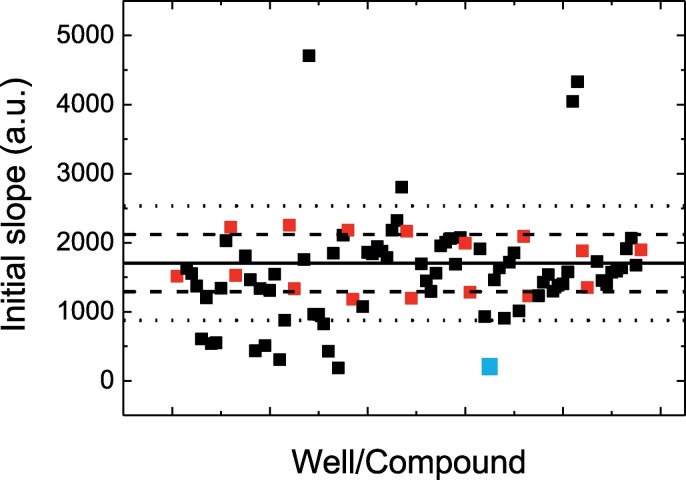
The continuous FRET assay for measuring the activity of 3CLpro was adapted to an in vitro experimental screening procedure. In a first step, the screening was performed using a small chemical library consisting of 150 compounds, which also included approved drugs with known therapeutic indications. Hits consisted of compounds reducing the enzymatic activity of 3CLpro below a specified threshold, defined as the average activity for the controls (enzyme with no compound) minus twice their corresponding standard deviation (Fig. 5 ). Several hits were selected and confirmed by the inhibition curve analysis.
Fig. 5.
Screening of a small chemical library. Enzyme activity, in the presence and the absence of compound, was quantitated as the initial slope of the fluorescence emission as a function of time. Control wells (red squares) contained enzyme and substrate in the presence of an identical concentration of DMSO as that in the wells with compounds (black squares). Hits were selected as those compounds lowering the activity below a certain threshold (average activity of controls minus twice the standard deviation of those controls, lower dotted line). The blue square corresponds to quercetin.
3.5. Quercetin inhibits SARS-CoV-2 3CLpro in vitro
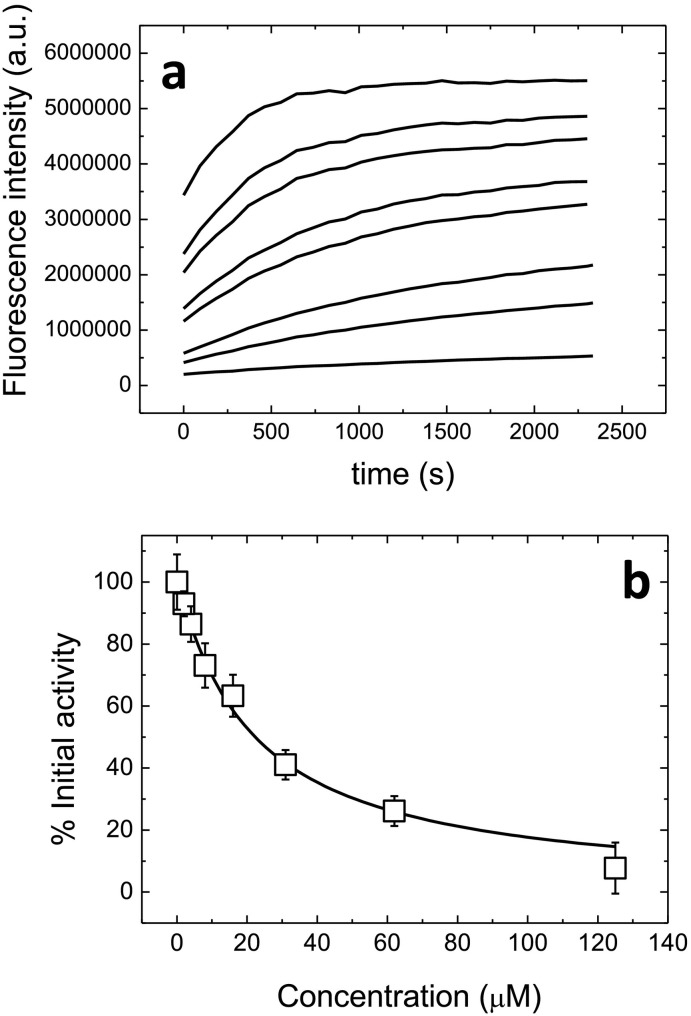
To assess the in vitro potency of the compounds selected through the small-scale screening, their inhibition constants were estimated from the inhibition curve. Inhibition curves were obtained by measuring the enzymatic activity, applying the previously described protocol for the continuous FRET assay, by fixing the enzyme and the substrate concentrations while varying the amount of compound (Fig. 6a). Enzymatic activity at each compound concentration was calculated as the initial slope in each curve, and a dose-dependent effect of the compound on the enzyme activity was observed (Fig. 6b). Non-linear regression analysis employing a simple inhibition model, according to Eq. (10) (see Materials & methods), allowed estimating an apparent inhibition constant, Ki app, of 21 μM for quercetin, the most active compound. Because it can be assumed this compound might bind to the active site, thus, functioning as a competitive inhibitor, this apparent inhibition constant can be transformed into an intrinsic inhibition constant according to: Ki = Ki app / (1 + [S] / Km), or, equivalently, the fitting can be carried out using Eq. (11), which accounts for competitive inhibition and the substrate concentration and Km appear explicitly (see Materials & methods). With a Km value of 11 μM (Fig. S3), an intrinsic inhibition constant Ki of 7.4 μM could be estimated for quercetin. This potency is similar to some of the first inhibitors found for SARS-CoV 3CLpro [30].
Fig. 6.
Hit confirmation through inhibition curve. (a) Substrate fluorescence emission as a function of time, fixing the enzyme concentration at 2 μM, the substrate concentration at 20 μM, and varying quercetin concentration from 0 to 125 μM (while maintaining constant the percentage of DMSO). The increase in quercetin concentration resulted in the decrease in the enzymatic activity. (b) Experimental inhibition data for quercetin (squares), together with the non-linear regression analysis fitting curve (black line) to a simple inhibition, from which the apparent inhibition constant (Kiapp = 21 μM) or the intrinsic inhibition constant (Ki = 9.6 μM) could be estimated.
3.6. Quercetin affects SARS-CoV-2 3CLpro thermal stability
In general, the interaction of a ligand with a protein results in a change in the stability of the protein against thermal or chemical denaturation. More precisely, the preferential interaction of the ligand with certain conformational state stabilizes that state, resulting in a net global stabilization effect if the preferential interaction occurs with the native state or a net destabilization if the preferential interaction occurs with the non-native state. Thermal shift assay (TSA), also called differential scanning fluorimetry, consists in detecting ligand-induced stability changes in proteins by fluorescence [31,32]. Using an extrinsic fluorescent probe reporting the progression of the protein unfolding process, it is possible to assess the thermal stability of the protein by measuring its apparent Tm in the presence of increasing concentrations of a given interacting compound.
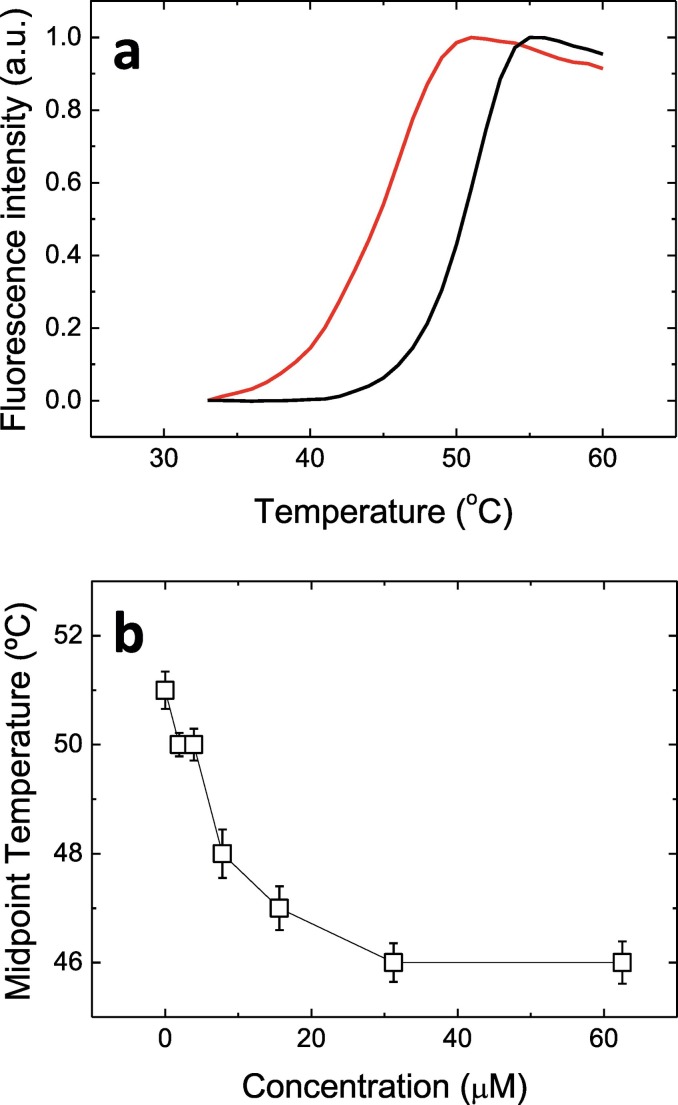
The identified compound, quercetin, altered the thermal stability of 3CLpro causing a destabilization (Fig. 7a). Although ligand-induced stabilizing effects on a protein are more common, it is not unusual to observe destabilizing effects [[33], [34], [35]]. In addition, the effect of the compound was concentration-dependent (Fig. 7b).
Fig. 7.
Hit confirmation through thermal shift assay (TSA). (a) Unfolding traces of the compound-free protein (black) and the protein in the presence of 62.5 μM quercetin (red). (b) Apparent unfolding temperature for the protein at different quercetin concentrations.
3.7. Quercetin interacts with SARS-CoV-2 3CLpro
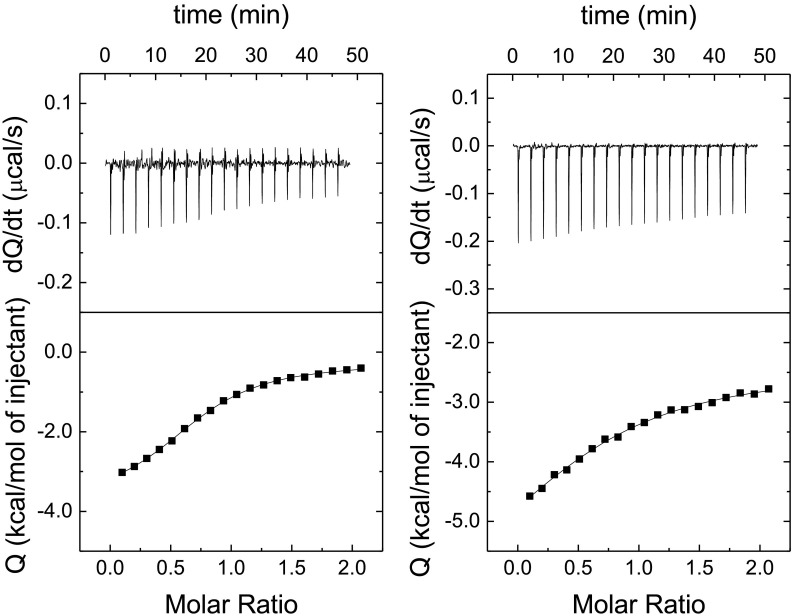
Isothermal titration calorimetry (ITC) is considered the gold-standard for binding affinity determination, as well as target engagement in drug discovery [36,37]. Careful experimental design in ITC provides invaluable and detailed information about the drug-target interaction [38]. ITC was employed to assess target engagement for quercetin and get a direct estimation of the dissociation constant for its interaction with 3CLpro. According to the experiments, quercetin interacts with 3CLpro with a dissociation constant of 2.7 μM in the absence of NaCl, and a dissociation constant of 10 μM in the presence of NaCl 150 mM (Fig. 8 ), in good agreement with the inhibition constant estimated through the inhibition curve (which was determined in the presence of NaCl 150 mM).
Fig. 8.
Hit conformation through isothermal titration calorimetry (ITC). Interaction of quercetin with 3CLpro assessed by isothermal titration calorimetry in sodium phosphate 50 mM, pH 8, without (left) and with (right) NaCl 150 mM. The upper plot shows the thermogram (thermal power required to maintain a null temperature difference between sample and reference cells as a function of time) and the lower plot shows the binding isotherm (ligand-normalized heat effect per injection as a function of the molar ratio, the quotient between the ligand and protein concentrations in the cell). The fitting curve corresponds to the single ligand binding site model (continuous line). According to the data analysis, in the absence of NaCl quercetin interacts with 3CLpro with favorable enthalpic (ΔH = −3.6 kcal/mol) and entropic (−TΔS = −4.0 kcal/mol) contributions to the Gibbs energy of binding (ΔG = −7.6 kcal/mol), corresponding to a dissociation constant Kd of 2.7 μM. In the presence of NaCl quercetin interacts with 3CLpro with favorable enthalpic (ΔH = −4.3 kcal/mol) and entropic (−TΔS = −2.5 kcal/mol) contributions to the Gibbs energy of binding (ΔG = −6.8 kcal/mol), corresponding to a dissociation constant Kd of 10 μM. In both cases, the percentage of active (or binding-competent) protein is 0.75.
3.8. Quercetin binds to SARS-CoV-2 3CLpro active site
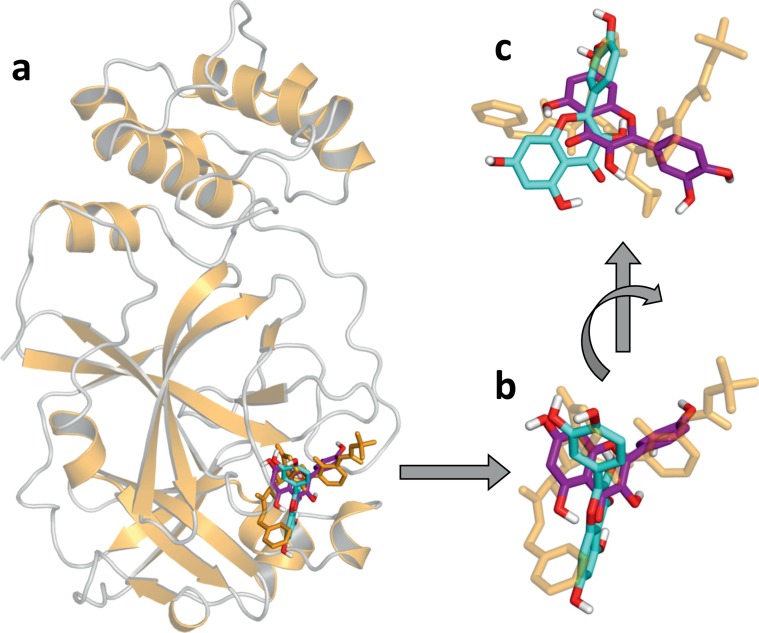
Molecular docking was used to clarify the interaction of quercetin with 3CLpro. Two crystallographic structures were considered, corresponding to the protein in unliganded form or with an inhibitor bound in the active site, reported as entry 6Y2E and 6Y2F in the PDB [22], respectively. Fig. 9 summarizes the overall findings obtained in the simulation with quercetin. For both molecular receptors, the most favorable binding modes predicted through a blind search carried out on the whole protein surface were found in the known protein active site. The binding affinity ranged from −7.5 to −7.2 kcal/mol, corresponding to a dissociation constant of ~3–5 μM, in reasonably good agreement with the value obtained experimentally (Kd of 2.7 μM). A variety of slightly different binding modes were obtained, all distributed in the same binding location. This observation is consistent with the pseudo-symmetric chemical structure of quercetin, with five hydroxyl groups determining a number of possibilities in the formation of hydrogen bonds as donors.
Fig. 9.
Molecular docking of quercetin to 3CLpro. Structure of (a) unliganded 3CLpro (from entry 6Y2E of the PDB; in ribbon representation, yellow) and α-ketoamide inhibitor bound in the active site (from entry 6Y2F; in stick representation, yellow) [22]. The liganded protein was superimposed by a least square fit on the Cα atoms of the liganded one, and only the latter is shown. (b) and (c) Detail of the 3CLpro active site, with the two best docking poses obtained using as receptor the structure 6Y2E (cyan) and 6Y2F (purple). Polar hydrogen atoms are explicitly shown (white) and oxygen atoms are also coloured (red).
As detailed in Fig. 9 (see insets b and c), the two binding modes of quercetin most distant from each other still occupied the same protein crevice. In particular, they mostly overlapped with the position of a single larger known ligand, the α-ketoamide inhibitor co-crystallized with 3CLpro in the PDB entry 6Y2F [22]. Interactions with a number of protein residues contribute to bind quercetin into the 3CLpro binding site. The key residue is Met165, which can form both hydrogen bonds and hydrophobic interactions with quercetin. Additional hydrogen bonds can be formed with the two residues Ser144 and Asn142. Energetic contributions are also due to supplementary hydrophobic interactions with Met49, Phe140 and Leu141, and to electrostatic interaction formed with the polar residue His164 and the charged residue Glu166.
4. Discussion
The need for pharmacological agents acting against SARS-CoV-2 is urgent. The development of drugs acting through different mechanisms provides a good starting point for a combined use in a therapy with low susceptibility to drug resistance. 3CLpro (or Mpro) is an appropriate target, given its high conservation among coronaviruses, as well as a suitable experimental tractability and druggability. Drug discovery can be initiated from experimental or computational screenings, by employing small or large, highly diverse or focused (target-derived) chemical libraries. On the basis of an experimental pipeline for drug screening successfully employed in the past [35,[39], [40], [41], [42], [43], [44], [45]], we have applied the same methodology to identify small molecules blocking the activity of SARS-CoV-2 3CLpro.
The first step in this screening program was to assess the stability and the in vitro activity of SARS-CoV-2 3CLpro. According to our results, 3CLpro is well folded in solution and exhibits a large kinetic stability (activation energy barrier ~ 200 kcal/mol), which allowed us to apply equilibrium models for unfolding. It has been established that monomeric 3CLpro is inactive [46] and the functional state of this protein is the homodimer. We observed that 3CLpro not only populates dimeric states in solution, but also tetrameric states, depending on the pH. Interestingly, even tetrameric and highly-active octameric states have been previously reported for 3CLpro [22,27]. In addition, the effect of pH on the structural stability parallels that of the enzymatic activity, suggesting some titratable residues in the range of pH 5–8 are instrumental to guarantee both these properties. Hydrodynamic radius measurements by dynamic light scattering showed a predominant population of particles compatible with dimeric 3CLpro (hydrodynamic radius of 3.89, 3.33, and 3.94 nm at pH 5, 7, and 8, respectively), with an expected smaller size around its isoelectric point (pI ~ 6) due to smaller net charge and lower electrostatic repulsions (Fig. S4). Additional experiments using size-exclusion chromatography were also in agreement with a predominantly dimeric 3CLpro (Fig. S4). It may be hypothesized that, along the thermal denaturations, temperature could trigger a transient dimerization of dimers into tetramers (tetrameric intermediate) at moderate temperatures, a temperature-driven oligomerization process that has been observed in other proteins [47,48], before reaching the unfolding temperature and ending in monomer unfolding/dissociation; work on this matter is in progress.
The screening test for 3CLpro could be based on detecting small molecules interfering with the enzymatic activity (which is fairly easy for a protease, by using a continuous enzymatic assay and a peptidic FRET substrate) or based on detecting the effect of small molecules on altering the thermal stability of the protein against thermal denaturation (by using fluorescence TSA). The TSA-based screening has the advantage of being quite general for any protein, but it has some disadvantages: 1) binding affinity does not correlate with stability changes; 2) false negatives are somewhat common, because ligands may bind to unfolded or non-native protein states besides binding to the native state; 3) stability changes are highly dependent on the binding enthalpy and binding heat capacity; and 4) the ligand binding does not always translates into an inhibitory effect [49]. In contrast, the techniques based on detection of the protein activity provide direct evidence for inhibition during the screening test, but the activity of the protein target must be measurable through a quick and appropriate procedure, which is not straightforward in general.
Through the application of a small-scale activity-based screening to SARS-CoV-2 3CLpro, quercetin was selected and further studied. Direct evidence for target engagement was gathered by different biophysical techniques, TSA and ITC, as well as through molecular docking simulations. Quercetin showed an inhibition constant of about 7 μM, which corresponds to a binding affinity sufficiently favorable to be considered as a good candidate for optimization or preclinical studies.
The fact that quercetin exerts a destabilizing effect may be related to its physico-chemical properties and the type of interaction with the protein target. The extent of the stabilization effect induced by a ligand depends on the specific set of conformational states, within the whole conformational landscape of the protein, that interact preferentially with the ligand. If the ligand interacts with most of the conformational states with similar binding affinities, the effect on the protein stability will be small. If the ligand interacts not only with the native state, but also with partially unfolded states (which could be the case for quercetin, due to its hydrophobic moiety), a destabilization effect could be expected if the affinity (or the binding stoichiometry) for the non-native states is higher. These are some of the caveats (described above) to be born in mind when TSA is applied for in vitro molecular screening. In addition, unlike in the case of isothermal inhibition assays, where only native-like states are accessible, through TSA it is possible to gradually explore non-native states close to the native state through a temperature-driven process, and binding to non-native states can be detected. In any case, regardless of whether the effect of the ligand on the protein was destabilizing or stabilizing, quercetin interacted and inhibited the activity of 3CLpro.
The influence of quercetin on the structural properties of 3CLpro was assessed (Fig. S5), and provided an additional piece of evidence for the interaction of quercetin with 3CLpro. The far-UV CD spectrum of the protein was not significantly distorted by quercetin, but considerable changes occurred in the near-UV CD and fluorescence spectra. In particular, a substantial decrease in tryptophan fluorescence could be observed in 3CLpro upon quercetin binding. Very likely these alterations in the tertiary structure of the protein are associated with the destabilizing effect of quercetin on the thermal stability of 3CLpro observed by TSA.
According to ITC experiments, quercetin binds with favorable enthalpic (ΔH = −3.6 kcal/mol) and entropic (−TΔS = −4.0 kcal/mol) contributions to the Gibbs energy of binding (ΔG = −7.6 kcal/mol). The hydrophobicity, the compactness and the rigidity of the molecule, and the ability to adopt multiple configurations within the binding site seem to be responsible for a large desolvation entropy gain and small conformational, configurational and roto-translational entropy losses upon binding, resulting in a global favorable entropy gain. Quercetin would be an appropriate scaffold to engineer new functional groups for developing new inhibitor compounds for SARS-CoV-2 3CLpro. The lower affinity at high ionic strength (Fig. 8) indicates that the binding of quercetin to 3CLpro is coupled to the release of salt ions.
Quercetin compares quite well to SARS-CoV-2 3CLpro inhibitors already reported. An intrinsic inhibition constant Ki of 0.19 μM could be estimated for an alpha-ketoamide inhibitor 13b reported by Zhang et al. [22]. In order to compare the inhibition potency of ligands, their molecular size must be considered. The ligand efficiency or binding efficiency index (BEI = pKi / MW) normalizes the inhibitor potency (pKi = −logKi) with its molecular weight (MW) [50]. Because quercetin and inhibitor 13b have a MW of 0.302 kDa and 0.598 kDa, respectively, their ligand efficiencies are 17.0 and 11.2, respectively. Thus, although the alpha-ketoamide inhibitor has a much lower inhibition constant, this is not surprising given its larger MW; in fact, quercetin is close to be considered as a fragment (MW < 300 Da) from a screening perspective. However, BEI is significantly higher for quercetin, suggesting it is a good drug candidate establishing favorable interactions with the target and possessing high potential for optimization and development.
It is worth to note that quercetin is a natural product with well-known pharmacokinetics and ADMET properties and some proposed therapeutic indications, which points to a potential repurposing for COVID-19 treatment. Interestingly, and in agreement with our blind assays, experimental evidences had been reported in the past for an inhibitory effect of quercetin on the 3CLpro from the coronavirus variant MERS-CoV [51]. The work presented here provides a direct evidence for quercetin ability to target SARS-CoV-2 3CLpro on solid experimental bases. Furthermore, it represents a successful starting point that guarantees the possibility to extend our experimental approach for identifying SARS-CoV-2 3CLpro drug candidates to larger chemical libraries, in order to find small molecules to be used (or further optimized) as potential COVID-19 drugs.
Author contributions
Conceptualization, O.A. and A.V.C.; Methodology, O.A., A.V.C. and B.R.; Software, O.A., A.V.C. and B.R.; Validation, O.A., A.V.C., D.O.A., A.J.A., L.C.L., S.V., B.R. and H.T.R.; Formal analysis, O.A., A.V.C., D.O.A., A.J.A., L.C.L., S.V., B.R. and H.T.R.; Investigation, D.O.A., A.J.A., L.C.L., S.V., H.T.R. and B.R.; Resources, O.A., A.V.C., B.R. and H.T.R.; Data curation, O.A., A.V.C., B.R. and H.T.R.; Writing – original draft preparation, O.A., A.V.C. and B.R.; Writing – review & editing, O.A., A.V.C., D.O.A., A.J.A., L.C.L., S.V., B.R. and H.T.R.; Visualization, O.A., A.V.C., D.O.A., A.J.A., L.C.L., S.V., and B.R.; Supervision, O.A., A.V.C.; Project administration, O.A. and A.V.C.; Funding acquisition, O.A. and A.V.C.
Acknowledgments
Acknowledgments
This work was supported by Fundación hna; Miguel Servet Program from Instituto de Salud Carlos III (CPII13/00017 to O.A.); Fondo de Investigaciones Sanitarias from Instituto de Salud Carlos III, and European Union (ERDF/ESF, ‘Investing in your future’) (PI18/00343 to O.A. and a FIS Research Contract to L.C.L.); Spanish Ministry of Economy and Competitiveness (BFU2016-78232-P to A.V.C. and SAF2017-83265-R to H.T.R.); Spanish Ministry of Science, Innovation and Universities (FPI Predoctoral Research Contract BES-2017-080739 to D.O.A.); the Spanish National Research Council (CSIC, project 202020E079); Diputación General de Aragón (Predoctoral Research Contract 2019 to A.J.A., ‘Protein Targets and Bioactive Compounds Group’ E45_20R to A.V.C., ‘Digestive Pathology Group’ B25_20R to O.A.); Centro de Investigación Biomédica en Red en Enfermedades Hepáticas y Digestivas (CIBERehd). The proteomic analysis was performed in the Proteomics Facility of The Spanish National Center for Biotechnology (CNB-CSIC) that belongs to ProteoRed, PRB3-ISCIII, supported by grant PT17/0019.
Declaration of competing interest
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2020.07.235.
Appendix A. Supplementary data
Supplementary material
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.W.H.O. (WHO) World Health Organization. Novel coronavirus (COVID-19) situation. https://covid19.who.int [DOI] [PMC free article] [PubMed]
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams J.G., Walls R.M. JAMA; United States: 2020. Supporting the Health Care Workforce During the COVID-19 Global Epidemic. [DOI] [PubMed] [Google Scholar]
- 5.Renu K., Prasanna P.L., Valsala Gopalakrishnan A. Coronaviruses pathogenesis, comorbidities and multi-organ damage - a review. Life Sci. 2020;255 doi: 10.1016/j.lfs.2020.117839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5(1):84. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke L. An introduction to economic studies, health emergencies, and COVID-19. J. Evid. Based Med. 2020;13(2):161–167. doi: 10.1111/jebm.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atzrodt C.L., Maknojia I., McCarthy R.D.P., Oldfield T.M., Po J., Ta K.T.L., Stepp H.E., Clements T.P. A guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020 doi: 10.1111/febs.15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pirrone V., Thakkar N., Jacobson J.M., Wigdahl B., Krebs F.C. Combinatorial approaches to the prevention and treatment of HIV-1 infection. Antimicrob. Agents Chemother. 2011;55(5):1831–1842. doi: 10.1128/AAC.00976-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., Norris A., Sanseau P., Cavalla D., Pirmohamed M. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 11.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata A.A. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020;9(4):1225. doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. 2003;100(23) doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muramatsu T., Kim Y.T., Nishii W., Terada T., Shirouzu M., Yokoyama S. Autoprocessing mechanism of severe acute respiratory syndrome coronavirus 3C-like protease (SARS-CoV 3CLpro) from its polyproteins. FEBS J. 2013;280(9):2002–2013. doi: 10.1111/febs.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu A., Wang Y., Zeng C., Huang X., Xu S., Su C., Wang M., Chen Y., Guo D. Prediction and biochemical analysis of putative cleavage sites of the 3C-like protease of Middle East respiratory syndrome coronavirus. Virus Res. 2015;208:56–65. doi: 10.1016/j.virusres.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. Eur. J. Chem. Biol. 2020;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63(9):4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 19.Khan S.A., Zia K., Ashraf S., Uddin R., Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1751298. [DOI] [PubMed] [Google Scholar]
- 20.Lee H., Mittal A., Patel K., Gatuz J.L., Truong L., Torres J., Mulhearn D.C., Johnson M.E. Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual and high-throughput screenings. Bioorg. Med. Chem. 2014;22(1):167–177. doi: 10.1016/j.bmc.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo C.J., Chi Y.H., Hsu J.T., Liang P.H. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem. Biophys. Res. Commun. 2004;318(4):862–867. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19(14):1639–1662. [Google Scholar]
- 25.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4(1):17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grande F., Rizzuti B., Occhiuzzi M.A., Ioele G., Casacchia T., Gelmini F., Guzzi R., Garofalo A., Statti G. Identification by molecular docking of homoisoflavones from Leopoldia comosa as ligands of estrogen receptors. Molecules. 2018;23(4) doi: 10.3390/molecules23040894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S., Zhong N., Xue F., Kang X., Ren X., Chen J., Jin C., Lou Z., Xia B. Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease. Protein Cell. 2010;1(4):371–383. doi: 10.1007/s13238-010-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shriver J.W., Edmondson S.P. Defining the stability of multimeric proteins. Methods Mol. Biol. 2009;490:57–82. doi: 10.1007/978-1-59745-367-7_3. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Ruiz J.M. Theoretical analysis of Lumry-Eyring models in differential scanning calorimetry. Biophys. J. 1992;61(4):921–935. doi: 10.1016/S0006-3495(92)81899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S.A., Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry. 2004;43(17):4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 31.Ericsson U.B., Hallberg B.M., Detitta G.T., Dekker N., Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 2006:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 32.Pantoliano M.W., Petrella E.C., Kwasnoski J.D., Lobanov V.S., Myslik J., Graf E., Carver T., Asel E., Springer B.A., Lane P., Salemme F.R. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 2001;6(6):429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 33.Cimmperman P., Baranauskiene L., Jachimoviciūte S., Jachno J., Torresan J., Michailoviene V., Matuliene J., Sereikaite J., Bumelis V., Matulis D. A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys. J. 2008;95(7):3222–3231. doi: 10.1529/biophysj.108.134973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.González A., Casado J., Chueca E., Salillas S., Velázquez-Campoy A., Sancho J., Lanas Á. Small molecule inhibitors of the response regulator ArsR exhibit bactericidal activity against Helicobacter pylori. Microorganisms. 2020;8(4) doi: 10.3390/microorganisms8040503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villanueva R., Romero-Tamayo S., Laplaza R., Martínez-Olivan J., Velázquez-Campoy A., Sancho J., Ferreira P., Medina M. Redox- and ligand binding-dependent conformational ensembles in the human apoptosis-inducing factor regulate its pro-life and cell death functions. Antioxid. Redox Signal. 2019;30(18):2013–2029. doi: 10.1089/ars.2018.7658. [DOI] [PubMed] [Google Scholar]
- 36.Lakowicz J.R., Jayaweera R., Szmacinski H., Wiczk W. Resolution of multicomponent fluorescence emission using frequency-dependent phase angle and modulation spectra. Anal. Chem. 1990;62(18):2005–2012. doi: 10.1021/ac00217a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward W.H., Holdgate G.A. Isothermal titration calorimetry in drug discovery. Prog. Med. Chem. 2001:309–376. doi: 10.1016/s0079-6468(08)70097-3. [DOI] [PubMed] [Google Scholar]
- 38.Claveria-Gimeno R., Vega S., Abian O., Velazquez-Campoy A. A look at ligand binding thermodynamics in drug discovery. Expert. Opin. Drug Discov. 2017;12(4):363–377. doi: 10.1080/17460441.2017.1297418. [DOI] [PubMed] [Google Scholar]
- 39.González A., Salillas S., Velázquez-Campoy A., Espinosa Angarica V., Fillat M.F., Sancho J., Lanas Á. Identifying potential novel drugs against Helicobacter pylori by targeting the essential response regulator HsrA. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-47746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santofimia-Castano P., Xia Y., Lan W., Zhou Z., Huang C., Peng L., Soubeyran P., Velazquez-Campoy A., Abian O., Rizzuti B., Neira J.L., Iovanna J. Ligand-based design identifies a potent NUPR1 inhibitor exerting anticancer activity via necroptosis. J. Clin. Invest. 2019:130. doi: 10.1172/JCI127223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neira J.L., Bintz J., Arruebo M., Rizzuti B., Bonacci T., Vega S., Lanas A., Velázquez-Campoy A., Iovanna J.L., Abián O. Identification of a drug targeting an intrinsically disordered protein involved in pancreatic adenocarcinoma. Sci. Rep. 2017;7 doi: 10.1038/srep39732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hidalgo J., Latorre P., Carrodeguas J.A., Velázquez-Campoy A., Sancho J., López-Buesa P. Inhibition of pig phosphoenolpyruvate carboxykinase isoenzymes by 3-mercaptopicolinic acid and novel inhibitors. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0159002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abian O., Vega S., Sancho J., Velazquez-Campoy A. Allosteric inhibitors of the NS3 protease from the hepatitis C virus. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0069773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pey A.L., Ying M., Cremades N., Velazquez-Campoy A., Scherer T., Thöny B., Sancho J., Martinez A. Identification of pharmacological chaperones as potential therapeutic agents to treat phenylketonuria. J. Clin. Invest. 2008;118(8):2858–2867. doi: 10.1172/JCI34355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cremades N., Velázquez-Campoy A., Martínez-Júlvez M., Neira J.L., Pérez-Dorado I., Hermoso J., Jiménez P., Lanas A., Hoffman P.S., Sancho J. Discovery of specific flavodoxin inhibitors as potential therapeutic agents against Helicobacter pylori infection. ACS Chem. Biol. 2009;4(11):928–938. doi: 10.1021/cb900166q. [DOI] [PubMed] [Google Scholar]
- 46.Shi J., Wei Z., Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 2004;279(23):24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campos L.A., Sharma R., Alvira S., Ruiz F.M., Ibarra-Molero B., Sadqi M., Alfonso C., Rivas G., Sanchez-Ruiz J.M., Romero Garrido A., Valpuesta J.M., Muñoz V. Engineering protein assemblies with allosteric control via monomer fold-switching. Nat. Commun. 2019;10(1):5703. doi: 10.1038/s41467-019-13686-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasimova M.R., Milstein S.J., Freire E. The conformational equilibrium of human growth hormone. J Mol Biol. 1998;1998:409–418. doi: 10.1006/jmbi.1997.1613. [DOI] [PubMed] [Google Scholar]
- 49.Velazquez-Campoy A., Sancho J., Abian O., Vega S. Biophysical screening for identifying pharmacological chaperones and inhibitors against conformational and infectious diseases. Curr. Drug Targets. 2016;17(13):1492–1505. doi: 10.2174/1389450117666160201110449. [DOI] [PubMed] [Google Scholar]
- 50.Abad-Zapatero C., Metz J.T. Ligand efficiency indices as guideposts for drug discovery. Drug Discov. Today. 2005:464–469. doi: 10.1016/S1359-6446(05)03386-6. [DOI] [PubMed] [Google Scholar]
- 51.Jo S., Kim H., Kim S., Shin D.H., Kim M.-S. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem. Biol. Drug Des. 2019;94(6):2023–2030. doi: 10.1111/cbdd.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material