Highlights
-
•
This study compared RT-qPCR sensitivity for SARS-CoV-2 detection.
-
•
False negatives were in the range of 2–39.8%.
-
•
The most sensitive solution (97.9% [92.8–99.7]) targeted the SARS-CoV-2 E-gene.
Keywords: COVID-19, SARS-CoV-2, Diagnosis, False negatives, Solution comparisons, Sensitivity
Abstract
Objectives
The ongoing COVID-19 pandemic continues to impose demands on diagnostic screening. In anticipation that the recurrence of outbreaks and the measures for lifting the lockdown worldwide may cause supply chain issues over the coming months, this study assessed the sensitivity of a number of one-step retrotranscription and quantitative polymerase chain reaction (RT-qPCR) solutions to detect SARS-CoV-2.
Methods
Six different RT-qPCR alternatives were evaluated for SARS-CoV-2/COVID-19 diagnosis based on standard RNA extractions. The one with best sensitivity was also assessed with direct nasopharyngeal swab viral transmission medium (VTM) heating; thus overcoming the RNA extraction step.
Results
A wide variability in the sensitivity of RT-qPCR solutions was found that was associated with a range of false negatives from 2% (0.3–7.9%) to 39.8% (30.2–50.2%). Direct preheating of VTM combined with the best solution provided a sensitivity of 72.5% (62.5–81.0%), in the range of some of the solutions based on standard RNA extractions.
Conclusions
Sensitivity limitations of currently used RT-qPCR solutions were found. These results will help to calibrate the impact of false negative diagnoses of COVID-19, and to detect and control new SARS-CoV-2 outbreaks and community transmissions.
1. Introduction
The ongoing pandemic of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) causing the coronavirus disease 2019 (COVID-19) has imposed an increasing demand on daily diagnostic screening. This is expected to perpetuate over the coming months, due to the recurrence of outbreaks and lifting of lockdown measures worldwide (Patel et al., 2020). Given the high sensitivity compared to serological testing (Cassaniti et al., 2020), standard diagnosis continues to rely on RNA extractions from respiratory or oral samples followed by one-step reverse transcription and real-time quantitative polymerase chain reaction (RT-qPCR) that entails one or several primer-probe sets for targeting SARS-CoV-2 sequences (Corman et al., 2020). While it has been shown that protocol modifications aiming to overcome supply chain issues and accelerate diagnosis affect assay sensitivity (Alcoba-Florez et al., 2020, Esbin et al., 2020), differences in target priming efficiencies and RT-qPCR kit components are also expected to account for dissimilarities in false negative results (Nalla et al., 2020).
This study aimed to evaluate the sensitivity of six different RT-qPCR solutions, including five marketed kits and one based on the World Health Organization (WHO) diagnostic assays with the best sensitivity (Corman et al., 2020, Vogels et al., 2020), using RNA extractions from nasopharyngeal swab viral transmission medium (VTM). To skip the RNA extraction step that has been described elsewhere, the alternative with the best sensitivity was also assessed by direct preheating of VTM samples (Alcoba-Florez et al., 2020).
2. Materials and methods
The study was conducted at the University Hospital Nuestra Señora de Candelaria (Santa Cruz de Tenerife, Spain) from March to June 2020. Six different RT-qPCR solutions were evaluated (Table 1 ): four based on three viral targets and two based on one viral target. Given the high specificity of the RT-qPCR (Alcoba-Florez et al., 2020), focus was on evaluating the rate of false negatives (FN) and assay sensitivity using the same 98 COVID-19 patient samples. The alternative with the best sensitivity was also assessed under an alternative procedure that skips the RNA extraction step described elsewhere (Alcoba-Florez et al., 2020).
Table 1.
Different RT-qPCR solutions evaluated for the detection of SARS-CoV-2 in nasopharyngeal swab samples from COVID-19 positive patients.
| Solution | #Targetsa | Target gene | Sensitivity, % (95% CI)b | FNc |
|---|---|---|---|---|
| TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific) combined with validated primer-probe setsd | 3 | E | 95.9 (89.9–98.9) | 4 |
| N | 75.5 (65.8–83.6) | 24 | ||
| RdRp | 77.6 (68.0–85.4) | 22 | ||
| LightMix® Modular SARS-CoV (COVID19) (TIB MOLBIOL) | 3 | E | 97.9 (92.8–99.7) | 2 |
| N | 78.6 (69.1–86.2) | 21 | ||
| RdRp | 89.8 (82.0–95.0) | 10 | ||
| SARS-COV-2 R-GENE (BioMérieux) | 3 | E | 65.3 (55.0–74.6) | 34 |
| N | 66.3 (56.1–75.6) | 33 | ||
| RdRp | 60.2 (49.8–70.0) | 39 | ||
| TaqPath COVID-19 CE-IVD RT-PCR Kit (Thermo Fisher Scientific) | 3 | ORF1ab | 65.3 (55.0–74.6) | 34 |
| S | 70.4 (60.3–79.2) | 29 | ||
| N | 76.5 (66.9–84.5) | 23 | ||
| Genesig Real-Time PCR COVID-19 kit (Primedesign Ltd.) | 1 | RdRp | 81.6 (72.5–88.7) | 18 |
| Real Accurate Quadruplex corona-plus PCR Kit (PathoFinder) | 1 | N | 83.7 (74.8–90.4) | 16 |
Specific primer-probes for SARS-CoV-2.
95% Confidence Interval.
False negative counts out of 98 patients.
Samples were collected in 2 mL of VTM (BioMérieux, Lyon, France). RNA extractions were conducted from 200 μL of VTM using the MagNA Pure Compact Nucleic Acid Isolation Kit I (Roche, Basel, Switzerland) or the STARMag Viral DNA/RNA 200C kit (Seegene, Seoul, Korea). The RT-qPCR was performed in 10 μL final volume reactions (5 μL of sample) using a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, California, USA) following the thermal cycling specifications of each solution. Positive and negative controls were included in all experiments, as described elsewhere (Alcoba-Florez et al., 2020). Sensitivity and 95% confidence intervals (95% CI) were calculated from the FN counts using MedCalc (MedCalc Software Ltd.).
3. Results
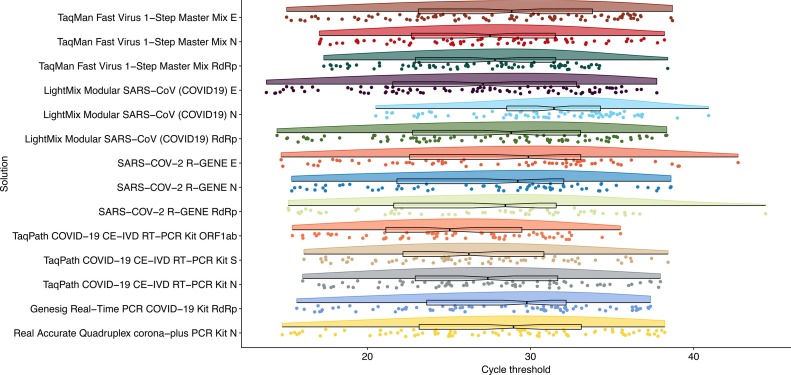
Since all samples were COVID-19 positive for at least one solution/viral target, results with threshold cycle (Ct) values >40 or those that remained undetected during the 45 cycles of the experiments were considered FN observations (Fig. 1 , Table 1). Attending to individual targets, it was found that the most sensitive solution was the LightMix® Modular SARS-CoV (COVID19) (TIB MOLBIOL, Berlin, Germany) used in combination with a primer-probe set for the E-gene (97.9% [92.8–99.7]) (Table 1). It was closely followed by the TaqMan Fast Virus 1-Step Master Mix kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA) combined with validated primer-probes for diagnosis (Corman et al., 2020) for the same viral gene (95.9% [89.9–98.9]). When combining at least two viral gene targets, it was found that the TaqMan Fast Virus 1-Step Master Mix kit with validated primer-probe sets targeting both E and RdRp genes (Corman et al., 2020) attained an equivalent sensitivity. The kit with the poorest performance for all three viral primer-probe sets was SARS-COV-2 R-GENE (BioMérieux) (range 60.2% [49.8–70.0] to 66.3% [56.1–75.6]). Its levels of sensitivity improved to those of all other kits when the E-gene primer-probe set was combined with those for N or the RdRp genes (71.4% [61.4–80.1] and 69.4% [59.3–78.3], respectively). The sensitivity of all other solutions did not benefit from combining the results of more than one primer-probe set.
Fig. 1.
Raincloud plot of the distribution of cycle threshold (Ct) values for the RT-qPCR solutions evaluated for the detection of SARS-CoV-2 in COVID-19 positive samples. Raw Ct data with the median and the interquartile range are also represented and overlaid on each distribution.
Finally, because the LightMix® Modular SARS-CoV (COVID19) kit with primer-probes for the E-gene showed the highest sensitivity, it was tested on samples that were preheated at 70 °C for 10 minutes in a substitution of the RNA extraction (Alcoba-Florez et al., 2020). Although this alternative decreased the kit sensitivity (72.5% [62.5–81.0]), the results were still comparable with other evaluated solutions (Table 1).
4. Discussion
RT-qPCR for selected target genes of SARS-CoV-2 has been key in the global response to the COVID-19 pandemic. Given the rapid spread of the virus at this time, it is likely that the RT-qPCR assays will continue to be a central tool for controlling COVID-19. However, as happened in the past due to supply chain issues, policy decisions and laboratory testing capacities (Alcoba-Florez et al., 2020), it is predictable that the diagnosis of COVID-19 will continue relying on a variety of solutions among laboratories and countries (Vogels et al., 2020).
The current results showed a wide variability in the sensitivity of RT-qPCR solutions for SARS-CoV-2 detection, which was associated with a proportion of FN ranging from 2% (0.3–7.9%) to 39.8% (30.2–50.2). Given that the same patient nasopharyngeal samples were assayed for different solutions, well-known factors affecting SARS-CoV-2 sensitivity (stage of infection and type of specimen) (Pan et al., 2020, Wölfel et al., 2020) were suitably controlled in the study, since all solutions were equally affected. Thus, the differences in sensitivity among solutions were due to their different components (i.e. primers-sets, buffers, enzymes, and reagent contents in general). These findings will help to assess the impact of the selected solution on FN diagnoses of COVID-19 (Ramdas et al., 2020) and to choose a solution that minimizes misdiagnoses of an active SARS-CoV-2 infection.
Authors’ contributions
JAF and CF designed the study. JAF, HGC and DGMA participated in data acquisition. JAF, LC and CF performed the analyses and data interpretation. LC, AVF, RGM, and CF wrote the draft of the manuscript. All authors contributed in the critical revision and final approval of the manuscript.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This research was funded by Cabildo Insular de Tenerife [grant number CGIEU0000219140]; the agreement with Instituto Tecnológico y de Energías Renovables (ITER) to strengthen scientific and technological education, training research, development and innovation in Genomics, Personalized Medicine and Biotechnology [grant number OA17/008]; Ministerio de Ciencia e Innovación [grant number RTI2018-093747-B-100 and RTC-2017-6471-1], co-funded by the European Regional Development Fund (ERDF); Lab P2+ facility [grant number UNLL10-3E-783], co-funded by the ERDF and “Fundación CajaCanarias”; and the Spanish HIV/AIDS Research Network [grant number RIS-RETIC, RD16/0025/0011], co-funded by Instituto de Salud Carlos III and by the ERDF. The funders had no role in the study design, collection, analysis and interpretation of data, in the writing of the manuscript or in the decision to submit the manuscript for publication.
Ethical approval
The University Hospital Nuestra Señora de Candelaria (Santa Cruz de Tenerife, Spain) review board approved the study (ethics approval number: CHUNSC_2020_24).
Acknowledgments
We deeply acknowledge the University Hospital Nuestra Señora de Candelaria board of directors and the executive team for their strong support and assistance in accessing diverse resources used in the study.
References
- Alcoba-Florez J., González-Montelongo R., Íñigo-Campos A., García-Martínez de Artola D., Gil-Campesino H., The Microbiology Technical Support Team Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples. Int J Infect Dis. 2020;97:66–68. doi: 10.1016/j.ijid.2020.05.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassaniti I., Novazzi F., Giardina F., Salinaro F., Sachs M., Perlini S. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol. 2020;30(March) doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;20:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalla A.K., Casto A.M., Huang M.W., Perchetti G.A., Sampoleo R., Shrestha L. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J Clin Microbiol. 2020;58:e00557–e620. doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R., Babady E., Theel E.S., Storch G.A., Pinsky B.A., St George K. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: Value of Diagnostic Testing for SARS-CoV-2/COVID-19. mBio. 2020;11:e00722–e820. doi: 10.1128/mBio.00722-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas K., Darzi A., Jain S. ‘Test, re-test, re-test’: using inaccurate tests to greatly increase the accuracy of COVID-19 testing. Nat Med. 2020;26:810–811. doi: 10.1038/s41591-020-0891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR primer-probe sets. MedRxiv. 2020 doi: 10.1101/2020.03.30.20048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]