Abstract
On-going pandemic pneumonia outbreak COVID-19 has raised an urgent public health issue worldwide impacting millions of people with a continuous increase in both morbidity and mortality. The causative agent of this disease is identified and named as SARS-CoV2 because of its genetic relatedness to SARS-CoV species that was responsible for the 2003 coronavirus outbreak. The immense spread of the disease in a very small period demands urgent development of therapeutic and prophylactic interventions for the treatment of SARS-CoV2 infected patients. A plethora of research is being conducted globally on this novel coronavirus strain to gain knowledge about its origin, evolutionary history, and phylogeny. This review is an effort to compare genetic similarities and diversifications among coronavirus strains, which can hint towards the susceptible antigen targets of SARS-CoV2 to come up with the potential therapeutic and prophylactic interventions for the prevention of this public threat.
Keywords: COVID-19, SARS-CoV2, Genomic comparison, ER stress, Target antigenic sites, Vaccine development
Abbreviations: NTD, N-terminal domain; S, Spike; E, envelope; M, membrane protein; N, nucleocapsid protein; Nsp, non-structure protein; ORF, open reading frame; PDB, protein data bank; RdRp, RNA-dependence RNA polymerase; 3CLpro, 3-chymotrypsin-like protease; ACE-2, angiotensin converting enzyme-2; HCoV, human coronavirus
1. Introduction
Coronaviruses (CoVs) are single-stranded, positive-sense RNA viruses belonging to the order Nidovirales, family Coronaviridae, and subfamily Coronavirinae (Spaan et al., 2012). CoVs possess the largest genomes among all RNA viruses ranging from 26 to 32 kilobases in length, with G + C contents varying from 32% to 43% (Woo et al., 2009; Woo et al., 2007; Woo et al., 2005). CoVs are predominantly associated with enteric and respiratory diseases in animals and humans (Cheng et al., 2004; Gélinas et al., 2001).
Subfamily coronavirinae further diverges into three major generas or groups -the alpha-CoVs (group 1), the beta-CoVs (group 2), and the gamma-CoVs (group 3) characterized by varying genetic makeup and antigenic cross-reactivity(Cleri et al., 2010; Gorbalenya et al., 2004; Khan et al., 2020; Woo et al., 2010). Delta-CoVs, representing a novel genus of Coronaviruses were also later found in birds and pigs. Out of these, only alpha-CoV strains (HCoV-229E and HCoV-NL63) along with beta-CoVs (HCoV-HKU1, HCoV-OC43, severe acute respiratory syndrome coronavirus [SARS-CoV], and Middle East respiratory syndrome coronavirus [MERS-CoV]) have been identified as human pathogenic strains (Cortellis, 2020). Swine acute diarrhea syndrome (SADS-CoV) (Zhou et al., 2018), porcine transmissible gastroenteritis virus(Brian and Baric, 2005), and porcine enteric diarrhea virus (PEDV) (Lin et al., 2016) are a few examples of alpha and beta-CoVs that hold the potential to pose heavy disease burden on livestock.
Currently, an ongoing pneumonia outbreak caused by a novel Coronavirus strain named SARS-CoV2 belonging to the beta-coronavirus genera is an emerging threat to public health all over the world. Ongoing extensive research has suggested that bats are the natural reservoir and pangolins are probably the potential intermediate host of the virus, as 99% homology was found between SARS-CoV2 and coronavirus strain isolated from pangolins(Zheng, 2020).
A plethora of research is being conducted globally on this novel coronavirus strain to gain knowledge about its origin, evolutionary history, and phylogeny. This review is an effort to compare genetic similarities and diversifications among coronavirus strains, which can hint towards the susceptible antigen targets of SARS-CoV2 to come up with the potential therapeutic and prophylactic interventions for the prevention of this public threat.
1.1. Genomic organization of the viral genome
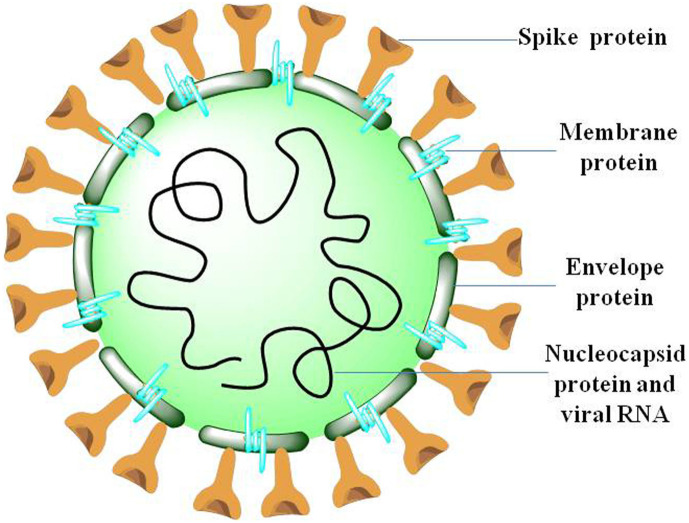
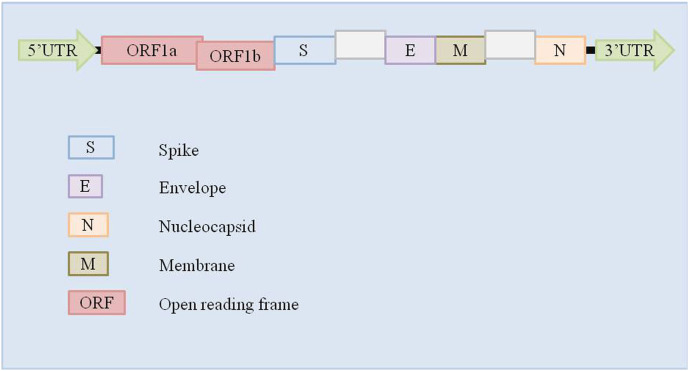
Virion structure of coronavirus is made up of four major structural proteins namely, spike, envelope, membrane, nucleocapsid (Fig. 1 ). Distinct CoV strains share a common genetic organization for the coding region encoding for a canonical set of genes in the order 5′ end- ORF1a/b replicase, spike, envelope, membrane, nucleocapsid-3′ end (Fig. 2 ), although the number and location of accessory ORFs present in different CoV species vary (6–11 ORFs)(Brian and Baric, 2005). Subgenomic (sg) mRNAs that are responsible for the translation of genes form a 5′ and 3′ co-terminal nested set with viral genome. A common 5′ leader sequence and a 3′ terminal sequence is present along with subgenomic mRNAs. Both 3′ and 5′ ends of the genome carry small untranslated regions (UTRs). Besides, several nonstructural proteins (nsps) including RNA-dependent RNA polymerase (RdRp), coronavirus main protease (3CLpro), and papain-like protease (PLpro) are also encoded by the viral genome (Anand Gaurav, 2020; Baez-Santos et al., 2014; Lai, 1990).
Fig. 1.
Coronavirus virion structure depicting structural proteins: S (spike), M (membrane), E(envelope) and N(Nucleocapsid).
Fig. 2.
Genomic organization of coronaviruses.
A typical CoV genomic and subgenomic sequence consists of at least six ORFs. The first ORF representing almost 67% of the entire viral genome (ORFa/b) encodes 16 nsps (non-structural proteins) (1–16 nsp), except gamma-CoVs that lack nsp1. A papain-like protease (PL2pro) in nsp3 and a 3C-like protease (3CLpro; also known as the “main protease”) in nsp5 are the two protease domains conserved in all types of CoVs which are encoded by ORF1a sequence in the genome (Van Boheemen et al., 2012). Other ORFs encodes for few accessory proteins and some major structural proteins including envelope (E), spike (S), membrane (M), and nucleocapsid (N) proteins (Van Boheemen et al., 2012; Czub et al., 2005; Woo et al., 2010).
1.2. Distinct characteristics of Novel Coronavirus SARS-CoV2
1.2.1. Receptor binding domain
The RBD sequence in the spike protein of a coronavirus is the most variable region and prone to mutations (Tao et al., 2020; Zhou et al., 2020ba). A total of six amino acid residues are suggested to be essential for binding to the human ACE-2 receptor (Yushun Wan, Jian Shang 1, Rachel Graham, Ralph S. Baric, 2020). As per the amino acid sequence coordinates of SARS-CoV, these are Y442, L472, N479, D480, T487, and Y4911, whereas in SARS-CoV2 these corresponds to L455, F486, Q493, S494, N501, and Y505 residues respectively (Walls et al., 2020; Yushun Wan, Jian Shang 1, Rachel Graham, Ralph S. Baric, 2020). Interestingly, studies have suggested that out of these six residues of SARS-CoV2 five does not share similarity to SARS-CoV counterparts and the change is probably due to mutations, deletions or insertions in the S1-S2 region of Coronaviruses (Walls et al., 2020; Wrapp et al., 2020; Yushun Wan, Jian Shang 1, Rachel Graham, Ralph S. Baric, 2020).
1.2.2. Polybasic furin cleavage site and O-linked glycans
Polybasic cleavage site (RRAR) is the second distinct characteristic of SARS-CoV2 located at the junction of two subunits i.e., S1 and S2 of Spike(Walls et al., 2020). Proteases like furin can effectively cleave this site. In SARS-CoV2, a leading proline is also inserted at the junction (between subunits S1 and S2 of Spike) that leads to the formation of a turn/stem-loop structure, which consequently results in the insertion of O-linked glycans to cleavage site residues S686, S673, and T678. The presence of O-linked glycans is very unique and different from other CoV variants (Chan et al., 2008; Vankadari and Wilce, 2020).
This polybasic proteolytic cleavage of S glycoprotein is responsible for determining the viral infectivity as well as host range because whether the virus can jump across species e.g., from bats to humans, is determined by this cleavage (Nao, 2017). The S protein isolated from Ugandan bats MERS-like CoV species, for instance, was found to duly cohere to human cells, but viral entry was not mediated. However, if during coherence of virus to the human cell, protease trypsin is inserted, it could cleave the S glycoprotein and allow the virus to enter into the cells (Menachery et al., 2019). This phenomenon demonstrates the importance of cleavage of S glycoprotein for mediating viral entry into human cells and thus determining viral infectivity and that restricting its cleavage by manipulating the expression of S protein or ACE-2 receptor binding motif might be a great therapy.
Although the protein sequence analysis has suggested the presence of furin cleavage sites in SARS-CoV2 protein, however, RaTG-13 (Zhou et al., 2020b), the most closely related bat CoV strain to SARS-CoV2 isolated from a bat in Yunnan in 2013 does not constitute this furin cleavage site (Zhou et al., 2020a). However, there are abundant furin proteases present in respiratory tracts of humans suggesting that S glycoprotein cleavage takes place while exiting from RBCs and thus infect other cells (Izaguirre, 2019). This furin cleavage site might have been acquired by RNA recombination and its presence in SARS-CoV2 might be responsible for infecting human cells. Also, this cleavage site might have allowed bat CoV to jump into humans and thus initiate the ongoing pneumonia outbreak COVID-19 (Andersen et al., 2020; WO, 2020; Wrapp et al., 2020; Zhou et al., 2020a).
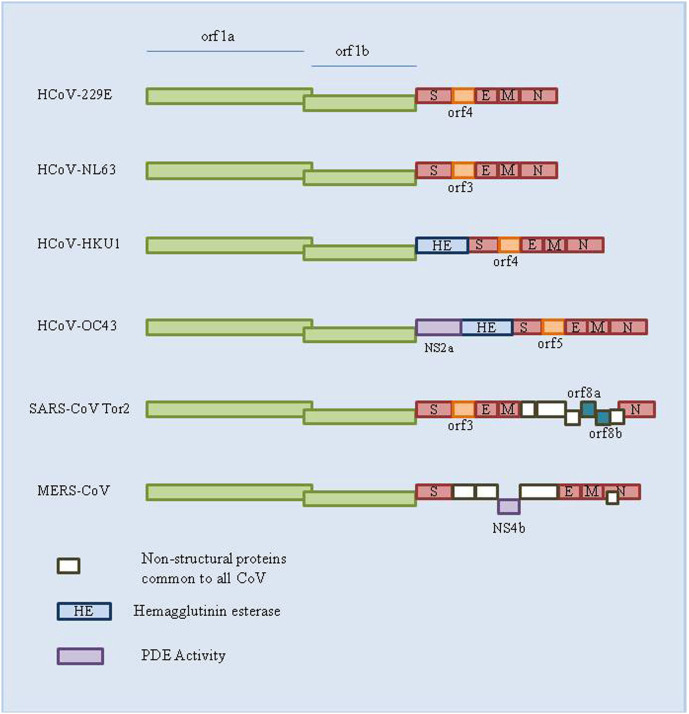
1.3. Human coronavirus serotypes
In 1965, B814 was described as the first human Coronavirus (HCoV) strain. In subsequent years, about 30 more types of HCoV strains have been identified. Out of these, seven HCoV strains i.e., two alpha-CoVs (HCoV-229E; HCoV-NL63), and five beta-CoVs (HCoV-OC43; HCoV-HKU1; MERS-CoV; SARS-CoV including novel SARS-CoV-2) (Table 1 ) (Fig. 3 ) are commonly circulating in the human population and predominantly responsible for cold symptoms and other respiratory diseases in healthy individuals. The Middle East respiratory syndrome CoVand Severe acute respiratory syndrome CoV are two zoonotic viruses that pose major public health threats as they are associated with severe lower respiratory tract infections, while most other HCoVs cause comparatively mild upper respiratory tract infections (common cold) (Jonsdottir and Dijkman, 2016).
Table 1.
The accession number, genus, abbreviation, name, and length (bps) for the seven most common human coronavirus strains.
| No. | Accession | Genera | Abbreviation | Genome | Length (bp) |
|---|---|---|---|---|---|
| 1 | NC_002645 | α | HCoV-229E | Human Coronavirus 229E | 27,317 |
| 2 | NC_005831 | α | HCoV-NL63 | Human Coronavirus NL63 | 27,553 |
| 3 | NC_006577 | β | HCoV-HKU1 | Human Coronavirus HKU1 | 29,926 |
| 4 | NC_006213 | β | HCoV-OC43 | Human Coronavirus OC43 | 30,741 |
| 5 | NC_019843 | β | MERS-CoV | Middle East respiratory syndrome Coronavirus | 30,119 |
| 6 | NC_004718 | β | SARS-CoV | Severe acute respiratory syndrome Coronavirus | 29,751 |
| 7 | NC_045512 | β | SARS-CoV2 | Severe acute respiratory syndrome Coronavirus-2 | 29,903 |
Fig. 3.
Genomes of different CoV strains.
1.3.1. Alpha coronavirus
This genus of CoVs contains two human pathogenic viral strains i.e., HCoV-229E and HCoV-NL63. HCoV-229E utilizes aminopeptidase N (APN) (Yeager et al., 1992) as its major receptor for entering into the host cell, like animal alpha-CoVs whereas; HCoV-NL63 utilizes angiotensin-converting enzyme-2 (ACE-2) (Hofmann et al., 2005) receptors to enter in the host cell, similar to SARS-CoV, which is otherwise a beta-CoV.
1.3.2. Beta coronavirus
MERS-CoV and SARS-CoV (Shults et al., 2012; Zaki et al., 2012) are two bat-viruses included in this genus. Novel COVID-2019 disease-causing virus is also classified as a beta-CoV and has been named as SARS-CoV2. The angiotensin-converting enzyme 2 (ACE2) (Gorbalenya et al., 2004; Woo et al., 2010) acts as one of the main receptors in SARS-CoV to enter the host cell, whereas, MERS-CoV, on the other hand, utilizes dipeptidyl peptidase 4 (DPP4, also known as CD26) (Song et al., 2014) as the primary receptor. Other than these, HCoV-OC43 and HCoV-HKU1, are two non-SARS CoV species included in this genus that probably utilize sialic acid residues as receptors and have hemagglutinin-esterase activity (Vlasak et al., 1988).
1.3.3. Gamma coronavirus
Mostly Avian Coronaviruses are included in this genus; the infectious bronchitis virus of chickens is the most prominent of all. This virus is responsible for causing respiratory and reproductive tract disease in chickens.
1.3.4. Delta coronavirus
This group represents a novel genus of Coronaviruses found in birds and pigs.
2. Origin and evolution of SARS-CoV and SARS-CoV2
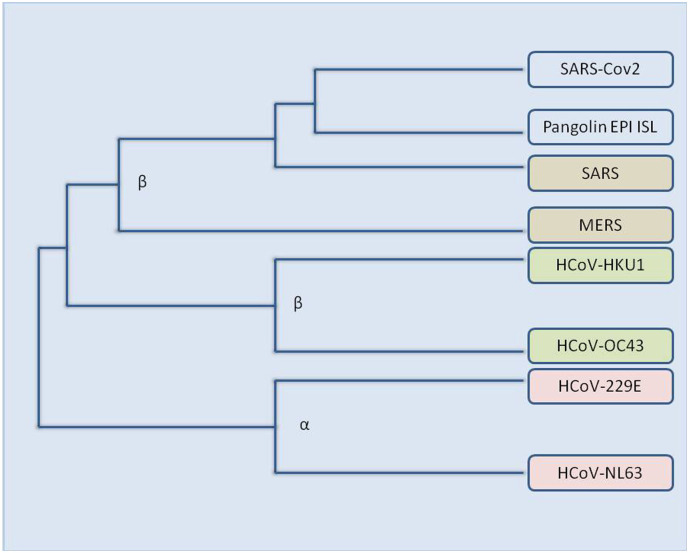
Current sequence database survey analysis shows an animal origin of all diverse sorts of human CoVs: HCoV-NL63, HCoV-229E, SARS-CoV and MERS-CoV are mulled over to have originated from bats, whereas, HCoV-OC43 and HCoV- HKU1 are considered to be evolved from rodents (Forni et al., 2017; Su et al., 2016) (Fig. 4 ). The transmission of the virus from natural hosts to humans requires an intermediate host; domesticated animals might serve as essential hosts for viral transmission, additionally, domestic animals themselves can endure the disease caused by bat-borne or closely associated CoVs. Of the various coronavirus strains found so far, most of them are considered to have evolved from bats, thus, bats make the major natural reservoirs of alpha and beta-CoVs. (Woo et al., 2012)
Fig. 4.
An Evolutionary tree of various coronavirus strains.
Initially, when the SARS epidemic originated, most of the diseased patients were found with a record of animal exposure before the disease development. In 2005, two groups independently documented that horseshoe bats (genus Rhinolophus) (Lau et al., 2005; Li et al., 2005) were the main natural reservoir of novel CoVs related to human SARS-CoV. Many other CoV strains which are phylogenetically related to SARS-CoV were also discovered to have bat-origin., isolated from various locations including diverse European, African and Southeast Asian countries and also from, China (Gouilh et al., 2011; Hu et al., 2017; Lacroix et al., 2017; Lau et al., 2005; Li et al., 2005; Rihtarič et al., 2010; Tong et al., 2009; Wacharapluesadee et al., 2015). As per studies, the SARS-CoV strains from Hipposideros bats (Tong et al., 2009) in Africa were found to be phylogenetically distant from those of Rhinolophus SARS-CoV strains suggesting that SARS-CoV have wide geographical spread and bats might have been a host of SARS-CoV from a very long period. A cave of Yunnan province, China, is designated as a diversity hot spot as this location contains a highly diverse SARSr-CoVs in bat populations (Ge et al., 2013; Hu et al., 2017; Wang et al., 2016). The SARSr-CoV in this location consists of all the genetic diversity of SARS-CoV found in all other places of china. All these pieces of evidence point towards an immensely diverse nature of SARS-CoV, additionally, despite 15 years of extensive searching, researchers are unable to locate the ultimate progenitor of SARS-CoV in bat populations. A process called viral RNA recombination is a major phenomenon involved in enhancing the diversity of RNA viruses, in this process, two non-segmented RNA genomes involve the exchange of genetic information and this phenomenon is found to be very frequent within Coronaviruses (Masters, 2006). So, it is anticipated that SARS-CoV emanated through recombination of bats SARSr-CoV or through another bat cave which is yet-to-be-identified.
The S gene and upstream oforf8 are the two regions of the viral genome suggested to be essential break-points for viral RNA recombination, where the former encodes the spike (S) protein containing the receptor-binding domain (RBD) whereas, later encodes an accessory protein (Hon et al., 2008; Hu et al., 2017; Wu et al., 2016). Frequent RNA recombination of CoVs and immense genetic diversity, as well as the prevalence of SARS-CoV, is likely to be one of the reasons for the emergence of novel coronavirus variant i.e., SARS-CoV2 and subsequently COVID-19 outbreak.
2.1. Natural selection
Andersen et al. (2020) have tried to explain the proximal origin of SARS-CoV2 by providing two hypotheses of its emergence. First, natural selection in an animal host prior to zoonotic transfer, and the second hypothesis is natural selection in humans after the zoonotic transfer (Andersen et al., 2020). Natural selection is a process that allows organisms to adapt to their environment by selectively reproducing suitable variations in their genotype or genetic constitution (Gregory, 2009).
2.1.1. Natural selection in an animal host prior to zoonotic transfer
The RBD region of S protein in SARS-CoV2 is optimized for binding to human-like ACE2 receptor, and the most probable reason for this change is natural selection (Tao et al., 2020; Zhang et al., 2020a; Zhou et al., 2020b). As bat SARS-CoV-like Coronaviruses share the closest homology to SARS-CoV2 therefore, bats are thought to be the natural reservoir of SARS-CoV2 (Tao et al., 2020). Additionally, the genome of Rhinolophus affinis bat (Zhou et al., 2020b) was found to share 96% similarity to the novel coronavirus with non-homologous regions belonging to the RBD region of S protein suggesting incompatibility of this strain with human ACE2 receptor (Wan et al., 2020). This suggests natural selection as the reason for the optimization of S protein in novel coronavirus.
Furthermore, no strain of bat beta-CoV is found to have a polybasic cleavage site as found in the genome of novel coronavirus SARS-CoV2 strain. Also, no direct progenitor of SARS-CoV2 is identified until now. All these shreds of evidence suggest that mutations, insertions, and deletions at the S1 and S2 junction of Spike protein of SARS-CoV2 are responsible for evolutionary changes (Andersen et al., 2020; Yamada and Liu, 2009).
A high population density of an animal host is a must for a precursor virus to achieve both mutations and polybasic cleavage site in S protein of SARS-CoV2 (Andersen et al., 2020).
2.1.2. Natural selection in humans after zoonotic transfer
Another hypothesis of adaptation of above mentioned genomic features and consequently the origin of SARS-CoV2 could be that a progenitor of SARS-CoV2 containing all the genomic characteristics (polybasic cleavage site and mutations in S protein) might have jumped into humans via yet unknown human to human transmission (Andersen et al., 2020; Tao et al., 2020; Zhou et al., 2020b).
Various strains of SARS-CoV2 sequenced so far carry similar genomic characteristics as described above, pointing towards a common progenitor from which they have adapted these features. The RBD region in SARS-CoV2 shows features very similar to the viral strains isolated from pangolins suggesting that the virus that jumped to humans probably consisted of this region and polybasic cleavage insertion might have occurred during human to human transmission (Andersen et al., 2020). This is true in the case of MERS-CoV where repeated jumps of the virus from dromedary camels were the reason for all human cases producing short transmission chains or single infections that without adapting to sustained transmission, eventually resolved (Dudas et al., 2018).
Retrospective serological studies and examination of banked human samples can be informative and assist in determining whether such cryptic spread has occurred or not (Andersen et al., 2020; Dudas et al., 2018; Wang et al., 2018).
3. RNA virus mutations
Most of the evolutionary changes in organisms occur as a consequence of mutations. The mutation is an alteration of the nucleotide sequence in the genome of an organism, these variations bring novelty that we observe in the course of evolution; also, natural selection can act upon these alterations (Baer, 2008). Usually, small count of these variations is beneficial, some are inconsequential (neutral) and most of them are harmful or non-beneficial for the organisms. Whether an organism has a low mutation rate or high, the count of harmful mutations always outnumbers beneficial mutation (Loewe and Hill, 2010).
The largest group of molecular parasites known to infect humans, animals, and plants are RNA viruses. In-depth study of different RNA virus genomes, as well as their replication pathways, suggests their heterogeneous nature in using a wide variety of strategies to enhance their stability and thus evolvability (Murphy et al., 2016; Steinhauer, 1987). Eradication of existing viruses and emergence or re-emergence of new viruses is likely to be balanced by this process of natural selection (Murphy et al., 2016). Environmental alterations along with genetic plasticity of RNA viruses favour the emergence of several new RNA viruses, consequently, new viral pathogens come in contact with potential hosts and facilitate host jumping (Morse and Schluederberg, 1990).
CoV mutations: RNA viruses usually have relatively high mutation rates than DNA viruses as well as a million times higher than their hosts; this is the reason for enhanced viral adaptability and evolvability (Duffy, 2018). Mutation rate or mis-insertion errors during RNA replication have been suggested to fall in the range of 10−3 to 10−5 substitutions per nucleotide (Domingo et al., 1988). Immensely high rates of mutations in RNA viruses results in a yield of offsprings that differ by 1–2 mutations from their parent, forming a diverse mutant cloud of descendants (Vignuzzi and Andino, 2012). Absence or lack of efficiency of proof-reading of RNA polymerases is one of the contributing factors of such high mutation rates in these viruses. In contrast, the RNA viruses with the largest known genome sizes i.e., coronaviruses, show relatively low rates of mutations than other RNA viruses. The slower mutation rates or preservation of such large genomes is probably associated with the exceptional characteristics of CoV RTC (replication-transcription complex) which contains 3′-5′ exoribonuclease activity that probably provides proof-reading function which is unique to CoV genomes among all other variants of RNA viruses (Chen et al., 2020; Minskaia et al., 2006).
The rate of RNA virus mutations usually lies in between the range of 10−6 to 10−4 substitutions per nucleotide site per cell infection,(Peck and Lauring, 2018) whereas, studies suggest that the rate of mutation of SARS-CoV falls in a range between 0.80 and 2.38 × 10−3 nucleotide substitutions per site per year, much lower than other RNA virus counterparts (Zhao et al., 2004).
4. Genomic comparison of CoV strains
The whole-genome sequence alignment of CoV revealed 54% identity among varying CoV strains, whereas, the genome sequence alignment of the nsp-coding region alone of CoV shows 58% similarity and that of the structural protein-coding region shows 43% similarity, suggesting that nsps (non-structural proteins) form the conserved region of the genome with high percent identity, contrarily, structural proteins in need of adaptation to new hosts are more diverse (Chen et al., 2020).
The mutation rates in the RNA virus replication are relatively much higher than DNA viruses. RNA virus genomes are usually small, with less than 10 kb length, however, the genome length of CoVs are comparatively larger with approx. ~30 kb length permitting easier accommodation and modification of genes (Vega et al., 2004). The slower mutation rates or preservation of such large genomes is probably associated with the exceptional characteristics of CoV RTC (replication-transcription complex) which contains 3′-5′ exoribonuclease activity of nsp14 as well as various other RNA processing enzymes (Lauber et al., 2013; Minskaia et al., 2006). The 3′-5′ exoribonuclease activity probably provides a proof-reading function that is unique to CoV genomes among all other variants of RNA viruses (Chen et al., 2020; Minskaia et al., 2006).
4.1. Genomic comparison of SARS-CoV2 (COVID-19) with other CoV strains
In pursuance of recent studies, the genome of SARS-CoV2 is believed to have originated from bats showing 96.2% homology to a bat SARS-related coronavirus (SARSr-CoV; RaTG13) collected in Yunnan province, China (Zhou et al., 2020b), besides, it was shown to possess 85% similarity to the genome of the SARS-like virus ZC45 (bat-SL-CoVZC45, MG772933.1), together these viruses along with SARS-like virus ZXC21 have been grouped into a unique Orthocoronavirinae subfamily (Ma et al., 2020). (Table 2 ).
Table 2.
Percent identity of novel coronavirus SARS-CoV2 strain with different CoV strains. (Chan et al., 2020; Chen et al., 2020; Gralinski and Menachery, 2020; Malik et al., 2020; Ren et al., 2020; Zaki et al., 2012; P. Zhou et al., 2020a).
| S·No | Viral strains | Genus | Percent identity |
|---|---|---|---|
| 1 | HCoV-229E | α | 65.04 |
| 2 | HCoV-NL63 | α | 65.11 |
| 3 | HCoV-HKU1 | β | 67.59 |
| 4 | HCoV-OC43 | β | 68.93 |
| 5 | MERS-CoV | β | 69.58 |
| 6 | SARS-CoV | β | 82.45 |
| 7 | bat-SL-CoVZC45 | β | 88 |
| 8 | bat-SL-CoVZXC21 | β | 88 |
| 9 | SARS-HCoV Tor2 | β | 82 |
| 10 | SARS-HCoV BJ01 | β | 82 |
| 11 | SARSr-CoV; RaTG13 | β | 96.2 |
4.1.1. SARS-CoV2 and SARS-CoV
The results of zpicture comparative genomic analyses of SARS-CoV2 and SARS-CoV have revealed extremely high homology between the two strains at the nucleotide level. Additionally, the genomes of these two strains differ from each other in six regions. The first three regions of difference belong to the partial coding sequences of ORF1a/b (448 nt, 55 nt, and 278 nt, respectively). The next two regions belong to the partial coding sequences of the S gene (315 nt and 80 nt, respectively) and the last region of difference is a part of the coding sequence of the orf7b and orf8 genes(214 nt) (Jiabao Xu, 2014). The spike gene of SARS-CoV2 shows more homology to bat-CoV, whereas two accessory genes 3a and 8b possess homology to SARS-CoV.
Proteomic similarity analyses of SARS-CoV and SARS-CoV2 have suggested that most of the proteins are highly homologous (95%–100%). RdRp and 3CLpro protease share over 95% of sequence similarity even though at the genome level these two strains share only 82% similarity (Chan et al., 2020; Dong et al., 2020; Lu et al., 2020; Morse et al., 2020). Additionally, both these strains share 76% of sequence similarity in their S proteins, a highly conserved receptor-binding domain (RBD), and a domain of S protein (Chan et al., 2020; Dong et al., 2020; Lu et al., 2020; Morse et al., 2020). Also, PLpro sequences of SARS-CoV and SARS-CoV2 share 83% similarity with a large number of similar active sites (Morse et al., 2020). All these pieces of evidence suggest a common evolutionary history of both these viral strains.
However, SARS-CoV2 possesses two proteins (orf8 and orf10) that share no homology to the SARS-CoV strain. SARS-CoV conserved sequences of orf8 are different from the amino acid sequence of orf8 derived from SARS-CoV2. Since these two proteins of SARS-CoV2 possess no homology to other CoV strains, it might be therapeutically beneficial to study the biological function of these two proteins i.e., orf8 and orf10 in SARS-CoV2 (Chan et al., 2020).
4.1.2. Structural comparison of SARS-CoV2 from SARS-CoV
Studies suggest a close relationship between SARS-CoV2 and SARS-CoV or SARS-like bat-CoV. Analysis of the structural and functional differences between SARS-CoV2 and SARS-CoV might be accomplished by shedding light on the amino acid substitutions in different proteins.
Altogether, 380 amino acid substitutions occurred between the amino acid sequences and corresponding consensus sequences of SARS-CoV. Envelope, matrix, or accessory proteins p6 and 8b, nsp7, and nsp13 revealed no change in amino acid sequences. Non-structural proteins nsp2 and nsp3 manifested single amino-acid substitution at positions 61 and 102 respectively (Wu et al., 2020). Additionally, spike protein was found to possess 27 amino acid substitutions with a length of 1273 amino acids, including amino acid region 357–528 in the RBD with six substitutions and another six at amino acid region 569–655 in the underpinning subdomain (SD). Furthermore, the C-terminal of the receptor-binding subunit S1 domain was found to have four substitutions (Q560L, S570A, F572T, and S575A) (Guo et al., 2004; Wu et al., 2020). The receptor-binding motifs that interact with the human ACE-2 receptor were found to be exactly similar to the SARS-CoV strain with no amino acid substitutions, whereas, six mutations occurred in the other RBD region (Ge et al., 2013). Due to limited knowledge about SARS-CoV2, the reasonable explanations for these amino acid substitutions are not known yet.
4.2. Genomic Divergence from other beta-CoV strains
Novel SARS-CoV2 is currently being researched the most around the globe to cope up with the current pandemic scenario affecting a great number of people worldwide for coming up with efficient vaccines and other therapeutic interventions. Globally a bunch of research groups are working to deduce an ample amount of information from the viral genome and to find out the most potential target for the development of the viral vaccine. The search for a potential vaccine has led the researchers to conduct SARS-CoV2 genomic comparison with the previous CoVs, to know if the vaccine targets for previous CoVs can be useful for developing a potential vaccine for the novel virus.
Ceraolo and Giorgi (2020) performed an interspecies genomic divergence investigation for all proteins encoded by SARS-CoV2 and its relatives by aligning all the sequences using MUSCLE (Ceraolo and Giorgi, 2020). They stated a close homology (>80%) of SARS-CoV2 with bat BCoV bat-SL-CoVZXC21 and the high conservation of E, M, and A structural proteins across all beta-CoV strains. As specified by the results of the above-stated research, we performed a multiple sequence alignment along with the assessment of the percent identity matrix using CLUSTAL OMEGA 1.2.4 and constructed phylogenetic trees using JALVIEW 2.10.5.
In compliance with the results stated by Ceraolo and Giorgi, 2020, we observed a very high genomic conservation between the Wuhan isolated SARS-CoV2 strain and SARS Bat coronavirus RaTG13 strain, with a percent identity of 97.71% and 85.41% in S protein and N protein respectively (Table 4). The ACE-2 RBD subdomain of S2 subunit of SARS-CoV2 spike protein displayed 74.41% of genomic similarity to bat SARS RaTG13 strain showing high conservation level in the RBD region.
Table 4.
Percent identity matrix of major proteins and domains of novel coronavirus SARS-CoV2 strain with other beta-CoVs obtained using CLUSTAL O (1.2.4).
| PROTEIN | SARS-CoV | MERS-CoV | HCoV HKU1 | HCoV OC43 |
|---|---|---|---|---|
| S (spike) | 97.71% | 32.79% | 30.50% | 31.26% |
| E (Envelope) | 96.00% | 36.00% | 28.00% | 20.00% |
| M (Membrane) | 89.59% | 39.27% | 35.29% | 38.74% |
| N (Nucleocapsid) | 85.41% | 48.47% | 34.28% | 35.20% |
| Receptor (ACE-2) binding domain | 74.41% | 18.75% | 24.44% | 22.83% |
| N-terminal domain | 52.55% | 21.67% | 21.49% | 20.26% |
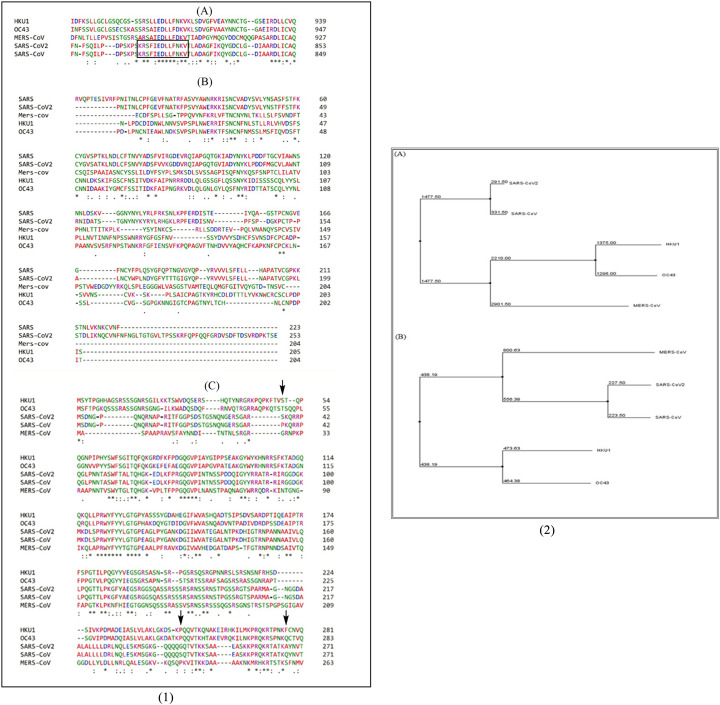
Furthermore, Robson, 2020 stated that subsequence KRSFIEDLLFNKV of the S2’ spike glycoprotein proteolytic cleavage site, is an important motif and is found to be responsible for initial binding of previous SARS-CoV to lung cells and their activation of the spike protein by proteolytic cleavage(Robson, 2020). So, we also carried sequence alignment for this particular region and found it to be completely conserved as we observed a 100% match between the SARS-CoV2 strain and SARS bat RaTG13 strain, whereas the motif differs at 3 amino acid positions in MERS strain. For further visual confirmation, of the level of genomic divergence and similarity between SARS-CoV2, SARS Bat coronavirus RaTG13, and other common beta CoV strains, via MSA, in the S and N structural proteins, please refer to Fig. 7.
Fig. 7.
(1). (A) MSA of KRSFIEDLLFNKV motif, (B) MSA of Receptor (ACE-2) binding domain of S1 subunit of spike protein, (C) MSA of Nucleocapsid (N) protein of various beta coronavirus strains representing sequences for SARS-CoV2, SARS, MERS, HCoV HKU1 and HCoV OC43. (2). Phylogenetic trees of (A) S protein and, (B) N protein of different beta CoVs. Both the trees demonstrate close homology of novel SARS CoV-2 with SARS-CoV.
5. CoV introduction into cell and its replication
The adherence of the host receptor to the virus via S protein marks the initiation of the viral infection. As soon as the virus adheres to the host receptor, the process of cleavage of viral S protein into two subunits i.e., S1 (N- terminal receptor binding domain) and S2 (C-terminal domain) are triggered by host proteases(Huang et al., 2006; Qiu et al., 2006; Yamada et al., 2009). The S1 subunit interaction with the host receptor plays a major role in determining the host range of CoVs (Kuo et al., 2000). Additionally, this interaction brings about a conformational change in the S2 subunit of S protein thus exposing the hidden fusion peptide which aids in viral entry into the host cellular membrane. The conformational change leads to the formation of a six-helix bundle fusion core that brings the virus and hosts cellular membrane nearby hence, fusing the lipid bilayers. This fusion ensures the entry of viral nucleocapsid into the host cellular cytoplasm (Fung and Liu, 2014; Masters, 2006).
Once the viral nucleocapsid is inside the host cell, it is uncoated thus revealing the viral genomic RNA. This genomic RNA now acts as mRNA for initiating translation of replicase polyprotein. Two open reading frames i.e., ORF1a and ORF1b are present in the replicase gene. Translation of these open reading frames i.e., ORF1a and ORF1b results respectively in the development of polyprotein 1a (pp1a) and a larger polyprotein 1ab (pp1ab) (Brierley et al., 1987). Newly synthesized pp1a and pp1ab undergo autoproteolytic cleavage. Cleavage of pp1a produces 11 non-structural proteins (nsps) i.e., nsp1– nsp11, and that of pp1ab results in the formation of 15 nsps (nsp1–nsp10andnsp12–nsp16). The functions of many nsps are not completely understood, however, the papain-like protease in nsp3 and main protease in nsp5 are responsible for this autoproteolytic cleavage of pp1a and pp1ab, whereas the RNA-dependent RNA polymerase (RdRp) is contained within nsp12 (Baker et al., 1993).
After translation, the replicase now utilizes the viral genomic RNA as a template to synthesize negative sense genomic RNAs which will later serve as a template for synthesis of progeny positive-sense genomic RNAs. Additionally, replicase synthesizes a nested set of subgenomic RNAs (sgRNAs) via discontinuous transcription process(Sawicki et al., 2007). The processes of replication and transcription of CoVs results in the formation of replication-transcription complexes (RTCs), membrane proteins nsp3, nsp4, and nsp6 aids in anchoring these complexes to the intracellular membrane (Oostra et al., 2007).
The translation of sgRNAs leads to the formation of transmembrane proteins (S, M, and E) as well as accessory proteins. After synthesis, the structural proteins are inserted into the endoplasmic reticulum, folded, and then conveyed to ER-Golgi intermediate compartment (ERGIC). Further, the formation of nucleocapsid takes place by encapsulation of progeny genomic RNA which is synthesized by translation of N proteins. With the aid of M proteins, protein-protein interactions occur inside ERGIC which consequently results in assembly of the complete virion (Masters, 2006). Secretory pathways present inside the smooth wall vesicles then help in transporting the newly formed virions and thus fusing them with the plasma membrane to release the mature virus particles. The S proteins of these viruses further infect the other uninfected host cells resulting in an escalation of viral growth inside the host body (Krijnse-Locker et al., 1994; Masters, 2006).
6. CoV infection and er stress
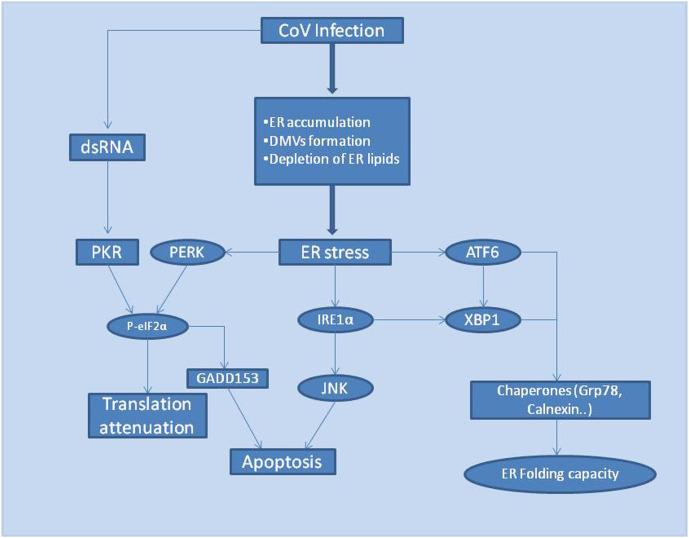
The endoplasmic reticulum is one of the main organelle involved in the synthesis and proper folding of newly synthesized proteins and thus the growth of viral infection (GM, 2000). However, under certain circumstances, the protein folding capacity of ER might not be adequate to carry out proper folding of all the newly synthesized proteins which can result in the accumulation of unfolded proteins in ER leading to ER stress. Unfolded protein response (UPR) is the signaling pathway evolved by the cells to combat ER stress and maintain cellular homeostasis (Shapiro et al., 2017; Ron and Walter, 2007). The accumulation of unfolded proteins in ER triggers the activation of three ER stress transducers, namely: PKR-like ER protein kinase(PERK), activating transcriptional factor-6 (ATF6), and inositol-requiring protein-1 (IRE1) (Tabas and Ron, 2011). The activation of these transducers further initiates UPR signaling. Once activated, this signaling pathway tries to regain cellular homeostasis by increasing the pace of proper protein folding and decreasing the protein synthesis inside ER and if the stage of homeostasis is impossible to achieve, UPR activates apoptotic pathways for the betterment of the organism (Fung and Liu, 2014; Tabas and Ron, 2011).
Research studies around the globe have proposed that SARS-CoV infected cells exhibited an up-regulated expression of genes associated with ER stress i.e., glucose-regulated protein 94 (GRP94) and glucose-regulated protein 78 (GRP78) (Jiang et al., 2003; Yeung et al., 2008). Moreover, current pieces of evidence suggest three chief phenomenons involved in the induction of ER stress by CoV namely: Formation of double-membrane vesicles, glycosylation of CoV structural proteins, and depletion of ER lipid (Fung and Liu, 2014).
-
1.
Formation of double-membrane vesicles (DMVs): CoV infected cells have been shown to induce the formation of DMVs. Cellular membrane modifications are suggested to occur during the replication of various plus-strand RNA viruses and CoV is one among them (David-Ferreira and Manakar, 1965).
As specified by the data gained using electron microscopy, DMVs are found to be located probably in the sites near RTCs in the vicinity of major CoV replicase proteins (Gosert et al., 2002; Snijder et al., 2006). The source of DMV formation is not yet clear but the late endosomes, autophagosomes, and the early secretory pathway are thought to be the originators of DMVs (Prentice et al., 2004; van der Meer et al., 1999; Verheije et al., 2008). A recent study by Reggiori et al., 2010, has suggested that to form DMVs, CoVs seize the EDEMosomes to obtain the ER membrane. The COPII-independent vesicles, EDEMosomes, are usually found in the ER and are responsible for maintaining the level of mannosidase alpha-like1 (EDEM1), a regulator of ER-linked degradation(Calì et al., 2008). So, the above pieces of evidence point towards ER-origin of CoV induced DMVs (Reggiori et al., 2010).
-
2.
Glycosylation of CoV structural proteins: During the process of CoV replication, transmembrane structural proteins (S, E, and M) are synthesized by ER in a tremendous amount to assembly progeny virions. The production, folding, and modifications of such a massive number of proteins put the immense workload on ER, thus enhancing the probability of generating ER stress (Shapiro et al., 2017; Fung and Liu, 2014).
Protein glycosylation is an integral part of protein folding in the ER. The reaction involves the addition of a carbohydrate moiety to the protein molecule (Roth et al., 2012). Excluding N proteins, all other structural proteins are synthesized in a massive amount by ER. In CoVs, based on amino-acid side-chain atoms to which glycans are attached, two types of protein glycosylation are found to take place in ER i.e., O-linked (in beta-CoVs) and N- linked (in alpha and gamma-CoVs) (Cavanagh, 2007; Jacobs et al., 1986; Nal et al., 2005). M protein is one of the most abundant proteins present in CoV and glycosylation of this protein is associated with induction of alpha interferon (IFN) function as well as in vivo tissue tropism (Charley and Laude, 1988; De Haan et al., 2003; Laude et al., 1992). S protein is also highly glycosylated(Masters, 2006), moreover, the glycans on S protein in SARS-CoV have been shown to interact with two alternative receptors of SARS-CoV (independent of major ACE-2 receptor) namely: L-SIGN (liver lymph node-specific intercellular adhesion molecule-3-grabbing non-integrin), and DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin) (Han et al., 2007).
Chaperones are the proteins present inside ER that assist in proper protein folding, maturation as well as assembly. S proteins are highly dependent on calnexin, an ER protein chaperone; moreover, studies have suggested that S2 subunit in S glycoproteins of SARS-CoV interact with calnexin and a decrease in the infectivity of pseudotype lentivirus carrying SARS-CoV S protein was observed after knocking out calnexin (Fukushi et al., 2012). So, striking out chaperones will hamper proper protein folding and consequently assembly of virions, thus decreasing viral spread and infectivity (Fung and Liu, 2014).
-
3.
Depletion of ER lipid: After synthesis and folding of proteins in the ER, they are transported to ERGIC for assembly of the virion. Mature virions are released by the process of exocytosis leaving behind an ER with depleted lipid levels (Fung and Liu, 2014).
The above-mentioned factors play a vital role in the generation of ER stress, which in turn activates the UPR pathway that brings stress response factors like PERK, IRE1, and ATF6 into action (Fig. 5 ). Although the detailed mechanism of molecular interactions between CoV S proteins and stress factors (PERK/IRE1/ATF6) has not been determined, but the layout of CoV induced UPR activation pathways are demonstrated below:
Fig. 5.
Flowchart displaying CoV induced generation of ER stress and activation of the UPR signaling cascade.
CoV induced activation of the PERK pathway: PERK is the first branch to get activated in the UPR pathway after ER stress (Szegezdi et al., 2006). The translation process entirely collapses as the activation of PERK (protein kinase RNA-like endoplasmic reticulum kinase) pathway results in phosphorylation of α-subunit ofeukaryoticinitiationfactor2 (eIF2α), that regulates the mRNA translation machinery, bringing a halt to protein translation (Ron and Walter, 2007). Additionally, the transcription factor GADD153 is responsible for the initiation of CoV induced apoptosis and it was found that the expression of GADD153 is up-regulated during late CoV infection stages (Fung and Liu, 2014; Marciniak et al., 2004; Puthalakath et al., 2007).
CoV induced activation of the IRE1 pathway: Inositol-requiring kinase 1 (IRE1), self-activates its free luminal domain by homodimerization and transautophosphorylation. The activated domain, in turn, activates the transcription factor XBP1 (Xbox binding protein) mRNA by splicing and removing a 26 bp intron (Calfon et al., 2002; Yoshida et al., 2001). Activated XBP1 then induces upstream UPR gene expression (Bechill et al., 2008; Versteeg et al., 2007). Besides, IRE1 also modulates JNK and Akt kinase activities that help infected cells escape virus-induced apoptosis (Fung and Liu, 2014; Urano et al., 2000).
CoV induced activation of the ATF6 pathway: Activating transcription factor (ATF6) is basic in nature consisting of leucine residues. As per studies, CoV infected cells lead to activation of ATF6 pathways which in turn results in the up-regulation of ER chaperone proteins to combat ER stress (Fung and Liu, 2014; Sung et al., 2009).
7. Key SARS-CoV2 targets for vaccine development
Recent pandemic pneumonia outbreak COVID-19, in Wuhan, China is spreading globally and has raised an urgent public health issue worldwide impacting millions of people with a continuous increase in both morbidity and mortality. The causative agent of this disease is identified and named as SARS-CoV2 because of its genetic relatedness to SARS-CoV species that was responsible for the 2003 coronavirus outbreak. The immense spread of the disease in a very small period demands urgent development of therapeutic and prophylactic interventions for the treatment of SARS-CoV2 infected patients.
A vaccine is a biological preparation designed to protect humans from viral and bacterial infections. Vaccines stimulate the production of antibodies inside the human body before disease generation, in the same manner as antibodies are produced after the individuals are exposed to the disease pathogen (Siegrist, 2013). In the last decade, vaccine development technology has evolved significantly, involving the formulation of cell-culture based vaccines (e.g., Flucelvax Tetra cell-based vaccine (Bühler and Ramharter, 2019)), licensed vectored vaccines ((e.g., Ervebo, a vesicular stomatitis virus [VSV]-vectored ebolavirus vaccine (Dumiak, 2019)), several DNA and RNA vaccine candidates and recombinant protein vaccines (e.g., a vaccine against hepatitis B virus (Huzair and Sturdy, 2017; Shang et al., 2020).
There is limited knowledge of how the immune system of humans reacts naturally against SARS-CoV2. The fact that SARS-CoV2 displays a high level of homology to SARS-CoV, the target epitopes for vaccine development of SARS-CoV might prove to be effective for SARS-CoV2 as well (Hoffmann et al., 2020; Letko et al., 2020; Lu et al., 2020). Despite the high level of similarity between both, there are certain genetic variations as well. Studies reveal that only 16% of B cell epitopes and 23% of T cell epitopes of the SARS-CoV map identically to SARS-CoV2 (Ahmed et al., 2020). Key SARS-CoV2 target antigenic sites for vaccine development (Table 3 ) are enlisted below:
Table 3.
Key target regions, accession, length and FASTA sequence of SARS-CoV2 for vaccine development.
| Target region | Accession | Length | FASTA Sequence |
|---|---|---|---|
| Spike protein S2 subunit | 6LXT_F | 132aa | GVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVESGGRGGPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYGG |
| Receptor Binding domain (RBD) | 6VW1_F | 217aa | RVVPSGDVVRFPNITNLCPFGEVFNATKFPSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLNAPATVCGPKLSTDLIK |
| N-Terminal Domain (NTD) | 6YI3_A | 140aa | GAMGLPNNTASWFTALTQHGKEDLKFPRGQGVPINTNSSPDDQIGYYRRATRRIRGGDGKMKDLSPRWYFYYLGTGPEAGLPYGANKDGIIWVATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGSRGGS |
| M protein | Q1Z97065 | 222aa | MADSNGTITVEELKKLLEQWNLVIGFLFLTWICLLQFAYANRNRFLYIIKLIFLWLLWPVTLACFVLAAVYRINWITGGIAIAMACLVGLMWLSYFIASFRLFARTRSMWSFNPETNILLNVPLHGTILTRPLLESELVIGAVILRGHLRIAGHHLGRCDIKDLPKEITVATSRTLSYYKLGASQRVAGDSGFAAYSRYRIGNYKLNTDHSSSSDNIALLVQ |
| E protein | QHU36866 | 75aa | MYSFVSEETGTLIVNSVLLFLAFVVFLLVTLAILTALRLCAYCCNIVNVSLVKPSFYVYSRVKNLNSSRVPDLLV |
| N protein | QIQ22768 | 419aa | MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSKQRRPQGLPNNTASWFTALTQHGKEDLKFPRGQGVPINTNSSPDDQIGYYRRATRRIRGGDGKMKDLSPRWYFYYLGTGPEAGLPYGANKDGIIWVATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGSRGGSQASSRSSSRSRNSSRNSTPGSSRGTSPARMAGNGGDAALALLLLDRLNQLESKMSGKGQQQQGQTVTKKSAAEASKKPRQKRTATKAYNVTQAFGRRGPEQTQGNFGDQELIRQGTDYKHWPQIAQFAPSASAFFGMSRIGMEVTPSGTWLTYTGAIKLDDKDPNFKDQVILLNKHIDAYKTFPPTEPKKDKKKKADETQALPQRQKKQQTVTLLPAADLDDFSKQLQQSMSSADSTQA |
7.1. Spike protein
Earlier vaccine development studies of SARS-CoV and MERS-CoV identified surface S glycoprotein of CoVs to be the most ideal target for vaccine development (Du et al., 2009; Schindewolf and Menachery, 2019). During host cell receptor interaction with the S glycoprotein of the CoV, S protein undergoes cleavage directed by host cell proteases and divides into two subunits: S1 and S2 (Huang et al., 2006; Kuo et al., 2000; Yamada et al., 2009). The S2 subunit then undergoes some conformational change to reveal the hidden fusion peptide that aids in fusing S protein to host cell membrane and thus making a path for viral RNA to enter the host cell cytoplasm (Masters, 2006). Since this surface glycoprotein (S protein) plays the most significant role in the initiation and spread of CoV infection inside an organism, therefore, S protein serves as the most vulnerable target for vaccine development of CoVs (Wrapp et al., 2020).
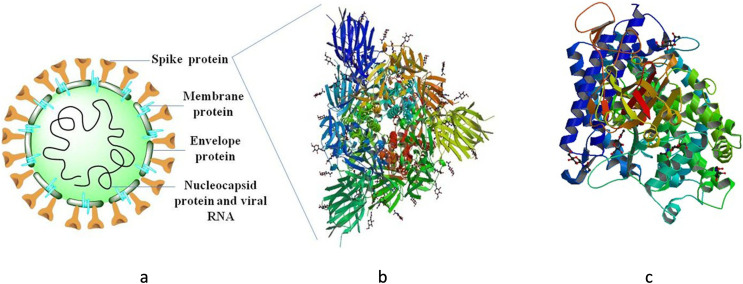
Developing vaccines against full length S protein is beneficial as full-length proteins can provide more target epitopes along with maintaining proper protein conformation and consequently higher immunogenicity (Pallesen et al., 2017; Zhang et al., 2020b). To date, a SARS-CoV-2S protein trimmer (S-Trimer) vaccine prepared using its patented Trimer-Tag© technology is reported to have been developed by Clover Biopharmaceuticals, which will be launched within next 4–5 weeks (Biopharmaceuticals, 2020). Other than full-length protein-based vaccines, specific regions of S protein such as S2 subunit, RBD, NTD, and FP also serve as potential targets for vaccine development (Fig. 6 ).
Fig. 6.
(a) Structure of coronavirus particle displaying different proteins. (b)The S protein is the major target for vaccine development. This picture depicts Electron microscopy obtained image of the Structure of SARS-CoV-2 spike glycoprotein (PDB ID: 6VXX) (c)Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 obtained by X-ray diffraction (PDB ID:6M0J).
7.1.1. Spike S2 subunit
Studies reveal that B cell epitopes in the S2 subunit derived from S protein of SARS-CoV2 map identically to SARS-CoV and thus epitopes of this region might prove to be promising candidates for induction of protective antibody response and thus vaccine development. Preliminary studies for confirming the compatibility of the S2 subunit for vaccine development have already been carried out and the results depict promising nature of these epitopes in generating cross-reactivity and neutralizing antibodies (Ahmed et al., 2020).
7.1.2. Spike Receptor Binding Domain (RBD)
RBD is present in the S1 subunit of SARS-CoV2 and aids in binding the host cell receptor and then fusing the membrane of the host cell to the virus with the help of S2 subunit. SARS-CoV2 binds to the same host cell receptor i.e., ACE-2 (angiotensin-converting enzyme-2) as SARS-CoV (Brielle et al., 2020; Lan et al., 2020). Therefore, like the S2 subunit of the spike, the RBD region also makes a promising candidate as a target region for the development of vaccines against SARS-CoV2. As per the findings from a recent study, the SARS-CoV RBD specific antibodies were observed to cross-react with SARS-CoV2 protein, additionally, SARS-CoV2 was found to be cross-neutralized by induction of RBD SARS-CoV antisera, suggesting that RBD-SARS CoV based vaccines have potential to prevent SARS-CoV2 infection (Tai et al., 2020). In addition, the manipulation of ACE-2 receptor binding domain will prove to be a great therapy as it will ensure preventing viral entry into the host cell.
7.1.3. Spike N-Terminal Domain (NTD)
In several CoV species, NTD like RBD is reported to constitute carbohydrate receptor binding activity. Thus, NTD also serves as a candidate target region for vaccine development. A study demonstrated that an antibody that binds to the MERS-CoV S1 subunit NTD region was found to cross-neutralize the wild-type strain EMC of MERS-CoV (2012) (Chen et al., 2017), thus, depicting the potential of spike NTD region as potential antigen target. However, so far there is not much knowledge available about the function of S1-NTD of SARS-CoV2, a gain of higher understanding might demonstrate its interaction with certain receptors and make it a potential vaccine development target (Zhang et al., 2020b).
7.2. M protein
Membrane protein is the most abundant protein present on the surface of SARS-CoV2 and is involved in virion assembly in the cells. As specified by immunogenic and structural analysis, M protein is found to harbour a T cell epitope cluster that holds the potential to generate a strong cellular immune response (van der Meer et al., 1999), thus depicting the potential of M protein to serve as a target for vaccine development (Jin et al., 2005; Li et al., 2020a; Oh et al., 2012; Zhang et al., 2020b).
7.3. E protein
Envelope glycoproteins are small proteins that are composed of 75 amino acids in SARS-CoV2. The E proteins of CoVs along with M proteins play a crucial role in virion morphogenesis and their assembly within the cell (Siu et al., 2008). As per studies, the potential of recombinant SARS and MERS-CoV with mutated E protein live attenuated vaccines have been explored earlier (Graham et al., 2013; Schoeman and Fielding, 2019; Shang et al., 2020). Moreover, E protein is believed to be a chief virulence factor as knock-out of this protein results in reduced secretion of major inflammatory factors i.e., IL-1, TNF, and IL-6 (Nieto-Torres et al., 2014; Zhang et al., 2020b).
7.4. N protein
Nucleocapsid protein is an integral protein of SARS-CoV2 that aids in the formation of ribonucleoprotein complex also known as capsid by packaging viral genome inside the viral envelope. The formation of the capsid is important for viral self-assembly and replication (Jin et al., 2005; Li et al., 2020b). As specified by the data obtained from a study, 89% of SARS infected patients were able to produce antibodies to this antigen, depicting high antigenicity of N protein (Leung et al., 2004). However, previous studies demonstrate very fluctuating observations regarding the potential of N protein as a target antigen for vaccine development. As per the results obtained from a previous study, the SARS-CoV N protein DNA vaccine was able to induce the production of antibodies in vaccinated C57BL/6 mice and was thus used for the treatment of vaccinia virus (Kim et al., 2004). In contrast, a recent study on SARS-CoV2 claims N protein to be unsuitable for vaccine development because the antibodies induced by the N protein of SARS-CoV2 were found to be incapable of providing immunity to SARS-CoV infection (Gralinski and Menachery, 2020; Shang et al., 2020).
8. Conclusion
Recent pandemic novel coronavirus emerged from Wuhan, China has caused widespread fear and concern and has turned on a global public health security alarm. The direct zoonotic origin of SARS-CoV2 is not confirmed yet, however, genetic sequence studies have revealed bat-origin of this virus as it shares 96.2% genetic homology to bat SARS-related coronavirus strain (SARSr-CoV; RaTG13) procured from Yunnan province, China. Despite the high level of genetic similarity of novel SARS-CoV2 virus to other CoV strains, it is revealed that the SARS-CoV2 genome contains a few distinct characteristics like mutations in the S1-S2 region of the receptor-binding domain, polybasic furin cleavage site, and the presence of o-linked glycans. Although, targeting the regions of difference for the development of drugs and vaccines could be of great help as these genomic variations might be the reason for increased infectivity and severity of the disease, but since the pandemic is causing immense public health damage, it is the need of the hour to come up with a prophylactic or therapeutic intervention at the earliest. So, drug/vaccine repurposing is the current hot research area and researchers are aiming for the use of potential target antigen sequences of previously known coronavirus strains to come up with a suitable vaccine for novel SARS-CoV2 strain.
Additionally, S glycoprotein is responsible for mediating viral entry into human cells and thus aids in determining host range as well as viral infectivity. Restricting its cleavage by manipulating the expression of S protein or ACE-2 receptor binding motif might be a great therapy. Moreover, S protein is the most potential target antigenic site suggesting that RBD-SARS CoV based vaccines have potential to prevent SARS-CoV2 infection. Therapeutics, Moderna Inovio Pharmaceuticals, Clover Biopharmaceuticals, Novavax, Johnson & Johnson, Codagenix, are a few among the various companies working for the development of SARS-CoV2 vaccines.
Although immense global research is being conducted on this novel coronavirus strain, based on the genetic comparison of various coronavirus strains we have enlisted a few potential antigen target sites of SARS-CoV2 for the development of vaccines. More research and in-depth understanding of pathogenic mechanisms and genomic variability of SARS-CoV2 will further help in coming up with more targets for better therapy of COVID-19.
Declaration of Competing Interest
Authors declare no conflicts of interest.
Acknowledgments
Acknowledgement
The authors duly acknowledge the support provided by the Indian Council of Medical Research (ICMR), New Delhi, India, for providing fellowship to Ms.Navpreet kaur (Grant number- 5/4-5/188/Neuro/2019-NCD-1) and Panjab University, Chandigarh, for providing essential research facilities.
References
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12 doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand Gaurav M.A.-N. 2020. Polymerases of Coronaviruses: Structure, Function, and Inhibitors. [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;2–4 doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer C.F. Does mutation rate depend on itself. PLoS Biol. 2008;6:0233–0235. doi: 10.1371/journal.pbio.0060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez-Santos Y., St. John S., Mesecar A. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2014;115 doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.C., Yokomori K., Dong S., Carlisle R., Gorbalenya A.E., Koonin E.V., Lai M.M. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 1993;67:6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechill J., Chen Z., Brewer J.W., Baker S.C. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J. Virol. 2008;82:4492–4501. doi: 10.1128/jvi.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biopharmaceuticals C. 2020. Clover Initiates Development of Recombinant Subunit-Trimer Vaccine for Wuhan Coronavirus (2019-nCoV) [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brielle E.S., Schneidman D., Linial M. The SARS-CoV-2 exerts a distinctive strategy for interacting with the ACE2 human receptor. bioRxiv. 2020 doi: 10.1101/2020.03.10.986398. 2020.03.10.986398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M.E., Binns M.M., Bilimoria B., Blok V.C., Brown T.D., Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler S., Ramharter M. Flucelvax tetra: a surface antigen, inactivated, influenza vaccine prepared in cell cultures. ESMO Open. 2019;4:2018–2019. doi: 10.1136/esmoopen-2018-000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Calì T., Galli C., Olivari S., Molinari M. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem. Biophys. Res. Commun. 2008;371:405–410. doi: 10.1016/j.bbrc.2008.04.098. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.-M., Woo P.C.Y., Lau S.K.P., Tse H., Chen H.-L., Li F. 2008. Spike Protein, S, of Human Coronavirus HKU1 : Role in Viral Life Cycle and Application in Antibody Detection; pp. 1527–1536. [DOI] [PubMed] [Google Scholar]
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charley B., Laude H. Induction of alpha interferon by transmissible gastroenteritis coronavirus: role of transmembrane glycoprotein E1. J. Virol. 1988;62:8–11. doi: 10.1128/jvi.62.1.8-11.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lu S., Jia H., Deng Y., Zhou J., Huang B., Yu Y., Lan J., Wang W., Lou Y., Qin K., Tan W. Erratum: a novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein (emerging microbes & infections (2017) 6 5 (e37)) Emerg. Microbes Infect. 2017;6:e60. doi: 10.1038/emi.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V.C.C., Hung I.F.N., Tang B.S.F., Chu C.M., Wong M.M.L., Chan K.H., Wu A.K.L., Tse D.M.W., Chan K.S., Zheng B.J., Peiris J.S.M., Sung J.J.Y., Yuen K.Y. Viral replication in the Nasopharynx is associated with Diarrhea in patients with severe acute respiratory syndrome. Clin. Infect. Dis. 2004;38:467–475. doi: 10.1086/382681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleri D.J., Ricketti A.J., Vernaleo J.R. Severe acute respiratory syndrome (SARS) Infect. Dis. Clin. N. Am. 2010;24:175–202. doi: 10.1016/j.idc.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellis . 2020. Disease Briefing: Coronaviruses. Dis. Brief. Coronaviruses; pp. 1–81. [Google Scholar]
- Czub M., Weingartl H., Czub S., He R., Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine. 2005;23:2273–2279. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro David J., Livezey Mara, Yu Liqun, Zheng Xiaobin. Anticipatory UPR activation: a protective pathway and target in cancer. Physiol. Behav. 2017;176:139–148. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Ferreira J.F., Manakar R.A. An electron microscope study of the development of a mouse Hepatitis virus in tissue culture cells. J. Cell Biol. 1965;24:57–78. doi: 10.1083/jcb.24.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haan C.A.M., De Wit M., Kuo L., Montalto-Morrison C., Haagmans B.L., Weiss S.R., Masters P.S., Rottier P.J.M. The glycosylation status of the murine hepatitis coronavirus M protein affects the interferogenic capacity of the virus in vitro and its ability to replicate in the liver but not the brain. Virology. 2003;312:395–406. doi: 10.1016/S0042-6822(03)00235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Holland J.J., Ahlquist P. Variability of RNA genomes. RNA Genetics. 1988 [Google Scholar]
- Dong N., Yang X., Ye L., Chen K., Chan E.W.-C., Chen S. Genomic and protein structure modelling analysis depicts the origin and pathogenicity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China. F1000Research. 2020;9:121. doi: 10.12688/f1000research.22357.2. [DOI] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas G., Carvalho L.M., Rambaut A., Bedford T. 2018. MERS-CoV Spillover at the Camel-Human Interface; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16:1–6. doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumiak M. Proven against Ebola, a vector shows its broader potential. IAVI Rep. 2019;23:4–9. [Google Scholar]
- Murphy E.A., Fauquet C.M., Ghabrial S.A., Jarvis A.W., Martelli G.P., Mayo M.A. Virus taxonomy classification and nomenclature of viruses. Encyclopedia of Parasitology. 2016 doi: 10.1007/978-3-642-27769-6_290-2. [DOI] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi M., Yoshinaka Y., Matsuoka Y., Hatakeyama S., Ishizaka Y., Kirikae T., Sasazuki T., Miyoshi-Akiyama T. Monitoring of S protein maturation in the endoplasmic reticulum by Calnexin is important for the infectivity of severe acute respiratory syndrome coronavirus. J. Virol. 2012;86:11745–11753. doi: 10.1128/jvi.01250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:1–13. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X. Lou, Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J., Luo C.M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.Y., Wang L.F., Daszak P., Shi Z.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gélinas A.M., Boutin M., Sasseville A.M.J., Dea S. Bovine coronaviruses associated with enteric and respiratory diseases in Canadian dairy cattle display different reactivities to anti-HE monoclonal antibodies and distinct amino acid changes in their HE, S and ns4.9 protein. Virus Res. 2001;76:43–57. doi: 10.1016/S0168-1702(01)00243-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GM C. 2nd edition. 2000. The Cell: A Molecular Approach. [Google Scholar]
- Gorbalenya A.E., Snijder E.J., Spaan W.J.M. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78:7863–7866. doi: 10.1128/jvi.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse Hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/jvi.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouilh M.A., Puechmaille S.J., Gonzalez J.P., Teeling E., Kittayapong P., Manuguerra J.C. SARS-coronavirus ancestor’s foot-prints in south-east Asian bat colonies and the refuge theory. Infect. Genet. Evol. 2011;11:1690–1702. doi: 10.1016/j.meegid.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Menachery V.D. Return of of the the coronavirus. Viruses. 2020;12:1–8. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory T.R. 2009. Understanding Natural Selection: Essential Concepts and Common Misconceptions; pp. 156–175. [DOI] [Google Scholar]
- Guo J.P., Petric M., Campbell W., McGeer P.L. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology. 2004;324:251–256. doi: 10.1016/j.virol.2004.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.P., Lohani M., Cho M.W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007;81:12029–12039. doi: 10.1128/jvi.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Krüger N., Müller M., Drosten C., Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. 2020 doi: 10.1101/2020.01.31.929042. 2020.01.31.929042. [DOI] [Google Scholar]
- Hofmann H., Pyrc K., Van Der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C.-C., Lam T.-Y., Shi Z.-L., Drummond A.J., Yip C.-W., Zeng F., Lam P.-Y., Leung F.C.-C. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008;82:1819–1826. doi: 10.1128/jvi.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Zeng L.P., Yang X. Lou, Ge X.Y., Zhang W., Li B., Xie J.Z., Shen X.R., Zhang Y.Z., Wang N., Luo D.S., Zheng X.S., Wang M.N., Daszak P., Wang L.F., Cui J., Shi Z.L. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:1–27. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bosch B.J., Li F., Li W., Kyoung H.L., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J.M., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huzair F., Sturdy S. Biotechnology and the transformation of vaccine innovation: the case of the hepatitis B vaccines 1968–2000. Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci. 2017;64:11–21. doi: 10.1016/j.shpsc.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre G. The proteolytic regulation of virus cell entry by Furin and other proprotein convertases. Viruses. 2019;11 doi: 10.3390/v11090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs L., van der Zeijst B.A., Horzinek M.C. Characterization and translation of transmissible gastroenteritis virus mRNAs. J. Virol. 1986;57:1010–1015. doi: 10.1128/jvi.57.3.1010-1015.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiabao Xu S.Z. SARS-CoV-2 and SARS-CoV; 2014. Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.-Y., Wek S.A., McGrath B.C., Scheuner D., Kaufman R.J., Cavener D.R., Wek R.C. Phosphorylation of the subunit of eukaryotic initiation factor 2 is required for activation of NF- B in response to diverse cellular stresses. Mol. Cell. Biol. 2003;23:5651–5663. doi: 10.1128/mcb.23.16.5651-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Xiao C., Chen Z., Kang Y., Ma Y., Zhu K., Xie Q., Tu Y., Yu Y., Wang B. Induction of Th1 type response by DNA vaccinations with N, M, and E genes against SARS-CoV in mice. Biochem. Biophys. Res. Commun. 2005;328:979–986. doi: 10.1016/j.bbrc.2005.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsdottir H.R., Dijkman R. Coronaviruses and the human airway: a universal system for virus-host interaction studies coronaviruses: emerging and re-emerging pathogens in humans and animals Susanna Lau emerging viruses. Virol. J. 2016;13:1–9. doi: 10.1186/s12985-016-0479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Kazmi S., Bashir A., Siddique N. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Lee J.H., Hung C.-F., Peng S., Roden R., Wang M.-C., Viscidi R., Tsai Y.-C., He L., Chen P.-J., Boyd D.A.K., Wu T.-C. Generation and characterization of DNA vaccines targeting the Nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:4638–4645. doi: 10.1128/jvi.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijnse-Locker J., Ericsson M., Rottier P.J.M., Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Godeke G.-J., Raamsman M.J.B., Masters P.S., Rottier P.J.M. Retargeting of coronavirus by substitution of the spike glycoprotein Ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix A., Duong V., Hul V., San S., Davun H., Omaliss K., Chea S., Hassanin A., Theppangna W., Silithammavong S., Khammavong K., Singhalath S., Greatorex Z., Fine A.E., Goldstein T., Olson S., Joly D.O., Keatts L., Dussart P., Afelt A., Frutos R., Buchy P. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia. Infect. Genet. Evol. 2017;48:10–18. doi: 10.1016/j.meegid.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. Coronavirus: organization, replication and expression of genome. Annu. Rev. Microbiol. 1990;44:303–333. doi: 10.1146/annurev.micro.44.1.303. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.L., Wong S.S.Y., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Goeman J.J., de Parquet M.C., Thi Nga P., Snijder E.J., Morita K., Gorbalenya A.E. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Gelfi J., Lavenant L., Charley B. Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J. Virol. 1992;66:743–749. doi: 10.1128/jvi.66.2.743-749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.T.M., Tam F.C.H., Ma C.H., Chan P.K.S., Cheung J.L.K., Niu H., Tam J.S.L., Lim P.L. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral Nucleocapsid. J. Infect. Dis. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Jonathan H., Wang H., Crameri Gary, Hu Z., Zhang H., Zhang J., Mceachern J., Field H., Daszak P., Eaton B.T., Zhang S., Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri Galy, Hu Z., Zhang H., Zhang J., Eaton B.T., Zhang S., Wang L. Bats Are Natural Reservoirs of SARS-like Coronaviruses Published by : American Association for the Advancement of Science Stable. 2005. http://www.jstor.org/stable/3842715 URL. (REFERENCES Linked references are available on JSTOR for this article : You may need. Science (80-. ). 310, 676–679) [DOI] [PubMed]
- Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Lin L., Wang H., Yin J., Ren Y., Zhao Z., Wen J., Zhou C., Zhang X. 2020. The Epitope Study on the SARS-CoV Nucleocapsid Protein Shuting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Saif L.J., Marthaler D., Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Hill W.G. The population genetics of mutations: good, bad and indifferent. Philos. Trans. R. Soc. B Biol. Sci. 2010;365:1153–1167. doi: 10.1098/rstb.2009.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Ph D., Wang D., Ph D., Xu W., Wu G., Gao G.F., Phil D., Tan W., Ph D. 2020. A Novel Coronavirus from Patients with Pneumonia in China, 2019; pp. 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak S.J., Yun C.Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H.P., Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;65:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer Y., Snijder E.J., Dobbe J.C., Schleich S., Denison M.R., Spaan W.J.M., Locker J.K. Localization of mouse Hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Dinnon K.H., Yount B.L., McAnarney E.T., Gralinski L.E., Hale A., Graham R.L., Scobey T., Anthony S.J., Wang L., Graham B., Randell S.H., Lipkin W.I., Baric R.S. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 2019 doi: 10.1128/jvi.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S.S., Schluederberg A. From the National Institute of Allergy and Infectious Diseases, the Fogarty International Center of the National Institutes of Health, and the Rockefeller University. Emerging viruses: the evolution of viruses and viral diseases. J. Infect. Dis. 1990;162:1–7. doi: 10.1093/infdis/162.1.1. [DOI] [PubMed] [Google Scholar]
- Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli sabelle, Crescenzo-Chaigne B., Escriou N., van der Wef S., Yuen K.Y., Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Nao N. Vol. 8. 2017. Genetic Predisposition To Acquire a Polybasic Cleavage Site for Highly Pathogenic Avian Influenza Virus Hemagglutinin Naganori; pp. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., DeDiego M.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Castaño-Rodriguez C., Alcaraz A., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein Ion Channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H.L.J., Gan S.K.E., Bertoletti A., Tan Y.J. Understanding the T cell immune response in SARS coronavirus infection. Emerg. Microbes Infect. 2012;1:0. doi: 10.1038/emi.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., te Lintelo E.G., Deijs M., Verheije M.H., Rottier P.J.M., de Haan C.A.M. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 2007;81:12323–12336. doi: 10.1128/jvi.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., Kong W.P., Andres E.L., Kettenbach A.N., Denison M.R., Chappell J.D., Graham B.S., Ward A.B., McLellan J.S. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck K.M., Lauring A.S. Complexities of viral mutation rates. J. Virol. 2018;92:1–8. doi: 10.1128/jvi.01031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H., O’Reilly L.A., Gunn P., Lee L., Kelly P.N., Huntington N.D., Hughes P.D., Michalak E.M., McKimm-Breschkin J., Motoyama N., Gotoh T., Akira S., Bouillet P., Strasser A. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Qiu Z., Hingley S.T., Simmons G., Yu C., Das Sarma J., Bates P., Weiss S.R. Endosomal proteolysis by Cathepsins is necessary for murine coronavirus mouse Hepatitis virus type 2 spike-mediated entry. J. Virol. 2006;80:5768–5776. doi: 10.1128/jvi.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S., Bernasconi R., De Haan C.A.M., Molinari M. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihtarič D., Hostnik P., Steyer A., Grom J., Toplak I. Identification of SARS-like coronaviruses in horseshoe bats (Rhinolophus hipposideros) in Slovenia. Arch. Virol. 2010;155:507–514. doi: 10.1007/s00705-010-0612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson B. 2020. COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles’ heel conserved region to minimize probability of escape mutations and drug resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Roth Z., Yehezkel G., Khalaila I. Review article identification and quantification of protein glycosylation. Int. J. Carbohydr. Chem. 2012;640923 doi: 10.1155/2012/640923. [DOI] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/jvi.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindewolf C., Menachery V.D. Middle east respiratory syndrome vaccine candidates: cautious optimism. Viruses. 2019;11 doi: 10.3390/v11010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W., Yang Y., Rao Y., Rao X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. npj Vaccines. 2020;5:2–4. doi: 10.1038/s41541-020-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shults R.A., Cen- N., Hepatitis V., Ruth A., Lindsey T., Highway N., Safety T., Services H., Safety H., Services H., J-p A., Force T. Severe Respiratory Illness Associated Coronavirus. Am. Med. Assoc. 2012;308 [Google Scholar]
- Siegrist C.-A. Vaccines - CH 2 how do vaccines mediate protection? Vaccines. 2013:16–23. doi: 10.1016/B978-1-4557-0090-5.00004-5. [DOI] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S.M., Bruzzone R., Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/jvi.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J.M., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/jvi.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Wang Y., Wang N., Wang D., Guo J., Fu L., Shi X. Identification of residues on human receptor DPP4 critical for MERS-CoV binding and entry. Virology. 2014;471:49–53. doi: 10.1016/j.virol.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Brian D., Cavanagh D., de Groot R., Enjuanes L., Gorbalenya A.E., Holmes K., Masters P.S., Rottier P., Taguchi F., Talbot P. Family Coronaviridae. Virus Taxon. Eighth Rep. Int. Comm. Taxon. Viruses. 2012:947–964. [Google Scholar]
- Steinhauer D. Rapid Evolution Of RNA Viruses. Annu. Rev. Microbiol. 1987;41:409–433. doi: 10.1146/annurev.micro.41.1.409. [DOI] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S.C., Chao C.Y., Jeng K.S., Yang J.Y., Lai M.M.C. The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology. 2009;387:402–413. doi: 10.1016/j.virol.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegezdi E., Logue S.E., Gorman A.M., Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I., Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z., Tian J., Pei Y., Yuan M., Zhang Y., Dai F. 2020. A new coronavirus associated with human respiratory disease in China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Conrardy C., Ruone S., Kuzmin I.V., Guo X., Tao Y., Niezgoda M., Haynes L., Agwanda B., Breiman R.F., Anderson L.J., Rupprecht C.E. Detection of novel SARS-like and other coronaviruses in bats from Kenya. Emerg. Infect. Dis. 2009;15:482–485. doi: 10.3201/eid1503.081013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F., Wang X.Z., Bertolotti A., Zhang Y., Chung P., Harding H.P., Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science (80-. ) 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Van Boheemen S., De Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D.M.E., Haagmans B.L., Gorbalenya A.E., Snijder E., Fouchier R.A.M. Genomic characterization of a newly discovered coronavirus. MBio. 2012;3:1–9. doi: 10.1128/mBio.00473-12.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankadari N., Wilce J.A. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega V.B., Ruan Y., Liu J., Lee W.H., Wei C.L., Se-Thoe S.Y., Tang K.F., Zhang T., Kolatkar P.R., Ooi E.E., Ling A.E., Stanton L.W., Long P.M., Liu E.T. Mutational dynamics of the SARS coronavirus in cell culture and human populations isolated in 2003. BMC Infect. Dis. 2004;4:1–9. doi: 10.1186/1471-2334-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Raaben M., Mari M., Te Lintelo E.G., Reggiori F., Van Kuppeveld F.J.M., Rottier P.J.M., De Haan C.A.M. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., van de Nes P.S., Bredenbeek P.J., Spaan W.J.M. The coronavirus spike protein induces endoplasmic reticulum stress and Upregulation of intracellular chemokine mRNA concentrations. J. Virol. 2007;81:10981–10990. doi: 10.1128/jvi.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M., Andino R. Closing the gap: the challenges in converging theoretical, computational, experimental and real-life studies in virus evolution. Curr. Opin. Virol. 2012;2:515–518. doi: 10.1016/j.coviro.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasak R., Luytjes W., Spaan W., Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U. S. A. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S., Duengkae P., Rodpan A., Kaewpom T., Maneeorn P., Kanchanasaka B., Yingsakmongkon S., Sittidetboripat N., Chareesaen C., Khlangsap N., Pidthong A., Leadprathom K., Ghai S., Epstein J.H., Daszak P., Olival K.J., Blair P.J., Callahan M.V., Hemachudha T. Diversity of coronavirus in bats from eastern Thailand emerging viruses. Virol. J. 2015;12:1–7. doi: 10.1186/s12985-015-0289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.N., Zhang W., Gao Y.T., Hu B., Ge X.Y., Yang X. Lou, Zhang Y.Z., Shi Z.L. Longitudinal surveillance of SARS-like coronaviruses in bats by quantitative real-time PCR. Virol. Sin. 2016;31:78–80. doi: 10.1007/s12250-015-3703-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Yang S.L.X., Zhang H.H.Y., Guo H., Luo C., Miller M., Zhu G., Chmura A.A., Hagan E., Zhang J.Z.Y., Peter L.W., Shi D.Z. Serological evidence of bat SARS-related coronavirus infection in humans , China. Virol. Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WO Z.G. 2020. Sequence Analysis Indicates that 2019-nCoV Virus Contains a Putative Furin Cleavage Site at the Boundary of S1 and S2 Domains of Spike Protein 1–9. [DOI] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Chu C.-m., Chan K.-h., Tsoi H.-w., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W.-k., Poon L.L.M., Wong S.S.Y., Guan Y., Peiris J.S.M., Yuen K.-y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/jvi.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Wang M., Lau S.K.P., Xu H., Poon R.W.S., Guo R., Wong B.H.L., Gao K., Tsoi H.-w., Huang Y., Li K.S.M., Lam C.S.F., Chan K.-h., Zheng B.-j., Yuen K.-y. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2007;81:1574–1585. doi: 10.1128/jvi.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lai K.K.Y., Huang Y., Lee P., Luk G.S.M., Dyrting K.C., Chan K.-H., Yuen K.-Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 2009;83:908–917. doi: 10.1128/jvi.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1805–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M., Zheng B.-J., Chan K.-H., Yuen K.-Y. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavi. J. Virol. 2012;86:3995–4008. doi: 10.1128/jvi.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.aax0902. (80-. ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yang L., Ren X., Zhang J., Yang F., Zhang S., Jin Q. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2016;213:579–583. doi: 10.1093/infdis/jiv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Liu D.X. Vol. 83. 2009. Proteolytic Activation of the Spike Protein at a Novel RRRR/S Motif Is Implicated in Furin-Dependent Entry , Syncytium Formation , and Infectivity of Coronavirus Infectious Bronchitis Virus in Cultured Cells; pp. 8744–8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Liu X.B., Fang S.G., Tay F.P.L., Liu D.X. Acquisition of cell-cell fusion activity by amino acid substitutions in spike protein determines the infectivity of a coronavirus in cultured cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager Curtis L., Ashmun Richard A., Williams Richard K., Cardellichio Christine B., Shapiro Linda H. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;359:710–713. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung Y.S., Yip C.W., Hon C.C., Chow K.Y.C., Ma I.C.M., Zeng F., Leung F.C.C. Transcriptional profiling of Vero E6 cells over-expressing SARS-CoV S2 subunit: insights on viral regulation of apoptosis and proliferation. Virology. 2008;371:32–43. doi: 10.1016/j.virol.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Wan Yushun, Shang Jian, Graham Rachel, Baric Ralph S. 2020. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. 2020. Pangolin homology associated with 2019-nCoV. [Google Scholar]
- Zhang J., Zeng H., Gu J., Li H., Zheng L., Zou Q. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines. 2020;8:153. doi: 10.3390/vaccines8020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Li H., Wu X., Zhong Y., Zhang K., Zhang Y.P., Boerwinkle E., Fu Y.X. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 2004;4:1–9. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. Vol. 16. 2020. SARS-CoV-2 : an Emerging Coronavirus that Causes a Global Threat. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X. Lou, Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–259. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X., Wang Xian-Guang, Hu B., Zhang L., Zhang W., Guo H., Jiang R., Liu M., Chen Y., Shen X., Wang Xi, Zhan F., Wang Y., Xiao G., Shi Z. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;2019 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., Hughes A.C., Bi Y., Shi W. A novel bat coronavirus reveals natural insertions at the S1/S2 cleavage site of the Spike protein and a possible recombinant origin of HCoV-19. bioRxiv. 2020 doi: 10.1101/2020.03.02.974139. 2020.03.02.974139. [DOI] [Google Scholar]