Erythroid cell lines are important tools for the study of erythropoiesis in health and disease, enabling manipulation of molecular targets. However, until recently, available continuous cell lines with erythroid characteristics were mainly derived from patients (K562, HEL) or mouse (MEL) with erythroleukaemia, and do not represent ‘normal’ erythroid cells or undergo terminal erythroid differentiation.
Although erythroid cells can be generated in vitro from peripheral blood stem cells, the approach is severely limited by their restricted expansion potential1 and thus the number of erythroid cells generated, with repeat collections required. Approaches to derive erythroid cells from more sustainable induced pluripotent stem cells are confounded by poor levels of erythroid cell expansion, aberrant or incomplete erythroid differentiation and predominantly fetal/embryonic rather than adult globin expression.2,3
Major improvements to the repertoire of human erythroid lines came about in 2013 with the inception of HUDEP-2,4 and in 2017 with BEL-A.5 We have reported improved erythroid differentiation and enucleation rates for BEL-A5,6 compared to those published for HUDEP-2,4,7 however the behavior and properties of these two lines have not been directly compared under the same conditions.
BEL-A and HUDEP-2 were generated using the same doxycycline-inducible HPV16-E6/E7 construct.4 However, BEL-A was created from adult5 and HUDEP-2 from cord blood CD34+ cells.4 In addition, CD34+ cells for BEL-A were cultured in an optimised erythroid culture medium prior to induction of immortalisation,5 whereas CD34+ cells for the HUDEP-2 line were cultured in a non-erythroid specific medium.4 Following induction of E6/E7 expression in early erythroblasts, both lines were transferred to the same expansion medium.4,5 A schematic of the protocols is shown in Figure 1A. Both established lines are maintained in the latter medium for expansion. Doubling time for BEL-A during expansion is faster than HUDEP-2, 18.7±2.0 hours versus 22.7±5.0 hours respectively (P<0.01).
Figure 1.
Improved erythroid differentiation of HUDEP-2 in Bristol erythroid culture system, and comparison of BEL-A and HUDEP-2 differentiation. (A) Schematic of approaches for establishing the BEL-A and HUDEP-2 cell lines.4,5 Erythroid 1° medium6 consists of IMDM with 3% human AB serum, 2% fetal bovine serum, insulin, heparin, holo-transferrin, SCF, IL-3 and EPO. SS: Stemspan™ SFEM; SCF: stem cell factor; IL-3: interleukin-3; EPO: erythropoietin; DEX: dexamethasone; DOX: doxycycline. (B) Cumulative fold expansion (i), % enucleation per total live cells (ii) and reticulocyte yield (iii) of BEL-A and HUDEP-2 in Bristol differentiation media and HUDEP-2 in EDM media. Live cell numbers were determined by trypan blue exclusion assay. Enucleation rates were calculated from cytospin counts (≥200 cells per slide). Reticulocyte yield was calculated by multiplying % enucleation by cumulative fold expansion for each time point. Results are mean ± standard deviation (SD) from at least three independent experiments. */♦P<O.05, **/♦♦P<0.01, ***/♦♦♦P<0.001, ****/♦♦♦♦P<0.0001 two-tailed Student t-test comparing HUDEP-2 in Bristol with HUDEP-2 in EDM media (♦), or BEL-A with HUDEP-2 in Bristol media (*). (C) Percentage of erythroid cell types present at day 0, 4, 6, 8 and 12 of differentiation for BEL-A and HUDEP-2 in Bristol differentiation media, and HUDEP-2 in EDM media. Cells were stained with Leishman’s and ≥200 cells counted per cytospin. Data show mean ± SD, n=3, and (D) representative images from cytospins of these cultures. Scale bar =20 μm. Arrow heads represent the following cell types: white, proerythroblast; blue, basophilic normoblasts; orange, polychromatic normoblasts; black, orthochromatic normoblasts; red, reticulocytes. (E) Relative proportion of erythroid cell types present at day 8 and 12 of differentiation for BEL-A and HUDEP-2 in Bristol differentiation media, and HUDEP-2 in EDM media, presented as extrapolated cell number. (F) Flow cytometry analysis of cell surface marker expression of BEL-A and HUDEP-2 in Bristol differentiation media. Cells were dual stained with anti-CD36 or anti-α4-integrin FITC conjugates (both Miltenyi Biotec) and anti-band 3 (BRIC71) or anti-glycophorin A (GPA) (BRIC256) primary antibodies (both IBGRL, Filton) used in conjunction with an IgG1 APC secondary (Biolegend). (G) Cell surface expression of band 3, α4-integrin, GPA and CD36 of BEL-A and HUDEP-2 nucleated cells (as determined by Hoechst staining) during erythroid differentiation, presented as median fluorescence intensity (MFI). Data show mean ± SD, n=4. (H) Cell surface expression of α4-integrin, CD36 and CD71 of BEL-A and HUDEP-2 orthochromatic normoblasts at day 11 of erythroid culture, presented as median fluorescence intensity (MFI). Data show mean ± SD, n=4.
We initially compared the performance of HUDEP-2 in the published, improved differentiation culture system for this line (EDM)7 and the improved version of our Bristol erythroid differentiation medium.6 As can be seen from Figure 1Bi, the expansion profile and rates were similar in both media. However, differentiation was slower in the EDM medium (Figure 1C-D) and, although cells progressed to orthochromatic normoblasts, enucleation rates (calculated as the percent reticulocytes per total number of live cells) were significantly lower at all time points analysed (Figure 1Bii, C-D). Increasing the culture duration did not result in further enucleation, the remaining orthochromatic normoblasts instead dying.
We next compared the performance of BEL-A and HUDEP-2 in Bristol medium. The expansion profile and rate for the two lines was not significantly different (Figure 1Bi), however the differentiation rate of BEL-A was faster than HUDEP-2, with a greater number of later stage erythroid cells present by day 6 of BEL-A cultures and at all time points thereafter (Figure 1C-D). The reduction in cell numbers after day 8 was predominantly due to apoptosis of orthochromatic normoblasts that fail to enucleate (more clearly seen in Figure 1E), as determined by Annexin V staining and flow cytometry, although we cannot exclude some progressive loss of reticulocytes.
For more detail, differentiation was analysed using the inverse expression of erythroid membrane proteins band 3 versus α4-integrin8 and GPA versus CD36; band3 and GPA are known to increase, and α4-integrin and CD36 to decrease during differentiation.8 The two lines showed a similar overall differentiation profile, but again an increased rate of differentiation was evident for BEL-A (Figure 1F). This is clearer when comparing mean fluorescence intensity (MFI) values for each protein individually (Figure 1G), with differentiating BEL-A cells having significantly higher levels of band 3 and GPA at all time points until maximum levels achieved, and significantly lower levels of CD36 from day 4 and α4-integrin from day 7 of culture. The level of CD36, α4-integrin, and also CD71, remained higher on HUDEP-2 than BEL-A orthochromatic normoblasts (Figure 1H) even with extended time in culture.
Enucleation rates were significantly higher for BEL-A than HUDEP-2 cultures at all time points analysed (Figure 1Bii). At day 12 there were 41±1% reticulocytes in BEL-A compared to 19±2% in HUDEP-2 cultures, with the remaining cells orthochromatic normoblasts in both (Figure 1Bii, C-D).
However, ‘enucleation rates’ are artificially inflated when viability of cultures decrease as values are calculated from the number of reticulocytes per total number of live cells, i.e. as more nucleated cells die the proportion of reticulocytes, and thus the ‘apparent’ enucleation rate increases. They do not therefore provide an accurate or transparent representation of true enucleation potential. For example, for HUDEP-2 the viability of cells at day 12 was decreased to ~67% (compared to ~81% for BEL-A), and when considered gave an enucleation rate of ~13% rather than ~19%. To overcome this discrepancy, we have therefore alternatively presented all data as reticulocyte yield, calculated as percent enucleation x cumulative fold expansion (live cell number relative to original input cell number) at a given time point, thus taking into account nucleated cell death. For lines, yield is calculated from transfer of cells to differentiation medium. We propose this method be adopted by others as it enables a far truer comparison of enucleation potential between cell lines and between different culture systems.
The reticulocyte yield was also significantly higher for BEL-A cultures at all time points analysed (Figure 1Biii), 4.3-fold higher than HUDEP-2 cultures at day 12. The yield of HUDEP-2 cultures in Bristol medium was higher than in EDM at all time points, but values only reached significance at day 8 (P<0.001).
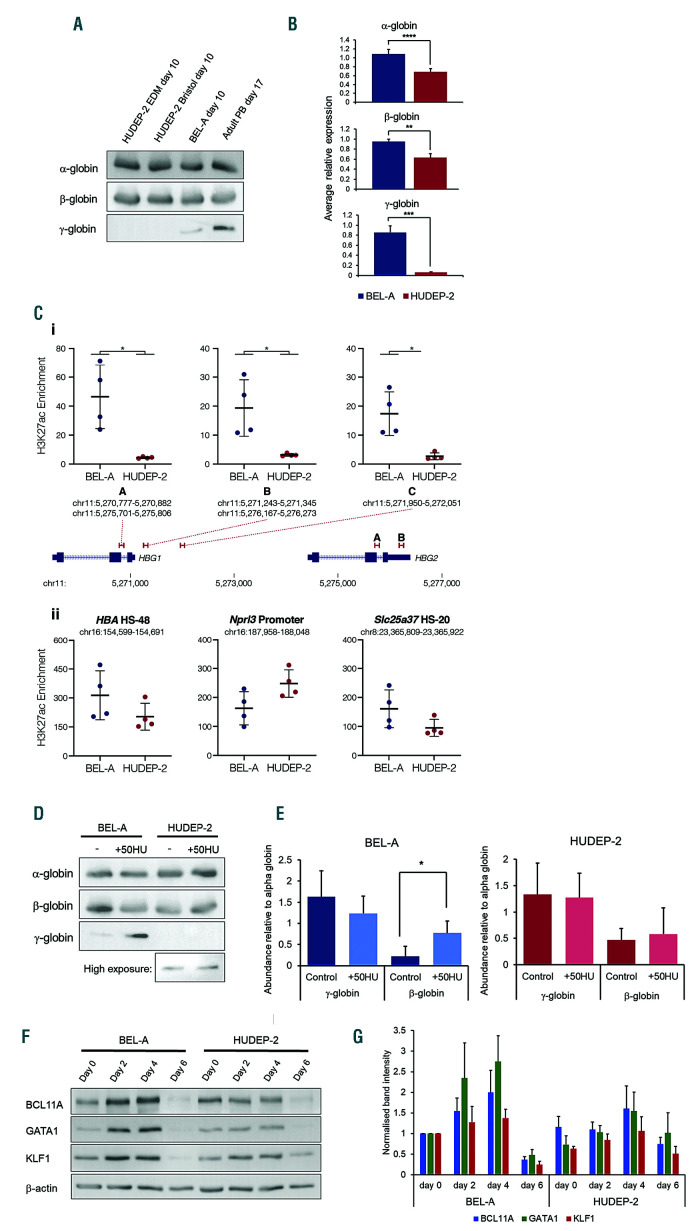
Although created from cord blood CD34+ cells, HUDEP-2 expresses β- rather than γ-globin. We were therefore interested to see whether globin profiles were equivalent to those of BEL-A, BEL-A having levels of adult globin equivalent to adult erythroid cells.5 When the level of α- and β-globin protein was compared by immunoblotting no difference was apparent (Figure 2A). However, when analysed by quantitative PCR (qPCR) significantly higher levels of both α- and β-globin transcripts were detected in BEL-A (Figure 2B), not due to differences in control PABPC1 levels. We have also previously shown that the level of γ-globin is lower in BEL-A than in cultured adult erythroid cells.5 However, the level of γ-globin was extremely low in HUDEP-2, significantly lower than in BEL-A (Figure 2A-B). Consistently, chromatin immunoprecipitation qPCR (ChIP-qPCR) showed significantly higher levels of the active histone mark histone-3 lysine-27 acetylation (H3K27ac) at the HBG1/2 promoters in the BEL-A line (Figure 2Ci); while the lines had similar levels of H3K27ac at control loci (Figure 2Cii). Such data may be important as there is much interest in the reactivation of fetal globin in adult erythroid cells as a treatment for hemoglobinopathies, with cell lines an important tool for evaluating novel drugs and reagents. However, although increased levels of γ-globin can be induced in HUDEP-2 via gene editing,9,10 the cells have been reported unable to respond to exogenous administration of drugs known to increase γ-globin in adult erythroid cells.11 In agreement, whereas addition of hydroxyurea to BEL-A cultures induced a significant increase (3.7-fold) in γ-globin levels, there was no significant effect detected in HUDEP-2 cells (Figure 2D-E). Analysis by flow cytometry showed the majority of BEL-A cells responded to hydroxyurea treatment.
Figure 2.
Globin subunit expression by BEL-A and HUDEP-2. (A). Western blots of lysates obtained from late stage (day 10) HUDEP-2 in EDM media, HUDEP-2 and BEL-A in Bristol media and adult peripheral blood (PB; day 17) erythroid cells incubated with antibodies to α-, β- and γ-globin (sc-514378; sc-21757; sc-21756; all Santa Cruz). (B) Analysis of α-, β- and γ-globin mRNA levels in BEL-A and HUDEP-2 in Bristol media on day 6 of culture by quantitative PCR (qPCR). Target gene expression was normalised to PABPC1 reference gene (using the 2-DDCt method) and compared to a BEL-A control sample. Results are means ± standard deviation (SD), n=4. TaqMan gene expression assays used were: HBA (Hs00361191_g1), HBB (Hs00747223_g1), HBG (Hs00361131_g1) and PABPC1 (Hs00743792_s1) (Fisher Scientific). (C) H3K27ac chromatin immunoprecipitation (ChIP) was carried out in 5×106 day 4 cells by fixing in 1% formaldehyde and performing immunoprecipitation (IP) with the ChIP Assay Kit (Merck Millipore) using 0.3 mg anti-H3K27ac (ab4729; Abcam). Enrichment at the HBG1/2 promoters (i) and control regions (ii) was determined using qPCR by comparing to a background control (hg19, chr16:6,002,736-6,002,834). Bars depict mean and standard deviation for IP from four separate differentiations (circles). *Mann-Whitney rank sum P=0.0286 (D-E) Response of BEL-A and HUDEP-2 cells to hydroxyurea treatment. Hydroxyurea (H8627; Sigma) was added to cells at 50 μM every two days from four days prior to the start of differentiation until harvesting of cells on day 10 of differentiation. (D) Representative Western blot of cell lysates from ± hydroxyurea (HU) cultures incubated with antibodies to α-, β- and γ-globin and (E) densitometric analysis of the bands obtained from at least three independent experiments normalised to α-globin expression (means ± SD). *P<0.05 two-tailed Student t-test. (F-G) Expression of BCL11A, GATA1 and KLF1 by BEL-A and HUDEP-2. β-actin was used as a protein loading control. (F) Representative western blots of lysates obtained from cells at day 0, 2, 4 and 6 of BEL-A and HUDEP-2 cultures incubated with BCL11A (ab19487; Abcam), GATA1 (sc-266; Santa Cruz), KLF1 (sc-14034; Santa Cruz) and β-actin (A1978; Sigma) antibodies, and (G) densitometric analysis of the bands obtained from two independent experiments normalised to β-actin expression and then compared to BEL-A day 0 (means ± SD).
The level of GATA1, KLF1 and BCL11A, key transcription factors for erythroid differentiation and β-globin expression, were overall lower in HUDEP-2 compared to BEL-A (Figure 2F-G), possibly contributing to differences in their differentiation and enucleation potential, and also the lower level of α- and β-globin transcripts in HUDEP-2.
Both BEL-A and HUDEP-2 are mixed clonal populations. The karyotype of HUDEP-2 has been reported previously, revealing a modal chromosome number of either 51 (range 49–5312) or 50,7 with a number of whole or partial trisomies and other chromosome anomalies, the slight variation between reports likely due to the number of metaphases (clones) analysed and methods used. Karyotype analysis of BEL-A by G-banding gave a modal chromosome number of 48, XX range 44-48 (50 metaphases). The most common trisomies were of chromosome 6 and 19 (46 and 45 metaphases respectively). Eleven metaphases had chromosome loss, most commonly monosomy of chromosome 8, 9, 12, 15, 17, 18 or 21. There was also a small number of partial chromosome losses.
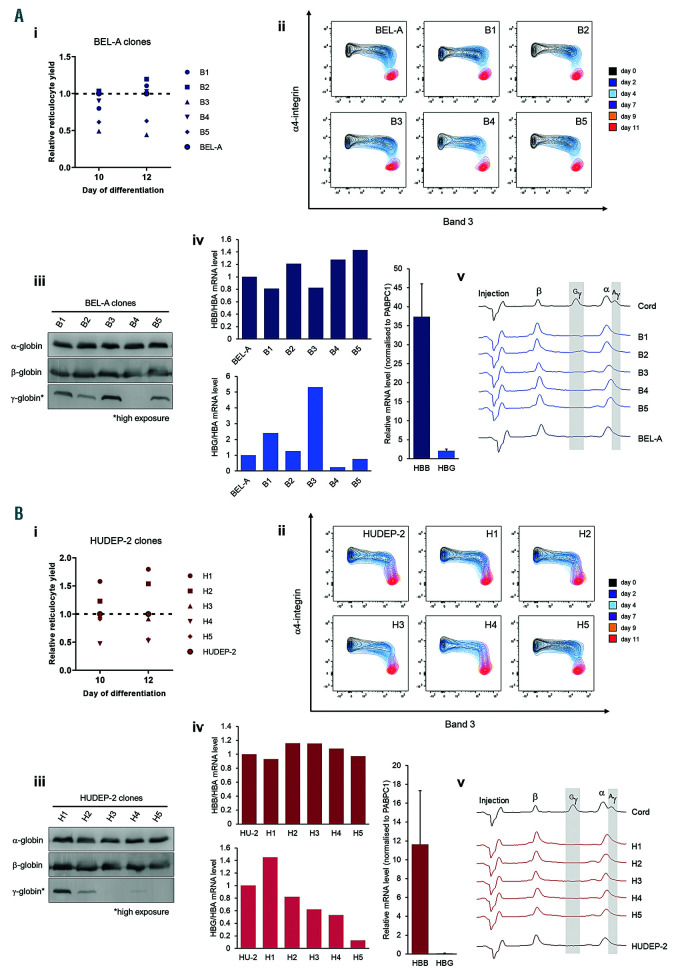
To investigate clonal variability we single cell sorted and analysed five individual clones selected at random of both BEL-A and HUDEP-2. There was slight variation in reticulocyte yield between clones compared to the respective unsorted BEL-A and HUDEP-2 population (Figure 3Ai and 3Bi). While the BEL-A clones all showed a similar differentiation profile (Figure 3Aii), HUDEP-2 clones H3 and H5 differentiated marginally more rapidly compared to the other HUDEP-2 clones (Figure 3Bii), but this did not equate with increased enucleation. The level of α- and β-globin was relatively consistent across respective BEL-A and HUDEP-2 clones (Figure 3 Aiii, iv, v and Biii, iv, v). Some variation in γ-globin level was observed between both BEL-A and HUDEP-2 clones by immunoblot and qPCR (Figure 3Aiii, iv and 3Biii, iv). However, these variations exist among a very low range of γ-globin protein and mRNA, which was barely detectable and not quantifiable by high performance liquid chromatography (HPLC) (Figure 3Av and Bv).
Figure 3.
Analysis of clonal variability in the BEL-A and HUDEP-2 cell lines. To generate clonal sub-populations of BEL-A and HUDEP-2 a BD FACSAria Cell Sorter was used to isolate single cells from the propidium iodide-negative population into 96-well plates. Single-cell clones were cultured for seven days before selecting five derived from each line at random for further characterisation (B1-B5 and H1-H5 from BEL-A [A] and HUDEP-2 [B] respectively). (i) Reticulocyte yield of BEL-A and HUDEP-2 clones compared to the respective unsorted cell line. (ii) Flow cytometry analysis of band 3 and α4-integrin expression during differentiation of BEL-A and HUDEP-2 cell lines and clonal populations. (iii-v) Globin subunit expression of BEL-A and HUDEP-2 clones. (iii) Western blots of lysates obtained from cells at day 10 of differentiation incubated with antibodies to α-, β- and γ-globin. (iv) qPCR at day 4 of differentiation with primers: HBA for GAGGCCCTGGAGAGGATGTTCC rev ACAGCGCGTTGGGCATGTCGTC; HBB for GCTCACCTGGACAACCTCAR rev CGTTGCCCAGGAGCCTGAA; HBG for TGGGTCATTTCACAGAGGAGR rev TAGACAACCAGGAGCCTTCC; PABPC1 for AGCTGTTCCCAACCCTGTAATC rev GGATAGTATGCAGCACGGTTCTG. Globin mRNA levels were normalised to respective PABPC1 mRNA level of sample. Upper and lower panels show relative HBB/HBA and HBG/HBA mRNA level to that of respective unsorted line. Right-hand panel shows average HBB and HBG mRNA levels for all BEL-A or HUDEP-2 samples. HU-2=HUDEP-2. (v) Reversed-phase high-performance liquid chromatography (RP-HPLC) of BEL-A and HUDEP-2 cells and clonal populations at day 10 of differentiation. The injection peak is identified along with peaks for β-globin (β), Gγ-globin (Gγ), α-globin (α) and Aγ-globin (Aγ); δ-globin expression is below the detection limit. A sample of erythroid cells differentiated from cord blood CD34+ at 21 day in culture is included as a positive control for γ-globin. Method was as Loucari et al.13 with the exception of the injection size (600,000 cells in 30 μL dH2O) and the LC column used (Jupiter 5 μm C18 300A, size 250×4.4 mm, protected with a Security Guard analytical guard system [KJ0-4282, Phenomenex]).
The data from this study corroborate BEL-A as a further advancement in the provision of human erythroid lines, having improved performance and more closely recapitulating adult erythropoiesis than HUDEP-2, possibly due to a more optimal erythroid phenotype at time of immortalisation. However, although not as efficient as BEL-A, we have demonstrated that HUDEP-2 can undergo improved differentiation and enucleation in our Bristol culture medium compared to that achieved in previous medium; useful information for researchers who have adopted this line.
Yield from adult erythroid cultures is less efficient than that in vivo, due to restricted expansion rates in vitro.1 In contrast the absolute reticulocyte yield from BEL-A is essentially unlimited as the progenitors divide indefinitely, restricted only by feasible culture volume (and cost). Nevertheless, significantly fewer BEL-A orthochromatic normoblasts enucleate than those differentiated from non-immortalised adult progenitors, an issue we are presently addressing.
Overall, these data provide an important resource, enabling researchers’ informed choice of the most appropriate line for their studies.
Acknowledgments
The authors would like to thank the Siriraj Central Cytogenetic Laboratory, Faculty of Medicine Siriraj Hospital, Mahidol University for performing karyotyping of BEL-A, the University of Bristol Proteomics Facility for use of the HPLC system and the University of Bristol Flow Cytometry Facility for use of equipment. We would also like to acknowledge the assistance of Dr. Andrew Herman and Lorena Sueiro Ballesteros of the Flow Cytometry Facility for cell sorting.
Footnotes
Funding: this study was supported by the National Institute for Health Research Blood and Transplant Research Unit (NIHR BTRU) in Red Cell Products (IS-BTU-1214-10032). The views expressed are those of the authors and not necessarily those of the National Health Service, NIHR, or the Department of Health and Social Care. DJD was supported by a Wellcome Trust Strategic Award (106130/Z/14/Z).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Anstee DJ, Gampel A, Toye AM. Ex-vivo generation of human red cells for transfusion. Curr Opin Hematol. 2012;19(3):163–169. [DOI] [PubMed] [Google Scholar]
- 2.Dias J, Gumenyuk M, Kang H, et al. Generation of red blood cells from human induced pluripotent stem cells. Stem Cells Dev. 2011;20(9):1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trakarnsanga K, Wilson MC, Griffiths RE, et al. Qualitative and quantitative comparison of the proteome of erythroid cells differentiated from human iPSCs and adult erythroid cells by multiplex TMT labelling and nanoLC-MS/MS. PLoS One. 2014;9(7):e100874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurita R, Suda N, Sudo K, et al. Establishment of immortalized human erythroid progenitor Ccell lines able to produce enucleated red blood cells. PLoS One. 2013;8(3):e59890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trakarnsanga K, Griffiths RE, Wilson MC, et al. An immortalized adult human erythroid line facilitates sustainable and scalable generation of functional red cells. Nat Commun. 2017;8(1):14750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawksworth J, Satchwell TJ, Meinders M, et al. Enhancement of red blood cell transfusion compatibility using CRISPR-mediated erythroblast gene editing. EMBO Mol Med. 2018;10(6):e8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vinjamur DS, Bauer DE. Growing and genetically manipulating human umbilical cord blood-derived erythroid progenitor (HUDEP) cell lines. Methods Mol Biol. 2018;1698:275–284. [DOI] [PubMed] [Google Scholar]
- 8.Hu J, Liu J, Xue F, et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood. 2013;121(16):3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoniani C, Meneghini V, Lattanzi A, et al. Induction of fetal hemoglobin synthesis by CRISPR/Cas9-mediated editing of the human beta-globin locus. Blood. 2018;131(17):1960–1973. [DOI] [PubMed] [Google Scholar]
- 10.Canver MC, Smith EC, Sher F, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Kurita R, Nakamura Y, Sheehan V. Screening fetal hemoglobin inducing compounds in human umbilical cord blood-derived erythroid progenitor cells. Blood. 2017;130(Suppl 1):2222. [Google Scholar]
- 12.Moir-Meyer G, Cheong PL, Olijnik A-A, et al. Robust CRISPR/Cas9 genome editing of the HUDEP-2 erythroid precursor line using plasmids and single-stranded oligonucleotide donors. Methods Protoc. 2018;1(3):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loucari CC, Patsali P, Van Dijk TB, et al. Rapid and sensitive assessment of globin chains for gene and cell therapy of hemoglobinopathies. Hum. Gene Ther Methods. 2018;29(1):60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]