Abstract
Platelets are key elements in thrombosis, particularly in atherosclerosis-associated arterial thrombosis (atherothrombosis), and hemostasis. Megakaryocytes in the bone marrow, differentiated from hematopoietic stem cells are generally considered as a uniform source of platelets. However, recent insights into the causes of malignancies, including essential thrombocytosis, indicate that not only inherited but also somatic mutations in hematopoietic cells are linked to quantitative or qualitative platelet abnormalities. In particular cases, these form the basis of thrombo-hemorrhagic complications regularly observed in patient groups. This has led to the concept of clonal hematopoiesis of indeterminate potential (CHIP), defined as somatic mutations caused by clonal expansion of mutant hematopoietic cells without evident disease. This concept also provides clues regarding the importance of platelet function in relation to cardiovascular disease. In this summative review, we present an overview of genes associated with clonal hematopoiesis and altered platelet production and/or functionality, like mutations in JAK2. We consider how reported CHIP genes can influence the risk of cardiovascular disease, by exploring the consequences for platelet function related to (athero)thrombosis, or the risk of bleeding. More insight into the functional consequences of the CHIP mutations may favor personalized risk assessment, not only with regard to malignancies but also in relation to thrombotic vascular disease.
Introduction
Atherosclerotic cardiovascular disease is a chronic inflammatory condition that frequently occurs in the aging population.1 Current understanding is that upon rupture or erosion of an atherosclerotic plaque, a thrombus is formed of aggregated platelets and fibrin which can become vaso-occlusive.2 Furthermore, platelets contribute to ensuing thrombo-inflammatory reactions through their multiple interactions with vascular cells, leukocytes and the coagulation system, thereby promoting disease progression.3
Platelets are formed from megakaryocytes in the bone marrow (BM) through a differentiation and maturation process known as megakaryopoiesis. Several transcription factors have been identified over the years that regulate megakaryopoiesis and platelet production, and understanding of key transcriptional regulators is still expanding. Mutations in genes encoding for these transcription factors, along with epigenetic regulators, are accompanied with quantitative and/or qualitative platelet abnormalities, causing thrombo-hemorrhagic complications.4 Multiple growth factors control megakaryopoiesis and platelet production, of which thrombopoietin and its binding to the thrombopoietin receptor plays a primary role.5 Megakaryocytes undergo endomitosis to become polyploid and during maturation extensive reorganization of cytoskeletal proteins is required for proplatelet formation and the budding of platelets.6
A number of recent studies stipulate that the incidence of cardiovascular disease (CVD), such as coronary artery disease, heart failure and ischemic stroke, is higher in patients with so-called somatic driver mutations in hematopoietic stem or progenitor cells, resulting in a clonal expansion of a subpopulation of blood cells.1 This process, referred to as clonal hematopoiesis of indeterminate potential (CHIP), was proposed to define individuals with somatic clonal mutations in genes related to hematologic malignancies with variant allele fractions of >2%, but without a known hematologic malignancy or other clonal disorder.7 This premalignant state is considered to be relatively frequent in the elderly population, where somatic mutations accumulate in a variety of genes controlling hematopoietic stem cell maintenance, expansion and survival. Although CHIP increases the risk of developing hematologic cancer, mostly myeloid neoplasms, the absolute risk is still small. Several excellent recent reviews describe in detail the etiology of clonal hematopoiesis and its relation with CVD.1,8,9 So far, attention has mainly been focused on proposed mechanisms of accelerated inflammation-driven atherosclerosis and increased thrombosis risk through altered function of innate immune cells.
In the present paper, we took a different approach. We confined our search to the current evidence on CHIP mutations that are directly or indirectly linked to qualitative or quantitative platelet traits. Starting from the Online Mendelian Inheritance in Man (OMIM) database complemented with recent literature, we selected and discussed genes that were linked to clonal hematopoiesis as well as to the platelet traits count and function. Since CHIP mutations appeared not to be only associated with increased platelet count and/or function, but also with decreases in these platelet traits, its potential relation to both (athero)thrombotic and hemostatic disorders is presented in this review.
Section 1: Clonal mutations in genes associated with increased platelet count and/or function
For several genes encoding for transcription regulators (ASXL1), epigenetic regulators (DNMT3A, IDH2) and cell signaling proteins (ABL1, BCR, BRAF, JAK2, SH2B3), clonal mutations are known that enhance platelet production, which can be accompanied by enhanced platelet functionality. Related effects are described for several genes with divergent roles (ABCB6, SF3B1) (Table 1 and Figure 1).
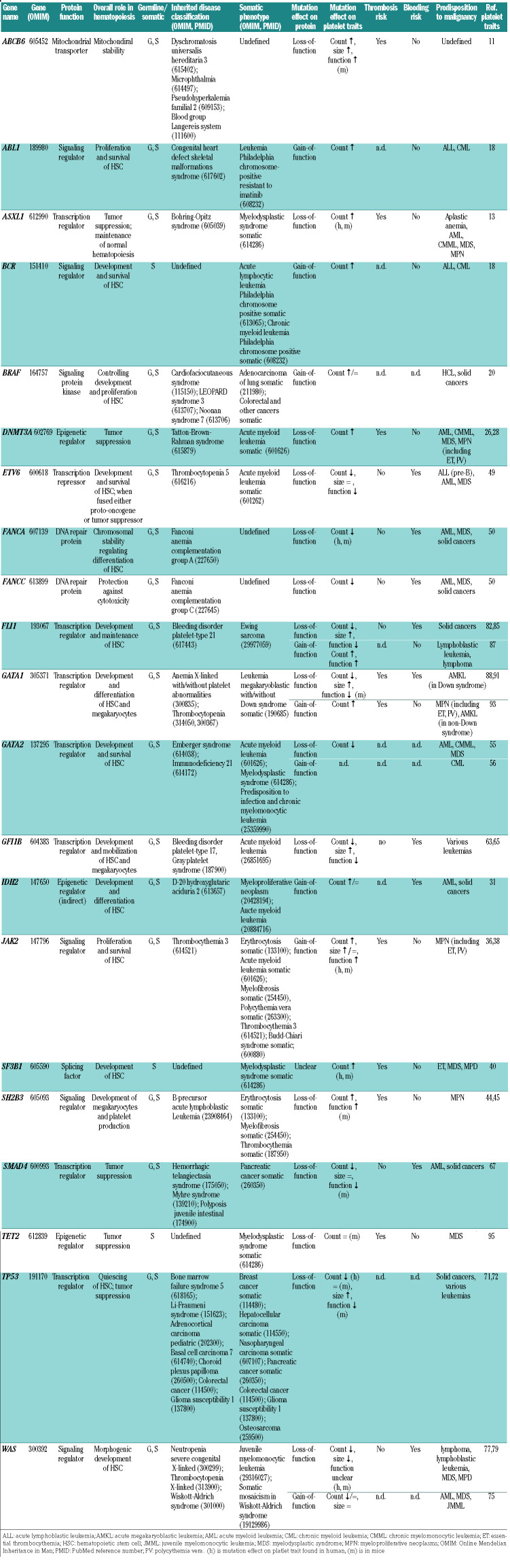
Table 1.
Relevant genes in clonal hematopoiesis with effects on platelet traits and disease.
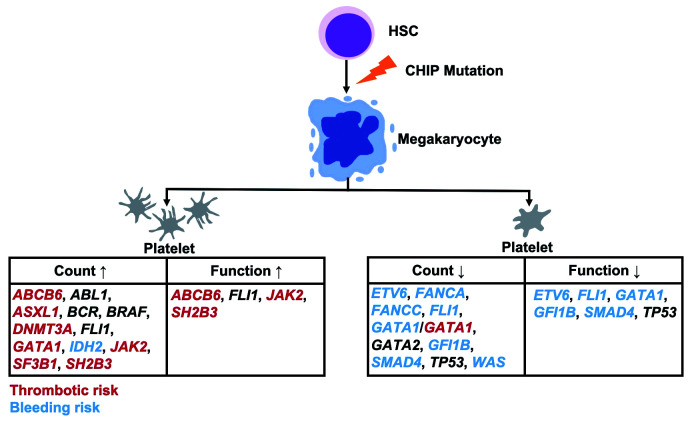
Figure 1.
Clonal hematopoiesis of indeterminate potential (CHIP)-related genes affecting platelet traits and the risk of thrombosis or bleeding. Mutations in genes associated with a thrombotic or bleeding risk are indicated in red and blue, respectively. For genes indicated in black, no such associations are known yet.
ABCB6
Multiple so-called ABC transporters play a role in lipid trafficking, and thus may contribute to atherosclerosis. However, the ABCB6 gene product (ATP binding cassette subfamily B member 6) facilitates the ATP-dependent import of porphyrins and heme into mitochondria.10 Markedly, germline mutations of ABCB6 are associated with several disease phenotypes, including dyschromatosis, microphthalmia and pseudo-hyperkalemia.
The gene ABCB6 is highly expressed in BM megakaryocyte progenitor cells and megakaryocytes, but only moderately in platelets. Evidence regarding CVD mainly comes from animal studies. In mice, deficiency in Abcb6 increased megakaryopoiesis and thrombopoiesis, resulting in an increased platelet count and larger platelet volume, effects that were explained by higher oxidative stress in the presence of accumulating porphyrins.11 The platelets produced in these mice were hyper-reactive and furthermore, against a high-lipid background, attracted leukocytes, thus enhancing atherosclerosis.10,11
In patients with acute promyelocytic or myeloid leukemia, RNA expression levels of ABCB6 are reduced, suggesting the occurrence of also acquired clonal mutations in this gene.12 However, so far no strong association with CHIP has been found.8
ASXL1
The transcriptional regulator Additional sex combs like 1 (ASXL1) is a chromatin-binding protein, which acts as tumor suppressor and is implicated in the maintenance of normal hematopoiesis. Somatic mutations of this gene are found in patients with a variety of myeloid malignancies, including acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), myelodysplastic syndrome (MDS), and myeloproliferative neoplasm (MPN).13 In particular, mutations in ASXL1 are detected in 10% of MDS patients and 40% of CMML patients.14 Hence, this gene is considered as a driver of leukemia and myelodysplasia. The majority of (somatic) mutations provoke a truncation of the C-terminus of the protein, resulting in a loss of transcription regulation. In addition, the truncated form can interact with other proteins to modulate cell proliferation.15 In mouse models, transgenic expression of a C-terminal truncated Asxl1 mutant resulted in age-dependent anemia, thrombocytosis, and morphological dysplasia.13 A similar type of thrombocytosis is seen in MDS-refractory anemia patients, carrying ASXL1 mutations.13 The prevalence of acquired hematopoietic mutations in ASXL1 in a healthy population of 60-69 years of age was estimated at 1.5%, and was associated with twice the risk of developing CVD.16
BCR and ABL1
The somatic gene effects of Breakpoint cluster region protein (BCR) and Abelson murine leukemia viral oncogene homolog 1 (ABL1) are highly related, if only because the two proteins share signaling pathways. The proto-oncogene ABL1 contains an auto-inhibitory SH3 domain which, when deleted, turns it into an oncogene. During a somatic reciprocal translocation of chromosomes 22 and 9, both genes can fuse together. The encoded BCR-ABL1 fusion protein is frequently detected in patients with chronic myeloid leukemia (CML) (90%) or acute lymphoblastic leukemia (ALL) (30%).17 While CML patients mostly carry the 210 amino-acid variant of BCR-ABL, in ALL patients also a shorter 185 amino-acid variant is present. Both fusion forms display constitutive protein tyrosine kinase activity.17
The current understanding is that aberrant roles of BCR and ABL1 in hematopoiesis are a consequence of fusion formation, although the main evidence comes from case studies. A fusion variant has been described, which is associated with an increased platelet count, although the mechanism is still unclear.18 In the few healthy adults carrying a BCR-ABL1 fusion mutation hematopoietic malignancies were not detected. On the other hand, BCR-ABL1 fusions can be considered as indicators for a premalignant state, while the absolute risk of developing CVD is smaller than for the JAK2 V617F mutation.19
BRAF
The serine-threonine protein kinase BRAF is an essential partner in the mitogenic RAS/RAF/MEK/ERK signaling pathway. The BRAF proto-oncogene is expressed in all tissues, where it controls cell proliferation, apoptosis, and differentiation. In addition, BRAF is necessary for embryonic development, as Braf-deficient embryos die because of disturbed blood vessel formation.20
Evidence on the role of BRAF in normal megakaryopoiesis comes from work mainly with immortalized human megakaryoblastic cell lines. Upon stimulation with thrombopoietin, differentiation and proliferation of the cells appeared to rely on BRAF-mediated signaling to ERK.21 Downregulation of BRAF thus lowered the number of megakaryocytic lineage cells, a phenomenon that was confirmed in vivo in chimeric mice.20
In the Noonan, LEOPARD and cardiofaciocutaneous syndromes, patients carry germline mutations in BRAF in regions distinct from those of somatic cancerous mutations. However, only limited changes in BRAF signaling are reported.22 On the other hand, somatic gain-of-function mutations in the BRAF gene accumulate in patients with AML, malignant lymphomas or solid cancers.23 Next to more common point mutations, rare chromosomal translocations are described for this gene.24
A frequently observed gain-of-function mutation (V600E) is the driver mutation present in different cancers, including melanomas, solid cancers and hairy cell leukemia (HCL). Patients who suffer from HCL have low blood cell counts, likely due to BM aberrations and splenomegaly.25 Whether megakaryopoiesis is altered due to a constitutively increased MEK/ERK signaling via BRAF still needs to be confirmed.
DNMT3A
Clonal mutations of three genes (DNMT3A, IDH2, TET2) have been reported which, directly or indirectly, affect histone methylation and hence these can be considered as epigenetic regulators.
The gene DNA methyltransferase 3α (DNMT3A) encodes for a DNA methylation enzyme that regulates gene imprinting, chromosome inactivation and tumor suppression. Genetic mutations in the DNMT3A gene occur in the rare Tatton-Brown-Rahman syndrome which, as far as is known, is not accompanied by hematopoietic aberrations.
In several acquired blood cancers, but especially in adults with AML, somatic mutations in DNMT3A have been reported.26 About a quarter of all AML patients with de novo disease carry variant forms of this protein, most commonly with R882H mutation. The loss-of protein-function in those patients resulted in chromosomal islands of hypomethylation.27 The same mutation, albeit less frequent, can occur in patients with CMML, MDS or MPN.26 It is considered that DNMT3A mutations in hematopoietic stem cells lead to a pre-leukemic state, waiting for additional mutations to induce leukemia. The time interval from first appearance of the mutation to disease is, however, unclear.26
In agreement with its relevance for clonal hematopoiesis, a recent report points to an increased incidence of acquired DNMT3A mutations in the elderly, with a prevalence of about 15% at 60-69 years of age.1 Combined with a JAK2 V617F mutation (see below), the mutated DNMT3A gene associates with essential thrombocythemia (ET) and polycythemia vera (PV).26 Current understanding is that first acquisition of a DNMT3A mutation followed by JAK2 will result in an ET phenotype. On the other hand, first appearance of the JAK2 mutation may result in a PV phenotype.26 Overall, DNMT3A mutations in AML patients are associated with higher platelet counts than patients with WT-DNMT3A; however, the absolute count is still low (<150×109/L).28
Regarding atherosclerosis development and atherothrombosis, studies report increased inflammation, linked to mutated DNMT3A, possibly due to a higher production of cytokines.29 Indeed, in patients carrying an acquired mutation of DNMT3A, the risk of CVD appears to be doubled.1 The higher platelet count observed in AML patients with DNMT3A mutations likely occurs secondary to the pro-inflammatory phenotype. So far, no mechanism has been found to link DNMT3A mutations directly to platelet traits.
IDH2
The enzyme isocitrate dehydrogenase NADP+ 2 (IDH2) localized in mitochondria generates NADPH from NADP+, whilst catalyzing the oxidative decarboxylation of isocitrate, ultimately producing D-2-hydroxy-glutarate. By producing NADPH, IDH2 regulates the mitochondrial redox balance, hence mitigating cellular oxidative damage.30 As expected, genetic mutations in IDH2 are described to be associated with metabolic diseases. On the other hand, somatic mutations of IDH2 are found in several cancers, including hematologic malignancies, sarcomas and colon cancer. This is compatible with a role of IDH2 as epigenetic regulator, although the direct evidence for epigenetic effects is still indirect.
The most frequent clonal mutations in IDH2, identified in patients with de novo AML, concern the protein arginine residues R140Q and R172K. These variants cause a gain-of-function resulting in an abnormal, damaging production of D-2-hydroxyglutaric acid, leading to a hypermethylated state of DNA and histones.30 In comparison to non-carriers, AML patients carrying the somatic IDH2 mutations showed a higher platelet count, although the absolute platelet count was still low (<150×109/L).31 The same trend for platelet count has also been found in MDS patients.32 Next to this, in primary myelofibrosis, IDH mutations can form a risk factor for leukemic transformation.33 No association with thrombotic events is known for these patient groups, but bleeding was more common in mutant-carrying patients. In the elderly, the percentage of individuals with clonal hemostasis driver mutations in IDH2 appeared to be rather low at around 1%.16 Together, this suggests only mild effects of somatic changes in this gene on clonal hematopoiesis, associated with a slight increase in platelet count by a so far unknown mechanism and an increased bleeding tendency.
JAK2
The non-receptor tyrosine kinase Janus kinase 2 (JAK2) is one of the general regulators of cell survival and proliferation, by controlling, for example, cytokine receptor signaling pathways. Also in hematopoiesis, JAK2 controls precursor cell maintenance and function.34 Inherited mutations of JAK2 are detected in patients with hereditary thrombocytosis,35 while somatic mutations of the gene link to various phenotypes including erythrocytosis.
A well-known acquired JAK2 variant is the mutation V617F, which is carried by the majority of patients with MPN, i.e. in nearly all PV patients and half of patients with ET or primary myelofibrosis. In general, the V617F mutation affects the proliferation of myeloid cells and leads to increased inflammatory responses.34 However, this somatic mutation as such is not considered to enhance the risk of thrombotic events in patients with ET or PV.36 Nevertheless, platelets from JAK2 V617F-positive patients demonstrated an enhanced activation status and procoagulant potential. In addition, the fraction of immature platelets, which can be more active than mature platelets, was higher in carriers of the JAK2 V617F mutation versus non-carriers.37
Transgenic mice have been generated carrying the human JAK2 V617F mutation in the megakaryocyte lineage.38 The JAK2 V617F megakaryocytes responded better to thrombopoietin, and displayed a greater migratory ability, proplatelet formation and increased ploidy. The produced platelets responded stronger to multiple agonists.
In aging healthy individuals, the prevalence of the JAK2 V617F variant is only 1%, but carriers have a 10-fold increased risk of CVD.16 Depending on the degree of mutation expansion, subjects may develop MPN instead of CHIP.8 How or under which conditions the thrombocytosis is linked to somatic JAK2 mutations aggravating CVD is still a matter of debate and requires further investigation.
SF3B1
The gene splicing factor 3B subunit 1 (SF3B1) encodes for a core component of the RNA spliceosome machinery.39 Somatic mutations in this gene, along with other genes of the spliceosome, have been identified in over half of MDS patients.39 Common mutations in the SF3B1 gene are those of K700E, K666N and R625C.40 To study the impact of the frequent K300E mutation, a conditional knock-in mouse model was developed, which revealed a RNA splicing defect similar as suggested in MDS patients harboring this mutation.39 Regarding the thrombotic risk, studies revealed that patients carrying a mutated SF3B1 gene had higher platelet counts and were more prone to develop CVD than patients without mutation,40 although the altered molecular players are unclear. Furthermore, a sequencing study identified SF3B1 mutations in 5% of ET patients.41 In the aging healthy population, clonal hematopoietic mutations of SF3B1 appear to be infrequent, ranging from 2 to 5%.42
SH2B3
The signaling adaptor protein Src homology 2 B3 (SH2B3, also named LNK) acts as an interactor of JAK2, and negatively regulates thrombopoietin-induced megakaryopoiesis. An associated inherited disease is B-precursor acute lymphoblastic leukemia. Somatic mutations in the SH2B3 gene are found in >5% of MPN patients. These concern frameshift and missense mutations throughout the whole gene, often co-existing with mutations in driver genes, including JAK2, CALR and MPL.43 The loss-of-function of SH2B3 can lead to a higher expansion of hematopoietic stem cells, acting by increased thrombopoietin signaling and megakaryopoiesis.44 The higher platelet and leukocyte counts may worsen atherosclerosis and the thrombotic risk.43 In Sh2b3 knockout mice, it was found that hyperlipidemia aggravated both atherosclerosis and thrombosis, likely due to positive platelet priming.45 Whether this priming event due to SH2B3 mutations also occurs in humans, is not known.
Section 2: Clonal mutations in genes associated with decreased platelet count and/or function
Several mutations in genes encoding for transcription regulators (ETV6, GATA2, GFI1B, SMAD4), cell signaling proteins (TP53, WAS), and other proteins (FANCA, FANCC) are linked to impaired hematopoiesis, causing thrombocytopenia of varying severity with evidence for concomitant platelet function defects (Table 1 and Figure 1).
ETV6
The transcriptional repressor E26 transformation-specific variant 6 (ETV6) serves to maintain the development of hematopoietic stem cells in the BM as a continuous survival signal. It acts by inhibiting other transcription factors, such as FLI1. However, it appears not to be required for embryonic stem cell expansion.46
ETV6 is known as a proto-oncogene, since it can be a fusion partner with over 30 other genes, but in case of truncating mutations it acts as a tumor suppressor gene. Depending on the fusion site, the fused protein can alter the transcription levels of ETV6 target genes, which may support the development of leukemia.46,47 On the other hand, germline heterozygous ETV6 mutations have been identified in some patients with dominantly inherited thrombocytopenia.48 Such patients seem to have a predisposition to hematologic malignancies, most commonly ALL, AML or MDS. Given that the complete loss of ETV6 is lethal, truncating or protein-inactivating mutations are mainly found as somatic events, and rarely as germline variants.46
As a transcriptional repressor, ETV6 has an established role in megakaryocyte and platelet (patho)physiology. Patients with germline ETV6 variants show a large expansion of immature megakaryocyte colony-forming units, accompanied by a reduced formation of proplatelets, thus explaining the thrombocytopenia. The mutant platelets are of normal size, although characterized by aberrant cytoskeleton organization, lower levels of small GTPases, and defective clot retraction.49 Evidence is lacking to link clonal variants of ETV6 to thrombotic phenotypes; however, a related bleeding tendency has been described.
FANCA, FANCC
The proteins Fanconi anemia complementation group A and C (FANCA/C) are repair factors after DNA damage or apoptosis. Inherited mutations in either gene are seen in the disorder Fanconi anemia, where patients in 60-70% of the cases show a mutation in FANCA and in 15% a mutation in FANCC. Such patients suffer from progressive bone marrow failure, pancytopenia and predisposition to cancer.50 Knockout mouse studies revealed that FANCA is needed for normal megakaryopoiesis and platelet production. Megakaryocytes in the deficient mice were found to be in a senescent state.50
In humans, heterozygous mutations of FANCA are observed in a proportion of patients with AML.51 However, carriers of such mutations do not seem to have a significant risk of developing cancer.52 On the other hand, FANCA deletion mutations, especially in combination with other germline mutations, might contribute to breast cancer susceptibility.53 The phenotype coupled to somatic mutations in FANCA and/or FANCC is probably linked to genomic instability caused by defective FANC proteins.51 How these somatic mutations contribute to CHIP-related CVD needs to be established.
GATA2
In immature hematopoietic stem cells, the transcription factor GATA-binding factor 2 (GATA2) is expressed earlier than GATA1, and becomes down-regulated upon differentiation.54 This has also been observed in Gata2-knockout mice, revealing that GATA2 is required for hematopoietic stem cell and progenitor cell development.55 In humans, congenital GATA2 deficiency is accompanied by a hypocellular and dysplastic bone marrow, resulting in low platelet counts.55 Furthermore, germline deletion mutations in the GATA2 gene are associated with an increased predisposition to infection, AML, CMML or MDS.
On the other hand, a somatic mutation (L359V) in GATA2 has been identified in approximately 10% of patients in the progression phase of CML.56 This concerns a gain-of-function resulting in increased transcription factor activity, in contrast to gene deletion. Reports indicate that in approximately 50% of patients with any GATA2 mutation, the megakaryocyte development is abnormal.57 Unlike GATA1, GATA2 regulates platelet GPIIb rather than GPIb expression.54 Variants of GATA2 have also been associated with increased susceptibility for coronary artery disease,58 linking this gene to CHIP.
GFI1B
Another transcription regulator crucial for erythroid and megakaryocytic differentiation is growth factor independent 1B transcription repressor (GFI1B). As a DNA-binding protein, it regulates the dormancy and mobilization of hematopoietic stem cells.59 Next to the full-size protein of 330 amino acids implicated in megakaryopoiesis, a shorter form is expressed that may rather regulate erythroid development.60 The longer protein modulates the expression of several proto-oncogenes and tumor suppressor genes.59 Accordingly, a functional disturbance of GFI1B can contribute to leukemia development. In mice, genetic deletion of Gfi1b resulted in early lethality, where the embryos showed failed megakaryocyte development.61
For human GFI1B, both germline and somatic mutations have been identified. These generally result in a truncated or a dysfunctional form of the protein, thereby reducing DNA binding and transcription repressor activity.62 In the inherited disorder gray platelet syndrome, patients with a GFI1B mutation display (macro)thrombocytopenia with platelets that are reduced in α-granules.63 The patient’s platelets were also found to be reduced in GPIb and GPIIb/IIIa expression, whereas that of the hematopoietic precursor marker CD34 was markedly increased. The suggestion that, in these and other patients with a truncating mutation of GFI1B, megakaryocyte development is impaired was recently supported by platelet proteome analysis.64 In mice, a megakaryocyte-specific deletion of Gfi1b enhanced the expansion of megakaryocytes, but resulted in severe thrombocytopenia.65 Here, the (tubulin) cytoskeleton appeared to be underdeveloped in the mutant megakaryocytes, explaining an inadequate proplatelet formation.
Whole-exome sequencing efforts have revealed the presence of alternative GFI1B splice variants, which is accompanied by impaired megakaryocyte differentiation and thrombopoiesis.60 However, in heterozygous carriers, platelet counts and function were in normal ranges. Little is known about clonal hematopoiesis. A somatic mutation of GFI1B has been identified in patients with AML.59
SMAD4
The ‘vascular’ transcription factor SMAD family member 4 (SMAD4) acts as a tumor suppressor, triggered by signaling pathways evoked by transforming growth factor-β or bone morphogenetic protein.66 Within the cellular nucleus, SMAD4 forms a complex with other SMAD isoforms to control gene expression. In mice, SMAD4 was found to play a role especially in vascular development.66 On the other hand, a megakaryocyte-specific deficiency of SMAD4 is described, causing mild thrombocytopenia with partially dysfunctioning platelets, likely as a consequence of altered promotor activities.67
In humans, both somatic and inherited mutations of SMAD4 are known. Inherited mutations of the gene present with distinct phenotypes, ranging from a vascular bleeding disorder (hereditary hemorrhagic telangiectasia) to gastro-intestinal and bone marrow abnormalities.68 Somatic mutations of the 358-515 amino-acid region are linked to pancreatic carcinoma.69 During the screening for somatic driver mutations linked to clonal hematopoiesis, a similar mutation of SMAD4 was found.16 Mutations in SMAD4 are related to a bleeding rather than thrombotic phenotype.
TP53
The tumor suppressor tumor protein p53 (TP53 or p53) is a critical player in cell cycle progression and apoptosis. Herein, TP53 maintains the quiescent state of hematopoietic stem cells, and controls DNA damage responses upon cellular stress.70 In megakaryocytic cells derived from Tp53 knockout mice, cell size and polyploidization were increased due to higher DNA synthesis and decreased apoptosis. In human cell cultures, TP53 knockdown affected the expression of platelet integrins, granule components and cytoskeletal proteins, which was accompanied by functional platelet defects.71
Regarding human disease, the TP53 deletion occurring in multiple myeloma is accompanied by a lowering in platelet count.72 In this context, mutant TP53 forms are considered to drive clonal hematopoiesis via the epigenetic regulator EZH2, leading to overmethylation of histone H3. This can down-regulated several genes associated with self-renewal and differentiation of hematopoietic stem cells.70 A common consequence is expansion of the affected hematopoietic cell clones. Markedly, the TP53 gene is top ranking in mutated genes found in CHIP.73 How mutated TP53 in hematopoietic cells contributes to CHIP-associated CVD still remains to be determined. Few studies have shown that there is higher expression of pro-inflammatory cytokines in p53-deficient murine leukocytes, which may accelerate the development of CVD.9 However, there is no evidence directly linking platelet traits to CVD development.
WAS
The Wiskott-Aldrich syndrome (WAS) protein is selectively expressed in hematopoietic cells, where it regulates actin cytoskeleton rearrangements. In the classical X-linked Wiskott-Aldrich syndrome, patients suffer from thrombocytopenia with smaller sized platelets and recurrent infections, due to an impaired functionality or availability of WAS.74 A milder phenotype is that of X-linked thrombocytopenia, in which patients only suffer from bleeding because of low platelet count.74 Rare inherited mutations that instead cause constitutive WAS activation are seen in patients with X-linked neutropenia, experiencing recurrent bacterial infections while having normal platelet count and size.75 In addition, these patients show an increased predisposition for AML or MDS.76
In classical Wiskott-Aldrich syndrome patients, the prevalence of malignancy is 13-22%, mostly due to development of lymphoma, but also to lymphoblastic leukemia, MDS or MPD.76 The thrombocytopenia is likely caused by increased platelet removal. In Was-deficient mice, platelet turnover was shortened, with proteomic evidence for alterations in proteins of metabolic and proteasomal pathways.77 Furthermore, in both the mutant mice and patients, there is evidence for a hyperactivation status of the platelets, thus explaining the higher elimination rate. Several groups reported on alterations in integrin activation in the patient’s platelets.78,79 However, one patient study concluded platelet activation properties were normal.80
There is limited evidence for the presence of somatic mutations in the WAS gene. This mainly concerns gain-of-function mutations, associated with poor outcome in patients with juvenile myelomonocytic leukemia.81 This suggested a clonal role of the gene in the pre-malignant state.
Section 3: Clonal mutations in other genes
For the genes FLI1 and GATA1 encoding for transcription regulators, whether the mutation is gain-of-function or loss-of-function likely determines its respective effect on platelet count and/or function. Mutations in TET2 have been associated with increased inflammation-induced atherosclerosis and thrombotic disease, although possible effects on platelets remain to be established (Table 1 and Figure 1).
FLI1
The protein Friend leukemia virus integration 1 (FLI1) is a member of the ETS transcription factor family, which is highly expressed in the hematopoietic lineage and endothelial cells. Due to a faulty vasculature, Fli1 knockout mice die during embryonic development, but heterozygous mice are viable without apparent phenotype.82 Detailed studies indicate that FLI1 plays an important role in both erythropoiesis and megakaryopoiesis by regulating the expression of multiple genes.83 It acts together with the transcription factor GABPA, especially in later phases of megakaryopoiesis. This is exemplified by the fact that, in Fli1 knockout mice, megakaryocytes are specifically reduced in the expression of late-stage genes, e.g. genes encoding for glycoprotein (GP)Ibα, GPIX and platelet factor 4.82
In humans, heterozygous mutations in FLI1 are commonly grouped together as ‘Bleeding disorder platelet-type 21’ (Phenotype MIM 617443). Examples are the Jacobsen syndrome and Paris-Trousseau syndrome, which are characterized by a heterozygous partial deletion of chromosome 11, encompassing the FLI1 gene. Such patients characteristically suffer from abnormal growth and mental retardation, accompanied by thrombocytopenia, most likely due to impaired megakaryopoiesis.84 In the Paris-Trousseau syndrome, platelets are enlarged and contain large fused α-granules.84 In patients with a mutated FLI1 gene, presenting with congenital macrothrombocytopenia, also an impaired agonist-induced platelet aggregation has been reported.85
In the case of somatic mutations, FLI1 can become fusion partner with the transcriptional repressing gene EWSR1, a condition known as Ewing sarcoma.86 The effect on platelets is unclear. On the other hand, in vitro studies have indicated that the overexpression of FLI1 in stem cells enhances megakaryopoiesis, thrombopoiesis, and platelet functionality.87 Furthermore, deregulated high levels of FLI1 are found in various types of cancer. In agreement with this, a predisposition to pre-T-cell lymphoblastic leukemia and lymphoma is described for transgenic mice over-expressing Fli1 in the hematopoietic progenitor cells.83 It remains to be established whether FLI1 is a main contributing gene in CHIP-related CVD.
GATA1
The transcription factor GATA-binding protein 1 (GATA1) controls the development and production of megakaryocytes, platelets and erythrocytes. In mouse studies, the loss of Gata1 in the megakaryocyte lineage resulted in smaller size megakaryocytes and a defect in proplatelet formation. The Gata1-deficient platelets were larger in size, showed an excess in rough endoplasmic reticulum, and contained fewer α-granules.88 Furthermore, the deficient mice were impaired in red blood cell development, and often died because of anemia.89 Consistent with this, GATA1 is highly expressed in human megakaryocytes and erythroid cells. Mutations in GATA1 can appear as germline or somatic. Inherited mutations associate with hematopoietic disorders, characterized by low blood cell counts. On the other hand, somatic mutations often result in the production of shorter GATA1 variants, for example, in cases of AMKL (acute megakaryoblastic leukemia) or Down syndrome.54 Here, platelets tend to be low in counts and display an atypical morphology.
A common consequence of germline GATA1 mutations in hematopoietic disorders is the altered interaction of GATA1 with its co-factor FOG1, i.e. a zinc finger protein co-operating with GATA1 to regulate cell differentiation. This has been reported for patients with X-linked thrombocytopenia, or other forms of macrothrombocytopenia, who experience bleeding diatheses.90 The patient’s megakaryocytes are abnormal in structure and the platelets show decreased numbers of α-granules.90 Patients with primary myelofibrosis, having upstream driver mutations resulting in low GATA1 levels in megakaryocytes, show an increased risk of both thrombosis and bleeding. Mice with Gata1low mutation resemble this phenotype, demonstrating similar megakaryocyte abnormalities, such as abnormal P-selectin localization, and thrombo-hemorrhagic events. The prothrombotic state was ascribed to increased platelet-leukocyte interactions through P-selectin.91
In cultured megakaryocytes, GATA1 has been shown to regulate the expression of GPIIb (fibrinogen receptor) and GPIb (von Willebrand factor receptor). Markedly, in GATA1-deficient megakaryocytes, expression levels of GPIIb can be maintained by GATA2 substitution, whereas those of GPIb are decreased.54 As expected, inherited mutations of GATA1 are accompanied by a bleeding phenotype rather than by an increased risk of thrombosis. On the other hand, high levels of GATA1 transcripts are found in patients with ET or PV.92 Overexpression of GATA1 in mice results in a similar phenotype.93 Regarding CHIP, somatic gain-of-function may increase the cardiovascular risk including atherothrombosis, whereas loss-of-function may be more associated with bleeding.
TET2
The protein Tet oncogene family member 2 (TET2) has a key role in DNA methylation, explaining how it functions as a tumor suppressor, maintaining normal hematopoiesis. The TET gene product in particular represses the transcription of inflammatory molecules, such as interleukin-6 and -8, which are known as proatherogenic mediators.1,94 This explains why somatic loss-of-function mutations in TET2 are associated with an increased inflammation tendency. Similarly, as described for DNMT3A, the mutations may increase the burden of atherosclerosis and arterial CVD.
Murine Tet2-null models are used to confirm that CHIP-like mutations lead to inflammation-driven cardiovascular pathologies,7,95 markedly without changes in blood cell counts. In the aging population, clonal hematopoietic mutations of TET2 have a prevalence of 2.5%.16 On the other hand, such mutations are found in approximately 25% of patients with myeloid neoplasms, and are then associated with an increased cardiovascular risk.7 It is still not clear to what extent the mutations affect megakaryopoiesis or platelet function, either directly or indirectly via enhanced inflammation.
Conclusions and perspectives
As outlined above, somatic mutations in multiple genes affecting hematopoiesis contribute as a risk factor to the development of CVD. So far, studies have focused on the effects of somatic and CHIP-linked mutations on blood cells, linking to increased inflammation, atherosclerotic disease and thrombosis risk. In this review, we provide evidence that many of the common CHIP genes are involved in quantitative (count) and/or qualitative (function) platelet traits, and therefore in this way can influence CVD, in particular triggered by thrombo-inflammatory mechanisms. On the other hand, insight is gained in a link between mutations in CHIP genes and impairment of hematopoiesis and hemostatic function.
Reactive (secondary) thrombocytosis, which is not due to a primary hematologic disorder but driven by inflammatory stimuli, trauma or acute bleeding, does not seem to increase the risk of thrombotic or hemorrhagic complications.96 In line with this, the degree of elevation in the platelet count does not correlate with the thrombosis risk in myeloproliferative disease, where clonal (primary) thrombocytosis has been demonstrated.97 This indicates that the platelet count as such is not the only determinant of the increased thrombosis risk in myeloproliferative disorders.98,99 Also, several CHIP mutations (e.g. DNMT3A mutations) can indirectly cause a rise in platelet count by inducing increased expression of inflammatory molecules that subsequently upregulate the thrombopoietin production by the liver. However, the combination of alterations in count and function may play an essential role in CHIP mutations related to thrombosis. So far, we have found evidence of seven CHIP-related genes (ABCB6, ASXL1, DNMT3A, GATA1, JAK2, SF3B1, SH2B3) with elevated platelet counts and an associated thrombotic risk (Figure 1). For the other genes, there is not enough evidence to make estimates of this kind; only for ABCB6, JAK2 and SH2B3 mutations is it known that the elevated platelet count is accompanied by a hyper-reactive platelet phenotype. Apparently, information regarding the functional status of platelets in the context of CHIP mutations is still scarce and further studies are needed to elucidate the contribution of platelets to the risk of thrombosis.
One of the most thoroughly investigated conditions, demonstrating the consequences of altered platelet traits due to somatic driver mutations, is essential thrombocythemia. Markedly, in these patients, there appears to be no direct correlation between platelet count and thrombosis. On the other hand, the JAK2 V617F mutation is known to increase the thrombosis risk in ET patients, when compared to patients without the mutation.100 The reported enhanced activation status of platelets in JAK2 V617F-positive patients provides a strong indication that platelet function changes induced by a CHIP mutation contribute to the risk of thrombosis, thus explaining part of the risk associations of CHIP mutations with CVD. Platelet reactivity also involves interactions with leukocytes, secretion of pro-inflammatory mediators and release of extracellular vesicles that may all contribute to CVD, like atherosclerosis and atherothrombosis. Given the increasing prevalence of CHIP mutations in the elderly who are prone to develop CVD (along with malignancies), more thorough investigation of platelet function linked to CHIP mutations would be worthwhile. Greater insight into the functional consequences of such acquired mutations may also favor personalized risk assessment, not only with regard to malignancies, but also in relation to thrombotic vascular disease.
Acknowledgments
AV is supported by the Dutch Landsteiner Foundation for Blood Transfusion Research (1711). IDS is supported by a joint PhD scholarship of Maastricht and Reading Universities, and by the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement n. 766118.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/8/2020.
References
- 1.Jaiswal S, Libby P. Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat Rev Cardiol. 2020;17(3):137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P, Pasterkamp G. Requiem for the ‘vulnerable plaque’. Eur Heart J. 2015;36(43):2984–2987. [DOI] [PubMed] [Google Scholar]
- 3.Van der Meijden PE, Heemskerk JW. Platelet biology and functions: new concepts and future clinical perspectives Nat Rev Cardiol. 2019;16(3):166–179. [DOI] [PubMed] [Google Scholar]
- 4.Daly ME. Transcription factor defects causing platelet disorders. Blood Rev. 2017;31(1):1–10. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi E, Norfo R, Pennucci V, et al. Genomic landscape of megakaryopoiesis and platelet function defects. Blood. 2016;127(10):1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machlus KR, Italiano JE., Jr The incredible journey: From megakaryocyte development to platelet formation. J Cell Biol. 2013; 201(6):785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steensma DP. Clinical consequences of clonal hematopoiesis of indeterminate potential. Blood Adv. 2018;2(22):3404–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haybar H, Shahrabi S, Ghanavat M, Khodadi E. Clonal hematopoiesis: genes and underlying mechanisms in cardiovascular disease development. J Cell Physiol. 2019;234(6):8396–8401. [DOI] [PubMed] [Google Scholar]
- 9.Sano S, Wang Y, Walsh K. Clonal Hematopoiesis and Its Impact on Cardiovascular Disease. Circ J. 2018;83(1):2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boswell-Casteel RC, Fukuda Y, Schuetz JD. ABCB6, an ABC transporter impacting drug response and disease. AAPS J. 2017;20(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy AJ, Sarrazy V, Wang N, et al. Deficiency of ATP-binding cassette transporter B6 in megakaryocyte progenitors accelerates atherosclerosis in mice. Arterioscler Thromb Vasc Biol. 2014;34(4):751–758. [DOI] [PubMed] [Google Scholar]
- 12.Abraham A, Karathedath S, Varatharajan S, et al. ABCB6 RNA expression in leukemias: expression is low in acute promyelocytic leukemia and FLT3-ITD-positive acute myeloid leukemia. Ann Hematol. 2014;93(3):509–512. [DOI] [PubMed] [Google Scholar]
- 13.Nagase R, Inoue D, Pastore A, et al. Expression of mutant Asxl1 perturbs hematopoiesis and promotes susceptibility to leukemic transformation. J Exp Med. 2018;215(6):1729–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbuccia N, Murati A, Trouplin V, et al. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23(11): 2183–2186. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Kurtenbach S, Guo Y, et al. Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood. 2018;131(3):328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acuna-Hidalgo R, Sengul H, Steehouwer M, et al. Ultra-sensitive sequencing identifies high prevalence of clonal hematopoiesis-associated mutations throughout adult life. Am J Hum Genet. 2017;101(1):50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo JQ, Wang JY, Arlinghaus RB. Detection of BCR-ABL proteins in blood cells of benign phase chronic myelogenous leukemia patients. Cancer Res. 1991;51(11): 3048–3051. [PubMed] [Google Scholar]
- 18.Yamagata T, Mitani K, Kanda Y, et al. Elevated platelet count features the variant type of BCR/ABL junction in chronic myelogenous leukaemia. Br J Haematol. 1996;94(2):370–372. [DOI] [PubMed] [Google Scholar]
- 19.Valent P, Kern W, Hoermann G, et al. Clonal hematopoiesis with oncogenic potential (CHOP): separation from CHIP and roads to AML. Int J Mol Sci. 2019;20(3):789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamata T, Kang J, Lee TH, et al. A critical function for B-Raf at multiple stages of myelopoiesis. Blood. 2005;106(3):833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia J, de Gunzburg J, Eychene A, et al. Thrombopoietin-mediated sustained activation of extracellular signal-regulated kinase in UT7-Mpl cells requires both Ras-Raf-1-and Rap1-B-Raf-dependent pathways. Mol Cell Biol. 2001;21(8):2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkozy A, Carta C, Moretti S, et al. Germline BRAF mutations in Noonan, LEOPARD, and cardiofaciocutaneous syndromes: molecular diversity and associated phenotypic spectrum. Hum Mutat. 2009;30(4):695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Wertheim G, Morrissette JJD, Bagg A. BRAF kinase domain mutations in de novo acute myeloid leukemia with monocytic differentiation. Leuk Lymphoma. 2017;58(3):743–745. [DOI] [PubMed] [Google Scholar]
- 24.Palanisamy N, Ateeq B, Kalyana-Sundaram S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16(7):793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falini B, Martelli MP, Tiacci E. BRAF V600E mutation in hairy cell leukemia: from bench to bedside. Blood. 2016;128(15):1918–1927. [DOI] [PubMed] [Google Scholar]
- 26.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15(3):152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Rodriguez B, Mayle A, et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell. 2016;29(6):922–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou HA, Kuo YY, Liu CY, et al. DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood. 2012;119(2):559–568. [DOI] [PubMed] [Google Scholar]
- 29.Leoni C, Montagner S, Rinaldi A, et al. Dnmt3a restrains mast cell inflammatory responses. Proc Natl Acad Sci U S A. 2017;114(8):E1490–E1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24(6):1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiNardo CD, Ravandi F, Agresta S, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol. 2015;90(8):732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiNardo CD, Jabbour E, Ravandi F, et al. IDH1 and IDH2 mutations in myelodysplastic syndromes and role in disease progression. Leukemia. 2016;30(4):980–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yonal-Hindilerden I, Daglar-Aday A, Hindilerden F, et al. The clinical significance of IDH mutations in essential thrombocythemia and primary myelofibrosis. J Clin Med Res. 2016;8(1):29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perner F, Perner C, Ernst T, Heidel FH. Roles of JAK2 in Aging, Inflammation, Hematopoiesis and Malignant Transformation. Cells. 2019;8(8):854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mead AJ, Rugless MJ, Jacobsen SE, Schuh A. Germline JAK2 mutation in a family with hereditary thrombocytosis. N Engl J Med. 2012;366(10):967–969. [DOI] [PubMed] [Google Scholar]
- 36.Fleischman AG, Tyner JW. Causal role for JAK2 V617F in thrombosis. Blood. 2013;122(23):3705–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbui T, Finazzi G, Falanga A. Myeloproliferative neoplasms and thrombosis. Blood. 2013;122(13):2176–2184. [DOI] [PubMed] [Google Scholar]
- 38.Hobbs CM, Manning H, Bennett C, et al. JAK2V617F leads to intrinsic changes in platelet formation and reactivity in a knock-in mouse model of essential thrombocythemia. Blood. 2013;122(23):3787–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mupo A, Seiler M, Sathiaseelan V, et al. Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia. 2017;31(3):720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin CC, Hou HA, Chou WC, et al. SF3B1 mutations in patients with myelodysplastic syndromes: the mutation is stable during disease evolution. Am J Hematol. 2014;89(8):E109–115. [DOI] [PubMed] [Google Scholar]
- 41.Tefferi A, Vannucchi AM, Barbui T. Essential thrombocythemia treatment algorithm 2018. Blood Cancer J. 2018;8(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKerrell T, Park N, Moreno T, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10(8):1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maslah N, Cassinat B, Verger E, et al. The role of LNK/SH2B3 genetic alterations in myeloproliferative neoplasms and other hematological disorders. Leukemia. 2017;31(8):1661–1670. [DOI] [PubMed] [Google Scholar]
- 44.Takizawa H, Eto K, Yoshikawa A, et al. Growth and maturation of megakaryocytes is regulated by Lnk/Sh2b3 adaptor protein through crosstalk between cytokine- and integrin-mediated signals. Exp Hematol. 2008;36(7):897–906. [DOI] [PubMed] [Google Scholar]
- 45.Wang W, Tang Y, Wang Y, et al. LNK/SH2B3 loss of function promotes atherosclerosis and thrombosis. Circ Res. 2016;119(6):e91–e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hock H, Shimamura A. ETV6 in hematopoiesis and leukemia predisposition. Semin Hematol. 2017;54(2):98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim CA, Phillips ML, Kim W, et al. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20(15):4173–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Paola J, Porter CC. ETV6-related thrombocytopenia and leukemia predisposition. Blood. 2019;134(8):663–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poggi M, Canault M, Favier M, et al. Germline variants in ETV6 underlie reduced platelet formation, platelet dysfunction and increased levels of circulating CD34+ progenitors. Haematologica. 2017;102(2):282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pawlikowska P, Fouchet P, Vainchenker W, et al. Defective endomitosis during megakaryopoiesis leads to thrombocytopenia in Fanca−/− mice. Blood. 2014;124(24):3613–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tischkowitz MD, Morgan NV, Grimwade D, et al. Deletion and reduced expression of the Fanconi anemia FANCA gene in sporadic acute myeloid leukemia. Leukemia. 2004;18(3):420–425. [DOI] [PubMed] [Google Scholar]
- 52.Garcia MJ, Benitez J. The Fanconi anaemia/BRCA pathway and cancer susceptibility. Searching for new therapeutic targets. Clin Transl Oncol. 2008;10(2):78–84. [DOI] [PubMed] [Google Scholar]
- 53.Solyom S, Winqvist R, Nikkila J, et al. Screening for large genomic rearrangements in the FANCA gene reveals extensive deletion in a Finnish breast cancer family. Cancer Lett. 2011;302(2):113–118. [DOI] [PubMed] [Google Scholar]
- 54.Crispino JD. GATA1 in normal and malignant hematopoiesis. Semin Cell Dev Biol. 2005;16(1):137–147. [DOI] [PubMed] [Google Scholar]
- 55.Ganapathi KA, Townsley DM, Hsu AP, et al. GATA2 deficiency-associated bone marrow disorder differs from idiopathic aplastic anemia. Blood. 2015;125(1):56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang SJ, Shi JY, Li JY. GATA-2 L359 V mutation is exclusively associated with CML progression but not other hematological malignancies and GATA-2 P250A is a novel single nucleotide polymorphism. Leukoc Res. 2009;33(8):1141–1143. [DOI] [PubMed] [Google Scholar]
- 57.Dickinson RE, Milne P, Jardine L, et al. The evolution of cellular deficiency in GATA2 mutation. Blood. 2014;123(6):863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Connelly JJ, Wang T, Cox JE, et al. GATA2 is associated with familial early-onset coronary artery disease. Plos Genet. 2006; 2(8):e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anguita E, Candel FJ, Chaparro A, Roldan-Etcheverry JJ. Transcription Factor GFI1B in Health and Disease. Front Oncol. 2017;7:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polfus LM, Khajuria RK, Schick UM, et al. Whole-exome sequencing identifies loci associated with blood cell traits and reveals a role for alternative GFI1B splice variants in human hematopoiesis. Am J Hum Genet. 2016;99(2):481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saleque S, Cameron S, Orkin SH. The zinc-finger proto-oncogene GFI1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 2002; 16(3):301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moroy T, Vassen L, Wilkes B, Khandanpour C. From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation. Blood. 2015;126(24):2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monteferrario D, Bolar NA, Marneth AE, et al. A dominant-negative GFI1B mutation in the gray platelet syndrome. N Engl J Med. 2013;370(3):245–253. [DOI] [PubMed] [Google Scholar]
- 64.Van Oorschot R, Hansen M, Koornneef JM, et al. Molecular mechanisms of bleeding disorder associated GFI1B (Q287*) mutation and its affected pathways in megakaryocytes and platelets. Haematologica. 2019;104(7):1460–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beauchemin H, Shooshtarizadeh P, Vadnais C, et al. GFI1b controls integrin signaling-dependent cytoskeleton dynamics and organization in megakaryocytes. Haematologica. 2017;102(3):484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mao X, Debenedittis P, Sun Y, et al. Vascular smooth muscle cell Smad4 gene is important for mouse vascular development. Arterioscler Thromb Vasc Biol. 2012;32(9): 2171–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Jiang L, Mo X, et al. Megakaryocytic Smad4 regulates platelet function through Syk and ROCK2 expression. Mol Pharmacol. 2017;92(3):285–296. [DOI] [PubMed] [Google Scholar]
- 68.Gallione CJ, Repetto GM, Legius E, et al. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4). Lancet. 2004;363(9412):852–859. [DOI] [PubMed] [Google Scholar]
- 69.Schutte M, Hruban RH, Hedrick L, et al. DPC4 gene in various tumor types. Cancer Res. 1996;56(11):2527–2530. [PubMed] [Google Scholar]
- 70.Chen S, Wang Q, Yu H, et al. Mutant p53 drives clonal hematopoiesis through modulating epigenetic pathway. Nat Commun. 2019;10(1):5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Apostolidis PA, Woulfe DS, Chavez M, et al. Role of tumor suppressor p53 in megakaryopoiesis and platelet function. Exp Hematol. 2012;40(2):131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shah V, Johnson DC, Sherborne AL, et al. Subclonal TP53 copy number is associated with prognosis in multiple myeloma. Blood. 2018;132(23):2465–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu Q, Zhang M, Blaese RM, et al. The Wiskott-Aldrich syndrome and X-linked congenital thrombocytopenia are caused by mutations of the same gene. Blood. 1995;86(10):3797–3804. [PubMed] [Google Scholar]
- 75.Devriendt K, Kim AS, Mathijs G, et al. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat Genet. 2001;27(3):313–317. [DOI] [PubMed] [Google Scholar]
- 76.Keszei M, Kritikou JS, Sandfort D, et al. Wiskott-Aldrich syndrome gene mutations modulate cancer susceptibility in the p53 +/- murine model. Oncoimmunology. 2018;7(9):e1468954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sereni L, Castiello MC, Villa A. Platelets in Wiskott-Aldrich syndrome: Victims or executioners? J Leukoc Biol. 2018;103(3):577–590. [DOI] [PubMed] [Google Scholar]
- 78.Shcherbina A, Cooley J, Lutskiy MI, et al. WASP plays a novel role in regulating platelet responses dependent on alphaIIbbeta3 integrin outside-in signalling. Br J Haematol. 2010;148(3):416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim H, Falet H, Hoffmeister KM, Hartwig JH. Wiskott-Aldrich syndrome protein (WASp) controls the delivery of platelet transforming growth factor-beta1. J Biol Chem. 2013;288(48):34352–34363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerrits AJ, Leven EA, Frelinger AL, et al. Effects of eltrombopag on platelet count and platelet activation in Wiskott-Aldrich syndrome/X-linked thrombocytopenia. Blood. 2015;126(11):1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coppe A, Nogara L, Pizzuto MS, et al. Somatic mutations activating Wiskott-Aldrich syndrome protein concomitant with RAS pathway mutations in juvenile myelomonocytic leukemia patients. Hum Mutat. 2018;39(4):579–587. [DOI] [PubMed] [Google Scholar]
- 82.Pang L, Xue HH, Szalai G, et al. Maturation stage-specific regulation of megakaryopoiesis by pointed-domain Ets proteins. Blood. 2006;108(7):2198–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y, Luo H, Liu T, et al. The ets transcription factor Fli-1 in development, cancer and disease. Oncogene. 2015;34(16):2022–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Favier R, Jondeau K, Boutard P, et al. Paris-Trousseau syndrome: clinical, hematological, molecular data of ten new cases. Thromb Haemost. 2003;90(5):893–897. [DOI] [PubMed] [Google Scholar]
- 85.Saultier P, Vidal L, Canault M, et al. Macrothrombocytopenia and dense granule deficiency associated with FLI1 variants: ultrastructural and pathogenic features. Haematologica. 2017;102(6):1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165. [DOI] [PubMed] [Google Scholar]
- 87.Vo KK, Jarocha DJ, Lyde RB, et al. FLI1 level during megakaryopoiesis affects thrombopoiesis and platelet biology. Blood. 2017;129(26):3486–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vyas P, Ault K, Jackson CW, et al. Consequences of GATA1 deficiency in megakaryocytes and platelets. Blood. 1999;93(9):2867–2875. [PubMed] [Google Scholar]
- 89.Fujiwara Y, Browne CP, Cunniff K, et al. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93(22):12355–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Freson K, Devriendt K, Matthijs G, et al. Platelet characteristics in patients with X-linked macrothrombocytopenia because of a novel GATA1 mutation. Blood. 2001;98(1):85–92. [DOI] [PubMed] [Google Scholar]
- 91.Zetterberg E, Verrucci M, Martelli F, et al. Abnormal P-selectin localization during megakaryocyte development determines thrombosis in the gata1low model of myelofibrosis. Platelets. 2014;25(7):539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lally J, Boasman K, Brown L, et al. GATA-1: A potential novel biomarker for the differentiation of essential thrombocythemia and myelofibrosis. J Thromb Haemost. 2019; 17(6):896–900. [DOI] [PubMed] [Google Scholar]
- 93.Yamaguchi Y, Zon LI, Ackerman SJ, et al. Forced GATA-1 expression in the murine myeloid cell line M1: induction of c-Mpl expression and megakaryocytic/erythroid differentiation. Blood. 1998;91(2):450–457. [PubMed] [Google Scholar]
- 94.Kaasinen E, Kuismin O, Rajamaki K, et al. Impact of constitutional TET2 haploinsufficiency on molecular and clinical phenotype in humans. Nat Commun. 2019;10(1):1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fuster JJ, MacLauchlan S, Zuriaga MA, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355(6327):842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vannucchi AM, Barbui T. Thrombocytosis and thrombosis. Hematology Am Soc Hematol Educ Program. 2007:363–370. [DOI] [PubMed] [Google Scholar]
- 97.Schafer AI. Thrombocytosis. N Engl J Med. 2004;350(12):1211–1219. [DOI] [PubMed] [Google Scholar]
- 98.Hengeveld PJ, Hazenberg MD, Biezeveld MH, Raphael MF. [Risk of thrombosis in reactive thrombocytosis]. Ned Tijdschr Geneeskd. 2018;162:D2697. [PubMed] [Google Scholar]
- 99.Scharf RE. Molecular complexity of the megakaryocyte-platelet system in health and disease. Hamostaseologie. 2016;36(3): 159–160. [PubMed] [Google Scholar]
- 100.Falanga A, Marchetti M. Thrombosis in myeloproliferative neoplasms. Semin Thromb Hemost. 2014;40(3):348–358. [DOI] [PubMed] [Google Scholar]