Significance
Human endogenous retroviruses (HERV) were incorporated into the genome over millions of years but are mostly inactive. HERV-K subtype HML-2 is the most recently incorporated and its incidental expression occurs in a variety of pathological conditions. However, its physiological role is not understood. We discovered that the envelope protein of this virus plays a critical role in early embryonic development. It is expressed at high levels on the surface of pluripotent stem cells and signals via direct binding to CD98HC, leading to activation of signaling pathways that regulate stem cell function. Down regulation of HML-2 env resulted in dissociation of the stem cell colonies and enhanced differentiation along neuronal pathways. Thus, HML-2 regulation is critical for human embryonic and neurodevelopment.
Keywords: endogenous retrovirus, neurogenesis, embryogenesis, stem cells, progenitor cells
Abstract
Stem cells are capable of unlimited proliferation but can be induced to form brain cells. Factors that specifically regulate human development are poorly understood. We found that human stem cells expressed high levels of the envelope protein of an endogenized human-specific retrovirus (HERV-K, HML-2) from loci in chromosomes 12 and 19. The envelope protein was expressed on the cell membrane of the stem cells and was critical in maintaining the stemness via interactions with CD98HC, leading to triggering of human-specific signaling pathways involving mammalian target of rapamycin (mTOR) and lysophosphatidylcholine acyltransferase (LPCAT1)–mediated epigenetic changes. Down-regulation or epigenetic silencing of HML-2 env resulted in dissociation of the stem cell colonies and enhanced differentiation along neuronal pathways. Thus HML-2 regulation is critical for human embryonic and neurodevelopment, while it’s dysregulation may play a role in tumorigenesis and neurodegeneration.
Human embryogenesis and brain development are precisely regulated by human-specific factors that differentiate us from other species. However, identification of these factors and the ensuing regulatory pathways has remained a mystery. A breakthrough in evolutionary biology has been the recognition that pervasive horizontal gene transfer between species—mediated by retroviruses—is one of the defining factors of evolution (1). If infection occurs in germline cells, the retroviral genes get transferred vertically from parent to offspring. In humans, over 8% of the genome consists of “relic” retrovirus genes (2). Named human endogenous retroviruses (HERVs), these genes were once considered “junk” because of the lack of known function in the human genome. However, it seems that some of the HERV genes have been adopted by their hosts for their own evolutionary benefits. As an example, the envelope protein of HERV-W, expressed in mammals as syncytin, has become an essential cellular protein for trophoblast and placental development (3, 4).
Among the HERVs, HERV-K subtype HML-2 is the most recently incorporated and the best-preserved family whose integration is unique in the human genome (5–7). The human genome has about 140 copies of HERV-K, some of which are specific to humans (8). Recently acquired HML-2 have nearly full-length viral sequences with ORFs for gag, pol, and env. The virus is classified as type 1 and 2. Only type 2 has an intact env gene (9). HML-2 activation has been observed in pluripotent stem cells (PSC) (10) and mesenchymal stem cells (11). Unregulated expression has been found in certain tumors (12) and overexpression in postmitotic neurons can cause neurodegeneration (13). Thus understanding the role of HML-2 in stem cell function and neuronal development could transform our understanding of human development and may have important consequences for studying disease pathogenesis and identifying new modes of treatment.
Results
PSC Express HML-2 Env, Which Is Down-Regulated During Neuronal Differentiation.
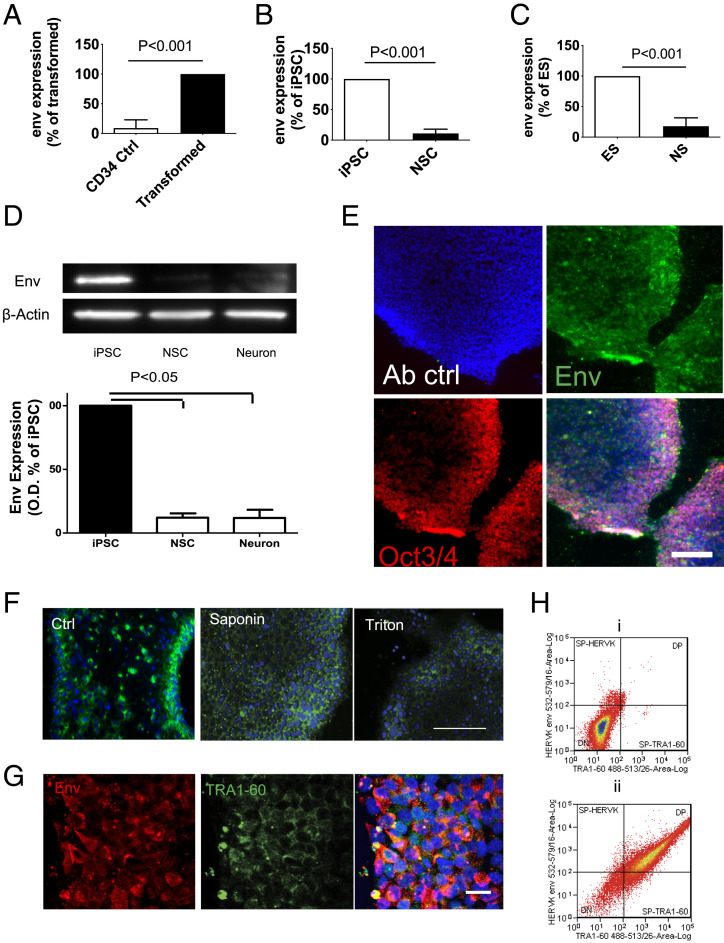
To study HML-2 Env expression during induced pluripotent stem cell (iPSC) transformation and neuronal differentiation, purified CD34 cells from peripheral blood of healthy donors were transformed into iPSC by transduction with Sendai virus containing transcriptional factors of Sox2, C-myc, Klf4, and Oct-3/4. The generated iPSCs had a normal karyotype and were characterized by immunostaining for stem cell markers (SI Appendix, Fig. S1 A and B). Five days after transduction they showed an increase in HML-2 env transcription (Fig. 1A). The iPSC lines were placed in neural induction medium for 7 d to generate neuronal stem cells (NSC) and in neural differentiation media for another 7 d to generate neurons. Three iPSC cell lines were used for these studies: ND2.0 (14), PAU, and ALEX (SI Appendix, Table S1). Neural induction was associated with a decrease in HML-2 env expression (Fig. 1B). A similar decrease in HML-2 env expression after neural differentiation was seen using the W9 embryonic stem (ES) cell line (Fig. 1C). HML-2 Env protein as monitored by Western blot analysis and immunostaining was expressed in iPSC but decreased in NSC and neurons (Fig. 1 D and E and SI Appendix, Fig. S1 C and E). However, unlike Env, Gag protein was expressed in iPSC and NSC but decreased only in neurons (SI Appendix, Fig. S1D). RNA-sequencing (RNA-seq) data (Gene Expression Omnibus [GEO]: GSE110267) showed that 4,305 HERV annotated regions (RepeatMasker) of the Human Genome (hg38) were expressed in at least one cell type profiled and 1,302 regions were exclusively in iPSC (SI Appendix, Fig. S2A). Of the HERV regions, 574 were differentially expressed between one or more cell types and most of the differential expressions were between iPSC and other cell types (SI Appendix, Fig. S2B). Principle component analysis (PCA) using expression for the 574 HERV regions revealed grouping within and between cell types, separating them by differentiation stage and time (SI Appendix, Fig. S2C).
Fig. 1.
Activation of HML-2 Env in PSC and silencing upon differentiation. (A) CD34+ hematopoietic stem cells were isolated from blood samples and infected with Sendai virus particles containing Yamanaka factors Oct-4, C-Myc, Klf-4, and Sox-2 to generate iPSC. Five days later, HML-2 env expression in the infected cells and control CD34 cells was monitored by qPCR. Data represent mean ± SEM from four independent experiments. (B) iPSC were differentiated into NSC using neural induction medium, HML-2 env expression in NSC was significantly decreased compared with iPSC. Data represent mean ± SEM from four independent experiments. (C) Similarly, ES cell line W9 had high levels of HML-2 env expression but declined upon differentiation to NSC. Data represent mean ± SEM from five independent experiments. (D) Western blot analysis showed that expression of HML-2 Env in the iPSC but no expression was seen at the NSC or neurons. Data are mean ± SEM from four independent experiments. (E) Immunostaining for HML-2 Env (green) shows HML-2 Env expression in iPSC (Env: green; stem cell marker Oct3/4: red; DAPI: blue). Antibody control (Ab ctrl) represents immunostaining without primary antibodies. Images are representative of three independent experiments. (Scale bar, 200 μm.) (F) Immunostaining of the iPSCs with an antibody of HML-2 Env (green) shows prominent immunostaining in the iPSC membrane, which decreased when the cells were treated with detergents, saponin, or triton after fixation with PFA. (Scale bar, 100 µm.) Images are representative of three independent experiments. (G) iPSC were immunostained for HML-2 Env (red) and TRA1-60 (green) directed against the iPSC membrane. (Scale bar, 20 µm.) DAPI (blue) was used to stain the nuclei. (H) Cell membrane staining of HML-2 Env and TRA1-60 on isolated live iPSCs was determined by flow cytometry. (i) No staining was observed in cells incubated with only secondary antibodies conjugated with fluorophores, (ii) Cells immunostained with antibodies against HML-2 and TRA1-16, showed that 92.3% of the cells were TRA1-60+, and 89% were also HML-2 Env+.
HERV-Env Is Expressed on the Cell Membrane of iPSC Cells.
Immunostaining showed that, HML-2 Env was expressed mainly on the cell surface of iPSC. Treatment of the cells with detergents resulted in loss of immunostaining (Fig. 1F), confirming that HML-2 Env was expressed on the cell membrane. Interestingly, there was increased HML-2 Env expression at the edge of some colonies, in the more compact cells. Costaining for a PSC marker TRA1-60 confirmed HML-2 Env expression in iPSC colonies (Fig. 1G). Dissociated single iPSC were immunostained and analyzed by flow cytometry. There was no staining of cells treated with secondary antibodies conjugated with fluorophores as a control (Fig. 1 H, i); 92.3% of cells were TRA1-60+, among which 89% were also HML-2 Env+, while only ∼10% of the TRA1-60− cells stained for HML-2 Env (Fig. 1 H, ii). Thus, HML-2 Env expression was not homogenous but predominantly on compact iPSCs and was integrated in the cell membrane.
Identification of Active HML-2 Env Loci in PSC.
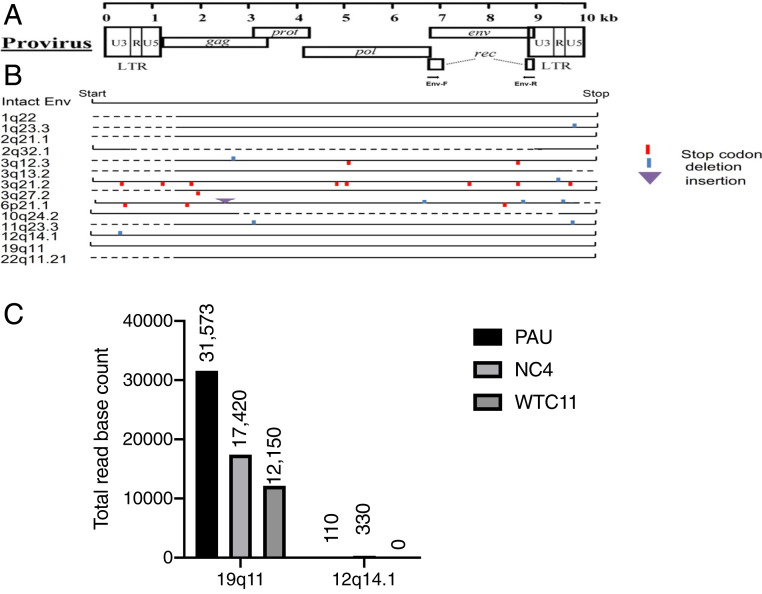
We amplified 1-kb transcripts of the HML-2 env derived from iPSC, cloned, and sequenced them. Fourteen loci were found to be transcriptionally active (Dataset S1), but only two loci, chromosomes 12q14.1 and 19q11, had complete ORFs (Fig. 2 A and B). The locus on 12q14.1 is also termed as K119, K(C12), K41, or ERVK-21 and the locus on 19q11 as K (C19) or ERVK-19 (15). To determine the predominant loci for the HML-2 env transcripts, RNA-seq data (GEO: GSE110497) from three iPSC lines were analyzed to generate the coverage values for the two env loci of interest using the “samtools” (www.htslib.org/). It showed that the env reads were predominantly from 19q11 (Chr 19: 27638539–27640638) (Fig. 2C). We next determined if HML-2 env expression at these loci was regulated by DNA methylation. Global DNA methylation (i.e., 5-methylcytosine) levels were determined by detecting LINE-1 methylation in iPSC and NSC. The methylation levels were significantly increased in NSC compared to the corresponding iPSC (SI Appendix, Fig. S3A). Conversely, when NSC cultures were treated with DNA methylation inhibitor 5-aza-2′-deoxycytidine, HML-2 env expression significantly increased (SI Appendix, Fig. S3B). Thus, DNA methylation was at least partially responsible for HML-2 env inhibition during neural differentiation from iPSC.
Fig. 2.
Determination the loci of HML-2 env expression. One-kilobase transcripts from the HML-2 env derived from iPSCs were amplified, cloned, and sequenced and 14 sites were found transcriptionally active. (A) Diagram shows where the HML-2 Env coding sequences and the PCR primers are. (B) Schematic of HML-2 env genes in the human genome expressed in iPSC are shown. Solid lines indicate the length of the transcript. The 12q14.1 env has a full-length transcript except for a 3-nt deletion. The 19q11 env has no deletion and no premature stop codon. (C) RNA-seq data from three iPSC lines—PAU, NC4 and WTC11—were used with “samtools” and the available command supported by that tool called “bedcov” to generate the coverage values for the two env loci. It showed that 19q11 predominantly produced the env reads.
We used a bisulfite conversion assay followed by sequencing to analyze the DNA methylation status of the HML-2 long terminal repeat (LTR) at chromosomes 12q14.1 and 19q11 from multiple iPSC lines, NSC, and neurons derived from the iPSC (SI Appendix, Table S1). At both loci, there was an increased trend in methylation as the cells differentiated from iPSC to NSC or to neurons (SI Appendix, Fig. S3C). However, the 19q11 LTR methylation was less than that of 12q14.1, supporting it is the originating locus for most of HML-2 Env expression.
HML-2 Env Is Critical for Maintaining Stemness of PSC.
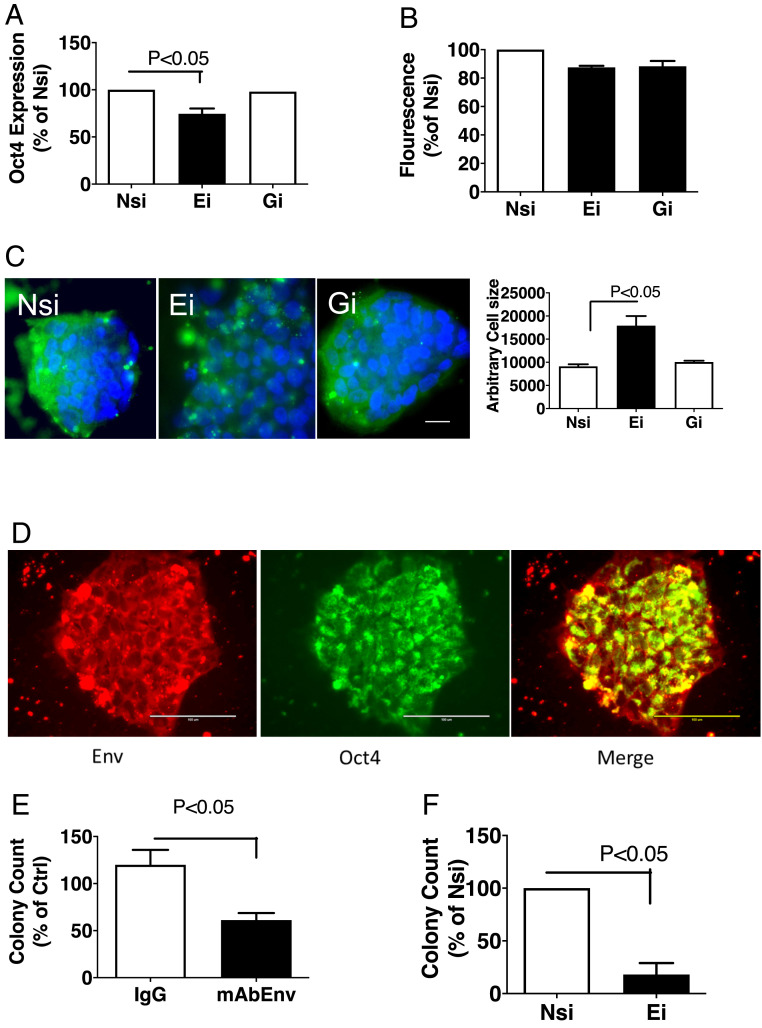
To study the role of HML-2 env in stem cell function, iPSC were transfected with small-interfering RNA (siRNA) targeting HML-2 env (Ei), a nontargeting control siRNA (Nsi), or a siRNA targeting HML-2 gag (Gi). Inhibition of HML-2 env transcript decreased expression of Oct-4/POU5F1 (Fig. 3A and SI Appendix, Fig. S4), which is a critical factor for the induction and maintenance of cellular pluripotency (16), but did not result in significant toxicity as reflected by estimation of cell numbers and mitochondrial function by Cellquanti blue assay (Fig. 3B). Ei also resulted in an increase in cell size, while Gi showed no significant changes compared to Nsi (Fig. 3C).
Fig. 3.
Inhibition of HML-2 env decreases stemness and stem cell colony formation. (A) Specific Ei and Gi were used to treat the iPSC. Oct-4 transcripts were decreased by Ei treatment in iPSC compared to control Nsi after 24 h. Data represents mean ± SEM from three independent experiments. (B) The effect on cell viability was monitored using cellquanti blue assay. No significant changes were observed in cell numbers following treatment with Ei or Gi compared to Nsi. Data represent mean ± SEM from three independent experiments. (C) After 48 h of treatment, cells were fixed and stained for HML-2 Env (green) and DAPI (blue) and imaged by a TIRF microscope. Decrease in HML-2 expression and an increase in cell size was observed in the cells treated with Ei compared with Nsi, while no significant effect of Gi treatment was observed. Data represent mean ± SEM from three independent experiments. (Scale bar, 10 µm.) (D) iPSCs were dissociated with ETDA treatment and replated on Matrigel-coated plates. After 48 h, iPSC colonies were observed which immunostained for HML-2 Env (red) and stem cell marker Oct4 (green). (Scale bars, 100 µm.) (E) iPSCs were similarly dissociated and cultured in the presence or absence of control IgG or monoclonal antibody to HML-2 Env (mAb Env), or (F) treated with Nsi or Ei. The number of stem cell colonies were counted and expressed as a percentage of control. Data represents mean ± SEM from three independent experiments.
To study the effect of HML-2 Env inhibition on colony formation in iPSC, iPSC were subcultured for 48 h in the presence of a monoclonal antibody to HML-2 Env and Oct-4+ colonies bigger than 100 µm were counted (Fig. 3D). Antibodies against HML-2 Env resulted in significantly fewer iPSC colonies, compared to control IgG (Fig. 3E). Furthermore, iPSCs transfected with siRNA against HML-2 env resulted in formation of significantly fewer colonies compared to Nsi (Fig. 3F). Thus HML-2 Env plays a role in maintaining of iPSC stemness and stem cell colony formation.
Induction of NSC and Differentiation to Neurons Is Regulated by HML-2 Env Expression.
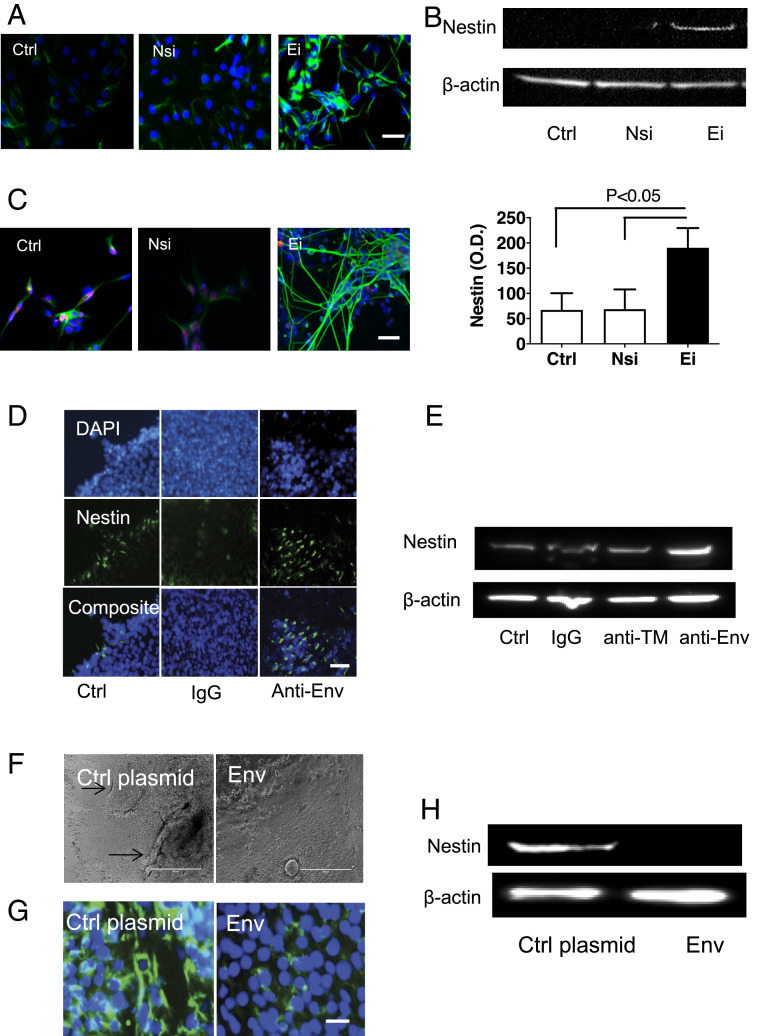
The effect of HML-2 Env during neural induction was studied by treating iPSC cultures with siRNA to HML-2 env and then culturing them in neural induction media. Within 3 d, the treated cells expressed nestin, an NSC marker, as detected by immunostaining and Western blot analysis (Fig. 4 A and B). After 7 d of further neuronal differentiation, the cells assumed a neuronal morphology and immunostained for β-III-tubulin (Fig. 4C). When iPSC were incubated with antibody against HML-2 Env in neural induction media, the iPSC colonies started to build up to make 3D structures within the center. At the same time, cells detached from the edge of the colony and assumed a neuronal morphology (Movie S1). These changes occurred at a much more rapid pace compared to untreated control cells similarly placed in neural induction media. Nestin expression was also more prominent in the cultures incubated with HML-2 Env antibody (Fig. 4 D and E), indicating that inhibition of HML-2 Env increased NSC induction from iPSC. Furthermore, when expression of HML-2 Env was forced by transfecting iPSC with a HML-2 env-containing plasmid (SI Appendix, Fig. S5), the neuronal induction process was inhibited as indicated by the maintenance of original morphology and inhibition of nestin expression in the cultures even after 7 d of induction (Fig. 4 F–H). Conversely, transfection of siRNA to HML-2 env in multiple iPSC lines resulted in the increased expression of nestin (SI Appendix, Fig. S4).
Fig. 4.
Inhibition of HML-2 Env promotes neuronal differentiation of stem cells. (A) Three days after iPSCs were transfected with Ei, and incubated in neural induction media, expression of NSC marker nestin was increased compared to control cells (Ctrl) and cells transfected with Nsi, as determined by immunostaining (blue: DAPI; green: nestin). (Scale bar, 50 µm.) (B) Corresponding Western blot analysis shows nestin in the Ei-treated cell extracts. Data are presented as mean ± SEM from three independent experiments. (C) After 7 d of further differentiation in neuronal medium, the cells assumed a neuronal morphology with immunostaining for β-III-tubulin (green). (Scale bars, 50 µm.) (D) iPSCs were treated with control IgG or antibody to HML-2 Env (Anti-Env). Increased immunostaining for nestin (green) was noted with anti-Env. (Scale bar, 50 µm.) (E) Corresponding Western blot analysis shows increased nestin with anti-Env treatment but not with control IgG or antibody to the transmembrane domain of the HML-2 Env (anti-TM). (F) Forced expression of HML-2 Env during iPSC neuronal differentiation by transfecting iPSC with a CMV promoter-driven Env plasmid resulted in delayed neuronal differentiation as shown by lack of 3D structure build-ups and (G) decreased expression of nestin expression by immunostaining (green for nestin, blue for nuclei DAPI staining), and (H) Western blot analysis. β-Actin was used as a loading control. (Scale bars, 400 µm in F and 25 µm in G.) Images are representative of three independent experiments.
HML-2 Env Interacts with CD98HC.
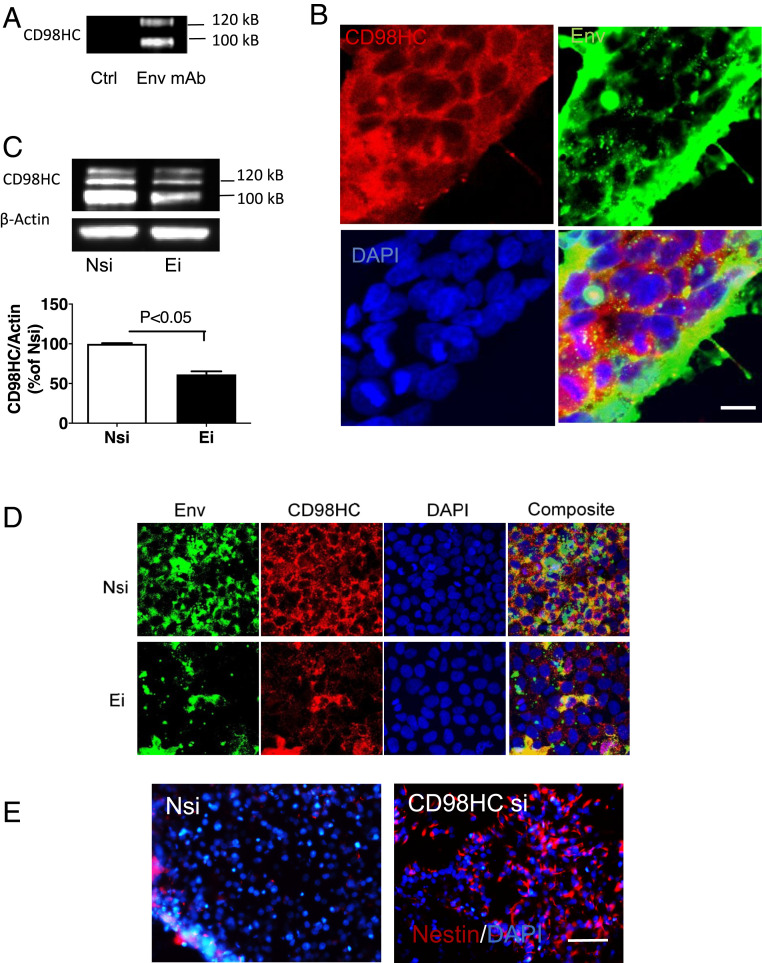
To determine the interactions between HML-2 Env and other cell membrane proteins, we prepared HML-2 Env immunobeads using recombinant HML-2 Env that matched to chromosome 19 and incubated them with iPSC lysates. The resulting protein complexes were analyzed by mass spectrometry. Fifty-five interacting proteins were found for which at least three or more peptides could be identified (Dataset S2). The vast majority of these were mitochondrial proteins, enzymes involved in energy metabolism, or nucleolar proteins. The protein complex also included histone H4 and lysophosphatidylcholine acyltransferase (LPCAT1). Only one cell membrane protein was isolated: 4F2 cell-surface antigen heavy chain, or CD98HC. The interaction between HML-2 and CD98HC was confirmed by immunoprecipitation of the cell extracts with an antibody to HML-2 Env followed by Western blot analysis using an antibody to CD98HC (Fig. 5A). Immunostaining showed colocalization of CD98HC and HML-2 Env in iPSCs (Fig. 5B). Interestingly, when cells were treated with siRNA against HML-2, CD98HC production was decreased compared to control, as shown by Western blot analysis (Fig. 5C) and immunostaining (Fig. 5D). When iPSCs were treated with siRNA targeting CD98HC and subsequently cultured in neural induction medium, the cells showed increased nestin expression (Fig. 5E). Thus HML-2 Env binds directly to CD98HC and may exert its stemness maintenance effect through CD98HC signaling.
Fig. 5.
Interaction between HML-2 Env and CD98HC. (A) iPSC lysates were immunoprecipitated using an antibody to HML-2 Env (anti-Env) and analyzed by Western blot using an antibody to CD98HC. Images are representative of three independent experiments (B) Immunostaining of stem cells showed colocalization of CD98HC (red) and HML-2 Env (green). (Scale bar, 10 µm.) (C) Forty-eight hours after transfection with Ei, CD98HC expression was decreased compared with Nsi. Data are presented as mean ± SEM from three independent experiments. (D) iPSCs were transfected with either Nsi or siRNA to HML-2 env. Inhibition of Env with Ei resulted in decreased expression of HML-2 Env (green) and CD98HC (red), as shown by immunostaining. (Scale bar, 20 µm.) (E) iPSCs were treated with either Nsi or siRNA to CD98HC (CD98HC si). Treatment with CD98HC si resulted in an increase in nestin+ cells (red) during neural induction as shown by immunostaining. (Scale bar, 100 µm.) (B, D, and E) DAPI (blue) was used to stain the nuclei. Images are representative of three independent experiments.
Molecular Pathways Underlying HML-2 Env Function in iPSC.
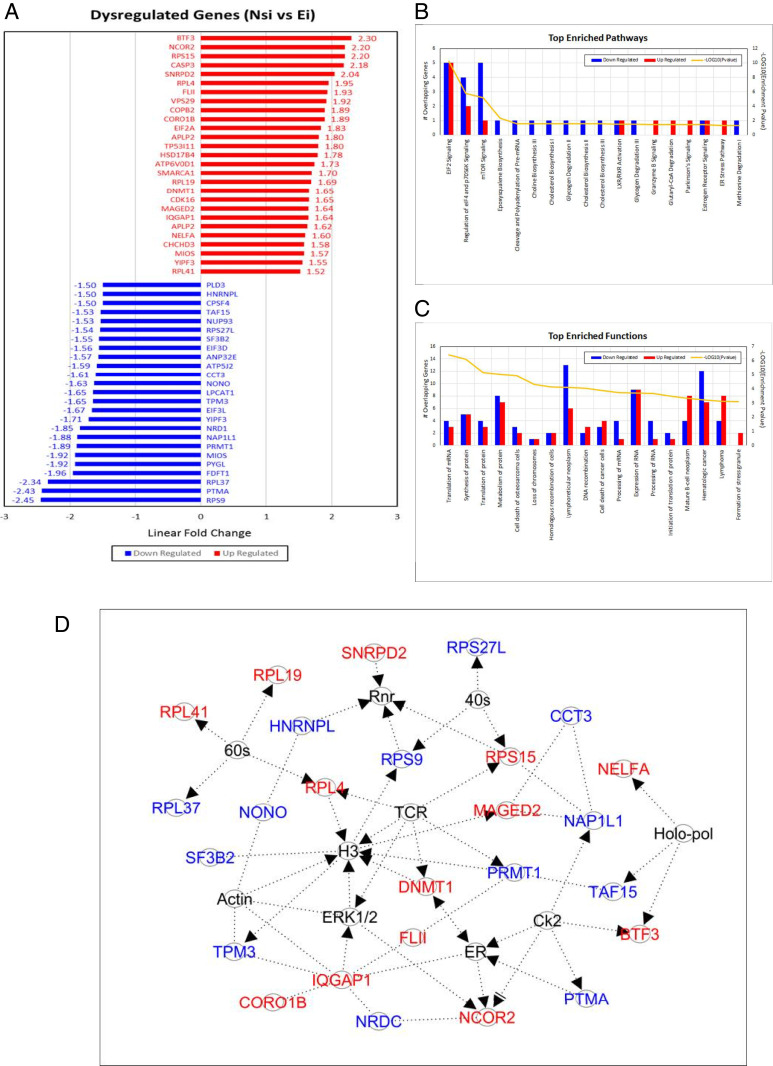
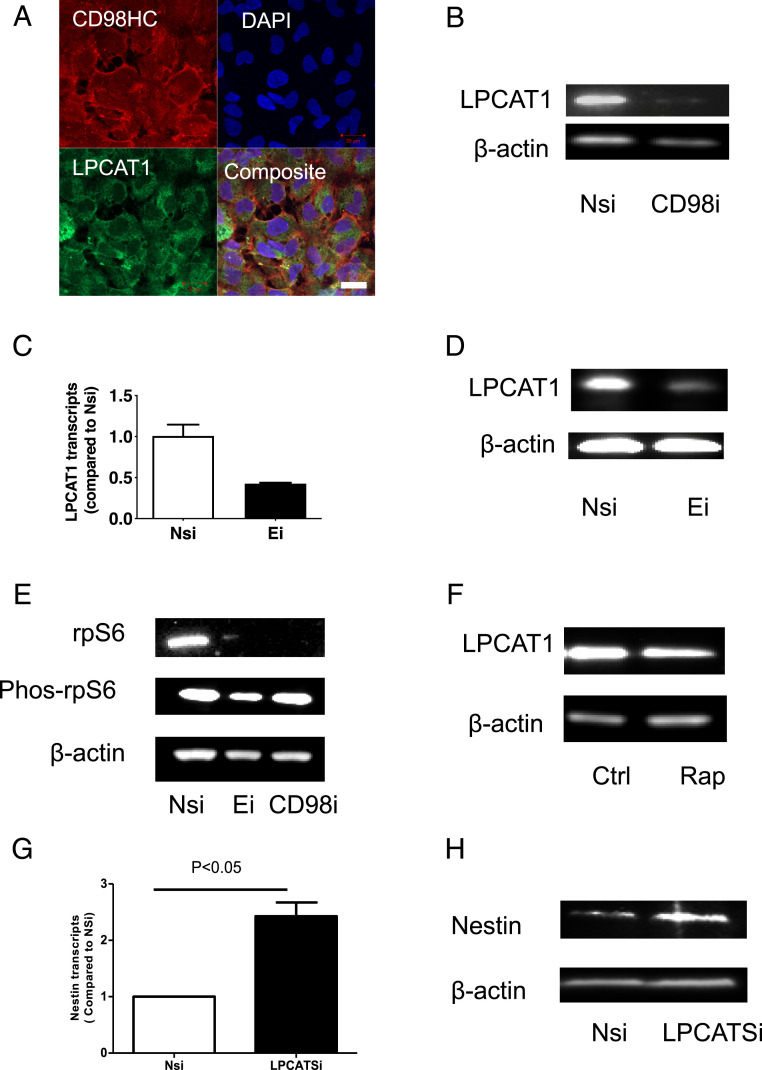
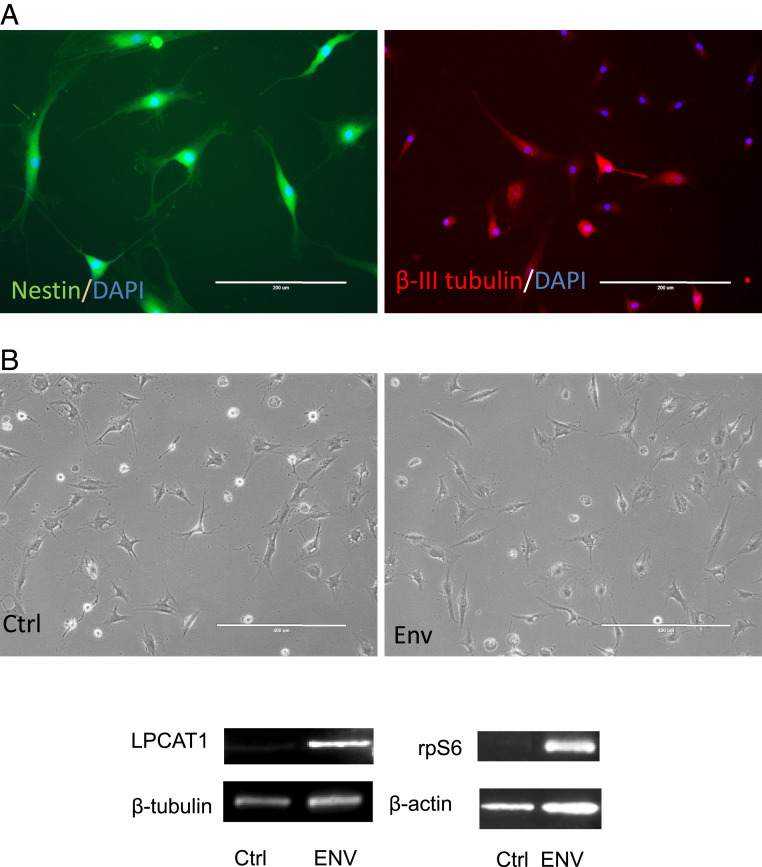
To determine the genes and pathways regulated by HML-2 Env, iPSC cell lines [PAU, NC 4.0, and WTC11 (17)] were treated with siRNAs targeting HML-2 Env and RNA-seq data (GEO: GSE110497) was analyzed. Twenty-seven genes were significantly increased, and 25 genes were significantly decreased after 24 h of siRNA treatment compared to Nsi (Fig. 6A and Dataset S3). The affected genes were involved in several pathways, including mammalian target of rapamycin (mTOR) signaling, DNA methylation, RNA transcriptional regulation, and others associated with stem cell functions (Fig. 6 B and C). Interestingly, 42 genes regulated by HML-2 env have also been associated with cancer, including LPCAT1 (Fig. 6A and Dataset S3). To determine if the interactions between HML-2 Env and CD98HC could signal via the LPCAT1 and mTOR pathway, we initially dual-stained the iPSC for CD98HC and LPCAT1. While CD98HC was localized to the cell membrane, LPCAT1 was predominantly cytoplasmic with some colocalization at the cell membrane (Fig. 7A). Inhibition of CD98HC using siRNA caused a reduction in LPCAT1 production (Fig. 7B), suggesting that the two molecules are functionally linked. Similarly, inhibition of HML-2 env using siRNA caused a reduction in LPCAT1 transcripts (Fig. 7C) and protein (Fig. 7D). To determine the relationship between these molecules and the mTOR pathway, we monitored the levels of ribosomal protein S6 (rpS6) following treatment with siRNA to HML-2 env or CD98HC. There was a reduction in both the nonphosphorylated and phosphorylated forms of rpS6 (Fig. 7E). LPCAT1 protein in iPSC was also decreased following treatment with rapamycin, which targets mTOR (Fig. 7F). When iPSC were treated with siRNA to LPCAT1, increased nestin transcripts (Fig. 7G) and protein expression were noted (Fig. 7H). This relationship was further confirmed in cells derived from rhesus macaques that do not have an endogenous HML-2. NSC were generated from rhesus fibroblasts (Fig. 8A). Transfection of these cells with HML-2 env plasmid resulted in a dramatic increase in expression of rpS6 and LPCAT1 (Fig. 8B). This suggests a tight relationship between HML-2 and the mTOR–LPCAT1 pathway and its critical role in embryogenesis.
Fig. 6.
HML-2 Env regulates gene expression in iPSCs. HML-2 Env expression was inhibited by treating iPSCs with siRNA targeting HML-2 env in three different lines. Total RNA was collected for RNA-seq analysis. (A) Vertical bar plot describes genes dysregulated between Nsi and Ei (uncorrected ANCOVA P < 0.05, linear fold-change ≥ 1.5×). Genes are ranked by linear fold-change (blue bars = down-regulated, red bars = up-regulated). (B) These genes were imported into the IPA tool (https://www.qiagen.com/us/) to identify the top enriched functions by Exact Test and (C) the top enriched pathways by Exact Test. Number of down-regulated genes and up-regulated genes overlapping per function and pathway are described (x axis) along with the corresponding enrichment P value (y axis). (D) The top-scoring molecular network.
Fig. 7.
Interactions among HML-2 Env, CD98HC, and LPCAT1. (A) iPSCs immunostained for CD98HC (red) and LPCAT1 (green) showed LPCAT1 expression in CD98HC+ cells. DAPI (blue) was used to stain the nuclei. (Scale bar, 20 μm.) (B) iPSC were treated with either Nsi or siRNA to CD98HC for 24 h. LPCAT1 expression was decreased by siRNA targeting CD98HC as determined by Western blot analysis. (C) iPSC were also treated with either Nsi or Ei. Inhibition of env with Ei resulted in decreased LPCAT1 expression, as detected by RT-PCR and (D) Western blot analysis. (E) Inhibition of HML-2 env or CD98HC using siRNA resulted in a decrease in rpS6 production as determined by a Western blot analysis. (F) Effect of rapamycin (Rap) on the LPCAT1 production in iPSC was determined using Western blot analysis. (G) iPSCs were treated with siRNA to LPCAT1 for 24 h which resulted in increased nestin transcripts and 72 h after neuronal induction, an increase in nestin protein was noted (H), as detected by Western blot assay. Data are presented as mean ± SEM and images are representative of three independent experiments.
Fig. 8.
Introduction of HML-2 Env expression in rhesus NSC activates mTOR and LPCAT1 pathway. (A) Induced rhesus NSC positive-stained with nestin (green) could be differentiated to β-III-tubulin+ (red) neurons. (B) The rhesus NSCs were transfected with HML-2 Env plasmid for 48 h, which resulted in the increased levels of rpS6 and LPCAT1, as determined by Western blot assay. β-Actin or β-tubulin was used as a loading control. Images are representative of three independent experiments.
Discussion
Emerging evidence shows that during human embryonic development and in PSC there is activation of retroviral and transposable elements, including LINE-1 and HERVs (18–20). The mechanisms by which they influence embryonic development are poorly understood. Some ERV families contain preserved splice sites that join the ERV segment with non-ERV exons in their genomic vicinity (21). Similarly, HERV-H activation is associated with stem cell pluripotency and cellular differentiation, where it functions as a nuclear noncoding RNA (22). We show that the HML-2 Env protein is expressed in the human PSC and regulates stemness and differentiation potential of the cells by interacting with cell-membrane molecules and cell-matrix networks, leading to the activation of signaling mechanisms implicated in cell proliferation.
HML-2 env transcripts increased during iPSC transformation. HML-2 env transcripts were maximally expressed in PSC but diminished as the cells differentiated into NSC and neurons. This silencing of HML-2 is critical for neuronal differentiation, since forced expression in stem cells prevents their differentiation along the neuronal pathway. This is akin to expression of retroviral elements in other dividing cells where they have been implicated in cancer (23). In fact, uncontrolled HML-2 expression has been found in teratocarcinoma, which is a germline or stem cell tumor (24). In contrast, forced expression of HML-2 Env protein in terminally differentiated neurons is toxic (13). The effects of HML-2 in stem cells were specific for the Env protein since silencing of gag transcripts had no effect on cell differentiation. The activated HML-2 env transcripts could be localized to several loci; however, only those on Chr12q14.1 and Chr19q11 have an intact ORF for HML-2 Env. These viral genes correspond to the HML-2 subgroup, which is the youngest and most active ERV. The locus on chromosome 12 is also termed HERV-K 119, K(C12), K41, or ERVK-21. The locus on chromosome 19 is termed HERV-K (C19) or ERVK-19 (15). These chromosomal loci have not been previously implicated in any physiological or pathological condition. The predominant HML-2 transcripts in teratocarcinomas originate from chromosome 22 (24) and, in patients with amyotrophic lateral sclerosis, were predominantly from chromosome 7 (13, 25).
Previous reports show that several HERV transcripts are increased during cell transformation and in human PSCs (10). These transcripts are tightly regulated and do not get activated by age alone (26). We found that the expression of HML-2 Env was regulated by epigenetic changes. iPSC showed a lower level of DNA methylation compared to NSC and nonspecific DNA methylation inhibitor 5-Aza-2′-deoxycitydine induced a significant increase of HML-2 env expression in NSC. LTR methylation is a well-known regulator of HML-2 transcription and the level of methylation throughout the LTR affects its expression (27). The methylation at the 3′ end of the LTR has a more significant effect on the locus transcription than the 5′ methylation (27). We observed that at loci 19q11 and 12q14.1, LTR methylation levels were decreased in iPSC compared to NSC and neurons. However, because the methylation at the 3′ end of the 19q11 LTR is much lower than 12q14.1, it is likely to be the locus from which most HML-2 transcripts are originating in these cells. Interestingly, many zinc finger genes exist in clusters in chromosome 19, which are known to drive the epigenetic repression of ERVs, including HML-2 through DNA methylation (28). Previous studies show that the Krueppel-associated box zinc finger protein ZFP809 targets KAP1 (TRIM28) and represses ERVs in neural progenitor cells (29).
We studied the function of HML-2 Env in PSC and in neuronal differentiation. In PSC, HML-2 Env expression was increased predominantly in smaller and more compact cells. Blocking of HML-2 Env using either siRNA or antibodies resulted in separation of cells and enlarged cell size, and fewer stem cell colonies formed after replating. Similarly, in another study excision of HML-2 env in a tumor cell line resulted in decreased cell proliferation (30). We observed that inhibition of HML-2 env by siRNA resulted in decreased Oct-4 expression, a critical marker for stemness. It also decreased expression of rpS6, a crucial effector of mTORC1 signaling pathway, and decreased novel protein synthesis. This is in agreement with previous reports that nonphosphorylated rpS6 is responsible for increased protein synthesis and smaller cell size (31) and that mTORC1 tunes ribosome concentration by regulating ribosome biogenesis and autophagy, subsequently affecting the biophysical properties of the cytosol, including viscosity (32), which are closely associated with cell size. Thus HML-2 env is critical for PSC function by maintaining cell-to-cell contact and cell adhesion to the extracellular matrix, which in turn regulate the cell functions through the evolutionary conserved mTOR pathway. There is an increased level of mTOR activation in human outer subventricular zone radial glia compared to nonhuman primates, possibly due to increased proliferation in human radial glia that function as NSC (33). We observed that rhesus NSC transfected with HML-2 Env resulted in high levels of rpS6 and LPCAT1 expression, further implying that HML-2 incorporation in the human genome caused the increased activity of mTOR in human stem cells and may have played a role in human evolution.
The function of HML-2 Env in stem cell differentiation was explored using a neuronal induction model. Inhibition of HML-2 Env using either siRNA or antibody to the Env resulted in enhanced NSC differentiation and production of neurons. In contrast, forced expression of HML-2 Env in the neural induction process resulted in slower differentiation and lower levels of nestin. This further supports the notion that HML-2 Env subtly modulates the stemness and differentiation potential of PSCs, as has also been reported for HERV-H (34). HERV-H is expressed as a noncoding RNA that regulates transcriptional factors. In contrast, HML-2 transcripts encode the Env protein, which interacts with other proteins on the cell membrane to mediate its effect. Studies in mouse ES cells have also implicated the role of ERVs in regulation of pluripotency and reprograming, however, have not addressed the role of viral protein expression (35).
Retroviral envelope proteins are known to facilitate cell-to-cell adhesion and in some cases cause cell fusion. For example, HERV-W syncytin leads to fusion of trophoblasts to form the placenta (3, 36). This syncytium formation is mediated via interactions between syncytin and the human sodium-dependent neutral amino acid transporter type 2 gene named SLC1A5 (37). We found that the HML-2 Env binds to 4F2 cell-surface antigen heavy chain, also named CD98HC or SLC3A2. Interestingly, treatment with siRNAs targeting HML-2 Env also decreased CD98HC expression, providing further evidence for functional interactions between the two proteins. CD98HC is a heterodimeric amino acid transporter that interacts with β1 integrin (38–40). CD98HC is well known for maintaining stem cell function and plays an important role in regulating cell morphology and has been implicated in the skin-healing process (39). CD98HC also enables fibronectin matrix assembly by interacting with integrins to support RhoA-driven contractility (38). Similarly, interactions between HML-2 Env and the CD98HC leads to activation of signaling pathways in PSC to maintain cell adhesion and stem cell morphology. These cell adhesions are indispensable for stem cell pluripotency (41). Interruption of these cell adhesions by inhibition of HML-2 expression results in inhibition of the RhoA pathway and differentiation of the cells along a neuronal pathway. Thus HML-2 Env may be another mechanism acquired by human stem cells to fine-tune the regulation between stemness and cell differentiation.
Overexpression of CD98HC has been associated with tumorigenesis (42–44) and inhibition of HML-2 env in the stem cells was found to modulate several genes associated with cancer. Among these genes was LPCAT1, which we found was regulated by mTOR and CD98HC. This interaction between mTOR and LPCAT1 has not been described previously. This enzyme catalyzes the conversion of lysophosphatidlycholine to phosphatidylcholine (45). LPCAT1 can also catalyze the palmitolysation of histone H4 (46) and our pull-down assay showed that histone H4 and LPCAT1 were part of the HML-2 Env interactome. This posttranslational modification of histone H4 is involved in epigenetic global gene regulation (46). These functions of LPCAT1 are calcium-dependent, suggesting that they are part of the signaling cascade. High levels of LPCAT1 and HML-2 have been reported in prostate cancer (47, 48), further suggesting an interaction between the two proteins in cells capable of renewing themselves. Together, these findings suggest a signaling pathway for HML-2 Env function in stem cells (SI Appendix, Fig. S8).
In summary, HML-2 expression in human pluripotent cells has functions associated with cell morphology and adhesion mediated by interactions with the membrane-bound protein CD98HC, resulting in activation of signaling pathways implicated in cell proliferation. The on and off nature of HML-2 Env expression may play an important role in the regulation of the delicate balance between stemness and neuronal differentiation and its dysregulation could potentially play a role in tumorigenesis and neurodegeneration.
Materials and Methods
Reagents and Supplies.
Details of the reagents and resources are provided in SI Appendix, Table S3.
Cell Cultures.
Culture media and components were purchased from Invitrogen, growth factors and cytokines were purchased from PeproTech, and chemicals were purchased from Sigma, if not specified. ES cell line WA09-FI was obtained from WiCell. iPSC, NC4, NCRM1, NCRM5 (49), and ND2.0 (50) were provided by the Center for Regenerative Medicine at the National Institutes of Health (NIH); WTC11 was kindly provided by Michael E. Ward, National institute of Neurological Disorders and Stroke, NIH, Bethesda, MD. Human PSCs were cultured in E8 medium on Matrigel-coated plates in a 5% CO2 incubator at 37 °C and subcultured using ethylene diamine tetra acetic acid (EDTA), following published protocols (50). Mouse embryonic fibroblasts were purchased from ThermoFisher and used as a negative control for HML-2 Env protein.
Human adult hematopoietic progenitor cells (CD34 cells) were enriched, cultured, and transformed following our previously published protocol (51). Briefly, blood from adult healthy donors was collected at the Transfusion Medicine Blood Bank of the NIH and signed informed consents were obtained in accordance with the NIH Institutional Review Board. CD34 cells were purified by negative selection using the human progenitor cell enrichment kit (Stemcell Technologies) and resuspended in StemSpan serum-free expansion medium (Stemcell Technologies) containing human thrombopoietin (100 ng/mL), fms-like tyrosine kinase 3 (Flt-3) ligand (100 ng/mL) and stem cell factor (100 ng/mL), IL-6 (20 ng/mL), and IL-7 (20 ng/mL) in a six-well plate (3 × 105 cells per well) at 37 °C in a 5% CO2 incubator. For transformation, 1 × 105 CD34 cells per well in 200 µL were seeded in a 24-well plate and infected with Sendai viral particles (CytoTune-iPS Reprogramming kit, Invitrogen) containing transcription factor constructs of hOct3/4, hSox-2, hKlf4, and hc-Myc at a multiplicity of infection of 15 in prewarmed media. Next, 500 µL of fresh medium was added after 24 h and half-media changes were performed every other day for 5 d. The cells were then transferred to murine embryonic fibroblasts (MEF feeder cells, Invitrogen) in E8 medium with daily medium change till the iPSC colonies emerged, usually after another 14 d. Generated iPSC were picked and expanded on Matrigel-coated six-well plates in E8 medium. Generated iPSC lines PAU and ALEX were confirmed by karyotyping, which showed normal chromosome pattern and by immunostaining for stem cell markers SSEA-3, Oct-4, and TRA1-60 (SI Appendix, Fig. S1 A and B). Additional iPSC lines 505, 506, and 516 generated from peripheral blood mononuclear cells from healthy human donors using a similar Sendai virus method by NIH National Heart, Lung, and Blood Institute iPSC core faculty were also used for cell differentiation and HML-2 LTR methylation studies.
Rhesus skin samples were collected and provided by the National Institute of Neurological Disorders and Stroke animal health care section based on the animal protocol (animal protocol number: 1196) approved by NIH Institutional Animal Care and Use Committee. The sterile collected skin samples were cut into 1-mm3 pieces and seeded onto a poly-d-lysine (PDL; Sigma) -coated six-well plate in DMEM/F12 medium containing 10% FBS (Gemini) and antibiotics at 37 °C in an incubator containing 5% CO2. After 2 wk, the fibroblasts growing out of the skin pieces were dissociated with trypsin/EDTA treatment and subcultured in a PDL-coated six-well plate. When the cells reaching 40% confluence (SI Appendix, Fig. S6A), Sendai virus containing Yamanaka factors were used to transduce the cells. After 5 d, the cells were dissociated with Accutase treatment and subcultured into a Matrigel-coated six-well plate and incubated with neural induction medium to select NSC. The resulting NSC (SI Appendix, Fig. S6B) were nestin+ and could differentiate to β-III-tubulin+ neurons spontaneously (Fig. 8A). The rhesus NSC within three passages were used for transfection using plasmids containing HML-2 Env with lipofectamine 3000 (ThermoFisher) (Fig. 8B).
Cell Differentiation.
NSC were differentiated from PSC following our previous protocol (51). Briefly, iPSC were seeded on a Matrigel-coated six-well plate in E8 medium. When ready for induction, the E8 medium was replaced with neural induction media (neural basal medium containing neural induction supplement, Invitrogen) for 3, 5, or 7 d, as needed for different experiments. The resulting NSC were characterized by nestin immunostaining and were further differentiated to neurons when subcultured on PDL-coated plates and incubated with neuronal differentiation medium (DMEM/F12) containing 1× N2 supplement, 1× B27 supplement, 300 ng/mL cyclic adenosine monophosphate (cAMP, Sigma), and 0.2 mM vitamin C (Sigma), 10 ng/mL brain-derived neurotrophic factor, and 10 ng/mL glial-derived neurotrophic factor for 14 d. The resulting neurons were confirmed by immunostaining for β-III-tubulin.
Immunocytochemistry.
Immunocytochemistry was performed following our previously published protocol (52). Briefly, cells were fixed in 4% paraformaldehyde (PFA) for 10 min at room temperature and then washed three times with PBS pH 7.4. After incubation in 0.1% Triton X-100 in PBS (PBS-T) for 10 min at room temperature, the cells were incubated with blocking buffer (PBS-T containing 4% goat serum and 1% glycerol [Sigma]) at room temperature for 20 min. Cells were immunostained with mouse monoclonal antinestin (1 µg/mL, Millipore Cat# MAB5326, RRID:AB_11211837), mouse monoclonal anti–β-III-tubulin antibodies (1 µg/mL, Promega Cat# G7121, RRID:AB_430874;1 µg/mL), rabbit anti-LPCAT1 (2 µg/mL; Proteintech Group Cat# 16112–1-AP, RRID:AB_2135554), mouse anti–TRA-1-60 antibody (10 µg/mL; Abcam Cat# ab16288, RRID:AB_778563), rat anti–SSEA-3 antibody (10 µg/mL; Millipore Cat# MAB4303, RRID:AB_177628), and the ready-to-use rabbit anti-Sox2 and anti–Oct-4, antibodies from the hES/iPS cell characterization kit (Applied Stemcell), followed by corresponding secondary antibodies (anti-mouse Alexa Fluor 488: ThermoFisher Scientific Cat# A-11001, goat anti-mouse IgM Alexa Fluor 488: 1 µg/mL; ThermoFisher Scientific Cat# A21042, RRID:AB_141357; Alexa RRID:AB_2534069, 1:400; anti-rabbit Alexa Fluor 594: ThermoFisher Scientific Cat# A-11012, RRID:AB_2534079, 1:400; Invitrogen) DAPI nuclear staining.
For immunostaining for HML-2 Env, the cells were fixed in PFA. Some samples were posttreated with either 0.1% Triton X-100 or 0.1% saponin for 30 min. The cells were then incubated with a monoclonal antibody to HML-2 Env (AMSBIO LLC Cat# HERM-1811–5, RRID:AB_10891119, 10 µg/mL; Austral Biologicals) overnight at 4 °C, followed by 1-h incubation with anti-mouse Alexa Fluor 488 (1:400) at room temperature. Images were acquired using a Zeiss LSM 510 META multiphoton confocal system (Carl Zeiss) or EVOS fluorescence microscope (AMG). Secondary antibody-only control was used to determine the specificity of the staining.
Validation of HML-2 Env Antibodies.
Additional rabbit anti–HML-2 Env and Gag antibodies were custom manufactured by GeneScript. Mouse anti-HML-2 Env mAb K06 GN was manufactured and provided by Geneuro. All of the HML-2 Env antibodies were further validated by immunoprecipitation of HML-2 Env from the cell lysates of HML-2 Env Flag Tag Plasmid transfected HEK 293 cells. The immunoprecipitated HML-2 Env was validated by Western blot analysis.
Flow Cytometry.
Accutase cell dissociation solution (ThermoFisher) was used to dissociate the iPSC. The dissociated cells were spun and resuspended at 1 million cells/mL in DPBS and incubated with anti–HML-2 Env mAb (K06 GN) and Tra1-60 mouse IgM antibody (Abcam, Cat# ab16288, RRID: AB_778563) at a final concentration 10 µg of antibody per 1 million cells in 1 mL of DPBS for 1 h at room temperature, followed by a wash step to remove excess antibody. The cells were subsequently stained using an Alexa Fluor 488-conjugated secondary antibody against mouse IgM and an Alexa Flour 546 anti-mouse IgG (1 µg antibody per 1 million cells in 1 mL DPBS) for 1 h at room temperature, followed by a wash step. At the same time, cells stained with secondary antibody alone were used as control. The cells were subsequently stained using 1 µM/mL DRAQ5 (ThermoFisher), a cell-permeable DNA dye, and 1 µg/mL DAPI (ThermoFisher), a cell impermeable DNA dye, to discriminate between live and dead cells and to enable single cell gating on the FACS machine (MoFlo Astrios cell sorter, Beckman Coulter). FACS gating strategy is shown in SI Appendix, Fig. S7. After the cells were aseptically passed through a 35-μm strainer to remove any remaining cell clumps, FACS analysis was performed by NIH Flow Cytometry Core facility.
Real-Time PCR (RT-PCR) Assay.
RNA was extracted and purified using a total RNA purification kit (Norgen) with on-column DNase I (Qiagen) treatment for 15 min following the manufacturer’s instructions. RT-PCR was conducted using one step SensiFAST SYBR kit (Bioline) and ViiA7 real-time PCR system (Life Technologies) according to the manufacturer’s instructions. HML-2 env and β-actin primers were manufactured by Qiagen, as previously published (13). Nestin and β-2-microglobin (B2M) primers were manufactured by Invitrogen and LPCAT1 primers were synthesized by Sigma. The sequences are: Nestin forward: 5′-CAGCGTTGGAACAGAGGTTGG, reverse: 5′-TGGCACAGGTGTCTCAAGGGTAG; B2M forward: 5′-GTATGCCTGCCGTGTGAAC, reverse: 5′-AAAGCAAGCAAGCAGAATTTGG; LPCAT1 forward: 5′-AGTATATACGGCCTGTGTTC, reverse: 5′-CATTATCTGTGGCCACTTTC. Ct values over 35 were considered undetectable. B2M and β-actin were used as internal control. Delta Ct value (ΔCt) was calculated according to the ViiA7 real-time PCR machine manual. PCR product for HML-2 env was also visualized and purified by running through SizeSelect Agarose Gels (E-gel, Life technologies) and then sequenced by Genewiz.
siRNA Synthesis and Treatment.
Consensus HML-2 (ERV-K6) env sequence was used as a template to design Env siRNA 1 and 2. When analyzed by nucleotide BLAST, Env siRNA2 also matched to HML-2 Env sequence on chromosome 22 of hg38. The DNA sequence on chromosome 22 (hg38_dna range = chr22:18945137–18946982) was used as a template for the design of other siRNAs using Dharmacon’s online tool. The resulting candidate sequences were further examined using nucleotide BLAST to avoid nonspecific matches. Three sequences (Ei 3, 4, and 5) were then chosen as they were also best matches to consensus HML-2 Env (ERV-K6) sequences. Sequences for siRNA targeting HML-2 gag were based on the published literature (53). Silencer select siRNAs based on these sequences as well as predesigned silencer select siRNAs targeting CD98HC (assay ID: s12944), LPCAT1 (assay ID: s36577), and a predesigned silencer select negative control siRNA were ordered from ThermoFisher. For siRNA transfection, iPSCs were cultured for 24 h and reached 30 to 50% confluence. TransIT-siQUEST transfection reagent (Mirus Bio) was mixed with siRNA (100-μM stock, 125-nM final concentration). After 30 min, the mixture was applied to the iPSC cultures.
Colony Formation Assay.
Colony formation was used to test the capability of stem cells to form new colonies after dissociation. To determine the effect of siRNA, iPSC cultures in 24-well plates at 40% confluence were transfected with siRNAs (SI Appendix, Table S2). Effect of siRNA on the inhibition of HML-2 env expression was determined after 24 h, which showed 40 to 60% inhibition. For assessment of colony formation, 48 h after transfection the cells were treated with EDTA dissociation solution (0.5 mM EDTA, 1.8 g/L NaCl in calcium/magnesium-free PBS) (54) for 5 min at room temperature and dissociated by vigorous pipetting in E8 medium. The dissociated cells were then seeded onto Matrigel-coated 24-well plates at a 1 to 10 dilution ratio. For determining the effect of antibodies, the replated iPSC were incubated with 1 µg/mL of each of the antibodies (monoclonal antibody against HML-2 Env, AMSBIO LLC Cat# HERM-1811-5, RRID:AB_10891119) and an isotype control antibody (ThermoFisher Scientific Cat# MA1-10418, RRID:AB_2536786). After 48 h of subculture, cells were fixed with 4% PFA and immunostained for Oct-4. To exclude nonspecific cell aggregates, only iPSC colonies with size of 100 µm or bigger were counted using an EVOS microscope by an investigator blinded to the treatment.
Cell Morphology and Cell-Size Determination.
Cells were cultured on Matrigel-coated 24-well plates and transfected with siRNAs, as described above. The cultures were live-imaged using FL-Auto microscopy (Life Technologies). After 48 h, cells were fixed with 4% PFA followed by immunostaining for HML-2 Env and Oct-4. Images of stem cell colonies were taken using an EVOS microscope and a Nikon Eclipse Ti total internal reflection fluorescence (TIRF) microscope equipped with a 488/561/405/647 LU4A Lasers System and an Andor iXon3 EMCCD camera by using a APO TIRF 100× NA 1.49 objective. Images were acquired with NIS-Elements AR software and processed with ImageJ to obtain composite images. Nuclear imaging was done through the DAPI channel of the TIRF microscope system, using fluorescence illumination supplied by an X-cite fluorescent lamp at 75% power (Lumen Dynamics). ImageJ was used to determine the area and total number of DAPI+ cells in the colonies. Average cell sizes were calculated by dividing the colony area by total cell numbers by an investigator blinded to the treatment.
Western Blot Analysis and Immunoprecipitation.
Cells were lysed with radio immunoprecipitation assay (RIPA) buffer (55) including the complete proteinase inhibitor mixture (Roche) or the phosphatase inhibitors mixture (Sigma). HML-2 Env magnetic beads were made by conjugating protein G magnet beads (50 μL; Life Technologies) with anti–HML-2 Env mAb (5 µg) for 30 min. The beads were then incubated with cell lysates for 1 h at room temperature. After washing three times with washing buffer, the pellets were collected and used for Western blot assay.
For Western blot analysis, protein (20 µg per lane) or pellets from immunoprecipitation were resolved by 4 to 10% Tris-glycine polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 5% nonfat milk or 5% BSA and incubated with monoclonal anti–HML-2 Env (2 µg/mL), rabbit anti–HML-2 Env (2 µg/mL), rabbit anti–HML-2 gag (2 µg/mL), monoclonal antinestin (1:1,000), rabbit anti-LPCAT1 (0.25 µg/mL), rabbit anti-S6 ribosomal protein (1:1,000; Cell Signaling Technology Cat# 2217, RRID:AB_331355), rabbit antiphospho-S6 ribosomal protein ([Ser235/236], 1:2,000; Cell Signaling Technology Cat# 2211, RRID:AB_331679), mouse monoclonal antipuromycin (1:5,000; Millipore Cat# MABE343, RRID:AB_2566826) or monoclonal mouse anti-CD98HC (3 µg/mL), and monoclonal mouse anti–β-actin (1:5,000, Sigma) antibodies overnight at 4 °C. After washing, membranes were incubated with peroxidase-linked anti-rabbit or anti-mouse IgG (1:5,000; GE Healthcare) for 1 h at room temperature. Amersham ECL Western blotting detection reagents (GE Healthcare) were used for detection with a FlourChem M imager (ProteinSimple). For HML-2 Env protein detection in Western blots, cell lysates from HEK 293 cells transfected with Env FlagTag plasmid (SI Appendix, Fig. S5) were used as a positive control and cell lysate of mouse embryonic fibroblasts was used as a negative control.
Transfection of HML-2 Env Plasmids.
HML-2 plasmids were designed (SI Appendix, Fig. S5) and used to transfect iPSC using Lipofectamine 3000 (Invitrogen) per the manufacturer’s instructions. Briefly, 30% confluent iPSC were seeded for 24 h before transfection. The cells were transfected with HML-2 plasmids or empty pcDNA as a control. After 48 h of transfection, the culture medium was replaced with neural induction medium to induce neural differentiation. The transfection efficiency was ∼30% as determined by a green fluorescent protein-expressing plasmid control (Lonza). HML-2 Env plasmid and the control empty plasmid were also used to transfect rhesus NSC with Lipofectamine 3000. The rhesus cells were collected 48 h after transfection for protein extraction and Western blot analysis.
Cloning and Sequencing of HML-2 Env RNA from iPSC.
Total RNA was extracted and purified from iPSC using RNeasy Plus Mini Kit (Qiagen). About 1 μg RNA was then reverse-transcribed with SuperScript III First-Strand Synthesis SuperMix kit (ThermoFisher). The resulting product was subjected to PCR amplification using Q5 High-Fidelity 2× Master Mix (New England Biolabs). The following PCR condition was used: Denaturing at 98 °C for 10 s, annealing at 55 °C for 20 s, and extension at 72 °C for 4 min. Total cycle number is 35. The PCR primers were used to amplify the full-length Env gene as follows: forward primer 5′- cccactagacatttgaagttctaca-3′, and reverse primer 5′- ggagtctcctatgtctacttcttt-3′. After separation and purification by agarose gel, the PCR product was cloned using TOPO TA cloning kit (ThermoFisher). Plasmids from 40 clones were prepared and sequenced using M13 forward primer. The sequencing results were blasted against Human genome sequence (hg38). The best hit from each clone was assigned to individual chromosomal locus. To determine what chromosome has active HML-2 expression in iPSC, percentage of clones from each chromosome was calculated for comparison.
LTR Methylation Analysis Using Bisulfite Treatment of DNA.
Genomic DNA was isolated from iPSC, NSC, and neuronal cell lines by standard protocol from a DNeasy Blood & Tissue Kit (Qiagen). Treatment of the denatured DNA (i.e., single-stranded DNA) with sodium bisulfite to determine CpG methylation levels was accomplished with an EpiTect Fast Bisulfite Conversion Kit (Qiagen). Genomic DNA was incubated at 95 °C for 10 min to induce fragmentation followed by sodium bisulfite conversion at 60 °C (pH 5–6) for 40 min. Desulphonation proceeded at 23 °C for 15 min. Two LTR loci were amplified from bisulfite-treated DNA by methylation-specific PCR using an EpiTect MSP Kit (Qiagen). Efficiency of bisulfite treatment was confirmed through standardized methylated and unmethylated controls provided by the EpiTect PCR Control DNA Set (Qiagen). Standardized controls and genomic DNA were treated simultaneously. PCR contained 2 μL of treated DNA (∼200 ng) with HotStarTaq d-Tect polymerase (Qiagen). PCR cycles were as follows: 10 min at 95 °C; 40 cycles, each cycle consisting 15 s at 94 °C, 30 s at 55 to 60 °C (dependent on primer set annealing temperature), and 30 s at 72 °C; and finally, 10 min at 72 °C. After gel electrophoresis, PCR products were eluted with a gel extraction kit (Qiagen). The various PCR primer sequences flanking the LTR of HML-2 loci at chromosomes 12q14.1 and 19q11 are shown in SI Appendix, Table S5B. PCR products were subsequently cloned into the pCR 4-TOPO TA vector with One Shot TOP10 Chemically Competent Escherichia coli (Invitrogen). Plasmid DNA from single colonies were purified using QIAprep Spin Miniprep Kit (Qiagen) and sequenced by Eurofins Genomics.
RNA-Seq and Pathway Analysis after HML-2 Env Inhibition.
iPSC seeded in Matrigel-coated 24-well plates were transfected with siRNA against Ei or Nsi for 24 h in E8 medium. RNA samples were collected from control and treated cells using a total RNA purification kit (Norgen) with on-column DNase I (Qiagen) treatment for 15 min following the manufacturer’s instructions. The inhibition effect of siRNA on HML-2 Env as well as on Oct4 and nestin expression was first determined using RT-PCR (SI Appendix, Fig. S4). The samples with confirmed HML-2 Env inhibition were used for RNA-seq. After CASAVA deplexing, .fastq files generated per library were imported into the CLCbio Genomics Workbench (https://digitalinsights.qiagen.com/) and quality control-inspected, adaptor-clipped, low quality-trimmed, filtered, then mapped. Per quality trimming, 15 bp from the 5′ end of each read was globally removed along with 1 bp from the 3′ end. Bases with a call accuracy <95% were trim-removed thereafter. Per filtering, reads having more than two ambiguities were discarded along with those having read length <15 bp postquality trimming. Per mapping, the “RNA-Seq” tool supported in the Workbench was used to locally align reads by library against the human genome (GRCh38/hg38) without masking, keeping only those mapped reads with ≥80% reference identity for ≥80% read length. Postmapping, the number of reads falling in known annotated transcripts (Ensembl) were enumerated and the counts converted into transcript per million mapped reads (TPM) expression values. TPM expression values were next imported into R (https://cran.r-project.org/), where TPM expression values were pedestalled by 2, log2-transformed, filtered to remove transcripts not having at least one transformed value >1, then quantile-normalized. Normalized values for remaining transcripts were then used to confirm the absence of library-level outliers via Tukey box plot, covariance-based PCA scatter plot, and Pearson correlation heat map. After, normalized expression was noise-modeled using LOWESS (CV∼mean) and transcripts not having at least one normalized value >3.25 were removed as noise-biased and noninformative. For transcripts not discarded, differential expression between Ei and Nsi treatment was tested for using ANOVA while correcting for iPSC line type. Transcripts having an uncorrected P < 0.05 by this test and an absolute difference of corrected means ≥1.5× were subsequently deemed to be differentially expressed between Ei and Nsi. Transcript annotations and corresponding enriched pathways and functions were obtained using the Ingenuity Pathway Analysis “IPA” tool (https://www.qiagen.com/us/). The sorted alignment files for control iPSC samples were also used to generate the coverage values for the two env loci 19q11 and 12q14.1, using the tool called “samtools” and the available command supported by that tool called “bedcov” (https://www.htslib.org).
mTOR Molecular Pathway Analysis.
Puromycin incorporation was used to study the effect of HML-2 Env on protein synthesis, iPSC were treated with siRNA targeting HML-2 env or CD98HC for 23 h, followed by incubation with puromycin (10 μg/mL; Sigma) for another hour. The cells were washed with PBS and then lysed with RIPA buffer for Western blot assay using antipuromycin antibody. Before transfer to the PVDF membrane, the gel was also stained with page blue protein staining buffer to compare the amounts of total protein loaded in the lanes. The effect of HML-2 env and CD98HC siRNA treatments on Rps6 and phospho-Rps6 were studied using Western-blot assay. The effect of mTOR pathway activation was also mimicked by treating iPSC with rapamycin (2 µM, Sigma) for 24 h and the cell lysates were collected for LPCAT1 protein production using Western blot assay.
Statistical Analysis.
For experiments that required immunostaining, the total number of cells and immunolabeled cells were counted in nine predetermined fields per coverslip or well using a fluorescence microscope with a 20× objective. Three coverslips per well were counted in each group. At least three independent experiments were performed. Mean and SE were calculated for each treatment group. Statistical analysis was performed using Prism v3.0. Differences were tested using one-way ANOVA followed by Bonferroni’s test for multiple groups and Student’s t test for two groups. Two-tailed values of P < 0.05 were considered significant. Details of statistical analysis are in SI Appendix, Table S4.
Code Availability.
Software used in this study include the Ingenuity Pathway Analysis “IPA” tool (https://www.qiagen.com/us/), R (https://cran.r-project.org/), CLCbio Genomics Workbench (https://digitalinsights.qiagen.com/), samtools (https://www.htslib.org/), Proteome Discoverer 2.1 (ThermoFisher Scientific), and a custom database constructed from the SwissProt portion of the UniProt database (release 2017_04). Additional information on RNA-seq analysis, global DNA methylation analysis, and liquid chromatography-mass spectrometry are provided in SI Appendix, SI Methods
Supplementary Material
Acknowledgments
We thank Dr. Hemali Phatnani and Dr. Isabel Hubbard, New York Genome Center ALS Consortium, for assistance with RNA-sequencing study. This work is funded by NIH intramural funds.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE110498). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier PXD019702 and 10.6019/PXD019702).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002427117/-/DCSupplemental.
Data Availability.
All relevant data supporting the key findings of this study are available within the article and SI Appendix. Two sets of RNA-seq data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus; the link to the superseries record is https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110498 (56). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (57) partner repository with the dataset identifier PXD019702 and 10.6019/PXD019702 (58).
References
- 1.Koonin E. V., Wolf Y. I., Evolution of microbes and viruses: A paradigm shift in evolutionary biology? Front. Cell. Infect. Microbiol. 2, 119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander E. S. et al.; International Human Genome Sequencing Consortium , Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Mi S., et al. , Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Dupressoir A., Lavialle C., Heidmann T., From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta 33, 663–671 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Hohn O., Hanke K., Bannert N., HERV-K(HML-2), the best preserved family of HERVs: Endogenization, expression, and implications in health and disease. Front. Oncol. 3, 246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbulescu M., et al. , Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9, 861–868 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Medstrand P., Mager D. L., Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 72, 9782–9787 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suntsova M., et al. , Molecular functions of human endogenous retroviruses in health and disease. Cell. Mol. Life Sci. 72, 3653–3675 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wildschutte J. H., et al. , Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc. Natl. Acad. Sci. U.S.A. 113, E2326–E2334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs N. V., et al. , Human endogenous retrovirus K (HML-2) RNA and protein expression is a marker for human embryonic and induced pluripotent stem cells. Retrovirology 10, 115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mareschi K., et al. , Human endogenous retrovirus-H and K expression in human mesenchymal stem cells as potential markers of stemness. Intervirology 62, 9–14 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Schmitt K., Reichrath J., Roesch A., Meese E., Mayer J., Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol. Evol. 5, 307–328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W., et al. , Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 7, 307ra153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ou W., Li P., Reiser J., Targeting of herpes simplex virus 1 thymidine kinase gene sequences into the OCT4 locus of human induced pluripotent stem cells. PLoS One 8, e81131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian R. P., Wildschutte J. H., Russo C., Coffin J. M., Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8, 90 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jerabek S., Merino F., Schöler H. R., Cojocaru V., OCT4: Dynamic DNA binding pioneers stem cell pluripotency. Biochim. Biophys. Acta 1839, 138–154 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Miyaoka Y., et al. , Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods 11, 291–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Hurk J. A. J. M., et al. , L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 16, 1587–1592 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Pi W., et al. , The LTR enhancer of ERV-9 human endogenous retrovirus is active in oocytes and progenitor cells in transgenic zebrafish and humans. Proc. Natl. Acad. Sci. U.S.A. 101, 805–810 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klawitter S., et al. , Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat. Commun. 7, 10286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Göke J., et al. , Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16, 135–141 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Lu X., et al. , The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol. 21, 423–425 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Cao M., et al. , Human endogenous retroviruses and cancer. Cancer Biol. Med. 13, 483–488 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhardwaj N., Montesion M., Roy F., Coffin J. M., Differential expression of HERV-K (HML-2) proviruses in cells and virions of the teratocarcinoma cell line Tera-1. Viruses 7, 939–968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douville R., Liu J., Rothstein J., Nath A., Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 69, 141–151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nevalainen T., et al. , Aging-associated patterns in the expression of human endogenous retroviruses. PLoS One 13, e0207407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavie L., Kitova M., Maldener E., Meese E., Mayer J., CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 79, 876–883 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukic S., Nicolas J. C., Levine A. J., The diversity of zinc-finger genes on human chromosome 19 provides an evolutionary mechanism for defense against inherited endogenous retroviruses. Cell Death Differ. 21, 381–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brattås P. L., et al. , TRIM28 controls a gene regulatory network based on endogenous retroviruses in human neural progenitor cells. Cell Rep. 18, 1–11 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Ibba G., Piu C., Uleri E., Serra C., Dolei A., Disruption by SaCas9 endonuclease of HERV-Kenv, a retroviral gene with oncogenic and neuropathogenic potential, inhibits molecules involved in cancer and amyotrophic lateral sclerosis. Viruses 10, 412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruvinsky I., et al. , Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 19, 2199–2211 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delarue M., et al. , mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174, 338–349.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollen A. A., et al. , Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743–756.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohnuki M., et al. , Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc. Natl. Acad. Sci. U.S.A. 111, 12426–12431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gautam P., Yu T., Loh Y.-H., Regulation of ERVs in pluripotent stem cells and reprogramming. Curr. Opin. Genet. Dev. 46, 194–201 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Blaise S., de Parseval N., Bénit L., Heidmann T., Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 13013–13018 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavillette D., et al. , The envelope glycoprotein of human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J. Virol. 76, 6442–6452 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Féral C. C., et al. , CD98hc (SLC3A2) participates in fibronectin matrix assembly by mediating integrin signaling. J. Cell Biol. 178, 701–711 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boulter E., et al. , CD98hc (SLC3A2) regulation of skin homeostasis wanes with age. J. Exp. Med. 210, 173–190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feral C. C., et al. , CD98hc (SLC3A2) mediates integrin signaling. Proc. Natl. Acad. Sci. U.S.A. 102, 355–360 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bedzhov I., et al. , Adhesion, but not a specific cadherin code, is indispensable for ES cell and induced pluripotency. Stem Cell Res. 11, 1250–1263 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Shishido T., et al. , Transformation of BALB3T3 cells caused by over-expression of rat CD98 heavy chain (HC) requires its association with light chain: Mis-sense mutation in a cysteine residue of CD98HC eliminates its transforming activity. Int. J. Cancer 87, 311–316 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Poettler M., et al. , CD98hc (SLC3A2) drives integrin-dependent renal cancer cell behavior. Mol. Cancer 12, 169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estrach S., et al. , CD98hc (SLC3A2) loss protects against ras-driven tumorigenesis by modulating integrin-mediated mechanotransduction. Cancer Res. 74, 6878–6889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harayama T., Shindou H., Shimizu T., Biosynthesis of phosphatidylcholine by human lysophosphatidylcholine acyltransferase 1. J. Lipid Res. 50, 1824–1831 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou C., et al. , Acyl-CoA:lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J. Biol. Chem. 286, 28019–28025 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X., et al. , The expression level of lysophosphatidylcholine acyltransferase 1 (LPCAT1) correlates to the progression of prostate cancer. Exp. Mol. Pathol. 92, 105–110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goering W., et al. , Human endogenous retrovirus HERV-K(HML-2) activity in prostate cancer is dominated by a few loci. Prostate 75, 1958–1971 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Efthymiou A., et al. , Functional screening assays with neurons generated from pluripotent stem cell-derived neural stem cells. J. Biomol. Screen. 19, 32–43 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Chen G., et al. , Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8, 424–429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang T., et al. , Derivation of neural stem cells from human adult peripheral CD34+ cells for an autologous model of neuroinflammation. PLoS One 8, e81720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang T., et al. , Activated T-cells inhibit neurogenesis by releasing granzyme B: Rescue by Kv1.3 blockers. J. Neurosci. 30, 5020–5027 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oricchio E., et al. , Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene 26, 4226–4233 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Beers J., et al. , Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc. 7, 2029–2040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alcaraz C., De Diego M., Pastor M. J., Escribano J. M., Comparison of a radioimmunoprecipitation assay to immunoblotting and ELISA for detection of antibody to African swine fever virus. J. Vet. Diagn. Invest. 2, 191–196 (1990). [DOI] [PubMed] [Google Scholar]
- 56.Johnson K. R., Human endogenous retrovirus-K in stem cell function and neuronal differentiation. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110498. Deposited 12 February 2018.
- 57.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kowalak J., Co-IP of fHERV-K env protein and interactors. PRIDE Database. www.ebi.ac.uk/pride/archive/projects/PXD019702. Deposited 11 June 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data supporting the key findings of this study are available within the article and SI Appendix. Two sets of RNA-seq data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus; the link to the superseries record is https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110498 (56). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (57) partner repository with the dataset identifier PXD019702 and 10.6019/PXD019702 (58).