Significance
Hepatitis delta virus (HDV) aggravates hepatitis B virus (HBV) infection of liver cells. Although the viruses are evolutionarily unrelated, HDV depends on HBV because it requires the HBV envelope protein for its transmission. HDV is only described in humans, which has triggered diverse hypotheses regarding its evolution and origins. Here we show that spiny rats (Proechimys semispinosus) carry a counterpart to HDV that surprisingly does not cause hepatitis and is not linked to HBV. The rodent deltavirus finding alone, but also taken together with the recent deltavirus findings in snakes and other vertebrates and invertebrates, suggests that a deltavirus precursor may have infected mammals before it acquired dependence on HBV as seen in humans.
Keywords: deltavirus, Proechimys semispinosus, hepadnavirus, coinfection, neotropical rodent
Abstract
Hepatitis delta virus (HDV) is a human hepatitis-causing RNA virus, unrelated to any other taxonomic group of RNA viruses. Its occurrence as a satellite virus of hepatitis B virus (HBV) is a singular case in animal virology for which no consensus evolutionary explanation exists. Here we present a mammalian deltavirus that does not occur in humans, identified in the neotropical rodent species Proechimys semispinosus. The rodent deltavirus is highly distinct, showing a common ancestor with a recently described deltavirus in snakes. Reverse genetics based on a tandem minus-strand complementary DNA genome copy under the control of a cytomegalovirus (CMV) promoter confirms autonomous genome replication in transfected cells, with initiation of replication from the upstream genome copy. In contrast to HDV, a large delta antigen is not expressed and the farnesylation motif critical for HBV interaction is absent from a genome region that might correspond to a hypothetical rodent large delta antigen. Correspondingly, there is no evidence for coinfection with an HBV-related hepadnavirus based on virus detection and serology in any deltavirus-positive animal. No other coinfecting viruses were detected by RNA sequencing studies of 120 wild-caught animals that could serve as a potential helper virus. The presence of virus in blood and pronounced detection in reproductively active males suggest horizontal transmission linked to competitive behavior. Our study establishes a nonhuman, mammalian deltavirus that occurs as a horizontally transmitted infection, is potentially cleared by immune response, is not focused in the liver, and possibly does not require helper virus coinfection.
The genus Deltavirus is an unclassified RNA virus taxon that currently includes only one species, the hepatitis delta virus (HDV). This virus is an important human pathogen and the only satellite virus known in animals. For transmission, HDV must acquire an envelope from a coinfecting hepatitis B virus (HBV; family Hepadnaviridae, genus Orthohepadnavirus). Following superinfection of HBV-infected individuals, HDV increases the severity of liver disease (1). Initially detected in 1977 (2), HDV is present in about 10% of chronically HBV-infected patients (3). The circular single-stranded RNA genome is ∼1,700 nt long and is packaged together with the small and large hepatitis delta antigens (S-HDAg and L-HDAg) into an HDV ribonucleoprotein particle that buds with the help of HBV envelope proteins from HDV/HBV-coinfected hepatocytes. During replication, a double rolling-circle scheme produces multimeric antigenomic RNAs that are self-cleaved into monomers by a viroid-like ribozyme structure. The ligated circular RNA (circRNA) is further transcribed into multimeric genomic RNAs which also undergo self-cleavage and ligation (4). Remarkably, HDV RNA replication is performed by cellular RNA polymerase II (Pol-II), with S-HDAg being essential in this process, as it coactivates Pol-II by possibly acting as a histone mimic (5). The RNA genome of HDV folds into an unbranched rod-like structure due to its high degree of self-complementarity (6). Budding and interaction with HBV envelope proteins require expression of L-HDAg, which is identical to S-HDAg except for a 19-amino acid extension at its C terminus, directed by cellular adenosine deaminase activity that edits the amber codon on the antigenomic RNA (7). Posttranslational farnesylation of the C terminus of L-HDAg is essential for successful interaction of the HDV ribonucleoprotein particle with HBV envelope proteins (8). HDV has one serotype and is divided into eight distinct genotypes which show only a moderately structured geographic distribution (9, 10). The distribution of HBV and HDV genotypes is also only slightly correlated (11). HDV can be experimentally transmitted to woodchucks that carry replicating woodchuck hepatitis virus, but has not been observed in feral animals (12, 13). HDV remains the only human RNA virus for which there is no known replicating counterpart in other animals.
Recently, deltavirus-like sequences have been found in transcriptome data derived from pooled oropharyngeal and cloacal samples of teals and ducks (14), tissues of boas (Boa constrictor) and a water python (Liasis mackloti) (15), and various vertebrate and invertebrate transcriptomes (16). The snake- and duck-associated sequences, although highly distinct from HDV, show predicted similarity in secondary structure to the HDV ribozyme element. Amino acid motifs known to have functional importance for HDV replication occur in both sequences. However, the sequences described by Chang et al. (16) lack the two ribozyme structural elements and some of the functional motifs, as well as experimental proof of the viral replication mechanism. Also, the natural history of disease, if transmissible, is not known for these putative viruses.
Here we describe a unique mammalian deltavirus from the neotropical rodent species Proechimys semispinosus, tentatively named rodent deltavirus (RDeV). We initially discovered the viral RNA following undirected next-generation sequencing of blood samples. Our study of 763 animals provides epidemiological and virological evidence for a transmissible and prevalent infection with a delta-like agent in a specific host. Surprisingly, and in striking contrast to human HDV, we neither find a liver-specific tropism of RDeV nor epidemiological or functional evidence of coinfection with a hepadnavirus in the animals studied.
Results
In an ongoing study in the Panama Canal area, focused on the effects of habitat disturbance on the infection rate with rodent Hepacivirus (SI Appendix, Fig. S1) (17), we subjected blood samples of 120 individuals of P. semispinosus to total RNA sequencing (RNA-seq) using the Illumina MiSeq platform. High bit-score matches to human HDV were obtained after comparing de novo assembled sequences with a viral reference library (18). Based on mapping against a reference alignment of all known HDV genotypes, we compiled HDV-related sequences covering the whole deltavirus genome from two individual animals. After aligning these initial genome assemblies with HDV reference genotypes, we designed an RT-PCR assay targeting a fragment in the coding region of the rodent deltavirus antigen (RDeAg). The assay was applied to pooled blood samples from 763 P. semispinosus individuals. Resolution of positive-testing pools resulted in 30 positive individuals (overall detection rate 3.9%; 95% CI 2.6 to 5.3%). Only adult animals were found positive. In addition to P. semispinosus, we examined blood samples from individuals (n = 183) from 11 other rodent and marsupial species. No species other than P. semispinosus yielded evidence of RDeV presence (SI Appendix, Table S1).
Because HDV has a circular genome, we used primer pairs whose 5′ ends are adjacent on reverse-complementary template strands, with 3′ ends facing in opposite directions (Fig. 1B and SI Appendix, Table S2). These primers can generate RT-PCR amplicons covering the near-full deltavirus genome if the template RNA molecule is circular. This was successful in three samples with high viral load (Fig. 1C). Genome circularity was further confirmed by mapping RNA-seq reads from the respective samples to the initial amplicon sequences, resulting in assemblies with protruding ends that are identical, matching both ends of the amplicon, and therefore derived from circular templates. Ten full circular RDeV genomes and 10 partial genome assemblies were subsequently obtained from the 30 initially RNA-positive samples by mapping reads from individual RNA-seq datasets. PCR products for the other 10 samples produced small fragments of less than 200 bases. The nucleotide sequences of all 10 full genomes are 97.5 to 99.6% identical. The sequences are available in GenBank under accession numbers MK598003 to MK598012.
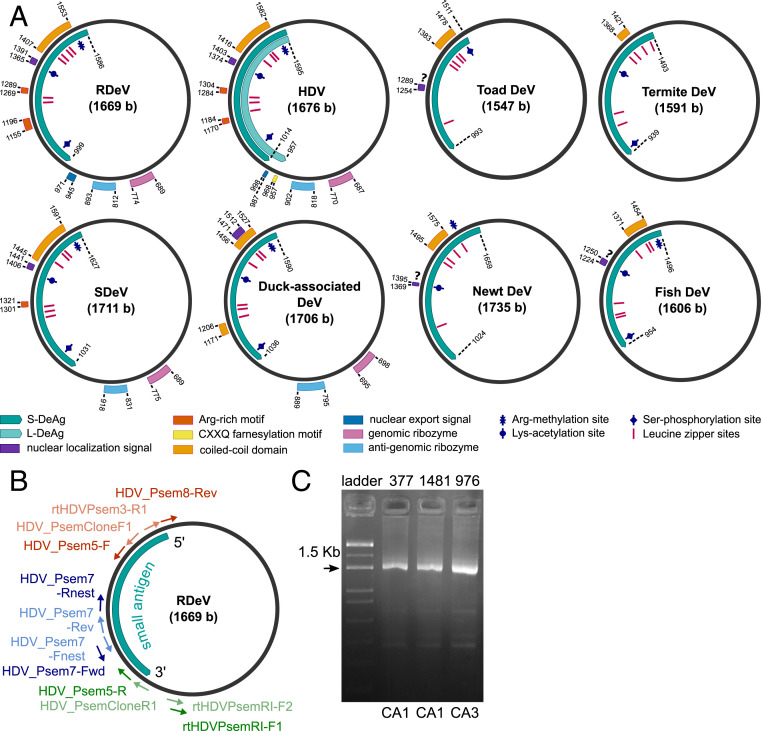
Fig. 1.
(A) Genome organization of human HDV, RDeV, SDeV, duck-associated DeV, newt DeV, toad DeV, fish DeV, and termite DeV. Delta antigen-coding regions, functional and structural motifs, as well as post-translational modification sites. (B) Primer-binding positions for the circularization assays shown in their locations on the RDeV genome. Primer names and sequences are listed in SI Appendix, Table S2. Blue, circularization assay 1 (CA1); red, circularization assay 2 (CA2); green, circularization assay 3 (CA3). External primers are shown in dark color, and nested primers in light color. The S-RDeAg ORF is shown for orientation. (C) Exemplary results from CA1 and CA3 obtained from total RNA blood extracts of three RDeV RNA-positive P. semispinosus. Fragment sizes are 1,608 bp for CA1 and 1,532 bp for CA3; label correspondence to RDeV GenBank accession numbers: 377, MK598006; 1481, MK598003; 976, MK598009.
Major structural and functional HDV domains are present in all recovered RDeV sequences (Fig. 1A and SI Appendix, Fig. S2 and Table S10), including a single open reading frame (ORF) representing the delta antigen. A 19-amino acid tail that differentiates the S-HDAg from the L-HDAg in human HDV is present in RDeV, though with a different amino acid composition. This tail is the result of antigenomic editing at a site corresponding to the amber stop codon in the S-HDAg ORF of human HDV (SI Appendix, Fig. S3). There is no experimental evidence of RNA editing or large-antigen expression in RDeV, as discussed below. In addition, the farnesylation motif CXXQ in the HDV L-HDAg C terminus, required to acquire the HBV-derived envelope (8), is absent in all P. semispinosus deltavirus sequences.
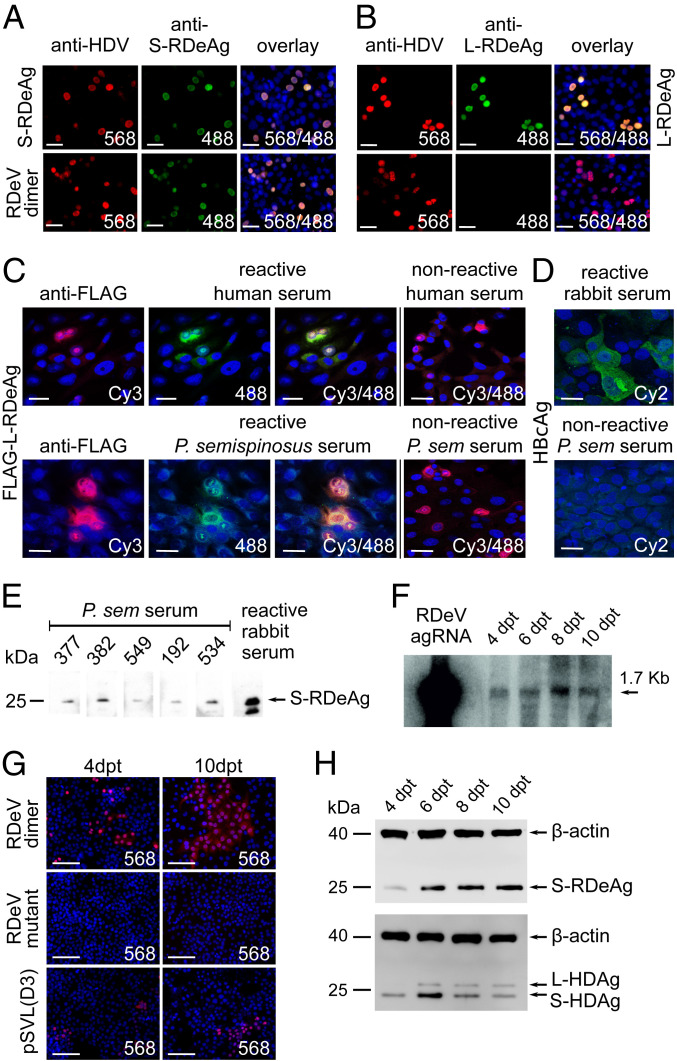
Predicted secondary structures of genomic and antigenomic ribozymes are highly similar between human and rodent deltavirus (SI Appendix, Fig. S4). The ribozyme active site is identical between rodent and human virus on both antigenomic and genomic strands. The nuclear export sequence motif and the nuclear localization signal of S-HDAg are also present in the S-RDeAg. Evidence for the existence and functionality of the nuclear localization signal is provided by the occurrence of the small delta antigen in the cell nuclei of HuH7 cells overexpressing S-RDeAg (Fig. 2A).
Fig. 2.
(A and B) Immunofluorescence staining of HuH7 cells transfected with plasmids expressing S-RDeAg (A), a stop codon mutant expressing a hypothetical L-RDeAg (B), as well as an intact dimer of the RDeV genome (both panels). Cell nuclei were stained with DAPI; delta antigens were detected with anti-HDAg immunoglobulin G (IgG) from an HDV/HBV-coinfected patient followed by an Alexa 568-labeled goat anti-human antibody (red); S- or L-RDeAg was identified using S- or L-RDeAg–specific peptide antisera from immunized rabbits, followed by an Alexa 488-labeled goat anti-rabbit antibody (green). Channels are overlaid for both panels. (C) Immunofluorescence staining of L-RDeAg expression in Vero B4 cells with HDAg-reactive human and P. semispinosus serum samples. Cell nuclei were stained with DAPI (blue); FLAG-tagged L-RDeAg was detected using a mouse anti-FLAG antibody followed by a goat anti-mouse Cy3-labeled antibody (red); reactivity of human and P. semispinosus serum against L-RDeAg is visualized by an Alexa 488-labeled goat anti-human and goat anti-guinea pig antibody, respectively (green). Nonreactive human and P. semispinosus serum samples are shown for comparison. (D) Immunofluorescence staining of HBcAg expression in HuH7 cells with anti-HBc–specific rabbit antiserum (Upper) and P. semispinosus serum (Lower) samples. Cell nuclei are stained with DAPI (blue); expression of HBcAg is visualized using a Cy2-labeled secondary antibody (green). Channels are merged for both panels. (Scale bars, 30 μm.) (E) Specific reactivity of P. semispinosus sera against S-RDeAg in a Western blot. Total protein extracts from HuH7 cells overexpressing S-RDeAg were separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis and blotted. The membranes were incubated with five P. semispinosus serum samples (IDs shown in figure) followed by a goat anti-guinea pig horseradish peroxidase (HRP) antibody. Anti-S-RDeAg–specific peptide antiserum from an immunized rabbit was used as positive control, with goat anti-rabbit HRP as secondary antibody. (F) Northern blot of antigenomic RDeV RNA from transfected HuH7 cells. The cells were transfected with an expression plasmid containing an intact dimer of the RDeV genome. Total RNA was isolated 4, 6, 8, and 10 d posttransfection (dpt) and subjected to a 1% denaturing formaldehyde agarose gel. After blotting, the RNA was hybridized with a single-stranded oligonucleotide probe labeled with [γ-32P]ATP and T4 polynucleotide kinase at the 5′ end. In vitro-transcribed antigenomic RDeV RNA was used as positive control. (G) Immunofluorescence staining of HuH7 cells transfected with expression plasmids containing an intact dimer of the RDeV genome, a mutant dimer of the RDeV genome with abrogated S-RDeAg expression, and a trimeric genome of human HDV genotype 1 [pSVL(D3)]. Cells (Left) were fixed on day 4 posttransfection. A 1:64 dilution of the cells (Left) were subjected to clonal expansion and fixed on day 10 posttransfection (Right). Cell nuclei were stained with DAPI (blue) and delta antigens were detected with anti-HDV IgGs followed by an Alexa 568-labeled goat anti-human antibody (red). (Scale bars, 100 μm.) (H) Detection of deltavirus antigens from HDV- and RDeV-transfected HuH7 cells by Western blot. HuH7 cells were transfected with a dimeric RDeV genome and a trimeric genome of human HDV genotype 1 [pSVL(D3)], and total protein was isolated on 4, 6, 8, and 10 d posttransfection. S-RDeAg (Upper) and S- and L-HDAg (Lower) were detected using cross-reactive human anti-HDV serum; beta-actin was detected by an anti-beta-actin antibody.
Deltavirus Phylogeny.
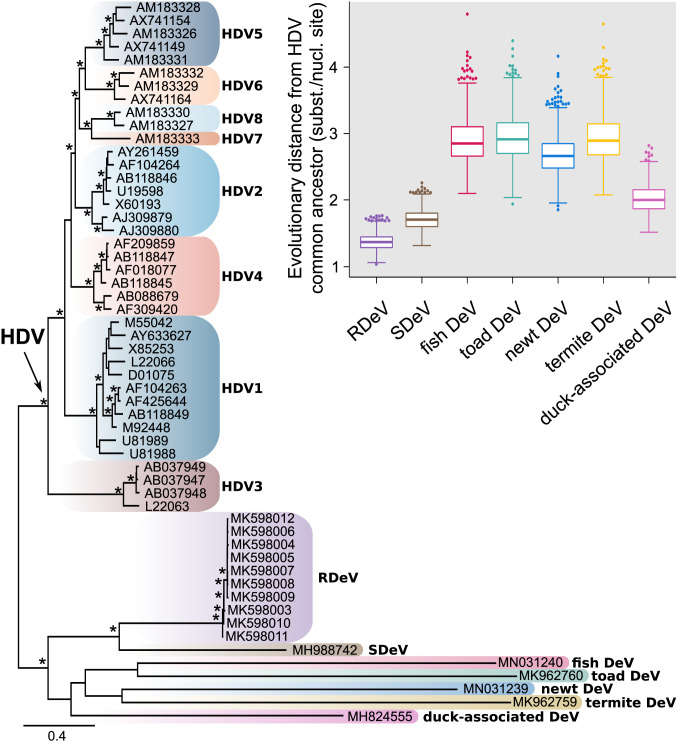
Nucleotide and amino acid sequences were compared as summarized in SI Appendix, Fig. S2 and Table S3. Phylogenies were calculated based on full-genome nucleotide sequence alignments, as well as translated small delta antigen-coding sequences. Both tree topologies are equivalent and the distinction of RDeV from human HDV is clear (the full-genome phylogeny is shown in Fig. 3). The nonhuman deltaviruses are highly diverged and therefore chosen to root the phylogeny. The snake deltavirus (SDeV) clusters with the rodent deltavirus, and both form a highly diversified and well-supported sister group to human HDV, with HDV genotype 3 being the most divergent among HDV genotypes. All other nonhuman deltaviruses group together consistently, although without significant bootstrap support among subclades, owing to their high sequence divergence. RDeV is most closely related to human HDV, as averaged over all 1,000 bootstrap replicate trees (Fig. 3).
Fig. 3.
Maximum-likelihood phylogeny based on a full-genome alignment of HDV representatives from the eight different genotype groups and RDeV, SDeV, duck-associated DeV, newt DeV, toad DeV, fish DeV, and termite DeV. The phylogenetic inference was done using RAxML-NG version 0.7.0 BETA (19). Asterisks indicate bootstrap support >80 and tree tips show GenBank accession numbers. The tree is rooted to the branch leading to the nonhuman DeVs. Distribution of evolutionary distances (branch lengths, extracted from 1,000 bootstraps) for all nonhuman DeVs relative to the common ancestor of HDV (arrow on tree) is shown in the boxplots. The boxplots show median, interquartile range, Tukey’s minimum and maximum (whiskers), and extreme outlier values (dotted).
Organ Distribution of RDeV RNA and Protein Detection.
To obtain insight into the infection pattern caused by the rodent deltavirus, we tested for viral RNA in organs of 18 animals that were found dead during sampling (note that we followed a noninvasive sampling approach and animals could not be purposefully euthanized during our study). One of these animals tested RDeV RNA-positive in all available organs (liver, kidney, lung, heart, and small intestine), but its organ-specific RNA concentrations did not suggest specific virus replication in the liver (SI Appendix, Table S4). Also, a commercial HDV antigen enzyme-linked immunosorbent assay that we found to cross-detect with RDeV did not identify organ-specific protein expression in the liver, lung, small intestine, heart, or kidney of this animal (SI Appendix, Fig. S5A). To further understand potential virus excretion, fecal samples from 822 individuals were tested by RT-PCR, 10 of which were positive for RDeV RNA (the numbers of individuals tested for each type of material are summarized in SI Appendix, Table S5). This 1.2% detection rate was significantly lower than that in blood samples (χ2 test, P < 10−4). The average virus concentration in fecal samples was 2.9 × 109 RNA copies per gram, with a maximum concentration of 2.2 × 1010 copies per gram. Average and maximum concentrations in blood were 1.8 × 108 and 2.1 × 109 copies per milliliter.
RDeV Genome Replication.
Rodent deltavirus genome replication was confirmed by Northern blot analysis, as well as by clonal expansion of RDeV-transfected cells. The full RDeV genome was cloned as a tandem head-to-tail fusion dimeric construct in genomic (minus-strand) orientation downstream of a cytomegalovirus (CMV) promoter (see the construct map in SI Appendix, Fig. S6) and transfected into HuH7 cells. Antigenomic RDeV RNA as a marker of ongoing RDeV replication was detected in transfected cells at all time points (Fig. 2F). The expression of antigenomic RDeV RNA was accompanied by a robust synthesis of genomic RDeV RNA (SI Appendix, Fig. S5D). Massive clonal expansion of RDeV-transfected cells verifies the autonomous replicating nature of RDeV, even exceeding HDV clonal expansion as determined by immunofluorescence staining (Fig. 2G).
Deltavirus Antigens and Editing of the Amber Codon.
One key feature of HDV is the expression of two viral proteins (S-HDAg and L-HDAg) from a single ORF. A 19-amino acid carboxyl-terminal extension differentiates the S-HDAg (195 codons) from the L-HDAg (214 codons) in human HDV. This extension is the result of antigenomic editing by cellular adenosine deaminase acting on RNA 1 (ADAR1) at a site corresponding to the amber stop codon in the S-HDAg ORF of the circular antigenomic HDV RNA, edited from a UAG to a UIG codon. Following HDV RNA replication, the UIG codon is converted into a tryptophan codon (UGG) (SI Appendix, Fig. S3) during subsequent HDV messenger RNA (mRNA) synthesis, allowing expression of the L-HDAg (20). Interestingly, a 19-amino acid carboxyl-terminal extension is present in RDeV, though different in 15 of the 19 amino acids. To investigate potential RDeV large antigen (L-RDeAg) expression, the full RDeV genome was cloned as a tandem head-to-tail fusion construct in genomic (minus-strand) orientation downstream of a CMV promoter (see the construct map in SI Appendix, Fig. S6) and transfected into HuH7 cells. As positive controls, S- or (hypothetical) L-RDeAg was expressed under the control of a CMV promoter. Specific S- or L-RDeAg expression was examined with antibodies from rabbits immunized with synthetic peptides against S-RDeAg (Fig. 2A) or the putative 19-amino acid extension of L-RDeAg (Fig. 2B). The tandem genome construct expressed S-RDeAg but not L-RDeAg (Fig. 2 A and B).
Corresponding results were obtained by Western blot analysis of RDeV dimer transfection experiments using a broadly cross-reactive human anti-HDV serum (Fig. 2 H, Upper). Evidence for the absence of RNA editing on the RDeAg ORF was also obtained from RNA-seq data. Mapping of reads from total RNA blood and organ extracts of infected animals and from transfected RDeV dimeric genome constructs in cell culture yielded only unedited reads, whereas both edited and unedited reads were detected for human HDV in a control experiment (SI Appendix, Fig. S3). In summary, there is no experimental evidence of RNA editing on the RDeV antigen ORF or of large-antigen expression during RDeV replication in vivo or in vitro.
Search for Signs of Hepadnaviral Coinfection.
Because human HDV transmission requires an envelope protein provided by an active coinfection with HBV, we subjected all 763 P. semispinosus blood samples to PCR screening for hepadnaviruses. The assay was designed to detect all mammalian orthohepadnaviruses, including those from rodents, bats, and primates (21). None of the samples tested positive. Specific reanalysis of all available transcriptome datasets from the present study did not yield any evidence for orthohepadnaviral genomes in a basic local alignment search tool (BLAST) analysis. All blood samples with sufficient volume, whether RNA-positive or -negative for RDeV (n = 68), were tested for antibodies against woodchuck HBV core antigen (WHcAg). This antigen was chosen because the woodchuck is most closely related to Proechimys among known HBV hosts. In previous studies, we demonstrated that anti-HBc antibodies broadly cross-react among HBVs from different hosts, even across mammalian orders (21), so it is likely that WHcAg would be bound by antibodies against a possible HBV in Proechimys. However, no such antibodies were found in any of the tested samples (Fig. 2D).
Investigation of Hepacivirus as Potential Cofactor.
Experimental evidence shows that apart from HBV, other viruses, including hepatitis C virus (genus Hepacivirus), can provide envelope proteins for human HDV (22). As we previously detected a high prevalence of hepacivirus in the rodents studied (17), this possibility was investigated. Overall, we found 4 out of 30 RDeV RNA-positive rodents in which hepacivirus was not detected by the tests initially applied (SI Appendix, Table S6). To exclude having missed hepacivirus detection, we applied RT-PCR assays specifically designed for the E1, NS3, and NS5B genes of P. semispinosus hepacivirus to those samples. All four animals were confirmed to be hepacivirus RNA-negative by these assays as well as by RNA-seq read mapping against P. semispinosus hepacivirus. Statistical analysis of the degree of dependency between deltavirus and hepacivirus infection in individuals was conducted, providing no support for a dependency between the two infections (SI Appendix, Table S6; χ2 test, P = 0.24). Also, analyses of sampling site-specific deltavirus and hepacivirus detection rates (SI Appendix, Table S7) did not reveal any correlation between deltavirus and hepacivirus (Pearson’s correlation coefficient, ρ = 0.15), arguing against a linear correlation. Logarithmic and linear regression fits did not yield significant associations either, suggesting that the two viruses follow different patterns of distribution. We conclude that RDeV infection in P. semispinosus does not depend on active hepacivirus infection.
Immune Reaction in RDeV-Infected Animals.
To determine whether RDeV causes an immune reaction in P. semispinosus, serum samples were subjected to an indirect immunofluorescence assay. L-RDeAg was cloned and transiently expressed in Vero B4 cells, as described (23). Fig. 2C shows the reactivity of antibodies from human sera against L-RDeAg, along with examples of reactive and nonreactive P. semispinosus sera. At a dilution of 1:100, 17 of 30 RDeV RNA-positive and 3 of 115 RDeV RNA-negative animals tested positive for anti-RDeAg antibodies (P < 10−6, χ2 test). To exclude RNA degradation in the case of antibody-positive samples for which RDeV RNA was not detected, we applied a real-time RT-PCR assay to detect RNA transcripts of a host housekeeping gene (TATA-binding protein) (SI Appendix, Table S8). The 20 positive sera were additionally tested at 1:1,000 dilution, testing positive in 5 cases. Western blots of these sera against expressed S-RDeAg confirmed the specificity of serum antibodies (Fig. 2E). The average RDeV RNA concentration in all anti-RDeAg-positive sera was 262,400 RNA copies per microliter of blood. The average RDeV RNA concentration in only those samples that tested antibody-positive up to a dilution of 1:1,000 was 8,500 RNA copies per microliter of blood (SI Appendix, Fig. S5C). These concentrations were significantly different (P < 0.05, Mann–Whitney U test), which suggests that an adaptive immune response can potentially limit or eliminate viral replication. Elimination is also suggested by the occurrence of three anti-RDeAg antibody-positive but RDeV RNA-negative animals.
Ecological Factors That Influence Rodent Deltavirus Infection.
The study area in the Barro Colorado Nature Monument consists of islands and peninsulas with different densities of populations of P. semispinosus, a generalist species that can adapt to environmental changes (24) and thus (relatively) competitively benefit from anthropogenic habitat disturbance. We have previously noted a positive correlation between site-specific P. semispinosus population density and the detection rate of hepacivirus infection (SI Appendix, Table S7) (17). For rodent deltavirus, a different site-specific distribution was observed (SI Appendix, Table S7). Highest detection rates were identified in continuous forest sites that have largely preserved the primordial habitat composition that existed before the flooding of the Gatún Lake area during the construction of the Panama Canal. Lower detection rates were found on forested islands, and minimal rates in forest fragments surrounded by agriculture. To identify environmental factors that might correlate with rodent deltavirus transmission, host- and habitat-specific properties were examined under two different logistic regression models (SI Appendix, Table S9). RDeV infection was found to be positively correlated with male sex and reproductive activity, as well as with inhabiting continuous forest sites with undisturbed habitat. In addition to the variables shown in SI Appendix, Table S9, other habitat-specific factors such as population density, canopy height and coverage, and understory density did not add explanatory power to the analysis. The inclusion of site-specific data on mosquito diversity narrowed the sample size to n = 686 individuals for sampling sites where these data were available. There was no detectable correlation between mosquito diversity and RDeV detection rate.
Discussion
The present results enhance our understanding of the evolution of hepatitis delta virus, a satellite virus to HBV and until recently the only human RNA virus with no known counterpart in other animals. The first question that must follow the discovery of novel deltavirus-like sequences in rodent, reptile (15), and putatively avian (14) and other vertebrate and invertebrate hosts (16) is whether these represent transmissible viruses as opposed to, for example, endogenous viral elements. Hetzel et al. (15) demonstrated protein expression in organs of those snakes in which viral sequences were detected, and Szirovicza et al. (25) showed that snake deltavirus can replicate in several cell lines. Our reverse genetic and clonal expansion experiments now prove the existence of mammalian nonhuman deltavirus genomes as self-replicating viral entities, in which genomic replication involves an internal initiation of replication. Moreover, correlations of serology and virus detection in a natural animal population of considerable size demonstrate that rodent deltavirus infection is acquired during the course of life and is likely to be cleared by an active immune response. Previous observations on the snake deltavirus corroborate this infection pattern, showing that although snake deltavirus infection was found in both maternal and offspring boas (15), not all offspring were infected, and snakes with no kinship to the mating pairs exhibited both the presence and absence of the virus. Horizontal transmission seems likely in both rodent and reptile deltaviruses. The significantly lower detection rate of viral RNA in stool samples compared with blood suggest blood-borne rather than fecal-oral transmission. The observed predominant infection of adult males is compatible with transmission through competitive behavior.
The novel deltaviruses associated with diverse hosts warrant considerations regarding evolutionary origins. Phylogeny suggests that fish- and amphibian-associated viruses are most distant from other known deltaviruses, while snake and rodent viruses are significantly closer to the HDV common ancestor. Considering the relationship of these hosts, the tree topology seems to disfavor virus-host codivergence. Besides, the duck-associated virus was not detected in an internal organ but in cloacal and oropharyngeal swabs, leaving open the possibility of contamination from alimentary sources (14). The phylogenetic branch pattern supports previous work on human HDV phylogeny, suggesting that HDV genotype 3 stems from the oldest common ancestor of human HDV genotypes (26). As this ancestor is more closely related to the rodent virus than to any of the nonmammalian deltaviruses, this would be compatible with the hypothesis that human and rodent deltaviruses have codiverged with mammalian lineages. Deltaviruses in other mammals may remain to be discovered, but equally may now be extinct. A hypothesis of codivergence would be corroborated by the discovery of at least one more mammalian virus whose phylogenetic position reflects that of its host in relation to humans and rodents. The clustering of rodent and snake viruses might then be interpreted as a sign of cross-species acquisition from mammals. It is noteworthy that the parental boas carrying the snake deltavirus stemmed from Panama, like the rodents studied here (15). Human HDV genotype 3 is also found in South and Middle America, but this alone should not be taken as support to claim cross-host transmission.
The dependence of HDV on HBV coinfection for transmission may have evolved only in a mammalian viral lineage leading to human HDV, given that hepadnaviruses occur in diverse mammalian species including rodents, bats, and particularly New World primates (21, 27, 28). Based on the lack of large-antigen expression and a farnesylation motif, both necessary for interaction with hepadnaviral envelope proteins, as well as on evidence from epidemiological and serological data, we can exclude the presence of HBV coinfection in deltavirus-infected P. semispinosus. Neither HBV DNA nor anti-HBc antibodies were detected in any animals infected with the deltavirus. In all other deltavirus studies, no sequences of coinfecting hepadnaviruses were found in transcriptome data. Other coinfecting viruses were identified in each of these studies, designating their envelope proteins as a potential source for an RDeV envelope (14–16). As very recently shown by Szirovicza et al. (25), snake deltavirus can produce infectious viral particles in vitro by utilization of envelope proteins from arenaviruses and orthohantaviruses.
In the present study, we investigated the potential influence of hepacivirus infection. Despite a high hepacivirus prevalence in the studied rodents [ca. 80% (17)], we found four deltavirus RNA-positive individuals lacking a hepacivirus coinfection. Human HDV has been found to produce infectious particles in vitro with the help of an envelope protein from hepatitis C virus, with the required presence of an L-HDAg farnesylation motif (22). Given the absence of this motif in the rodent delta antigen and the lack of a hepacivirus coinfection in some individuals carrying deltavirus RNA, we do not consider deltavirus infection to depend on hepacivirus infection in these rodents. In addition, we screened individual RNA-seq datasets from 120 animals and did not identify any other infectious viral agent in any deltavirus RNA-positive P. semispinosus. There are multiple alternatives that may have resulted in rodent deltavirus transmission. One possibility is the presence of a currently unknown virus, bacterium, or integrated sequence that provides an envelope protein to rodent deltavirus. Furthermore, recent advances in cellular cross-communication have revealed the exchange of extracellular vesicles containing mRNA, circRNA, and other noncoding RNA molecules (29). It should be investigated whether rodent deltavirus might employ such pathways for transmission.
In their study of snake organ samples, Hetzel et al. (15) found two bands of ca. 20 and 27 kDa in Western blots using an anti-S-SDeAg serum. While the larger band might correspond to a larger protein variant, expression of an actual L-SDeAg was not confirmed by specific anti-L-SDeAg immunoblots, analyses of RNA editing, or experimental studies of gene expression. The present data, based on all these approaches, show that the related rodent virus does not express a large delta antigen, suggesting that the use of L-HDAg may have evolved only in connection with the utilization of the HBV envelope. In this scenario, the encounter with hepadnaviruses may have selected for viral variants with a farnesylation signal in an L-HDAg tail that is expressed by an initially low-level editing activity or stop codon readthrough, allowing these viruses to exploit a novel transmission opportunity by acquisition of an HBV envelope. The sequence information from the present study can now be used in directed searches for deltaviruses in other mammalian species, even in the absence of HBV infection.
Alternative hypotheses of HDV origin have suggested either a viroid origin (perhaps via an insect vector) or derivation from a host DIPA gene (reviewed in ref. 11). A viroid origin is supported by the structural similarity of the HDV ribozyme to plant viroid ribozymes, the genome circularity of both HDV and viroids, and the interaction with common cellular proteins (30). Derivation from the human genome has been suggested based on the incomplete set of genes essential for replication, the occurrence of HDV-like ribozyme elements in the human genome (31), and the interaction of HDV with a human-derived protein (DIPA) (32). The deltavirus findings do not increase or decrease support for the viroid origin hypothesis but force us to consider the DIPA hypothesis from a wider perspective. The rodent deltavirus ORF shows 30% nucleotide and 22% amino acid identity when compared with the DIPA ORF in mice and rats, at which levels similarity due to chance must be considered as a likely explanation (33). In addition to these caveats, the present results suggest that other mammalian deltaviruses existed before the origin of human HDV, and that precursors to these mammalian viruses existed in other vertebrates (14–16). Alternate scenarios, specifically the derivation of nonhuman deltaviruses from human HDV precursors, seem unlikely, considering the genetic distances between these viruses, the relatively short time span available for the evolution of such diversity in the case of a human origin, and the requirement that the human virus infect several nonhuman hosts. It appears instead that deltavirus originated as an exogenous infectious agent that established acute infection in mammals. The present study provides a basis for the evolution of HDV, confirming the general understanding that RNA virus diversity evolved along the major evolutionary lineages of their hosts (34).
Methods
Animals were captured using Sherman traps in the Barro Colorado Nature Monument, Panama (17). RNA-seq experiments were done on an Illumina MiSeq instrument. RT-PCR followed standard protocols, including for circular genome amplification. Immunofluorescence assay (IFA) serology was based on cells overexpressing the L-RDeAg or woodchuck WHc antigen under the control of a CMV promoter. Viral full RDeV genome complementary DNA was cloned as a monomer or head-to-tail dimer under the control of a CMV promoter. A full description of methods is provided in SI Appendix, Materials and Methods.
All fieldwork was carried out with full ethical approval (Smithsonian Institutional Animal Care and Use Committee protocols 2013-0401-2016-A1-A7 and 2016-0627-2019-A2) and samples were exported to Germany with permission from the Panamanian government (SEX/A-21-14, SE/A-69-14, SEX/A-22-15, SEX/A-24-17, SEX/A-120-16, and SEX/A-52-17).
Supplementary Material
Acknowledgments
We thank Stefan Brändel for arranging research permits and Tobias Bleicker for technical assistance. This work was supported by grants from the German Research Foundation (DFG), B08/SFB 1021/2 (to D.G.), SPP 1596 Grants SO 428/9-1 and 2 (to S. Sommer), and DR772/8-22 (to C.D.). The National Reference Centre for Hepatitis B Viruses and Hepatitis D Viruses is supported by the German Ministry of Health via the Robert Koch Institute. Additional support for sequencing work was provided by the European Commission’s COMPARE project.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Sequences reported in this paper have been deposited in GenBank (accession numbers MK598003–MK598012).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006750117/-/DCSupplemental.
Data Availability.
The rodent deltavirus complete genome sequences described here are available in GenBank under accession numbers MK598003 to MK598012.
References
- 1.Rizzetto M. et al., Chronic hepatitis in carriers of hepatitis B surface antigen, with intrahepatic expression of the delta antigen. An active and progressive disease unresponsive to immunosuppressive treatment. Ann. Intern. Med. 98, 437–441 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Rizzetto M. et al., Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut 18, 997–1003 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H.-Y. et al., Prevalence and burden of hepatitis D virus infection in the global population: A systematic review and meta-analysis. Gut 68, 512–521 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Branch A. D., Robertson H. D., A replication cycle for viroids and other small infectious RNA’s. Science 223, 450–455 (1984). [DOI] [PubMed] [Google Scholar]
- 5.Abeywickrama-Samarakoon N. et al., Hepatitis delta virus histone mimicry drives the recruitment of chromatin remodelers for viral RNA replication. Nat. Commun. 11, 419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K.-S. et al., Structure, sequence and expression of the hepatitis delta (δ) viral genome. Nature 323, 508–514 (1986). [DOI] [PubMed] [Google Scholar]
- 7.Wong S. K., Lazinski D. W., Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. U.S.A. 99, 15118–15123 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto J. C., Casey P. J., The hepatitis delta virus large antigen is farnesylated both in vitro and in animal cells. J. Biol. Chem. 271, 4569–4572 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Jackson K. et al., Epidemiology and phylogenetic analysis of hepatitis D virus infection in Australia. Intern. Med. J. 48, 1308–1317 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Le Gal F. et al., Eighth Major Clade for Hepatitis Delta Virus. Emerging Infectious Diseases 12, 1447–1450 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor J., Pelchat M., Origin of hepatitis δ virus. Future Microbiol. 5, 393–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Negro F. et al., Hepatitis delta virus (HDV) and woodchuck hepatitis virus (WHV) nucleic acids in tissues of HDV-infected chronic WHV carrier woodchucks. J. Virol. 63, 1612–1618 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freitas N. et al., Hepatitis delta virus infects the cells of hepadnavirus-induced hepatocellular carcinoma in woodchucks. Hepatology 56, 76–85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wille M. et al., A divergent hepatitis D-like agent in birds. Viruses 10, 720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hetzel U. et al., Identification of a novel deltavirus in boa constrictors. MBio 10, e00014-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang W.-S. et al., Novel hepatitis D-like agents in vertebrates and invertebrates. Virus Evol. 5, vez021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmid J. et al., Ecological drivers of Hepacivirus infection in a neotropical rodent inhabiting landscapes with various degrees of human environmental change. Oecologia 188, 289–302 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Goodacre N., Aljanahi A., Nandakumar S., Mikailov M., Khan A. S., A reference viral database (RVDB) to enhance bioinformatics analysis of high-throughput sequencing for novel virus detection. MSphere 3, e00069-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozlov A. M., Darriba D., Flouri T., Morel B., Stamatakis A., RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo G. X. et al., A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64, 1021–1027 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drexler J. F. et al., Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 110, 16151–16156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Vargas J. et al., Enveloped viruses distinct from HBV induce dissemination of hepatitis D virus in vivo. Nat. Commun. 10, 2098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller M. A. et al., Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: A nationwide, cross-sectional, serological study. Lancet Infect. Dis. 15, 629 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Adler G. H., The island syndrome in isolated populations of a tropical forest rodent. Oecologia 108, 694–700 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Szirovicza L. et al., Snake deltavirus utilizes envelope proteins of different viruses to generate infectious particles. MBio 11, e03250-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Gal F. et al., Eighth major clade for hepatitis delta virus. Emerg. Infect. Dis. 12, 1447–1450 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Carvalho Dominguez Souza B. F. et al., A novel hepatitis B virus species discovered in capuchin monkeys sheds new light on the evolution of primate hepadnaviruses. J. Hepatol. 68, 1114–1122 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Rasche A., Sander A.-L., Corman V. M., Drexler J. F., Evolutionary biology of human hepatitis viruses. J. Hepatol. 70, 501–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K. M., Abdelmohsen K., Mustapic M., Kapogiannis D., Gorospe M., RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 8, e1413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores R., Ruiz-Ruiz S., Serra P., Viroids and hepatitis delta virus. Semin. Liver Dis. 32, 201–210 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Webb C.-H. T., Lupták A., HDV-like self-cleaving ribozymes. RNA Biol. 8, 719–727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brazas R., Ganem D., A cellular homolog of hepatitis delta antigen: Implications for viral replication and evolution. Science 274, 90–94 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Long M., de Souza S. J., Gilbert W., Delta-interacting protein A and the origin of hepatitis delta antigen. Science 276, 824–825 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y.-Z., Wu W.-C., Shi M., Holmes E. C., The diversity, evolution and origins of vertebrate RNA viruses. Curr. Opin. Virol. 31, 9–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The rodent deltavirus complete genome sequences described here are available in GenBank under accession numbers MK598003 to MK598012.