Significance
The ability to culture human spermatogonial stem cells (SSCs) in vitro is critical to develop SSC therapeutic approaches to treat male infertility. To achieve this goal, it is important to define molecular pathways in human SSCs. Toward this end, we developed an approach to purify human primitive undifferentiated spermatogonia (uSPG) highly enriched in SSCs. Comparative RNA-sequencing analysis of these primitive uSPG versus differentiating SPG identified differentially expressed genes encoding key components in several signaling pathways. Leveraging this information, coupled with single-cell RNA-sequencing analysis, we studied the effects of modulating signaling pathways on human SPG fate in vitro. Inhibition of AKT signaling favored primitive uSPG fate, which can be applied to culture SSCs for therapeutic applications.
Keywords: human spermatogonia, in vitro culture, testis, RNA sequencing, PLPPR3
Abstract
Spermatogonial stem cells (SSCs) are essential for the generation of sperm and have potential therapeutic value for treating male infertility, which afflicts >100 million men world-wide. While much has been learned about rodent SSCs, human SSCs remain poorly understood. Here, we molecularly characterize human SSCs and define conditions favoring their culture. To achieve this, we first identified a cell-surface protein, PLPPR3, that allowed purification of human primitive undifferentiated spermatogonia (uSPG) highly enriched for SSCs. Comparative RNA-sequencing analysis of these enriched SSCs with differentiating SPG (KIT+ cells) revealed the full complement of genes that shift expression during this developmental transition, including genes encoding key components in the TGF-β, GDNF, AKT, and JAK-STAT signaling pathways. We examined the effect of manipulating these signaling pathways on cultured human SPG using both conventional approaches and single-cell RNA-sequencing analysis. This revealed that GDNF and BMP8B broadly support human SPG culture, while activin A selectively supports more advanced human SPG. One condition—AKT pathway inhibition—had the unique ability to selectively support the culture of primitive human uSPG. This raises the possibility that supplementation with an AKT inhibitor could be used to culture human SSCs in vitro for therapeutic applications.
Spermatogonial stem cells (SSCs) support spermatogenesis throughout adult life by balancing self-renewal and differentiation (1, 2). SSC transplantation provides a possible future therapeutic option to treat human infertility, which afflicts >100 million men world-wide. Such “SSC therapy” has potentially broad clinical value for treating various forms of human infertility, including as a means to restore fertility to prepubescent males (which, by definition, do not yet make sperm) that have been rendered infertile by cytotoxic chemotherapy (3).
While many markers have been reported to label human SSCs—including UTF1, UCHL1, ITGA6, SALL4, and GPR125—these markers have uncertain specificity for human SSCs vs. human progenitors; and many of them also clearly label differentiating (d) spermatogonia (SPG) (2, 4, 5). Furthermore, only some of the reported “human SSC markers” have been functionally tested by the only assay currently available to judge human SSC activity: Germ-cell xenograft transplantation analysis. In this assay, human testicular cell fractions are transplanted into the testes of immune-deficient mice that have been chemically or genetically depleted of germ cells, followed 2 mo later by determination of the number of spermatogonial colonies (6, 7). This assay has shown that cells positive for specific cell-surface proteins (ITGA6 or EpCAMlow), or combinations of cell-surface proteins (EpCAMlow/HLA-ABC−/CD49e− or HLA-ABC−/CD9+) have 3- to 12-fold more spermatogenic colonies (indicative of 3- to 12-fold higher SSC activity) relative to nonenriched cells (6, 8, 9). Using cell number (not colony number) as a readout, other studies have shown that the cell-surface protein SSEA4 enriches rhesus monkey and human testicular cells for SSC activity by ∼5- and ∼40-fold, respectively (10, 11).
A powerful approach to define cell types and cell type-specific gene markers is single-cell RNA-sequencing (scRNA-seq) analysis. Thus, several groups, including ours, have recently used scRNA-seq analysis to investigate human spermatogenesis (12–15), as reviewed in refs. 16 and 17. A key outcome of the scRNA-seq studies on adult human testes was the discovery of a cell cluster composed of highly undifferentiated (u) SPG (12, 14). This cell cluster—referred to as “state 0” or “SSC-1B” (12, 14)—was defined as the most uSPG subset, based on a plethora of gene markers and pseudotime trajectory analysis. We will refer to these highly undifferentiated cells as “primitive uSPG.” Whether there are SSCs in this primitive uSPG subset has not yet been functionally determined.
One of the genes shown—through scRNA-seq analysis—to be preferentially expressed in primitive human uSPG is PLPPR3 (14), a member of lipid phosphate phosphatase-related protein (LPPR) gene family. All five LPPR family members encode six-transmebrane proteins that are expressed in neurons and have neural functions, including neuronal plasticity and excitatory efficacy (18–20). Less is known about PLPPR3 than most other family members. PLPPR3 has been shown to be both necessary and sufficient for neurons to generate axon filopodia and branches (21); it has also been found to interact with PLPPR1 in a neural cell line to promote S6 ribosomal protein phosphorylation, an event known to elevate protein synthesis (22). We found that PLPPR3 is not only expressed in the nervous system, but also the testis (14). Immunofluorescence (IF) analysis showed that PLPPR3 is expressed in germ cells in the periphery of human seminiferous tubules where it labels a subset of SPG positive for the broad uSPG marker proteins, UTF1, raising the possibility that PLPPR3 is a human SSC marker (14). In this communication, we demonstrate that PLPPR3 is selectively expressed in primitive uSPG and that its expression on the surface allows for purification of highly enriched human SSCs. Comparative transcriptome analysis of PLPPR3+ cells with KIT+ cells (enriched for dSPG) identified thousands of significantly differentially expressed genes (DEGs), including genes critical for several signaling pathways. Using this information, we tested agents known to impact several of these signaling pathways, allowing us to identify conditions favorable for culture of human primitive uSPG. These findings have important potential implication for future studies on human SSCs, including their characterization, culture, and expansion for clinical application.
Results and Discussion
PLPPR3 Labels Human SSCs.
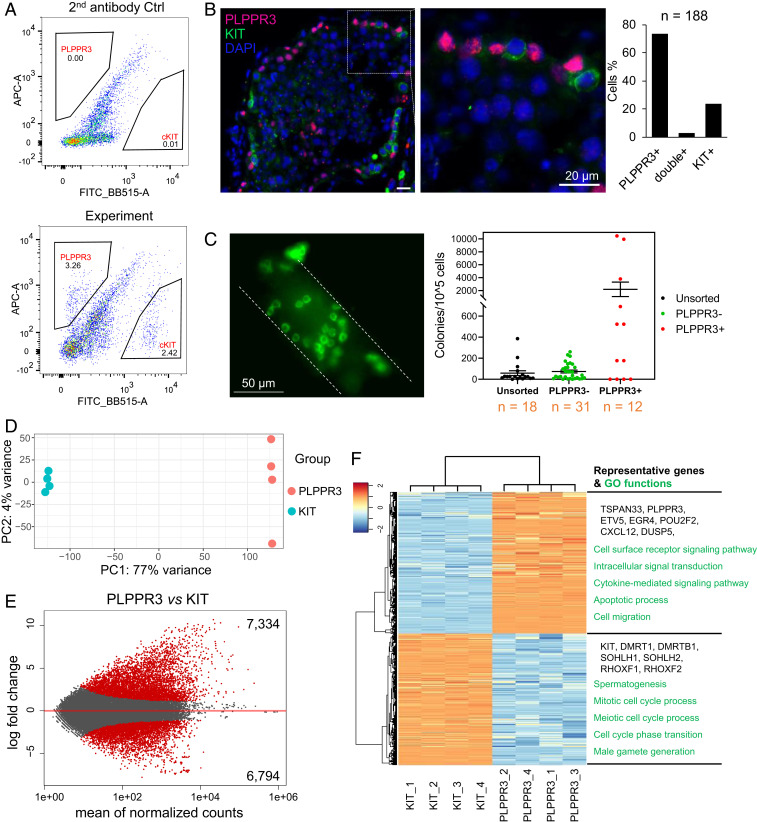
As described in the introductory paragraphs, we previously showed that the PLPPR3 gene is primarily expressed in the most primitive uSPG subset, as determined by scRNA-seq analysis (14). This raised the possibility that its encoded protein also displays this specificity. PLPPR3 is a transmembrane protein expressed on the cell surface (14, 22), allowing us to test its specificity by FACS analysis. We compared PLPPR3+ cells with KIT+ cells, as KIT is a well-established dSPG cell-surface marker (5). FACS analysis showed that PLPPR3+ cells and KIT+ cells are distinct cell populations, with very few double-positive cells (Fig. 1A). This was confirmed by IF analysis of adult human testicular cross-sections (Fig. 1B).
Fig. 1.
Transcriptome profiling of human primitive uSPG and dSPG. (A) FACS plot of human adult testicular cells stained with antisera against PLPPR3 and KIT, followed by labeled secondary antisera (Lower) or secondary antisera alone (Upper). (B, Left) IF analysis of a human adult testis section costained with antisera against PLPPR3 and KIT. Cell nuclei were stained with DAPI (blue). (Scale bar, 20 µm.) (Right) Quantification of the positive cells that costain or not as indicated. n, number of cells counted. (C) Germ-cell xenograft-transplantation analysis. (Left) Representative image of human germ cell colonies after xenograft transplantation. Cells are stained against primate testis cell-specific antisera. (Right) Quantification of colonies in each group (colonies contain greater than four cells). The number of recipient mice tested are shown below. Unfractionated cells and PLPPR3− cells were used as control conditions. (D) PCA of RNA-seq datasets from PLPPR3+ and KIT+ cells. (E) MA plot of DEGs in PLPPR3+ vs. KIT+ cells. Red points indicate genes with q < 0.01 and log2 fold-change >1 or <1. The number genes significantly enriched in PLPPR3+ and KIT+ cells are shown on the top and bottom, respectively. (F) Heatmap of DEGs from PLPPR3+ vs. KIT+ cells (the four biological replicates from each are shown). (Right) Representative genes and biological processes in PLPPR3+ and KIT+ cells, are shown (Upper and Lower, respectively).
PLPPR3 protein is known to not only be expressed on the cell surface, but intracellularly (14, 22), raising the possibility that its expression, as detected by IF analysis of fixed cells, differs from its surface expression. To test this, we permeabilized testicular cells prior to FACS analysis in order to detect both intracellular and cell-surface PLPPR3. This revealed that twice as many permeabilized testicular cells are PLPPR3+ (SI Appendix, Fig. S1A) than are viable testicular cells (Fig. 1A) (6% vs. 3%, respectively). This indicated that only one-half of PLPPR3+ cells express PLPPR3 on the cell surface. Coupled with our finding that the PLPPR3 gene is primarily expressed in primitive uSPG (14), this raised the possibility that cell-surface PLPPR3 is a highly selective primitive uSPG marker that could be used to purify human SSCs. To test this, we performed germ-cell xenograft germ-cell transplantation analysis (23). While human SPG transplanted into mice testes are unable to fully progress through spermatogenesis, the colonies that form must migrate to the SSC niche in seminiferous tubules and are long-term, suggesting that xenograft germ-cell transplantation analysis is a reasonable assay for measuring human SSC activity (2). Using this assay, we found that PLPPR3+ SPG were ∼38-fold enriched for SSC activity (as assayed by the number of colonies that formed) compared with unfractionated cells (Fig. 1C). PLPPR3− cells did not display elevated colony formation relative to unfractionated cells, but did generate some colonies, suggesting that a low frequency of PLPPR3− cells are SSCs. Together, these results demonstrate that cell-surface PLPPR3 is a marker of primitive human uSPG that greatly enriches for SSCs, and thus this marker is a potentially powerful tool for understanding the molecular events involved in human SPG development.
Purification and Transcriptome Analysis of Human uSPG and dSPG.
Given the specificity of PLPPR3 for primitive human uSPG, coupled with its ability to enrich for human SSCs, we elected to use PLPPR3 as a vehicle to purify primitive human uSPG by FACS for bulk RNA-seq analysis. For comparison, we performed RNA-seq analysis on FACS-purified KIT+ cells in parallel.
Principal component analysis (PCA) showed that the PLPPR3+ cells purified from four independent donors clustered together, indicative of a highly correlated pattern of gene expression (Fig. 1D). The same was observed for KIT+ cells purified from these four donors (Fig. 1D). In contrast, the PLPPR3+ and KIT+ cells plotted distant from each other (Fig. 1D), indicating these two cell populations have dramatically different patterns of gene expression. Indeed, we found that 14,128 genes were differentially expressed between PLPPR3+ and KIT+ cells (when using a stringent statistical cutoff: q < 0.01, fold-change > 2) (Fig. 1E). The genes significantly more highly expressed in PLPPR3+ cells included many known uSPG marker genes, such as TSPAN33, ETV5, EGR4, POU2F2, CXCL12, and DUSP5 (Dataset S1). In contrast, the genes significantly more highly expressed in KIT+ cells included many known dSPG marker genes, such as DMRT1, DMRTB1, SOHLH1, SOHLH2, RHOXF1, and RHOXF2 (Dataset S1). This verified that PLPPR3+ and KIT+ cells are enriched in uSPG and dSPG, respectively.
As a further validation, we compared the DEGs from PLPPR3+ and KIT+ cells with genes exhibiting developmentally regulated expression in SPG subsets defined by scRNA-seq analysis (SI Appendix, Fig. S1B). In this scRNA-seq study, we performed Monocle pseudotime analysis to align the developmental progression of human SPG from the primitive uSPG stage to the dSPG stage (14). Here, we used this dataset to identify genes that significantly change their expression along this trajectory (SI Appendix, Fig. S1C). These genes fell into three groups that had peak expression in immature, intermediate maturity, and the most advanced SPG, respectively, confirmed by known marker genes (SI Appendix, Fig. S1C). We then compared the genes in these three groups with the genes exhibiting enriched expression in PLPPR3+ and KIT+ cells. The PLPPR3+-enriched genes overlapped the most with group-1 genes, consistent with the PLPPR3+ population being enriched in primitive uSPG (SI Appendix, Fig. S1D). The KIT+-enriched genes overlapped most with group-3 genes, consistent with the KIT+ population being enriched in dSPG (SI Appendix, Fig. S1D). These PLPPR3+- and KIT+-enriched genes exhibiting stage-specific expression during SPG development are candidates to mediate stage-specific functions during human SPG development.
The Human uSPG Transcriptome.
The genes displaying enriched expression in PLPPR3+ cells (relative to KIT+ cells) are candidates to have roles in human SSCs. Consistent with this, some of the PLPPR3+-enriched genes—including LMO2, FLI1, ETV6, ASCL2, FOXO3, and KLF4 (Fig. 1F and Dataset S1)—have known roles in other adult stem cell systems, including hematopoietic stem cells, intestinal epithelial stem cells, and neural stem cells (24–29). It will be interesting to determine whether these factors have similar roles in human SSCs. Gene ontology (GO) analysis revealed that PLPPR3+ cells preferentially express genes involved in “cell-surface receptor signaling,” “intracellular signal transduction,” and “cytokine-mediated signaling pathway” (Fig. 1F). GO analysis showed that other genes enriched in PLPPR3+ cells—including ATM, BCL6, CCR2, PRDM1, CSF1, IGF1, IRF1, and CDKN1A (Dataset S1)—are involved in cell proliferation, and thus may mediate the proliferative expansion of human SPG progenitors. Also enriched in PLPPR3+ cells are genes involved in cell migration (e.g., CXCL12, MMP9, MYO1C, NOTCH1, and VIL1), differentiation (e.g., WNT4, XBP1, CHD7, CDK6, EGR1, and FOXP1), metabolism (e.g., ADAM8, ASTL, CAMK2D, CCND1, DUSP5, and PDGFB), and apoptotic processes (e.g., BAX, BCL2, ADAR, AGO4, CFLAR, and MCL1) (Dataset S1).
As a complementary approach to address enrichment for signaling pathways, we used Ingenuity pathway analysis, which showed that PLPPR3+ cells selectively express genes in the EIF2, integrin, JAK/STAT, germ-Sertoli cell junction, and CXCR4 signaling pathways (SI Appendix, Fig. S1E). Consistent with this, several of these signaling pathways—including the integrin, JAK/STAT, and germ-Sertoli cell junction signaling pathways—have been previously shown to be important for SSCs (30–32).
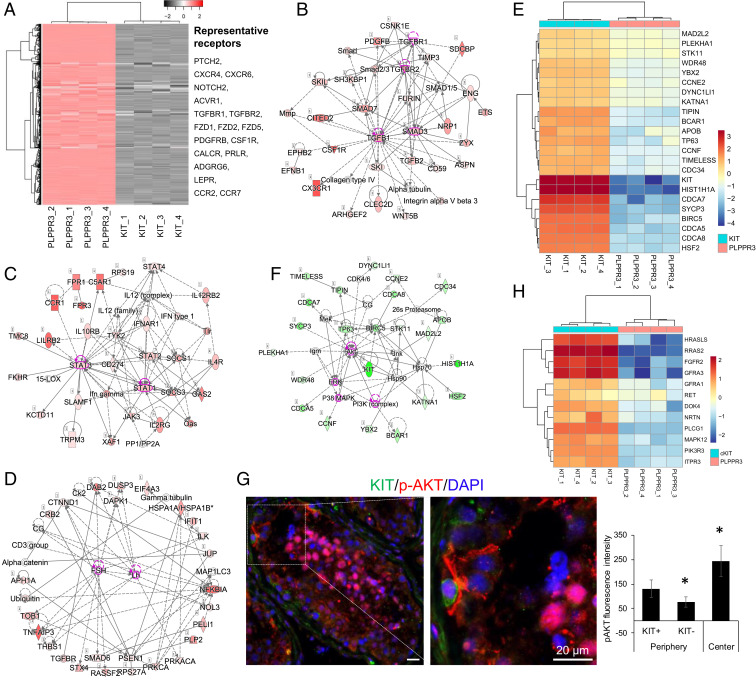
SPG are known to express cell-surface receptors that bind to many factors made by testicular somatic cells (33, 34). Thus, we examined whether PLPPR3+ cells express genes encoding components of cell-surface signaling pathways. GO analysis revealed that PLPPR3+ cells preferentially express a remarkably large number of genes (844) associated with cell-surface receptor signaling (Fig. 2A and Dataset S1). Ingenuity pathway analysis revealed that many of these genes encode receptors regulated by the TGF-β and JAK-STAT signaling pathways, as well as hormones such as FSH and LH (Fig. 2 B–D). The finding that many signaling pathways are enriched in the PLPPR3+ subset raised the possibility that one or more of these signaling pathways could be leveraged as a means to propagate human uSPG in vitro, which we tested, as described below.
Fig. 2.
Signaling pathways enriched in human primitive uSPG and dSPG. (A) Heatmap of genes encoding cell-surface proteins enriched in PLPPR3+ cells. (Right) Representative examples. (B–D) Regulatory networks composed of cell-surface receptor-associated factors encoded by genes exhibiting enriched expression in PLPPR3+ cells (from A). The intensity of the red color represents the degree of increased expression in PLPPR3+ vs. KIT+ cells. (E) Heatmap of genes encoding AKT signaling pathway components. KIT+ cells more highly express these genes than PLPPR3+ cells (all four biological replicates from each are shown). (F) Regulatory networks composed of AKT signaling pathway-associated factors encoded by genes exhibiting enriched expression in KIT+ cells (from E). The intensity of the green color represents the degree of increased expression in KIT+ vs. PLPPR3+ cells. (G) IF analysis of human adult testis section costained with antisera against p-AKT and KIT. (Scale bars, 20 µm.) (Right) Quantification of average pAKT fluorescence intensity per unit area within the region of interest, as assessed using ImageJ software (NIH), as described previously (66); 67 cells were quantified. The periphery of the seminiferous tubule contains SPG, while the center contains later-stage germ cells. (H) Heatmap of genes encoding GDNF signaling pathway components. KIT+ cells more highly express these genes than PLPPR3+ cells (all four biological replicates from each are shown).
The Human dSPG Transcriptome.
The genes that display enriched expression in KIT+ cells (relative to PLPPR3+ cells) are candidates to have roles in dSPG, including their differentiation. In agreement with this, we identified many KIT+ cell-enriched genes involved in cell differentiation, including DMRTC2, DMC1, SPO11, TBPL1, CHD5, and MAST2 (Fig. 1F and Dataset S1). GO analysis revealed that KIT+ cells exhibit enriched expression of genes involved in “cell-cycle phase transition” (Fig. 1F), consistent with the fact that dSPG tend to be proliferating (35). Another enriched category is “spermatogenesis” (Fig. 1F), consistent with the fact that dSPG are responsible for undergoing the initial steps of spermatogenesis (35). Interestingly, KIT+ cells also up-regulate the expression (relative to PLPPR3+ cells) of many genes important for later stages of spermatogenesis, suggesting that germ cells prepare for later events at the dSPG stage (Dataset S1). These up-regulated genes include SYCP3, which is involved in meiosis in spermatocytes; DOT1L, SMYD2, SETD3, SUZ12, KDM1A, KMT5B, PRMT1, and PRMT5, all of which catalyze the addition of specific histone methylation marks that are known to shift during spermatocyte development (36); and PRM1, PRM2, and TNP1, which encode proteins that replace histones when spermatids elongate (37).
Ingenuity pathway analysis of the genes selectively expressed in KIT+ cells revealed a striking enrichment for genes involved in AKT signaling (Fig. 2E). The protein products of these enriched genes can be aligned in a complex gene network with AKT as its central node (Fig. 2F). This finding led us to test whether human SPG have active AKT signaling. Using an antisera against phospho (p)-AKT, the active form of this kinase (38), we observed that SPG do indeed exhibit staining with this antisera (Fig. 2G). An even stronger signal was observed in spermatocytes and spermatids (Fig. 2G), indicating that AKT signaling occurs not only in SPG, but throughout human spermatogenesis.
Another signaling pathway whose components are enriched in dSPG is the glial cell line-derived neurotrophic factor (GDNF) pathway (Fig. 2H). This was surprising, as GDNF is known to instead act in uSPG in mice, where it promotes SSC self-renewal over differentiation (39), and is widely used to expand mouse germline stem cells in vitro (40). Among the GDNF pathway genes more highly expressed in KIT+ cells than PLPPR3+ cells are the GDNF receptor genes—GFRA1 and RET—as shown by RNA-seq analysis of all four biological replicates (Fig. 2H) and verified by qPCR analysis (SI Appendix, Fig. S2A). This is consistent with recent scRNA-seq studies showing that GFRA1 is infrequently expressed in cells with characteristics of SSCs, and instead is mainly expressed in later-stage SPG, including progenitors (12, 14, 15). To determine whether this is also the case for the GFRA1 protein, we performed IF analysis on adult human testes. This showed that ∼77% of GFRA1+ cells also express NANOS3 protein (SI Appendix, Fig. S2B), a well-established progenitor marker (14, 41). In addition, ∼38% of KIT+ cells and ∼46% of PRPPL3+ cells coexpress GFRA1 protein (SI Appendix, Fig. S2B), demonstrating that GFRA1 marks some dSPG and primitive uSPG as well. This expression pattern suggests that GDNF may act broadly in human SPG, driving not only the proliferation of a subset of SSCs, but most progenitors as well as many dSPG.
Identification of Signaling Pathways That Alter SPG Fate In Vitro.
Having identified signaling pathway components that are differentially expressed in uSPG vs. dSPG, we next tested whether manipulating the corresponding signaling pathways modified the properties of human SPG in vitro cultures. Our primary goal was to identify signaling conditions that favor primitive uSPG, as such cultures would likely be enriched in human SSCs. We used the workflow depicted in Fig. 3A, in which dissociated cells derived from human testicular biopsies were enriched for SPG using the cell-surface protein, integrin-α6 (ITGA6) and then cultured for 2 wk. We chose ITGA6 because it is a conserved SPG marker that enriches for SSCs in rodents, monkeys, and human (10, 42, 43), allowing us to test the properties of human SPG in vitro cultures. The cells were cultured in a previously described SSC culture feeder-free basal media—IMDM/SFM (44)—in combination with signaling pathway factors described below. We first tested the role of FGF2, as 1) it is widely used for mouse SSC culture (40), 2) it promotes self-renewal of cultured mouse SSCs/uSPG through activation of the transcription factors ETV5 and BCL6B (45) by a GDNF-independent mechanism (46), and 3) the gene encoding its receptor, FGFR3, is broadly expressed in human uSPG (12, 14). In support of its critical role, we found that FGF2 supplementation to the basal media dramatically improved cell survival (SI Appendix, Fig. S2C). Therefore, FGF2 was included as a standard component of our culture media.
Fig. 3.
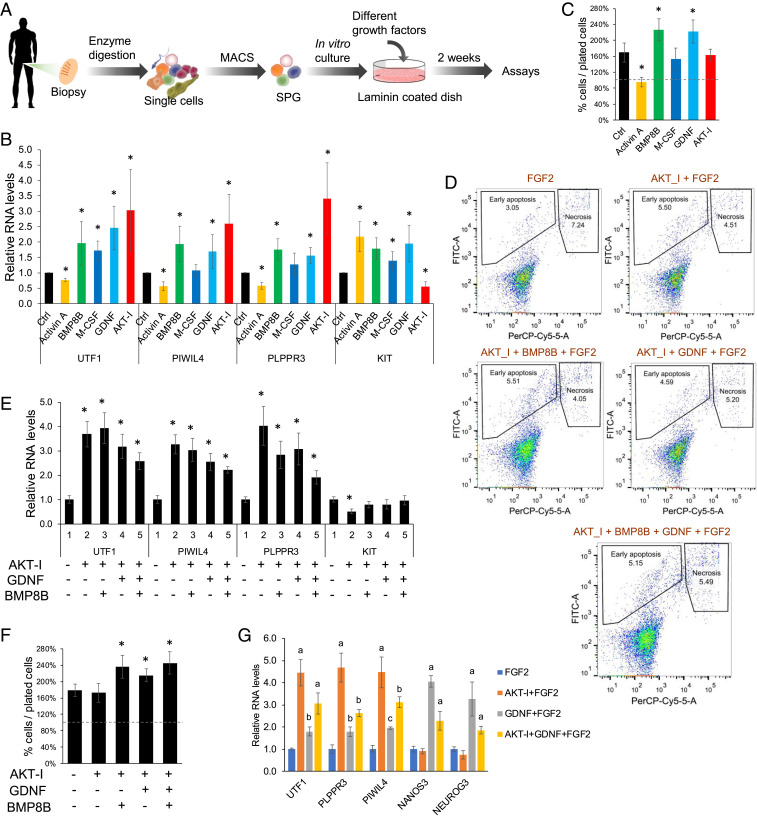
Identification of signaling pathways that alter human SPG fate. (A) Schematic illustration of the human SPG culture experimental workflow. (B) qPCR analysis of MACS-purified ITGA6+ cells cultured for 2 wk with basal media + FGF2 + the indicated signaling factor or modulator. The values shown are relative to FGF2 alone (labeled as “Ctrl”) culture condition (mean ± SD from five biological replicates). (C) Percentage of cells after 2-wk culture, relative to the number of cells initially plated (dashed line). FGF2 alone is labeled as “Ctrl;” other cultures have FGF2 + the indicated factor. (D) Apoptosis/necrosis analysis of ITGA6+ cells cultured for 2 wk with the agents shown. Annexin V-FITC+ labels cells undergoing early apoptosis, while cells labeled by both Annexin V-FITC+ and PI+ are necrotic. (E) qPCR analysis of cells cultured as in B with the agents shown. The values shown are relative to cells incubated with FGF2 alone (mean ± SD from four biological replicates). (F) Percentage of cells after 2-wk culture, relative to the number of cells initially plated (dashed line). (G) qPCR analysis of the indicated markers expressed in MACS-purified ITGA6+ cells cultured for 4 wk with the factors indicated. The values shown are relative to our basal media, which contains FGF2 (mean ± SD from two biological replicates). *P < 0.05.
We then compared cells incubated with FGF2 alone vs. cells incubated with FGF2 in combination with the following signaling factors that our studies indicated were candidates to favor culture of human uSPG: Activin A, GDNF, M-CSF, BMP8B, and the AKT pathway inhibitor MK-2206 2HCl. We chose to test the TGF-β–signaling ligand, activin A, because: 1) The activin A receptor gene, ACVR, exhibits enriched expression in PLPPR3+ cells (Fig. 2A and Dataset S1); 2) activin A is necessary for the maintenance of pluripotency in human embryonic stem cells (47); and 3) many TGF-β–signaling genes are enriched in PLPPR3+ cells (Fig. 2B and Dataset S1). We chose BMP8B because: 1) Many BMP signaling component genes—including JUN, KRAS, NFKB1, MRAS, SMAD6, BMP2, SMAD7, and GRB2—exhibit enriched expression in PLPPR3+ cells (Dataset S1); 2) BMP4, BMP7, and BMP8B has been shown to induce the generation of germ cells from human embryonic stem cells (48); and 3) BMP8B is required for the resumption of mouse germ-cell proliferation in early puberty in mice (49). M-CSF (also called CSF1) was chosen because: 1) The gene encoding its receptor, CSF1R, is enriched in PLPPR3+ uSPG (Fig. 2A and Dataset S1); 2) CSF acts through the JAK-STAT pathway (50), which we found is likely to be up-regulated in uSPG (Fig. 2C); and 3) addition of CSF1 significantly enhances SSC self-renewal in heterogeneous Thy1+ mouse spermatogonial cultures (51). We chose to test GDNF because: 1) It regulates the self-renewal and differentiation of mouse uSPG in a dosage-dependent manner (39) and 2) it is widely used for mouse SSCs culture (40). Finally, we examined the effect of the AKT pathway inhibitor MK-2206 2HCl (which we will refer to as “AKT-I”) because: 1) AKT signaling component genes are highly enriched in dSPG where they form an elaborate network (Fig. 2 E and F and Dataset S1) and 2) we obtained evidence that AKT signaling increases during SPG differentiation (Fig. 2G).
The effect of these signaling factors on SPG cultures was determined by quantifying the expression of the well-established uSPG and dSPG markers, UTF1 and KIT, respectively (12, 14, 52). We also quantified the expression of the scRNA-seq–defined primitive uSPG markers, PLPPR3 and PIWIL4, both of which were validated by follow-up studies (12, 14). qPCR analysis of these four marker genes indicated that the signaling pathway factors we tested fell into four categories: Category 1, minimal effect on either uSPG and dSPG; category 2, broadly supportive of SPG; category 3, preferentially supportive of dSPG; and category 4, preferentially supportive of uSPG (Fig. 3B).
M-CSF had only a modest effect on UTF1 and KIT expression and had no significant effect on PIWIL4 and PLPPR3 expression, and thus we placed M-CSF in category 1. In contrast, GDNF and BMP8B significantly increased expression of all four SPG marker genes we tested, indicative of these factors belonging to category 2. Consistent with GDNF being assigned in category 2, its receptor, GFRA1, is broadly expressed in human SPG, including progenitors (SI Appendix, Fig. S2B). To directly test whether GDNF supports progenitors in our culture system, we examined the progenitor markers, NANOS3 and NEUROG3, and found that both were greatly elevated in response to GDNF supplementation (SI Appendix, Fig. S2D). We placed activin A in category 3, as it increased the expression of the dSPG marker KIT, but significantly decreased expression of all three uSPG markers, suggesting that activin A drives dSPG differentiation. Finally, the the AKT-I strongly increased the expression of all three uSPG markers and decreased KIT expression, indicative of the AKT-I being in category 4. Because it decreased KIT expression, this raised the possibility that the AKT-I acts by inhibiting the differentiation of cultured human uSPG, thereby enriching for uSPG, a possibility verified by scRNA-seq analysis, as described below.
While the primary goal of our study was to identify signaling factors that favor culture of uSPG, not their long-term proliferative expansion, nonetheless, we also measured the effect of the different signaling pathway conditions on cell number. As shown in Fig. 3C, the two factors that supported the culture of both uSPG and dSPG—GDNF and BMP8B—also significantly increased the number of cells in culture, relative to FGF2 alone, over the 2-wk culture period. This is consistent with the known growth-promoting effects of GDNF on mouse uSPG (39). In contrast, the other factors did not significantly increase cell number relative to FGF2 alone. Indeed, activin A modestly decreased cell number (Fig. 3C), consistent with our finding that activin A favored dSPG over uSPG markers (Fig. 3B) and thus might promote SPG differentiation. We considered the possibility that the AKT-I did not further increase cell number relative to FGF2 alone because it reduced cell viability, based on the previous finding that high concentrations (≥3 μM) of the AKT inhibitor we used induces cell apoptosis in many human carcinoma cell lines (53, 54). However, we found that the mild dose we used (100 nM) did not significantly increase either apoptosis or necrosis, when compared with FGF2 treatment alone (Fig. 3D).
The finding that AKT-I supplementation strongly up-regulated three of three uSPG markers and down-regulated the dSPG marker, KIT, suggested that this agent could prove useful for culturing highly enriched human uSPG. Thus, we next tested the effect of the AKT-I in combination with other signaling factors. This analysis showed that while most factor combinations increased uSPG marker expression relative to FGF2 alone, it was not higher than in response to AKT-I + FGF2 (Fig. 3E). Furthermore, only AKT-I + FGF2 significantly decreased KIT expression relative to FGF2 alone (Fig. 3E). Thus, from this perspective, AKT-I + FGF2 was the most effective combination favoring uSPG over dSPG. However, cell-count analysis revealed that triple-combination treatment (with either GNDF + AKT-I + FGF2 or BMP8 + AKT inhibitor + FGF2) significantly increased cell counts relative to FGF2 alone, whereas AKT-I + FGF2 did not (Fig. 3F). Quadruple-combination treatment (BMP8 + GNDF + AKT-I + FGF2) also significantly increased cell number compared to FGF2 alone (Fig. 3F). None of the combinations we tested had a significant effect on either apoptosis or necrosis (Fig. 3F). We conclude that the AKT-I selectively favors the culture of uSPG, and when it is used in combination with other factors, it is able to support proliferative expansion of uSPG, even during short-term culture.
To further test the efficacy of the AKT-I in promoting the culture of primitive uSPG, we extended the culture period to 1 mo. This revealed that AKT-I supplementation increased the expression of primitive uSPG markers (by approximately fourfold, when compared with FGF2 alone) (Fig. 3G), indicative of expansion of primitive uSPG over the 1-mo culture period. The increase in primitive uSPG markers was more modest when the cultures were also supplemented with GDNF or supplemented with GDNF alone (Fig. 3G). AKT-I supplementation also permitted cell expansion over a 1-mo period (SI Appendix, Fig. S2E). While cell expansion was no better in response to the AKT-I than in response to FGF2 alone (SI Appendix, Fig. S2E), this is not surprising for two reasons. First, given that our evidence suggests that the AKT-I acts by suppressing SSC differentiation, one would not expect this inhibitor to necessarily also promote growth. Second, SSCs normally divide slowly (presumably to reduce the mutational load to the male germline), and thus if the AKT inhibitor acts to prevent conversion of SSCs to form more rapidly dividing progenitors, this predicts that cultures supplemented with the AKT-I would proliferate slowly. Indeed, when we instead supplemented the cultures with GDNF, there was a greater increase in cell number than in response to the AKT-I (SI Appendix, Fig. S2E). This is consistent with our finding that GDNF favors progenitors over SSCs.
scRNA-Seq Analysis Reveals That the AKT-I Elicits a SPG Subset Switch.
To scrutinize—in detail—the effects of the AKT-I on human SPG subsets, we turned to scRNA-seq analysis. We compared human SPG cultured with FGF2 alone vs. AKT-I + FGF2, as we found the former is essential for cell survival (SI Appendix, Fig. S2C), and the latter is the only condition that preferentially favors primitive uSPG over dSPG (Fig. 3 B–E). We purified ITGA6+ testicular cells from two fertile adults (aged 32 and 37 y) and cultured them under these two conditions for 2 wk, followed by scRNA-seq analysis. After filtering out poor-quality cells, 8,916 cells remained for downstream analysis. Quality parameters are summarized in SI Appendix, Fig. S3A and Table S1. The two replicates had highly correlated expression patterns (SI Appendix, Fig. S3B).
We identified cell clusters corresponding to the major cell types in the cultures using well-established cell type-specific gene markers (Fig. 4A and SI Appendix, Fig. S3C and Table S2). As expected, the major cell population was SPG: 51% and 60% of all cells in the FGF2 group and AKT-I + FGF2 group, respectively. There were also small numbers of later-stage germ cells (spermatocytes and spermatids), as well as stromal cells and blood cells. No cell clusters corresponding to Sertoli cells, Leydig cells, or peritubular myoid cells were observed.
Fig. 4.
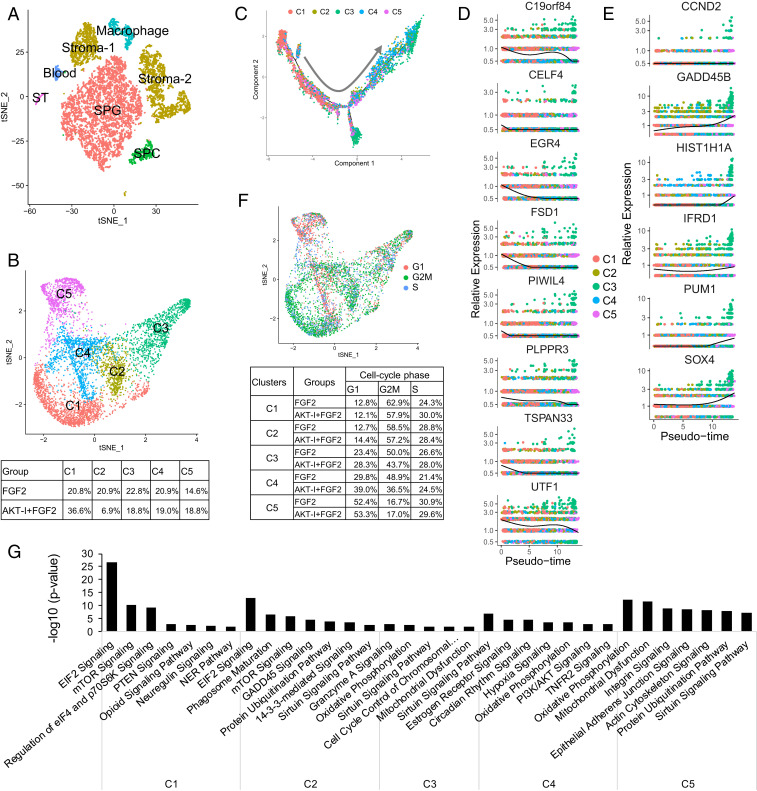
scRNA-seq analysis elucidates the constituents in human ITGA6+ cell cultures. (A) t-Distributed stochastic neighbor-embedding (tSNE) plot showing the cell clusters present in testicular ITGA6+ cells cultured for 2 wk with the agents shown. (B) Reclustering of the SPG cell cluster defined in A reveals five subclusters. (Lower) The percentage of cells in each subcluster. (C) Monocle trajectory analysis of subclusters defined in B. Arrow shows the pseudotime direction. (D) The expression pattern of genes preferentially expressed in the C1 subcluster, as shown on the pseudotime axis defined in C. (E) The expression pattern of differentiation-associated genes plotted along the pseudotime axis defined in C. (F, Upper) tSNE plot inferring the cell-cycle phase of the cells in the five subclusters defined in B, based on expression of a large set of G1-, G2/M-, and S-phase genes (67). (Lower) Percentage of cells in the cell-cycle phases indicated. (G) Enriched signaling pathways in each cell cluster defined in B.
To determine the impact of these two culture conditions on SPG subsets, we reclustered only the SPG. This revealed the existence of five distinct SPG subclusters: C1 to C5 (Fig. 4B). All five subclusters expressed uSPG marker genes, albeit at varying levels (SI Appendix, Fig. S3D and Dataset S1), suggesting they represent distinct uSPG states, as described below. Consistent with this, none of the subclusters were enriched for the dSPG marker KIT (SI Appendix, Fig. S3D). Indeed, scRNA-seq analysis only detected 198 KIT+ cells of a total of 5,001 SPG (4.0%). The notion that these in vitro cultures had few dSPG was further substantiated by the finding that another well-established dSPG marker, STRA8 (55), was detectably expressed in <1% of the cultured SPG cells (SI Appendix, Fig. S3D). There were modestly fewer KIT+ cells in the AKT-I + FGF2 cultures than FGF2 cultures (3.8% vs. 4.3%, respectively), which is consistent with the reduction in KIT mRNA expression measured by qPCR analysis of the bulk cultures (Fig. 3B). The KIT mRNA Ct values in these bulk cultures were high (∼30 to 33 in FGF2 alone and AKT-I + FGF2 cultures, respectively), consistent with few KIT+ cells, as detected by our scRNA-seq analysis. One explanation for why there are few dSPG in these cultures is that they tend to die in culture; dead dSPG would not have been detected as we removed dead cells prior to scRNA-seq analysis.
Interestingly, we observed that the two conditions we tested dramatically altered the proportion of cells in the C1 and C2 subclusters. The AKT-I + FGF2 condition yielded a much greater proportion of C1 cells than did FGF2 alone (36.6% vs. 20.8%, respectively) (Fig. 4B). Conversely, FGF2 alone yielded a much greater proportion of C2 cells than did AKT-I + FGF2 (20.9% vs. 6.9%, respectively) (Fig. 4B). To understand the significance of this C1/C2 shift, we investigated the nature of the C1 and C2 subclusters.
C1 is primarily composed of primitive uSPG, based on several lines of evidence. First, several human primitive uSPG markers recently defined by scRNA-seq analysis—PIWIL4, TSPAN33, PLPPR3, UTF1, EGR4, FSD1, CELF4, and C19orf84 (12, 14)—were preferentially expressed in C1 as compared to the other subsets (Fig. 4D and Dataset S1). Second, GO analysis revealed that genes preferentially expressed in the C1 subcluster overlapped with those enriched in primitive uSPG obtained directly from human testes (14). Overlapping biological functions include “establishment of protein localization to endoplasmic reticulum,” “protein targeting to membrane,” “transcription,” “RNA catabolic process,” and “peptide biosynthetic process” (Dataset S1). A third line of evidence came from Monocle trajectory analysis, which showed that C1 is at one end of the trajectory, while the other end is primary composed of cells from the more differentiated C3 and C4 subclusters (Fig. 4C). C1 preferentially expresses primitive uSPG markers (Fig. 4D), while C3 and C4 preferentially expresses genes associated with differentiation, including CCND2, GADD45B, HIST1H1A, IFRD1, PUM1, and SOX4 (Fig. 4E).
The C2 subcluster has a differentiation status intermediate between C1 and C3/C4/C5, based on the following lines of evidence: 1) Several stem cell self-renewal and maintenance genes preferentially expressed in C1 are down-regulated in C2 (SI Appendix, Fig. S3E); 2) several genes preferentially expressed in C2 are associated with cell differentiation (SI Appendix, Fig. S3F); and 3) C3, C4, and C5 are primarily composed of more differentiated cells than C1 and C2, based on their preferential expression of SOHLH1, SMC1A, SMC3, H3F3B, and SOX4 (Dataset S1), which encode proteins involved in SPG differentiation, mitosis, meiosis, and spermatocyte development (56–58). Fig. 4G provides a list of enriched functions in each of the subclusters, as defined by Ingenuity pathway analysis.
That the addition of the AKT-I to the SPG cultures increased the proportion of C1 cells and decreased the proportion of C2 cells (Fig. 4B) is consistent with the finding that the AKT-I increases the expression of uSPG markers (Fig. 3B) and it provides a mechanism by which primitive uSPG are favored in response to AKT signaling inhibition. In particular, our scRNA-seq results suggest that AKT pathway inhibition reduces the ability of C1 to differentiate into C2, resulting in an accumulation of C1 subcluster cells. While it is also possible that the AKT-I acts by promoting the proliferation of C1 cells, we regard this as unlikely, as the AKT-I did not increase the expression of cell-cycle genes (Fig. 4F), a reliable means to infer cell cycle status (14, 59, 60). Coupled with our evidence that the AKT-I also does not act by promoting SPG survival (Fig. 3D), our data instead favor that the AKT-I increases the proportion of primitive uSPG by inhibiting their differentiation. This is interesting in light of the many studies suggesting that AKT inhibition has the opposite role in mice, where it prevents SSC self-renewal and thus drives SPG differentiation (61, 62). This raises the possibility that mouse and human SPG fundamentally differ in their response to AKT signaling.
Our scRNA-seq analysis also allowed us to compare the cell-cycle frequency of our in vitro SPG cultures vs. SPG in vivo. To achieve this, we compared our in vitro and in vivo scRNA-seq datasets described herein and in Sohni et al. (14), respectively, for the expression of a large array of cell-cycle genes. This analysis indicated that the cells in our in vitro cultures supplemented with AKT-I + FGF2 are actively cycling more often than uSPG in vivo (SI Appendix, Table S3). Thus, while the growth rate of our cultures supplemented with the AKT-I is relatively slow relative to immortalized cell lines (SI Appendix, Fig. S2E), the cells are proliferating as rapidly as can be expected for cultures enriched for primitive uSPG.
Conclusions
Previous studies have reported conditions for culturing human SSCs from either dissociated cells obtained from whole testes or from purified testicular cell populations (63–65). While these studies were promising, they defined SSCs based on morphology and a limited number of broadly expressed SPG markers. Furthermore, the culture conditions necessary to favor different SPG stages, including SSCs, progenitors, and dSPG was not explored in these past studies.
In our study, we identified the cell-surface protein PLPPR3 as a tool to purify primitive human uSPG highly enriched for SSC activity. Using RNA-seq, which has far greater read depth than scRNA-seq, we identified the full complement of genes expressed in these enriched human SSCs, as well as thousands of genes up- and down-regulated as these cells progress to form dSPG. The datasets we generated will be valuable for the field to define the molecular mechanisms underlying SSC self-renewal and differentiation.
Among the statistically enriched categories of genes differentially expressed in primitive uSPG vs. dSPG were several signaling pathways. This prompted us to test the functional role of these signaling pathways, which led to the identification of signaling pathway manipulations that alter the fate of SPG in vitro. Most significantly, we found that the AKT-inhibitor MK-2206 2HCl significantly increases the proportion of primitive uSPG in culture, as defined by both conventional approaches and scRNA-seq analysis. To our knowledge, this defining of conditions that favor the culture of human primitive uSPG has the potential to be used to culture human SSCs for clinical applications.
Materials and Methods
Human Testis Samples Preparation.
The experiments with human material were approved by the University of California, San Diego Human Research Protections Program council. Informed consent was obtained from all of the human subjects. Testicular biopsies were obtained from 29 fertile men aged between 30 and 50 y, undergoing vasectomy reversal at the University of California, San Diego Medical Center, following Institutional Review Board-approved protocol #120471.
The biopsies were transported to the research laboratory on ice in Minimum Essential Medium Alpha Medium (αMEM) + 10% FBS. The samples were then immediately cut into smaller portions and cryopreserved using freezing media composed of 10% DMSO + 40% αMEM + 50% FBS under controlled cooling conditions in a freezing container (Thermo Fisher Scientific) at −80 °C. The samples were subsequently transferred to liquid nitrogen storage until use.
Single testicular cells were isolated using a two-step enzymatic digestion protocol described previously (14, 42). In brief, testicular tissue was mechanically disrupted and enzymatically digested with 1 mg mL−1 collagenase type IV (Worthington Biochemical) in HBSS (Gibco) at 37 °C. The tubules were sedimented and washed with HBSS and digested in 0.25% Trypsin-EDTA (ThermoFisher) and DNase I (Worthington Biochemical). The suspension was triturated vigorously 10 times, incubated at 37 °C for 5 min, followed by repeat trituration and incubation. The digestion was stopped by adding the same volume of αMEM + 10% FBS medium and the cells were size-filtered through 70-μm and 40-μm strainers (ThermoFisher) and pelleted by centrifugation at 300 × g for 5 min.
Additional Experimental Procedures.
The procedures for xenograft germ-cell transplantation analysis, immunofluorescence analysis, bulk RNA-seq, human SPG culture, Annexin V/PI staining and flow cytometry, qRT-PCR, and 10× Genomics scRNA-seq and analysis are presented in SI Appendix.
Supplementary Material
Acknowledgments
We thank the University of California, San Diego Institute for Genomics Medicine Genomics Center for sequencing analysis of our RNA-sequecing and single-cell RNA-sequencing experiments; the University of California, San Diego Tissue Technology Shared Resource for generating human testes sections; and the San Diego Supercomputer Center for providing data analysis resources. This work was supported by NIH Grants T32 HD087194 (to S.M.), R01 HD092084 (to K.E.O.), and R01 GM119128 (to M.F.W.); and the Lalor Foundation (K.T.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE144085).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000362117/-/DCSupplemental.
Data Availability.
The RNA-seq and scRNA-seq data generated in this study has been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus database under the accession no. GSE144085.
References
- 1.de Rooij D. G., The nature and dynamics of spermatogonial stem cells. Development 144, 3022–3030 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Fayomi A. P., Orwig K. E., Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. (Amst.) 29, 207–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadri-Ardekani H., Atala A., Testicular tissue cryopreservation and spermatogonial stem cell transplantation to restore fertility: From bench to bedside. Stem Cell Res. Ther. 5, 68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dym M., Kokkinaki M., He Z., Spermatogonial stem cells: Mouse and human comparisons. Birth Defects Res. C Embryo Today 87, 27–34 (2009). [DOI] [PubMed] [Google Scholar]
- 5.von Kopylow K., Spiess A. N., Human spermatogonial markers. Stem Cell Res. (Amst.) 25, 300–309 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Dovey S. L., et al. , Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J. Clin. Invest. 123, 1833–1843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermann B. P., Sukhwani M., Hansel M. C., Orwig K. E., Spermatogonial stem cells in higher primates: Are there differences from those in rodents? Reproduction 139, 479–493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zohni K., Zhang X., Tan S. L., Chan P., Nagano M., CD9 is expressed on human male germ cells that have a long-term repopulation potential after transplantation into mouse testes. Biol. Reprod. 87, 27 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Nickkholgh B., et al. , Enrichment of spermatogonial stem cells from long-term cultured human testicular cells. Fertil Steril 102, 558–565.e5 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Maki C. B., et al. , Phenotypic and molecular characterization of spermatogonial stem cells in adult primate testes. Hum. Reprod. 24, 1480–1491 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Izadyar F., et al. , Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum. Reprod. 26, 1296–1306 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Guo J., et al. , The adult human testis transcriptional cell atlas. Cell Res. 28, 1141–1157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermann B. P., et al. , The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 25, 1650–1667.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sohni A., et al. , The neonatal and adult human testis defined at the single-cell level. Cell Rep. 26, 1501–1517.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M., et al. , Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell 23, 599–614.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Tan K., Wilkinson M. F., Human spermatogonial stem cells scrutinized under the single-cell magnifying glass. Cell Stem Cell 24, 201–203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki S., Diaz V. D., Hermann B. P., What has single-cell RNA-seq taught us about mammalian spermatogenesis? Biol. Reprod. 101, 617–634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bräuer A. U., Nitsch R., Plasticity-related genes (PRGs/LRPs): A brain-specific class of lysophospholipid-modifying proteins. Biochim. Biophys. Acta 1781, 595–600 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Brindley D. N., Lipid phosphate phosphatases and related proteins: Signaling functions in development, cell division, and cancer. J. Cell. Biochem. 92, 900–912 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Strauss U., Bräuer A. U., Current views on regulation and function of plasticity-related genes (PRGs/LPPRs) in the brain. Biochim. Biophys. Acta 1831, 133–138 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Brosig A., et al. , The axonal membrane protein PRG2 inhibits PTEN and directs growth to branches. Cell Rep. 29, 2028–2040.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu P., et al. , Cooperative interactions of LPPR family members in membrane localization and alteration of cellular morphology. J. Cell Sci. 128, 3210–3222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagano M., Patrizio P., Brinster R. L., Long-term survival of human spermatogonial stem cells in mouse testes. Fertil. Steril. 78, 1225–1233 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Cleveland S. M., et al. , Lmo2 induces hematopoietic stem cell-like features in T-cell progenitor cells prior to leukemia. Stem Cells 31, 882–894 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruse E. A., et al. , Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proc. Natl. Acad. Sci. U.S.A. 106, 13814–13819 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hock H., et al. , Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 18, 2336–2341 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuijers J., et al. , Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell 16, 158–170 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Renault V. M., et al. , FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5, 527–539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bialkowska A. B., Yang V. W., Mallipattu S. K., Krüppel-like factors in mammalian stem cells and development. Development 144, 737–754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanatsu-Shinohara M., et al. , Homing of mouse spermatogonial stem cells to germline niche depends on beta1-integrin. Cell Stem Cell 3, 533–542 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Kopera I. A., Bilinska B., Cheng C. Y., Mruk D. D., Sertoli-germ cell junctions in the testis: A review of recent data. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1593–1605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrera S. C., Bach E. A., JAK/STAT signaling in stem cells and regeneration: From Drosophila to vertebrates. Development 146, dev167643 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolci S., Pellegrini M., Di Agostino S., Geremia R., Rossi P., Signaling through extracellular signal-regulated kinase is required for spermatogonial proliferative response to stem cell factor. J. Biol. Chem. 276, 40225–40233 (2001). [DOI] [PubMed] [Google Scholar]
- 34.De Rooij D., “The spermatogonial stem cell niche in mammals” in Sertoli Cell Biology, Griswold M. D., Ed. (Elsevier, ed. 2, 2015), pp. 99–121. [Google Scholar]
- 35.Stukenborg J. B., et al. , Male germ cell development in humans. Horm. Res. Paediatr. 81, 2–12 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Güneş S., Kulaç T., The role of epigenetics in spermatogenesis. Turk. J. Urol. 39, 181–187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward W. S., Function of sperm chromatin structural elements in fertilization and development. Mol. Hum. Reprod. 16, 30–36 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao Y., Hung M. C., Physiological regulation of Akt activity and stability. Am. J. Transl. Res. 2, 19–42 (2010). [PMC free article] [PubMed] [Google Scholar]
- 39.Meng X., et al. , Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287, 1489–1493 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Kanatsu-Shinohara M., et al. , Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 69, 612–616 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Suzuki H., Sada A., Yoshida S., Saga Y., The heterogeneity of spermatogonia is revealed by their topology and expression of marker proteins including the germ cell-specific proteins Nanos2 and Nanos3. Dev. Biol. 336, 222–231 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Valli H., et al. , Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrich human spermatogonial stem cells. Fertil Steril 102, 566–580.e7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shinohara T., Avarbock M. R., Brinster R. L., beta1- and alpha6-integrin are surface markers on mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. U.S.A. 96, 5504–5509 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanatsu-Shinohara M., et al. , Improved serum- and feeder-free culture of mouse germline stem cells. Biol. Reprod. 91, 88 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Ishii K., Kanatsu-Shinohara M., Toyokuni S., Shinohara T., FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development 139, 1734–1743 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Takashima S., et al. , Functional differences between GDNF-dependent and FGF2-dependent mouse spermatogonial stem cell self-renewal. Stem Cell Reports 4, 489–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James D., Levine A. J., Besser D., Hemmati-Brivanlou A., TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 132, 1273–1282 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Kee K., Gonsalves J. M., Clark A. T., Pera R. A., Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev. 15, 831–837 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Zhao G. Q., Deng K., Labosky P. A., Liaw L., Hogan B. L., The gene encoding bone morphogenetic protein 8B is required for the initiation and maintenance of spermatogenesis in the mouse. Genes Dev. 10, 1657–1669 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Rawlings J. S., Rosler K. M., Harrison D. A., The JAK/STAT signaling pathway. J. Cell Sci. 117, 1281–1283 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Oatley J. M., Oatley M. J., Avarbock M. R., Tobias J. W., Brinster R. L., Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development 136, 1191–1199 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Persio S., et al. , Spermatogonial kinetics in humans. Development 144, 3430–3439 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Hirai H., et al. , MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol. Cancer Ther. 9, 1956–1967 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Jiao P., et al. , MK-2206 induces cell cycle arrest and apoptosis in HepG2 cells and sensitizes TRAIL-mediated cell death. Mol. Cell. Biochem. 382, 217–224 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Endo T., et al. , Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 112, E2347–E2356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki H., et al. , SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev. Biol. 361, 301–312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jan S. Z., et al. , Unraveling transcriptome dynamics in human spermatogenesis. Development 144, 3659–3673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuen B. T., Bush K. M., Barrilleaux B. L., Cotterman R., Knoepfler P. S., Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development 141, 3483–3494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nestorowa S., et al. , A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, e20–e31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan K., Song H. W., Wilkinson M. F., Single-cell RNAseq analysis of testicular germ and somatic cell development during the perinatal period. Development 147, dev183251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee J., et al. , Akt mediates self-renewal division of mouse spermatogonial stem cells. Development 134, 1853–1859 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Oatley J. M., Avarbock M. R., Brinster R. L., Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J. Biol. Chem. 282, 25842–25851 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadri-Ardekani H., et al. , Propagation of human spermatogonial stem cells in vitro. JAMA 302, 2127–2134 (2009). [DOI] [PubMed] [Google Scholar]
- 64.He Z., Kokkinaki M., Jiang J., Dobrinski I., Dym M., Isolation, characterization, and culture of human spermatogonia. Biol. Reprod. 82, 363–372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goharbakhsh L., et al. , Isolation and culture of human spermatogonial stem cells derived from testis biopsy. Avicenna J. Med. Biotechnol. 5, 54–61 (2013). [PMC free article] [PubMed] [Google Scholar]
- 66.Tan K., et al. , Actin disorganization plays a vital role in impaired embryonic development of in vitro-produced mouse preimplantation embryos. PLoS One 10, e0130382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kowalczyk M. S., et al. , Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860–1872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and scRNA-seq data generated in this study has been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus database under the accession no. GSE144085.