Significance
Essential genes control the most fundamental processes of bacterial life but are missing from standard comprehensive mutant screens because loss-of-function mutants are absent from the libraries examined. We employed a method for generating insertion mutations at genome-saturation scale in naturally competent bacteria. The insertions can be immediately detected by sequencing, making it possible to follow their loss from the population during subsequent growth. The procedure allowed us to order essential processes based on how rapidly inactivation blocks DNA replication and growth. Processes whose inactivation most immediately blocks replication represent attractive antibiotic targets, since even partial inhibition should curtail growth.
Keywords: essential gene, Acinetobacter baylyi, natural transformation, Tn-seq, TFNseq
Abstract
Mutant phenotype analysis of bacteria has been revolutionized by genome-scale screening procedures, but essential genes have been left out of such studies because mutants are missing from the libraries analyzed. Since essential genes control the most fundamental processes of bacterial life, this is a glaring deficiency. To address this limitation, we developed a procedure for transposon insertion mutant sequencing that includes essential genes. The method, called transformation transposon insertion mutant sequencing (TFNseq), employs saturation-level libraries of bacterial mutants generated by natural transformation with chromosomal DNA mutagenized heavily by in vitro transposition. The efficient mutagenesis makes it possible to detect large numbers of insertions in essential genes immediately after transformation and to follow their loss during subsequent growth. It was possible to order 45 essential processes based on how rapidly their inactivation inhibited growth. Inactivating ATP production, deoxyribonucleotide synthesis, or ribosome production blocked growth the fastest, whereas inactivating cell division or outer membrane protein synthesis blocked it the slowest. Individual mutants deleted of essential loci formed microcolonies of nongrowing cells whose sizes were generally consistent with the TFNseq ordering. The sensitivity of essential functions to genetic inactivation provides a metric for ranking their relative importance for bacterial replication and growth. Highly sensitive functions could represent attractive antibiotic targets since even partial inhibition should reduce growth.
Essential genes underlie the most basic activities of life, with a set of about 300 required for growth in most bacterial species under most conditions (1). Although it has become straightforward to identify essential gene sets for different bacteria (2–5), it has been challenging to evaluate the relative importance of different essential genes for growth under differing conditions. Indeed, essential gene mutants are conspicuously absent from genome-scale phenotyping studies employing transposon mutant pools or arrayed libraries of individual mutants, with the notable exceptions of recent work employing CRISPR interference libraries (2, 6–9). Including essential functions in genotype–phenotype studies should help reveal how the most fundamental processes of cell physiology support growth under a given condition.
In addition, although essential functions have been viewed as a potential source of new antibiotic targets, Projan (10) has pointed out that essentiality alone is flawed as a criterion because it represents the best-case, never-achieved scenario of full inhibition of the corresponding function. Essential gene products whose inactivation most rapidly block growth should be potentially attractive targets since partial inhibition by drugs should also be effective at inhibiting growth.
We sought to assess the relative importance of different essential genes for sustaining growth by evaluating how rapidly their inactivation blocks it. Essential gene analysis has traditionally relied on conditional activity or expression alleles, for example those leading to temperature-sensitive gene products or regulated transcription (4, 8, 11). It is problematic to use such conditional alleles to compare essential processes to each other because individual alleles differ both in how much they reduce activity from wild type under permissive conditions and how completely they eliminate activity under nonpermissive conditions. For example, CRISPR interference alleles vary in both basal and maximal levels of repression (6, 7, 12, 13). Mutant differences may thus reflect the characteristics of the conditional alleles examined as much or more than the processes affected. An ideal analysis of how rapidly inactivation of different essential genes blocks growth would compare cells directly after wild-type genes had been fully inactivated by mutation.
To achieve this objective, we developed an approach in which inactivating mutations are generated genome-wide by natural transformation with transposon-mutagenized chromosomal DNA. The relative abundance of different essential gene mutants in the population during subsequent growth is then monitored to assess how rapidly their growth arrests.
The work employs Acinetobacter baylyi, a naturally competent bacterium in the same taxonomic class as Escherichia coli (Gammaproteobacteria), which is closely related to the notorious nosocomial pathogen Acinetobacter baumannii (14, 15). The bacterium has been developed as a model for genetic analysis and synthetic biology (14, 16–19). In the study reported here, we found that it was possible to order the essential processes of A. baylyi based on how rapidly their genetic inactivation blocks replication and growth.
Results
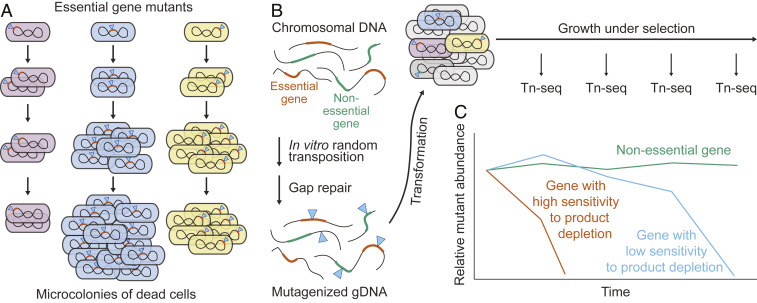
We wished to evaluate the growth of bacteria immediately following the inactivation of essential genes (e.g., due to transposon insertion). Initially, an essential gene product would be present at wild-type levels and should support further mutant replication. However, as growth and division without new synthesis dilute the product, it will eventually be depleted to a level insufficient for further growth, leading to a microcolony of nongrowing cells on solidified medium (Fig. 1A). The time it takes for growth to stop (as reflected in the size of a microcolony) should depend on how critical the corresponding essential process is for continued replication, how much in excess over the minimum level required for replication the essential product is at the time of mutation, and how rapidly the product turns over. This property, which we refer to as “depletion sensitivity,” should thus differ for different essential genes. To evaluate depletion sensitivity of essential genes at a comprehensive scale, we developed a highly efficient transposon mutagenesis technique to expand the capabilities of traditional transposon insertion mutant sequencing (Tn-seq) (20) and make it possible to analyze the behavior of mutations inactivating all genes—essential and nonessential. The approach is called transformation Tn-seq (TFNseq).
Fig. 1.
Distinguishing essential gene mutant growth arrest rates. (A) Essential genes are expected to differ in how fast depletion of their products blocks growth following mutation, resulting in different terminal microcolony sizes. (B) Transformation Tn-seq (TFNseq) overview. Chromosomal DNA is mutagenized in vitro by insertion of a Tn5-based transposon, and the resulting single-stranded gaps are repaired by gap filling and ligation. Bacteria are then transformed with the mutagenized DNA and grown under selection for the transposon. At different times, population samples are analyzed by Tn-seq. (C) Essential genes are distinguished from one another by their relative kinetics of mutant depletion.
TFNseq.
TFNseq is based on natural transformation of A. baylyi with genomic DNA that has been heavily mutagenized in vitro by insertion of a transposon Tn5 derivative (Fig. 1B) (Materials and Methods). Such transformations could generate cultures with a high fraction of mutagenized bacteria (up to 2%), with saturation-level coverage of essential and nonessential genes (≥5,000 independent mutants per average gene) (SI Appendix, Table S1). It was thus possible using Tn-seq to detect large numbers of insertions in the mutagenized DNA and in bacteria soon after transformation and to follow the loss of different essential gene insertions during subsequent growth (i.e., the decreases in fractional abundance in the mixed populations of essential and nonessential gene mutants) (Fig. 1C).
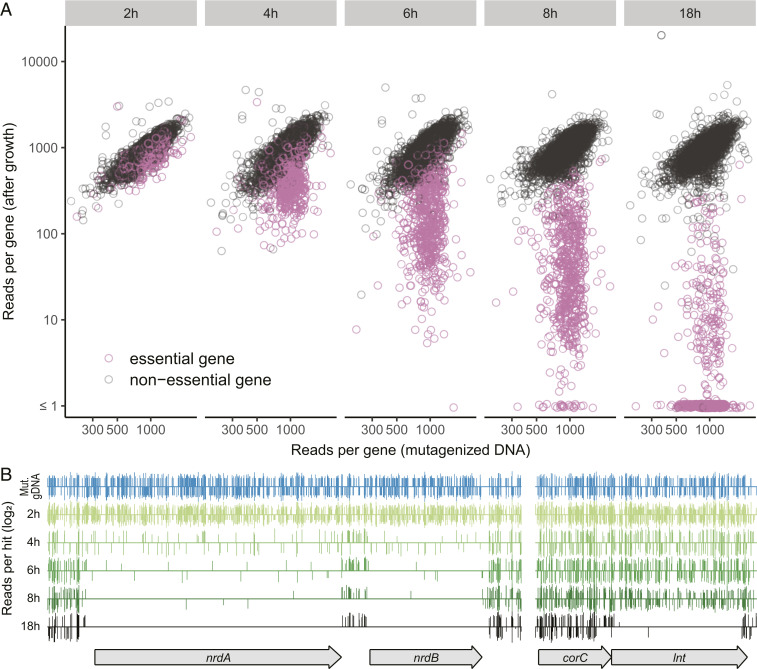
Mutants in different essential genes were gradually lost during growth over 18 h (nonessential mutant population doubling time 37 min) (Fig. 2A). The reduction of insertion sequence reads in four genes illustrates different depletion sensitivities (Fig. 2B). Insertion reads in two genes needed for deoxyribonucleotide synthesis (nrdAB, encoding subunits of ribonucleotide reductase) were largely gone by 4 h of growth, reflecting severe depletion sensitivity. In contrast, insertions in an essential gene needed for lipoprotein production (lnt, encoding an N-acyltransferase) were only lost after 18 h, reflecting more modest depletion sensitivity. Insertion reads in a nonessential gene (corC, encoding a divalent cation efflux protein) showed little reduction with growth.
Fig. 2.
Evaluating essential gene mutant speed of growth arrest. (A) Recovery of mutants in essential and nonessential genes (Tn-seq sequence reads per gene normalized for gene length) relative to mutagenized DNA used for transformation. The results show the progressive loss of insertions in essential genes. Wild-type doubling time was 37 min. (B) Examples of essential genes with different rates of mutant loss. The positions and read counts of transposon insertions at two loci are shown for the mutagenized DNA and samples collected at different times following transformation and growth. The vertical bars above and below each horizontal line represent forward- and reverse-oriented transposon insertions, respectively, with a maximum vertical axis value of ∼10. Insertions in a nonessential gene upstream of lnt (corC) showed little orientation bias, indicating they are not polar on lnt. However, there was strong orientation bias for insertions situated between nrdA and nrdB and at the 3′ end of corC at later time points, indicating that reverse-oriented insertions were polar, perhaps because the missing orientation interferes with downstream translation initiation and leads to death.
We note two likely sources of heterogeneity in our assays of depletion sensitivities. First, since the bacteria transformed are in the exponential phase of growth, a fraction will carry multiple copies of target genes, and mutant phenotypes will only be expressed after segregation of wild-type copies from such cells. Second, the transformation process itself was carried out for 150 min, so that cells that obtained insertion mutations early in the period would experience more rapid depletion of the corresponding product than those that obtained them at the end of the period.
We addressed two technical issues as part of the study. First, to simplify the analysis, we wished to reduce potential polar effects of transposon insertions on the expression of downstream genes in operons. Accordingly, the transposon we constructed for TFNseq (T33) promotes downstream transcription in one insertion orientation (Materials and Methods). However, we found little evidence of polarity in either orientation in most cases. Among 73 nonessential genes immediately upstream of essential genes in operons, ∼70% exhibited approximately equal numbers of insertions in both orientations, for example corC (Fig. 2B). The findings indicate that polarity was not a significant complicating factor in the analysis. Second, we suspected that transforming DNA that had not recombined into the genome could contribute to insertions detected by sequencing. To help evaluate this possibility, we mutagenized a heterologous genomic DNA (from E. coli), which should be unable to recombine into the A. baylyi genome, and analyzed it after transformation by TFNseq. Although the DNA did indeed fail to form stable kanamycin-resistant recombinants in A. baylyi, insertions could be detected by sequencing at an early time point. Two hours after transformation ∼25% of the E. coli insertions could be detected relative to admixed A. baylyi control DNA. The finding indicates that TFNseq detects unrecombined DNA, which could thus contribute to an overestimation of A. baylyi mutant representation, particularly at early time points. The fastest-depleting genes would be most affected by this overestimation (and thus represent maximum values), but the order of depletion times for different genes should not be affected. In addition, in verification studies using qPCR to assay constructed deletion mutants, we also obtained results indicating that unrecombined DNA was not a significant determinant of the order of depletion sensitivity observed by TFNseq (discussed below).
Inactivation Sensitivities of Different Essential Processes.
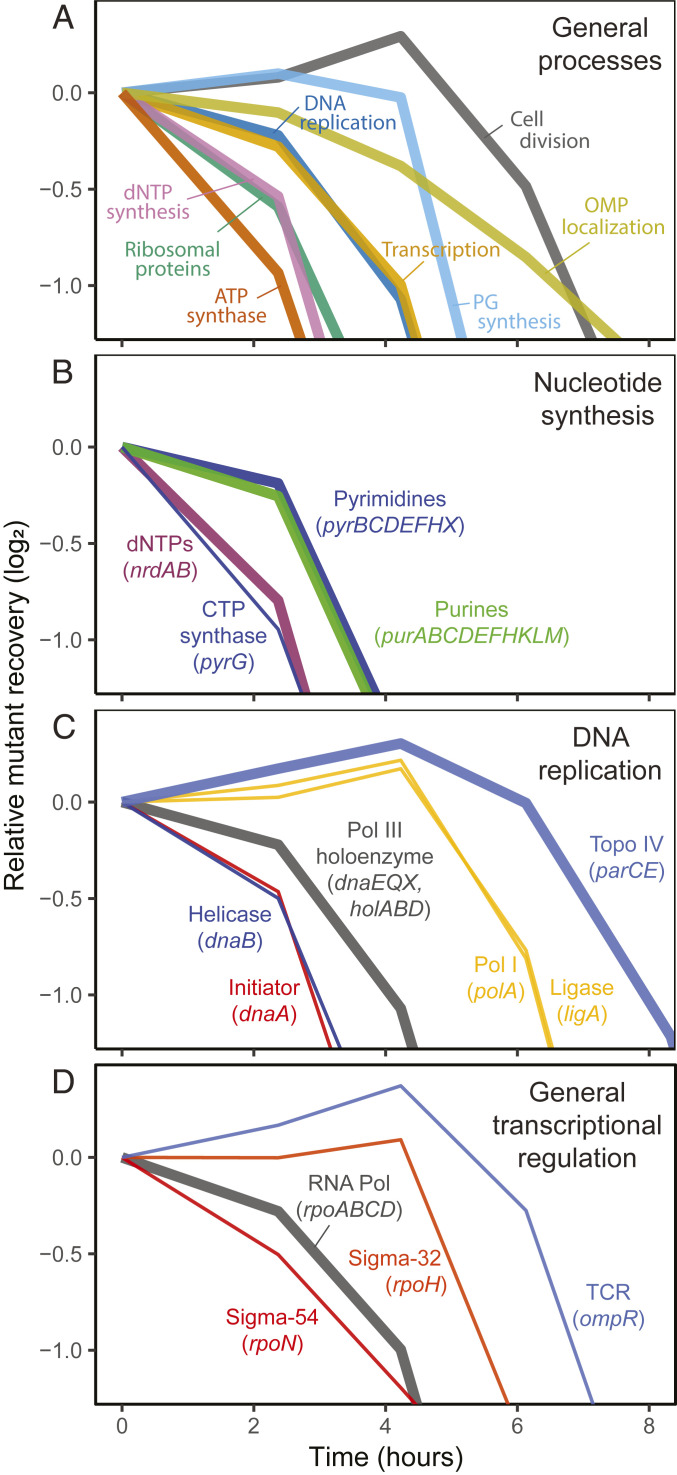
We defined 565 candidate essential genes by TFNseq based on the loss of their insertions after outgrowth on a minimal medium (Materials and Methods and Dataset S1). The set includes most (432/499; 86.6%) of the essential genes identified previously by single-gene deletion (21). Essential gene mutants were lost from the population at different rates during outgrowth following mutagenesis (Fig. 2A), and the results were reproducible in independent experiments (Dataset S1 and SI Appendix, Fig. S1A). Mutants could be grouped according to the essential processes they affected and 45 such processes ordered by how rapidly inactivation blocked growth (Figs. 3A and 4). For some processes, the loss of mutants from the population was relatively abrupt (e.g., peptidoglycan synthesis) (Fig. 3A), suggesting that growth is relatively normal until the corresponding products fall below a threshold. For others, the loss was more gradual (e.g., outer membrane protein localization), suggesting that depletion may slow replication gradually before stopping it altogether. It seems likely that for most of the processes other than small-molecule biosynthesis, growth arrest is irreversible and corresponds effectively to death of the mutant cells.
Fig. 3.
Mutant depletion kinetics. The loss of mutants with growth for selected essential processes is shown. (A) General processes. A subset of general processes is shown. (B) Nucleotide synthesis. (C) DNA replication. (D) General transcriptional regulation. The rpoN gene was nonessential in a set of deletion mutants (21), a result we confirmed. The essentiality seen in TFNseq may thus be due to a difference in the selective condition, such as created by the high density of plating. Thick lines represent average values from the multiple genes indicated, while thin lines represent individual genes. dNTP, deoxyribonucleotide triphosphate; PG, peptidoglycan; OMP, outer membrane protein; Pol III, DNA polymerase III; Pol I, DNA polymerase I; Topo IV, DNA topoisomerase IV; RNA Pol, RNA polymerase holoenzyme; TCR, two-component regulator.
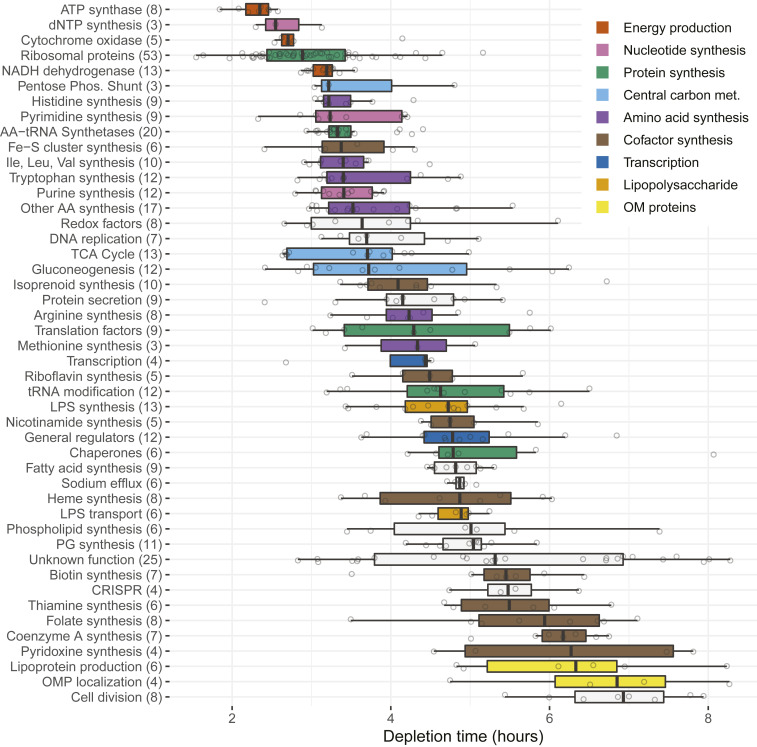
Fig. 4.
Mutant depletion times for different essential processes. The median and interquartile ranges of times required for twofold mutant depletion relative to the input mutagenized DNA are shown for the processes indicated. The central essential genes included for each process are listed in SI Appendix, Table S2, and the numbers of genes included for each process are indicated in parentheses. Colors indicate functionally related processes. Essential genes were assigned to biological processes primarily using KEGG2 Pathway Maps (https://www.genome.jp/kegg/pathway.html) and PGAT (22). dNTP, deoxyribonucleotide; LPS, lipopolysaccharide; PG, peptidoglycan; OMP, outer membrane protein.
Mutations blocking energy production arrested growth most rapidly and included genes encoding adenosine 5′-triphosphate (ATP) synthase, cytochrome oxidase, and reduced nicotinamide-adenine dinucleotide (NADH) dehydrogenase (Fig. 4). Among the genes needed for small molecule synthesis, the approximate order of mutant loss was nucleotides > amino acids > fatty acids > cofactors. Mutations in genes responsible for deoxyribonucleotide synthesis (ribonucleotide reductase and thymidylate synthetase) were particularly fast-acting. In cofactor synthesis, mutations blocking Fe–S cluster production led to faster depletion than those affecting other cofactors. Among functions needed for protein synthesis, mutations inactivating ribosomal proteins and aminoacyl-tRNA (transfer RNA) synthetases blocked growth faster than those affecting auxiliary factors (e.g., initiation factors), tRNA modification functions, and chaperones. In central carbon metabolism, mutations inactivating pentose shunt functions depleted faster than those affecting the tricarboxylic acid cycle and gluconeogenesis. Central replication and transcription functions depleted at intermediate rates, while mutants in functions needed for envelope synthesis (phospholipid, peptidoglycan, and outer membrane protein production) and cell division depleted slowly.
The depletion times of essential genes of unknown function varied over a wide range, presumably reflecting the fact that they affect a variety of different processes. The depletion times of mutants in individual genes should help identify the processes they affect. For example, the fastest-depleting such gene (ACIAD2193), with a depletion time of 2.9 h, is more likely to affect a rapidly depleting process like energy production or ribosome synthesis than a slowly depleting process like envelope synthesis or cell division. The opposite assignments are likely for the slowest-depleting gene of unknown function (ACIAD3039), which exhibited a depletion time of 8.2 h.
In the genes needed for purine and pyrimidine synthesis, there was an outlier (pyrG, encoding CTP synthetase) whose mutants depleted earlier than those for the other pyrimidine and purine biosynthetic genes (Fig. 3B). Perhaps a metabolic imbalance caused by the pyrG mutation is toxic. There was also heterogeneity in the inactivation sensitivities of DNA replication functions, with mutations affecting functions needed for replication fork formation and movement (dnaA, dnaB, and DNA polymerase holoenzyme genes) blocking growth faster than those needed for repair of lagging strand Okazaki fragments (polA, encoding DNA polymerase I, and ligA, encoding DNA ligase) and to decatenate replicated genomes (parCE, encoding topoisomerase IV) (Fig. 3C). Among general transcriptional regulators, mutations in rpoN (the nitrogen limitation sigma factor) blocked growth at about the same time as the vegetative RNA polymerase holoenzyme (rpoABCD), whereas mutations inactivating the heat shock sigma factor (rpoH) and an essential two-component regulator involved in outer membrane protein synthesis (ompR) were slower (Fig. 3D).
Validation of TFNseq Results with Deletion Mutants.
To help verify TFNseq findings, we examined a sample of 40 essential gene deletion mutations using qPCR. The mutations were generated by natural transformation of PCR fragments that replaced essential loci with a kanamycin resistance determinant, and the qPCR employed primers situated outside of the transformed DNA fragments to eliminate amplification signals due to unrecombined DNA (Materials and Methods). The mutant depletion times we observed using this assay agreed well with those found by TFNseq (SI Appendix, Fig. S1B). The results thus help validate the TFNseq results and indicate that sequences derived from unrecombined DNA do not seriously affect the order of depletion sensitivity.
Deletion Mutant Microcolonies.
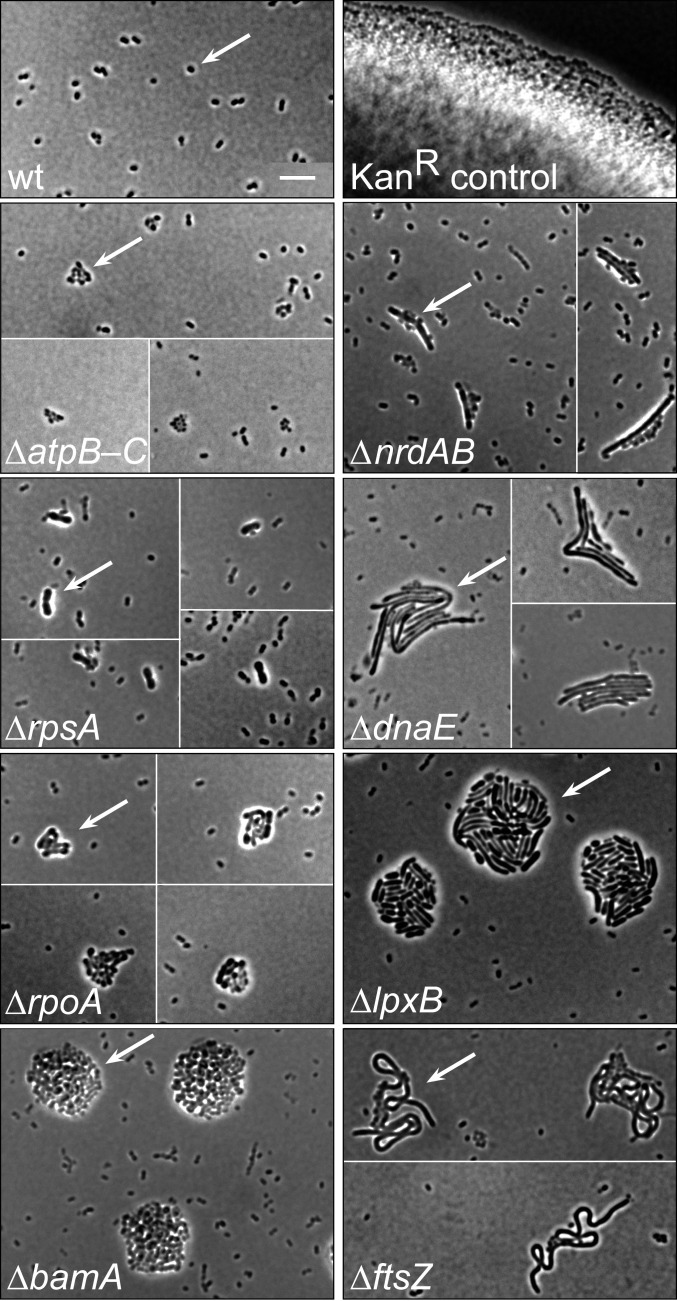
TFNseq assays changes in the relative amount of mutant DNA during growth, a measure of replication. As a complementary assay, we examined the microscopic appearance of microcolonies of dead cells formed by bacteria deleted of different essential genes (16). The sizes of such microcolonies reflect the increase in cell mass that occurs before loss of the targeted essential functions prevents further growth. To generate the mutant microcolonies, we transformed wild-type cells with PCR fragments designed to create deletion mutations marked with kanamycin resistance and plated them on medium containing the antibiotic (Materials and Methods). The untransformed wild type showed mostly singlets and doublets of unelongated cells due to rapid inhibition of growth by kanamycin, and a kanamycin-resistant control formed large colonies (Fig. 5). The essential gene mutants formed microcolonies whose sizes and cell morphologies varied, and for most of the genes microcolony sizes were consistent with depletion sensitivities seen by TFNseq (SI Appendix, Fig. S2). Mutations inactivating the ATP synthase (∆atpB-C), ribonucleotide reductase (nrdAB), and a ribosomal protein (rpsA) led to small microcolonies made up of few cells, while those inactivating the other genes led to larger microcolonies. The DNA polymerase (dnaE) mutants formed microcolonies that were somewhat larger than expected, perhaps because cells continued to elongate after DNA synthesis was blocked (23). A lipopolysaccharide biosynthesis mutant (∆lpxB) also formed somewhat larger colonies than expected from its depletion behavior. Overall, the results indicate that depletion sensitivities based on replication (TFNseq) are broadly consistent with those based on mutant cell mass increase (microcolony size) visualized microscopically.
Fig. 5.
Essential gene deletion mutant microcolonies. The microcolonies formed by deletion mutants generated by transformation with mutagenic PCR fragments and plating on selective (kanamycin-containing) medium are shown. Arrows indicate examples of microcolonies of each mutant. Panels showing the growth of wild type (kanamycin sensitive) and a kanamycin-resistant nonessential mutant (∆ACIAD0937) are shown for reference. The mutants are shown in order of increasing depletion times of the affected processes (Fig. 4): ∆atpB-C, ATP synthesis; ∆nrdAB, deoxyribonucleotide synthesis; ∆rpsA, ribosome synthesis; ∆dnaE, DNA replication; ∆rpoA, transcription; ∆bamA; outer membrane protein localization; ∆ftsZ, cell division. Note that a background of presumptive untransformed cells is seen in most images of mutants. Most mutants exhibited distinctive cell morphologies, and in many cases there appeared to be wild-type cells associated with mutants (e.g., ∆nrdAB). These cells may have segregated from transformants with multiple target genes, only one of which acquired the marked deletion. Deletion mutants of rplL and rplJKAL grew into colonies similar in size to that of rpsA, and microcolonies of all three deletions were similar in size whether marked with kanamycin resistance or ceftazadime resistance. In some cases, images of different fields were juxtaposed due to sparseness of microcolonies. ∆atpB-C corresponds to ∆atpBEFHAGDC. (Scale bar, 10 µm.)
Discussion
The principle finding of this study is that it is possible to order the essential processes of a bacterium based on how rapidly inactivation of the underlying genes blocks replication and growth. This order of “depletion sensitivity” presumably reflects how critical a process is for replication under a particular condition, how much in excess of the minimum required the underlying genes are expressed, and how rapidly the essential product turns over. It may be possible to help specify the relative importance of these contributions using complementary approaches, such as the analysis of temperature-sensitive alleles that rapidly inactivate functions of interest. We suspect that some elements of the depletion sensitivity order will be relatively general, and others will change with growth conditions, just as the sets of essential genes themselves do (4, 24). The procedures developed here thus provide a general approach to include essential genes in genome-scale mutant phenotyping studies.
Three of the most depletion-sensitive processes we identified are required for the generation of metabolic energy: ATP synthase, cytrochrome oxidase, and NADH dehydrogenase. Since A. baylyi is a nonfermenting obligate aerobe fully dependent on proton motive force and ATP synthase for producing ATP, the extreme inactivation sensitivities of these mutants presumably reflect the critical need for ATP replenishment for ongoing replication and growth. Indeed, metabolic models for A. baylyi identify ATP synthase and cytochrome oxidase as two functions exhibiting very high flux during growth under aerobic conditions (25), suggesting that this property could help account for their rapid mutant depletion times (SI Appendix, Table S3). Succinate dehydrogenase, which is required both for entry of succinate into central metabolism and as a major electron donor for respiration, also showed predicted high flux and fast mutant depletion.
The extreme inactivation sensitivities of functions needed for deoxyribonucleotide production (ribonucleotide reductase and thymidylate synthetase) likewise implies that these nucleotides rapidly deplete when new synthesis is reduced, inhibiting DNA replication. The findings support the proposal that ribonucleotide reductase activity is limiting for replication (26). The fast depletion phenotypes of mutants inactivating ribosomal proteins are more difficult to rationalize, although they do not appear to be due to the resistance marker employed (to kanamycin, which acts on ribosomes) since similar results were found with a β-lactam resistance marker. One interesting possibility is that ribosomes lacking individual proteins can be toxic to cells. The slow inactivating phenotypes of envelope and division mutants indicate that their inactivation only slowly feeds back on replication. The finding fits with the observation that filaments formed by inhibition of division have multiple genomes, indicative of continued replication in cells that fail to divide (11).
The results of the TFNseq screens were validated in two ways. In the first, a sample of deletion mutations of essential genes generated by transformation of PCR fragments was analyzed by qPCR in a manner that only detected integrated fragments. The order of depletion sensitivity agreed with that seen for the corresponding mutants in TFNseq. The results indicate that the TFNseq analysis was not compromised because it analyzed transposon mutants (as opposed to deletions) or due to its inability to distinguish recombined from unrecombined mutations after transformation. In the second validation experiments, the microcolonies of nongrowing cells formed by individual deletion mutants were examined. Although the size of the microcolonies reflects cell mass rather than DNA replication as a measure of growth, we found reasonable agreement between the microcolony and TFNseq assay results. Although not included this study, it should be feasible to quantify DNA amounts in individual mutant cells after staining (27).
TFNseq and other procedures we employed in this study relied on the efficient natural transformation competence of A. baylyi. Since natural competence is widely distributed in bacteria, including important pathogens like Vibrio cholera and Streptococcus pneumoniae (28, 29), it should be possible to extend approaches developed here to other species.
Finally, gene products whose inactivation most rapidly blocks growth under multiple conditions represent attractive antibiotic targets since partial pharmacological inhibition should also be effective at inhibiting growth. The order of depletion sensitivity thus provides a useful parameter to include in prioritizing potential targets.
Materials and Methods
Strains and Media.
A. baylyi ADP1 (15) was used throughout. Growth medium was M9 minimal medium supplemented with 15 mM Na succinate and 1 µM FeSO4. Solidified medium contained 1.5% Bacto-agar (Difco).
Transposon T33.
Transposon T33 is a 988-bp (base pair) transposon Tn5 derivative constructed for in vitro mutagenesis of ADP1 genomic DNA. The transposon carries the A. baumannii AB5075-UW aphA6 gene (encoding aminoglycoside 3′-phosphotransferase) and promoter and confers resistance to kanamycin and other aminoglycosides (30). Transposon T33 was constructed without the aphA6 transcription terminator so that transcription would extend into genes 3′ of insertion sites and minimize polar effects on downstream gene expression. We constructed T33 after observing that mutants produced by the commercially available transposon <KAN-2> (Lucigen) displayed strong orientation bias in their pattern of insertion into the ADP1 genome, such that it appeared that target gene promoters likely contributed to nptII expression. Transposon T33 exhibits no such coorientation bias in ADP1. The nucleotide sequence of transposon T33 is available at https://www.gs.washington.edu/labs/manoil/sequences.htm.
In Vitro Transposon Mutagenesis.
The ADP1 genomic DNA used for in vitro transposon mutagenesis was isolated by DNeasy Blood and Tissue Kit (Qiagen). Transposon T33 DNA was synthesized by amplifying the transposon by PCR using 5′-phosphorylated primers that corresponded to the exact transposon ends, followed by purifying the product by polyethylene glycol precipitation (31).
For mutagenesis, purified transposon T33 DNA in TE (Tris–ethylenediaminetetraacetic acid) buffer was mixed with 800 ng purified ADP1 genomic DNA at a molar ratio of ∼400:1 in the presence of 1× reaction buffer and one unit of EZ-Tn5 Transposase (Lucigen) in a 30-µL reaction volume, incubated for 2 h at 37 °C, and terminated by the addition of 1/10 volume 10× Stop Solution (Lucigen) and incubation at 70 °C for 10 min.
The mutagenesis reaction leaves short single-stranded gaps adjacent to the 5′ ends of the transposon in insertion products (32). In natural transformation, only one of the two DNA strands is used in recombination, and the other is degraded (28). Without repair, the gaps in the insertion products would thus eliminate genome homology on the 5′ side of the inserted transposon and prevent the formation of recombinants carrying transposon insertions. Indeed, we confirmed that, as expected, transformation by unrepaired product was highly inefficient at generating insertion mutants. To repair the gaps left by in vitro transposition, mutagenized genomic DNA (800 ng) was ethanol-precipitated and incubated for 2 h at 37 °C in a 50-µL reaction mixture with 1× ThermoPol Buffer (NEB), 1 mM dNTPs (NEB), 20× NAD+ (NEB), 0.8 µL of a 1:5 dilution (in NEB Dilution Buffer A) of Full Length Bst DNA Polymerase (NEB), and 1.6 µL of Taq DNA ligase (NEB). The mixture was stored at −20 °C prior to transformation.
Natural Transformation.
Transformation of ADP1 employed a previously described procedure (16). An overnight culture (optical density at 600 nm ≥1.2) of bacteria grown in M9 at 30 °C was diluted fivefold in the same media and incubated 60 to 70 min at 30 °C with agitation. A 260-µL aliquot of the resulting culture was then incubated with 100 to 300 ng transforming DNA for 150 min (for T33-mutagenized genomic DNA) or 70 min (for the mixture of deletion mutant constructs used for qPCR analysis and for individual deletion mutant constructs used for microscopy) at 30 °C with agitation. Glycerol was added to 10% (weight:volume) and the mixture was stored at −80 °C prior to plating.
Assay of Transposon Mutant Depletion during Growth.
To evaluate the kinetics of depletion of essential gene mutants from transposon mutant pools, aliquots of ADP1 transformed with T33-mutagenized genomic DNA were thawed and plated on M9 agar containing 20 µg/mL kanamycin. Platings corresponded to ∼450,000 kanamycin-resistant colonies per plate. The plates were incubated at 30° and at ∼2, 4, 6, and 8 h, and bacteria were harvested into M9–succinate medium. At these times, the population of mutant cells had doubled ∼0.5, 3.5, 6.5, and 10 times, respectively (SI Appendix, Table S1). Many more plates were harvested at early time points than at later points in order to collect sufficient mutant bacteria for saturation-level Tn-seq coverage, since at early time points mutant cells had multiplied minimally and represented only a small fraction of the total cellular population (SI Appendix, Table S1). Two 18-h mutant pools were generated from a lower plating density (∼2% of that used for the timed depletion assays) (SI Appendix, Table S1). Harvested bacteria were pelleted by centrifugation and the pellets frozen at −80 °C for later Tn-seq analysis.
We also analyzed by Tn-seq a sample of in vitro T33 mutagenized and repaired genomic DNA analogous to that used for transformation (SI Appendix, Table S1).
Tn-seq.
Tn-seq analysis employed the TdT method with minor modifications (31, 33). DNA purified from cell pellets by DNeasy Blood and Tissue Kit (Qiagen) was sheared to an average length of 300 bp using an M220 focused ultrasonicator (Covaris) and end-repaired with the NEBNext End Repair Module (NEB). Following purification (Qiagen MinElute), the DNA was C-tailed using terminal deoxynucleotidyl transferase (Promega) in the presence of 950 µM dCTP (Affymetrix) and 50 µM ddCTP (Affymetrix) by incubation for 60 min at 37 °C and 20 min at 75 °C then purified by Performa DTR gel filtration columns (Edge Biosystems). The purified C-tailed DNA served as template in a first round of PCR using primers olj376 (0.6 µM) and T33-66 (GGGGCGAATTCAAACATGAGGTGCGACA) (0.2 µM) and Kapa HiFi Hotstart ReadyMix (Kapa Biosystems) (cycling conditions: 2 min at 95 °C, then 22 cycles of 20 s at 98 °C, 25 s at 64 °C and 95 s at 72 °C). The first-round PCR product (1 to 3 µL) served as a template in a second round of real-time PCR using primer T33_ILMN_AmpF-4 (AATGATACGGCGACCACCGAGATCTACACTCTTTGAGGTGCGACAGTTTCAAAAGTGATGGTTGAGATGTGTATAAGAG) and one TdT_i8 primer per sample (for multiplexing) (31). The reaction conditions were the same as for the first round (with the addition of SYBR Green I [Invitrogen]), except that amplifications were stopped after a cycle in which at least 20% of full amplification had been achieved, but before the rate of exponential amplification had noticeably declined (by real-time fluorescence signal) (usually between cycles 5 and 10). Samples were combined as necessary for sequencing and fragment sizes ∼250 bp to 700 bp were isolated by acrylamide gel electrophoresis then purified by incubation of the gel slices in TE (56 °C for at least 2 h) and centrifugation though NanoSep MF 0.2 µm columns (Pall Life Sciences). Samples were sequenced on a MiSeq (Illumina) with a PhiX spike-in (Illumina) using 50-bp reads (and a 6-bp indexing read) and custom sequencing primer mix of T33_SEQ-6 (GAGGTGCGACAGTTTCAAAAGTGATGGTTGAGATGTGTATAAG) and primer PE_Read1_SEQ for reading the PhiX fragments. The 3′ end of T33_SEQ-6 corresponds to the sixth base before the end of transposon T33 in the amplified transposon–genome junctions, allowing sequence confirmation of the insertion junction in authentic amplicons. For additional primer sequences, see ref. 31.
Tn-seq Data Analysis.
Custom bioinformatics scripts (discussed below) were used for the following sequence read processing and analysis steps.
Reads counts per gene were calculated by summing reads from all sites within each gene’s “functional window,” defined for >97% of genes as the complete open reading frame (ORF) except the 5′ 5% (maximum 45 bases) and the 3′ 10% (maximum 90 bases). For ∼3% of genes that displayed negative selection within portions of the ORF (e.g., within a putative domain), we manually adjusted the functional window based on the pattern of hits in the primary 18-h mutant pool sample. We used hidden Markov model analysis of total read counts per 10-bp segments of the chromosome (34) to help define the essential or growth defective regions within each gene.
To compare samples quantitatively (i.e., the individual time points, the 18-h outgrown samples, and the pretransformation DNA), we normalized each sample’s read counts using a method akin to one used for RNA-sequencing analysis (35): We generated a “pseudo reference sample” by calculating the geometric mean of reads per gene per gene length for all genes with nonzero read counts within their functional windows in all samples (>85% of genes). We calculated a size factor for each sample as the median of the ratios of the sample gene read counts to the pseudo reference sample gene read counts for the same subset of genes. We then normalized the gene and insertion site read counts for each sample by dividing by the sample’s size factor.
To quantify mutant depletion rates and to define essential genes, we defined a metric called “delta log reads,” computed for each gene and sample as the difference in log2 (normalized reads per gene per gene length) relative to the pretransformation sample (Dataset S1). Depletion times were defined as the interpolated time at which the delta log reads fell permanently below a value of −1 (i.e., a twofold depletion relative to the pretransformation sample).
Essential genes (including 17 genes with essential domains corresponding to less than 60% of the length of the ORF) were defined as those whose transposon insertion sequence reads were significantly depleted within their functional windows (discussed above) in the 8-h time points in both time course assays and in both 16-h outgrown samples. Significant depletion corresponded to one-sided P values (without correction for multiple comparisons) of less than 10−5 for a normal curve fit to the major peak in the distribution of the delta log reads values for all genes. The normal curve was defined by the mean and SD of all values that fell within histogram bins (of 201 bins between values of −3 and 1) with frequency of at least 10% of the average bin frequency for the top 2% of bins, excluding the bin for zero reads. These constraints were intended to exclude bins for nongrowing and slow-growing mutants. For most samples, a P value of 10−5 corresponded to a delta log reads value close to −1. The analysis defined 571 genes. We further excluded six genes (ACIAD1220, ACIAD1322, ACIAD2029 [prmB], ACIAD2432 [rplI], ACIAD3368 [mraW] and ACIAD3570 [ribB1]) that were immediately upstream of essential genes and which displayed strong depletion only for reverse-oriented insertions, implying that their “essentiality” was due to polar effects of these insertions on the downstream essential genes.
Analysis of Deletion Mutant Depletion Rates by qPCR.
ADP1 was transformed with a mixture of targeted deletion constructs representing 41 essential and 2 nonessential loci (SI Appendix, Table S4). Each construct consisted of the aphA6 resistance cassette (promoter plus aphA6 coding sequence) flanked by ∼1,000-bp sequences representing the chromosomal sequences flanking the target gene or genes. Frozen transformation mixtures were thawed, plated on M9 agar containing 15 µg/mL kanamycin, and grown and harvested as described above for the transposon mutant depletion assays (with harvest time points at 2, 3, 4, 5.5, 7, and 9 h). A portion of the combined transformation mixes was also kept as the “inoculum” sample). For the inoculum sample and for each time point, the quantity of each essential-locus deletion allele relative to that of a nonessential control locus (d143; SI Appendix, Table S4) was determined by qPCR. The locus-specific amplification primer used in each qPCR reaction was designed to anneal to a chromosomal location outside of the ∼1000-bp region used as flanking sequence in the corresponding deletion construct used for transformation. The other primer in each reaction, T33alt1-28 (TGATGGTTGAGATGTGTATAAGTCAGTC), annealed within the aphA6 cassette. Thus, the qPCR detected only deletion alleles that had recombined into the chromosome and not input PCR fragments used to generate the deletions. Each qPCR reaction was performed in duplicate or triplicate using a Bio-Rad MiniOpticon instrument, a standard curve was performed for quantification of each unique amplicon, and standard methods were used to calculate initial template quantities.
The rate of mutant depletion for each targeted deletion was calculated as the rate of change of the log ratio of the allele of interest to the control allele by least-squares linear regression.
Microcolony Microscopy.
Bacterial microcolonies were routinely imaged after growth on minimal-succinate agar supplemented with 120 µg/mL kanamycin in 15- × 60-mm-diameter Petri plates under bright field illumination using a Nikon Eclipse 90i with an ELWD 20× objective equipped with 2× digital zoom.
Supplementary Material
Acknowledgments
We thank Pradeep Singh and Carrie Harwood for helpful comments on the manuscript. This work was supported by NIH Grants 1R01AI136904 and 1R01AI148208.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The Tn-seq sequence reads analyzed in this study are available at the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA600030). The bioinformatic scripts used for TFNseq read processing and analysis are available in GitHub at https://github.com/lg9/TFNseq_2020.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001507117/-/DCSupplemental.
Data Availability.
The Tn-seq sequence reads analyzed in this study are available at the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) (PRJNA600030) (36). The bioinformatic scripts used for TFNseq read processing and analysis are available at https://github.com/lg9/TFNseq_2020.
References
- 1.Kong X. et al., ePath: An online database towards comprehensive essential gene annotation for prokaryotes. Sci. Rep. 9, 12949 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rousset F. et al., Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. PLoS Genet. 14, e1007749 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodall E. C. A. et al., The essential genome of Escherichia coli K-12. MBio 9, e02096-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S. A. et al., General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112, 5189–5194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price M. N. et al., Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557, 503–509 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Peters J. M. et al., A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165, 1493–1506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caro F., Place N. M., Mekalanos J. J., Analysis of lipoprotein transport depletion in Vibrio cholerae using CRISPRi. Proc. Natl. Acad. Sci. U.S.A. 116, 17013–17022 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X. et al., High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol. Syst. Biol. 13, 931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T. et al., Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat. Commun. 9, 2475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Projan S. J., Whither antibacterial drug discovery? Drug Discov. Today 13, 279–280 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Hirota Y., Mordoh J., Jacob F., On the process of cellular division in Escherichia coli. 3. Thermosensitive mutants of Escherichia coli altered in the process of DNA initiation. J. Mol. Biol. 53, 369–387 (1970). [DOI] [PubMed] [Google Scholar]
- 12.Vento J. M., Crook N., Beisel C. L., Barriers to genome editing with CRISPR in bacteria. J. Ind. Microbiol. Biotechnol. 46, 1327–1341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui L. et al., A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 9, 1912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott K. T., Neidle E. L., Acinetobacter baylyi ADP1: Transforming the choice of model organism. IUBMB Life 63, 1075–1080 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Barbe V. et al., Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32, 5766–5779 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey J. et al., Essential gene deletions producing gigantic bacteria. PLoS Genet. 15, e1008195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin L., Ringel P. D., Vettiger A., Durr L., Basler M., DNA uptake upon T6SS-dependent prey cell lysis induces SOS response and reduces fitness of acinetobacter baylyi. Cell Rep. 29, 1633–1644.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Cuff L. E. et al., Analysis of IS1236-mediated gene amplification events in Acinetobacter baylyi ADP1. J. Bacteriol. 194, 4395–4405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng P., Leonard S. P., Mishler D. M., Barrick J. E., Synthetic genome defenses against selfish DNA elements stabilize engineered bacteria against evolutionary failure. ACS Synth. Biol. 8, 521–531 (2019). [DOI] [PubMed] [Google Scholar]
- 20.van Opijnen T., Camilli A., Transposon insertion sequencing: A new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Berardinis V. et al., A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 4, 174 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brittnacher M. J. et al., PGAT: A multistrain analysis resource for microbial genomes. Bioinformatics 27, 2429–2430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirota Y., Ryter A., Jacob F., Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harb. Symp. Quant. Biol. 33, 677–693 (1968). [DOI] [PubMed] [Google Scholar]
- 24.Baba T. et al., Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006 0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durot M. et al., Iterative reconstruction of a global metabolic model of Acinetobacter baylyi ADP1 using high-throughput growth phenotype and gene essentiality data. BMC Syst. Biol. 2, 85 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrick J., Sclavi B., Ribonucleotide reductase and the regulation of DNA replication: An old story and an ancient heritage. Mol. Microbiol. 63, 22–34 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Nonejuie P., Burkart M., Pogliano K., Pogliano J., Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. U.S.A. 110, 16169–16174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston C., Martin B., Fichant G., Polard P., Claverys J. P., Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Dalia A. B., McDonough E., Camilli A., Multiplex genome editing by natural transformation. Proc. Natl. Acad. Sci. U.S.A. 111, 8937–8942 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher L. A. et al., Resources for genetic and genomic analysis of emerging pathogen acinetobacter baumannii. J. Bacteriol. 197, 2027–2035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallagher L. A., Methods for Tn-Seq analysis in acinetobacter baumannii. Methods Mol. Biol. 1946, 115–134 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Reznikoff W. S., Transposon Tn5. Annu. Rev. Genet. 42, 269–286 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Klein B. A. et al., Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics 13, 578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeJesus M. A., Ioerger T. R., A Hidden Markov Model for identifying essential and growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinformatics 14, 303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallagher L. A., Essential genes TFNseq. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/PRJNA600030. Deposited 9 January 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Tn-seq sequence reads analyzed in this study are available at the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) (PRJNA600030) (36). The bioinformatic scripts used for TFNseq read processing and analysis are available at https://github.com/lg9/TFNseq_2020.