Significance
Here, we discover prototypical pacemaker neurons in the ancient cnidarian Hydra and provide evidence for a direct interaction of these neurons with the commensal microbiota. We uncover a remarkable gene-expression program conservation between the Hydra pacemaker neurons and pacemaker cells in Caenorhabditis elegans and the mammalian gut. We suggest that prototypical pacemaker cells emerged as neurons using components of innate immunity to interact with the microbial environment and ion channels to generate rhythmic contractions. The communication of pacemaker neurons with the microbiota represents a mechanistic link between the gut microbiota and gut motility. Our discoveries improve the understanding of the archetypical properties of the enteric nervous systems, which are perturbed in human dysmotility-related conditions.
Keywords: pacemaker neuron, Hydra, ion channel, microbiome, antimicrobial peptide
Abstract
Pacemaker neurons exert control over neuronal circuit function by their intrinsic ability to generate rhythmic bursts of action potential. Recent work has identified rhythmic gut contractions in human, mice, and hydra to be dependent on both neurons and the resident microbiota. However, little is known about the evolutionary origin of these neurons and their interaction with microbes. In this study, we identified and functionally characterized prototypical ANO/SCN/TRPM ion channel-expressing pacemaker cells in the basal metazoan Hydra by using a combination of single-cell transcriptomics, immunochemistry, and functional experiments. Unexpectedly, these prototypical pacemaker neurons express a rich set of immune-related genes mediating their interaction with the microbial environment. Furthermore, functional experiments gave a strong support to a model of the evolutionary emergence of pacemaker cells as neurons using components of innate immunity to interact with the microbial environment and ion channels to generate rhythmic contractions.
The enteric nervous system (ENS) coordinates the major functions of the gastrointestinal tract (1). In all extant animals, the structurally conserved ENS is a diffuse nerve net located within the wall of the gastrointestinal tract. In prebilaterian animals, such as Hydra, the nervous system is structurally simple and thus has great potential to inform us about evolutionary ancient basic structural and functional principles of neural circuits (2) (Fig. 1A). The principal function of the ENS is coordination of the rhythmic intestine motility, known as peristalsis, that occurs ubiquitously in the animal kingdom (Fig. 1A) and is driven by rhythmic electrical pulses generated by pacemaker cells (3). In mammals, proper functioning of the interstitial cells of Cajal (ICC), that serve as pacemakers in the muscular wall of the gastrointestinal tract (4–6), is essential for normal gut motility (7–9). Dysfunction of the pacemaker system contributes to functional gastrointestinal disorders, such as irritable bowel syndrome (IBS), chronic constipation, and intestinal pseudo-obstruction (9–12). The Ca2+-activated Cl−-channel Anoctamin-1 (encoded by the ANO1 gene) and the voltage-gated Na+-channel Nav1.5 (SCN5A gene) represent molecular markers of the interstitial pacemaker cells in human and mice (13–15), and DNA variants in the corresponding genes have been shown to associate with increased risk of IBS (16–18). Ion channel dysfunction (channelopathy) appears to be a plausible pathogenetic mechanism in functional gastrointestinal disorders (19), as the transient receptor potential cation channel TRPM8 (known as the cold and menthol receptor) and other ion channels have also been implicated in IBS susceptibility and gut dysmotility (20–23). Spontaneous contractile gut activities are not only affected by microbes. In fact, there is evidence that bacterial population dynamics themselves are affected by the periodic stimulation (24). Previous studies in Hydra suggested that the rhythmic peristaltic movements of the body column are dependent on neurons (25) and that they are modulated by the host-associated microbiota since germ-free (GF) animals display reduced and less-regular contraction frequencies (26). Single-cell RNA sequencing (scRNA-seq) uncovered neuron-specific transcriptional signatures and the presence of distinct neuronal subtypes (27). Little, however, is known about the nature of the neurons that generate peristaltic movement in a prebilaterian animal and how such prototypical neurons engage with the resident microbiota.
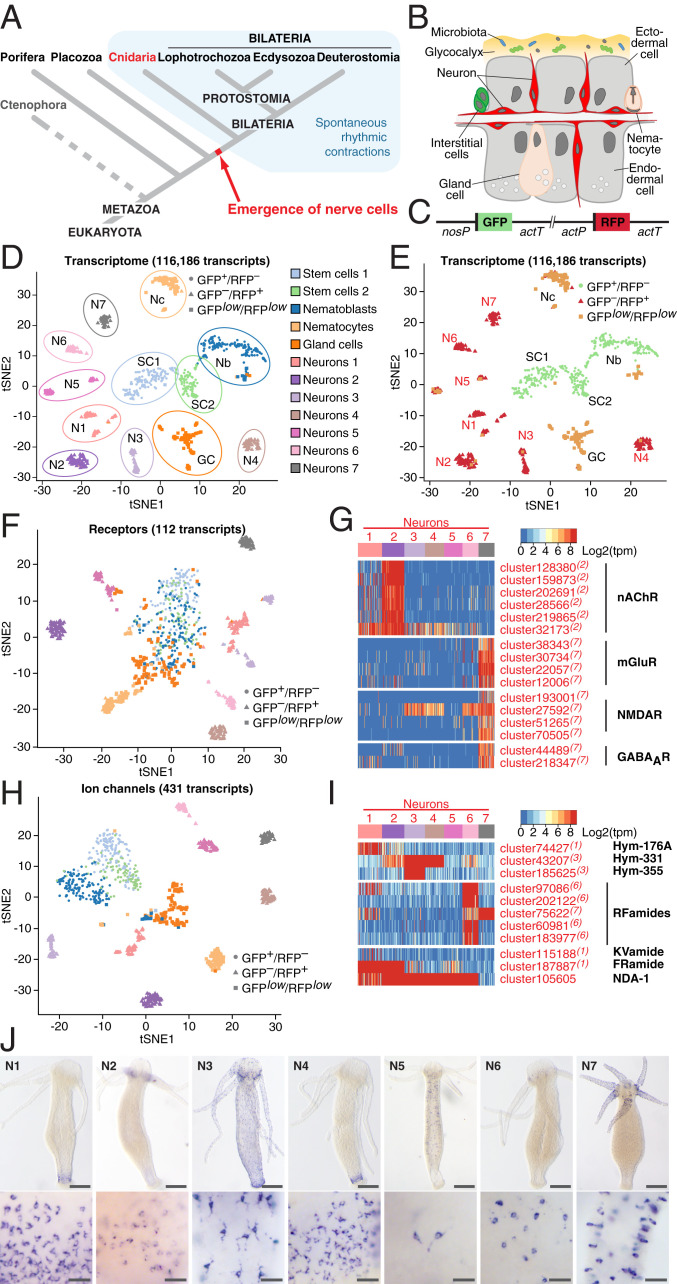
Fig. 1.
Single-cell transcriptome profiling uncovers the molecular anatomy of Hydra nervous system. (A) Emergence of the first nerve cells preceded the divergence of Cnidaria and Bilateria. Cnidarians possess structurally simple nervous systems and offer a great potential to reveal the fundamental structural and functional principles of neural circuits. Spontaneous rhythmic contractions are ubiquitously observed in Eumetazoa. (B) The Hydra body is made of three cell lineages: The ectodermal and endodermal epithelia separated by the extracellular matrix and the lineage of interstitial cells. The outer surface of the ectoderm is covered by a glycocalyx that serves as a habitat for symbiotic bacteria. The endoderm lining the gastric cavity is free of glycocalyx and stable microbiota. Two nerve nets made of sensory and ganglion neurons are embedded within both epithelia. (C) Genetic construct used to generate transgenic Hydra polyps and differentially label cells within the interstitial lineage by a combination of two fluorescent proteins: GFP expressed under a stem cell-specific nanos promoter (nosP), and RFP driven by the actin promoter (actP) active in terminally differentiated neurons. Both cassettes are flanked by the actin terminator (actT). (D) t-Distributed stochastic neighbor embedding (t-SNE) map constructed by dimensionality-reduction principal component analysis defined by highly covariable genes (Materials and Methods). A total of 928 cells were partitioned in 12 clusters and colored by their cell-type identities inferred from expressed proliferation and cell-type–specific marker genes (Datasets S5 and S6). (E) t-SNE map based on analysis of the entire transcriptome made of 116,186 transcripts segregates 12 clusters, including 7 subpopulations of neurons. Cells are color-coded by their phenotype captured by FACS upon sorting. (F) t-SNE map based on expression analysis of 112 transcripts coding for putative neurotransmitter receptors (Dataset S7). Seven neuronal populations are clearly segregated, indicating that each neuronal population is characterized by a specific set of receptors. (G) Heatmap illustrating expression of genes coding for putative nAChR, mGluR and NMDAR, and GABAAR within seven neuronal populations. Expression within the entire interstitial lineage is presented in SI Appendix, Fig. S3. Transcripts specifically up-regulated in the neurons are labeled red; superscript numbers indicate the nerve cell cluster (N1–N7) where the transcripts are significantly (adjusted P < 0.05) enriched. (H) t-SNE map constructed by expression analysis of 431 transcripts coding for putative ion channels (Dataset S8). Seven neuronal populations are clearly segregated, suggesting that each neuronal population is characterized by a specific set of channels. (I) Heatmap illustrates expression of 11 genes coding for main known neuropeptides in Hydra. Each neuronal population expresses a unique combination of neuropeptides. (J) In situ hybridization for marker genes strongly enriched in each of seven nerve cells clusters (N1–N7) (SI Appendix, Fig. S6) reveals that seven neuronal subpopulations reside in spatially restricted domains along the body column of Hydra (Scale bars, 100 µm upper panel, 25 µm lower panel.)
Here, we provide a definition of prototypical pacemaker cells, which integrates marker genes discovered in human dysmotility patients with the recent discovery that spontaneous contractile activities are affected by microbes. The functional experiments connected the rhythm generation and interactions with microbes at the level of this specific neuronal population. These findings shed new light on the evolution of pacemaker neurons, emphasize the role of the microbial environment in dysmotility, and underscore the importance of cross-species comparisons in tracking cell type evolution.
Results
Identification of Pacemaker Cells in Hydra Using Human Orthologous Genes.
Previous studies in Hydra suggested that a subpopulation of neurons located in the head region (28–30) might have properties of pacemaker cells to control the regularity of spontaneous body contractions. To gain insight into this specific cell population, we first assessed the molecular and functional diversity of the neuronal populations by scRNA-seq using red fluorescent protein (RFP)-labeled neurons (Fig. 1 B–J and SI Appendix, Figs. S1 and S2). Similar to the analysis of the entire transcriptome, the expression profile of 112 transcripts coding for putative neurotransmitter receptors clearly identified seven distinct clusters of neurons and separated them from the stem cells and nonneuronal cells (Fig. 1 D–F). This indicates that each neuronal subpopulation is characterized by a specific set of neurotransmitter receptors (Fig. 1G and SI Appendix, Fig. S3). For example, most transcripts coding for putative nicotinic acetylcholine receptors (nAChRs) were expressed almost exclusively in neuronal subpopulation N2 (Fig. 1G and SI Appendix, Fig. S3), while diverse homologs of muscarinic and N-methyl-D-aspartate glutamate receptors (mGluRs and NMDARs), and γ-aminobutyric acid type-A receptors (GABAARs) were enriched in the neuronal subpopulation N7 (Fig. 1G and SI Appendix, Fig. S3). Similarly, the expression profiles of 431 transcripts coding for ion channels clearly segregated GFP+/RFP– stem cells from the GFPlow/RFPlow nonneuronal cells and the seven remarkably distinct clusters of GFP–/RFP+ neurons (Fig. 1H). In addition, each neuronal subpopulation was found to express a unique combination of neuropeptides (Fig. 1I and SI Appendix, Fig. S4). The Hym-355 neuropeptide precursor gene, for example, was exclusively expressed in the neuronal subpopulation N3, while Hym-176A transcripts were discovered predominantly in the N1 subpopulation. Most of RFamide precursor transcripts were found in subpopulation N6 (Fig. 1I and SI Appendix, Fig. S4). Genes coding for some other peptides, including Hym-331 and FRamide, were expressed in two or more neuronal subpopulations. Taken together, these observations indicate that the molecular identity of different subpopulations of neurons is determined by the specific expression of ion channels, neurotransmitter receptors, and neuropeptides.
Interestingly, based on the expression profiles of 364 transcripts coding for putative transcription factors (TFs), all seven subpopulations of neurons express a common set of TFs, which separates them from stem cells and nonneuronal cell types (SI Appendix, Fig. S5). The neuron-specific TF signature consists mainly of Zn-finger, homeodomain and helix–loop–helix DNA-binding proteins, including the Achaete-scute homologous TFs. Furthermore, each neuronal population is characterized by a combinatorial expression of few genes encoding other TFs, such as the homologs of Aristales, NeuroD, and Orthopedia (SI Appendix, Fig. S5 B and C and Dataset S1).
In situ hybridization for selected marker genes enriched in each of the seven neuronal subpopulations (Fig. 1J and SI Appendix, Fig. S6A) showed that the neuronal subpopulations in Hydra reside in spatially restricted domains along the body column. Neuronal clusters N1 were found confined to the foot region of the polyp (Fig. 1J and SI Appendix, Fig. S6B). Cluster N2 was represented by a population of neurons located in the base of tentacles. N3-specific neurons were spread in the ectoderm along the entire Hydra body, whereas neurons of subpopulations N4 and N5 were found in the endodermal epithelial layer (Fig. 1J and SI Appendix, Fig. S6B). Neurons from the cluster N6 were confined to the hypostome of a polyp, whereas neurons of the cluster N7 were restricted to the tentacles. Notably, most of the seven spatially restricted neuronal subpopulations contain both sensory and ganglion cells (Fig. 1J). Analysis of marker gene expression (SI Appendix, Fig. S7) indicated a clear correspondence between the neuronal clusters identified by us and the subtypes reported by Siebert et al. (27).
To identify the pacemaker cells among the neurons in Hydra, we focused on a few human orthologs known to be either restricted in their expression to human ICCs (ANO1 and SCN5A) (13, 14) or mechanistically involved in the control of gut motility via circular smooth muscle cell contractions (menthol sensitive Ca2+ channels such as TRPM8) (31) (Fig. 2A). Homology search and phylogenetic analysis uncovered three Hydra genes coding for SCN-like sodium channels, six homologs of ANO1-like chloride channels, and four homologs encoding TRPM-like cation channels, which are remarkably similar to their human counterparts (Fig. 2 B–D and SI Appendix, Figs. S8–S10). Analysis of the single-cell transcriptome (Fig. 2E and SI Appendix, Fig. S11) revealed that the expression of genes encoding SCN-, ANO1-, and TRPM-like channels overall was very weak, often restricted to only few cells. However, several of the transcripts coding for SCN- and ANO1-homologs were more expressed in neurons (Figs. 2E and SI Appendix, Fig. S11), with some of the transcripts specific for neuronal subpopulation N2, which is located at the base of tentacles (Figs. 1J and 2E and SI Appendix, Fig. S11). Real-time PCR confirmed (Fig. 2F) that most of the SCN-, ANO1-, and TRPM-like ion channel genes are up-regulated in the head region. Transcripts of two of these genes, cluster2505 and cluster30856, coding for ANO1-like and SCN-like channels, could be detected by in situ hybridization at the base of tentacles (Fig. 2 G and H). Immunohistochemical analysis using specific antibodies raised against synthetic peptides confirmed the presence of ANO1-like and SCN-like channel proteins at the base of the tentacles (Fig. 2 I–K) in the subpopulation N2 domain (Fig. 1J). High-magnification confocal microscopy identified the cells expressing SCN and ANO1 channels as neurons (Fig. 2 L and M). Taken together, these observations indicate that the genes encoding SCN-, and ANO1-, and TRPM-like channels are expressed in a population of nerve cells resident in the subpopulation N2 at the base of tentacles (Fig. 2N).
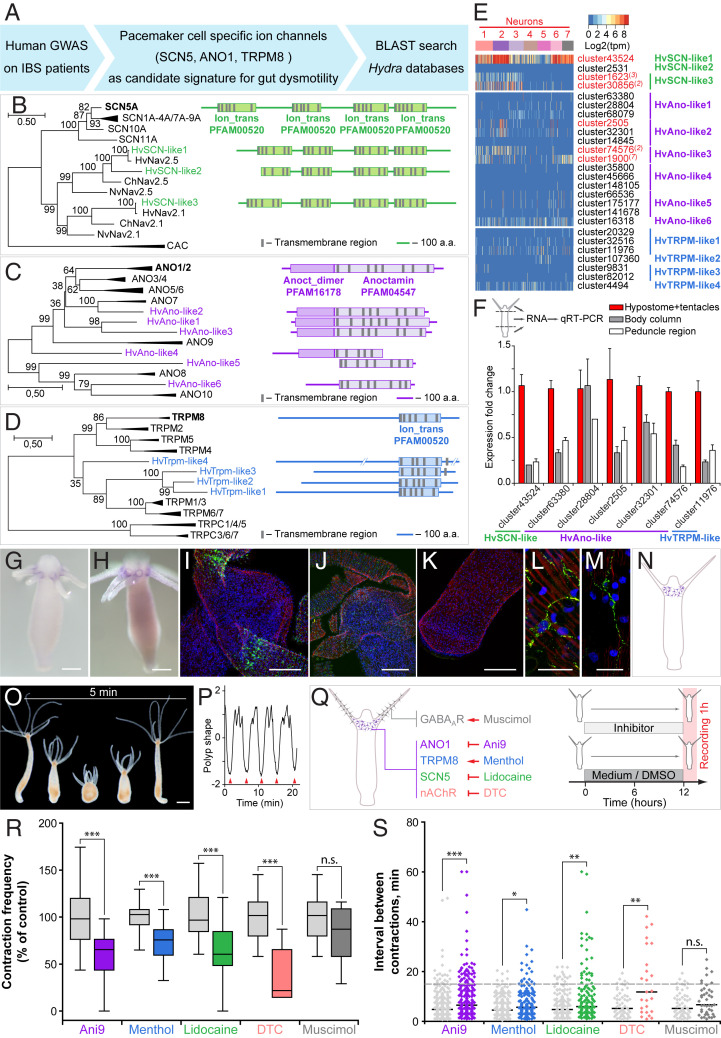
Fig. 2.
Identification of Hydra pacemaker cells using orthologs of human ion channels. (A) Genome-wide association studies on patients with gut motility disorders, such as IBS identified ion channels SCN5, ANO1, and TRPM8 that are expressed in human pacemaker cells (ICCs) and found to be essential for gut motility control. A BLAST search was used to identify the homologous genes in Hydra. (B–D) Pacemaker-specific ion channels are highly conserved in Hydra. Phylogenetic tree and domain structure of human SCN (B), ANO1 (C), and TRPM (D) channels and the orthologs from Hydra (Hv). Additionally, sequences from other cnidarians, Nematostella vectensis (Nv) and Clytia hemisphaerica (Ch) are included into the phylogenetic analysis. Noncollapsed trees are presented in SI Appendix, Figs. S8–S10. The topology and domain structure of three Hydra SCN-like sodium channels, six ANO1-like chloride channels and four homologs of TRPM-like cation channels are remarkably similar to their human counterparts. (E) Expression of genes encoding SCN-, ANO1-, and TRPM-like channels in Hydra single-cell dataset is overall very weak and restricted to only few cells. However, several transcripts coding for SCN and ANO1 homologs are significantly up-regulated in neurons (red) with some of the transcripts specifically enriched in the neuronal subpopulation N2 (red superscript). (F) Expression levels of most transcripts coding for SCN-, ANO1-, and TRPM-like channels are highest in the head region of Hydra, including the hypostome and tentacles, as revealed by real-time PCR analysis. Mean ± SEM, n = 3 to 6. (G and H) In situ hybridization with two probes specific for the transcripts cluster2505 (coding for an ANO1-like channel) and cluster30856 (SCN-like), confirms their localized expression in the base of tentacles. (Scale bars, 100 µm.) (I–M) Immunohistochemical analysis confirmed the presence of ANO1-like (I and L) and SCN-like (J and M) channels in the neurons of the subpopulation N2 at the base of tentacles. No signal can be detected in the peduncle region of Hydra (K). (Scale bars, 20 µm in L and M and 50 µm in I, J, and K.) (N) Taken together, the expression of gut dysmotility-associated ion channels identifies the nerve cell population N2 resident at the base of tentacles as putative pacemakers in Hydra. Pharmacological interference experiments corroborate the essential role of ANO-, SCN-, and TRPM-like channels and nicotinic acetylcholine receptors expressed in the neuronal population N2 in pacemaker activity in Hydra. (O) Hydra demonstrates spontaneous rhythmic contractions followed by body extensions that occur on average every 5 min. (Scale bar, 200 µm.) (P) The contraction pattern was video-recorded and transformed into a diagram of polyp shape to assess the contraction frequency, defined as the number of full body contractions (red arrowheads) that occurred within 1 h, and the time intervals between consecutive contractions. (Q) Polyps were exposed to chemicals specifically modulating the activity of the ANO1-, SCN-, and TRPM-like ion channels and nAChR expressed in the neuronal population N2. Experimental design: Hydra polyps were treated for 12 h prior to a 1-h recording of contractions. Polyps incubated in 0.16% DMSO-supplemented (for Ani9) or pure (other chemicals) Hydra medium served as control. (R) Contraction frequency is reduced in the presence of all chemicals targeting the channels expressed on the pacemaker population N2, but not affected in the presence of muscimol, which likely interferes with the population N7. (S) Modulating the activity of the pacemaker-specific ANO1-, SCN-, and TRPM-like channels and nAChRs in Hydra also disturbs the rhythmicity of spontaneous contractions, since the intervals between contractions become longer and less regular. Sampling size: n = 10 to 49 animals (contraction frequency), n = 25 to 283 intervals (interval length), *P < 0.05; **P < 0.005; ***P < 0.0005; n.s., P > 0.05.
ANO1-, SCN-, and TRPM-like Channels Are Essential for Pacemaker Activity in Hydra.
We next tested the role of the neuronal subpopulation N2-specific ANO1-, SCN-, and TRPM-like channels in controlling the pacemaker-driven rhythmic spontaneous contractions in Hydra (Fig. 2 O–Q). Exposing polyps to Ani9, a potent inhibitor of ANO1 channels (32), resulted in both a twofold reduction of the contraction frequency (Fig. 2 Q and R) and also in less regular contractions compared to controls (Fig. 2S). Similar results were obtained upon treatment of polyps with menthol, which activates TRPM8 channels in vertebrates (33), and lidocaine, which interferes with SCN-like ion channels (Fig. 2 Q–S). The results show that modulating the activity of the neuron-specific ANO1-, SCN-, and TRPM-like channels in Hydra greatly disturbs the rhythmicity of spontaneous contractions. Since neuronal subpopulation N2 is also characterized by the expression of putative nAChRs (Fig. 1G and SI Appendix, Fig. S3), we next tested the effects of tubocurarine (DTC), known as a potent antagonist of nAChRs (34, 35), on the frequency and rhythmicity of the spontaneous contractions in Hydra (Fig. 2Q). We found that the presence of DTC strongly reduced the frequency and affected the regularity of the spontaneous contractions (Fig. 2 R and S). Other reflexes dependent on neural circuits, such as the feeding reflex (36), were not affected by most of the channel-specific inhibitors (SI Appendix, Fig. S12), indicating a specific role of the ion channels expressed in the N2 neurons in controlling rhythmic body contractions. Inhibiting GABAA receptors specifically expressed in neurons of subpopulation N7 and absent in the subpopulation N2 (Figs. 1G and 2Q and SI Appendix, Fig. S3) had no effect onto the contraction pattern (Fig. 2 R and S) but strongly influenced the feeding response (SI Appendix, Fig. S12).
Together with the previous pharmacological findings (36), our observations unequivocally identify the pacemaker population N2 as cholinergic, and the neuronal population N7 controlling the feeding response as predominantly GABAergic. Notably, all cells within neuronal population N2 homogeneously expressed high levels of one of the innexin genes, cluster41630 (SI Appendix, Fig. S13). Innexins are the only components of gap-junction complexes known in Hydra (37, 38). Homotypic gap junctions established between neurons of the N2 subpopulation therefore may electrically couple these neurons and allow generation of a neural network with pacemaker properties. In a similar way, a net of neurons present in the peduncle of Hydra is likely coupled by gap junctions (39), and the epithelial cells of both the ectoderm and endoderm are electrically coupled by gap junctions (40–42). In sum, these data suggest that neurons of the N2 subpopulation express marker genes for gut dysmotility (Fig. 2 A–N) and that inhibition of these ion channels disturbs spontaneous and regular body contractions (Fig. 2 O–S). Therefore, neurons of the N2 subpopulation may act as pacemaker cells controlling the spontaneous contraction pattern. This is consistent with earlier electrophysiological recordings in Hydra (30) and the observations that removal of the head region results in loss of spontaneous contractile activity (28).
Pacemaker Neurons in Hydra Are Immunocompetent Cells.
We have shown previously that neurons in Hydra secrete antimicrobial peptides (AMPs) to shape the resident microbiome (43). Neurons express a rich repertoire of peptides, including the previously characterized antimicrobial peptide NDA-1 and the dual-function neuropeptides RFamide III, Hym-370, and Hym-357, which previously were found to have strong activity against gram-positive bacteria (43) (Fig. 3A). In addition, neurons express homologs of Kazal2 and Arminin proteins that have been previously characterized (44–46) as antimicrobial peptides in epithelial or gland cells in Hydra (Fig. 3A). Notably, the N2 pacemaker population also expresses multiple AMP molecules (Fig. 3A), indicating that these neurons, in addition to governing peristaltic motion, are also directly interacting with the resident microbiota. To determine if interaction with bacteria is a general characteristic of neurons in Hydra, we analyzed the seven neuronal subpopulations for expression of immune-related genes (Fig. 3 B–D). Neurons express virtually all components of the Toll-like receptor (TLR/MyD88) pathway (47) (Fig. 3B and SI Appendix, Fig. S14A; see also SI Appendix, Supplementary Results), as well as many C-type lectin (CTL) receptors (Fig. 3C and SI Appendix, Fig. S14C) and intracellular NACHT and NB-ARC domain-containing NOD-like receptors (48) (Fig. 3D and SI Appendix, Fig. S14 B and C). Overall, these observations indicate that neurons in Hydra are immunocompetent cells, equipped with receptors, signal transducers, and effector molecules to interact with bacteria.
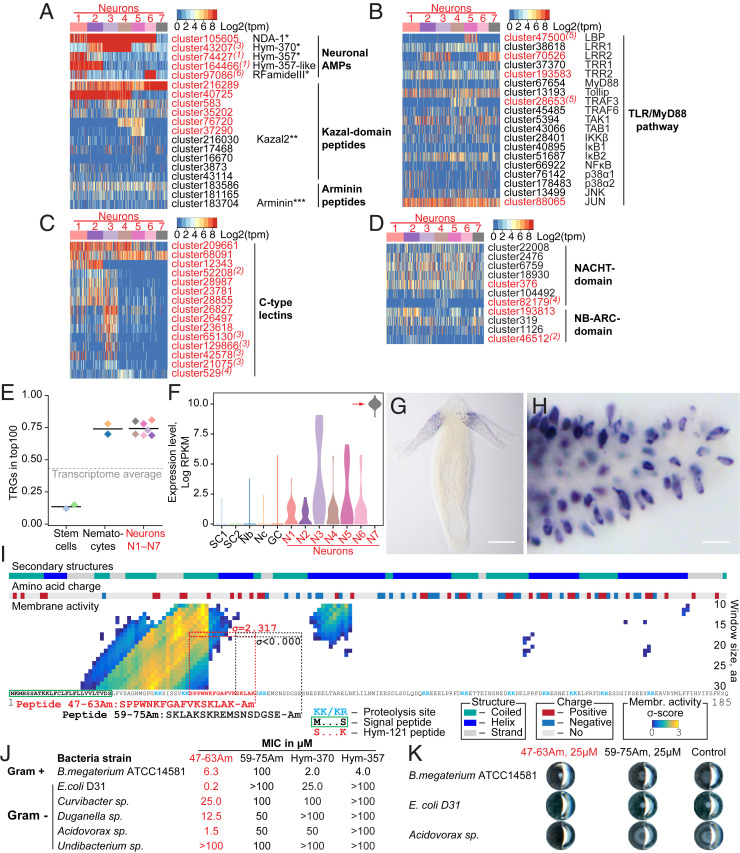
Fig. 3.
Neurons in Hydra are immunocompetent cells. (A) Neurons express a rich set of peptides that have been previously characterized as antimicrobial peptides or their homologs. *ref. 43; **ref. 44; ***ref. 46. (B) Heatmap illustrates expression of transcripts coding for components of the TLR/MyD88-dependent immune pathway. Most components are present in the neurons, and five of them are significantly enriched in the neuronal population (red). Superscript numbers indicate the nerve cell cluster (N1 to N7), where the transcripts are significantly (adjusted P < 0.05) enriched. (C) Heatmap illustrates expression of some transcripts coding for NACHT and NB-ARC domain-containing NOD-like receptors that have immune function. (D) Multiple C-type lectin receptors that might recognize bacterial products are strongly expressed in the neurons. (E) Over 70% of top 100 transcripts specifically expressed in each of seven neuronal subpopulations (N1 to N7) is represented by genes that have no homologs outside of Cnidaria, and thus are considered as TRGs. In contrast, among the top 100 transcripts specifically enriched in the interstitial stem cells, only 15% are identified as TRGs. (F) Transcripts of TRG cluster62692 are strongly up-regulated in the neuronal subpopulation N7, weakly expressed in other neurons, and absent from nonneuronal cells of the interstitial lineage. (G and H) In situ hybridization provides evidence that the TRG cluster62692 is expressed exclusively in the sensory neurons of the tentacles. (Scale bars, 100 µm in G and 10 µm in H.) (I) Moving-window small-peptide scan prediction map for the peptide encoded by TRG cluster62692 with residue charge and secondary structure annotations. The heat map reflects the peptide’s probability (σ-score) of being membrane active as predicted by the machine learning classifier (49). High σ-scores (yellow) suggest that cluster62692 peptide is a potent antimicrobial peptide. N-terminal signal peptide, putative proteolysis sites, and a sequence identical to a previously described peptide Hym-121 (50) are found within the cluster62692 peptide, providing evidence that a preprohormone cluster62692 is processed and gives rise to a secreted active peptide. The 17-aa-long peptide corresponding to amino acids 47 to 63 (SPPWNKFGAFVKSKLAK = Hym-121) with high membrane activity score (σ = 2.317) and control peptide amino acids 59 to 75 (SKLAKSKREMSNSDGSE) with no membrane activity (σ = −1.878) were synthesized, C terminally amidated, and tested for antimicrobial activity in a MIC assay. (J) The peptide 47–63 Am is a potent antimicrobial peptide that shows selective growth inhibiting activity against gram-positive and -negative bacteria. Control peptide 59–75 Am demonstrates no antimicrobial activity. Consistently with previous observations (43), dual-function neuropeptides Hym-370 and Hym-357 show some antibacterial activity, yet weaker and more restricted than the peptide 47–63 Am. (K) Representative wells from plates of MIC assay. At concentration 25 µM, the peptide 47–63 Am inhibits growth of Curvibacter sp. and Acidovorax sp. and affects colony morphology of B. megaterium. The growth in the presence of control peptide (59–75 Am, 25 µM) is not different from that in the pure medium (control).
In addition to a conserved toolbox of immune genes (Fig. 3 B–D), Hydra neurons also employ some of their nonconserved, taxonomically restricted genes (TRGs) to interact with bacteria. Nonconserved genes comprise over 70% of the cell-type–specific genes in transcriptomes of the seven neuronal subpopulations (N1–N7) (Fig. 3E, SI Appendix, Fig. S15, and Dataset S2). The majority of the neuron-specific TRGs code for short peptides (<200 aa) with an N-terminal signal peptide sequence, but with no detectable structural domains (Dataset S2). A machine learning-based approach (49) identified that some of these novel genes code for putative peptides with high membrane-destabilizing activity, indicating strong antimicrobial activity (SI Appendix, Fig. S16 and Dataset S3). Surprisingly, the neuronal cluster N2 that contains the pacemakers was one of the populations most enriched in secreted peptides with putative antimicrobial activity (SI Appendix, Fig. S16).
To provide direct evidence for functional relevance of TRGs in immune reactions in neurons, we characterized TRG cluster62692 that is specifically expressed in neuronal subpopulation N7 in the tentacles (Fig. 3 F–H) and encodes a 185-aa-long peptide (Fig. 3I). The N-terminal signaling peptide is followed by a stretch of mostly positively charged residues, which is predicted to have a high membrane-destabilizing activity (Fig. 3I). Screening a Hydra peptide database (50) uncovered a 17-aa amidated peptide, referred to as Hym-121, that is identical to amino acids 47 to 63 within the cluster62692 polypeptide (Fig. 3I), thus providing an evidence for translation and proteolytic processing of the preprohormone encoded by TRG cluster62692. To test whether this peptide may serve as an antimicrobial peptide, we synthetized the amidated amino acids 47 to 63 peptide and subjected it to a minimum inhibitory concentration (MIC) assay against diverse bacteria. We found the peptide to inhibit growth of Escherichia coli at concentrations as low as 0.2 μM (Fig. 3 J and K). The peptide also displayed a strong growth inhibiting activity (MIC 1.5 μM) against Acidovorax, a gram-negative commensal microbe found on Hydra (51). The growth of other two resident microbes, Curvibacter and Duganella, was impaired only at peptide concentrations 12.5 to 25 μM. Finally, the growth of Undibacterium was not inhibited by the 47–63 peptide even at 100 μM, indicating a high selectivity of the peptide’s antimicrobial activity (Fig. 3J). Surprisingly, while the peptide did not inhibit the growth of Bacillus megaterium, it drastically affected the colony morphology at concentrations as low as 6.3 μM (Fig. 3 J and K). This suggests that the peptide modulates the motility, swarming behavior, or spore formation of B. megaterium, thus implying a novel mechanism of AMP activity. Taken together, our data suggest that in addition to a conserved toolbox of immune genes (Fig. 3 B–D), neurons also employ some of their TRGs to interact with bacteria, supporting the view (52) that the nervous system, with its rich repertoire of neuropeptides, is involved in controlling resident beneficial microbes.
Pacemaker Gene Expression Is Dependent on Microbes.
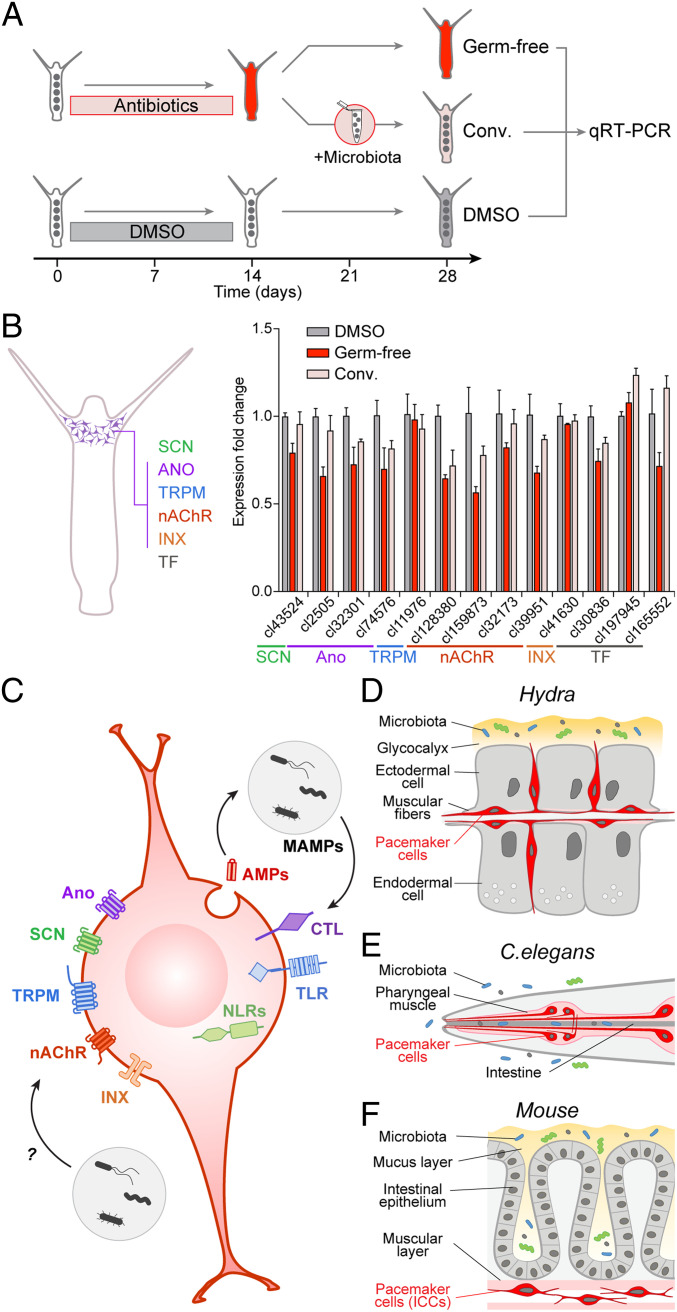
To gain further insights into the immune function of the pacemaker cells, we compared the expression of pacemaker-specific genes in Hydra polyps harboring normal microbiota, in GF polyps lacking microbiota, and in GF polyps recolonized with the natural microbiota, referred to as conventionalized polyps (Fig. 4A). In GF polyps, virtually all genes were substantially down-regulated compared to control (Fig. 4B). In the conventionalized polyps, the expression level of most of the genes was restored to values close to control ones. These observations provide a direct evidence that expression of pacemaker genes is dependent on the presence of symbiotic microbes. These findings are also consistent with our previous observation that the presence of microbes is required for the functionality of pacemaker cells to control regular body column contractions (26): The contraction frequency in GF polyps was reduced by 50% compared to control and was partially restored in conventionalized polyps. Taken together, these observations provide a strong evidence for the link between the microbiota and the pacemaker activity in Hydra.
Fig. 4.
Prototypical pacemaker neurons interact with microbiota. (A) To test the immune function of the pacemaker neurons in Hydra, GF animals were generated by antibiotic treatment and then recolonized with the natural Hydra microbiota to obtain conventionalized (Conv.) polyps. Polyps treated with DMSO solvent were used as control. Total RNA was extracted from polyps for gene-expression analysis by qRT-PCR. (B) Analysis of expression level of 13 genes specific for the pacemaker neurons in Hydra (scheme, Left) in GF, conventionalized (conv.) and control (DMSO) polyps. Average fold-changes (mean ± SEM; n = 3) are shown. Most of the genes are substantially down-regulated in GF polyps compared to control, and their expression level is restored to values close to control in conventionalized polyps. Thus, expression of pacemaker genes in Hydra is dependent on the presence of symbiotic microbes. (C–F) The molecular anatomy of the pacemaker cell. The gene-expression program characteristic for pacemaker cells is highly conserved and is present in the neuronal population N2 that controls spontaneous contractions in Hydra (D), in the neuro-muscular pacemaker complex of C. elegans pharynx (E), and in the ICCs driving the gut motility in mammals (F). This evolutionary conserved signature of a pacemaker cell is composed of ANO-, SCN-, and TRPM-like ion channels, nAChRs, and innexin gap junction. It also includes receptors such as Toll- and NOD-like receptors and C-type lectins that are capable of recognizing bacterial products (MAMPs). Bacteria-derived products may have profound effects onto the gene-expression program of the pacemakers via immune pathways or might directly target the pacemaker ion channels or neuromediator receptors (C).
Murine Intestinal Pacemaker Cells Also Are Immunocompetent Cells.
To search for commonalities between Hydra and mice pacemaker neurons, we screened the transcriptome of murine intestinal pacemakers, the ICCs (15), for the presence of transcripts coding for immune receptors and pathways. Surprisingly, almost the entire signal-transducing cascade of the TLR/MyD88 pathway is present in the ICCs (SI Appendix, Fig. S17 and Dataset S4). ICCs also express antimicrobial peptides, such as Defensin-8 (Dataset S4), as well as components of the NLR and CTL pathways, including NOD1, NOD2, and multiple CTL-domain receptors, which are essential for detecting bacteria (SI Appendix, Figs. S18 and S19 and Dataset S4). This strongly suggests that ICCs in mice, similar to pacemaker neurons resident in subpopulation N2 in Hydra, are capable of interacting with bacteria.
Molecular Architecture of Pacemaker Cells Is Highly Conserved in Evolution.
The similarity in the gene-expression profile of Hydra pacemaker neurons and murine ICCs suggests that the molecular architecture of pacemaker cells is highly conserved in evolution. To support that view, we expanded our analysis by using the single-cell transcriptional profiling of the nematode Caenorhabditis elegans (53). We surveyed the expression of the C. elegans homologs of the genes that comprise the pacemaker signature in both Hydra and mouse (coding for SCN, ANO, and TRPM ion channels, nAChR receptors, and gap-junction proteins) in 27 cell types identified by Cao et al. (53) (SI Appendix, Fig. S20). Although this did not lead to an immediate identification of a certain neuronal cell type as the pacemaker, three C. elegans cell types drew our attention: The pharyngeal neurons, pharyngeal muscle cells, and interneurons. These three cell types express to some extent all of the pacemaker-specific genes, suggesting that they might function as pacemakers. This is consistent with the observations that the pharyngeal system represents a neuro-muscular complex that, as a whole, possesses a pacemaker activity and drives regular contractions of the pharynx that assist food ingestion and movement along the intestine. A body of functional data provide evidence that both the neurons and epithelio-muscular cells, as well as a number of pacemaker-relevant genes (egl-19, gtl-1 and -2, lev-8, eat-2, and inx-6 and -16) (SI Appendix, Fig. S20) are functionally indispensable for the pacemaker function of the pharynx in C. elegans (54–58). Taken together, our findings point to the presence of a specific expression program in the pacemakers of the roundworm, and thus support the view that the molecular architecture of pacemaker cells (Fig. 4 C–F) is highly conserved in evolution.
Discussion
We have generated a comprehensive molecular profile of the neural subpopulations in Hydra and proposed a conceptual framework for phenotypic diversification of one of the simplest nervous systems in the animal kingdom. This framework builds upon seven spatially and functionally segregated neural subpopulations, allowing for the emergence of diverse and complex behaviors within a morphologically simple nerve net structure.
Our data establish the neurons of the N2 population as a major contributor to controlling the rhythmicity of spontaneous body contractions in Hydra. These prototypical pacemaker cells reside in the head region and specifically express ANO1-, SCN5-, and TRPM-like ion channels that characterize human gut pacemakers. Consistently, the experimental inhibition of these channels greatly disturbs peristalsis in Hydra, which is consistent with our finding that these neurons are coupled by gap junctions into a network exhibiting pacemaker activity. The high degree of gene-expression program conservation between the Hydra pacemaker subpopulation N2, the pharyngeal pacemaker complex of C. elegans, and murine pacemaker cells (Fig. 4 C–F) supports that peristaltic motor activity of the gut is an evolutionary ancient archetypical property necessary to sustain life (59, 60), and that the cells with recurrent spontaneous electric activity have evidently emerged as early as the nervous system itself.
Finally, we discovered that Hydra pacemaker cells express a rich set of immune genes, including antimicrobial peptides providing a mechanism for direct interference with resident microbes. The finding that many neuron-specific novel genes also encode antimicrobial peptides underlines the important function of neurons in interacting with microbes. Experimental interfering with microbiome in Hydra has a profound effect on the gene-expression program of the pacemaker neurons (Fig. 4B) and disrupts the rhythmic peristaltic activity of the polyps (26). Similarly, disturbances of gut microbiota in humans (61–63) result in changes in pacemaker rhythmicity and abnormal peristalsis. Because the Hydra pacemaker neurons can directly mediate the interaction with the microbiome, it might be plausible that human pacemakers in the gut similarly communicate with microbial communities. In fact, emerging data on mice provide first evidence for direct interactions between the gut microbiota, enteric neurons, and the intestinal motility (64–66). The evolutionary similarity or dissimilarity of the molecular toolkit used for such communication should become a subject of further investigations.
Altogether, our findings will improve the understanding of the archetypical properties of net nerve systems with pacemakers including the human enteric nervous system, which is perturbed in human dysmotility-related conditions affecting a large portion of the general population worldwide. We therefore presume that the principles identified here are relevant far beyond Hydra.
Materials and Methods
Experimental Design.
Experiments were carried out using Hydra vulgaris strain AEP. Animals were maintained under constant conditions, including the culture medium, food, and temperature (18 °C) according to standard procedures (67). Experimental animals were chosen randomly from clonally growing asexual Hydra cultures. The animals were typically fed three times a week; however, they were not fed for 24 h prior to pharmacological interference experiments, or for 48 h prior to RNA isolation, immunohistochemical staining, and in situ hybridization.
Generation of Transgenic Hydra Strains.
To facilitate the FACS-mediated molecular profiling of single cells, we developed a transgenic Hydra line expressing two reporter proteins. The enhanced GFP (eGFP) was cloned downstream from the previously reported cnnos1 promoter (68) and flanked by the actin terminator, and the codon-optimized DsRED2 protein was driven by the actin promoter sequence and flanked by the actin terminator region. After cloning into the LigAF vector (69), the transgenic construct was propagated in E. coli DH5α strain and microinjected into zygotes of H. vulgaris strain AEP, as previously described (67, 69). Founder mosaic transgenic animals were clonally propagated, screened, and enriched for transgenic cells until all interstitial stem cells were transgenic. The transgenic animals show no developmental abnormalities and are maintained in the laboratory for over 5 y.
FACS Isolation of Cells.
To isolate cells of the interstitial stem cell lineage from the transgenic Hydra by FACS (SI Appendix, Fig. S1A), the polyps were disintegrated into a cell suspension, as previously described (68). The single cells were sorted according to FSC, SSC, eGFP, and RFP fluorescence using FacsAria III cell sorting system (BD Biosciences), directly into 382-well plates containing lysis buffer, RNase inhibitor, oligo-dT30VN primer and dNTP mix, and snap-frozen. Sorting procedures are described in detail in SI Appendix, Supplementary Methods). In total, 1,152 individual cells were harvested: 384 GFP–/RFP+ neurons, 384 GFP+/RFP– stem cells, and 384 GFPlow/RFPlow cells.
Smart-Seq2 Library Preparation and Sequencing.
To generate cDNA libraries from the isolated cells, a previously described Smart-seq2 protocol (70) was implemented with minor modifications (SI Appendix, Supplementary Methods). The libraries were pooled and paired-end sequenced on Illumina HiSeq2500 instrument. Raw sequences and quality scores for all clusters were extracted using CASAVA software.
scRNA-Seq Data Processing, Quality Control, and Hierarchical Clustering.
Raw data from scRNA-seq were processed using a snakemake scRNA-seq pipeline. Cells passing quality control (i.e., 1,016) were analyzed using the R package Seurat_2.3.4. To map the reads, we used the previously described (71) reference transcriptome of H. vulgaris strain AEP (accession no. SRP133389). We included genes detected in at least three cells and cells that contained at least 5,000 transcripts. This resulted in a total of 928 cells and 166,186 transcripts. In the initial phase of the project, we generated in total 14 alternative clustering maps that segregated 928 cells into 9 to 22 clusters (SI Appendix, Fig. S21). Since the partitioning into 12 clusters, including 7 populations of neurons, was most consistent with our previous observations and the literature data, and resulted in clusters composed of at least 40 cells each, we further used only this clustering scheme. Specific parameters for the different input gene sets and input genes for each gene set are presented in SI Appendix, Supplementary Methods and Datasets S1, S7, and S8.
In Situ Hybridization.
To map the seven neuronal clusters populations in the Hydra body, we performed in situ hybridization with a set of genes strongly enriched in either of the seven neuronal subpopulations. Expression patterns were detected in the whole-mount Hydra preparations by in situ hybridization with antisense digoxigenin-labeled RNA probes, as previously described (72). A DIG-labeled sense-probe was used as a control. Signal was developed using anti-DIG antibodies conjugated to alkaline phosphatase (1:2,000; Roche) and NBT/BCIP staining solution (Roche). Images of the in situ preparations were captured on a Zeiss Axioscope with Axiocam camera.
Pharmacological Interference Assays.
To investigate the role of specific ANO1-, SCN-, and TRPM-like channels in the pacemaker activity in Hydra, we exposed normal H. vulgaris AEP polyps to different pharmacological agents, recorded, and quantified their behavior. Polyps were treated with 25 µM Ani9 (Sigma, Cat. No. SML1813), 200 µM menthol (Sigma, Cat. No. 15785), 100 µM lidocaine (Sigma, Cat. No. L5647), 1 mM tubocurarine (DTC, Sigma, Cat. No. 93750), or 100 µM muscimol (Sigma, Cat. No.M1523) for 1 or 12 h at 18 °C. Control polyps were incubated either in Hydra-medium or in the medium supplemented with 0.16% DMSO (for Ani9, which has been dissolved in 100% DMSO to stock concentration 15 mM). The spontaneous contractions were video-recorded and quantified, as previously described (26). We recorded the behavior for 90 min with a frequency 20 frames per minute. For further analysis, we excluded first 30 min of the recorded sequence, and quantified number of full-body contractions and their periodicity using a custom ImageJ plugin (26). The contraction frequencies were normalized to the average frequency of contractions in corresponding control polyps. To examine the effects of the modulators on the feeding reflex, Hydra polyps were pretreated with the pharmacological agents, their feeding reflex was elicited by 10 µM reduced glutathione (GSH, Sigma, Cat. No. G4251), and the duration of feeding response was recorded as described by Lenhoff (73).
Statistical Analysis.
The sample size (n) reported in the figure legends is the total amount of animals used in each treatment. Each animal employed was assigned to only one treatment and was recorded only once. Treatment of the polyps with pharmacological substances and evaluation of the behavioral parameters (contraction frequency, intervals between contractions, and duration of feeding response) was blinded. Differences in contraction frequency, interval between contractions, and feeding response duration between the treatments (i.e., Ani9, lidocaine, menthol, DTC, and muscimol) and corresponding control (i.e., Hydra-medium or DMSO) were analyzed using unpaired t test.
MIC Determination of Antimicrobial Activity.
To test whether peptides encoded in TRGs may have an antimicrobial function, two 17-aa-long peptides corresponding to amino acids 47 to 63 (SPPWNKFGAFVKSKLAK) and amino acids 59 to 75 (SKLAKSKREMSNSDGSE) of the prepropeptides encoded by the cluster62692 were synthesized (up to 5 mg), C-terminally amidated, and purified to a purity of >95% (GenScript), and their antimicrobial activity was estimated in an MIC assay, as previously described (43). The following bacterial strains were used in MIC assays: B. megaterium ATCC14581, E. coli D31, and four isolates from the natural H. vulgaris strain AEP microbiota: Curvibacter sp., Duganella sp., Acidovorax sp., and Undibacterium sp. (46). Microdilution susceptibility assays were carried out in 96-well microtiter plates that were precoated with sterile 0.1% bovine serum albumin (BSA). After removal of BSA the wells were filled with a twofold dilution series of either amino acids 47 to 63 or amino acids 59 to 75 peptide. We also tested the neuropeptides Hym-370 (KPNAYKGKLPIGLW-amide) and Hym-357 (KPAFLFKGYKP-amide) that has been previously identified as putative antimicrobial substances (43). Lyophilized peptides were dissolved in ultrapure water to stock concentration of 10 mg/mL. Incubation with an inoculum of ∼100 CFU per well was performed in phosphate buffered saline (PBS) buffer (pH 6.2) overnight at 37 °C for B. megaterium and E. coli, or in R2A media for 3 to 4 d at 18 °C for four isolates of Hydra bacteria. The MIC was determined as the lowest serial dilution showing absence of a bacterial cell pellet. Experiments were carried out in triplicates.
Phylogenetic Analysis of Ion Channel Genes.
To uncover whether Hydra has homologs of the ion channel genes whose expression is known to be either restricted to mammalian ICCs or essential for gut motility, we performed a BLAST search (tblastn) using full-length amino acid sequences of ANO1 (UniProt accession no. Q5XXA6), SCN5A (UniProt accession no. Q14524), and TRPM8 (UniProt accession no. Q7Z2W7) proteins from Homo sapiens against the genome of H. vulgaris (38) (available at https://research.nhgri.nih.gov/hydra/) and the reference transcriptome of H. vulgaris strain AEP. Matches with expectation e-value <10e-10 were considered as signs of homolog presence and were verified by manual domain composition analysis using SMART (74), transmembrane domain prediction with TMHMM (75), and reciprocal BLAST against the UniProt database. Maximum-likelihood phylogenetic trees of ANO1, SCN5A, and TRPM8 homologs from Hydra, human, African clawed frog Xenopus laevis, and zebrafish Danio rerio were built using full-length amino acid sequences aligned using MUSCLE (76) with 1,000 bootstrap iterations.
Generation of GF and Conventionalized Polyps.
GF polyps were generated by treating control Hydra polyps for 2 wk with an antibiotic mixture containing rifampicin, ampicillin, streptomycin, and neomycin in final concentrations of 50 µg/mL each and spectinomycin at 60 µg/mL, as previously described (26). Since rifampicin stock is dissolved in DMSO, control polyps were incubated in the corresponding 0.1% DMSO concentration for the same period of time. Antibiotic solution and control medium were replaced every 48 h. After 2 wk of treatment, the animals were transferred into sterile Hydra-medium that was further replaced every 48 h before isolation of total RNA. Conventionalized Hydras were generated by incubating GF polyps with tissue homogenates of control animals. Previously, we demonstrated that recolonization of GF polyps in this way results in the establishment of a bacterial community similar to that of an intact control Hydra polyp (26). Generation of GF polyps and recolonization were repeated in triplicates. The GF status of the polyps and success of recolonization were tested by plating a macerated polyp on R2A agar, which supports growth of Hydra microbiota. Absence of colonies following 3 d of incubation at 18 °C confirmed the GF status. Polyp macerates were also analyzed by 16S rDNA amplification, using universal primers Eub-27F and Eub-1492R. Absence of amplification product confirmed the GF status.
Quantitative Real-Time PCR Gene-Expression Analysis.
To test whether the genes coding for ANO1, SCN5A, and TRPM8 homologs in Hydra are differentially expressed in the polyp along the oral-aboral axis, we performed quantitative real-time PCR. We dissected polyps into three body sections: Head (hypostome area with tentacles), body column, and foot (peduncle). Each total RNA was extracted from body fragments obtained from 50 polyps, and converted into the cDNA as previously described (72). For each body part, we made three to five biological replicates.
To examine whether the gene-expression profile in the pacemaker cells of Hydra is dependent on the presence of specific microbiota, we compared expression of genes coding for ANO1, SCN5A, and TRPM8 homologs, as well as nAChR, innexin gap-junction proteins, and three pacemaker-specific transcription factors using real-time PCR. Total RNA was extracted from 100 normal, GF, or conventionalized polyps and converted into cDNA. For each condition, we made three biological replicates. Real-time PCR was performed using GoTaq qPCR Master Mix (Promega) and oligonucleotide primers specifically designed to amplify the homologs of ANO1, SCN5A, and TRPM8 ion channel genes, nAChR, innexins, and transcription factors, as well as the ef1a (translation elongation factor 1α) and actin genes as equilibration references (SI Appendix, Table S1). The data were collected by ABI 7300 Real-Time PCR System (Applied Biosystems) and analyzed by the conventional ΔΔCt method.
Generation of Antibodies and Immunohistochemistry.
To localize the expression of ANO1-like and SCN5A-like ion channels in Hydra using immunocytochemistry, polyclonal antibodies were raised against synthetic peptides in rabbits. The peptides that correspond to intracellular loops located between transmembrane domains of the ion channels (hySCN5-like: SRSKPKMFKDYKPE; hyANO-like: ETRRPIADRAQD) were synthetized, purified, and N terminally conjugated with KLH prior to injection (GenScript). Polyclonal antibodies were affinity-purified on the antigen and concentrated to 1.5 mg/mL. Serum harvested from the rabbits prior to their immunization was used as control.
Immunohistochemical detection in whole-mount Hydra preparations was carried oud as described previously (77). Briefly, polyps were relaxed in urethane, fixed in paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS, incubated in blocking solution for 1 h, and incubated further with primary antibodies diluted to 1.0 μg/mL in blocking solution at 4 °C. AlexaFluor488-conjugated goat anti-rabbit antibodies (Invitrogen) were diluted to 4 μg/mL and incubations were done for 1 h at room temperature. Rhodamin-phalloidin (Sigma) and TO-PRO3-iodide-AlexFluor633 (Invitrogen) counterstaining was conducted as described previously (77). Confocal laser-scanning microscopy was done using a TCS SP1 laser scanning confocal microscope (Leica).
Supplementary Material
Acknowledgments
We thank Vassilis Pachnis for critically reading the manuscript and for sharing unpublished observations in mouse enteric neurons; Eva Herbst (Kiel University), Hans-Heinrich Oberg (University Medical Center Schleswig-Holstein, Kiel), Kiran Sedimbi (SciLifeLab, Stockholm), Simone Picelli (Institute of Molecular and Clinical Opthalmology Basel), and The Eukaryotic Single Cell Genomics facility at SciLifeLab for technical support; the Uppsala Multidisciplinary Center for Advanced Computational Science for providing computational infrastructure; Jan Taubenheim (Heinrich Heine University, Düsseldorf) and Tatiana Chontorotzea (Medical University of Vienna) for help in analyzing the data; and Toshitaka Fujisawa (Center for Organismal Studies, Heidelberg) for providing access to the Hydra Peptide database. This work was supported in part by grants from the Deutsche Forschungsgemeinschaft and the CRC 1182 (“Origin and Function of Metaorganisms”) (to T.C.G.B.). T.C.G.B. appreciates support from the Canadian Institute for Advanced Research. I.A. was supported by Swedish Research Council and European Research Council Consolidator Grant STEMMING-FROM-NERVE. A.K. was supported by the Alexander von Humboldt Foundation. S.G. and Å.B. were financially supported by the Knut and Alice Wallenberg Foundation as part of the National Bioinformatics Infrastructure Sweden at SciLifeLab (grant to I.A. and A.K.). S.G. was also financially supported by Formas Grant 2017-01066_3.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Single-cell RNA-sequencing data generated in this study have been deposited in the National Center for Biotechnology Information Sequence Read Archive database (BioProject accession codes PRJNA614614 and PRJNA614611). Expression count matrices, corresponding metadata, and analytical pipelines are available as Mendeley dataset DOI: 1017632/ctdfn57ds2.1. Custom scripts are available at https://github.com/stefaniagiacomello/Hydra_scRNA-seq.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920469117/-/DCSupplemental.
Data Availability.
scRNA-seq data generated in this study have been deposited on the National Center for Biotechnology Information Sequence Read Archive database under the BioProject accession codes PRJNA614614 and PRJNA614611. Expression count matrices, corresponding metadata, and analytical pipelines are available as Mendeley dataset DOI: 1017632/ctdfn57ds2.1. Custom scripts are available at https://github.com/stefaniagiacomello/Hydra_scRNA-seq.
References
- 1.Sasselli V., Pachnis V., Burns A. J., The enteric nervous system. Dev. Biol. 366, 64–73 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Bosch T. C. G. et al., Back to the basics: Cnidarians start to fire. Trends Neurosci. 40, 92–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olsson C., Holmgren S., The control of gut motility. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 128, 481–503 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Barajas-López C., Berezin I., Daniel E. E., Huizinga J. D., Pacemaker activity recorded in interstitial cells of Cajal of the gastrointestinal tract. Am. J. Physiol. 257, C830–C835 (1989). [DOI] [PubMed] [Google Scholar]
- 5.Sanders K. M., Ward S. M., Koh S. D., Interstitial cells: Regulators of smooth muscle function. Physiol. Rev. 94, 859–907 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomsen L. et al., Interstitial cells of Cajal generate a rhythmic pacemaker current. Nat. Med. 4, 848–851 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Farrugia G., Interstitial cells of Cajal in health and disease. Neurogastroenterol. Motil. 20 (suppl. 1), 54–63 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Huizinga J. D. et al., W/kit gene required for interstitial cells of Cajal and for intestinal pacemaker activity. Nature 373, 347–349 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Yamataka A. et al., A lack of intestinal pacemaker (c-kit) in aganglionic bowel of patients with Hirschsprung’s disease. J. Pediatr. Surg. 30, 441–444 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Cogliandro R. F. et al., Patient-reported outcomes and gut dysmotility in functional gastrointestinal disorders. Neurogastroenterol. Motil. 23, 1084–1091 (2011). [DOI] [PubMed] [Google Scholar]
- 11.De Giorgio R., Sarnelli G., Corinaldesi R., Stanghellini V., Advances in our understanding of the pathology of chronic intestinal pseudo-obstruction. Gut 53, 1549–1552 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellow J. E., Phillips S. F., Miller L. J., Zinsmeister A. R., Dysmotility of the small intestine in irritable bowel syndrome. Gut 29, 1236–1243 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Pinilla P. J. et al., Ano1 is a selective marker of interstitial cells of Cajal in the human and mouse gastrointestinal tract. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G1370–G1381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strege P. R. et al., Sodium current in human intestinal interstitial cells of Cajal. Am. J. Physiol. Gastrointest. Liver Physiol. 285, G1111–G1121 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Lee M. Y. et al., Transcriptome of interstitial cells of Cajal reveals unique and selective gene signatures. PLoS One 12, e0176031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beyder A. et al., Loss-of-function of the voltage-gated sodium channel NaV1.5 (channelopathies) in patients with irritable bowel syndrome. Gastroenterology 146, 1659–1668 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strege P. R. et al., Irritable bowel syndrome (IBS) patients have SCN5A channelopathies that lead to decreased NaV1. 5 current and mechanosensitivity. Am. J. Physiol. Liver Physiol. 314, G494–G503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzone A. et al., Direct repression of anoctamin 1 (ANO1) gene transcription by Gli proteins. FASEB J. 33, 6632–6642 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beyder A., Farrugia G., Ion channelopathies in functional GI disorders. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G581–G586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henström M. et al., TRPM8 polymorphisms associated with increased risk of IBS-C and IBS-M. Gut 66, 1725–1727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ek W. E. et al., Exploring the genetics of irritable bowel syndrome: A GWA study in the general population and replication in multinational case-control cohorts. Gut 64, 1774–1782 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Bonfiglio F. et al., A GWAS meta-analysis from 5 population-based cohorts implicates ion channel genes in the pathogenesis of irritable bowel syndrome. Neurogastroenterol. Motil. 30, e13358 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Bonfiglio F. et al., Female-specific association between variants on chromosome 9 and self-reported diagnosis of irritable bowel syndrome. Gastroenterology 155, 168–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiles T. J. et al., Host gut motility promotes competitive exclusion within a model intestinal microbiota. PLoS Biol. 14, e1002517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell R. D., Josephson R. K., Schwab W. E., Rushforth N. B., Excitability of nerve-free hydra. Nature 262, 388–390 (1976). [DOI] [PubMed] [Google Scholar]
- 26.Murillo-Rincon A. P. et al., Spontaneous body contractions are modulated by the microbiome of Hydra. Sci. Rep. 7, 15937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siebert S. et al., Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 365, eaav9314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Passano L. M., McCullough C. B., Co-ordinating systems and behaviour in Hydra: I. Pacemaker system of the periodic contractions. J. Exp. Biol. 41, 643–664 (1964). [Google Scholar]
- 29.Passano L. M., McCullough C. B., Pacemaker hierarchies controlling the behaviour of hydras. Nature 199, 1174–1175 (1963). [DOI] [PubMed] [Google Scholar]
- 30.Kass-Simon G., Longitudinal conduction of contraction burst pulses from hypostomal excitation loci in Hydra attenuata. J. Comp. Physiol. 80, 29–49 (1972). [Google Scholar]
- 31.Amato A., Liotta R., Mulè F., Effects of menthol on circular smooth muscle of human colon: Analysis of the mechanism of action. Eur. J. Pharmacol. 740, 295–301 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Seo Y. et al., Ani9, a novel potent small-molecule ANO1 inhibitor with negligible effect on ANO2. PLoS One 11, e0155771 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peier A. M. et al., A TRP channel that senses cold stimuli and menthol. Cell 108, 705–715 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Willcockson I. U. et al., Orientation of d-tubocurarine in the muscle nicotinic acetylcholine receptor-binding site. J. Biol. Chem. 277, 42249–42258 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Karpen J. W., Hess G. P., Acetylcholine receptor inhibition by d-tubocurarine involves both a competitive and a noncompetitive binding site as determined by stopped-flow measurements of receptor-controlled ion flux in membrane vesicles. Biochemistry 25, 1786–1792 (1986). [DOI] [PubMed] [Google Scholar]
- 36.Pierobon P., Coordinated modulation of cellular signaling through ligand-gated ion channels in Hydra vulgaris (Cnidaria, Hydrozoa). Int. J. Dev. Biol. 56, 551–565 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Alexopoulos H. et al., Evolution of gap junctions: The missing link? Curr. Biol. 14, R879–R880 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Chapman J. A. et al., The dynamic genome of Hydra. Nature 464, 592–596 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takaku Y. et al., Innexin gap junctions in nerve cells coordinate spontaneous contractile behavior in Hydra polyps. Sci. Rep. 4, 3573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hufnagel L., Kass-Simon G., “The ultrastructural basis for the electrical coordination between epithelia of Hydra” in Coelenterate Ecology and Behavior, Mackie G. O., Ed. (Springer, 1976), pp. 695–704. [Google Scholar]
- 41.Fraser S. E., Green C. R., Bode H. R., Gilula N. B., Selective disruption of gap junctional communication interferes with a patterning process in hydra. Science 237, 49–55 (1987). [DOI] [PubMed] [Google Scholar]
- 42.Hand A. R., Gobel S., The structural organization of the septate and gap junctions of Hydra. J. Cell Biol. 52, 397–408 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Augustin R. et al., A secreted antibacterial neuropeptide shapes the microbiome of Hydra. Nat. Commun. 8, 698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Augustin R., Siebert S., Bosch T. C. G., Identification of a kazal-type serine protease inhibitor with potent anti-staphylococcal activity as part of Hydra’s innate immune system. Dev. Comp. Immunol. 33, 830–837 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Augustin R. et al., Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob. Agents Chemother. 53, 5245–5250 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franzenburg S. et al., Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl. Acad. Sci. U.S.A. 110, E3730–E3738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franzenburg S. et al., MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc. Natl. Acad. Sci. U.S.A. 109, 19374–19379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lange C. et al., Defining the origins of the NOD-like receptor system at the base of animal evolution. Mol. Biol. Evol. 28, 1687–1702 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Lee E. Y., Fulan B. M., Wong G. C. L., Ferguson A. L., Mapping membrane activity in undiscovered peptide sequence space using machine learning. Proc. Natl. Acad. Sci. U.S.A. 113, 13588–13593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujisawa T., Hydra peptide project 1993–2007. Dev. Growth Differ. 50 (suppl. 1), S257–S268 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Fraune S., Bosch T. C. G., Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. U.S.A. 104, 13146–13151 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klimovich A. V., Bosch T. C. G., Rethinking the role of the nervous system: Lessons from the Hydra holobiont. BioEssays 40, e1800060 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Cao J. et al., Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trojanowski N. F., Raizen D. M., Fang-Yen C., Pharyngeal pumping in Caenorhabditis elegans depends on tonic and phasic signaling from the nervous system. Sci. Rep. 6, 22940 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raizen D. M., Lee R. Y., Avery L., Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics 141, 1365–1382 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schüler C., Fischer E., Shaltiel L., Steuer Costa W., Gottschalk A., Arrhythmogenic effects of mutated L-type Ca 2+-channels on an optogenetically paced muscular pump in Caenorhabditis elegans. Sci. Rep. 5, 14427 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franks C. J., Holden-Dye L., Bull K., Luedtke S., Walker R. J., Anatomy, physiology and pharmacology of Caenorhabditis elegans pharynx: A model to define gene function in a simple neural system. Invert. Neurosci. 6, 105–122 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Harris T. W. et al., WormBase: A modern model organism information resource. Nucleic Acids Res. 48, D762–D767 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huizinga J. D. et al., The origin of segmentation motor activity in the intestine. Nat. Commun. 5, 3326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spencer N. J., Dinning P. G., Brookes S. J., Costa M., Insights into the mechanisms underlying colonic motor patterns. J. Physiol. 594, 4099–4116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quigley E. M. M., Microflora modulation of motility. J. Neurogastroenterol. Motil. 17, 140–147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dey N. et al., Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell 163, 95–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ge X. et al., Potential role of fecal microbiota from patients with slow transit constipation in the regulation of gastrointestinal motility. Sci. Rep. 7, 441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Obata Y., et al. , Neuronal programming by microbiota enables environmental regulation of intestinal motility. bioRxiv:10.1101/579250 (posted 15 March 2019).
- 65.Cryan J. F., Dinan T. G., Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Lai N. Y. et al., Gut-innervating nociceptor neurons regulate Peyer’s patch microfold cells and SFB levels to mediate Salmonella host defense. Cell 180, 33–49.e22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klimovich A., Wittlieb J., Bosch T. C. G., Transgenesis in Hydra to characterize gene function and visualize cell behavior. Nat. Protoc. 14, 2069–2090 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Hemmrich G. et al., Molecular signatures of the three stem cell lineages in hydra and the emergence of stem cell function at the base of multicellularity. Mol. Biol. Evol. 29, 3267–3280 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Wittlieb J., Khalturin K., Lohmann J. U., Anton-Erxleben F., Bosch T. C. G., Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 6208–6211 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Picelli S. et al., Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Mortzfeld B. M. et al., Temperature and insulin signaling regulate body size in Hydra by the Wnt and TGF-beta pathways. Nat. Commun. 10, 3257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Domazet-Lošo T. et al., Naturally occurring tumours in the basal metazoan Hydra. Nat. Commun. 5, 4222 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Lenhoff H. M., Activation of the feeding reflex in Hydra littoralis. I. Role played by reduced glutathione and quantitative assay of the feeding reflex. J. Gen. Physiol. 45, 331–344 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Letunic I., Bork P., 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L., Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 76.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klimovich A. et al., Non-senescent Hydra tolerates severe disturbances in the nuclear lamina. Aging (Albany NY) 10, 951–972 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data generated in this study have been deposited on the National Center for Biotechnology Information Sequence Read Archive database under the BioProject accession codes PRJNA614614 and PRJNA614611. Expression count matrices, corresponding metadata, and analytical pipelines are available as Mendeley dataset DOI: 1017632/ctdfn57ds2.1. Custom scripts are available at https://github.com/stefaniagiacomello/Hydra_scRNA-seq.