Significance
The therapeutic use of drugs against microbial pathogens and cancer is currently undergoing a paradigm shift from traditional therapies toward adaptive therapies, which attempt to minimize the risk of the evolution of drug resistance. Adaptive therapies treat pathogens with short bouts of drug application followed by periods without treatment to allow resistant cells to be naturally outcompeted by susceptible cells. However, the fitness cost of drug resistance is frequently too small for resistant cells to be rapidly outcompeted by susceptible cells, compromising the effectiveness and the future of adaptive therapies. In this study, we show that this problem can be potentially solved by amplifying the fitness cost of drug resistance to encourage the natural competition between resistant and susceptible cells.
Keywords: adaptive therapy, therapeutic resistance, evolutionary trade-offs, protein synthesis, mistranslation
Abstract
Antibiotic resistance frequently evolves through fitness trade-offs in which the genetic alterations that confer resistance to a drug can also cause growth defects in resistant cells. Here, through experimental evolution in a microfluidics-based turbidostat, we demonstrate that antibiotic-resistant cells can be efficiently inhibited by amplifying the fitness costs associated with drug-resistance evolution. Using tavaborole-resistant Escherichia coli as a model, we show that genetic mutations in leucyl-tRNA synthetase (that underlie tavaborole resistance) make resistant cells intolerant to norvaline, a chemical analog of leucine that is mistakenly used by tavaborole-resistant cells for protein synthesis. We then show that tavaborole-sensitive cells quickly outcompete tavaborole-resistant cells in the presence of norvaline due to the amplified cost of the molecular defect of tavaborole resistance. This finding illustrates that understanding molecular mechanisms of drug resistance allows us to effectively amplify even small evolutionary vulnerabilities of resistant cells to potentially enhance or enable adaptive therapies by accelerating posttreatment competition between resistant and susceptible cells.
The therapeutic use of drugs against microbial pathogens and cancer is currently undergoing a paradigm shift from traditional therapies toward adaptive therapies, in which disease is treated as an evolutionary process to minimize the risk of drug-resistance evolution (1–5).
With traditional therapy, the goal is to eradicate the disease by eliminating every pathogenic cell in the human body via the long-term administration of a drug at the maximum tolerable dose. A notorious drawback of this approach is that, after an initially effective treatment stage, it frequently results in the development of drug resistance during later treatment stages. In contrast, adaptive therapies aim to manage the disease without necessarily eradicating it. Evidence from studies of adaptive therapies suggest that—rather than forcing pathogenic cells into evolving elaborate forms of drug resistance during the long-term administration of antimicrobial or anticancer drugs—pathogens should be treated using repeated, short-term bouts of drug application that are interrupted by periods without treatment (3, 5). The rationale for this approach is that, during these periods without treatment, resistant cells will be naturally outcompeted by nonresistant cells (or cells with lower levels of resistance) either due to genetic drifts or because the resistant cells are frequently less fit than nonresistant cells in the absence of drug (1, 2).
Multiple studies have shown that integrating this evolutionary principle into clinical treatment protocols can drastically improve disease progression in patients suffering from advanced forms of infectious disease or cancer (1, 3–6). For example, adopting this approach dramatically improved the effectiveness of abiraterone-mediated treatment of metastatic forms of prostate cancer, where just 1 patient of 11 developed therapeutic resistance during a course of adaptive therapy, compared with 14 of 16 patients who developed therapeutic resistance following traditional therapy (5). Therefore, it is becoming increasingly important to learn whether we can reduce the periods without treatments in order to combat therapeutic resistance by encouraging natural competition between resistant and nonresistant cells.
In this study, we sought to exploit the evolutionary trade-off associated with drug-resistance evolution as a means to promote competition between resistant and nonresistant cells. Our aim was to capitalize on the fact that drug resistance frequently evolves as a trade-off in which the very same genetic mutations that alter a drug target and decrease the affinity of a target for the drug can also compromise the biological activity of the target. This compromised activity leads to growth defects in drug-resistant cells in the absence of the drug—a scenario that is especially common for synthetic drugs that act on a single target in the cell (7–12). For example, numerous studies showed that evolution of drug resistance in bacterial and cancer cells can lead to collateral sensitivity (a phenomenon in which resistance to one drug makes cells more sensitive to another drug), making it possible to exploit these evolutionary trade-offs to manage populations of drug-resistant cells after drug application (13–17).
In this study, we tested our hypothesis that drug-resistant cells can be selectively inhibited by enhancing toxicities associated with drug resistance, i.e., by placing them under conditions in which the molecular defects associated with drug resistance are especially fitness-costly.
For our drug model, we used tavaborole (also known as AN2690), a synthetic small-molecule inhibitor of protein synthesis (18–20). In 2014, this drug was approved by the Food and Drug Administration to treat onychomycosis (nail fungus) (21). Numerous studies have indicated that tavaborole and its chemical derivatives may also be of potential use against microbial pathogens, including Plasmodium falciparum (22, 23), Toxoplasma gondii (24), Trypanosoma brucei (25–29), Cryptosporidium parvum (24), Staphylococcus aureus (30), Mycobacterium tuberculosis (31–34), Streptococcus pneumoniae (35), and multidrug-resistant strains of Pseudomonas aeruginosa and Escherichia coli (20).
Tavaborole inhibits cell growth by inactivating leucyl-tRNA synthetase (LeuRS), an essential enzyme that attaches leucine to its corresponding tRNA (tRNALeu) to produce leucyl-tRNALeu, a substrate used in protein synthesis. LeuRS has two catalytic centers: the synthesis site, which is responsible for leucyl-tRNALeu formation, and the editing site, which is responsible for quality control during leucyl-tRNALeu formation. When the synthesis site makes occasional errors by attaching amino acids other than leucine to tRNALeu, the editing site detaches these amino acids from tRNALeu, thereby preventing errors in protein synthesis (36). Tavaborole inhibits LeuRS by targeting its editing domain, where tavaborole covalently binds with tRNALeu and prevents the dissociation of tRNALeu from LeuRS, leading to the inhibition of both protein synthesis and cell growth (19).
Previous studies in the model organism Saccharomyces cerevisiae have suggested that resistance to tavaborole can evolve via mutations in the leuS gene (the gene that codes for leucyl-tRNA synthetase): these mutations occur in the editing domain of LeuRS and frequently impair the protein’s editing activity (19, 37–39). A similar resistance mechanism was observed in clinical isolates of tavaborole-treated multidrug-resistant Escherichia coli (40) and in S. aureus that were treated with a tavaborole derivative, AN3365, the phase II clinical trials of which have been suspended due to the rapid development of AN3365 resistance (39). In the present study, therefore, we endeavored to determine if it is feasible to use this development of resistance, which likely arises at the expense of LeuRS editing, as a weakness via which tavaborole-resistant cells could be inhibited.
Results
We first tested how frequently tavaborole resistance originates from mutations in LeuRS. For this purpose, we evolved tavaborole-resistant E. coli. We used six initially identical populations of E. coli. Having determined the minimal inhibitory concentration (MIC) of tavaborole being 4 µg/ml in our assay, we then cultured these populations over 8 d, with the daily transfer of 5% of each culture into fresh media supplemented tavaborole, gradually increasing the tavaborole concentration from sub-MIC (3 µg/mL) to doses above the MIC (160 µg/mL) (SI Appendix, Fig. S1). By day 8, all populations were able to rapidly grow in the presence of tavaborole, indicating the development of tavaborole resistance (SI Appendix, Fig. S1).
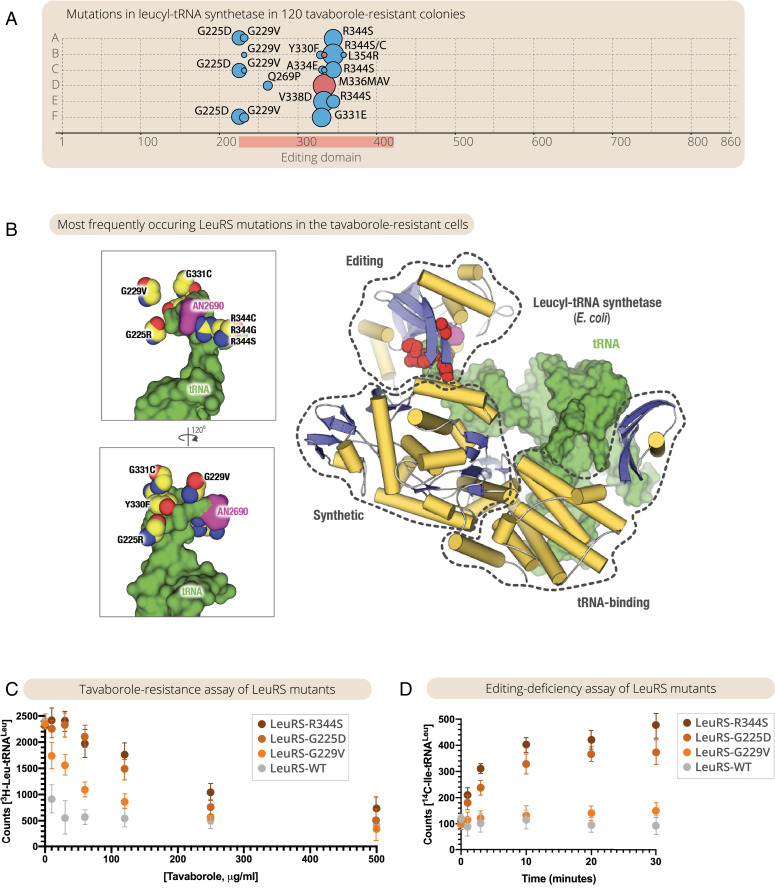
We next tested whether the resistant cells had mutations in the leuS gene. First, we sequenced the leuS gene in 120 tavaborole-resistant colonies (20 colonies selected at random from each of the six evolved populations) and found that one or two mutations in leuS were present in organisms from each colony. All these mutations were located in the leuS segment corresponding to the editing domain of LeuRS (Fig. 1A and SI Appendix, Table S1 and Supplementary Data 1). We next tested whether it was these mutations (and not other mutations possibly present in the E. coli genome) that conferred tavaborole resistance. We selected five of the tavaborole-resistant colonies that contained one of the five most frequently observed mutations, including G225D, G229V, Y330F, G331S, and R344S. In each of these colonies, we replaced the mutated leuS gene with the wild-type leuS gene and found that all of the derived clones had lost their tavaborole resistance, indicating that tavaborole resistance was conferred by mutations in the leuS gene (and not by other mutations that may possibly be present elsewhere in the genome) (SI Appendix, Fig. S2A). In a complementary experiment, we used wild-type E. coli and mutated their leuS gene by introducing one of the five frequently observed LeuRS mutations, G225D, G229V, Y330F, G331S, or R344S. We found that each of these mutations conferred resistance to tavaborole (10 µg/mL) upon their insertion into the wild-type E. coli genome, illustrating that each of these mutations on its own is sufficient to endow E. coli with a high level of resistance to tavaborole (SI Appendix, Fig. S2B). Finally, we performed time-resolved whole-population genome sequencing of the two evolving populations (lineages A and B) and found that mutations in the leuS gene were the only detectable mutations the frequency of which gradually increased during the course of the experiment and which were present in the majority of cells by the end of the experiment (SI Appendix, Fig. S3 and Supplementary Data 3). Taken together, our data showed that not only may leuS mutations confer resistance to tavaborole, as previously observed (19, 37–39), but also, at least in our experimental conditions, they appear to be the most preferential route for tavaborole resistance evolution.
Fig. 1.
Impairing LeuRS editing activity is the primary route of tavaborole-resistance evolution in continuous E. coli cultures. (A) The panel summarizes mutations in the leuS gene that were observed in 120 tavaborole-resistant colonies (20 colonies from each of the six tavaborole-resistant lineages of E. coli). The size of the circles indicates the frequency of the observed mutations, including amino acid substitutions (blue circles) and repeat insertions (red circles). As the panel shows, all of the observed mutations were located in the editing domain segment of the leuS gene, with one or two mutations per colony. (B) The crystal structure of LeuRS in complex with tRNALeu and tavaborole shows the location of tavaborole-resistant mutations in LeuRS synthetase. Remarkably, all of the observed mutations are located not at the interface between LeuRS and tavaborole but at the interface between LeuRS and tRNALeu, suggesting that these mutations prevent tavaborole inhibition via impaired editing activity. (C) A tavaborole-resistance assay shows IC50 measurements, in which LeuRS activity (assessed by monitoring leucyl-tRNALeu formation) was measured in the presence of tavaborole (0 to 500 μg/mL). (D) An editing activity assay shows that, unlike the wild-type LeuRS, the LeuRS-resistant mutants can erroneously charge tRNALeu with isoleucine, leading to the formation of an erroneous reaction product, isoleucyl-tRNALeu (Ile-tRNALeu). Collectively, the figure illustrates not only that tavaborole resistance may sometimes evolve through mutations in the editing domain of LeuRS, as previously observed in individual clones (19, 37–39), but also that these mutations represent the primary route of the resistance evolution, with the most frequently observed mutants being the most editing-defective.
We next tested whether the tavaborole-resistance mutations affected the editing activity of LeuRS. First, we mapped the observed mutations onto the previously determined structure of LeuRS synthetase bound to tavaborole-modified tRNALeu (Fig. 1B). Remarkably, none of the mutations were located at the interface between LeuRS and tavaborole (Fig. 1B). Instead, the mutations were clustered at the interface between tRNALeu and LeuRS, suggesting that tavaborole resistance is likely acquired not via the direct prevention of drug binding but because LeuRS mutants cannot properly bind tRNALeu within the editing site. This could be due either to steric clashes between tRNALeu and LeuRS (as seems to be the case for G225D, G229V, and G331C mutants) or to disrupted tRNA-LeuRS contact (as seems to be the case for R344C/S and Y330F mutants) (Fig. 1B and SI Appendix, Fig. S4). We then purified LeuRS mutants carrying the most frequently observed mutations (G225D, G229V, and R344S) and tested their activity in vitro by measuring kinetics of tRNALeu aminoacylation with leucine (to assess the synthetic activity of LeuRS) or isoleucine (to assess the editing activity of LeuRS). Consistent with the structural observations, the in vitro measurements showed that all mutants tested exhibited high levels of resistance to tavaborole (Fig. 1C), but at the same time were also editing-defective (Fig. 1D). Thus, we found not only that some of the tavaborole-resistance mutations may impair the editing activity of LeuRS, as observed previously in individual resistant clones (19, 37, 38), but also that the most frequently occurring mutations are the ones that impair the editing activity the most, with the majority of resistant cells in the evolving populations being editing-defective.
We next tested if we could exploit these editing defects in LeuRS to inhibit tavaborole-resistant cells. Previously, laboratory-engineered E. coli strains with editing-deficient LeuRS were shown to be hypersensitive to the toxicity of chemical analogs of leucine (including norleucine, norvaline, homocysteine, and homoserine); in these strains, and in some naturally occurring parasites (41, 42), the absence of editing activity caused leucine analog incorporation into protein sequences, resulting in protein misfolding and cell growth arrest (43, 44) as a result of mistranslation (45). We therefore anticipated that tavaborole-resistant cells would also be hypersensitive to the toxicity of leucine analogs.
To test this hypothesis, we first assessed norvaline sensitivity of the evolved E. coli clones with mutations G225D, G229V, and R344S in the editing domain of LeuRS (SI Appendix, Fig. S5). For this purpose, we isolated clones with these mutations from the experimentally evolved populations A and B and compared growth rates of these clones in the presence or absence of norvaline (1 mM), using daily serial dilutions for 8 d. We found that, compared to the wild-type E. coli, these clones showed hypersensitivity to norvaline toxicity: their growth was arrested by day 5, compared to day 7 for wild-type E. coli (SI Appendix, Fig. S5). Notably, the R344S mutant, which conferred the highest degree of tavaborole resistance among the evolved clones, showed the strongest hypersensitivity to norvaline toxicity (SI Appendix, Fig. S5).
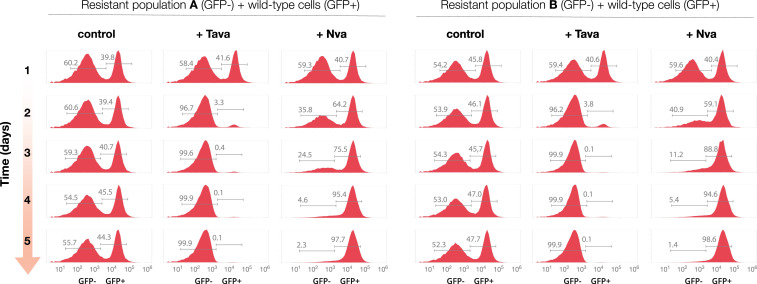
We then tested if this hypersensitivity to norvaline can be used to encourage the natural competition between resistant and nonresistant cells. For this purpose, we used a competition assay in which we mixed tavaborole-resistant cells with wild-type E. coli expressing GFP, at a ratio of ∼1:1, and incubated the cell mixture in a microturbidostat for 5 d (SI Appendix, Fig. S6). We observed that, in the absence of leucine analogs, the ratio between the two populations remained largely unaltered throughout the experiment, indicating comparable growth rates of both the resistant and nonresistant E. coli populations (Fig. 2). However, when the growth media was supplemented with norvaline (0.5 mM), the tavaborole-resistant population rapidly declined to less than 3% of the total cell count (Fig. 2). Thus, this experiment illustrated that tavaborole-resistant E. coli are indeed vulnerable to the toxicity of leucine-like amino acids, showing that we can capitalize on the genetic defects of tavaborole-resistant populations to create the additional burden (negative selective pressure) associated with antibiotic-resistance mutations and selectively inhibit antibiotic-resistant cells.
Fig. 2.
Tavaborole-resistant cells can be rapidly and selectively suppressed under conditions that require LeuRS editing activity. Flow-cytometry analysis of a competition assay in which two E. coli populations of E. coli were mixed at a ratio of ∼1:1 and grown for 5 d either in chemically defined media (“control”) or in the same media supplemented with tavaborole (“+Tava,” 1 μg/mL) or norvaline (“+Nva,” 0.5 mM). The competing populations were 1) wild-type E. coli that expressed genome-encoded GFP (corresponding to the GFP-positive peaks) and 2) populations of the evolved tavaborole-resistant E. coli (corresponding to the GFP-negative peaks and representing samples of population A or population B that were collected on the final day of the evolutionary experiment shown in Fig. 1A and SI Appendix, Table S1).
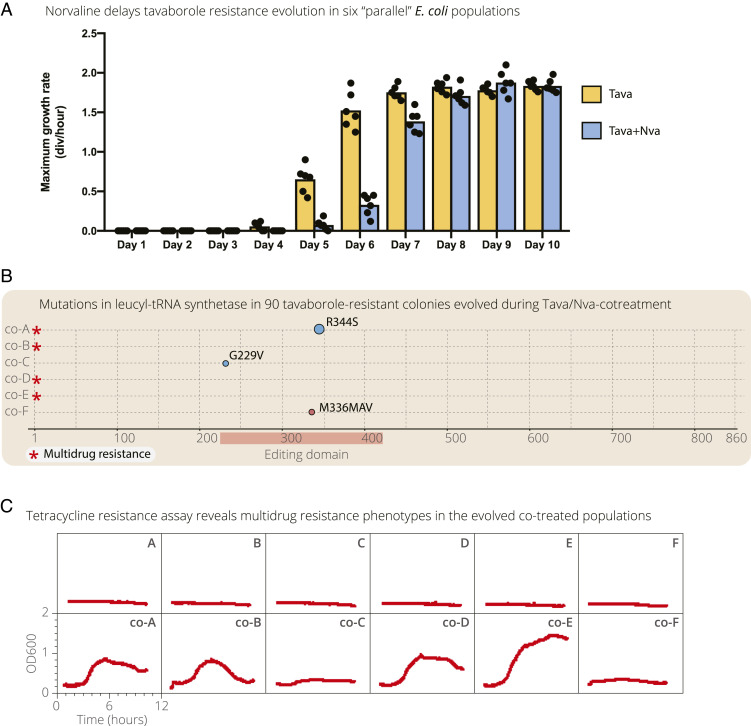
We finally set up an evolutionary experiment in which wild-type E. coli were simultaneously treated with tavaborole and norvaline. This experiment was meant to compare our consecutive application of tavaborole and norvaline with a more traditional approach in which multiple drugs/small molecules are applied simultaneously. The idea of this approach is to maximize fitness costs to prevent antibiotic assistance evolution in the first place, rather than to manage resistant cells after the first drug application using application of the second drug, small molecule, or environmental factor (this strategy is reviewed in ref. 46). For this purpose, we repeated our evolutionary experiment with six initially identical E. coli organisms with the only difference that, in addition to tavaborole, cells were continuously treated with norvaline (0.3 mM). We found that, compared to the initial evolutionary experiment, it took E. coli ∼1 to 2 d longer to evolve detectable levels of tavaborole resistance, consistent with the idea that norvaline can attenuate tavaborole resistance evolution (Fig. 3A and SI Appendix, Fig. S7). Sequencing the leuS gene in 90 random colonies (15 colonies per each population) revealed that only 6 colonies had a mutated leuS gene, corresponding to R344S, G229V, and M336MAV mutations in the editing domain of LeuRS (Fig. 3B). These sequencing data suggest that tavaborole-resistance evolution was acquired through a more general mechanism than alteration of the drug target. We therefore tested if the evolved populations are resistant to other drugs, using a tetracycline growth assay (Fig. 3C). We found that four of the evolved populations could rapidly grow in the presence of tetracycline (10 μg/mL), indicating the multidrug resistance phenotype (Fig. 3C). By contrast, the six E. coli populations that were evolved in the presence of tavaborole alone could not grow in the presence of tetracycline (Fig. 3C). Thus, although the simultaneous treatment delayed tavaborole resistance evolution, it also provoked multidrug resistance evolution in several lineages, consistent with the idea that excessive selective pressure can compromise subsequent management of resistant populations (1, 2).
Fig. 3.
Simultaneous treatment of E. coli with tavaborole and norvaline slows down rates of tavaborole resistance evolution but increases the risk of multidrug resistance. (A) The plot shows gradual acquisition of tavaborole resistance in the evolving E. coli populations: six populations that evolved in the presence of tavaborole (as shown in Fig. 1 and SI Appendix, Fig. S1) and six populations that evolved in the presence of tavaborole and norvaline (0.3 mM). For this experiment, aliquots of each of the evolving populations were taken at days 1 to 10 of the experiment, regrown overnight, diluted to OD600 = 0.1, and regrown in the presence of tavaborole (10 μg/mL). A illustrates that, in the presence of tavaborole and norvaline, it took the E. coli populations ∼1 to 2 d longer to evolve detectable tavaborole resistance compared to the experiment in which E. coli were treated with tavaborole only. (B) Targeted sequencing of the leuS gene in 90 random colonies (15 colonies per each of the 6 populations) shows that most of the E. coli colonies that evolved during norvaline/tavaborole cotreatment do not have mutations in the leuS gene, indicating that tavaborole resistance in these cells was acquired through mechanisms other than mutations in the editing domain of LeuRS. (C) The growth curves illustrates tetracycline resistance in the E. coli populations (collected at day 8 of the experiment) that evolved in the presence of tavaborole and norvaline (growth curves A–F) or tavaborole alone (growth curves co-A–co-F). Four of the six populations evolved in the presence of tavaborole, and norvaline could also grow in the presence of tetracycline (10 μg/mL), indicating a multidrug resistance phenotype.
Discussion
Overall, our case study illustrates that the natural competition between resistant and nonresistant cells can be drastically enhanced by exploiting the burden associated with those mutations that confer antibiotic resistance. In the case of tavaborole, the fitness cost associated with LeuRS editing deficiency can be amplified by adding norvaline to the growth media, making LeuRS editing defects highly disadvantageous for E. coli growth and leading to the rapid and selective suppression of drug-resistant E. coli populations.
Given that resistance to many synthetic drugs evolves primarily through alterations of a drug’s target (11), it is entirely possible that a conceptually similar approach could be applied to develop antiresistance strategies for other drugs. By encouraging the natural competition between resistant and nonresistant cells during the “off” phases of the “on–off” cycles of adaptive therapy (e.g., by administration of norvaline following the cycle of tavaborole administration), we can potentially accelerate adaptive therapies by significantly shortening their drug-free phases. Thus, our study provides an important opportunity and adds to the current arsenal of strategies [including collateral sensitivity, spatial competition constrains (47), containment strategy (48), and others (49)] of how to use a drug to minimize the threat of drug-resistance evolution.
Can this approach help resume therapeutic testing of drugs that were suspended due to rapid evolution of therapeutic resistance? In regard to tavaborole derivatives, can this approach give a second chance to drugs like AN3365 (39, 50)? Because administration of norvaline is not fully feasible in clinical settings, the answer to this question will depend on the future studies in which norvaline will be replaced by other agents for amplified mistranslation. One potential alternative to norvaline is aminoglycosides, a class of antibiotics that are used to treat various bacterial infections and that amplify mistranslation by targeting ribosomes and rendering them error-prone (51–54). Thus, it will be important to test if, in clinical settings, aminoglycosides can be used instead of tavaborole to manage the evolution of therapeutic resistance.
Finally, it is important to note that, unlike laboratory-evolved resistant cells, clinical isolates typically have smaller fitness costs of resistance, as was shown in the cases of Mycobacteria and other human pathogens (9, 55–58). This is because, due to longer periods of drug application (among other factors), clinical isolates frequently accumulate secondary, compensatory mutations. It is therefore plausible that successful off-phases of adaptive therapy may require shorter courses of drug administration to lower risks of acquisition of secondary mutations and preserve the strongest growth defects in the emerging populations of resistant cells.
Materials and Methods
Evolution of Tavaborole Resistance in E. coli.
The tavaborole-resistant E. coli were evolved by growing six initially identical populations of BL21(DE3) in LB media at 37 °C using 12-well plates (Corning, no. 353047) that were incubated in a Synergy|HTX multimode plate reader (BioTek). Each population was growing in 1.5 mL of lysogeny broth (LB) media, and, each day, 5% of each of the evolving cell cultures was transferred onto a new plate where each culture was diluted with 1.5 mL of fresh LB medium. During the 8 d of the experiment, the tavaborole concentration was gradually increased from the initial of 2.5 μg/mL to the final of 180 μg/mL (SI Appendix, Fig. S1). The cell growth during the experiment was continuously assessed by making one OD600 measurement per minute for each of the evolving cultures. Maximum growth rates were calculated by using the package for growth rate analysis described in ref. 59.
Sequencing of leuS Gene in Tavaborole-Resistant E. coli Colonies.
To determine the sequence of leuS gene in tavaborole-resistant cells, each of the evolved E. coli populations was plated on an LB-agar plate supplemented with tavaborole (60 μg/mL) and grown overnight at 37 °C. Then 20 individual colonies were picked at random from each of the six plates. Each of these colonies was then used as a DNA template for PCR amplification of leuS sequence. To amplify the leuS gene, we used Phusion High-Fidelity DNA Polymerase (NEB) and primers 1 and 2 (here and below we refer to the primers according to the SI Appendix, Table S2, and SI Appendix, Supplementary Data 1). The amplified gene was analyzed by Sanger sequencing (The Keck DNA Sequencing Facility at Yale School of Medicine) using each of the following primers for sequencing: 1, 3, and 4.
E. coli Genetics to Alter the leuS Sequence.
To introduce tavaborole-resistance mutations in the leuS gene into the E. coli genome, we used the previously published protocol for recombineering, a homologous recombination-based method of genetic engineering (60). In brief, the editing domain-coding segment of the leuS gene was PCR-amplified by using Phusion High-Fidelity DNA Polymerase (NEB), primers 1 and 2, and the tavaborole-resistant colonies as DNA template. The PCR product was used for transformation of the electro-competent E. coli (BL21[DE3]) that were provided with λ Red recombination genes by using the pSIM6 plasmid, as described in “Provision of the λ Red recombination genes” section of ref. 60 (60). The transformed cells were then grown for 1 h in LB medium at 37 °C, transferred onto LB-agar plates supplemented with tavaborole (10 μg/mL), and grown overnight at 37 °C. The resulting clones were grown in LB media, analyzed by Sanger sequencing of the PCR-amplified leuS gene (using primers 3 and 4), and used for growth assessment at 37 °C in LB media using the Synergy|HTX multimode plate reader (BioTek).
To replace the mutated leuS gene in tavaborole-resistant cells with the wild-type leuS, we fused the PCR-amplified leuS gene (using primers 3 and 4 and an aliquot of the wild-type E. coli as a DNA template) with the Neo-selection gene using the In-Fusion HD Cloning kit (Takara) and integrated the resulting leuS-Neo DNA into genomes of the tavaborole-resistant cells by using the scarred method for insertion of a nonselectable DNA fragment recombineering protocol with the Neo gene as a selectable marker (60). To introduce the genomic T247V and T248V mutations into the leuS gene, we used the identical protocol except that we first used primers 14 and 15 (SI Appendix, Table S2) to mutate leuS gene in the LeuRS-coding plasmid leuS-pBAD28 (SI Appendix, Supplementary Data 3) using a QuikChange II Site-directed mutagenesis kit (Agilent) and then used primers 3 and 4 to amplify the plasmid-encoded mutated leuS gene for its integration into the E. coli genome.
Library Preparation and Whole-Genome Sequencing.
Genomic DNA from the evolving E. coli populations was purified by using 1 mL of each E. coli sample that was treated by using the QIAamp DNA Mini Kit (Qiagen). For the sequencing library construction, the genomic DNA was fragmented by ultrasonication to a size between 50 and 800 bp. The DNA fragments (from 100 to 300 bp) were selected and further capped with the flanking dATP ends. The dTTP tailed adapters were ligated to both ends of the DNA fragments. The ligation product was then amplified and subjected to the single-strand circularization process. The remaining linear molecule was digested with the exonuclease to generate a single-strand circular DNA library. Sequencing was conducted according to the standard BGISEQ-500 protocol. Zebracall (a base-calling software) was used to make raw reads.
Sequencing Quality Control and Mapping.
Quality control of sequencing reads was performed before mapping. Reads with adapters shorter than 100 bp were removed. Reads containing more than 1% of unknown base and containing more than one base with a Phred score lower than 10 were removed. The quality control step resulted in cleaned paired-end reads with a more than 1,000-fold sequencing depth of the reference genome. The reads after filtration were mapped to reference sequences using BWA 0.5.6 according to the standard settings (61). For each alignment result, local realignment was performed with GATK 2.7 built-in RealignerTargetCreator and IndelRealigner tools to clean up mapping artifacts due to reads mapping on the edges of indels (62). The resulting files in BAM format were then prepared for initial single-nucleotide variations (SNV)/indel calling.
Identification of SNVs and Indels and Data Deposition.
The GATK 2.7 pipeline (62) was utilized to identify the SNVs and indels using default parameters. The variants were filtered by the following criteria: QUAL < 50 or FS > 3 or BaseQRankSum > 3 or MQRankSum > 3 or ReadPosRankSum > 3 or MQ < 10 or DP < 10. Next, annotation was performed for observed variant types including synonymous type, nonsynonymous type, frameshifts, and variants outside the coding region. The sequencing data were then deposited to the China National GeneBank CNSA CNGB Sequence Archive (https://db.cngb.org/search/project/CNP0000842; accession number CNP0000842), and the links to the datasets are provided in SI Appendix, Supplementary Data 1–6.
Preparation of the tRNALeu Transcript for Biochemical Assays.
To produce the tRNALeu transcript for enzymatic assays, we used the tRNALeu sequence for tRNALeuCAA-1–1 from the E. coli K-12 MG1655 strain (http://gtrnadb.ucsc.edu/genomes/bacteria/Esch_coli_K_12_MG1655/genes/tRNA-Leu-CAA-1-1.html). We first produced the DNA template for T7 transcription using PCR with Taq polymerase (NEB) and primers 11, 12, and 13. We then used the HiScribe T7 High Yield RNA Synthesis Kit (NEB) to produce tRNALeu. For this purpose, 4 μg of the DNA template were added to the reaction mixture (the total volume of 40 μL), containing 10 mM ATP, 10 mM CTP, 10 mM GTP, 10 mM UTP, and 200 mM GMP (to prevent the presence of phosphates at the tRNALeu 5′-end) and 4 μL of T7 RNA polymerase solution. The reaction mixture was incubated at 40 °C for 15 h and treated with 2 units of DNase I (NEB) at 37 °C for 1 h followed by DNase I heat inactivation. The resulting mixture was diluted to 100 μL with water, mixed with 100 μL of acid phenol (pH 4.5):chloroform:IAA (125:24:1). After the extraction, the aqueous phase was used for size-exclusion chromatography by using a disposable PD SpinTrap G-25 column (GE Healthcare Life Sciences). The purified tRNALeu was then ethanol precipitated, dissolved in water, analyzed by 12% polyacrylamide gel/7 M urea, and stored at 2 μg/μL concentration at −20 °C.
Purification of LeuRS and Its Mutants for Biochemical Assays.
To purify recombinant LeuRS synthetase, the leuS gene and its mutants were PCR-amplified by using primers 3 and 4 and genomic DNA from either wild-type or tavaborole-resistant E. coli colonies as the template. The PCR products were cloned into pBAD28 vector (ATCC 87400) by using In-Fusion HD Cloning kit (Takara) by using primers 7 and 8 to amplify the pBAD28 vector (SI Appendix, Table S2 and Supplementary Data 3). To express LeuRS, E. coli BL21(DE3) cells were transformed with LeuRS-coding plasmids, and the synthesis of recombinant proteins was induced at 16 °C by the addition of arabinose (to the final concentration of 0.2% wt/vol) to the growth medium. The proteins were then purified by the kit of Ni-affinity purification (BioRad), followed by ammonium sulfate protein fractionation and size-exclusion chromatography on a Superdex Increase 200 10/300 GL column (Pharmacia Biotech) in the aminoacylation buffer containing 60 mM Tris⋅HCl, pH 7.5, 10 mM MgCl2, and 1 mM dithiothreitol. Then, the proteins were concentrated to 10 mg/mL using Amicon Ultracel centrifugal filters (molecular weight cutoff, 30 kDa) and immediately used in biochemical assays.
LeuRS Aminoacylation Activity Assay.
To assess LeuRS aminoacylation activity, we used the previously described protocol monitoring synthesis of [14C]-labeled leucyl-tRNAL (63). In brief, we measured LeuRS activity in the aminoacylation buffer supplemented with 8 μM tRNALeu transcript, 40 μM L-leucine, 2 μM L-[3H]-leucine (PerkinElmer, 144 Ci/mmol), tavaborole (0 to 500 μg/mL), and a recombinant LeuRS from E. coli (the final concentration 20 nM). The reaction was initiated by the addition of LeuRS and terminated by the addition of trichloroacetic acid (TCA) to a 5% vol/vol concentration. In the meantime, we prepared Whatman 3 MM filter pads by adding 100 μL of 5% TCA onto each pad and drying each pad before taking measurements. Then, 10 μL of each reaction mixture was transferred to the filter, and the filters were immediately soaked in 15 mL of 5% ice-cold TCA. Each filter was washed 3 × 20 min by using 15 mL of ice-cold 5% TCA to remove the free amino acid. Then each filter was washed by using 15 mL of 96% ethanol, dried, mixed with 5 mL of scintillation fluid (MP Biomedicals), and used for measurements by using LS 6500 scintillation counter (Beckman Coulter).
LeuRS Editing Activity Assay.
To assess LeuRS editing activity, we measured the ability of LeuRS and its mutants to produce the misaminoacylated tRNALeu, Ile-tRNALeu, as described previously (64). In brief, we measured LeuRS activity in the aminoacylation buffer supplemented with 8 μM tRNALeu transcript, 40 mM L-isoleucine (99%, W527602; Sigma-Aldrich), 2 μM L-[14C]-isoleucine (PerkinElmer, 322 mCi/mmol), tavaborole (0 to 500 μg/mL), and a recombinant LeuRS from E. coli (final concentration is 2 μM). The reaction was initiated by the addition of LeuRS and terminated by the addition of TCA to 5% concentration. The amount of Ile-tRNALeu was then assessed by using aliquot transfer on Whatman 3 MM filter pads, as described for the aminoacylation activity assay.
Construction of GFP-Encoding E. coli.
The electrocompetent E. coli cells (BL21[DE3]) were transformed with the pOSIP-CT (TetR, P21) integration plasmid coding for superfolder GFP (sfGFP) (SI Appendix, Supplementary Data 4). The transformed cells were grown at 37 °C for 1 h in LB media and then for 12 h in LB media supplemented with tetracycline (50 μg/mL). Then, the GFP-positive cells were sorted by using a BD FACS Aria III Cell Sorter (BD Biosciences). The sorted cells were plated on a 100-mm LB-agar petri dish and grown at 37 °C for 16 h. Next, individual cell colonies were regrown in LB media at 37 °C, collected at approximately OD600 = 1, and stored at −80 °C in LB media supplemented with 50% glycerol.
Competition Assay in a Microbioreactor.
Before the experiment, the milliliter-scale microbioreactor chips (the total volume of 2 mL) (SI Appendix, Fig. S5) were γ-irradiated and sealed as part of the standard preinoculation sterile protocol (65). The medium bottles and feed lines were autoclaved separately, and 0.22-µm filters were installed between the bottles and the lines to prevent microbial contamination of the microbioreactor. Before each experiment, both the antibiotic-resistant populations and the sfGFP-positive nonresistant E. coli were grown separately for ∼12 h in the chemically defined media (New Minimal Media [NMM]) containing 7.5 mM (NH4)2SO4, 8.5 mM NaCl, 22 mM KH2PO4, 50 mM K2HPO4, 1 mM MgSO4, 20 mM D-glucose, 50 mg/L of 20 canonical amino acids, 1 µg/mL each of Ca2+ and Fe2+, 0.01 µg/mL each of Cu2+, Zn2+, Mn2+, and Mo2+, 10 µg/mL of thiamine, and 10 µg/mL biotin) to achieve the exponential growth phase for each of the populations. Then, the cell cultures were mixed at an ∼1:1 ratio and immediately injected into a microbioreactor (the total volume of 2 mL) for the competition assay. Three microbioreactors were used simultaneously to support cell growth in 1) NMM media, 2) NMM media supplemented with tavaborole (1 µg/mL), and 3) NMM media supplemented with norvaline (0.5 mM). Each of the microbioreactors was operating in the turbidostatic mode, maintaining the OD600 of mixed populations at 1, and the fermentation temperature was controlled at 37 ± 0.1 °C throughout the experiment. Each day, 100 µL of E. coli populations (5% of the total population size) was ejected from the each of the microbioreactors and used for flow cytometry analysis to immediately assess the ratio between sfGFP-positive and sfGFP-negative cells.
Flow Cytometry.
Analysis of live cells by flow cytometry was carried out on the Attune NxT flow cytometer (Invitrogen). For each measurement, we used 100,000 cells. FlowJo v10 software was used to analyze the flow cytometry data.
Supplementary Material
Acknowledgments
We thank the following scientists for insightful discussions and perceptive comments on various drafts of this manuscript and for bringing to our attention gaps in our knowledge and holes in our logic: Jessica Cunningham Reynolds (Moffitt Cancer Center); Alita Burmeister and Richard Prum (Department of Ecology and Evolutionary Biology, Yale); Jeffrey Tharp, Jon Fischer, Kazuaki Amikura, and Oscar Vargas-Rodriguez (Department of Molecular Biophysics and Biochemistry, Yale); Antonia van den Elzen (Department Cellular and Molecular Physiology, Yale); Haissi Cui (The Scripps Research Institute); and Ya-Ming Hou and her laboratory members (Thomas Jefferson University). We also thank the laboratory of Alanna Schepartz (University of California, Berkeley) for providing access to the flow cytometer and Kenneth Nelson (Yale Flow Cytometry Core Facility) for his technical assistance. This work was supported by the Guangdong Provincial Key Laboratory of Genome Read and Write Grant 2017B030301011 (to Y.S.) and NIH Grant R35GM122560 (to D.S.).
Footnotes
Competing interest statement: X.F., J.-T.Z., and Y.S. are employees of BGI. J.S. is an employee of Thermo Fisher Scientific. K.L. and H.L. are employees of Erbi Biosystems.
Data deposition: The sequencing data were deposited to the China National GeneBank CNSA CNGB Sequence Archive (accession number CNP0000842), and the links to the datasets are provided in SI Appendix, Supplementary Data 1–6. Supplementary Data 1–6 were deposited in the Figshare Data Repository and can be accessed at https://doi.org/10.6084/m9.figshare.11886288.v2.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003132117/-/DCSupplemental.
Data Availability.
Supplementary Data 1–6 were deposited in the Figshare Data Repository and can be accessed at https://doi.org/10.6084/m9.figshare.11886288.v2 (66).
References
- 1.Staňková K., Resistance games. Nat. Ecol. Evol. 3, 336–337 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Cunningham J. J., A call for integrated metastatic management. Nat. Ecol. Evol. 3, 996–998 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Thomas F. et al., Is adaptive therapy natural? PLoS Biol. 16, e2007066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hochberg M. E., An ecosystem framework for understanding and treating disease. Evol. Med. Public Health 2018, 270–286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J., Cunningham J. J., Brown J. S., Gatenby R. A., Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nat. Commun. 8, 1816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham J. J., Gatenby R. A., Brown J. S., Evolutionary dynamics in cancer therapy. Mol. Pharm. 8, 2094–2100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maharjan R., Ferenci T., The fitness costs and benefits of antibiotic resistance in drug-free microenvironments encountered in the human body. Environ. Microbiol. Rep. 9, 635–641 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Melnyk A. H., Wong A., Kassen R., The fitness costs of antibiotic resistance mutations. Evol. Appl. 8, 273–283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson D. I., Hughes D., Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Lenski R. E., Bacterial evolution and the cost of antibiotic resistance. Int. Microbiol. 1, 265–270 (1998). [PubMed] [Google Scholar]
- 11.Spratt B. G., Resistance to antibiotics mediated by target alterations. Science 264, 388–393 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Munita J. M., Arias C. A., Mechanisms of antibiotic resistance. Microbiol. Spectr. 4, 1–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S., Lieberman T. D., Kishony R., Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc. Natl. Acad. Sci. U.S.A. 111, 14494–14499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichol D. et al., Steering evolution with sequential therapy to prevent the emergence of bacterial antibiotic resistance. PLOS Comput. Biol. 11, e1004493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuentes-Hernandez A. et al., Using a sequential regimen to eliminate bacteria at sublethal antibiotic dosages. PLoS Biol. 13, e1002104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao B. et al., Exploiting temporal collateral sensitivity in tumor clonal evolution. Cell 165, 234–246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mira P. M. et al., Rational design of antibiotic treatment plans: A treatment strategy for managing evolution and reversing resistance. PLoS One 10, e0122283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker S. J. et al., Discovery of a new boron-containing antifungal agent, 5-fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (AN2690), for the potential treatment of onychomycosis. J. Med. Chem. 49, 4447–4450 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Rock F. L. et al., An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316, 1759–1761 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Zhang P., Ma S., Recent development of leucyl-tRNA synthetase inhibitors as antimicrobial agents. MedChemComm 10, 1329–1341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jinna S., Finch J., Spotlight on tavaborole for the treatment of onychomycosis. Drug Des. Devel. Ther. 9, 6185–6190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manhas R. et al., Leishmania donovani parasites are inhibited by the benzoxaborole AN2690 targeting leucyl-tRNA synthetase. Antimicrob. Agents Chemother. 62, e00079-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonoiki E. et al., Antimalarial benzoxaboroles target Plasmodium falciparum leucyl-tRNA synthetase. Antimicrob. Agents Chemother. 60, 4886–4895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palencia A. et al., Cryptosporidium and Toxoplasma parasites are inhibited by a benzoxaborole targeting leucyl-tRNA synthetase. Antimicrob. Agents Chemother. 60, 5817–5827 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin W. et al., Design and synthesis of α-phenoxy-N-sulfonylphenyl acetamides as Trypanosoma brucei Leucyl-tRNA synthetase inhibitors. Eur. J. Med. Chem. 185, 111827 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Zhang F. et al., Discovery of N-(4-sulfamoylphenyl)thioureas as Trypanosoma brucei leucyl-tRNA synthetase inhibitors. Org. Biomol. Chem. 11, 5310–5324 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Ding D. et al., Discovery of novel benzoxaborole-based potent antitrypanosomal agents. ACS Med. Chem. Lett. 1, 165–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs R. T. et al., SCYX-7158, an orally-active benzoxaborole for the treatment of stage 2 human African trypanosomiasis. PLoS Negl. Trop. Dis. 5, e1151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding D. et al., Design, synthesis, and structure-activity relationship of Trypanosoma brucei leucyl-tRNA synthetase inhibitors as antitrypanosomal agents. J. Med. Chem. 54, 1276–1287 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Si Y. et al., Antibacterial activity and mode of action of a sulfonamide-based class of oxaborole leucyl-tRNA-synthetase inhibitors. ACS Infect. Dis. 5, 1231–1238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X. et al., Discovery of a potent and specific M. tuberculosis leucyl-tRNA synthetase inhibitor: (S)-3-(Aminomethyl)-4-chloro-7-(2-hydroxyethoxy)benzo[c][1,2]oxaborol-1(3H)-ol (GSK656). J. Med. Chem. 60, 8011–8026 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Palencia A. et al., Discovery of novel oral protein synthesis inhibitors of Mycobacterium tuberculosis that target leucyl-tRNA synthetase. Antimicrob. Agents Chemother. 60, 6271–6280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gudzera O. I. et al., Identification of Mycobacterium tuberculosis leucyl-tRNA synthetase (LeuRS) inhibitors among the derivatives of 5-phenylamino-2H-[1,2,4]triazin-3-one. J. Enzyme Inhib. Med. Chem. 31, 201–207 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Gudzera O. I. et al., Discovery of potent anti-tuberculosis agents targeting leucyl-tRNA synthetase. Bioorg. Med. Chem. 24, 1023–1031 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Hu Q. H. et al., Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase. Sci. Rep. 3, 2475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palencia A. et al., Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 19, 677–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao H. et al., Analysis of the resistance mechanism of a benzoxaborole inhibitor reveals insight into the leucyl-tRNA synthetase editing mechanism. ACS Chem. Biol. 10, 2277–2285 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Sarkar J., Mao W., Lincecum T. L. Jr., Alley M. R., Martinis S. A., Characterization of benzoxaborole-based antifungal resistance mutations demonstrates that editing depends on electrostatic stabilization of the leucyl-tRNA synthetase editing cap. FEBS Lett. 585, 2986–2991 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta A. et al., A polymorphism in leuS confers reduced susceptibility to GSK2251052 in a clinical isolate of Staphylococcus aureus. Antimicrob. Agents Chemother. 60, 3219–3221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Dwyer K. et al., Bacterial resistance to leucyl-tRNA synthetase inhibitor GSK2251052 develops during treatment of complicated urinary tract infections. Antimicrob. Agents Chemother. 59, 289–298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L. et al., Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl. Acad. Sci. U.S.A. 108, 9378–9383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melnikov S. V. et al., Error-prone protein synthesis in parasites with the smallest eukaryotic genome. Proc. Natl. Acad. Sci. U.S.A. 115, E6245–E6253 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karkhanis V. A., Mascarenhas A. P., Martinis S. A., Amino acid toxicities of Escherichia coli that are prevented by leucyl-tRNA synthetase amino acid editing. J. Bacteriol. 189, 8765–8768 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cvetesic N. et al., Proteome-wide measurement of non-canonical bacterial mistranslation by quantitative mass spectrometry of protein modifications. Sci. Rep. 6, 28631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ribas de Pouplana L., Santos M. A., Zhu J. H., Farabaugh P. J., Javid B., Protein mistranslation: Friend or foe? Trends Biochem. Sci. 39, 355–362 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Baym M., Stone L. K., Kishony R., Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351, aad3292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bacevic K. et al., Spatial competition constrains resistance to targeted cancer therapy. Nat. Commun. 8, 1995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen E., Karslake J., Woods R. J., Read A. F., Wood K. B., Antibiotics can be used to contain drug-resistant bacteria by maintaining sufficiently large sensitive populations. PLoS Biol. 18, e3000713 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen E., Woods R. J., Read A. F., How to use a chemotherapeutic agent when resistance to it threatens the patient. PLoS Biol. 15, e2001110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwon N. H., Fox P. L., Kim S., Aminoacyl-tRNA synthetases as therapeutic targets. Nat. Rev. Drug Discov. 18, 629–650 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Krause K. M., Serio A. W., Kane T. R., Connolly L. E., Aminoglycosides: An overview. Cold Spring Harb. Perspect. Med. 6, a027029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi Y., Igarashi M., Destination of aminoglycoside antibiotics in the ‘post-antibiotic era’. J. Antibiot. (Tokyo), 4–14 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Arenz S., Wilson D. N., Blast from the past: Reassessing forgotten translation inhibitors, antibiotic selectivity, and resistance mechanisms to aid drug development. Mol. Cell 61, 3–14 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Arenz S., Wilson D. N., Bacterial protein synthesis as a target for antibiotic inhibition. Cold Spring Harb. Perspect. Med. 6, a025361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Böttger E. C., Springer B., Pletschette M., Sander P., Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat. Med. 4, 1343–1344 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Gagneux S. et al., The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312, 1944–1946 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Comas I. et al., Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 44, 106–110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacLean R. C., Vogwill T., Limits to compensatory adaptation and the persistence of antibiotic resistance in pathogenic bacteria. Evol. Med. Public Health 2015, 4–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mira P., Barlow M., Meza J. C., Hall B. G., Statistical package for growth rates made easy. Mol. Biol. Evol. 34, 3303–3309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharan S. K., Thomason L. C., Kuznetsov S. G., Court D. L., Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKenna A. et al., The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Francklyn C. S., First E. A., Perona J. J., Hou Y. M., Methods for kinetic and thermodynamic analysis of aminoacyl-tRNA synthetases. Methods 44, 100–118 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Splan K. E., Musier-Forsyth K., Boniecki M. T., Martinis S. A., In vitro assays for the determination of aminoacyl-tRNA synthetase editing activity. Methods 44, 119–128 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perez-Pinera P. et al., Synthetic biology and microbioreactor platforms for programmable production of biologics at the point-of-care. Nat. Commun. 7, 12211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melnikov S., Söll D., Supplementary data for "Exploiting evolutionary trade-offs for management of drug-resistant populations." Figshare. 10.6084/m9.figshare.11886288.v2. Deposited 18 May 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary Data 1–6 were deposited in the Figshare Data Repository and can be accessed at https://doi.org/10.6084/m9.figshare.11886288.v2 (66).