Significance
Reproductive success in placental mammals relies on proper development of the trophoblast lineage. In particular, a proper balance in self-renewal vs. differentiation of trophoblast progenitors is critical for establishment of pregnancy. A defect in this process causes early pregnancy loss. Here, we showed that the Hippo signaling effector, TEAD4, is essential for trophoblast progenitor self-renewal in early postimplantation mammalian embryos. Using genetic mouse models and human TSCs, including TSCs from patients with recurrent pregnancy losses, we identified a TEAD4-dependent, evolutionarily conserved gene expression program that promotes stemness and cell proliferation in trophoblast progenitors and ensures in utero survival of the developing fetus.
Keywords: placenta, trophoblast progenitor, Hippo signaling, TEAD4, recurrent pregnancy loss
Abstract
Early pregnancy loss affects ∼15% of all implantation-confirmed human conceptions. However, evolutionarily conserved molecular mechanisms that regulate self-renewal of trophoblast progenitors and their association with early pregnancy loss are poorly understood. Here, we provide evidence that transcription factor TEAD4 ensures survival of postimplantation mouse and human embryos by controlling self-renewal and stemness of trophoblast progenitors within the placenta primordium. In an early postimplantation mouse embryo, TEAD4 is selectively expressed in trophoblast stem cell–like progenitor cells (TSPCs), and loss of Tead4 in postimplantation mouse TSPCs impairs their self-renewal, leading to embryonic lethality before embryonic day 9.0, a developmental stage equivalent to the first trimester of human gestation. Both TEAD4 and its cofactor, yes-associated protein 1 (YAP1), are specifically expressed in cytotrophoblast (CTB) progenitors of a first-trimester human placenta. We also show that a subset of unexplained recurrent pregnancy losses (idiopathic RPLs) is associated with impaired TEAD4 expression in CTB progenitors. Furthermore, by establishing idiopathic RPL patient-specific human trophoblast stem cells (RPL-TSCs), we show that loss of TEAD4 is associated with defective self-renewal in RPL-TSCs and rescue of TEAD4 expression restores their self-renewal ability. Unbiased genomics studies revealed that TEAD4 directly regulates expression of key cell cycle genes in both mouse and human TSCs and establishes a conserved transcriptional program. Our findings show that TEAD4, an effector of the Hippo signaling pathway, is essential for the establishment of pregnancy in a postimplantation mammalian embryo and indicate that impairment of the Hippo signaling pathway could be a molecular cause for early human pregnancy loss.
Human reproduction is an inefficient process as the majority of conceptions do not lead to live birth (1, 2). Many of these losses occur prior to blastocyst implantation, and in many women conceptions are lost within 3 to 4 wk of implantation, leading to infertility. However, ∼15% of all conceptions are associated with clinical early pregnancy losses where pregnancy is confirmed but does not progress beyond 20 wk of gestation (1–5). Although a majority of early pregnancy losses are sporadic, 1 to 3% of pregnant women suffer from recurrent pregnancy losses (RPLs, defined as the occurrence of two or more consecutive pregnancy losses) (1, 3–5). While fetal chromosomal abnormalities are responsible for a majority (∼70%) of sporadic miscarriages, only a smaller fraction of RPLs is associated with chromosomal abnormalities. Furthermore, almost half of the RPLs are idiopathic (of unknown origin) and therefore difficult to counsel; thus, treatment is empirical. Early pregnancy losses are devastating to patients, and a better understanding of underlying mechanisms is necessary for better diagnosis, appropriate counseling, and development of patient-specific treatment options.
Studies in mutant mouse models showed that failure in placentation often leads to in utero embryonic death (6, 7). Therefore, impaired placentation due to defective development or function of trophoblast cell lineages is considered one of the major underlying causes of early pregnancy loss. Disruptions of trophoblast progenitor differentiation and defective placentation have also been implicated as probable causes of pregnancy-associated complications, such as preeclampsia, intrauterine growth restriction, and preterm birth (8–17). These pathological conditions cause significant morbidity and mortality for both mother and fetus. Furthermore, defective trophoblast development and placentation could lead to alteration in fetal developmental programming, leading to poor postnatal health or susceptibility to adult diseases (18–20). However, regulatory mechanisms that are critical to maintain the stemness and self-renewal of trophoblast progenitors within a placenta primordium or promote their differentiation during subsequent stages of placentation are poorly defined.
Development of the trophoblast cell lineage begins with the establishment of the trophectoderm (TE) during blastocyst maturation. Proper development of the TE is essential for embryo implantation (21–24). In a postimplantation mouse embryo, multipotent trophoblast stem cell–like progenitor cells (TSPCs) arise from the TE. The TSPCs undergo extensive self-renewal to develop the placenta primordium, consisting of the extraembryonic ectoderm (ExE)/ectoplacental cone (EPC) and the chorionic ectoderm (25–27). Subsequently, lineage-specific trophoblast progenitors arise from TSPCs, which differentiate to specialized trophoblast subtypes, leading to successful placentation (27). Although human placentation differs significantly from that of the mouse, very early stages of human placentation are associated with development of columns of proliferating cytotrophoblast (CTB) progenitors, which, similar to mouse TSPCs, undergo extensive self-renewal and differentiation to form a villous placenta (26, 28). Furthermore, expressions of several key trophoblast stem state factors, such as GATA-binding proteins 2 and 3 (GATA2 and GATA3), transcription factor AP-2 gamma (TFAP2C), and E74-like ETS transcription factor 5 (ELF5), are conserved in both mouse TSPCs and human CTBs (29–31), indicating that these conserved factors could establish common regulatory programs in both mouse TSPCs and human CTBs to regulate their stemness vs. differentiation balance.
In an earlier study, we showed that expression of TEA domain transcription factor 4 (TEAD4) is conserved in the TE lineages of multiple mammalian blastocysts, including mouse and human (32). Furthermore, using mouse models, our laboratory (32, 33) and others (34–36) have defined that TEAD4, in association with its cofactor, YAP1, regulates a global gene expression program to establish TE lineage development in preimplantation mouse embryos. Intriguingly, along with the TE lineages in preimplantation embryos, Tead4 messenger RNA (mRNA) expression has been detected within the TSPCs of a postimplantation mouse embryo (34). However, the importance of TEAD4 for trophoblast development after embryo implantation is unknown. Despite TEAD4 expression being conserved in CTB progenitors of a first-trimester human placenta, the functional importance of TEAD4 in the context of human trophoblast lineage has not been tested. Therefore, in this study, we investigate the importance of TEAD4 in the development and function of trophoblast progenitors of early postimplantation mouse and human embryos. We used conditional knockout mouse models and human trophoblast stem cells (TSCs) to evaluate TEAD4-dependent molecular mechanisms involved in trophoblast progenitor self-renewal. In addition, using patient-specific human TSCs, we tested whether alteration of those mechanisms is associated with idiopathic RPL. Our analyses revealed an evolutionarily conserved mechanism in which TEAD4 maintains the stemness and promotes self-renewal of trophoblast progenitors in postimplantation embryos to ensure placentation and progression of pregnancy.
Results
TEAD4 Is Selectively Expressed in Self-Renewing Trophoblast Progenitors during Early Postimplantation Mammalian Development.
Earlier studies established that TEAD4 and its cofactor YAP1 orchestrate the establishment of the TE/TSC-specific transcriptional program (34–36) by directly regulating expression of several key trophoblast genes (32). However, the importance of TEAD4 for trophoblast development after blastocyst implantation is unknown. So we tested TEAD4 and YAP1 expression in the developing mouse placenta after embryo implantation. We found that in an embryonic day 7.5 (E7.5) mouse embryo TEAD4 protein is abundantly expressed within the trophoblast progenitors of the ExE/EPC region (Fig. 1A). Furthermore, gene expression analyses of TEAD4 positive cells within the ExE/EPC region revealed that they highly express mouse trophoblast stem cell (mouse TSC)-specific genes, such as caudal type homeobox 2 (Cdx2), Eomesodermin (Eomes), and Elf5 (SI Appendix, Fig. S1 A and B). Thus, we concluded that the TEAD4-postive cells within the ExE/EPC region are the TSPC population. We found that similar to TEAD4, the YAP1 protein is also highly expressed within the TSPCs of an E7.5 mouse embryo (SI Appendix, Fig. S1C). Interestingly, we also noticed that cells within the developing embryo proper region lack TEAD4 expression (Fig. 1A), an observation also reported at the mRNA expression level in an earlier study (34).
Fig. 1.
TEAD4 is selectively expressed in trophoblast progenitors within early postimplantation mammalian embryos. (A) Immunofluorescence images showing selective expression of the TEAD4 protein in TSPCs of an E7.5 mouse embryo. Note the absence of TEAD4 expression within cells of the developing embryo proper. (B) Immunostaining image showing TEAD4 protein expression in a first-trimester (week 8) human placental villous. TEAD4 is highly expressed in vCTBs (green arrows) and in CCTs (blue arrows) at the base of the trophoblast column. However, TEAD4 is absent in STBs and in nascent EVTs that arise at the distal part of the trophoblast column. (C) scRNA-seq analyses in first-trimester human placentae. The t-distributed stochastic neighbor embedding (t-SNE) plot of the aggregate of the hierarchical clustering of two first-trimester human placental samples (one week 7 and one week 8) identified 22 different cell clusters. (D) Clusters of trophoblast and nontrophoblast cells are labeled by KRT7 and HLA-A expressions, respectively, on a t-SNE plot. Lack of KRT7 expression and induction of HLA-A expression identified clusters 19, 21, and 22 as nontrophoblast cells. (E) Expressions of specific genes were monitored to identify clusters of different cell types. ELF5 and TP63 expressions identified cells in clusters 1 and 12 and at the tip of cluster 4 as CTB progenitors. HLA-G expression identified clusters 18 and 10 and part of clusters 2 and 9 as cells which are differentiating to EVTs. CGB3 expression identified cluster 4 and part of cluster 9 as cells undergoing STB differentiation. Interestingly, TEAD4 and YAP1 show expression patterns similar to ELF5 and TP63, respectively.
We also tested TEAD4 and YAP1 protein expressions in first-trimester human placenta (6 to 8 wk of gestation). A first-trimester human placental villous contains two different layers of trophoblast cells: 1) a layer of villous CTB progenitors (vCTBs; Fig. 1B, green arrows), close to the stroma, and 2) the postmitotic syncytiotrophoblast (STB; Fig. 1B, red arrows) layer, overlaying the vCTBs (Fig. 1B). In addition, a column of proliferating CTBs, known as column CTBs (CCTs; Fig. 1B, blue arrows), arises where placental villi anchor to the maternal decidua (37). Eventually, CCTs differentiate to the invasive extravillous trophoblasts (EVTs). We found that in a first-trimester human placenta, TEAD4 is predominantly expressed within the vCTBs and in CCTs, which are at or near the base of the trophoblast column of an anchoring villous. However, the differentiated STBs or the nascent EVTs, which arise at the distal part of the trophoblast column from the CCTs, do not express TEAD4 (Fig. 1B). These observations support earlier studies (37, 38), which also reported selective expression of TEAD4 in CTB progenitors. Also, similar to TEAD4, within first-trimester placental villi YAP1 protein expression is confined to the vCTBs and in CCTs, which are at the base of the trophoblastic column (SI Appendix, Fig. S1D).
To further compare expression of TEAD4 with other regulators of CTB progenitors in a first-trimester human placenta, we performed single-cell RNA sequencing (scRNA-seq). Based on gene expression patterns, the entire cell populations of 6 to 8 wk euploid placentae were distributed into 22 cell clusters (Fig. 1C). These cell clusters were further analyzed for mRNA expression of human cytokeratin 7 (KRT7) and human leukocyte antigen-A (HLA-A) to distinguish between trophoblast cell clusters and nontrophoblast cells. We found that the majority of cell clusters consist of KRT7-expressing trophoblast cells (Fig. 1 D, Upper), whereas the HLA-A-expressing nontrophoblast cells were divided into three clusters (clusters 19, 21, and 22, Fig. 1 D, Lower). Next, we tested mRNA expression of ELF5 and TP63, which are indicated to mark the proliferating CTBs within a first-trimester human placenta (30, 39). ELF5 expression was detected mainly in cells of clusters 1 and 12 (Fig. 1 E, Upper Left) and within a few cells of cluster 4. TP63 expression was detected in all ELF5-expressing cell clusters. In addition, TP63 expression was also detected in some cells of clusters 7 and 8 (Fig. 1 E, Lower Left). Thus, we concluded that Elf5, TP63-expressing cells within clusters 1 and 12, and a population of cluster 4 cells represent the undifferentiated CTB progenitors. We also tested mRNA expression of HLA-G and CG beta subunit 3 (CGB3) to detect cells, which are differentiating to the EVTs and STBs, respectively. We noticed that cells within clusters 18 and 10 and part of cluster 2 express high levels of HLA-G, indicating that those cells are differentiating to the EVT lineage clusters (Fig. 1 E, Upper Right). We identified cells in cluster 4, which do not express ELF5 and TP63, most abundantly express CGB3 (Fig. 1 E, Lower Right), indicating that they constitute the differentiated STB population or cells undergoing STB differentiation. We also noticed that cells in cluster 9 express both HLA-G and CGB3, indicating a differentiating cell population, which might have both STB and EVT differentiation potential. Interestingly, none of these CTB, EVT, and STB markers are expressed in a large population of KRT7-expressing cells within clusters 3, 5, 6, 2, and 14, indicating that those clusters could contain the CTB population, which are transitioning from the progenitor state and are poised for differentiation.
Next, we tested mRNA expression of TEAD4; its cofactors YAP1, VGLL1, and WWTR1; and other TEAD family transcription factors, TEAD1 to TEAD3, in different cell clusters. Intriguingly, we found that only TEAD4 (Fig. 1 E, Upper Middle) and YAP1 (Fig. 1 E, Lower Middle) are selectively expressed in the ELF5- and TP63-expressing undifferentiated CTBs. VGLL1 showed a broader expression pattern and is expressed in all HLA-A-negative cells (SI Appendix, Fig. S1E), including cells in clusters 3, 5, 6, 2, and 14 that are ELF5 and TP63 negative and poised for differentiation. TEAD1 and TEAD3 also showed a broader expression pattern; however, they express at a low level in undifferentiated CTBs (SI Appendix, Fig. S1E). TEAD1 is more prominently expressed in all differentiated cell clusters, and TEAD3 is prominently expressed in CGB3-expressing clusters 4 and 9 (SI Appendix, Fig. S1E). In contrast, WWTR1, TEAD1, and TEAD2 mRNAs are mainly expressed in differentiating/differentiated trophoblast cells (SI Appendix, Fig. S1E). Thus, expression analyses revealed that TEAD4 is selectively expressed within TSPCs of an early postimplantation mouse embryo, and the expression pattern is conserved in CTB progenitors of a first-trimester human placenta.
TEAD4 Function in Trophoblast Progenitors Is Essential for Placentation and Embryonic Development in a Postimplantation Mouse Embryo.
Genetic ablation of Tead4 in mouse embryo results in embryonic death before the blastocyst stage due to impairment of a TE/TSC-specific transcriptional program (8–12). However, the importance of TEAD4 in trophoblast development after implantation is still unknown. As TEAD4 is selectively expressed in TSPCs of an early postimplantation mouse embryo (Fig. 1A), we tested its importance during early postimplantation mouse development. For this study, we used a mouse model (Tead4fl/fl:UBC-Cre/ERT2) in which Tead4 could be conditionally deleted with synthetic estrogen receptor ligand, 4-hydroxytamoxifen (tamoxifen).
We crossed Tead4fl/fl:UBC-Cre/ERT2 males with Tead4fl/fl females to confine Tead4 deletion only within developing embryos. We induced Tead4 gene deletion starting at ∼E5.5 and monitored embryonic development on or prior to E9.5. We started tamoxifen treatment at ∼E5.5, as the presence of tamoxifen on or prior to E4.5 affects the implantation process (40–42). Given the fact that expression of TEAD4 is restricted within the TSPCs of an early postimplantation mouse embryo (Fig. 1A), this inducible gene knockout system allowed us to study the importance of TSPC-specific TEAD4 function during early mouse development.
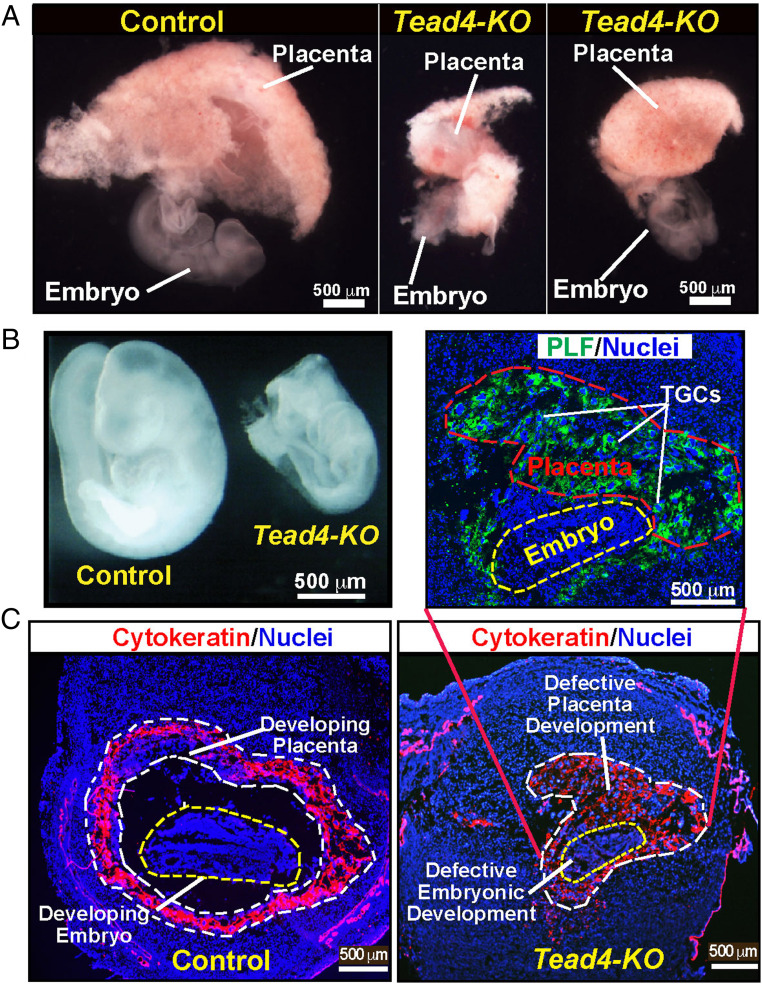
We found that deletion of Tead4 starting at ∼E5.5 resulted in embryonic death/loss at implantation sites. Most of the Tead4 knocked-out (Tead4-KO) embryos did not develop beyond the gastrulation stage and were associated with impaired placentation (Fig. 2A). Only a few Tead4-KO embryos apparently developed to Theiler stage 12 (∼E8.5, Fig. 2B). However, all of the Tead4-KO embryos died prior to E9.5 (SI Appendix, Fig. S2 A and B). Also, in comparison to Tead4fl/fl controls, placentae in Tead4-KO embryos were significantly smaller and contained only trophoblast giant cells (TGCs) (Fig. 2C). Furthermore, trophoblast stem state–specific genes, such as Esrrb, Tfap2C, and Gata3, were repressed, whereas TGC-specific genes, such as prolactin-2C2 (Prl2c2, also known as proliferin-1) and prolactin-3D1 (Prl3d1, also known as placental lactogen-I), were induced in Tead4-KO placentae (SI Appendix, Fig. S2C), indicating impaired trophoblast cell lineage development in the absence of TEAD4.
Fig. 2.
Loss of TEAD4 function in TSPCs of an early postimplantation mouse embryo impairs placentation, leading to embryonic death. (A) Conditional Tead4 alleles were deleted in TSPCs of a developing mouse placenta, and embryonic and placental developments were analyzed at E9.5 in control (Tead4fl/fl) and Tead4-KO embryos. Representative images of one control and two Tead4-KO conceptuses are shown. Embryonic and placentation defects were prominent in Tead4-KO conceptuses. (B) Images of developing control and Tead4-KO embryos at ∼E9.5. The Tead4-KO embryo represents one of the few that developed approximately to Theiler stage 12. (C) Placentation at control and Tead4-KO implantation sites were analyzed at ∼E9.5 via immunostaining with anti-pan-cytokeratin antibody (red, trophoblast marker). The developing Tead4-KO placenta is smaller and mainly contains TGCs, which can be identified via proliferin (PLF) expression (Green, expanded Inset).
TEAD4 Orchestrates Gene Expression to Maintain TSPC Self-Renewal in a Mouse Placenta Primordium.
As trophoblast lineage development was impaired in Tead4-KO developing placentae, we hypothesized that TEAD4 fine-tunes gene expression programs in TSPCs to balance their self-renewal vs. differentiation. To test this hypothesis, we tested the importance of the TEAD4-mediated gene expression program in both mouse TSCs and primary TSPCs within a placenta primordium.
In an earlier study, using chromatin immunoprecipitation along with deep sequencing (ChIP-seq), we identified TEAD4 binding regions on mouse TSC chromatin (32). However, how TEAD4 orchestrates the global gene expression program in mouse TSCs and the importance of TEAD4-regulated gene expression in TSC self-renewal are not well characterized.
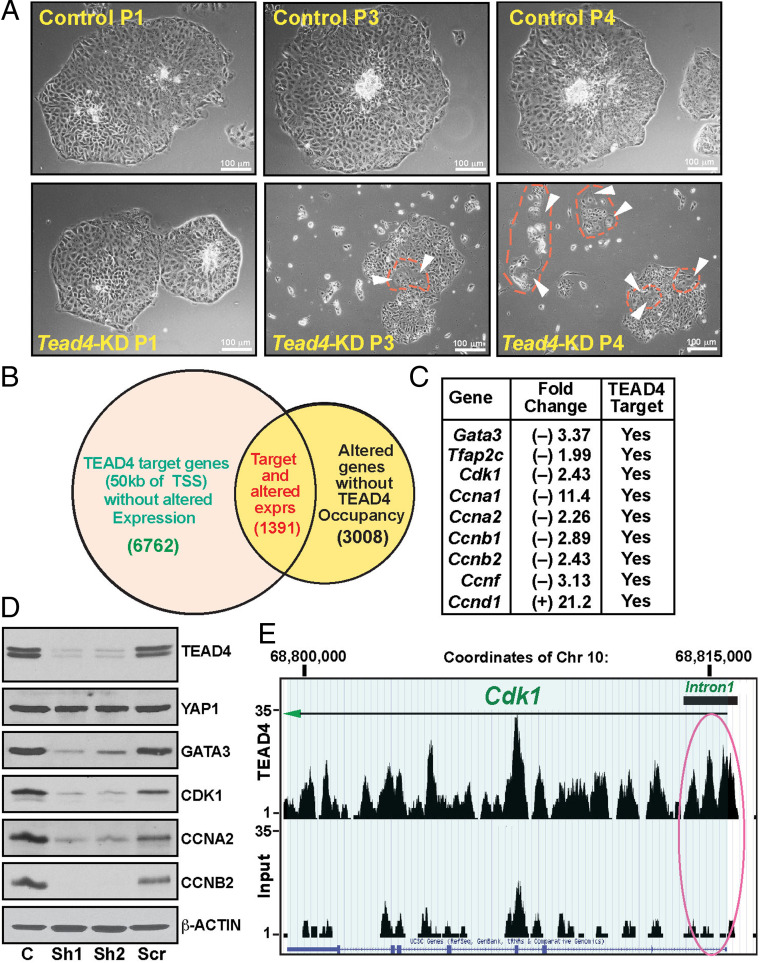
To test whether TEAD4 regulates mouse TSC self-renewal, we specifically depleted TEAD4 in mouse TSCs (Tead4-KD mouse TSC) via short hairpin RNA (shRNA)-mediated RNA interference (RNAi) (SI Appendix, Fig. S3A). We confirmed the specificity of RNAi by testing mRNA expression of Tead1 and Yap1, which remained unaltered in Tead4-KD mouse TSC (SI Appendix, Fig. S3A). Our in vitro cell culture studies in TSC stem state culture conditions (with FGF4 and heparin) showed that loss of TEAD4 induces mouse TSC differentiation, leading to loss of stem state colony morphology (Fig. 3A) over multiple passages of cell culturing. Interestingly, we also noticed significant inhibition of cell proliferation in Tead4-KD mouse TSC (Fig. 3A and SI Appendix, Fig. S3B). These studies indicated that TEAD4 is essential to maintain optimum self-renewal ability and stemness in mouse TSCs.
Fig. 3.
Loss of TEAD4 impairs mouse TSC self-renewal. (A) Micrographs show cell and colony morphologies of control and Tead4-KD mouse TSCs when cultured for multiple passages in standard mouse TSC culture conditions. Unlike wild-type control TSCs, Tead4-KD mouse TSCs gradually lost stem state colony morphology. (B) Venn diagrams showing the number of genes that are direct targets of TEAD4 and also showing significant changes in their mRNA expression in Tead4-KD mouse TSCs (TSS; transcription start site). (C) The table shows the alteration of mRNA expression of both trophoblast regulators (Gata3 and Tfap2c) and cell cycle regulators in Tead4-KD mouse TSCs. (D) Western blot analyses showing loss of altered protein expressions of cell cycle regulators in Tead4-KD mouse TSCs (C, wild-type control TSC; Sh1 and Sh2, mouse TSCs with TEAD4 RNAi using two distinct shRNAs; and Scr, mouse TSCs expressing a sham shRNA). (E) TEAD4 ChIP-seq peak shown in a region of mouse chromosome 10, which contains the Cdk1 gene.
To validate that TEAD4 directly regulates the gene expression program to maintain proliferation and stemness in mouse TSCs, we performed unbiased RNA-seq analyses in control vs. Tead4-KD mouse TSCs. Global transcriptome analyses in Tead4-KD mouse TSCs identified altered expression of 4,399 genes, of which 2,218 genes were up-regulated and 2,181 genes were down-regulated (Dataset S1). We noticed repression of TSC regulatory genes Gata3 and Tfap2c and induction of TGC-specific genes Prl2c2 and prolactin-3B1 (Prl3b, also known as placental lactogen-II) in Tead4-KD mouse TSCs (SI Appendix, Fig. S3C). However, expressions of mouse STB-specific retroviral genes, syncytin A (SynA), syncytin B (SynB), and glial cell missing 1 (Gcm1), were not significantly altered in Tead4-KD mouse TSCs (SI Appendix, Fig. S3C).
Comparative analyses of RNA-seq data with TEAD4 target genes (32) identified 1,391 genes that are direct targets of TEAD4 in mouse TSCs and either up-regulated or down-regulated in Tead4-KD mouse TSCs (Fig. 3B and Dataset S2). Interestingly, we found that mRNA expression of several cell cycle regulators (43), including cyclin-dependent kinase 1 (Cdk1), cyclin A1 (Ccna1), cyclin A2 (Ccna2), cyclin B1 (Ccnb1), cyclin B2 (Ccnb2), cyclin E2 (Ccne2), and cyclin F (Ccnf), was down-regulated in Tead4-KD mouse TSCs (Fig. 3C, SI Appendix, Fig. S3D, and Dataset S1). However, mRNA expression of cyclin D1 (Ccnd1), a known regulator of the G1/S phase (44), was up-regulated. Western blot analyses also validated the mRNA expression patterns (Fig. 3D). Furthermore, analyses of ChIP-seq data confirmed that all these cell cycle regulator genes are direct targets of TEAD4 in mouse TSCs (Fig. 3E, SI Appendix, Fig. S3E, and Dataset S2).
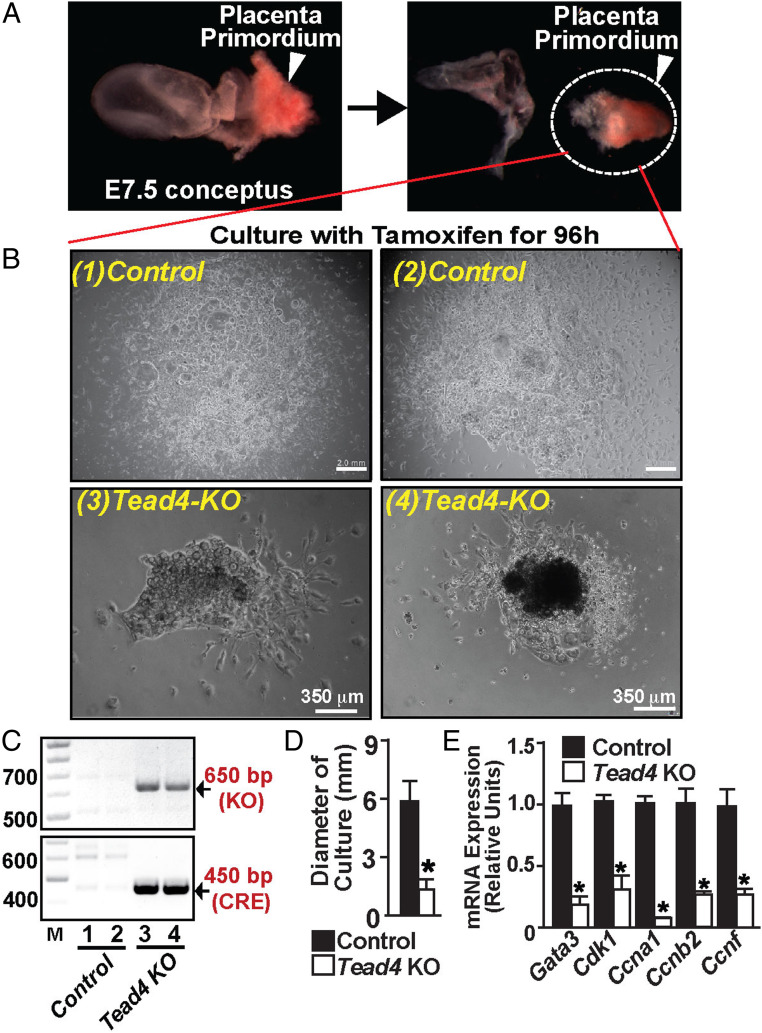
To further confirm TEAD4-mediated regulation of the trophoblast stem/progenitor state during mouse placentation, we tested primary TSPC self-renewal upon Tead4 deletion. We established ex vivo cultures of placenta primordium containing the developing ExE/EPC regions from E7.5 mouse embryos (Fig. 4A). Earlier, we showed that the ExE/EPC explant cultures contain nearly pure (≥97%) primary TSPCs (24), which could be maintained in the self-renewing stem/progenitor state with FGF4/heparin-containing mouse TSC culture conditions. Therefore, we cultured Tead4fl/fl (control) and Tead4fl/fl:UBC-Cre/ERT2 (Tead4-KO) explants in TSC culture conditions and treated them with tamoxifen. We also tested mRNA expression of cell cycle regulators in primary TSPCs. We found that, similar to Tead4-KD mouse TSCs, loss of TEAD4 impairs expansion of primary TSPCs (Fig. 4 B–D) and represses mRNA expression of cell cycle regulators, including Cdk1, Ccna1, Ccnb2, and Ccnf (Fig. 4E). Collectively, our studies of Tead4-KD mouse TSCs and Tead4-KO primary TSPCs strongly indicated that TEAD4 ensures gene expression balance to promote self-renewal and expansion of TSPCs during early postimplantation mammalian development.
Fig. 4.
Loss of TEAD4 impairs self-renewal in primary mouse TSPCs. (A) E7.5 mouse placenta primordia containing the ExE/EPC regions were isolated for testing the importance of TEAD4 in primary TSPC self-renewal. (B) Tead4fl/fl (control) and Tead4fl/fl:UBC-Cre/ERT2 (Tead4-KO) ExE/EPC explants were cultured with tamoxifen in FGF4/heparin-containing mouse TSC culture medium. Micrographs show a severe defect in expansion of primary TSPCs upon Tead4 deletion. (C) Confirmation of Tead4 deletion and the presence of Cre recombinase in explant cultures, which are shown in B. (D) Quantitative analyses of diameters of explant cultures of ExE/EPC regions from control and Tead4-KO embryos (mean ± SE; n = 5 ExE/EPC cultures for each condition, *P ≤ 0.05). (E) Quantitative mRNA expression analyses (mean ± SE; n = 5, *P ≤ 0.001) showing repression of cell cycle regulators in Tead4-KO TSPCs.
TEAD4 Orchestrates a Gene Expression Program to Promote Human TSC Self-Renewal.
TEAD4 is highly expressed in undifferentiated CTBs of human first-trimester placentae (Fig. 1B), and its expression is maintained in CTB-derived human TSCs (45). So we tested the importance of TEAD4 in human TSCs via loss-of-function analyses. We depleted TEAD4 in human TSCs (TEAD4-KD human TSC) by RNAi using lentiviral-mediated transduction of shRNAs (Fig. 5A). Depletion of TEAD4 strongly reduced human TSC proliferation (Fig. 5B) and promoted loss of stem state colony morphology in a culture condition that promotes human TSC stem state (Fig. 5C).
Fig. 5.
Loss of TEAD4 impairs human TSC self-renewal. (A) TEAD4 and YAP1 protein expressions were tested in human TSCs without (control) and with TEAD4 depletion by RNAi [shTEAD4 (1) and shTEAD4(2)]. (B) Plot showing reduced cell proliferation in TEAD4-KD human TSCs (mean ± SE; n = 3, *P ≤ 0.005). (C) Equal numbers of control and TEAD4-KD human TSCs were plated and cultured in the stem state culture condition. Micrographs confirm reduced cell proliferation and loss of stem state colony morphology (white circle) in TEAD4-depleted TSCs. (D) Confocal images of organoids generated with human TSCs. Immunostaining shows expressions of E-cadherin (CDH1, green) in the outer cell layer and human CG-β subunit (HCG-β, ρεδ) in the inner cell TSC organoids, confirming that the organoids develop an inside-out pattern of CTB vs. STB populations with respect to the first-trimester placental villi. (E) Micrographs show inefficient organoid formation by TEAD4-KD human TSCs. (F) Venn diagrams showing the number of genes that are direct targets of TEAD4 and also showing significant changes in their mRNA expression in Tead4-KD human TSCs. (G) The table shows alteration of mRNA expression of cell cycle regulators and STB and EVT markers in TEAD4-KD human TSCs. (H) Western blot analyses showing loss of protein expressions of cell cycle regulators in TEAD4-KD human TSCs. (I) Venn diagrams showing the number of unique and common TEAD4-regulated genes in mouse and human TSCs. (J) Ingenuity pathway analysis showing major biofunctions that are associated with genes regulated by TEAD4 in both mouse and human TSCs.
We also tested human TSC self-renewal by assessing their ability to form self-renewing three-dimensional trophoblast organoids (TSC organoids). Similar to undifferentiated CTBs (46, 47), human TSCs readily form trophoblast organoids (Fig. 5D). Also, similar to CTB-derived trophoblast organoids and in contrast to the architecture of the first-trimester placental villous epithelium, the human TSC organoids show an inverse organization of undifferentiated and differentiated cells. The outer layer of the human TSC organoids contained undifferentiated, E-cadherin-expressing mononuclear stem cells, whereas the center of the human TSC organoids contained the multinuclear, E-cadherin-negative and HCG-producing STB population (Fig. 5D). Our analyses of human TSC self-renewal using the TSC organoids confirmed the importance of TEAD4 in human TSC self-renewal. Unlike the control human TSCs, TEAD4-KD human TSCs showed severe impairment in organoid formation (Fig. 5E). Control human TSCs formed large organoids with prolonged culture (8 to 10 d) and could be dissociated and reorganized to form secondary organoids, indicating the self-renewing ability. In contrast, TEAD4-KD human TSCs either did not organize into an organoid structure or formed much smaller organoids. Thus, the human TSC organoid formation assay complemented the finding with the two-dimensional stem state culture assay and strongly indicated the importance of TEAD4 in promoting human TSC self-renewal.
To better understand how TEAD4 regulates human TSC self-renewal, we performed RNA-seq and ChIP-seq analyses to identify the TEAD4-regulated gene expression program. Depletion of TEAD4 significantly altered the expression of 2,238 genes (689 genes down-regulated and 1,549 genes up-regulated) in human TSCs (Dataset S3). ChIP-seq analyses showed that among these 2,238 genes, 750 genes are TEAD4 targets as TEAD4 occupancy is detected at their chromatin loci (Datasets S4 and S5 and Fig. 5F). The combinatorial analyses of RNA-seq and ChIP-seq data showed that, similar to Tead4-KD mouse TSCs, depletion of TEAD4 in human TSCs strongly reduced mRNA expression of cell cycle regulators, CDK1, CCNA2, CCNB1, CCNB2, CCNE2, and CCNF (Fig. 5G and SI Appendix, Fig. S4A). Interestingly, we also noticed that many of the STB-specific genes, CG A (CGA), CG B isoforms (CGBs), pregnancy-specific beta-1 glycoproteins (PSGs), and endogenous retroviral encoded gene ERVW1, are up-regulated in TEAD4-KD human TSCs (Fig. 5G and Datasets S3 and S5). Interestingly, HLA-G and HLA-F genes, which encode trophoblast-specific HLAs and are mainly expressed in an invasive EVT lineage, were identified as TEAD4 targets in human TSCs, and their mRNA expression was also up-regulated upon TEAD4 depletion (Fig. 5G and Datasets S3 and S5). In contrast, mRNA expression of achaete-scute family BHLH transcription factor 2 (ASCL2) and placenta-associated 8 (PLAC8), which are implicated in trophoblast invasion (48, 49), were down-regulated in TEAD4-KD human TSCs (Fig. 5G). Nevertheless, the unbiased genomics analyses indicated that loss of TEAD4 promoted expressions of differentiated trophoblast markers, especially differentiated STB-specific genes, in human TSCs.
Similar to our findings in Tead4-KD mouse TSCs, depletion of TEAD4 down-regulated both mRNA (Fig. 5G) and protein expressions (Fig. 5H) of conserved cell cycle regulators in human TSCs. Therefore, we performed comparative genomics analyses to identify whether TEAD4 establishes a common gene expression program in these cells to regulate cell proliferation. Our analyses identified 98 conserved genes that are directly regulated by TEAD4 in both mouse and human TSCs (Fig. 5I and Dataset S6). Among these 98 genes, 24 genes were down-regulated, and 41 genes were up-regulated in both cell types upon TEAD4 depletion (SI Appendix, Fig. S4B). In silico analyses revealed that, along with cell cycle regulation, the major biofunctions of these TEAD4-regulated genes include cell proliferation, cell survival, DNA replication, and lipid metabolism (Fig. 5J). Collectively, our genomics analyses revealed that TEAD4 fine-tunes gene expression programs in human TSCs to promote cell proliferation and to inhibit STB differentiation to maintain a self-renewing stem state.
Idiopathic Recurrent Pregnancy Losses, Which Are Associated with Defective Placental Villi Formation, Are Associated with Loss of TEAD4 Expression in Trophoblast Progenitors.
Nearly 3% of pregnant women suffer from RPL, and almost half of the RPLs are considered idiopathic, as they do not have deviations in clinical, environmental, and lifestyle risk factors, including parental chromosomal anomalies and maternal thrombophilic, anatomic, endocrine, and immunological disorders (3–5, 50). Based on our findings in Tead4-KO mouse embryos and TEAD4-depleted mouse and human TSCs, we hypothesized that some of these idiopathic RPLs might be associated with defective development of trophoblast progenitors due to impaired TEAD4 function. Therefore, we collected placentae from patients with idiopathic RPLs. We ensured that the collected placental tissues were not necrotic. Intriguingly, we noticed a major defect in placental villi formation (Fig. 6A) in a subset of idiopathic RPLs (7 out of 82 idiopathic RPL placenta samples). Ultrastructure analyses of those placentae showed defective formation of the CTB/STB bilayer, characterized by the presence of patches of a single trophoblast layer or lack of trophoblast cells (Fig. 6 B, Upper Right), as well as impaired trophoblastic column formation (SI Appendix, Fig. S5A). In addition, we also noticed strong reduction in TEAD4 expression in probable vCTBs (Fig. 6 B, Lower Right) and CCTs (SI Appendix, Fig. S5B) in those idiopathic RPL placentae.
Fig. 6.
A subset of idiopathic RPLs is associated with reduced TEAD4 expression in CTB progenitors. (A) Micrographs show defective villi formation in a first-trimester (7 wk) placenta from a patient suffering from idiopathic RPL. A 7 wk placenta from elective pregnancy termination was used as a control. (B) Immunofluorescence images show defective trophoblast bilayer formation and strong reduction of TEAD4 expression in CTBs in idiopathic RPL placenta, shown in A. (C) Patient-specific human TSCs were established from CTBs, which were isolated from placentae, associated with idiopathic RPL. Micrographs show efficient establishment of TSC colonies from an idiopathic RPL, which was not associated with defective placental villi formation (RPL 10, Left). In contrast, TSC establishment was inefficient from an idiopathic RPL (RPL 7), which was characterized by defective placental villi formation. We could obtain small TSC colonies only in the presence of feeder mesenchymal stromal cells isolated from first-trimester human placenta.
To further understand the correlation of reduced TEAD4 expression with the idiopathic RPLs, we isolated CTBs from RPL placentae and established patient-specific human TSC lines (RPL-TSCs). We established 22 RPL-TSC lines (DatasetS7). Among these 22 TSC lines, 15 lines were established from RPL placentae without any prominent defect in placental villi. These 15 RPL-TSC lines were easily established (Fig. 6 C, Left) and can be maintained in a self-renewing stem state for multiple passages without affecting their self-renewal ability (SI Appendix, Fig. S6A) or without inducing genomic instability (SI Appendix, Fig. S6B). They can also be efficiently differentiated to both SynTB and EVT lineages (SI Appendix, Fig. S6 C and D).
We were able to establish TSC lines from five of the seven RPL placentae, which showed a prominent defect in placental villi formation (RPL-TSC lines 4, 7, 15, 17, and 19, Dataset S7). However, during establishment, due to extremely slow proliferation, we needed to culture them in the presence of placental mesenchymal stromal feeder cells during the initial three passages (Fig. 6 C, Right). When transferred to the feeder-free culture condition, one of the lines (RPL-TSC 7) could not be maintained (Dataset S7).
We maintained RPL-TSC lines 4, 15, 17, and 19 in a feeder-free culture condition for multiple passages. Despite being euploid (Fig. 7A), RPL-TSCs showed a strong reduction in their proliferation rate (Fig. 7B). We confirmed reduced expression of TEAD4 mRNA (Fig. 7C) and protein expression in those TSCs (Fig. 7D). They also showed a higher propensity to lose stem state cellular morphology (Fig. 7E and SI Appendix, Fig. S6E; yellow circles) and were inefficient in forming TSC organoids (Fig. 7 F, Middle). In addition, they showed reduced mRNA expression of cell cycle regulators (plot of cell cycle regulator cell proliferation [SI Appendix, Fig. S6G]) and rescue of TEAD4 expression promoted stem state morphology (Fig. 7 E, Bottom Right) and increased the efficiency to form self-renewing TSC organoids (Fig. 7 F, Right). Furthermore, gene expression analyses showed that rescue of TEAD4 expression rescued mRNA expression of cell cycle regulator genes, such as CDK1 and CCNB1, in RPL-TSCs (SI Appendix, Fig. S6G).
Fig. 7.
TEAD4 expression rescues self-renewal in human TSCs, which are derived from defective placentae, associated with idiopathic RPL. (A) RPL-TSCs (line 15), which were derived from CTBs, associated with idiopathic RPL and defective placental villi formation, were cultured for five passages, and chromosome content was analyzed by karyotyping, and a euploid, XY karyotype was confirmed. (B) The plot shows reduced cell proliferation in RPL-TSCs, associated with defective placental villi formation. Three human TSC lines, established from first-trimester (weeks 7 to 9) vCTBs by Okae et al. (45), were used as a control (mean ± SE; n = 3, *P ≤ 0.005). (C) Quantitative RT-PCR analyses (mean ± SE; n = 4, *P ≤ 0.001) showing reduced expression of TEAD4 mRNA in human TSCs, derived with isolated CTBs from idiopathic RPLs, which are associated with defective villi formation. TEAD4 mRNA expression was compared with two control human TSC lines, one line generated by Okae et al. (45) and the RPL-TSC (line 10), which did not show any defect in self-renewal ability (shown in SI Appendix, Fig. S6A). (D) The Western blot shows reduced endogenous TEAD4 protein expression in RPL-TSCs (line 15) and rescue of TEAD4 expression after transduction with a lentiviral construct containing cloned human TEAD4 complementary DNA. (E) Micrographs show colony morphologies of RPL-TSCs (line 15) with or without ectopic TEAD4 expression. The Okae TSCs were used as a control. (F) Micrographs show organoid formation with RPL-TSCs (line 15) with or without ectopic TEAD4 expression. The Okae TSCs were used as a control.
We also tested mRNA expression of STB-specific genes (PSG4, ERVW1. and CGB) and EVT-specific genes (ASCL2, PLAC8. and HLA-G) in RPL-TSCs before and after rescue of TEAD4 expression. These genes were differentially expressed in TEAD4-KD human TSCs (data shown in Fig. 5G). We found that similar to that in TEAD4-KD human TSCs, ASCL2 and PLAC8 mRNA expression was reduced in the RPL-TSCs, and rescue of TEAD4-expression rescued their expression (SI Appendix, Fig. S6G). Also, similar to that in TEAD4-KD human TSCs, expression of PSG4 was induced in RPL-TSCs. However, rescue of TEAD4 expression did not reduce PSG4 expression. We also noticed that mRNA expression of the retroviral gene ERVW1 was not significantly altered in RPL-TSCs. In contrast to that in TEAD4-KD TSCs, expressions of CGB and HLA-G were reduced in RPL-TSCs, and rescue of TEAD4 expression only partially induced CGB and HLA-G expressions (SI Appendix, Fig. S6G). Nevertheless, the rescue of mRNA expression of cell cycle regulators, induced cell proliferation, and formation of self-renewing trophoblast organoids upon rescue of TEAD4 expression in RPL-TSCs strongly indicated that TEAD4 promotes CTB self-renewal in a developing human placenta, and impaired TEAD4 function could be one of the molecular causes of idiopathic RPL.
Discussion
Extensive self-renewal of CTB progenitors, which begins at the peri-implantation stage and continues throughout the first trimester of pregnancy, is essential for reproductive success in humans (26, 28). Eventually, establishment of differential gene expression programs in CTBs instigate STB and EVT differentiation, thereby leading to formation of the matured villous placenta. However, the mechanisms that fine-tune gene transcription to promote self-renewal in CTBs or instigate specific differentiation program are poorly understood. Our findings in this study provide strong evidence that Hippo signaling effector TEAD4 is essential to maintain self-renewal in CTB progenitors as well as in TSPCs of a developing mouse placenta. Meinhardt et al. (51) also provides evidence that the TEAD4 cofactor, YAP1, is pivotal to promote the stemness in CTB progenitors during early human placentation. Our studies implicate the Hippo signaling components, TEAD4/YAP1, which are conserved in placental mammals, in establishing pregnancy and progression of postimplantation mammalian development.
Our scRNA-seq analyses showed that TEAD4 and YAP1 are specifically expressed in CTB progenitors. Similarly, TEAD4 is selectively induced in TSPCs of a mouse placenta primordium. Along with this specific expression pattern, the strong phenotype in TSPC-specific Tead4-KO mouse embryos and loss of self-renewal in TEAD4-depleted RPL-TSCs emphasize the importance of TEAD4/YAP1-mediated gene regulation in trophoblast progenitor stemness. The rescue of self-renewal in RPL-TSCs upon ectopic TEAD4 expression further supports this concept. These findings also raise the question, What are the functions of other TEAD factors and TEAD cofactors during human trophoblast lineage development? As other TEAD factors, TEAD1, TEAD2, TEAD3, and cofactor WWTR1, are specifically induced during CTB differentiation, we predict that they are important for the development of the differentiated STBs and/or EVTs. However, it is possible that VGLL1, which is also highly expressed in undifferentiated CTBs (SI Appendix, Fig. S1E), possesses functional redundancy with YAP1 in undifferentiated CTBs. Interestingly, an earlier study noted that, unlike YAP1, VGLL1 expression is not prominent in mouse trophoblast progenitors (38). Thus, we propose that the TEAD4/YAP1 establishes a conserved gene expression program in both mouse TSPCs and human CTBs, whereas the TEAD4/VGLL1 complex uniquely regulates genes that are specific to human CTBs.
Our “omics” studies revealed that TEAD4 directly regulates CDK1 and several CYCLINs in both mouse and human TSCs. TEAD4-regulated CYCLINs are known to regulate various stages of the cell cycle, including the G1/S phase transition (CCNE2) (44), progression of the S phase (CCNA2) (52), and the G2/M phase (CCNB1/CCNB2) transition (43). CDK1, which is also regulated by TEAD4, is essential for the G2/M phase transition (53, 54). Thus, the lack of cell proliferation in TEAD4-depleted TSCs could be mainly due to the dysregulation of cell cycle regulators. Interestingly, an earlier study (55) involving scRNA-seq analyses showed that CCNB1 and CDK1 expression marks the most proliferative CTBs in a first-trimester human placenta. As both of these genes are directly regulated by TEAD4 in human TSCs, we propose that TEAD4 is the master regulator for the maintenance of proliferating CTB populations in a developing human placenta.
Our study indicated that along with promoting cell proliferation, TEAD4 maintains human TSC/CTB stemness by repressing transcriptions of STB-associated genes, such as CGA, CGBs, PSGs, and ERVW1 (Fig. 5G). Along with these STB genes, several hundred other genes were also up-regulated in TEAD4-KD human TSCs. This raises the question about the TEAD4-dependent mechanism that instigates gene repression vs. gene induction. In a different cellular context, it was shown that TEAD4 forms a complex with VGLL4 and C-terminal-binding protein 2 (CtBP2) to repress gene transcription (56). Thus, it is possible that a TEAD4-VGLL-CtBP2 complex represses transcription of STB-specific genes in human TSC as well as CTB progenitors.
The specific expression of TEAD4 in CTB progenitors raises two important questions: 1) Which cellular mechanism induces and maintains TEAD4 expression in undifferentiated CTBs? 2) Why is TEAD4 expression repressed in CTBs of certain idiopathic RPL placentae? Studies involving establishment of human TSCs and CTB organoids implicate activation of the epidermal growth factor and Wnt signaling pathways to maintain self-renewal in CTBs and CTB-derived human TSCs (45–47). Thus, it is possible that these signaling pathways are required to promote TEAD4 expression in CTB progenitors and suppression of these signaling pathways leads to TEAD4 repression in idiopathic RPL CTBs. However, there exist other possibilities. For example, we have shown that in the TE lineage and in mouse TSCs, TEAD4 autoregulates its own expression by binding to cis-elements located at the 5′ UTR region (32). Thus, one possible mechanism of loss of TEAD4 expression in idiopathic RPLs could be acquired mutation at the regulatory cis-element, leading to impaired TEAD4 gene transcription. Considering the rate of idiopathic RPL increases with maternal aging (57, 58), our findings in this study will provide an excellent platform for future studies to test mechanisms that are involved in TEAD4 regulation in human CTB progenitors during early human placentation.
Experimental Procedures
Animal and Tissue Collection.
To generate a Tead4fl/fl:UBC-Cre/ERT2 mouse, we crossed Tead4fl/fl mice (B6;129-Tead4tm1Bnno, MMRC) and UBC-Cre/ERT2 mice [B6;129S-Tg(UBC-Cre/ERT2)1Ejb/J, Jackson Laboratory], in which Cre recombinase is constitutively expressed from the human ubiquitin C (UBC) promoter (24) and is fused to a modified human estrogen receptor, thereby allowing deletion of Tead4 conditional alleles with only a synthetic ligand, tamoxifen. All experiments with mouse models were performed after obtaining approval from the Institutional Animal Care and Use Committee at the University of Kansas Medical Center.
Human Placental Sample Analysis.
Discarded, deidentified first-trimester placental tissues from elective termination were obtained from Mount Sinai Hospital, Toronto. Placental tissues from patients with idiopathic RPLs were obtained at the University of Kansas Medical Center with consent from patients. The institutional review board (IRB) at Mount Sinai Hospital and the University of Kansas IRB approved all collections and studies. Fresh placental tissues were embedded in optimum cutting temperature compound and cryosectioned or used for scRNA-seq analyses.
Establishment of Human TSC Lines and Culture.
Human TSC lines were derived from first-trimester CTBs and cultured following the protocols described previously (45). Although multiple lines were derived from idiopathic RPLs that were without apparent defect in placental villi formation, the data presented in this paper were generated using only one such line (RPL-TSC 10). Human TSCs were cultured in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12- supplemented with Hepes and l-glutamine along with a mixture of inhibitors. We followed the established protocol (45) to induce STB and EVT differentiation.
Statistical Significance.
Statistical significances were determined for expansion of ex vivo cultures of mouse ExE/EPC regions, quantitative RT-PCR analyses for mRNA expression, and cell proliferation analyses. We performed at least n = 3 experimental replicates for all of those experiments. For statistical significance of generated data, statistical comparisons between two means were determined with Student’s t test, and significantly altered values (P ≤ 0.05) are highlighted in figures by an asterisk. Although in a few figures studies from multiple groups are presented, the statistical significances were tested by comparing data of two groups, and significantly altered values (P ≤ 0.05) are highlighted in figures. RNA-Seq data were generated with n = 3 experimental replicates per group. The statistical significance of altered gene expression (absolute fold change ≥ 1.5 and false discovery rate q value ≤ 0.05) was initially confirmed with a right-tailed Fisher’s exact test. For ChIP-seq, we extended sequence reads in silico at their 3′ end to 200 bps. We used the Model-based Analysis of ChIP-seq algorithm (59) with a P value cutoff of 1e−3 for peak detection.
Supplementary Material
Acknowledgments
This research was supported by NIH Grants HD062546, HD0098880, and HD079363 and bridging grant support under the Kansas Idea Network of Biomedical Research Excellence (K-INBRE, Grant P20GM103418) to S.P. A pilot grant under the NIH Centers of Biomedical Research Excellence program (COBRE, Grant P30GM122731) supported P.H. This study was also supported by various core facilities, including the Genomics Core, the Transgenic and Gene Targeting Institutional Facility, the Imaging and Histology Core, and the Bioinformatics Core of the University of Kansas Medical Center. We thank Dr. Hiroaki Okae and Dr. Takahiro Arima of Tohoku University Graduate School of Medicine, Japan, for sharing human TSC lines. We thank Ms. Brandi Miller for critical comments on the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Data have been deposited in the Gene Expression Omnibus (GEO) database: RNA-seq data in mouse TSCs (accession no. GSE86809), RNA-seq data in human TSCs (accession no. GSE144809), TEAD4 ChIP-seq data in human TSCs (accession no. GSE144807), and single-cell RNA-seq in first-trimester human placenta (accession no. GSE145036).
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002449117/-/DCSupplemental.
Data Availability.
All raw data for RNA-seq analyses have been deposited in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/gds, accession no. GSE144809 for human TSCs and accession no. GSE86809 for mouse TSCs). The raw data for ChIP-seq analyses in human TSCs have been deposited in the GEO database (www.ncbi.nlm.nih.gov/gds, accession no. GSE144807). We used published (32) TEAD4 ChIP-seq data (accession no. GSE37350) for comparative analysis of TEAD4 targets in mouse TSCs. The raw data for single-cell RNA-seq in first-trimester human placenta are also available in the GEO database (accession no. GSE145036). Additional details of experimental procedures are mentioned in SI Appendix.
References
- 1.Larsen E. C., Christiansen O. B., Kolte A. M., Macklon N., New insights into mechanisms behind miscarriage. BMC Med. 11, 154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macklon N. S., Geraedts J. P., Fauser B. C., Conception to ongoing pregnancy: The “black box” of early pregnancy loss. Hum. Reprod. Update 8, 333–343 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Rull K., Nagirnaja L., Laan M., Genetics of recurrent miscarriage: Challenges, current knowledge, future directions. Front. Genet. 3, 34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison J. L., Schust D. J., Recurrent first trimester pregnancy loss: Revised definitions and novel causes. Curr. Opin. Endocrinol. Diabetes Obes. 16, 446–450 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Ford H. B., Schust D. J., Recurrent pregnancy loss: Etiology, diagnosis, and therapy. Rev. Obstet. Gynecol. 2, 76–83 (2009). [PMC free article] [PubMed] [Google Scholar]
- 6.Perez-Garcia V. et al., Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature 555, 463–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossant J., Cross J. C., Placental development: Lessons from mouse mutants. Nat. Rev. Genet. 2, 538–548 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Red-Horse K. et al., Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J. Clin. Invest. 114, 744–754 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaufmann P., Black S., Huppertz B., Endovascular trophoblast invasion: Implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol. Reprod. 69, 1–7 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Pijnenborg R., Vercruysse L., Hanssens M., The uterine spiral arteries in human pregnancy: Facts and controversies. Placenta 27, 939–958 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Velicky P., Knöfler M., Pollheimer J., Function and control of human invasive trophoblast subtypes: Intrinsic vs. maternal control. Cell Adhes. Migr. 10, 154–162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brosens I., Pijnenborg R., Vercruysse L., Romero R., The “great obstetrical syndromes” are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 204, 193–201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Resnik R., Intrauterine growth restriction. Obstet. Gynecol. 99, 490–496 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Pardi G., Marconi A. M., Cetin I., Placental-fetal interrelationship in IUGR fetuses–A review. Placenta 23, S136–S141 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Genbacev O., Joslin R., Damsky C. H., Polliotti B. M., Fisher S. J., Hypoxia alters early gestation human cytotrophoblast differentiation/invasion in vitro and models the placental defects that occur in preeclampsia. J. Clin. Invest. 97, 540–550 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y. M. et al., Failure of physiologic transformation of the spiral arteries in patients with preterm labor and intact membranes. Am. J. Obstet. Gynecol. 189, 1063–1069 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Kim Y. M. et al., Failure of physiologic transformation of the spiral arteries in the placental bed in preterm premature rupture of membranes. Am. J. Obstet. Gynecol. 187, 1137–1142 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Gluckman P. D., Hanson M. A., Cooper C., Thornburg K. L., Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 359, 61–73 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godfrey K. M., Barker D. J., Fetal nutrition and adult disease. Am. J. Clin. Nutr. 71 (suppl. 5), 1344S–1352S (2000). [DOI] [PubMed] [Google Scholar]
- 20.Funai E. F. et al., Long-term mortality after preeclampsia. Epidemiology 16, 206–215 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Rossant J., Lineage development and polar asymmetries in the peri-implantation mouse blastocyst. Semin. Cell Dev. Biol. 15, 573–581 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Zernicka-Goetz M., Cleavage pattern and emerging asymmetry of the mouse embryo. Nat. Rev. Mol. Cell Biol. 6, 919–928 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Kunath T., Strumpf D., Rossant J., Early trophoblast determination and stem cell maintenance in the mouse–A review. Placenta 25, S32–S38 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Home P. et al., Genetic redundancy of GATA factors in the extraembryonic trophoblast lineage ensures the progression of preimplantation and postimplantation mammalian development. Development 144, 876–888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts R. M., Fisher S. J., Trophoblast stem cells. Biol. Reprod. 84, 412–421 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemberger M., Hanna C. W., Dean W., Mechanisms of early placental development in mouse and humans. Nat. Rev. Genet. 21, 27–43 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Knott J. G., Paul S., Transcriptional regulators of the trophoblast lineage in mammals with hemochorial placentation. Reproduction 148, R121–R136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knofler M. et al., Human placenta and trophoblast development: Key molecular mechanisms and model systems. Cell. Mol. Life Sci. 76, 3479–3496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul S., Home P., Bhattacharya B., Ray S., GATA factors: Master regulators of gene expression in trophoblast progenitors. Placenta 60 (suppl. 1), S61–S66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hemberger M., Udayashankar R., Tesar P., Moore H., Burton G. J., ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum. Mol. Genet. 19, 2456–2467 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Lee C. Q. et al., What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Reports 6, 257–272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Home P. et al., Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc. Natl. Acad. Sci. U.S.A. 109, 7362–7367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar R. P. et al., Regulation of energy metabolism during early mammalian development: TEAD4 controls mitochondrial transcription. Development 145, dev162644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yagi R. et al., Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827–3836 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Ralston A. et al., Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Nishioka N. et al., Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 125, 270–283 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Haider S. et al., Notch1 controls development of the extravillous trophoblast lineage in the human placenta. Proc. Natl. Acad. Sci. U.S.A. 113, E7710–E7719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soncin F. et al., Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development 145, dev156273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y. et al., BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a ΔNp63+ cytotrophoblast stem cell state. Development 140, 3965–3976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dao B., Vanage G., Marshall A., Bardin C. W., Koide S. S., Anti-implantation activity of antiestrogens and mifepristone. Contraception 54, 253–258 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Pugh D. M., Sumano H. S., The anti-implantation action of tamoxifen in mice. Arch. Toxicol. Suppl. 5, 209–213 (1982). [DOI] [PubMed] [Google Scholar]
- 42.Bloxham P. A., Pugh D. M., Tamoxifen inhibition of an in vitro oestradiol-induced surface coat change on mouse blastocysts. Br. J. Pharmacol. 60, 517–519 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satyanarayana A., Kaldis P., Mammalian cell-cycle regulation: Several cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28, 2925–2939 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Bertoli C., Skotheim J. M., de Bruin R. A., Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 14, 518–528 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okae H. et al., Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Haider S. et al., Self-renewing trophoblast organoids recapitulate the developmental program of the early human placenta. Stem Cell Reports 11, 537–551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turco M. Y. et al., Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 564, 263–267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bogutz A. B. et al., Transcription factor ASCL2 is required for development of the glycogen trophoblast cell lineage. PLoS Genet. 14, e1007587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang W. L. et al., PLAC8, a new marker for human interstitial extravillous trophoblast cells, promotes their invasion and migration. Development 145, dev148932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saravelos S. H., Li T. C., Unexplained recurrent miscarriage: How can we explain it? Hum. Reprod. 27, 1882–1886 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Meinhardt G. et al., Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta. Proc. Natl. Acad. Sci. U.S.A. 117, 13562–13570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woo R. A., Poon R. Y., Cyclin-dependent kinases and S phase control in mammalian cells. Cell Cycle 2, 316–324 (2003). [PubMed] [Google Scholar]
- 53.Lohka M. J., Hayes M. K., Maller J. L., Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proc. Natl. Acad. Sci. U.S.A. 85, 3009–3013 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diril M. K. et al., Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc. Natl. Acad. Sci. U.S.A. 109, 3826–3831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y. et al., Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 28, 819–832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang W. et al., The TEA domain family transcription factor TEAD4 represses murine adipogenesis by recruiting the cofactors VGLL4 and CtBP2 into a transcriptional complex. J. Biol. Chem. 293, 17119–17134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li T. C. et al., An analysis of the pattern of pregnancy loss in women with recurrent miscarriage. Fertil. Steril. 78, 1100–1106 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Sauer M. V., Reproduction at an advanced maternal age and maternal health. Fertil. Steril. 103, 1136–1143 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Feng J., Liu T., Qin B., Zhang Y., Liu X. S., Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data for RNA-seq analyses have been deposited in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/gds, accession no. GSE144809 for human TSCs and accession no. GSE86809 for mouse TSCs). The raw data for ChIP-seq analyses in human TSCs have been deposited in the GEO database (www.ncbi.nlm.nih.gov/gds, accession no. GSE144807). We used published (32) TEAD4 ChIP-seq data (accession no. GSE37350) for comparative analysis of TEAD4 targets in mouse TSCs. The raw data for single-cell RNA-seq in first-trimester human placenta are also available in the GEO database (accession no. GSE145036). Additional details of experimental procedures are mentioned in SI Appendix.