Significance
The presence of a dominant male-determining locus (M-locus) in one of a pair of autosomes establishes the male sex in the dengue fever mosquito Aedes aegypti. The Ae. aegypti M-locus contains 30 genes, including Nix, a previously reported male-determining factor. Here we show that the Nix transgene alone was sufficient to convert females into fertile males, which continued to produce sex-converted progeny. We also show that a second M-locus gene named myo-sex was needed for male flight. Nix-mediated sex conversion was 100% penetrant, heritable, and stable, indicating great potential for developing mosquito-control strategies to reduce vector populations by female-to-male conversion. This work also sheds lights into the molecular basis of the function of the M-locus.
Keywords: mosquito, sex-determination, M factor, infectious diseases, Aedes aegypti
Abstract
A dominant male-determining locus (M-locus) establishes the male sex (M/m) in the yellow fever mosquito, Aedes aegypti. Nix, a gene in the M-locus, was shown to be a male-determining factor (M factor) as somatic knockout of Nix led to feminized males (M/m) while transient expression of Nix resulted in partially masculinized females (m/m), with male reproductive organs but retained female antennae. It was not clear whether any of the other 29 genes in the 1.3-Mb M-locus are also needed for complete sex-conversion. Here, we report the generation of multiple transgenic lines that express Nix under the control of its own promoter. Genetic and molecular analyses of these lines provided insights unattainable from previous transient experiments. We show that the Nix transgene alone, in the absence of the M-locus, was sufficient to convert females into males with all male-specific sexually dimorphic features and male-like gene expression. The converted m/m males are flightless, unable to perform the nuptial flight required for mating. However, they were able to father sex-converted progeny when presented with cold-anesthetized wild-type females. We show that myo-sex, a myosin heavy-chain gene also in the M-locus, was required for male flight as knockout of myo-sex rendered wild-type males flightless. We also show that Nix-mediated female-to-male conversion was 100% penetrant and stable over many generations. Therefore, Nix has great potential for developing mosquito control strategies to reduce vector populations by female-to-male sex conversion, or to aid in a sterile insect technique that requires releasing only non-biting males.
Highly diverse primary signals serve as the master switches to initiate sex-determination in insects (reviewed in ref. 1). In some species the initiating signals are female-determining factors that trigger female development. For example, a double dose of the X-linked signal elements in the fruit fly Drosophila melanogaster instigates female development in XX embryos (2); in honey bees the heterozygosity of the complementary sex determiner (csd) gene initiates female development in diploid embryos produced by fertilized queen bees (3); and a W chromosome-linked Piwi-interacting RNA gene determines female sex in ZW silkworms (4). In contrast, a dominant male-determining factor (M factor) serves as the primary signal that triggers male development in many other insects, including mosquitoes and other non-Drosophila flies, beetles, and true bugs (1, 5–8). The M factor is located either on a Y chromosome or within a sex locus named the M-locus on a homomorphic sex-determining chromosome, both of which are repeat-rich and thus difficult to study. Facilitated by recent advances in bioinformatics and genetic technologies, the M factor has been discovered in five dipteran insects, including Nix in the yellow fever and dengue fever mosquito Aedes aegypti (9), gYG2/Yob in the African malaria mosquito Anopheles gambiae (10, 11), Guy1 in the Asian malaria mosquito Anopheles stephensi (12), Mdmd in the housefly Musca domestica (13), and MoY in the Medfly Ceratitis capitata (14). None of these M factors are apparently related to each other, indicating frequent turnover of the initiating signals for sex-determination. However, through a cascade of events, these highly divergent primary signals are eventually transduced as sex-specific isoforms of conserved transcription factors doublesex (DSX) and fruitless (FRU) that program sexual differentiation. Thus, diverse primary signals in different species regulate the alternative, sex-specific splicing of dsx and fru pre-mRNAs, leading to sex-specific DSX and FRU protein isoforms.
Sex conversion resulting from a loss-of-function mutant male or a gain-of-function female would strongly indicate that an M factor is the master switch for sex determination. Such evidence has been presented only in species for which sex chromosome dosage compensation is not necessary or where the M factor may not be involved in dosage compensation (9, 13). We have previously shown that somatic knockout of Nix in male embryos resulted in feminized Ae. aegypti adults with developing ovaries, and female embryos injected with a plasmid that contains the Nix ORF driven by a strong constitutive promoter (polyUb) developed into partially masculinized adults (9). However, full phenotypic sex conversion was not observed, presumably due to somatic mosaicism, the transient nature of Nix expression, or the use of a different promoter. Moreover, the Ae. aegypti M-locus contains four other protein-coding and 25 long-noncoding RNA (lncRNA) genes, several of which showed highly enriched expression in the testes and male accessory glands (15). Thus, it remains unclear whether Nix alone is sufficient for conferring complete male sexually dimorphic traits and fertility.
Here, we present molecular and genetic characterizations of Nix-transgenic lines and show that the Nix transgene alone, in the absence of the M-locus, is sufficient to convert females into fertile males with all male-specific sexually dimorphic features. We show that a second M-locus gene named myo-sex is needed for male flight. We also show that Nix-mediated sex conversion is 100% penetrant, heritable, and highly stable, indicating great potential for developing mosquito control strategies to reduce vector populations by female-to-male sex conversion. This work will also inform future investigations into homomorphic sex chromosomes that are found in other insects, vertebrates, and plants.
Results
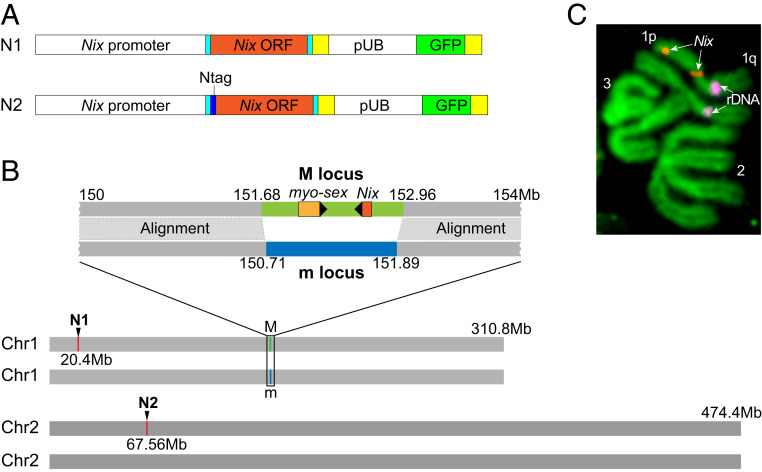
Generation of Nix Transgenic Lines.
Initial efforts failed to produce transgenic lines when Nix was under the control of the polyUb promoter. Thus, we sought to make transgenic mosquitoes that stably express a Nix transgene under the control of its native promoter. We isolated a 2.5-kb region upstream of the Nix gene, which was used to express the Nix transgene in the plasmid (Fig. 1A). In addition to the native Nix ORF (N1), we also designed a second plasmid that contains a Strep II tag (16) at the N terminus (N2) to facilitate future biochemical studies and to distinguish between the endogenous Nix and the Nix transgene (Fig. 1A). Three transgenic lines were obtained (SI Appendix, Tables S1–S3) using the plasmids shown in Fig. 1A. In subsequent analyses we focused on two of these (one line derived from each of the two constructs), hereto referred to as N1 and N2. The transgene insertion sites were identified by inverse PCR (SI Appendix, Fig. S1; primers are shown in SI Appendix, Table S4). Bioinformatic mapping of the sequences flanking the N2 transgene revealed an insertion on chromosome 2 (Fig. 1B), while the N1 transgene was present near the telomere on the 1p arm of chromosome 1 (Fig. 1B), distantly linked to the native M-locus, as confirmed by chromosomal fluorescence in situ hybridization (FISH) (Fig. 1C). To determine whether the native Nix promoter fragment recapitulated Nix expression in the context of these new genomic locations, we determined the transcription profile of the N2 transgene using primers that readily distinguish the N2 Nix transcript from the endogenous Nix transcript (SI Appendix, Fig. S2 and Table S4). Like the endogenous Nix, N2 Nix transcription was observed from the onset of embryonic development starting 2 to 4 h after egg deposition throughout all developmental stages. The N2 Nix transcript was detected in transgenic m/m pupae and adults but not in wild-type male pupae or adults. Also as expected, we did not observe endogenous Nix transcripts in transgenic m/m pupae or adults. Thus, the 2.5-kb Nix promoter that we isolated appears to function similarly to the native Nix promoter in the M-locus in its temporal expression pattern.
Fig. 1.
Transgenic lines that stably express the Nix transgene. (A) Plasmid constructs used to create transgenic lines that stably express the Nix transgene. N1 was designed to express Nix from its own promoter. A 3.7-kb Nix sequence containing the ∼2.5-kb promoter, the 5′UTR (light blue), the Nix ORF (orange), and 3′UTR (light blue) is followed by the SV40 polyadenylation signal (yellow). This Nix expression cassette is followed by the transformation marker cassette, GFP driven by the Ae. aegypti polyubiquitin promoter. These two cassettes are flanked by the Mos1 transposon arms, which are not shown (37). The N2 construct is identical to N1 except that a Strep-tag II (16) was added to the N terminus of the Nix ORF. (B) A schematic drawing showing the insertion sites in the N1 and N2 transgenic lines, respectively. The relative position of M and m loci and the content of known genes in the M-locus are also shown. (C) Chromosomal in situ hybridization using the N1 plasmid as a probe showed a signal (red) on the p arm of chromosome 1 in addition to a signal in the known M-locus (15). The rDNA signal (magenta) was used as a landmark for the q arm of chromosome 1. Image was taken using 1000× magnification.
Transgenic Nix Causes Female-to-Male Sex Conversion.
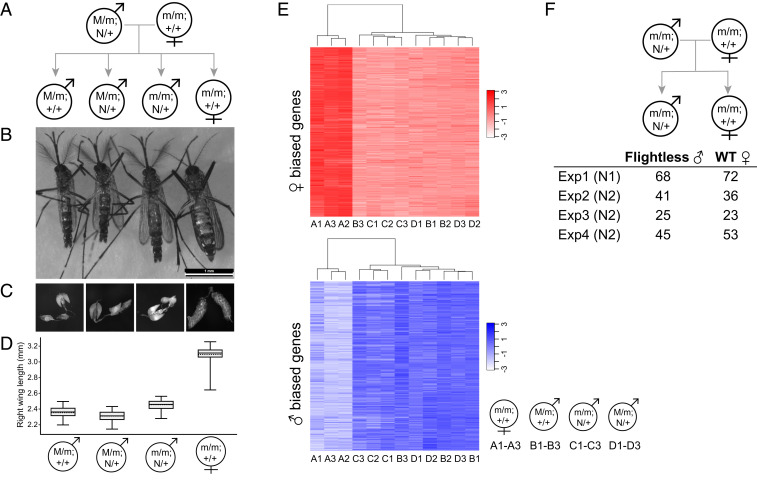
We next determined the phenotype of the genetic females (as indicated by the lack of the M-locus including myo-sex and the native Nix gene) (SI Appendix, Fig. S3) that contained the N1 or N2 Nix transgene. Four different genotypes are possible from crosses between transgenic males and wild-type females (Fig. 2A). They can be distinguished by the presence or absence of the EGFP transgenic marker (N/+ vs. +/+) and the presence or absence of the endogenous M-locus (M/m vs. m/m) (SI Appendix, Fig. S3). All genetic females with the N2 transgene showed conversion to complete male sexually dimorphic features, including plumose antennae, external genitalia showing male gonocoxite and gonostylus, and internal sex organs such as testes and male accessory glands (Fig. 2 B and C and SI Appendix, Fig. S3). The same female-to-male conversion phenotype was also observed in the N1 line (SI Appendix, Fig. S4). Thus, the Nix transgene alone converted females into phenotypic males. The sex-converted males showed a slightly larger body size compared to the wild-type and transgenic M/m males, as indicated by their wing-length measurement (Fig. 2D and SI Appendix, Fig. S5 and Table S5), which is used as a reliable proxy for body size (17). It is possible that subtle differences in Nix transgene expression or other factors in the M-locus may contribute to the wing-length difference between sex-converted m/m males and M/m males. We performed RNA-sequencing (RNA-seq) analysis of three biological replicates of pooled individuals of the four genotypes (National Center for Biotechnology Information [NCBI] BioProject number PRJNA625258). The overall transcription profile of the sex-converted m/m males was highly similar to wild-type M/m and transgenic M/m males, but clearly different from the wild-type m/m females (Fig. 2E and SI Appendix, Tables S6–S8). In addition, dsx and fru splicing was shifted toward the male isoforms, as confirmed by digital-droplet RT-PCR (ddPCR) (SI Appendix, Fig. S6).
Fig. 2.
The Nix transgene alone is sufficient to convert genetic females into fertile males with high penetrance and stability. (A) Pedigree showing the four genotypes of progeny from a cross between transgenic (N/+) males (M/m) and wild-type (+/+, or nontransgenic) females (m/m). N denotes the N1 or N2 Nix transgene. In this particular mating scheme, N2 transgenic males were used and produced progeny shown in B–D. (B) Representative individuals showing the phenotypes of the four genotypes shown in A. Genotyping was performed as shown in SI Appendix, Fig. S3. (C) Reproductive organs from individuals of the four genotypes. Images were taken using the LAS v4.5 software suite with 10× (whole body) or 40× (dissected tissues) magnification. (D) Length of the right wing of individuals of the four groups. Thirty individuals within each group were measured from the same cohort. Box plot, starting from bottom, shows minimum values, first quartile, median, third quartile, and maximum values using horizontal solid lines, with the mean indicated by a horizontal dashed line. As shown in SI Appendix, Table S5, all pairwise comparisons were significant (P < 0.0001), except for (M/m; +/+) vs. (M/m; N2/+) males (P = 0.1982). (E) RNA-seq of biological triplicates of each of the four genotypes from a cross between N1 transgenic males and wild-type females. The log2(FPKM+1) expression level heatmap of female-biased (red) and male-biased (blue) genes are shown and clustering of the samples was based on the transcription profile. (F) Mating of the sex-converted flightless m/m males with cold-anesthetized wild-type females produces wild-type females and m/m flightless males in four independent experiments. Similar results were obtained for both N1 and N2 (indicated in parentheses) transgenic m/m fathers. Genotype of the converted flightless males was confirmed (SI Appendix, Fig. S7).
Males Converted from Females by Nix Are Flightless.
Interestingly, all genetic females that were converted to phenotypic males due to the ectopic expression of the N1 or N2 transgene could not fly (Table 1; see also a video link at https://www.youtube.com/watch?v=fwUqN5iKTi0&feature=youtu.be). The majority of flightless males were not able to completely fold their wings; they could walk and sometimes jump but could never sustain flight. Wild-type males, N1 or N2 M/m males, and wild-type females displayed normal flight phenotypes. To assess the stability of these phenotypes, we screened for the four genotypes over multiple generations by crossing either (M/m; N1/+) or (M/m; N2/+) males with wild-type females. Between the two lines, we found 0 transgenic individuals that developed as females, while scoring 4,541 transgenic males and 2,215 flightless transgenic males (Table 1). As expected, the percentage of flightless males were higher in the N2 line than the N1 line because N1 is linked to the M-locus while N2 is on a separate chromosome. However, the numbers of the four genotypes did not strictly follow the expected Mendelian segregation ratios in the N2 lines in some generations (SI Appendix, Table S3). It is possible that some N2 male converts may have died prior to adult emergence.
Table 1.
Total number of progeny from crosses between transgenic males and wild-type females
| Line | Transgenic ♂ | Nontransgenic ♂ | Transgenic ♀ | Nontransgenic ♀ | Transgenic flightless ♂ |
| N1 | 2,696 | 829 | 0 | 2,186 | 902 |
| N2 | 1,845 | 2,079 | 0 | 2,027 | 1,313 |
The latest screening was done for G13 and G15 for N1 and N2 lines, respectively. Detailed numbers from each generation are provided in SI Appendix, Tables S2 and S3.
Males Converted from Females by Nix Are Fertile and the Sex-Conversion Phenotype Is Highly Penetrant and Heritable.
We then tested whether N1 or N2 m/m males were fertile, despite the absence of the M-locus. As these converted males were flightless and flying is required during mating (18), they could not mate with females that are alert. However, in four independent experiments that include both N1 and N2 lines, the sex-converted males fathered viable progeny when placed with cold-anesthetized wild-type females in a confined space (Fig. 2F). As there is no M-locus at all in these sex-converted transgenic males (m/m; N/+), when mated with wild-type females (m/m; +/+), only two genotypes are possible in their progeny: (m/m; N/+) and (m/m; +/+), which will manifest as flightless sex-converted males and wild-type females, respectively. Indeed, flightless males and wild-type females were observed at an approximately 1:1 ratio in all four experiments, and genotyping results confirmed that these flightless males were indeed sex-converted genetic females (SI Appendix, Fig. S7). Thus, sex-converted phenotypic males are fertile and continued to produce sex-converted progeny, indicating that the Nix transgene alone is sufficient to convert females into fertile males and that this sex-conversion is heritable. These results, together with the fact that not a single transgenic individual developed a female phenotype over a combined 28 generations with thousands of individuals screened (Table 1), suggest that the female-to-male conversion conferred by the Nix transgene is highly penetrant and stable.
The Myosin Heavy-Chain Gene Myo-Sex Is Required for Male Flight.
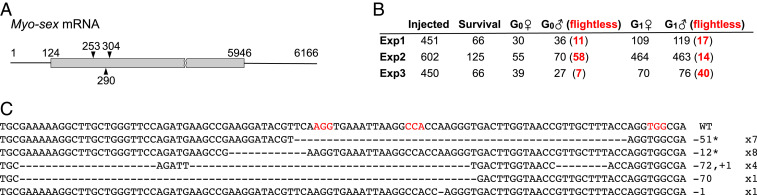
In addition to Nix, a myosin heavy-chain gene, myo-sex (19) is also located in the M-locus (15). Myo-sex and its closest autosomal paralog (AAEL005656) show sex-specific expression in males and females, respectively (19–21), and could be involved in sex-specific muscle functions. Therefore, we hypothesized that the flightless phenotype observed in N1 (m/m) and N2 (m/m) males is caused by the lack of the myo-sex gene associated with the M-locus. To test this hypothesis, we performed CRISPR/Cas9-mediated knockout using single-guide RNAs (sgRNAs) designed to specifically target the hemizygous myo-sex gene (Fig. 3). Following injection of preblastoderm embryos with a Cas9/sgRNA mixture, a significant portion of the surviving G0 males were flightless in three independent experiments. Despite the fact that only flying G0 males produced progeny when wild-type females were provided for mating, a significant portion of the G1 male offspring were also flightless in all three independent experiments (Fig. 3). We sequenced plasmids cloned from PCR products from 21 flightless G1 males and identified mutations in the myo-sex gene in all of them. Thus, we conclude that the M-locus gene myo-sex is required specifically for male flight.
Fig. 3.
Myo-sex knockout results in flightless males. (A) Three sgRNAs, starting at positions 253, 290, and 304, were used to target the myo-sex coding region, which spans positions 124 to 5,946. (B) CRISPR/Cas9-mediated myo-sex knockout produced flightless males in both the G0 and G1 generations in three independent experiments. G1 progeny were produced only from crosses between wild-type virgin females and sgRNA+Cas9-injected flying G0 males. (C) Sequence analysis of mutations in CRISPR/Cas9-edited flightless G1 progeny. Top line denotes the wild-type sequence; subsequent lines show various mutant sequences. The number of deleted (−) and inserted (+) bases and their occurrence are indicated to the right; in-frame mutations are indicated by an asterisk; deleted bases are denoted by dashes. The PAM sites for the three sgRNAs are highlighted in red.
Transgenic and Nontransgenic Males Are Not Significantly Different in Reproductive Fitness under Laboratory Conditions.
We performed competition assays by setting up six replicate cages, each containing 20 transgenic males (M/m; N/+) and 20 nontransgenic sibling males (M/m; +/+) competing for 10 virgin Liverpool females. We recorded the number of transgenic and nontransgenic progeny of each female from all six cages for the N1 and N2 lines, respectively (SI Appendix, Table S9). Overall, the percent of females that produced broods that contained transgenic progeny was 51.9% and 55.8% for the N1 and N2 lines, respectively (SI Appendix, Table S10). However, this does not necessarily mean that the transgenic males are more competitive in mating than their nontransgenic siblings because we cannot assume that females mate only once (22). To investigate transgenic fitness, we focused on testing whether the proportion of transgenic progeny deviated from the expected 25% under the assumption of equal reproductive fitness between the hemizygous transgenic and nontransgenic males (Table 2). We note that this test does not assume that females mate only once or use sperm from just one male. Overall, the N1 cages produced 21.5% (603 of 2,802, 3.5% lower than expected) transgenic progeny, while the N2 cages produced 31.3% (721 of 2,303, 6.3% higher than expected) transgenic progeny. However, none of these differences were statistically significant.
Table 2.
Proportion of transgenic progeny
| Replicate | + | − | Percent+, % |
| N1 replicate | |||
| 1 | 72 | 372 | 16.2 |
| 2 | 104 | 394 | 20.9 |
| 3 | 84 | 439 | 16.1 |
| 4 | 66 | 362 | 15.4 |
| 5 | 170 | 268 | 38.8 |
| 6 | 107 | 364 | 22.7 |
| Total | 603 | 2,199 | 21.5 |
| N2 replicate | |||
| 1 | 57 | 279 | 17.0 |
| 2 | 129 | 378 | 25.4 |
| 3 | 154 | 329 | 31.9 |
| 4 | 148 | 214 | 40.9 |
| 5 | 108 | 118 | 47.8 |
| 6 | 125 | 264 | 32.1 |
| Total | 721 | 1,582 | 31.3 |
Generalized linear mixed-effect model analyses were performed to test the null hypothesis that 25% of the progeny will be transgenic under the assumption of equal reproductive fitness between the hemizygous transgenic and nontransgenic males. No statistically significant departure from the 25% expectation was detected for either N1 or N2 (P > 0.05). Power analysis by 1,000 simulations show that the probability to correctly reject the null hypothesis (at α = 0.05) is 98.50% (95% confidence interval: 97.54 to 99.16%) and 99.10% (95% confidence interval: 98.30 to 99.59%) for N1 and N2, respectively. Data for each individual brood are provided in SI Appendix, Table S9. +, Number of transgenic (positive) progeny; −, number of nontransgenic (negative) progeny.
Discussion
The generation and analyses of Nix-expressing transgenic lines (Fig. 1) provided insights unattainable from previous transient experiments. Here, we have shown that Nix alone fulfills the role of a dominant master switch for male determination in Ae. aegypti, converting females into fertile males (Fig. 2). The fact that Nix is transcribed at the onset of maternal-to-zygotic transition (9) and that Nix expression does not require any other factors in the M-locus, as shown by the transcription of the Nix transgene in m/m females (SI Appendix, Fig. S2), indicate that Nix is the primary signal in the sex-determination pathway. Therefore, we conclude that Nix is indeed the M factor for sex-determination in Ae. aegypti.
The Ae. aegypti M-locus is a 1.3-Mbp repeat-rich region that contains a total of 30 annotated genes (15). These include Nix, four other protein-coding genes, and 25 lncRNA genes, several of which showed highly enriched expression in the testes or male accessory glands (15). Although they are apparently not necessary for the sex conversion observed in this study, it is reasonable to speculate that some of these genes may be involved in male reproductive biology or male-specific behavior. We note that our mating assay is a test of the minimal requirement of male fertility. Therefore, our results should not be regarded as indication of a lack-of-function of other M-locus genes. In fact, the converted m/m males could not fly and thus mating with cold-anesthetized wild-type females was necessary. We show that knockout of myo-sex, another gene located in the M-locus, results in a flightless male phenotype and may explain why Nix alone cannot convert females to flying males. Although it remains unknown whether Nix and myo-sex together can transform genetic females into both fertile and flying males, this study defined the roles of two key protein-coding genes in the sex locus of a unique, homomorphic, sex-determining chromosome in an insect species having significant medical importance. This work will inform future investigations of homomorphic sex chromosomes that are found in other insects, vertebrates, and plants.
The high penetrance and stability of the Nix transgene make it an attractive candidate for manipulation to reduce the number of biting and egg-laying females. Nix transgenic males do not appear to be significantly different in reproductive fitness compared to their nontransgenic sibling males under laboratory conditions. To further assess the fitness-related parameters important for genetic applications, tests need to be performed with more replicates, larger sample sizes, and field-like conditions. Multiple transgenic lines and various populations should also be tested for position effect and population-specific parameters. Converting genetic females into flightless males by expressing an endogenous mosquito gene, Nix, already successfully removes those females from the progeny. Although conceptually similar to female-killing approaches, the Nix transgenic males need not endure potentially leaky expression of a toxin designed to target females. To convert females into reproductively competitive males, a sex-conversion unit that includes both Nix and myo-sex, and possibly other M-locus genes, will be necessary. This may be achieved either by engineering a large sex-conversion cassette or by linking the necessary components in a region of suppressed recombination, which is not difficult to find in Ae. aegypti (15, 23, 24). Modeling suggests that the release of homozygous males carrying such a dominant sex-conversion unit is more effective than the sterile insect technique and the female-killing approaches in suppressing pest populations (25). The hemizygous Nix transgene in the current N1 and N2 males (M/m; N/+) is transmitted to only half of their progeny. The use of a conditional expression method, such as the tetracycline-repressible system (26), could allow for the development of homozygous strains carrying one or more copies of a Nix transgene or a sex-conversion unit. Such transgenic lines would produce male-only progeny and may be used as a population-suppression method (25, 27) or to significantly reduce the cost and augment the scale of other mosquito-control strategies that require the separation of males from females (28).
Nix-based female-removal or sex-conversion approaches could also be adapted to a homing-based gene-drive strategy (29) that would enable the inheritance of the transgene unit in almost all offspring, resulting in the removal or conversion of potentially all females. To mitigate resistance to homing, the Nix-containing homing cassette will need to target an essential gene (30–32). Here, sex-conversion is again much more powerful than female-removal. The outcome of releasing a sex-ratio distorter that is linked to a gene drive depends on the strength of the drive and the fitness of the transgenic individual (33, 34). Therefore, the successful design of a homing strategy for a sex-conversion unit requires balancing the need to transform females into sufficiently fit males using a multicomponent sex-conversion unit, and the ability to faithfully replicate that unit. Alternatively, the sex-conversion unit may be selected based on its linkage to a recoded essential gene that is no longer sensitive to a homing endonuclease that disrupts the endogenous copy of the essential gene (35).
Materials and Methods
Identification of the Nix Promoter Region.
To isolate the promoter/upstream sequence of Nix, PCR was performed using two primers (SI Appendix, Table S4), one of which contained the Nix ORF and the other was from ∼3 kb upstream. PCR was performed with an Arktik Thermal Cycler (Thermo Fisher Scientific) using Ae. aegypti (Liverpool) genomic DNA and the Q5 DNA Polymerase (New England Biolabs) according to the manufacturer’s protocol, generating a single amplicon of ∼3 kb. PCR products were cloned using the PCR-Blunt II-TOPO Cloning Kit (Thermo Fisher Scientific) and JM109 Competent Cells (Promega). A 2,575-bp sequence upstream of the ORF, a consensus derived from six clones, was used as the promoter. The ORF and the 5′ UTRs and 3′UTRs of Nix were previously determined (9).
Transgenic Constructs.
As shown in Fig. 1A, the N1 construct was designed to express Nix from its own promoter. A 3.7-kb sequence containing the 2,575-bp promoter described above (including the 102-bp 5′ UTR), the 864-bp Nix ORF, and the 19-bp Nix 3′ UTR, followed by the SV40 polyadenylation/termination signal sequence was synthesized (Epoch Life Science), and cloned into the pM2_pUB_EGFP vector (36) using the PstI/AsiSI restriction sites. The pM2_pUB_EGFP vector consists of Mos1 transposase substrate arms and the EGFP transformation marker driven by the Ae. aegypti polyubiquitin promoter (36). The N2 construct was designed to allow NIX protein purification and identification via an N-terminal Strep-tag II (Trp-Ser-His-Pro-Gln_phe-Glu-Lys) (16), (Fig. 1A). The N2 construct is identical to N1 except that the Strep-tag II was added to the N terminus of the Nix ORF (synthesized by Epoch Life Science).
Mosquito Rearing and Mos1-Mediated Transformation.
Ae. aegypti mosquitoes (Liverpool strain) were maintained at 28 °C and 60 to 70% humidity, with a 14/10-h day/night light cycle. Adult mosquitoes were maintained on 10% sucrose and blood-fed using artificial membrane feeders and defibrinated sheep’s blood (Colorado Serum Company). Donor plasmid was coinjected at 0.5 μg/μL with the Mos1 helper plasmid, pGL3-PUbMos1 at 0.3 μg/μL into 1-h-old embryos (37). Surviving G0 females were mated to Liverpool males in pools of 20 to 25. G0 males were mated individually to 5 Liverpool females and then merged into pools of 15 to 20 males. G1 larvae were screened for GFP fluorescence using a Leica M165 FC fluorescence microscope. Positive G1 individuals were outcrossed to Liverpool females to ensure that all transgene cassettes were stably inherited to the G2 generation. Images were taken of pupae and mature adults (7 to 10 d postemergence) at 10× (adults, pupae) or 40× (heads, genitalia, dissected tissues) magnification using the LAS v4.5 software suite and the following settings: Gain 1, Gamma 1, greyscale (white light), or pseudocolor (509 nm).
Inverse PCR for Transgene Insertion Site Determination.
Inverse PCR was used to determine the insertion site of the transgenic cassette for the N1 (generation 10) and N2 (generation 7) transgenic mosquito lines. Genomic DNA was isolated from three male adult individuals for each line, using the Quick-DNA Miniprep Kit (Zymo Research) according to the manufacturer’s protocol, and with an elution volume of 50 μL H2O. Three restrictions enzymes (Thermo Fisher), HpaI, MspI, and Bsp143I, were used to digest genomic DNA from each individual of each transgenic line. Approximately 1 μg DNA was digested for 8 h using 4 μL enzyme in a 60-μL reaction volume. Digested DNA was purified using the illustra GFX PCR DNA and Gel Band Purification Kit (GE Helathcare) with an elution volume of 50 μL H2O. Approximately 500 ng DNA was ligated in a 400-μL reaction volume with 1 μL T4 DNA Ligase (3 Weiss Units; Promega) overnight at 16 °C. DNA was purified using the illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare) with an elution volume of 30 μL H2O. PCR was performed using 1 μL of the purified DNA in a 50-μL reaction volume with Q5 DNA Polymerase (New England Biolabs), in a T100 Thermal Cycler (Bio-Rad), with primers (SI Appendix, Table S4) specific for the Mos1 transposable element right hand arm, which is located at the 5′ terminus of the transgenic cassette (see Fig. 1A for transgenic constructs). One specific PCR product was observed for each of the three enzymes used, for both lines. Sequencing results for all PCR products were the same for all individuals within each line, consistent with a single insertion site for each of the transgenic lines.
Chromosome FISH.
Slides of mitotic chromosomes were prepared from imaginal discs of fourth instar larvae from the N1 transgenic line following published protocols (38). FISH was performed using the N1 plasmid construct (Fig. 1A) as a probe and 18S rDNA as a landmark for the q arm of chromosome 1. The N1 plasmid probe was labeled by nick translation (Invitrogen), with Cy3-deoxyuridine 3-triphosphate (dUTP; Enzo Life Sciences). 18S rDNA was labeled by PCR (Bioline) with Cy5-dUTP (Enzo Life Sciences). Chromosomes were counterstained with Oxasole yellow (YOYO-1) iodide and mounted in Prolong Gold Antifade (Invitrogen). Slides were analyzed using a Zeiss LSM 510 Laser Scanning Microscope (Carl Zeiss Microimaging) at 1,000× magnification.
Endogenous and Transgenic Nix Transcription Profile.
Transgenic Nix GFP+ males from line N2 (M/m; N2/+) were crossed with Liverpool strain females and blood fed. To collect aged embryos ∼20 to 30 females were placed into 50-mL conical tubes with a wet cotton ball and a disk of filter paper at the bottom. Females were allowed to lay over the interval time and then removed from the tubes and eggs allowed to mature to the desired age. Embryos were transferred from the filter paper to a 1.5-mL tube with a fine paint brush and snap-frozen in liquid nitrogen at the appropriate time. Embryos (n ≥ 100) were collected for the following time points: 0 to 1 h, 2 to 4 h, 4 to 8 h, 8 to 12 h, 12 to 24 h, and 24 to 36 h. More eggs were collected from the same cage and hatched in order to collect siblings at all four larval stages (n = 50). One- and 2-d-old pupae, as well as 2- to 4-d postemergence adults were snap-frozen individually in 1.5-mL tubes so they could be genotyped by PCR before further processing (SI Appendix, Fig. S3). RNA was isolated using Quick-RNA Miniprep (Zymo Research). cDNA was then synthesized using the SuperScript RT kit (Life Technologies). RT-PCR was performed using Phire II DNA polymerase (Thermo Fisher Scientific) and primers are listed in SI Appendix, Table S4. We took advantage of the Strep-tag II inserted between the 5′UTR and the ORF of the Nix in the N2 construct to design primers that only amplify cDNA from the endogenous Nix transcript (SI Appendix, Table S4). Similarly, primers were also designed to only amplify the N2 transgenic Nix transcript (SI Appendix, Table S4).
Wing-Length Measurement and Statistical Analysis.
Wing-length values were measured for 30 individuals from line N2, eclosed from the same cohort of G14 larvae. Right wings were detached from 1-d postemergence adults, and mounted on a slide. A photograph was taken of each wing using a Leica DFC3000 G camera mounted on a Leica M165 FC Fluorescent Stereo Microscope. A 2-mm scale bar was photographed to standardize size measurements. ImageJ was used to measure the wing length as the distance from the anal lobe to the wing tip (17, 39) (SI Appendix, Fig. S5). Wing length values for each phenotype are reported in box plots (Fig. 2D) and shown in SI Appendix, Table S5. One-way ANOVA was performed using all samples, resulting in a P value of <0.0001. The Tukey simultaneous test for difference of means was performed resulting in adjusted P < 0.0001 for all pairwise comparisons except for transgenic males vs. wild-type males (P = 0.1982). A test for equal variances was performed using both Levene’s method and the multiple comparisons method.
RNA-Seq.
Two- to 4-d-postemergence adult siblings from the N1 transgenic line were snap-frozen in liquid nitrogen and preserved at −80 °C until the time of extraction. RNA was extracted using the Quick-RNA Miniprep kit (Zymo Research). Triplicate RNA-seq libraries were prepared for each of the four genotypes (Fig. 2E) using the NEBNext Ultra RNA Library Prep Kit for Illumina with the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs) and multiplexed into one lane of a HiSEq. 2500.
RNA-Seq Data Analysis.
RNA-seq reads from the wild-type and transgenic mosquito samples were aligned using Tophat2 (v2.1.1) to the Ae. aegypti L5 reference genome. The resulting BAM files were sorted and indexed. MarkDuplicates from the Picard tool kit v1.119 was used to identify and remove PCR duplicates (broadinstitute.github.io/picard/). Cufflinks v2.2.1 was used to assemble transcripts and estimate the relative abundances of the transcripts (40). Transcription levels were estimated as fragments per kilobase per million mapped reads (FPKM). The reference transcript file (AaegL5.0.gtf) was downloaded from VectorBase (vectorbase.org). Cuffdiff, a component of Cufflinks was used to normalize and compare the transcript expression levels between samples (41). The log2(FPKM+1) expression-level heatmaps (Fig. 2E) were generated using the heatmap.2 function in R’s gplots package (https://www.rdocumentation.org/packages/gplots/). Both columns (samples) and rows (genes) were clustered using the default hierarchical clustering settings. Row scaling was applied and the row z-score values were used for the color scale.
ddPCR for Doublesex and Fruitless Isoforms and Data Analysis.
Total RNA was isolated from each of the four genotypes using the ZymoResearch Quick-RNA MiniPrep Kit according to the manufacturer’s protocol. Three adult individuals (biological replicates) were used for each sample and all were siblings from generation 9 of the N1 transgenic line. cDNA was synthesized in a 20-μL reaction volume with ∼500 ng total RNA and random hexamers, using the Invitrogen SuperScript III First-Strand Synthesis Super Mix according to the manufacturer’s protocol, and the 20-μL completed cDNA reaction was diluted to a total of 60 μL with H2O. cDNA quality was checked by PCR using primers specific for ribosomal protein S7 (Rps7). These primers span a 110-nt intron, which should yield a 501- or 611-bp PCR product for cDNA or a genomic DNA template, respectively. Primers were ordered from Sigma Aldrich. Ae. aegypti (Liverpool) male genomic DNA and H2O was used as templates for positive and negative controls, respectively. PCR was performed using Phire HS II DNA polymerase from Thermo Fisher Scientific according to the manufacturer’s protocol. One microliter of the cDNA reaction was used in a total of 20-μL PCR volume and run for 30 cycles on a Bio-Rad MyCycler thermal cycler. Products were size-separated on a 1% agarose gel by electrophoresis, using a 100-bp step ladder from Promega. ddPCR was performed using 1 μL of the cDNA reaction with the Bio-Rad QX100 ddPCR machine according the manufacturers protocols. Taqman assays were designed to detect sex-specific doublesex and fruitless isoforms (SI Appendix, Table S4). The gene AAEL002401 was used as an internal reference for gene expression (9, 42). Probes were ordered from Biosearch Technologies and primers were ordered from Sigma Aldrich. Expression values are reported as the mean ± the SEM (SI Appendix, Fig. S6). One-way ANOVA was performed using all samples for each assay and the Tukey simultaneous test for difference of means was performed for all pairwise comparisons (SI Appendix, Fig. S6). A test for equal variances was performed using both Levene’s method and the multiple-comparisons method.
Mating Conditions for Flightless Males.
Five-day-old virgin Liverpool females were bloodfed and immediately placed in the refrigerator for 20 to 30 min so that they became immobile. Five of these cold-anesthetized females were placed in a 50-mL plastic conical tube together with five-to-eight flightless m/m males. The cotton ball covering the tube was pushed down close to the bottom to confine the mosquitoes to a small space to induce mating. The flightless males only had a few minutes to mate before the females became active. Mosquitoes were transferred to a 16-oz soup cup after half an hour. An egg cup was added to the soup cup after 2 d to collect eggs and embryos were hatched 5 d later.
CRISPR/Cas9-Mediated Myo-Sex Knockout.
Genomic regions of the myo-sex gene were manually searched for the presence of NGG (PAM), where N is any nucleotide. Selected target sites were 5′-GAAGCCGAAGGATACGTTCAAGG-3′, 5′-GTAACCGTTGCTTTACCAGGTGG-3′, and 5′-GGTTACCAAGTCACCCTTGGTGG-3′, with PAM sites underlined. sgRNAs were generated as previously described (43) using primers listed in SI Appendix, Table S4, in vitro-transcribed using MEGAscript T7 kit, and purified using the MEGAclear kit (Thermo Fisher Scientific). RNAs were aliquoted and stored at −80 °C. Cas9 mRNA was in vitro-transcribed, as previously described (44), and used for the first two knockout experiments (Fig. 3B). For the third replicate, Cas9 (ARCA) mRNA (Trilink Biotechnologies) was used. Three rounds of injections were performed with Cas9 mRNA at 0.6 µg/µL and all three sgRNAs at 0.1 µg/µL each for ∼500 embryos per replicate experiment. G0 males displayed a noticeable phenotype and were grouped by ability to fly, then crossed with Liverpool females. Mutations in the flightless male G1 were verified using the Phire II Animal Tissue Direct PCR kit (Thermo-Fisher Scientific), following the dilution procedure utilizing a single leg for the DNA template. Primers used for mutation screening are listed in SI Appendix, Table S4. PCR amplicons were purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) and sequenced.
Male Competitiveness Assay.
To mitigate the effect of genetic background and rearing condition, transgenic and their nontransgenic siblings were sexed, screened, and collected at the pupal stage from the same cohort and allowed to emerge separately. Six replicate cages were set up and assayed for each line as described below. Within 48 h after emergence, 20 flying transgenic males and 20 of their nontransgenic male siblings were placed in a 44-oz cage to commingle for 3 d before 10 5-d-old wild-type Liverpool virgin females were added and allowed to mate for another 2 d before blood feeding. Females were inspected individually to ensure that they were fully engorged. Two days later each female was transferred into an egg-laying tube, which is a 50-mL plastic conical tube with a hardened 50-mm wet paper disk on top of a water-soaked cotton ball at the bottom. Three to four days after egg laying, the eggs from individual females were hatched. The hatched larvae were fed with one crushed pellet of fish food and reared in sanitary, but nutrient-rich water at an optimal density, changing water or adding food as needed. Larvae were screened at L3–L4 using the GFP fluorescence marker and the numbers of GFP+ (transgenic) and GFP− (nontransgenic) were recorded.
Statistical Analysis of the Proportion of Transgenic Progeny.
Statistical analyses were performed using generalized linear mixed-effects models with transgenic and nontransgenic progeny counts as the independent variables (R package lme4). These variables were treated as fixed-effects and individual broods nested within cages/replicates were treated as random-effects. We employed a binomial regression in the analysis to model independent variables represented as proportions. The data in each test was assessed for overdispersion using the Pearson χ2 statistic and all assumptions of the tests were met. Statistical power of the generalized linear mixed-model tests was estimated using the simr package in R, with a number of iterative simulations set at 1,000. We used R v3.6.2 (45) to perform these analyses.
Materials and Data Availability.
All data and associated protocols are either described in the paper or deposited at the National Center for Biotechnology BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA625258). Transgenic mosquitoes and cloned DNA will be made promptly available on request by qualified researchers for their own use.
Supplementary Material
Acknowledgments
We thank Kate Morton, Camden Delinger, and Clare Morris for mosquito care and screening; Clemont Vinauger for generalized linear mixed-model analysis and producing one of the video clips; Karthikeyan Chandrasegaran for advice on wing-length measurement and for generalized linear mixed-model analysis; Brantley Hall and Giuseppe Saccone for comments; and Janet Webster and Jean Clarke for editorial suggestions. This work is supported by NIH Grants AI123338 and AI121853 and the Virginia Agriculture Experimental Station.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data have been deposited in the National Center for Biotechnology BioProject database (accession no. PRJNA625258).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001132117/-/DCSupplemental.
References
- 1.Bachtrog D. et al.; Tree of Sex Consortium , Sex determination: Why so many ways of doing it? PLoS Biol. 12, e1001899 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salz H., Erickson J. W., Sex determination in Drosophila: The view from the top. Fly (Austin) 4, 60–70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasselmann M. et al., Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454, 519–522 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Kiuchi T. et al., A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509, 633–636 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Baker R. H., Sakai R. K., Triploids and male determination in the mosquito, Anopheles culicifacies. J. Hered. 70, 345–346 (1979). [DOI] [PubMed] [Google Scholar]
- 6.Willhoeft U., Franz G., Identification of the sex-determining region of the Ceratitis capitata Y chromosome by deletion mapping. Genetics 144, 737–745 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla J. N., Palli S. R., Production of all female progeny: Evidence for the presence of the male sex determination factor on the Y chromosome. J. Exp. Biol. 217, 1653–1655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charlesworth D., Mank J. E., The birds and the bees and the flowers and the trees: Lessons from genetic mapping of sex determination in plants and animals. Genetics 186, 9–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall A. B. et al., SEX DETERMINATION. A male-determining factor in the mosquito Aedes aegypti. Science 348, 1268–1270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall A. B. et al., Radical remodeling of the Y chromosome in a recent radiation of malaria mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 113, E2114–E2123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzywinska E., Dennison N. J., Lycett G. J., Krzywinski J., A maleness gene in the malaria mosquito Anopheles gambiae. Science 353, 67–69 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Criscione F., Qi Y., Tu Z., GUY1 confers complete female lethality and is a strong candidate for a male-determining factor in Anopheles stephensi. eLife 5, e19281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma A. et al., Male sex in houseflies is determined by Mdmd, a paralog of the generic splice factor gene CWC22. Science 356, 642–645 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Meccariello A. et al., Maleness-on-the-Y (MoY) orchestrates male sex determination in major agricultural fruit fly pests. Science 365, 1457–1460 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Matthews B. J. et al., Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563, 501–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt T. G. M., Skerra A., The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2, 1528–1535 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Gleiser R. M., Urrutia J., Gorla D. E., Body size variation of the floodwater mosquito Aedes albifasciatus in Central Argentina. Med. Vet. Entomol. 14, 38–43 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Cator L. J., Arthur B. J., Ponlawat A., Harrington L. C., Behavioral observations and sound recordings of free-flight mating swarms of Ae. Aegypti (Diptera: Culicidae) in Thailand. J. Med. Entomol. 48, 941–946 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall A. B. et al., Insights into the preservation of the homomorphic sex-determining chromosome of Aedes aegypti from the discovery of a male-biased gene tightly linked to the M-locus. Genome Biol. Evol. 6, 179–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Leary S., Adelman Z. N., Disrupting female flight in the vector Aedes aegypti. bioRxiv:10.1101/862300 (2 December 2019).
- 21.Jiang X., Biedler J. K., Qi Y., Hall A. B., Tu Z., Complete dosage compensation in Anopheles stephensi and the evolution of sex-biased genes in mosquitoes. Genome Biol. Evol. 7, 1914–1924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helinski M. E. et al., Evidence of polyandry for Aedes aegypti in semifield enclosures. Am. J. Trop. Med. Hyg. 86, 635–641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudchenko O. et al., De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juneja P. et al., Assembly of the genome of the disease vector Aedes aegypti onto a genetic linkage map allows mapping of genes affecting disease transmission. PLoS Negl. Trop. Dis. 8, e2652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schliekelman P., Ellner S., Gould F., Pest control by genetic manipulation of sex ratio. J. Econ. Entomol. 98, 18–34 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Fu G. et al., Female-specific flightless phenotype for mosquito control. Proc. Natl. Acad. Sci. U.S.A. 107, 4550–4554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adelman Z. N., Tu Z., Control of mosquito-borne infectious diseases: Sex and gene drive. Trends Parasitol. 32, 219–229 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papathanos P. A. et al., A perspective on the need and current status of efficient sex separation methods for mosquito genetic control. Parasit. Vectors 11 (suppl. 2), 654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gantz V. M. et al., Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. U.S.A. 112, E6736–E6743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noble C., Olejarz J., Esvelt K. M., Church G. M., Nowak M. A., Evolutionary dynamics of CRISPR gene drives. Sci. Adv. 3, e1601964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Champer J. et al., A toxin-antidote CRISPR gene drive system for regional population modification. Nat. Commun. 11, 1082 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Champer J., et al. , Resistance is futile: A CRISPR homing gene drive targeting a haplolethal gene. bioRxiv:10.1101/651737 (27 May 2019). [Google Scholar]
- 33.Simoni A. et al., A male-biased sex-distorter gene drive for the human malaria vector Anopheles gambiae. Nat. Biotechnol., 10.1038/s41587-020-0508-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaghton A., Beaghton P. J., Burt A., Gene drive through a landscape: Reaction-diffusion models of population suppression and elimination by a sex ratio distorter. Theor. Popul. Biol. 108, 51–69 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Burt A., Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270, 921–928 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson M. A., Gross T. L., Myles K. M., Adelman Z. N., Validation of novel promoter sequences derived from two endogenous ubiquitin genes in transgenic Aedes aegypti. Insect Mol. Biol. 19, 441–449 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coates C. J., Jasinskiene N., Miyashiro L., James A. A., Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 95, 3748–3751 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timoshevskiy V. A., Sharma A., Sharakhov I. V., Sharakhova M. V., Fluorescent in situ hybridization on mitotic chromosomes of mosquitoes. J. Vis. Exp., e4215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Handel E., Day J. F., Correlation between wing length and protein content of mosquitoes. J. Am. Mosq. Control Assoc. 5, 180–182 (1989). [PubMed] [Google Scholar]
- 40.Roberts A., Trapnell C., Donaghey J., Rinn J. L., Pachter L., Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12, R22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C. et al., Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu W., Tu Z. J., Functional analysis of the promoter of an early zygotic gene KLC2 in Aedes aegypti. Parasit. Vectors 11 (suppl. 2), 655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bassett A. R., Tibbit C., Ponting C. P., Liu J. L., Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220–228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basu S. et al., Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 112, 4038–4043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R version 3.6.2. https://www.r-project.org. Accessed 8 July 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.