Significance
In mouse models of triple-negative breast cancer, we show that the oxidation of tumor cells prior to lysate generation, coupled with their compartmentalization in the core of liposomal spherical nucleic acids (SNAs) comprised of adjuvant DNA, yields a powerful immunotherapeutic that significantly inhibits tumor growth, dramatically extends survival, and promotes a tumoricidal immune cell population within the tumor microenvironment. Specifically, this work points toward the importance of properly packaging and presenting the adjuvant and antigens such that biodistribution, dendritic cell activation, and therapeutic efficacy can be controlled.
Keywords: spherical nucleic acids, cancer vaccines, oxidized tumor lysates, cancer immunotherapy
Abstract
Highly heterogenous cancers, such as triple-negative breast cancer (TNBC), remain challenging immunotherapeutic targets. Herein, we describe the synthesis and evaluation of immunotherapeutic liposomal spherical nucleic acids (SNAs) for TNBC therapy. The SNAs comprise immunostimulatory oligonucleotides (CpG-1826) as adjuvants and encapsulate lysates derived from TNBC cell lines as antigens. The resulting nanostructures (Lys-SNAs) enhance the codelivery of adjuvant and antigen to immune cells when compared to simple mixtures of lysates with linear oligonucleotides both in vitro and in vivo, and reduce tumor growth relative to simple mixtures of lysate and CpG-1826 (Lys-Mix) in both Py230 and Py8119 orthotopic syngeneic mouse models of TNBC. Furthermore, oxidizing TNBC cells prior to lysis and incorporation into SNAs (OxLys-SNAs) significantly increases the activation of dendritic cells relative to their nonoxidized counterparts. When administered peritumorally in vivo in the EMT6 mouse mammary carcinoma model, OxLys-SNAs significantly increase the population of cytotoxic CD8+ T cells and simultaneously decrease the population of myeloid derived suppressor cells (MDSCs) within the tumor microenvironment, when compared with Lys-SNAs and simple mixtures of oxidized lysates with CpG-1826. Importantly, animals administered OxLys-SNAs exhibit significant antitumor activity and prolonged survival relative to all other treatment groups, and resist tumor rechallenge. Together, these results show that the way lysates are processed and packaged has a profound impact on their immunogenicity and therapeutic efficacy. Moreover, this work points toward the potential of oxidized tumor cell lysate-loaded SNAs as a potent class of immunotherapeutics for cancers lacking common therapeutic targets.
Mobilizing the immune system against tumors is a central goal of personalized cancer treatments. Indeed, the identification of tumor-associated antigens (TAAs) and the advent of cell-based therapies represent significant progress toward achieving this aim (1–5). However, these approaches, including the use of dendritic cell (DC) vaccines (6) and chimeric antigen receptor T cell therapies (7), are expensive and labor intensive, as they require the extraction of immature immune cells from patients, expansion of cells ex vivo, incubation with TAAs, and reinfusion to the patient. Furthermore, these therapies are restricted to the subset of patients whose tumors express known TAAs (8), and raising an immune response with single-antigen vaccines may ultimately have limited efficacy due to tumor heterogeneity and loss of antigen expression over time (9–11).
An attractive alternative to single-antigen vaccines is to use lysates isolated from a patient’s own tumor as the TAA source (12–18). Exploiting tumor cell lysates as antigens broadens the set of proteins that can be processed and targeted by immune cells—in principle, the entire tumor proteome can be accessed (18). Therefore, this also addresses several potential limitations of using a finite set of well-defined TAAs, including 1) the challenge of identifying immunogenic epitopes from tumors, 2) epitope restriction by only one of the major histocompatibility complexes (MHC class I or class II), and 3) loss of targeted antigens in tumors. However, direct vaccination using tumor lysates has been met with limited success, due to low cellular uptake and bioavailability after injection, resulting in minimal immunogenicity (19). Oxidizing tumor cells prior to lysate isolation and preparation significantly increases immunogenicity when the lysates are utilized as the antigen source in DC vaccines (19–21). Importantly, protein chlorination by hypochlorous acid (HOCl), an oxidant produced by neutrophils as part of the adaptive immune response, increases the immunogenicity of antigens severalfold (22), potentially due to their increased proteolytic susceptibility (23). In addition, HOCl oxidation generates aldehyde-modified antigens that are more immunogenic than their unmodified counterparts (24). However, the underlying mechanism of how these changes facilitate antigen presentation and alter the tumor microenvironment (TME) remains unclear. Moreover, a major challenge in immunotherapeutic development is the selection of the appropriate vehicle for delivering both adjuvant and antigen (25), as the way components are formulated can significantly influence delivery to the immune system and thus activation of immunostimulatory pathways (26, 27). Nanoscale therapeutics have shown promise in this regard, by enhancing antigen-presenting cell (APC) activation over mixtures of adjuvant and antigen (28).
Spherical nucleic acids (SNAs) are a class of nucleic acids that exhibit completely different behavior from their linear analogs (29), including rapid cellular uptake without the use of ancillary transfection reagents (30). The SNA architecture is defined by a dense, highly oriented packing of nucleic acids into a spherical morphology, which imparts new chemical, biological, and physical properties to the nucleic acids from which SNAs are derived. To date, SNAs have been formed from a variety of nanoparticle cores, including gold and other inorganic nanoparticles (29–36), liposomes (37–41), polymers (42–44), and proteins (45). Liposomes are an especially attractive scaffold for SNA templating because the resulting systems are biodegradable and biocompatible, and liposomes are a validated, Food and Drug Administration-approved nanoscale formulation for drug delivery (46). In addition, the hollow core of liposomal SNAs can encapsulate TAAs and other cargo. Liposomal SNAs have been previously observed to initiate antigen presentation, activate immune cells, and induce production of proinflammatory cytokines for cancer treatment and other applications (39, 40, 41, 47, 48). In many of these examples, the sequence of the oligonucleotide shell comprises an unmethylated cytosine−guanosine sequence called CpG-1826. CpG-1826 mimics microbial genomes and acts as a pathogen-associated molecular pattern (49), which is recognized by Toll-like receptor 9, a component of the innate immune system located in the endosome of APCs, including DCs (50).
Given these facts, we reasoned that SNAs containing tumor cell lysates could be used to develop potent nanoscale immunotherapeutics for the treatment of highly heterogenous cancers, such as triple-negative breast cancer (TNBC). Accounting for the majority of all breast cancer-related mortality (51, 52), TNBC is a highly heterogeneous and aggressive disease that lacks functional expression of both estrogen and progesterone receptors and no overexpression of human epidermal growth factor receptor 2 protein (51–54). Paradoxically, TNBC primary tumors often initially respond well to chemotherapy; yet, there is a high incidence of relapse and metastasis. The early and aggressive nature of TNBC recurrence is exemplified by significantly decreased rates of progression-free and 3-y overall survival vs. other breast cancer subtypes (52, 55, 56), necessitating the development of new and effective treatment options. In an effort to explore the potential of SNAs as a therapeutic for treating TNBC, we synthesized liposomal SNAs that encapsulate lysates derived from TNBC cell lines in their core and present CpG-1826 on their surfaces (Lys-SNAs; Fig. 1), as well as analogs that contain lysates from TNBC cells that were oxidized with HOCl prior to lysis (OxLys-SNAs), and evaluated their immunomodulatory activity and antitumor properties in syngeneic, orthotopic mouse models of TNBC.
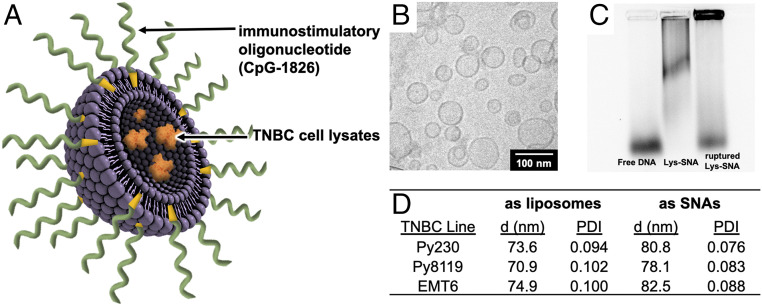
Fig. 1.
A lysate-loaded, immunostimulatory SNA (Lys-SNA). (A) A schematic of a Lys-SNA. TNBC lysates (orange) from either oxidized or nonoxidized TNBC cells are encapsulated in the core of a liposome (purple), which is functionalized with cholesterol-modified nucleic acids (green) to generate the SNA. (B) Cryo-TEM of Lys-SNAs. (C) Gel electrophoresis of free CpG-1826 (left lane), Lys-SNAs (middle lane), and Lys-SNAs after exposure to Triton-X to break apart liposomes (right lane). (D) Hydrodynamic diameter of lysate-loaded liposomes and SNAs, as measured by DLS, where “d” represents hydrodynamic diameter and “PDI” represents polydispersity index.
Results
Lysates from TNBC Cell Lines Can Be Compartmentalized in Lys-SNAs.
To assess the feasibility of using TNBC lysates as antigen sources, we utilized three murine mammary carcinoma cell lines to recapitulate the heterogeneity of TNBC (57, 58). Toward this end, we utilized Py230, a luminal cell line (59–61), and Py8119, a basal cell line (59–61), derived from the mouse mammary tumor virus−polyoma middle tumor antigen mouse model of breast cancer, which loses expression of estrogen and progesterone as it progresses (62). We chose the EMT6 cell line as a third model, as this syngeneic line has been recently recognized as a valuable model to study immune response in TNBC (63, 64). Cells were grown to confluency in monolayer cell culture, dissociated and subjected to several freeze−thaw cycles to induce cell necrosis and rupture cell membranes, and centrifuged to remove cellular debris, producing “whole” tumor lysate (65). The lysate was encapsulated in ∼70-nm liposomes prepared from 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC). After purification to remove unencapsulated lysate, the liposomes were incubated with 3′-cholesteryl-modified CpG-1826 (SI Appendix, Fig. S1) to generate Lys-SNAs (Fig. 1A), whose single-lamellar spherical morphologies were validated by cryogenic transmission electron microscopy (cryo-TEM; Fig. 1B). The average ratio of protein to DNA was determined to be 1.1 ± 0.7 mg of protein per micromole of DNA from three independent batches of EMT6 Lys-SNAs. Analysis via gel electrophoresis (Fig. 1C) and the observed increase in hydrodynamic diameter, as measured by dynamic light scattering (DLS; Fig. 1D), are consistent with DNA functionalization and SNA generation.
Lys-SNAs Increase Codelivery of Lysates and CpG DNA to DCs In Vitro and In Vivo.
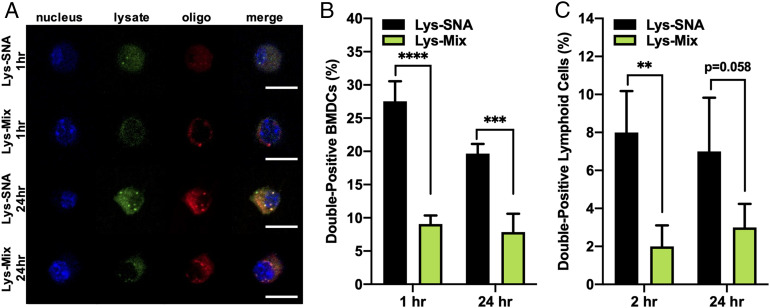
Codelivery of antigen and adjuvant to the same APC is vital for maximal antigen processing and presentation, as well as inducing the most potent antigen-specific immune response (66). Thus, we investigated the codelivery of lysate and DNA to APCs in vitro and in vivo. Toward this end, Lys-SNAs containing fluorophore-labeled lysate (fluorescein and Oregon Green 488) and CpG-1826 (Cy5) were synthesized. Purified lysates were incubated with an Oregon Green 488 succinimidyl ester (OR488-NHS) dye and fluorescein isothiocyanate (FITC)-5-maleimide dye to label both the free amines and thiols in the bulk protein solution. Following removal of any unreacted dye, FITC/OR488-labeled lysates and Cy5-modified DNA were used to generate dual fluorophore-labeled Lys-SNAs. Bone marrow-derived DCs (BMDCs) were isolated from C57BL/6 mice and incubated with either fluorophore-labeled Lys-SNAs or a mixture of fluorophore-labeled lysate and CpG-1826 (Fig. 2) at the same protein and DNA concentrations as Lys-SNAs. At set time points, cells were collected and analyzed by both confocal microscopy (Fig. 2A) and flow cytometry (Fig. 2B) to determine the number of cells positive for both FITC/OR488 and Cy5. At all time points, Lys-SNAs showed higher codelivery to immune cells in vitro than the simple mixture of lysate with CpG-1826 (Lys-Mix), showing a threefold and 2.5-fold enhancement in codelivery after 2 and 24 h incubation, respectively. To evaluate codelivery of antigen and adjuvant in vivo, C57BL/6 mice were subcutaneously (s.c.) administered Lys-SNA or Lys-Mix containing fluorophore-labeled lysate and CpG-1826 (n = 3 per group). At 2 and 24 h postinjection, animals were killed, and inguinal lymph nodes were isolated and dissociated into single-cell suspensions, and then analyzed by flow cytometry (Fig. 2C). In agreement with the in vitro data, codelivery to CD11c+ immune cells in vivo was enhanced fourfold when lysates and adjuvant DNA were formulated as Lys-SNAs at 2 h postinjection, with over 2.3-fold enhancement still observed at 24 h.
Fig. 2.
Delivery of FITC-labeled lysate within Cy5-labeled SNAs. (A) Confocal microscopy images of BMDCs incubated with dual fluorophore-labeled Lys-SNA and Lys-Mix for 1 and 24 h. (Scale bar, 10 µm.) (B) Codelivery of lysate and DNA by Lys-SNA (n = 3, black bars) and Lys-Mix (n = 3, green bars) to BMDCs in vitro after 1- and 24-h incubations as assessed via flow cytometry. (C) Codelivery of lysate and DNA by Lys-SNA (n = 3, black bars) and Lys-Mix (n = 3, green bars) to lymphoid cells in vivo 2 and 24 h following s.c. injection. Lymph nodes were isolated, and CD11c+ lymphoid cells were analyzed by flow cytometry. Statistical analysis was performed using an ordinary one-way ANOVA, where “**” represents a P value of <0.01, “***” represents a P value of <0.001, and “****” represents a P value of <0.0001.
Lys-SNAs Show Antitumor Activity in Multiple TNBC Models.
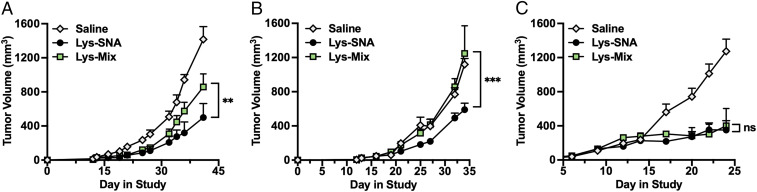
To assess the antitumor activity of Lys-SNAs in vivo, we established orthotopic syngeneic models of TNBC by inoculating mice with ∼106 TNBC cells (Py230, Py8119, or EMT6) in the left inguinal mammary fat pad. At days 6, 10, and 15 postinoculation, animals were peritumorally administered Lys-SNA, Lys-Mix, or saline as a negative control at a dose of 10 nmol of CpG-1826 and 20 μg of lysate. In the Py230 and Py8119 models, animals administered Lys-SNAs showed a 42% and 53% reduction in tumor volume, respectively, relative to animals administered Lys-Mix at day 30 of the study (Fig. 3 A and B), suggesting that packaging lysates and adjuvant DNA into an SNA architecture increases the observed antitumor efficacy, as compared to administration of mixtures of individual components. In the EMT6 model, administration Lys-SNA stalled tumor growth relative to saline, showing a 73% reduction in tumor growth at day 25; however, no significant difference in tumor growth was observed between animals administered Lys-SNA and Lys-Mix (Fig. 3C). Based on previous reports of using tumor lysates as antigen sources (22–24), we hypothesized that the lysates generated in the EMT6 model were poorly immunogenic, thus leading to suboptimal T cell priming and subsequent antitumor activity. Therefore, we investigated the use of lysates derived from oxidized tumor cells in this model.
Fig. 3.
Antitumor effects of Lys-SNA and Lys-Mix in vivo. (A) Antitumor efficacy of mice bearing orthotopic syngeneic (A) Py230 tumors, (B) Py8119 tumors, or (C) EMT6 tumors when administered Lys-SNA (black circles), Lys-Mix (green squares), or saline (white diamonds). Treatment initiation began at day 6 postinoculation and was repeated on days 10 and 15. Statistical analysis was performed using an ordinary one-way ANOVA, where “**” represents a P value of < 0.01, “***” represents a P value of < 0.001, and “n.s.” represents a P value > 0.05, which is considered not significant.
Oxidizing Tumor Cells prior to Lysis Increases Observed Immunogenicity.
Since it has been reported that oxidation increases the immunogenicity of tumor lysates, we sought to determine whether lysates derived from oxidized tumor cells could be utilized as potent antigen sources following incorporation into SNAs. We generated oxidized tumor cell lysates by incubating EMT6 cells in 60 μM HOCl for 1 h to ensure complete cell death (67). Oxidized lysates were then prepared by subjecting the cells to several freeze−thaw cycles and centrifugation to remove cellular debris. The total amount of protein lysate collected from oxidized tumor cells (herein referred to as “oxidized lysate”) was similar to that of protein lysate collected from nonoxidized tumor cells (herein referred to as “nonoxidized lysate”); however, the bulk protein population of oxidized lysate differed from that of nonoxidized lysate, with larger protein bands appearing in the oxidized sample (SI Appendix, Fig. S2).
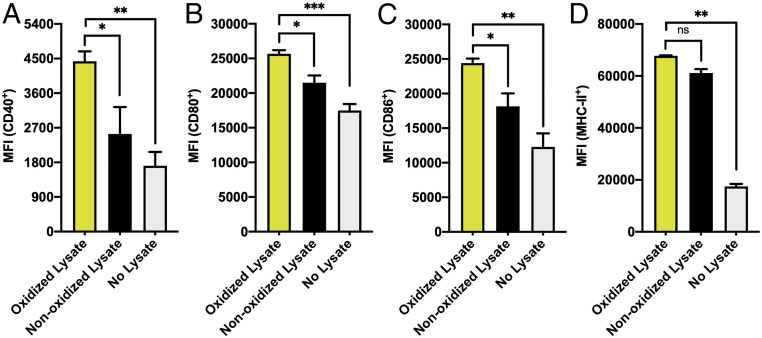
To determine whether oxidation prior to lysate generation increases the immunogenicity of the isolated lysate, we incubated BMDCs (Fig. 4 and SI Appendix, Fig. S3) with CpG-1826 (0.1 nmol) and either nonoxidized lysate or oxidized lysate at equivalent protein concentrations (1 µg of total protein). The expression of maturation markers CD40, CD80, and CD86 was significantly elevated in BMDCs incubated with oxidized lysates (Fig. 4 A–C) relative to treatment with nonoxidized lysates alone, showing a 75%, 20%, and 34% increase in CD40, CD80, and CD86 marker expression over nonoxidized lysates, respectively, as well as a 160%, 47%, and 98% increase over CpG-1826 alone. Furthermore, expression of the MHC-II was elevated nearly fourfold in BMDCs incubated with oxidized lysate as compared to those incubated with CpG-1826 alone (Fig. 4D).
Fig. 4.
Activation of BMDCs in vitro following incubation. Cells isolated from C57BL6 mice were purified and cocultured with CpG-1826 and oxidized lysates (yellow bars), nonoxidized lysates (black bars), or no lysate (gray bars). After 2 d, DC activation was measured by flow cytometry for expression levels of (A) CD40, (B) CD80, (C) CD86, and (D) MHC-II. Statistical analysis was performed using an ordinary one-way ANOVA, where “*” represents a P value of <0.05, “**” represents a P value of <0.01, “***” represents a P value of <0.001, and “n.s.” represents a P value of >0.05, which is considered not significant. “MFI” denotes median fluorescence intensity.
OxLys-SNAs Significantly Inhibit Tumor Growth and Extend Survival In Vivo.
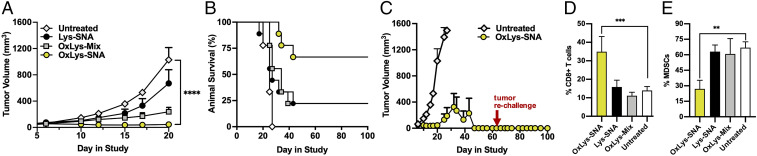
To evaluate the function of OxLys-SNAs as cancer immunotherapeutics, the in vivo antitumor activity of OxLys-SNAs was compared to Lys-SNAs and mixtures of oxidized lysate with CpG-1826 (OxLys-Mix) in the EMT6 model of TNBC. Balb/C mice were inoculated with ∼106 tumor cells in the left inguinal mammary fat pad, and, at day 6 postinoculation, treatment was initiated via peritumoral s.c. injection of OxLys-SNA, Lys-SNA, or OxLys-Mix (n = 9 per group) at a dose of 5 nmol of DNA (50% dose as compared to the studies shown in Fig. 3) and 20 µg of protein. Saline-treated animals were used as a negative control. Injections were repeated at days 10 and 15. Tumor mass and animal survival were monitored for 100 d postinoculation. Animals administered OxLys-SNA responded remarkably well to treatment as compared to all other treatment groups (Fig. 5A), with seven out of nine OxLys-SNA treated animals experiencing complete tumor remission at day 20 (SI Appendix, Fig. S4A). Furthermore, animal survival was significantly extended when administered OxLys-SNA (Fig. 5B), with six out of nine OxLys-SNA−treated animals surviving beyond day 100 postinoculation. Indeed, the first OxLys-SNA−treated animal to succumb to tumor burden (tumor volume exceeding 1,500 mm3) survived longer than all saline-treated animals (SI Appendix, Fig. S4B). To assess the ability of OxLys-SNAs to induce an immunological memory response, a small cohort of animals administered OxLys-SNAs were rechallenged by implanting ∼106 EMT6 cells in the inguinal mammary fat pad on day 60 following initial tumor implantation (Fig. 5C). All animals (n = 3) remained tumor free following rechallenge with EMT6 cells, indicating that vaccination with OxLys-SNAs not only eradicates existing tumors but also prevents new tumors from forming.
Fig. 5.
OxLys-SNA in vivo analyses. (A) Antitumor efficacy and (B) corresponding survival curve of BALB/c mice (n = 9 per group) bearing orthotopic syngeneic EMT6 tumors when administered OxLys-SNA (yellow circles), OxLys-Mix (gray squares), Lys-SNA (black circles), or saline (white diamonds). Treatment initiation began at day 6 postinoculation and was repeated on days 10 and 15. (C) Full tumor growth curve of animals administered OxLys-SNA (yellow circles) or saline (white diamonds). A subset of animals that had been administered OxLys-SNAs (n = 3) were rechallenged by inoculation with ∼106 EMT6 cells in the right inguinal mammary fat pad at day 65 (red arrow), and tumor growth was monitored out to day 100. Population of (D) cytotoxic CD8+T cells and (E) MDSCs isolated from the TME of EMT6-bearing mice at day 11 postinoculation, following treatment on days 6 and 15 with OxLys-SNA (yellow bars), Lys-SNA (black bars), OxLys-Mix (gray bars), or saline (white bars). Statistical analysis was performed using an ordinary one-way ANOVA, where “**” represents a P value of <0.01, “***” represents a P value of <0.001, and “****” represents a P value of <0.0001.
OxLys-SNAs Alter the Immune Cell Population within the TME.
To determine the effect of OxLys-SNA administration on the immune cell population at the tumor site, BALB/c mice were inoculated with EMT6 cells in the left inguinal mammary fat pad. At days 6 and 10 postinoculation, animals were administered OxLys-SNA, Lys-SNA, OxLys-Mix, or saline (n = 3 per group) via peritumoral s.c. injection. On day 11, animals were killed. Tumors were dissociated into single-cell suspensions and split into two fractions. One cell fraction was incubated with antibodies against CD45, CD3, and CD8 to identify CD8+ (cytotoxic) T cells present in the TME. The second cell fraction was incubated with CD45, CD11b, and Gr1 to identify myeloid-derived suppressor cells (MDSCs) present in the TME. Following antibody incubation, cells were fixed and analyzed via flow cytometry. Excitingly, the population of CD8+ T cells at the tumor site was significantly elevated in animals administered OxLys-SNAs (Fig. 5D) relative to all other treatment groups, showing a 2.3-fold increase relative to saline-treated controls. Concurrently, the population of MDSCs in animals administered OxLys-SNAs (Fig. 5E) was decreased 2.5-fold relative to saline-treated controls.
Discussion
Encapsulating tumor cell lysates into the core of liposomal SNAs increases the codelivery of lysate and adjuvant DNA to immune cells relative to simple mixtures, both in vitro and in vivo. This highlights the importance of structural arrangement in the design of immunotherapeutics, as the maximal immune response is achieved when both adjuvant and antigen are delivered to the same target cell (66). Indeed, formulation of lysate and CpG-1826 into SNAs leads to higher codelivery of both immunomodulatory components to the same CD11c+ cells in the draining lymph node in vivo at 2 h postinjection, as compared to a simple mixture of linear CpG-1826 and bare liposomes containing lysate. Importantly, the percent of CD11c+ cells staining positive for both Cy5-labeled DNA and fluorophore-labeled lysate is maintained up to 24 h after in vivo delivery with Lys-SNAs, whereas the percentage of double-positive cells increased when treated with the simple mixture, but not to the level of Lys-SNAs.
Furthermore, generating lysates from cells that have been subjected to oxidative stress with HOCl to induce cell death results in protein populations of higher molecular weight than those isolated from cells that have not been subjected to oxidative stress. This finding confirms that cellular oxidation prior to lysate generation changes the available antigen pool, which is in agreement with published studies on the use of oxidized lysates in DC vaccines (20, 68). When incubated with BMDCs in vitro, oxidized lysates enhance DC maturation over both nonoxidized lysates and CpG-1826. This finding suggests that inducing cell death via oxidation leads to lysate populations that have elevated adjuvant behavior as well as antigenic behavior, as DC maturation is primarily dictated by activation with adjuvants (69).
In the EMT6 model of TNBC, OxLys-SNAs show significant antitumor activity, with six out of nine animals experiencing complete tumor remission by day 20. Intriguingly, the first animal administered OxLys-SNA does not succumb to tumor burden (day 32 postinoculation) until 5 d after the last saline-treated animal (day 27 postinoculation). Thus, OxLys-SNAs act as potent immunotherapeutics, with superior performance to both Lys-SNAs and OxLys-Mix. Excitingly, animals who received OxLys-SNAs resisted rechallenge with EMT6 cells at 60 d postinoculation, indicating that this treatment regime has the potential to offer protective immunity. Moreover, OxLys-SNAs significantly increase the populations of cytotoxic CD8+ T cells and simultaneously decrease the population of MDSCs in the TME, relative to all other treatment groups. High levels of cytotoxic T cells in the microenvironment of breast tumors have been shown to be correlated with positive antitumor effects (70, 71), while high levels of MDSCs promote immune evasion (72). Therefore, this finding provides insight on the mechanism responsible for the observed antitumor efficacy of OxLys-SNAs.
This work is important because it describes a class of potent vaccines based upon the compartmentalization of antigens in the form of oxidized lysate within a nanotherapeutic. These structures, in three TNBC tumor models, show very promising activity with respect to codelivery of lysate and adjuvant and DNA, antitumor efficacy, extended animal survival, and alteration of the immune cell population within the TME. Importantly, this is only one way to compartmentalize lysates in SNA-type architectures, and, when one considers recent work that shows single antigen presentation on the outside of SNAs can increase vaccine performance (73), there is a path forward for identifying new structures with vaccine performance superior to those presented herein. Indeed, if one could present the lysate components on the outside of such structures, even better responses may be possible. Moreover, owing to the modularity and programmability of the SNA architecture, the rapid development of translationally relevant systems may be possible through the incorporation of human TNBC lysates and human-specific CpG adjuvants, such as CpG-7909 (40). Taken together, these results show that the method in which tumor cell lysates are generated, as well as the way adjuvant and antigen(s) are packaged and delivered to the immune system, has a profound impact on the resulting antitumor efficacy. Therefore, these results have important implications in the development of personalized, translationally relevant immunotherapies for TNBC and other cancers.
Materials and Methods
Generation and Characterization of Cell Lysates.
All animal protocols were approved by the Institutional Animal Care and Usage Committee at Northwestern University. EMT6 cell lines were obtained from ATCC and grown in Minimum Essential Media supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin−streptomycin. Py230 and Py8119 were grown in F-12K Medium (ATCC), 5% FBS, 0.1% MITO+ serum extender (Corning), 2.5 μg/mL amphotericin B (Gibco), and 50 μg/mL gentamycin (Gibco). All lysates were prepared from cells under passage number six. For lysate preparation, cells were trypsinized, washed, collected, and resuspended at 106 cells per milliliter in Dulbecco’s phosphate-buffered saline (DPBS), then subjected to five freeze−thaw cycles in liquid nitrogen and a 37 °C water bath. Cellular debris was removed by centrifugation at 10,000 relative centrifugal force (RCF) units for 10 min, and the supernatant was then collected as the protein lysate. Total protein concentration was measured using the bicinchoninic acid (BCA) assay with albumin as the protein standard (Pierce, ThermoFisher Scientific). Protein content was characterized using 4 to 12% sodium dodecyl sulfate (SDS)/ polyacrylamide gel electrophoresis at 100 V for 1 h and loading 20 µg of total protein.
To prepare oxidized lysates, EMT6 cells were grown to confluence in a Petri dish. Cells were washed with DPBS (3×) and then incubated with 60 μM HOCl in DPBS for 1 h at 37 °C. Following the incubation, cells were collected and washed with DPBS to remove any unreacted HOCl. Following centrifugation at 500 RCF for 5 min, cells were resuspended in DPBS at a density of 1 × 107 cells per mL. Cells were subjected to five freeze−thaw cycles using liquid nitrogen and a 37 °C water bath, followed by centrifugation for 10 min at 10,000 RCF. The soluble fraction was collected as the oxidized lysate.
Purified cell lysates were fluorophore-labeled for in vitro and in vivo uptake experiments. One milligram each of Oregon Green 488-NHS (Thermo Fisher) and FITC-maleimide (Thermo Fisher) dye were incubated with 1 mg/mL lysate in PBS (pH 7.5) for 16 h at 4 °C. The unreacted dye was removed by washing the lysate with 10 mL of PBS 10 times using 50 kDa cutoff centrifugation filters (4,000 × g, 10 min). Fluorophore-labeled lysate was subsequently used to make lysate-SNAs.
DNA Synthesis.
Cholesteryl-modified CpG-1826 (5′-TCC ATG ACG TTC CTG ACG TT (Sp18)2 Chol-3′) and Cholesteryl-Cy5-modified CpG-1826 (5′-TCC ATG ACG TTC CTG ACG TT -Cy5-(Sp18)2 Chol-3′) were synthesized with phosphorothioate backbones via automated solid-phase DNA synthesis using an MerMade 12 Synthesizer (Bioautomation), using 4,5-dicyanoimidazole as an activator and 3-((Dimethylamino-methylidene)amino)-3H-1,2,4-dithiazole-3-thione as the sulfurizing agent. Following synthesis, DNA strands were cleaved from solid support via overnight incubation with 30% ammonium hydroxide at room temperature (RT). Excess ammonia was removed through evaporation under nitrogen, and oligonucleotides were purified using an Agilent high-pressure liquid chromatography system using a C4 or C18 column, using a gradient of triethylammonium acetate and acetonitrile (10 to 100% acetonitrile) over 30 min. Purified oligonucleotides were collected and lyophilized. Powdered oligonucleotides were reconstituted in 5 mL of acetic acid and incubated at RT for 1 h, then extracted with ethyl acetate (7 mL, 3×). The purified, deprotected DNA was then lyophilized, resuspended in 1 mL of deionized water, and analyzed by matrix-assisted laser desorption ionization time-of-flight and native gel electrophoresis.
Synthesis of Lysate-Loaded SNAs.
Tumor cell lysate (either oxidized or not) was encapsulated within DOPC liposomes using the thin-film rehydration method (55). Solutions were adjusted to 1 mg/mL (with respect to protein concentration) in PBS, which was used to rehydrate 5 mg of DOPC for 1 h at RT. After the rehydration period, liposomes were formed through five freeze−thaw cycles, using liquid nitrogen and sonication in a 37 °C water bath. Liposomes were then diluted with PBS such that the highest concentration of lipid was no greater than 2 mg/mL lipid for extrusion, as measured by the commercially available phosphotidylcholine (PC) assay (Sigma). Liposome size was controlled through sequential high-pressure extrusion using polycarbonate filters (T&T Scientific) with pore sizes of 200, 100, 80, and 50 nm. Liposomes were passed through each filter size 10 times. Following the final extrusion, tangential flow filtration with a pore size of 500 kDa (Spectrum) was used to remove any nonencapsulated proteins, and the sample was repeatedly washed with PBS until no protein was detected in the flow-through, as monitored by measuring the absorption of the flow-through at 280 nm with ultraviolet visible (UV-vis) spectroscopy (Cary) and BCA assay. The amount of protein encapsulated within the liposomes was measured using the BCA assay after disruption of the liposome with 1% SDS to release encapsulated protein. The phospholipid concentration was measured using a commercially available PC assay kit.
To form SNAs, cholesterol-terminated oligonucleotides (3′) were embedded into the outer membrane of the liposomes by mixing 20 µM oligonucleotides with a solution of liposomes at 1.63 mM lipid at 25 °C overnight. The oligonucleotide concentration was determined by measuring the absorption at 260 nm with UV-vis. The resulting SNAs (both OxLys-SNAs and Lys-SNAs) were then concentrated to 20 µM by DNA using centrifugation filter units (Millipore), which also removed any unbound DNA. The resulting structures were analyzed by zeta potential (Malvern Zetasizer), gel electrophoresis, and DLS (Fig. 1).
Characterization of Lysate-Loaded SNAs.
Lysate-loaded SNAs were characterized using cryo-TEM, gel electrophoresis, DLS (Fig. 1D), and zeta potential (SI Appendix, Table S1). Cryo-EM samples were prepared by FEI Vitrobot Mark III by dropping 4 μL on a 200-mesh copper TEM grid with lacey carbon film, blotted for 5 s and then plunged into liquid ethane before transfer to a cryo-holder and storage in liquid nitrogen prior to imaging. Cryo-EM imaging was performed using a Hitachi HT7700 transmission electron microscope with a Gatan cryo-transfer holder under 120-kV accelerating voltage, and images were taken with a Gatan imaging camera at 30,000× magnification. Confirmation of DNA loading was done using gel electrophoresis, DLS, and zeta potential measurements. Cy5-labeled, cholesteryl-modified oligonucleotides, Cy5-labeled Lys-SNAs, and Cy5-labeled Lys-SNAs that had been incubated with Triton-X to dissociate liposomes (50 pmol each) were loaded into a 1% agarose gel on ice and run at 100 V for 45 min (Fig. 1C).
Uptake of Lys-SNAs and Lys-Mix by BMDCs In Vitro.
Bone marrow was isolated from femurs of Balb/C or C57BL/6 mice and cultured in Roswell Park Memorial Institute (RPMI)-1640 media, supplemented with 10% heat-inactivated FBS, 1% penicillin−streptomycin, and 20 ng/mL granulocyte-macrophage colony-stimulating factor. Media was replenished on day 3, and cells were harvested on day 6. SNAs were prepared by encapsulating AlexaFluor488-labeled lysate within the liposomal core and functionalizing Cy5-labeled DNA using the same methods as described above. Fluorophore-labeled SNAs were used to measure the uptake of particles in BMDCs. BMDCs were added to 24-well plates at 500,000 cells per well, and immediately treated with 1 µM Cy5-labeled DNA and AlexaFluor488-labeled lysate, or doubly labeled SNAs, for 1 or 24 h. Cells were washed, fixed with 4% paraformaldehyde for 10 min at RT, and either resuspended in DPBS for flow cytometry (BD LSRFortessa) or stained with 4′,6-diamidino-2-phenylindole and imaged with confocal microscopy (Zeiss LSM 800) using a 63× objective.
Uptake of Lys-SNAs and Lys-Mix by Lymphoid Cells In Vivo.
Female C57BL/6 mice (n = 3, 8 wk to 10 wk old) were injected s.c. (flank) with 200 µL of 50 µM fluorophore-labeled Lys-SNAs or a mixture of CpG-1826 and lysate (Lys-Mix). Mice were killed 2 h or 24 h postinjection, and draining lymph nodes were excised. Lymph nodes were dissociated into single cells using a cell strainer. Single-cell suspensions were stained with antibodies for CD11c (PE-Cy7) as well as a live/dead stain, and analyzed using flow cytometry.
Lys-SNA Antitumor Efficacy.
Female mice (age 8 wk to 10 wk) were inoculated with 1 × 106 TNBC cells (C57BL/6 mice for Py230 and Py8119; Balb/C mice for EMT6) via s.c. injection into the right inguinal mammary fat pad. On days 6, 10, and 15, animals were administered Lys-SNA, Lys-Mix, or saline (n = 5 per group) via peritumoral injection (50 µM, 200 µL). Tumor volumes were calculated by measuring the length and width with calipers and applying the formula V = L × W × W/2. Studies were stopped and animals killed when tumor burden of saline-treated animals exceeded 1,200 mm3.
OxLys-SNA Antitumor Efficacy.
Female Balb/C mice (age 8 wk to 10 wk) were inoculated with 1 × 106 EMT6 cells via s.c. injection into the right inguinal mammary fat pad. On days 6, 10, and 15, animals were administered OxLys-SNA, OxLys-Mix, Lys-SNA, or saline (n = 9 per group) via peritumoral at a dose of 5 nmol DNA and 20 µg of protein. Tumor volumes were calculated by measuring the length and width with calipers and applying the formula V = L × W × W/2. Animal survival was monitored up to 100 d, and animals were killed when tumor burden exceeded 1,500 mm3. At day 60 postinoculation, a subset of surviving OxLys-SNA animals (n = 3) was rechallenged by inoculation with ∼106 EMT6 cells in the right inguinal mammary fat pad, and monitored for evidence of tumor growth for an additional 40 d.
BMDC Activation.
BMDCs were isolated and cultured as described above. On day 6, BMDCs were harvested and incubated (100,000 BMDCs/sample) with CpG-1826 (0.1 nmol) plus oxidized lysates, nonoxidized lysates, or saline (1 µg of total protein) to induce BMDC maturation. After 48 h incubation, cells were washed with DPBS (3×) and stained with antibodies against CD40, CD80, CD86, and MHC-II, as well as with a live/dead stain, by incubation with the appropriate antibodies for 20 min at RT. Cells were then washed with DPBS (3×) and fixed with paraformaldehyde prior to analysis using flow cytometry. Immune cells were identified by gating for CD11b+/CD11c+ double-positive cells, followed by gating for the appropriate marker (CD40, CD80, CD86, or MHC-II).
Analysis of Immune Cell Populations at EMT6 Tumor Site.
Female Balb/C mice (age 8 wk to 10 wk) were inoculated with ∼106 EMT6 cells via injection into the right inguinal mammary fat pad. At days 6 and 10 postinoculation, animals (n = 3 per group) were administered OxLys-SNA, OxLys-Mix, Lys-SNA, or saline via peritumoral injection. On day 11, animals were killed, and tumors were harvested for immune cell population analysis. Tumors were washed with DPBS and dissociated into single-cell suspensions using a cell strainer. Tumor cells were then split into two samples. One sample was stained with antibodies against CD45, CD3, and CD8 to identify CD8+ cytotoxic T cells. The second sample was incubated with CD45, CD11b, and Gr1 to identify MDSCs. After incubation for 20 min at RT, cells were washed with DPBS (3×) and fixed with paraformaldehyde prior to analysis via flow cytometry. CD8+ T cells were identified by first gating for CD45+ cells, followed by gating for CD3+/CD8+ double-positive cells (SI Appendix, Fig. S5). MDSCs were identified by first gating for CD45+ cells, followed by gating for CDllb+/Gr1+ double-positive cells (SI Appendix, Fig. S6).
Data Availability Statement.
All data are available in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the NIH under Awards U54CA199091 and R01CA208783. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. It was also supported by the Prostate Cancer Foundation and the Movember Foundation under Award 17CHAL08, the Lefkofsky Family Foundation, and the Vannevar Bush Faculty Fellowship program sponsored by the Basic Research Office of the Assistant Secretary of Defense for Research and Engineering and funded by the Office of Naval Research through Grant N00014-15-1-0043. C.E.C. acknowledges support from the Eden and Steven Romick Post-Doctoral Fellowship through the American Committee for the Weizmann Institute of Science. L.E.C. acknowledges support from Northwestern University’s Cancer Nanotechnology Training Program Award T32CA186897. Z.H. was supported, in part, by the Northwestern University Graduate School Cluster in Biotechnology, Systems, and Synthetic Biology, which is affiliated with the Biotechnology Training Program. D.H. was supported by the Lynn Sage Foundation as Lynn Sage Scholar. The cryo-TEM experiment made use of the BioCryo facility of Northwestern University’s NUANCE Center, which has received support from the SHyNE Resource (NSF ECCS-1542205), the International Institute for Nanotechnology, and Northwestern’s Materials Research Science and Engineering Center program (NSF DMR-1720139).
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005794117/-/DCSupplemental.
References
- 1.Ali S. A. et al., T cells expressing an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of multiple myeloma. Blood 128, 1688–1700 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maude S. L. et al., Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neelapu S. S. et al., Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 377, 2531–2544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park J. H. et al., Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med. 378, 449–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuster S. J. et al., Chimeric antigen receptor T cells in refractory B-cell Lymphomas. N. Engl. J. Med. 377, 2545–2554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timmerman J. M., Levy R., Dendritic cell vaccines for cancer immunotherapy. Annu. Rev. Med. 50, 507–529 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Brown C. E., Mackall C. L., CAR T cell therapy: Inroads to response and resistance. Nat. Rev. Immunol. 19, 73–74 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Kuai R., Ochyl L. J., Bahjat K. S., Schwendeman A., Moon J. J., Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat. Mater. 16, 489–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang J. et al., A multi-antigen vaccine in combination with an immunotoxin targeting tumor-associated fibroblast for treating murine melanoma. Mol. Ther. Oncolytics 3, 16007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slingluff C. L., Jr., The present and future of peptide vaccines for cancer: Single or multiple, long or short, alone or in combination? Cancer J. 17, 343–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balkwill F. R., Capasso M., Hagemann T., The tumor microenvironment at a glance. J. Cell Sci. 125, 5591–5596 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Kawahara M., Takaku H., A tumor lysate is an effective vaccine antigen for the stimulation of CD4+ T-cell function and subsequent induction of antitumor immunity mediated by CD8+ T cells. Cancer Biol. Ther. 16, 1616–1625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González F. E. et al., Tumor cell lysates as immunogenic sources for cancer vaccine design. Hum. Vaccin. Immunother. 10, 3261–3269 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moyer J. S., Maine G., Mulé J. J., Early vaccination with tumor-lysate-pulsed dendritic cells after allogeneic bone marrow transplantation has antitumor effects. Biol. Blood Marrow Transplant. 12, 1010–1019 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Win S. J. et al., Secretomes from metastatic breast cancer cells, enriched for a prognostically unfavorable LCN2 Axis, induce anti-inflammatory MSC actions and a tumor-supportive premetastatic lung. Br. J. Cancer 106, 92–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields R. C., Shimizu K., Mulé J. J., Murine dendritic cells pulsed with whole tumor lysates mediate potent antitumor immune responses in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 95, 9482–9487 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J., Mooney D. J., In vivo modulation of dendritic cells by engineered materials: Towards new cancer vaccines. Nano Today 6, 466–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang C. L., Coukos G., Kandalaft L. E., Whole tumor antigen vaccines: Where are we? Vaccines (Basel) 3, 344–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang C. L.-L., Benencia F., Coukos G., Whole tumor antigen vaccines. Semin. Immunol. 22, 132–143 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant M. L. et al., Combining dendritic cells and B cells for presentation of oxidised tumour antigens to CD8+ T cells. Clin. Transl. Immunology 6, e149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang C. L. et al., A dendritic cell vaccine pulsed with autologous hypochlorous acid-oxidized ovarian cancer lysate primes effective broad antitumor immunity: From bench to bedside. Clin. Cancer Res. 19, 4801–4815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcinkiewicz J., Neutrophil chloramines: Missing links between innate and acquired immunity. Immunol. Today 18, 577–580 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Marcinkiewicz J., Chain B. M., Olszowska E., Olszowski S., Zgliczyński J. M., Enhancement of immunogenic properties of ovalbumin as a result of its chlorination. Int. J. Biochem. 23, 1393–1395 (1991). [DOI] [PubMed] [Google Scholar]
- 24.Allison M. E. D., Fearon D. T., Enhanced immunogenicity of aldehyde-bearing antigens: A possible link between innate and adaptive immunity. Eur. J. Immunol. 30, 2881–2887 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Coulie P. G., Van den Eynde B. J., van der Bruggen P., Boon T., Tumour antigens recognized by T lymphocytes: At the core of cancer immunotherapy. Nat. Rev. Cancer 14, 135–146 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Frey S., Castro A., Arsiwala A., Kane R. S., Bionanotechnology for vaccine design. Curr. Opin. Biotechnol. 52, 80–88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irvine D. J., Hanson M. C., Rakhra K., Tokatlian T., Synthetic nanoparticles for vaccines and immunotherapy. Chem. Rev. 115, 11109–11146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemp J. A., Shim M. S., Heo C. Y., Kwon Y. J., “Combo” nanomedicine: Co-delivery of multi-modal therapeutics for efficient, targeted, and safe cancer therapy. Adv. Drug Deliv. Rev. 98, 3–18 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Mirkin C. A., Letsinger R. L., Mucic R. C., Storhoff J. J., A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Cutler J. I., Auyeung E., Mirkin C. A., Spherical nucleic acids. J. Am. Chem. Soc. 134, 1376–1391 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Seferos D. S., Prigodich A. E., Giljohann D. A., Patel P. C., Mirkin C. A., Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett. 9, 308–311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lytton-Jean A. K. R. et al., Highly cooperative behavior of peptide nucleic acid-linked DNA-modified gold-nanoparticle and comb-polymer aggregates. Adv. Mater. 21, 706–709 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J.-S., Lytton-Jean A. K. R., Hurst S. J., Mirkin C. A., Silver nanoparticle-oligonucleotide conjugates based on DNA with triple cyclic disulfide moieties. Nano Lett. 7, 2112–2115 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell G. P., Mirkin C. A., Letsinger R. L., Programmed assembly of DNA functionalized quantum dots. J. Am. Chem. Soc. 121, 8122–8123 (1999). [Google Scholar]
- 35.Cutler J. I., Zheng D., Xu X., Giljohann D. A., Mirkin C. A., Polyvalent oligonucleotide iron oxide nanoparticle “click” conjugates. Nano Lett. 10, 1477–1480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young K. L. et al., Hollow spherical nucleic acids for intracellular gene regulation based upon biocompatible silica shells. Nano Lett. 12, 3867–3871 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banga R. J., Chernyak N., Narayan S. P., Nguyen S. T., Mirkin C. A., Liposomal spherical nucleic acids. J. Am. Chem. Soc. 136, 9866–9869 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sprangers A. J., Hao L., Banga R. J., Mirkin C. A., Liposomal spherical nucleic acids for regulating long noncoding RNAs in the nucleus. Small 13, 1602753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meckes B., Banga R. J., Nguyen S. T., Mirkin C. A., Enhancing the stability and immunomodulatory activity of liposomal spherical nucleic acids through lipid-tail DNA modifications. Small 14, 1702909 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radovic-Moreno A. F. et al., Immunomodulatory spherical nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 112, 3892–3897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skakuj K. et al., Conjugation chemistry-dependent T-cell activation with spherical nucleic acids. J. Am. Chem. Soc. 140, 1227–1230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z., Zhang Y., Fullhart P., Mirkin C. A., Reversible and chemically programmable micelle assembly with DNA block-copolymer amphiphiles. Nano Lett. 4, 1055–1058 (2004). [Google Scholar]
- 43.Calabrese C. M. et al., Biocompatible infinite-coordination-polymer nanoparticle-nucleic-acid conjugates for antisense gene regulation. Angew. Chem. Int. Ed. Engl. 54, 476–480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu S., Xing H., Gordiichuk P., Park J., Mirkin C. A., PLGA spherical nucleic acids. Adv. Mater. 30, e1707113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brodin J. D., Sprangers A. J., McMillan J. R., Mirkin C. A., DNA-mediated cellular delivery of functional enzymes. J. Am. Chem. Soc. 137, 14838–14841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bulbake U., Doppalapudi S., Kommineni N., Khan W., Liposomal formulations in clinical use: An updated review. Pharmaceutics 9, 12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guan C. et al., RNA-based immunostimulatory liposomal spherical nucleic acids as potent TLR7/8 modulators. Small 14, e1803284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Z. N., Cole L. E., Callmann C. E., Wang S., Mirkin C. A., Sequence multiplicity within spherical nucleic acids. ACS Nano 14, 1084–1092 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vollmer J. et al., Characterization of three CpG oligodeoxynucleotide classes with distinct immunostimulatory activities. Eur. J. Immunol. 34, 251–262 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Kumagai Y., Takeuchi O., Akira S., TLR9 as a key receptor for the recognition of DNA. Adv. Drug Deliv. Rev. 60, 795–804 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Gross B. P., Wongrakpanich A., Francis M. B., Salem A. K., Norian L. A., A therapeutic microparticle-based tumor lysate vaccine reduces spontaneous metastases in murine breast cancer. AAPS J. 16, 1194–1203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z., Qiu Y., Lu W., Jiang Y., Wang J., Immunotherapeutic interventions of triple negative breast cancer. J. Transl. Med. 16, 147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stagg J., Allard B., Immunotherapeutic approaches in triple-negative breast cancer: Latest research and clinical prospects. Ther. Adv. Med. Oncol. 5, 169–181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders C., Carey L. A., Understanding and treating triple-negative breast cancer. Oncology (Williston Park) 22, 1233–1239. Discussion in: Oncology (Williston Park)22, 1239–1240, 1243 (2008). [PMC free article] [PubMed] [Google Scholar]
- 55.André F., Zielinski C. C., Optimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agents. Ann. Oncol. 23 (suppl. 6), vi46-vi51 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Anders C. K., Carey L. A., Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin. Breast Cancer 9 (suppl. 2), S73–S81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fedele M., Cerchia L., Chiappetta G., The epithelial-to-mesenchymal transition in breast cancer: Focus on basal-like carcinomas. Cancers (Basel) 9, 134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su Y. et al., Epigenetic reprogramming of epithelial mesenchymal transition in triple negative breast cancer cells with DNA methyltransferase and histone deacetylase inhibitors. J. Exp. Clin. Cancer Res. 37, 314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meade K. J. et al., Secretomes from metastatic breast cancer cells, enriched for a prognostically unfavorable LCN2 axis, induce anti-inflammatory MSC actions and a tumor-supportive premetastatic lung. Oncotarget 10, 3027–3039 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ellies L. G., Kuo C., PyVmT luminal and EMT cell lines exhibit characteristic patterns of orthotopic and tail vein metastasis. Cancer Res. 73, C91 (2013). [Google Scholar]
- 61.Biswas T., Yang J., Zhao L., Sun L., Abstract P2-04-07: Modeling orthotopic and metastatic progression of mammary tumors to evaluate the efficacy of TGF-β inhibitors in a pre-clinical setting. Cancer Res. 72, P2-4-7-P02-4–7 (2012). [Google Scholar]
- 62.Christenson J. L. et al., MMTV-PyMT and derived met-1 mouse mammary tumor cells as models for studying the role of the androgen receptor in triple-negative breast cancer progression. Horm. Cancer 8, 69–77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ouzounova M. et al., Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat. Commun. 8, 14979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barnes S., “EMT-6 syngeneic breast tumor model—A powerful tool for immuno-oncology studies” (Covance, 2018).
- 65.Liu L. N., Shivakumar R., Allen C., Fratantoni J. C., Delivery of whole tumor lysate into dendritic cells for cancer vaccination. Methods Mol. Biol. 423, 139–153 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Krishnamachari Y., Salem A. K., Innovative strategies for co-delivering antigens and CpG oligonucleotides. Adv. Drug Deliv. Rev. 61, 205–217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prokopowicz Z. M. et al., Hypochlorous acid: A natural adjuvant that facilitates antigen processing, cross-priming, and the induction of adaptive immunity. J. Immunol. 184, 824–835 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Chiang C. L. et al., A dendritic cell vaccine pulsed with autologous hypochlorous acid-oxidized ovarian cancer lysate primes effective broad antitumor immunity: From bench to bedside. Clin. Cancer Res. 19, 4801–4815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ho N. I., Huis In ’t Veld L. G. M., Raaijmakers T. K., Adema G. J., Adjuvants enhancing cross-presentation by dendritic cells: The key to more effective vaccines? Front. Immunol. 9, 2874 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maimela N. R., Liu S., Zhang Y., Fates of CD8+ T cells in tumor microenvironment. Comput. Struct. Biotechnol. J. 17, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim P. S., Ahmed R., Features of responding T cells in cancer and chronic infection. Curr. Opin. Immunol. 22, 223–230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lesokhin A. M., Merghoub T., Wolchok J. D., Myeloid-derived suppressor sells and the efficacy of CD8+ T-cell immunotherapy. OncoImmunology 2, e22764 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang S. et al., Rational vaccinology with spherical nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 116, 10473–10481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript and SI Appendix.