Significance
Orexin receptors belong to the superfamily of G-protein–coupled receptors (GPCRs) which represent the largest class of drug targets in humans. Despite the recent progress in structural biology, the development of subtype-selective orexin receptor and GPCR ligands in general remains challenging, due to the high sequence similarity among individual receptor subtypes. However, subtype-selective molecules are key to discerning the individual contributions of receptor subtypes to (patho-)physiology. Starting from the clinically used, non–subtype-selective orexin receptor antagonist suvorexant, we demonstrate how docking, crystallography, medicinal chemistry, and in vitro pharmacology can be combined to exploit single amino acid sequence differences for the discovery of subtype-selective probes. Such compounds will help to better understand orexin receptor pharmacology and develop promising drug candidates.
Keywords: GPCR, orexin receptor, drug design, subtype selectivity, crystal structure
Abstract
Orexins are neuropeptides that activate the rhodopsin-like G protein-coupled receptors OX1R and OX2R. The orexin system plays an important role in the regulation of the sleep-wake cycle and the regulation of feeding and emotions. The nonselective orexin receptor antagonist suvorexant has been the first drug on the market targeting the orexin system and is prescribed for the treatment of insomnia. Subtype-selective OX1R antagonists are valuable tools to further investigate the functions and physiological role of the OX1R in vivo and promising lead compounds for the treatment of drug addiction, anxiety, pain or obesity. Starting from the OX1R and OX2R crystal structures bound to suvorexant, we exploited a single amino acid difference in the orthosteric binding site by using molecular docking and structure-based drug design to optimize ligand interactions with the OX1R while introducing repulsive interactions with the OX2R. A newly established enantiospecific synthesis provided ligands showing up to 75-fold selectivity for the OX1R over the OX2R subtype. The structure of a new OX1R antagonist with subnanomolar affinity (JH112) was determined by crystallography in complex with the OX1R and corresponded closely to the docking-predicted geometry. JH112 exhibits high selectivity over a panel of different GPCRs, is able to cross the blood–brain barrier and acts as slowly diffusing and insurmountable antagonist for Gq protein activation and in particular β-arrestin-2 recruitment at OX1R. This study demonstrates the potential of structure-based drug design to develop more subtype-selective GPCR ligands with potentially reduced side effects and provides an attractive probe molecule and lead compound.
While G-protein–coupled receptors (GPCRs) are outstanding therapeutic targets (1), many GPCR drugs lack selectivity (2). An example is suvorexant, a Food and Drug Administration-approved drug targeting the orexin system. The orexin system consists of two rhodopsin-like GPCRs, named orexin-1 (OX1R) and orexin-2 receptor (OX2R), and their endogenous peptide ligands orexin A and orexin B (3), which are also known as hypocretin-1 and -2 (4). Both orexin neuropeptides are exclusively expressed in hypothalamic neurons, which, however, project widely in the brain and are involved in many different regulation mechanisms in the central nervous system (5). Since OX1R is selectively activated in cholinergic and noradrenergic systems as well as in the amygdala and in the ventromedial hypothalamic nucleus, it is, among other functions, responsible for the regulation of emotions, pain, feeding, and addiction (5–7). In contrast, the OX2R is predominantly activated in neurons in the paraventricular nucleus and in histaminergic neurons (e.g., in the tuberomammillary nucleus), which are responsible for the regulation of the sleep–wake cycle and arousal (5, 6, 8–11). Suvorexant and the very recently approved lemborexant (12, 13) are nonselective orexin receptor antagonists prescribed for the treatment of insomnia. A few subtype-selective OX2R antagonists are currently in clinical development, e.g., seltorexant, which has been developed for the treatment of insomnia and major depressive disorders and is currently being investigated in phase II clinical trials (14). The pharmaceutical potential of selective OX1R antagonists for treatment of obesity (15), anxiety (16), and drug addiction (17–19) has been shown in several in vivo experiments.
Structure-based drug design enabled the development of new probes with improved selectivity (20, 21) and activities (22) due to the multitude of novel high-resolution structures. The high-resolution crystal structures of suvorexant bound to both OX1R (Protein Data Bank [PDB] number: 4ZJ8) (23) and OX2R (PDB number: 4S0V) (24) provide detailed insight into the highly conserved orthosteric binding sites of the two receptor subtypes and have been the basis for our work. Although still challenging, a structural understanding of the binding pocket enables a focus on the few differences that exist between the two subtypes. We aimed to develop selective OX1R antagonists exploiting a single Ala/Thr difference in the orthosteric binding pockets of the two receptors. Starting with the crystal structures of the OX1R and OX2R bound to the nonselective antagonist suvorexant, we have used a cycle of structure-based design, synthesis, binding assays, and crystallography to find compounds based on the structure of suvorexant with subnanomolar OX1R affinity and up to 75-fold selectivity over the OX2R subtype.
Results
Structure-Based Design.
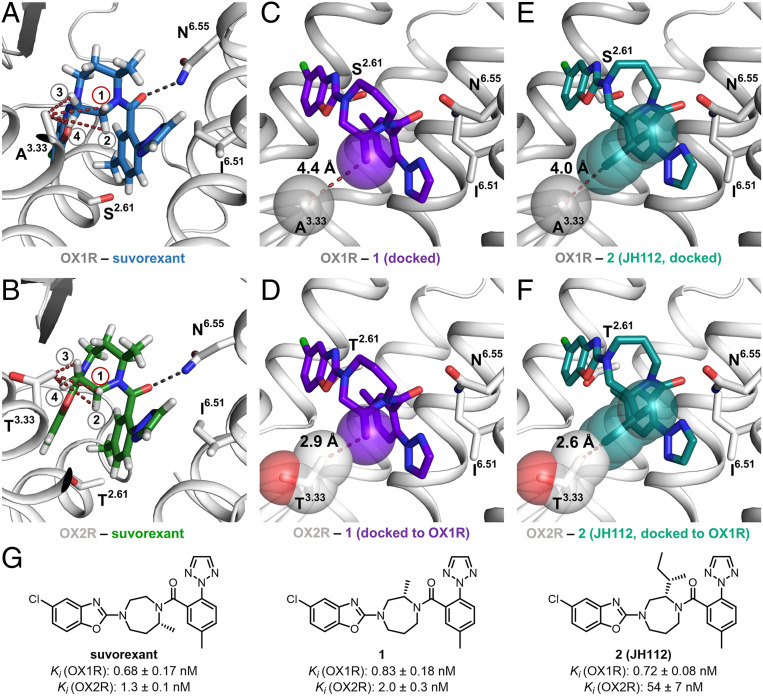
Suvorexant adopts a horseshoe-like conformation in the binding pockets of both orexin receptors, putatively leading to face-to-face interaction between the two aromatic ring systems (Fig. 1 A and B) (25). From a drug-optimization perspective, it is interesting that the interactions between the receptor and the ligands are predominantly hydrophobic. Very recently, solving the crystal structures of thermostabilized OX1R and OX2R in complex with various ligands, a study by Rappas et al. revealed that high-affinity binding to orexin receptors is dependent on hydrophobic interactions and, ideally, replacement of high-energy water molecules within the binding pockets of OX1R and OX2R (26). The only polar interaction between suvorexant and the orexin receptors is a hydrogen bond between the carbonyl oxygen of the amide bond of suvorexant and the side-chain amide nitrogen of Asn3186.55 of OX1R [numbers in superscript refer to the GPCRdb (Ballesteros-Weinstein) enumeration scheme (27)]. There are only two sequence differences in the orthosteric binding sites within 4 Å of the ligand, conferring that the binding site of the OX2R subtype is ∼30 Å3 smaller than the binding site of OX1R. The residue Ser1032.61 of OX1R is exchanged for Thr1112.61 in the OX2R. In addition, Ala1273.33 of OX1R is replaced by the larger Thr1353.33 for OX2R. Interestingly, the side chain in position 3.33 is located in close proximity to the ethylene bridge of the central homopiperazine ring of suvorexant (Fig. 1 A and B). The homopiperazine moiety links the two terminal heteroarene moieties via a longer propylene and a shorter ethylene bridge, the former of which has a methyl substituent. Because the methyl group is located at the propylene bridge, the substituent does not point toward a nonconserved residue and therefore does not have an influence on subtype selectivity.
Fig. 1.
Comparison of the orthosteric binding sites of OX1R and OX2R and development of compound 2 (JH112). (A and B) The orthosteric binding pocket of OX1R and OX2R with conserved features of ligand recognition and binding affinities of the cocrystallized ligand suvorexant (hydrogens displayed). The only nonconserved residues in the two binding pockets are located in TM2 (OX1R: Ser2.61; OX2R: Thr2.61) and TM3 (OX1R: Ala3.33; OX2R: Thr3.33). (A) Distances between hydrogen 1 to 4 and the carbon of the Ala3.33 side chain: H1: 4.4 Å; H2: 5.3 Å; H3: 3.9 Å; H4: 4.3 Å. (B) Distances between hydrogen 1 to 4 and the terminal carbon of the Thr3.33 side chain: H1: 3.4 Å; H2: 4.5 Å; H3: 3.0 Å; H4: 3.2 Å. (C) Docking pose of compound 1 indicating that a methyl substituent in position H1 of the suvorexant points toward the nonconserved Ala3.33 of the OX1R. Distance between the carbon of the methyl substituent and Cβ of the Ala3.33 side chain: 4.4 Å. (D) Alignment of OX2R to OX1R with docked compound 1 in the binding pocket, indicating a steric clash between the methyl substituent of compound 1 and the nonconserved Thr3.33 of the OX2R. Distance between methyl substituent and Thr3.33: 2.9 Å. (E) Docking pose of compound JH112 indicating that the sec-butyl substituent fits into OX1R’s binding site (minimum distance between the sec-butyl substituent and Ala3.33 side chain: 4.0 Å). Compound JH112 adopts a similar binding conformation as suvorexant. (F) Alignment of OX2R to OX1R with docked compound JH112 in the binding pocket indicating a steric clash between the sec-butyl substituent of JH112 and the nonconserved Thr3.33 of OX2R. Minimum distance between sec-butyl substituent and Thr3.33: 2.6 Å. (G) Chemical structures of the compounds and binding affinity.
Guided by the crystal structures of OX1R and OX2R bound to suvorexant, we intended to relocate the methyl group from the propylene to the ethylene bridge. Having identified the position and stereochemistry, molecular docking guided us to optimize the size and nature of the substituent for OX1R affinity and subtype selectivity over OX2R. Our design aimed to find compounds with a group that points directly to the nonconserved residue in TM3, resulting in optimized ligand interactions with OX1R, while suffering from repulsive interactions with OX2R at the Thr1353.33 residue (Fig. 1 A and B). Determination of the distances between the four hydrogens attached to the ethylene bridge and the terminal carbons of Ala1273.33 or Thr1353.33 indicated that hydrogen 3 has the shortest distance for both subtypes (OX1R: 3.9 Å, OX2R: 3.0 Å). However, replacement of hydrogen 3 by a methyl group will cause repulsive intramolecular interactions with the axially oriented hydrogen at the central CH2 of the propylene bridge resulting in conformational changes of the ligand and reduction of binding affinity for both receptor subtypes. Replacement of hydrogens 2 and 4 by an alkyl substituent was considered ineffectual because these hydrogens are not pointing toward the nonconserved residue. Hydrogen 1 appeared to be very promising because of its proximity (OX1R: 4.4 Å, OX2R: 3.4 Å) and orientation to the nonconserved residue. Thus, we focused on the development of subtype-selective compounds by exchanging hydrogen 1 with alkyl substituents.
According to our molecular docking studies, the methyl-substituted derivative 1 fits well into the OX1R-binding pocket with a distance of 4.4 Å between the substituent and Ala1273.33 (Fig. 1C). Alignment of the OX2R to the docking pose of compound 1 in the OX1R revealed a steric clash between the methyl substituent of the ligand and the nonconserved Thr1353.33 of OX2R (distance 2.9 Å; Fig. 1D), suggesting reduced OX2R-binding affinity. Chemical synthesis of the new ligand 1 was performed using the natural amino acid (S)-alanine as a chiral building block. The reaction sequence included a Michael-type addition of acrylonitrile to the amino group, lactamization, and chemical reduction to give an N-protected homopiperazine, which was functionalized with the two heteraromatic moieties by sequential N-arylation and N-acylation (SI Appendix). A radioligand-binding assay with membranes from transiently transfected HEK 293T cells showed Ki values of compound 1 in the single-digit nanomolar range for both subtypes (OX1R: Ki = 0.83 ± 0.18 nM, OX2R: Ki = 2.0 ± 0.3 nM, mean ± SEM), indicating that the newly installed methyl group was indeed tolerated at the OX1R (Fig. 1G). However, selectivity was low (2.4-fold) (SI Appendix, Table S1), suggesting that a clash was ameliorated in the OX2R by movement of ligand or protein. Molecular docking of candidates with sterically more demanding methyl surrogates (SI Appendix, Figs. S1 and S2) led us to compounds including the (S,S)-sec-butyl substituted analog 2 (JH112). The docking pose of JH112 at the OX1R (Fig. 1E) was similar to the position of suvorexant in the crystal structure. The newly installed substituent in JH112 points toward Ala1273.33, but has a slightly shifted position (distance: 4.0 Å), thus avoiding repulsion. As a consequence, the sec-butyl substituent is highly complementary to its environment (Fig. 1E). Alignment of the OX2R to the docking pose of JH112 in the OX1R clearly predicted repulsive interactions because the minimum heavy-atom distance between the ligand and Thr1353.33 is only 2.6 Å (Fig. 1F). In fact, chiral pool synthesis of enantiomerically pure JH112 from natural isoleucine, which was done following the reaction sequence established for the alanine derivative 1, and radioligand-binding experiments showed subnanomolar binding affinity to the OX1R (Ki = 0.72 ± 0.08 nM), while the affinity for the OX2R was 75-fold lower (Ki = 54 ± 7 nM). To find out whether ligand association or dissociation were affected by our chemical modification, we compared kinetic binding of JH112 with the binding of suvorexant at OX2R (SI Appendix, Fig. S3 and Table S2). Importantly, dissociation of JH112 from OX2R was 17-fold faster (0.5576 ± 0.1835 min−1) than for suvorexant (0.0334 ± 0.0065 min−1) whereas a decrease of the association rate could not be observed. In detail, JH112 showed even slightly faster association (2.543 ± 0.804⋅106⋅min−1⋅M−1 and 0.485 ± 0.062⋅106⋅min−1⋅M−1, for JH112 and suvorexant, respectively). We suggest that a repulsive interaction between the sec-butyl group of JH112 and Thr1353.33 provokes the substantial reduction in dissociation half-life (1.2 and 20.7 min, respectively).
To further evaluate whether the selectivity of JH112 reflects the design for preferential binding to Ala1273.33 and interference with Thr1353.33 of OX1R and OX2R, respectively, we explored the effect of reciprocal residue substitutions in the OX1R and OX2R backgrounds (SI Appendix, Fig. S4). In fact, we observed an increase of affinity for the OX1R selective ligand JH112 at OX2R_T135A (4.9-fold, Ki 11 ± 3 nM), while its affinity dropped 21-fold at the reciprocal construct OX1R_A127T (Ki 15 ± 3 nM). In comparison, the affinity of the dual orexin receptor antagonist suvorexant was slightly reduced for both mutants OX1R_A127T (2.4-fold, Ki 1.6 ± 0.2 nM) and OX2R_T135A (6.2-fold, Ki 8.1 ± 1.8 nM). The mutational analysis supports our structure-based design, although the OX2R_T135A mutation did not improve the affinity of JH112 to the OX1R wild-type level. This indicates additional contributions from other amino acids that may result from a slightly modified binding pose.
Similar to suvorexant, compound JH112 showed an excellent selectivity profile with a 10,000-fold higher affinity for the OX1R compared to 20 aminergic and peptidergic GPCRs (SI Appendix, Fig. S5 and Table S3).
Crystallography.
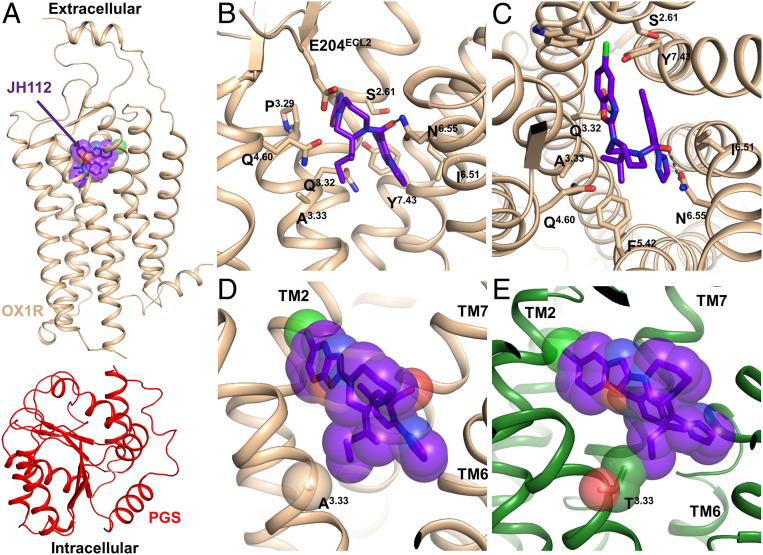
To verify the model on which the design of these ligands has been based, and to provide a template for future drug design, the structure of the OX1R in complex with JH112 was determined by X-ray crystallography. A Pyrococcus abysii glycogen synthase (PGS) fusion domain was introduced in the third intracellular loop (ICL3) of the OX1R in a similar manner as previously carried out for OX1R and OX2R (23, 24). We purified the receptor construct in the presence of JH112, and crystals were grown in lipid cubic phase (28). We obtained a 3.5 Å dataset from eight crystals and solved the structure by molecular replacement (Fig. 2A and SI Appendix, Table S4). Compound JH112 binds to OX1R in the predicted orientation (heavy atom rmsd to docked pose 0.66 Å) with the (S,S)-sec-butyl substituent oriented toward Ala1273.33 (Fig. 2 B and C and SI Appendix, Fig. S6). Interestingly, compound JH112 binds to OX1R in a slightly distinct orientation compared to suvorexant; the whole molecule swings “upward,” which prevents the (S,S)-sec-butyl substituent from clashing with Ala1273.33. The triazole moiety of JH112 rotates counterclockwise as it would otherwise intramolecularly clash with the (S,S)-sec-butyl substituent. Fig. 2 D and E shows that compound JH112 fits well into the OX1R’s binding site, while it would clash with Thr1353.33 of OX2R.
Fig. 2.
Crystal structure of OX1R in complex with the selective antagonist JH112. (A) Overall structure of the OX1R-PGS-JH112 complex. (B and C) Binding pocket of OX1R interacting with JH112. (D and E) Interaction of JH112 with the nonconserved position in TM3 of OX1R and OX2R. (D) The crystal structure shows the (S,S)-sec-butyl substituent of JH112 pointing directly to the nonconserved Ala3.33 in OX1R without leading to a steric clash between the ligand and the receptor. (E) Superimposition of the OX2R structure on the OX1R-JH112 structure indicates repulsive interactions between Thr3.33 of the OX2R and the (S,S)-sec-butyl substituent of JH112.
Molecular Dynamics Simulations.
Molecular dynamics simulations of suvorexant and JH112 in the OX1R and OX2R were carried out to investigate the atomistic reasons for the OX1R-selectivity of JH112. In all these simulations, the conformations of the ligands show little fluctuation below 1.5 Å RMSD, and the two residues in the immediate vicinity of the bound ligand that differ between OX1R and OX2R (Ser103/Thr1112.61 and Ala127/Thr1353.33) are mostly stable (SI Appendix, Fig. S7). Ser103/Thr1112.61 fluctuate more because they do not directly interact with the ligand. Interestingly, amino acid Asn3186.55, the residue entertaining the only direct hydrogen bond interaction with the ligands, shows a less stable interaction in the OX2R-JH112 complex (34 ± 20% SD of all frames) compared to the OX1R-JH112 complex (51 ± 7% SD) (SI Appendix, Fig. S8). For suvorexant, no substantial differences were observed for this interaction between the two receptor subtypes (17 ± 8% SD and 21 ± 13% SD for OX1R and OX2R, respectively) (SI Appendix, Fig. S8). Interestingly, the numbers for the suvorexant complex are considerably lower, indicating that hydrophobic interactions and possibly entropic contributions play a crucial role, too. Additional simulations of suvorexant and JH112 in water (without the receptors) showed that JH112 adopts two different equally stable conformations (two clusters containing 53.7 and 44.7% of all conformations of JH112 of five clusters with an equal average distance between clusters). In contrast, suvorexant’s second conformation is less frequent (the two most frequent clusters contain 85 and 9.5%, respectively, of all conformations) (SI Appendix, Figs. S9–S11). Comparing the conformations in solution with their respective conformations in the complexes shows that JH112 needs to limit itself to one of the conformations in the process of binding to the receptor (SI Appendix, Fig. S10), whereas suvorexant’s most populated conformation in solution is already very close to its bound conformation (SI Appendix, Fig. S11). Given that the conformations of JH112 and suvorexant in the OX1R and OX2R are similar, one can hypothesize that during binding JH112 loses more degrees of freedom than suvorexant, but can compensate for this in the OX1R by forming a more frequent hydrogen bond interaction. The fact that the association rate is very similar between JH112 and suvorexant, as shown by the kinetic measurements, points to a rather small energetic barrier between the two conformations of JH112, as the conversion does not seem to be the rate-limiting step.
Fractional native contact analysis of JH112 provides a descriptor of the number of interactions in the binding pocket. The average percentage of native contacts along the simulation trajectory is 28 ± 3% SD in the OX1R. In contrast, suvorexant in the OX1R forms these contacts much less frequently with 8 ± 8% SD on average (SI Appendix, Fig. S12). This indicates that JH112 occupies the binding pocket in a more stable fashion.
Functional Investigation.
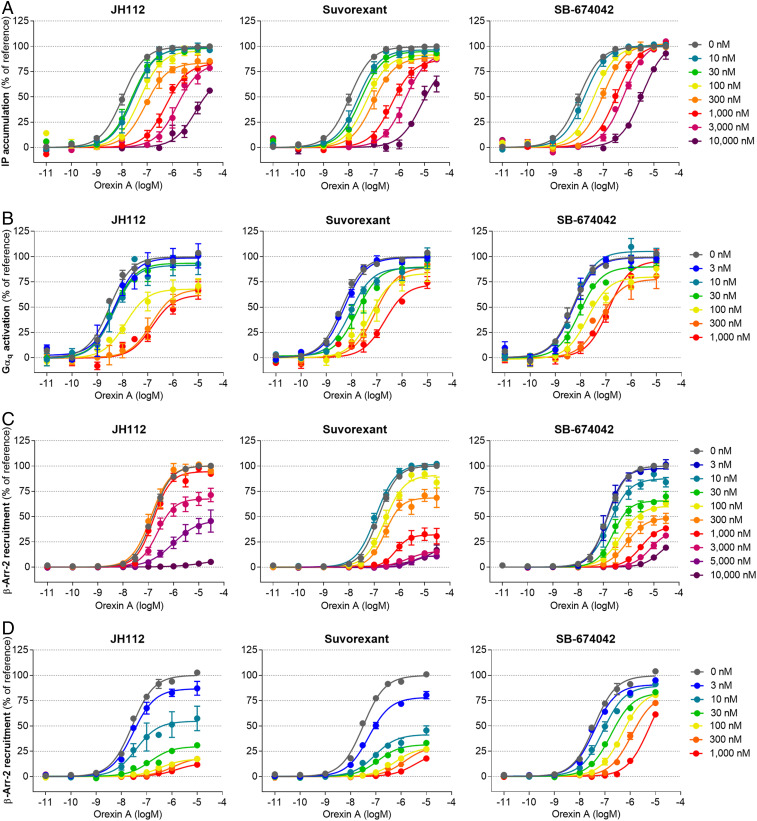
Stimulation of OX1R is known to trigger a broad variety of intracellular responses promoted by recruitment and activation of heterotrimeric Gq proteins and β-arrestins (29, 30). Initially, we evaluated the effect of compound JH112 on OX1R-induced Gαq protein activation in the presence or absence of the endogenous agonist orexin A by measuring the level of the second messenger inositol phosphate (31) (IPOne assay, Cisbio). Stimulation of OX1R-expressing HEK 293T cells with orexin A resulted in a sigmoidal concentration-response curve with a potency in the nanomolar range (EC50 = 15 ± 4 nM). Incubation with JH112, suvorexant, or SB-674042 did not induce inositol phosphate accumulation (SI Appendix, Fig. S13A), but compound JH112 was able to suppress the effect of orexin A (used at a concentration corresponding to its EC80) in a concentration-dependent manner. The observed IC50 of 18 ± 3 nM for JH112 was lower than those of suvorexant and SB-674042 (IC50 100 ± 24 nM and 56 ± 9 nM, SI Appendix, Fig. S14A). When concentration-response curves of orexin A were collected after preincubation with the antagonists, different effects were observed for SB-674042, suvorexant, and JH112 (Fig. 3A). While the inhibitory effects of 10 µM SB-674042 could be surmounted by orexin A, JH112 and suvorexant did not allow orexin A to reach its maximum response. To account for individual kinetic effects of the antagonists, we complemented the functional IPOne assay by a proximity-based experiment, allowing us to add the antagonist after incubation with orexin A. SI Appendix, Figs. S13B and S14B, show that concentration-response curves of orexin A and the antagonists obtained employing a Gαq-RlucII/Gγ-GFP10 BRET biosensor (32, 33) were highly similar to the results from inositol phosphate accumulation (EC50 orexin A 5.0 ± 1.8 nM). Fig. 3B shows concentration-response curves of orexin A obtained in the presence of the antagonists, confirming insurmountable properties. In fact, JH112, suvorexant, and, in this case, also the reference agent SB-674042 diminished the Emax of orexin A to 65 to 75%. In addition to Gq activation, β-arrestin coupling has been described for OX1R upon stimulation with orexin A. To investigate potential inhibition by JH112, suvorexant, and SB-674042, the recruitment of β–arrestin-2 was studied using two different assays employing enzyme fragment complementation (DiscoverX Pathhunter) or bystander BRET between β–arrestin-2-RLucII and a GFP-fused plasma membrane marker (rGFP-CAAX) (34). In both cases, we could detect a concentration-dependent recruitment of β–arrestin-2 for orexin A with potencies that were slightly inferior to those observed for G-protein activation (EC50 160 ± 20 nM and 37 ± 6 nM for the Pathhunter and BRET assay, respectively), but no activation with the antagonists (SI Appendix, Figs. S13 C and D and S14 C and D). Exploiting fragment complementation, all three ligands substantially diminished, if not abolished, the orexin A-mediated recruitment of β–arrestin-2 to OX1R (Fig. 3C). A similar trend was observed in the β–arrestin-2 BRET assay, which does not require a preincubation period with antagonist (Fig. 3D).
Fig. 3.
Inhibition of OX1R activation by JH112, suvorexant and SB-674042. Inhibition of the orexin A effect was determined in (A) an IP1 accumulation assay (Cisbio), (B) a Gαq-RlucII/Gγ-GFP10 biosensor BRET assay, (C) a β-arrestin recruitment assay based on enzyme fragment complementation (DiscoverX), and (D) a BRET assay for the recruitment of β–arrestin-2 to the plasma membrane. In each case, orexin A concentration-response curves were collected in the presence of a given concentration of the antagonist. While SB-674042 mostly leads to a decrease of the orexin A potency (rightward shift of EC50), JH112 and suvorexant additionally depress the maximum response (Emax) of orexin A, especially for the recruitment of β–arrestin-2. Curves represent the mean ± SEM from 3 to 16 individual experiments, each performed in duplicate (A and C) or triplicate (B and D).
The results clearly demonstrate that JH112 exerts insurmountable antagonism (35–37), a phenomenon that has been previously described for other small-molecule OX1R antagonists (38–40). For JH112, the effect is particularly pronounced for the inhibition of β–arrestin-2 recruitment when the Emax of orexin A is reduced to below 20%. Analysis of the functional data employing an operational hemi-equilibrium model for competitive antagonism (35, 37, 41) yielded a dissociation half-life of 8 to 89 h for suvorexant and at least 37 h, if not pseudoirreversible antagonism, for JH112. In contrast, significantly faster dissociation rates were calculated for SB-674042 (maximum half-life 12 h), pointing toward a kinetic mechanism behind the observed insurmountable antagonism of JH112 (SI Appendix, Table S5 and Fig. S15).
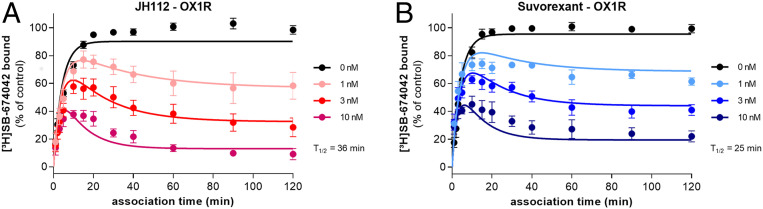
Kinetic analysis of binding studies with the radioligand [3H]SB-674042, suvorexant, and JH112 revealed dissociation rates ranging from 0.1383 ± 0.0094 min−1 to 0.01915 ± 0.00367 min- 1 (Fig. 4 and SI Appendix, Table S6). Although the resulting dissociation half-lives of 5.1 min for SB-674042, 25 min for suvorexant, and 36 min for JH112 are considerably shorter than those calculated from the functional assays, both methodologies lead to the same kinetic rank order of the ligands. Our observation is in agreement with previous functional investigations on suvorexant at the OX2R in the presence of orexin A (35), which indicated an up to 15-fold longer dissociation half-life compared to kinetic binding experiments. In its canonical binding pose, orexin A binds bitopically with crucial contacts extending deep into the TM bundle and specific interactions engaging the amphiphilic helix located at the N terminus of OX1R (23). These two sites are located far away from each other and connected by a conformationally flexible sequence of seven amino acids. The difference in dissociation rate constants determined by hemi-equilibrium functional assays in the presence of high concentrations of orexin A and kinetic binding may confer an allosteric binding mode of the native hormone if a small molecule antagonist is bound. Hence, if JH112 occupies the site resolved within the crystal structure, orexin A may still be able to bind the amphiphilic helix at the top of the binding site and thereby slow down the dissociation of the antagonist.
Fig. 4.
Ligand-binding kinetics of JH112 and suvorexant at OX1R. Kinetic binding experiments with the radioligand [3H]SB-674042 and membranes from HEK293T cells expressing OX1R reveal a dissociation half-life of (A) 36 min for JH112 and (B) 25 min for suvorexant. Data represent mean ± SEM and the global fit from 5 to 10 individual experiments.
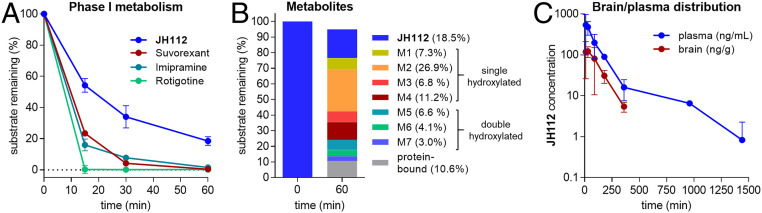
We investigated the in vitro metabolic stability of JH112 in a rat liver microsome assay (Fig. 5 A and B). JH112 shows a significantly increased stability (t1/2 = 25.3 ± 2.7 min, intrinsic clearance 55.5 ± 5.5 µL⋅min−1⋅mg−1) compared to suvorexant (t1/2 = 7.32 ± 0.49 min, intrinsic clearance 190 ± 13 µL⋅min−1⋅mg−1). High metabolic stability is usually associated with a long duration of action, which is favorable for potential pharmacological applications in the fields of addiction, anxiety, pain, or obesity. The observed metabolites of JH112 are either single- or double-hydroxylated analogs. Preliminary pharmacokinetic studies of JH112 revealed the compound being brain-penetrant with a brain-to-plasma concentration ratio of 0.3 (Fig. 5C).
Fig. 5.
Phase I metabolism and brain/plasma distribution of compound JH112. (A) Exposure of suvorexant and JH112 to rat liver microsomes over 60 min. Rotigotine and imipramine serve as positive controls for extensive phase I metabolism. Data represent mean ± SEM of n = 3 independent experiments. (B) Quantitative analysis of metabolite pool composition after 60 min by HPLC-MS reveals predominantly hydroxylated metabolites (M1 to M7). Total amount of JH112 and metabolite pool after 60 min is slightly lower than 100%, reflecting cumulative error in liquid chromatography–mass spectrometry analysis. (C) Pharmacokinetic studies of JH112 in female CD-1 mice (n = 3 animals for each time point) show central nervous system penetration of the compound, with a peak level of 122 ng of JH112 per gram of brain tissue after 30 min, with a maximum plasma concentration of 479 ng/mL after 10 min. All data are mean ± SEM.
Structure–Activity Relationship Studies.
We have carried out molecular docking calculations, chemical synthesis, and binding experiments with a set of further suvorexant analogs revealing an informative structure–activity relationship profile (SI Appendix, Figs. S1 and S2). Hence, elongation of the substituent from methyl to ethyl leads to only a weak increase (1.8-fold) of subtype selectivity, as the substituent is conformationally flexible and thus able to avoid a steric clash with Thr1353.33 of OX2R. Going from methyl (compound 1) to ethyl (compound 3) and propyl (compound 4) substituents, the docking poses in the OX1R are conserved, and the side chains can be accommodated close to Ala1273.33 (SI Appendix, Fig. S1). The binding poses of these molecules in the OX2R are similar. However, the increase of the side chain becomes sterically more demanding, and compound 4 does not interact with Asn3246.55 but shifts its polar contact to His3507.39. Overall, homologization has been beneficial, resulting in 16-fold subtype selectivity for the propyl analog 4, indicating repulsive interactions of the terminal CH3 position in the OX2R, even in a low-energy conformational state (SI Appendix, Fig. S1).
Compounds with branched substituents including isopropyl (compound 5), cyclopropyl (compound 6), isobutyl (compound 7) and cyclopropylmethyl (compound 8) showed higher subtype selectivity (26- to 52-fold) and single-digit nanomolar binding affinity for the OX1R (Ki = 1.1 ± 0.2 nM to 6.1 ± 0.7 nM) as a result of an increased bulkiness (SI Appendix, Figs. S1 and S2). Whereas the impact on subtype selectivity was higher when the branch was closer to the homopiperazine ring, compounds with a branch more distant showed a more favorable binding affinity. The docking poses of the increasingly bulky side chains of compounds 5 to 8 showed a similar trend as the n-alkyl substituted molecules. Compound 8 is an exception, in the docking pose of which the side chain flips over to helix 6, relinquishing the contact with helix 3. In the OX2R, these branched molecules behave similarly to their nonbranched counterparts: Compound 5, with its isopropyl group as a branched n-ethyl chain, does not interact with Asn318/3246.55 anymore and forms an interaction with His344/3507.39 instead, according to docking. This is even enhanced for the longer branched molecules, which could explain the high selectivity of these compounds.
We have also synthesized the enantiomer (compound 9) as well as two regioisomers (compounds 10 and 11) of the isopropyl-substituted compound 5 (SI Appendix, Fig. S1). Changing the stereochemistry of the substituent drastically decreases the binding affinities to both receptor subtypes by a factor of 25 to 55. By formally moving the isopropyl substituent to the neighboring carbon atom of the homopiperazine ring closer to the benzoxazole, we obtained two regioisomers of compound 5 with opposite stereochemistry (compounds 10 and 11). The OX1R-binding affinities of those compounds were also lower compared to compound 5 (compound 10: ∼10-fold; 11: ∼40-fold), which corroborates our choice of the position of substituents at the homopiperazine ring.
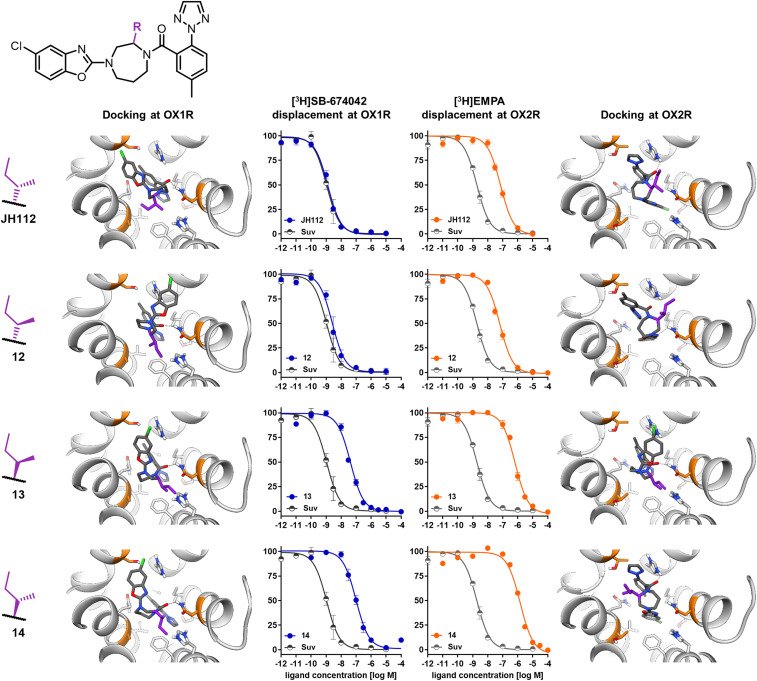
Compound 12, a stereoisomer of JH112, which has an inverted configuration at the sec-butyl substituent, shows an about 3-fold lower OX1R affinity and reduced selectivity over the OX2R (28-fold), proving the high complementarity between receptor and JH112 (Fig. 6). As expected, the two stereoisomers of JH112, which have an inverted configuration at the homopiperazine ring system resulting from the formal replacement of hydrogen 2 by a sec-butyl group, show significantly lower affinity with Ki values in the high nanomolar range. Docking of the stereoisomers of JH112 shows that only the (S,S)-sec-butyl pose fits into the space around Ala1273.33 without any clashes. All of the other stereoisomers are either unable to stabilize the interaction with Asn318/3246.55 (compound 12) or, in the case of compounds 13 and 14, the side chain points away from helix 3 and engages helix 5 instead. In the OX2R docking, a similar trend can be observed, i.e., that the molecules can interact only with His344/3507.39 (Fig. 6).
Fig. 6.
Receptor-binding curves and docking poses of compounds JH112 and 12 to 14 in OX1R and OX2R. The competition curve obtained with suvorexant (in gray) is displayed as reference. Data from competition binding experiments were normalized to total (100%) and unspecific binding (0%) and are displayed as mean ± SEM from four to six independent experiments.
Docking of derivatives with larger substituents (compounds 15 and 16) predicted that a phenyl or cyclohexyl substituent, respectively, is not well tolerated by the receptors, which was confirmed by decreased binding affinities (SI Appendix, Fig. S2).
We also investigated the importance of the ring size of those suvorexant analogs (SI Appendix, Fig. S2). The desmethyl-suvorexant analog (compound 17) shows binding affinities comparable to suvorexant and no subtype selectivity. By opening the seven-membered ring at the longer propylene bridge, we obtained compound 18, which showed dramatically lower binding affinities to both receptor subtypes in the single-digit micromolar range probably due to an entropic penalty. Reducing the ring size to the less flexible piperazine analog (compound 19) is not well tolerated by the receptors and resulted in micromolar binding affinities, as previously described (38). Interestingly, enlargement of the ring size leads to the very flexible 1,5-diazocane derivative 20 which showed only slightly reduced binding affinities (∼30- to 60-fold) compared to the homopiperazine analog (compound 17).
Discussion
Two observations are particularly worth highlighting. First, the newly developed orexin receptor antagonist JH112 shows subnanomolar binding affinity and 75-fold subtype selectivity for OX1R over the OX2R. Because OX1R is responsible for the regulation of addiction, pain, obesity, and anxiety, JH112 may have important clinical applications. The clinically approved drugs targeting the orexin system, suvorexant and lemborexant, are nonselective dual orexin receptor antagonists used for the treatment of insomnia. Despite several approaches toward the development of subtype-selective OX1R antagonists, none of them has resulted in clinical approval yet (42). Ligands with high OX1R selectivity like SB-674042 have been described, but were not pursued further due to a number of liabilities including poor solubility, limited plasma exposure, hydrolytic instability, or off-target affinities (43). Interestingly, first-in-human studies with the selective OX1R antagonist JNJ-61393215 as a potential treatment of mood and anxiety disorders have been initiated (44, 45). JH112 exhibits potent and insurmountable antagonism for Gq activation and in particular β–arrestin-2 recruitment at OX1R, putatively resulting from very slow receptor dissociation when the endogenous agonist orexin A is noncanonically bound to the amphiphilic helix at the N-terminus of the receptor. Moreover, JH112 is able to cross the blood–brain barrier, is slowly metabolized in a phase I liver microsomal stability assay, and possesses an excellent selectivity profile over 20 other GPCRs. Thus, JH112 could be a promising candidate for the treatment of disorders associated with OX1R activation with less off-target effects related to the OX2R, such as sleep–wakefulness disorders.
Second, structure-based drug design has been proven successful for the differentiation between highly related GPCR subtypes (20, 46). The herein presented cycle based on docking, enantiospecific synthesis, molecular pharmacology, and crystallography starting from the orexin receptor crystal structures bound to suvorexant led to the subtype-selective OX1R antagonist JH112. Selectivity for OX1R over OX2R could be achieved by exploiting a single amino acid difference in the binding pocket of the two receptors. A crystal structure of the OX1R in complex with JH112 and mutational studies confer that JH112 occupies the space provided by Ala1273.33 in OX1R while producing repulsive interactions with Thr1353.33 in OX2R, consistent with the docking-derived design hypothesis. Kinetic binding studies showed that JH112 has a shorter half-life time at OX2R compared to suvorexant, suggesting that the steric repulsion between Thr1353.33 and the sec-butyl substituent of JH112 provokes an increase of ligand dissociation rather than slowing down association. Based on this understanding, the resolved crystal structure of OX1R bound to JH112 will be a useful template for further lead-structure optimization studies.
Materials and Methods
The data that support the findings of this study are available in this paper and/or in SI Appendix. Atomic coordinates and structure factors of the JH112-OX1R complex have been deposited in the Protein Data Bank (https://www.rcsb.org/) under accession code 6V9S.
Ligand Design.
The design of selective OX1R ligands was guided by the inactive state crystal structures of OX1R (PDB ID 4ZJ8) (23) and OX2R (PDB ID 4S0V) (24) bound to the dual selective antagonist suvorexant. Ligand conformations were generated using OMEGA (47) and docked with OpenEye’s FRED tool (48, 49). Further details and the synthetic procedures leading to ligands 1 to 20 are provided in SI Appendix.
Characterization of Ligands.
Synthesized ligands were characterized in binding studies with the radioligands [3H]SB-674042 and [3H]EMPA and membranes from HEK 293T cells transiently transfected with the complementary DNAs coding for OX1R and OX2R. The pharmacologic properties of JH112 were assessed in G-protein activation and β-arrestin recruitment assays in transiently transfected HEK 293T cells, and its microsomal stability and pharmacokinetics were determined as described in SI Appendix. If not stated otherwise, data are indicated as mean ± SEM.
Structure Determination.
The structure of OX1R bound to JH112 was determined by lipid cubic phase (LCP) crystallography. Data collection was performed at beamline 23ID-D (GM/CA-CAT), Advanced Photon Source, Argonne National Laboratory. The structure was solved by molecular replacement using the previously reported structures of PGS (PDB ID 2BFW) and hOX1R in complex with suvorexant (PDB ID 4ZJ8) as independent search models. Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 6V9S. Further details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Michel Bouvier (Institute for Research in Immunology and Cancer, University of Montreal, Canada) for providing the BRET biosensor plasmids and Stefania Monteleone and Tobias Hüfner for helpful discussions. This research was supported by the German Research Foundation Deutsche Forschungsgemeinschaft Heisenberg Professorship KO4095/4-1 and KO4095/5-1 and Research Grants KO4095/3-1 (to P.K.) and GRK1910 (to D.W. and P.G.); NIH Grant R01NS103939 (to D.M.R.); and the Welch Foundation Grant I-1770 (to D.M.R.). D.W., P.K., and P.G. participate in European Cooperation in Science and Technology (COST) Action CA18133 European Research Network on Signal Transduction (ERNEST).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Data have been deposited in the Protein Data Bank (accession number 6V9S).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002704117/-/DCSupplemental.
References
- 1.Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee S.-M., Booe J. M., Pioszak A. A., Structural insights into ligand recognition and selectivity for classes A, B, and C GPCRs. Eur. J. Pharmacol. 763, 196–205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakurai T. et al., Orexins and orexin receptors: A family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 92, 573–585 (1998). [DOI] [PubMed] [Google Scholar]
- 4.de Lecea L. et al., The hypocretins: Hypothalamus-specific peptides with neuroexcitatory activity. Proc. Natl. Acad. Sci. U.S.A. 95, 322–327 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcus J. N. et al., Differential expression of orexin receptors 1 and 2 in the rat brain. J. Comp. Neurol. 435, 6–25 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Trivedi P., Yu H., MacNeil D. J., Van der Ploeg L. H. T., Guan X.-M., Distribution of orexin receptor mRNA in the rat brain. FEBS Lett. 438, 71–75 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Razavi B. M., Hosseinzadeh H., A review of the role of orexin system in pain modulation. Biomed. Pharmacother. 90, 187–193 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Sakurai T., The neural circuit of orexin (hypocretin): Maintaining sleep and wakefulness. Nat. Rev. Neurosci. 8, 171–181 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Lin L. et al., The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98, 365–376 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Thannickal T. C. et al., Reduced number of hypocretin neurons in human narcolepsy. Neuron 27, 469–474 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roecker A. J., Cox C. D., Coleman P. J., Orexin receptor antagonists: New therapeutic agents for the treatment of insomnia. J. Med. Chem. 59, 504–530 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Beuckmann C. T. et al., In vitro and in silico characterization of Lemborexant (E2006), a novel dual orexin receptor antagonist. J. Pharmacol. Exp. Ther. 362, 287–295 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Scott L. J., Lemborexant: First approval. Drugs 80, 425–432 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Zisapel N., Current Phase II investigational therapies for insomnia. Expert Opin. Investig. Drugs 24, 401–411 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Perez-Leighton C. E., Butterick-Peterson T. A., Billington C. J., Kotz C. M., Role of orexin receptors in obesity: From cellular to behavioral evidence. Int. J. Obes. 37, 167–174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steiner M. A. et al., Discovery and characterization of ACT-335827, an orally available, brain penetrant orexin receptor type 1 selective antagonist. ChemMedChem 8, 898–903 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Sakurai T., Orexin: A link between energy homeostasis and adaptive behaviour. Curr. Opin. Clin. Nutr. Metab. Care 6, 353–360 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Zhou L., Smith R. J., Do P. H., Aston-Jones G., See R. E., Repeated orexin 1 receptor antagonism effects on cocaine seeking in rats. Neuropharmacology 63, 1201–1207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris G. C., Wimmer M., Aston-Jones G., A role for lateral hypothalamic orexin neurons in reward seeking. Nature 437, 556–559 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Liu H. et al., Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists. Proc. Natl. Acad. Sci. U.S.A. 115, 12046–12050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S. et al., D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science 358, 381–386 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manglik A. et al., Structure-based discovery of opioid analgesics with reduced side effects. Nature 537, 185–190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin J. et al., Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors. Nat. Struct. Mol. Biol. 23, 293–299 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Yin J., Mobarec J. C., Kolb P., Rosenbaum D. M., Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature 519, 247–250 (2015). [DOI] [PubMed] [Google Scholar]
- 25.McGaughey G. et al., Shaping suvorexant: Application of experimental and theoretical methods for driving synthetic designs. J. Comput. Aided Mol. Des. 28, 5–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rappas M. et al., Comparison of orexin 1 and orexin 2 ligand binding modes using X-ray crystallography and computational analysis. J. Med. Chem. 63, 1528–1543 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballesteros J. A., Weinstein H., “Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors” in Methods in Neurosciences, Sealfon S. C., Ed. (Academic Press, 1995), Vol. 25, pp. 366–428. [Google Scholar]
- 28.Caffrey M., Cherezov V., Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kukkonen J. P., Leonard C. S., Orexin/hypocretin receptor signalling cascades. Br. J. Pharmacol. 171, 314–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C. et al., The orexin/receptor system: Molecular mechanism and therapeutic potential for neurological diseases. Front. Mol. Neurosci. 11, 220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trinquet E. et al., D-myo-inositol 1-phosphate as a surrogate of D-myo-inositol 1,4,5-tris phosphate to monitor G protein-coupled receptor activation. Anal. Biochem. 358, 126–135 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Galés C. et al., Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nat. Struct. Mol. Biol. 13, 778–786 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Breton B. et al., Multiplexing of multicolor bioluminescence resonance energy transfer. Biophys. J. 99, 4037–4046 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Namkung Y. et al., Monitoring G protein-coupled receptor and β-arrestin trafficking in live cells using enhanced bystander BRET. Nat. Commun. 7, 12178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mould R., Brown J., Marshall F. H., Langmead C. J., Binding kinetics differentiates functional antagonism of orexin-2 receptor ligands. Br. J. Pharmacol. 171, 351–363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treiber A. et al., The use of physiology-based pharmacokinetic and pharmacodynamic modeling in the discovery of the dual orexin receptor antagonist ACT-541468. J. Pharmacol. Exp. Ther. 362, 489–503 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Kenakin T., Jenkinson S., Watson C., Determining the potency and molecular mechanism of action of insurmountable antagonists. J. Pharmacol. Exp. Ther. 319, 710–723 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Heidmann B. et al., Discovery of highly potent dual orexin receptor antagonists via a scaffold-hopping approach. ChemMedChem 11, 2132–2146 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Williams J. T. et al., Discovery and optimisation of 1-acyl-2-benzylpyrrolidines as potent dual orexin receptor antagonists. MedChemComm 6, 1054–1064 (2015). [Google Scholar]
- 40.Perrey D. A. et al., The importance of the 6- and 7-positions of tetrahydroisoquinolines as selective antagonists for the orexin 1 receptor. Bioorg. Med. Chem. 23, 5709–5724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riddy D. M. et al., Label-free kinetics: Exploiting functional hemi-equilibrium to derive rate constants for muscarinic receptor antagonists. Mol. Pharmacol. 88, 779–790 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Perrey D. A., Zhang Y., Therapeutics development for addiction: Orexin-1 receptor antagonists. Brain Res. 1731, 145922 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lebold T. P., Bonaventure P., Shireman B. T., Selective orexin receptor antagonists. Bioorg. Med. Chem. Lett. 23, 4761–4769 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Salvadore G. et al., F13. Safety, tolerability, pharmacokinetic and pharmacodynamic properties of the selective orexin-1 receptor antagonist JNJ-61393215: Results from the first-in-human and multiple ascending dose studies. Biol. Psychiatry 85, S217–S218 (2019). [Google Scholar]
- 45.Janssen L., Research & development: A study of JNJ-61393215 in the treatment of depression (2019). https://ClinicalTrials.gov/show/NCT04080752. Accessed 27 May 2020.
- 46.Chevillard F. et al., Binding-site compatible fragment growing applied to the design of β2-adrenergic receptor ligands. J. Med. Chem. 61, 1118–1129 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Hawkins P. C. D., Skillman A. G., Warren G. L., Ellingson B. A., Stahl M. T., Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 50, 572–584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGann M., FRED pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 51, 578–596 (2011). [DOI] [PubMed] [Google Scholar]
- 49.McGann M., FRED and HYBRID docking performance on standardized datasets. J. Comput. Aided Mol. Des. 26, 897–906 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.