Significance
DNA mismatch repair (MMR) proteins are essential for correcting base incorporation errors that occur during replication, greatly enhancing genomic stability. In all organisms, MutS and MutL homologs initiate MMR repair and are involved in several other DNA transactions, including DNA-damage–induced apoptosis and homologous recombination. Our study reveals that MutS mobile clamps hydrolyze ATP while remaining bound to DNA and frequently revisit the mismatch. We also find that MutL converts MutS mobile clamps into immobile MutS–MutL complexes. These immobile, multimeric complexes present a striking contrast to current MMR initiation models that envision MutS–MutL sliding freely on the DNA, effectively diffusing away from the mismatch. Our results support mechanisms that localize repair complexes to the vicinity of the mismatch.
Keywords: smFRET, MMR, mismatch repair, MutS
Abstract
DNA mismatch repair (MMR), the guardian of the genome, commences when MutS identifies a mismatch and recruits MutL to nick the error-containing strand, allowing excision and DNA resynthesis. Dominant MMR models posit that after mismatch recognition, ATP converts MutS to a hydrolysis-independent, diffusive mobile clamp that no longer recognizes the mismatch. Little is known about the postrecognition MutS mobile clamp and its interactions with MutL. Two disparate frameworks have been proposed: One in which MutS–MutL complexes remain mobile on the DNA, and one in which MutL stops MutS movement. Here we use single-molecule FRET to follow the postrecognition states of MutS and the impact of MutL on its properties. In contrast to current thinking, we find that after the initial mobile clamp formation event, MutS undergoes frequent cycles of mismatch rebinding and mobile clamp reformation without releasing DNA. Notably, ATP hydrolysis is required to alter the conformation of MutS such that it can recognize the mismatch again instead of bypassing it; thus, ATP hydrolysis licenses the MutS mobile clamp to rebind the mismatch. Moreover, interaction with MutL can both trap MutS at the mismatch en route to mobile clamp formation and stop movement of the mobile clamp on DNA. MutS’s frequent rebinding of the mismatch, which increases its residence time in the vicinity of the mismatch, coupled with MutL’s ability to trap MutS, should increase the probability that MutS–MutL MMR initiation complexes localize near the mismatch.
DNA mismatch repair (MMR) plays a major role in an organism’s ability to avoid mutations, including correcting DNA replication errors, modulating cellular responses to DNA damaging agents, and preventing recombination between diverged sequences. MMR is initiated by MutS and MutL proteins (1, 2). Mutations in these proteins cause Lynch syndrome, the most common hereditary cancer (3–6). In addition, they are linked to sporadic cancers and cause resistance to the cytotoxic effects of DNA-damaging agents used often in cancer treatment (7–9).
MutS initiates repair by binding to a mismatch or insertion-deletion loop and then undergoing ATP-dependent conformational changes to form a clamp that moves along the DNA (2, 10). This ATP-activated state of MutS also promotes its interaction with one or more MutL proteins (2, 11, 12). Subsequently, the MutS–MutL complex, or MutL that has been activated by MutS (13), interacts with the strand discrimination signal to promote nicking and excision of the error-containing daughter strand. In eukaryotes and most bacteria, strand discrimination appears to be achieved by interaction of the MutS–MutL complex with the mobile DNA replication processivity clamp (PCNA in eukaryotes, β-clamp in prokaryotes) (2, 14, 15). This interaction activates the latent endonuclease activity of MutL to nick the daughter strand. In contrast, in Escherichia coli, which employs methyl-directed MMR, hemimethylated GATC sites serve as the strand discrimination signal. In this case, MutS–MutL interacts with and activates MutH endonuclease to nick the unmethylated strand at a hemimethylated GATC site up to 1 kB from the mismatch. The mechanisms by which MutS and MutL interact with each other and with the strand-discrimination signal to initiate downstream repair events after mismatch recognition remain unresolved and controversial, with several disparate models currently under debate in the field. Although ongoing studies of MMR in various organisms could lead to reconciliation, it is also possible that fundamental differences in strand discrimination after mismatch recognition and mobile clamp formation result in divergence of E. coli (methyl-directed) versus MutH-independent (nonmethyl-directed) repair mechanisms.
Studies of E. coli MMR have led to the proposal that ATP-bound, freely diffusing MutS clamps recruit MutL to form MutS–MutL clamps that remain mobile on DNA (16, 17). However, other studies using proteins from a variety of species suggest that MutL can trap MutS at the mismatch (18) and that multiple MutL proteins associate with MutS in complexes (12, 18–22). Given that the strand discrimination signal (PCNA\β-clamp) is mobile in organisms using nonmethyl-directed MMR, signaling at a distance between the MutS–MutL complex and the strand identity marker is not essential (unlike the situation in E. coli methyl-directed repair), and a mobile MutS–MutL clamp or MutS-activated mobile MutL clamp may not be necessary in nonmethyl-directed MMR. For decades, it has been known that in the absence of MutL, MutS forms a mismatch- and ATP-dependent mobile clamp in all organisms. Despite its prevalence, the function of this postrecognition MutS clamp and its interactions with MutL remain poorly understood even though they play a critical role in initiating repair.
In this work, we used single-molecule FRET (smFRET) to investigate the properties of the MutS mobile clamp and its interactions with MutL, focusing on proteins from Thermus aquaticus (Taq), a MutH-independent MMR model system. smFRET with Taq MMR proteins is a powerful tool for studying transient interactions that initiate MMR because the proteins are amenable to fluorophore labeling in specific, well-defined positions and are well-behaved in vitro. On interrogating individual MutS proteins in the long-lived mismatch- and ATP-dependent mobile clamp state on end-blocked DNA, we unexpectedly found that the MutS clamp undergoes frequent cycles of mismatch rebinding (or revisiting) and mobile clamp formation without releasing DNA. Furthermore, this mismatch rebinding requires ATP hydrolysis by MutS. Ensemble measurements of ATPase activity confirm that the MutS mobile clamps hydrolyze ATP while bound to doubly end-blocked DNA. These results contrast with current MMR models that suggest ATP hydrolysis occurs after MutS dissociates from DNA (10, 17, 23–27). We also found that MutL stops movement of the MutS clamp on DNA. Repetitive rebinding of the mismatch by a MutS mobile clamp likely helps corral MutS–MutL complexes in the vicinity of the mismatch, thereby localizing subsequent excision activity for more efficient DNA repair.
Results
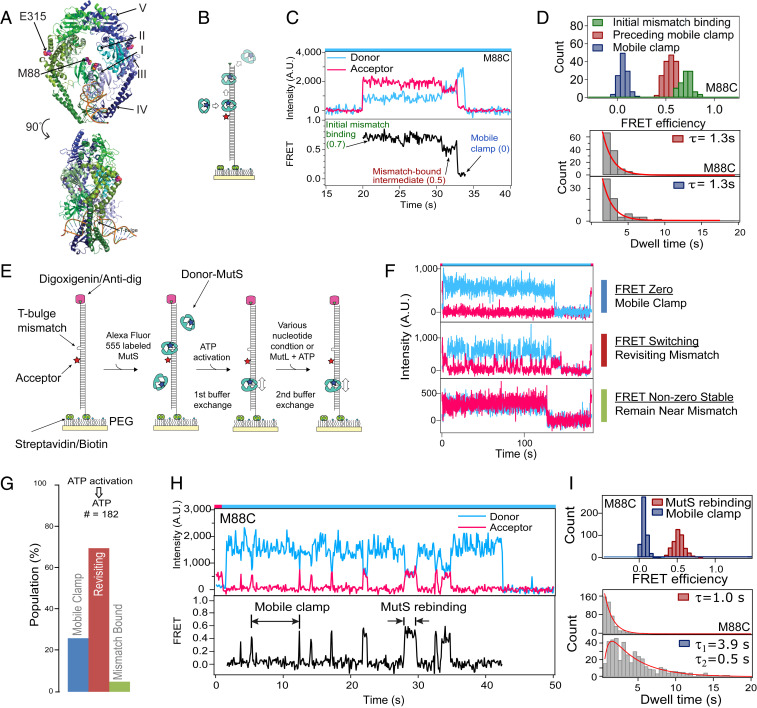
In previous experiments, we characterized conformational changes in Taq MutS–T-bulge DNA complexes (Fig. 1A) as MutS transitions from mismatch recognition to a mobile clamp in the absence and presence of MutL (18, 28). We used a 550-base pair, surface-tethered DNA substrate containing a central T-bulge and an acceptor fluorophore 9 bases away (Fig. 1B). Addition of MutS containing a donor fluorophore on DNA binding domain I (M88C) (Fig. 1A) and ATP results in a sequence of readily distinguishable, distinct FRET efficiency levels (0.7 → 0.5 → 0 → no fluorescence) (Fig. 1 C and D). These FRET values represent mismatch binding by MutS (0.7), a conformational change in the protein at the mismatch (0.5), the clamp moving away from the mismatch (0), and sliding off the free DNA end (no fluorescence). This cycle can repeat when MutS from solution binds to a mismatch freed after a previous mobile clamp leaves and, if both DNA ends are blocked, multiple MutS mobile clamps can load onto the DNA (10, 17, 18, 24). These studies also showed that MutL can trap MutS in an intermediate state at the mismatch if MutL arrives before MutS moves away (18). However, these studies did not address the fate and function of the postmismatch recognition MutS mobile clamp, a long-standing question in the field that is key to understanding the initiation of MMR.
Fig. 1.
MutS mobile clamps revisit a DNA mismatch on end-blocked DNA. (A) Crystal structure of Taq MutS–T-bulge complex (PDB ID code 1EWQ) (64), highlighting the domains and label positions. (B) Schematic of experiments with unblocked T-bulge DNA, 2 mM ATP, and 5 nM MutS (dimer). (C) Example time trace of donor and acceptor intensities (Upper) and FRET (Lower) between AF555-MutS (M88C) donor and Cy5–T-bulge DNA acceptor reports MutS binding a mismatch (FRET 0.7), transitioning to an intermediate state at the mismatch (FRET 0.5), converting to a mobile clamp that slides away (FRET 0), and falling off the DNA end (loss of donor signal). This behavior is not observed in the absence of a mismatch or ATP. (D) FRET histograms and dwell-time distributions for unblocked DNA experiments. Single-exponential fits to the dwell-time data are in red. Distributions are shown for the initial binding state (0.7), the preceding mobile (0.5), and the mobile clamp states (0.0). (E) Schematic of experiments with end-blocked DNA: Addition of MutS without nucleotide, first buffer exchange introducing ATP (no additional MutS), and second buffer exchange introducing the nucleotide to be tested. (F) Single AF555-MutS mobile clamps on end-blocked Cy5–T-bulge DNA exhibit three types of behaviors of donor (blue) and acceptor (red) intensities before the signals disappear, likely due to donor photobleaching given the ∼10-min lifetime of mobile clamps on DNA (24): Continuous zero FRET (Top; mismatch bypassing clamp), FRET switching between 0 and 0.5 for at least one mismatch rebinding event (Middle; mismatch revisiting clamp), and constant FRET 0.5 (Bottom; mismatch-bound MutS). The zero FRET population (Top) includes clamps that remain continuously mobile and any MutS colocalized with DNA in a mismatch-independent manner (e.g., nonspecific adsorption to the surface or interaction with DNA end-blocking proteins streptavidin or anti-dig) (Materials and Methods). (G) Fractions of MutS colocalized on DNA after the second buffer exchange with ATP, exhibiting the three behaviors described above. Color bars and state names are as defined in F. (H) Zoom-in of a trace with MutS revisiting the mismatch. (I) Histograms of FRET values from many traces with revisiting events show peaks at 0.5 and 0 (Upper). Dwell time distribution for the FRET 0.5 state fit with a single exponential (red) yields a 1.0 s mismatch rebinding lifetime. Fitting the dwell-time distribution for the FRET 0 state (lowest panel) with a two-step kinetic model (red) (SI Appendix, Supporting Methods) yields lifetimes 3.9 s and 0.5 s for the mobile clamp states between revisits (Lower). Color bars on top of the intensity vs. time graphs indicate laser illumination color (red = 640 nm; blue = 532 nm).
MutS Mobile Clamps Revisit a Mismatch.
In vivo, MutS will not encounter DNA ends; therefore, we blocked the free end of our DNA substrate to prevent the clamp from sliding off and monitored the mismatch- and ATP-activated MutS mobile state over an extended time period. To characterize the properties of single MutS mobile clamps without complications from the signals of multiple MutS loading on the same DNA substrate, we optimized a protocol that results in no more than one MutS bound per DNA in most cases (Materials and Methods, Fig. 1E, and SI Appendix, Fig. S1). This protocol starts with a flow chamber where well-spaced T-bulge DNA molecules are tethered to the surface. We first incubated donor-labeled MutS in the acceptor-labeled DNA chamber in buffer without ATP, which results in one MutS dimer bound stably at the mismatch, blocking additional binding. The chamber was then washed with buffer containing ATP, which removes unbound MutS from solution and activates conformational changes in mismatch-bound MutS for conversion into mobile clamps (Fig. 1E, first buffer-exchange step). This protocol results in significant fractions of DNAs with only one MutS bound or no protein bound, and a very small fraction with multiple MutS bound (Materials and Methods and SI Appendix, Table S1); the analysis includes only DNAs colocalized with exactly one MutS. The majority of MutS in these complexes exhibits behavior consistent with mobile clamps (FRET 0), with only a small fraction (5%) remaining at the mismatch (FRET 0.5) (Fig. 1 F, Bottom trace; Fig. 1G, green bar). As reported previously (24), the lifetime of a MutS mobile clamp on a blocked DNA substrate is ∼10 min.
To our surprise, only a small fraction of the ATP-induced mobile MutS clamps (26%) exhibit constant zero FRET (Fig. 1 F, Top trace; Fig. 1G, blue bar), and the majority (69%) alternate between FRET 0 and FRET 0.5 without dissociating from DNA (Fig. 1 F, Middle trace; Fig. 1G, red bar; Fig. 1H). For these alternating MutS molecules, the dwell-time distribution of the FRET 0 events, which represent the process of the mobile clamp rebinding the mismatch, fits a two-step model with lifetimes of 3.9 s and 0.5 s, indicating a hidden kinetic step that does not result in a FRET change (18, 29, 30). The two-step kinetics indicate that after formation, the mobile clamp must undergo some subsequent conformational transition for it to become capable of rebinding the mismatch. The dwell-time distribution of the FRET 0.5 events fit a single exponential with lifetime 1.0 s (Fig. 1I). The FRET efficiency of 0.5 indicates that MutS is located at/near the mismatch, and the width of the FRET distribution is consistent with a single static state (Fig. 1I) (31–34). Notably, we also observed a FRET 0.5 state as an intermediate on the path from the initial mismatch recognition (FRET 0.7) to mobile clamp formation (FRET 0) (Fig. 1 C and D, initial FRET 0.5 state, red histogram). These similarities suggest that the pre- and postmobile clamp FRET 0.5 states may represent the same (or similar) mismatch-bound conformation of MutS, and analysis of the kinetic properties of the observed FRET states (Fig. 1H) further supports this idea. Specifically, the dwell-time distributions of the pre- and postmobile clamp FRET 0.5 states exhibit similar lifetimes (1.3 s and 1.0 s, respectively) (compare red histograms in Fig. 1 D and I) (18, 28). Taken together, these observations indicate that MutS repeatedly cycles between a mobile clamp (FRET 0) and a mismatch-bound state (FRET 0.5) without dissociating from the DNA.
Our findings differ from a previous study of MutS, which was labeled on the domain IV dimer interface (T469C) (24). That study did not detect an intermediate MutS state during initial clamp formation nor the MutS clamp revisiting the mismatch. We considered the possibility that fluorophore location on MutS might influence the results and therefore repeated the smFRET experiments with MutS labeled at another position (E315C on connector domain III in Fig. 1A; see also SI Appendix, Fig. S2) (30). We observed mismatch rebinding with this labeled version of MutS as well. Furthermore, both the M88C- and the E315C-labeled MutS proteins exhibit similar mismatch-rebinding kinetics (compare Fig. 1I to SI Appendix, Fig. S2D) (MutS [E315C] FRET 0.5 lifetime is 1.5 s and the two-step FRET 0 lifetimes are 4.7 s and 0.6 s). Together, these results strongly support our conclusion that MutS mobile clamps repeatedly revisit the mismatch.
Although we observed mismatch rebinding, previous studies have provided evidence that MutS clamps can bypass the mismatch (17, 24, 35). Our studies are also consistent with these observations. Specifically, given the fast diffusivity of a MutS clamp (28, 36), the clamp is expected to slide over the mismatch hundreds of times during its ∼4-s lifetime between mismatch release and rebinding on our 550-base pair-blocked DNA (Fig. 1I and SI Appendix, Supporting Methods and Fig. S2D) (35). Importantly, the ability of MutS mobile clamps to both bypass and rebind the mismatch indicates the need for a switch in the clamp conformation that makes MutS competent to bind the mismatch again.
MutS Revisiting the Mismatch Requires ATP Hydrolysis.
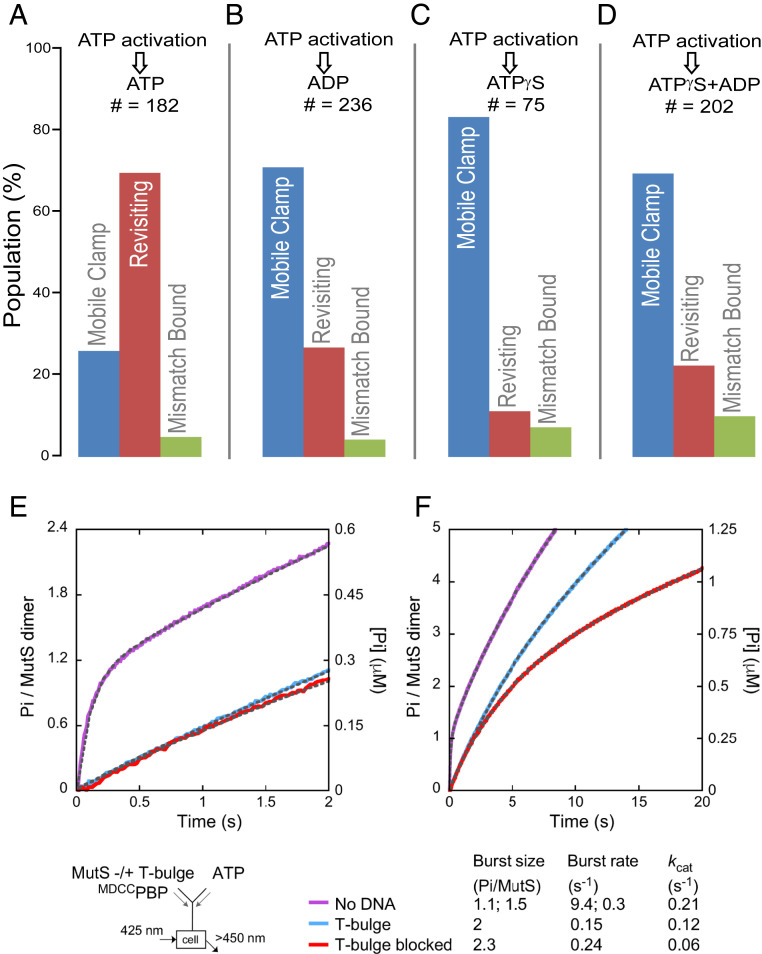
Most current models of MMR signaling, with the notable exception of the translocation model from Modrich and coworkers (37–39), posit that mismatch- and ATP-activated MutS maintains a hydrolysis-independent mobile clamp state that does not rebind the mismatch, and only hydrolyzes or exchanges nucleotides after dissociating from DNA, essentially remaining unchanged as an ATP-bound, freely diffusing clamp while on DNA (10, 17, 23–27). Our observation that MutS mobile clamps can both bypass and rebind a mismatch sharply contrasts these models, and it implies that MutS adopts more than one conformation on DNA. To determine if this conformational switching is linked to its ATPase activity, we examined the effects of different nucleotides on preformed MutS mobile clamps (Fig. 2 A–D). First, we formed mobile clamps as described above (Fig. 1E, up to first buffer exchange), and then performed another buffer-exchange step with varying nucleotides (Fig. 1E, second buffer exchange). In a control experiment to establish a baseline, we exchanged the ATP-containing buffer with the same buffer (Fig. 2A), which resulted in 69% of the clamps revisiting the mismatch (switching between FRET 0 and FRET 0.5) and 26% remaining mobile (constant FRET 0). When the second exchange buffer contains ADP instead of ATP (Fig. 2B), there is a dramatic reduction in the revisiting fraction, with 70% of MutS remaining in the mobile clamp state, bypassing the mismatch. Similarly, in exchange buffers containing slowly hydrolysable ATP-γ-S or a mixture of ADP+ATP-γ-S (Fig. 2 C and D), most of the mobile clamps do not revisit the mismatch (note that a small revisiting fraction in these conditions may be due to residual ATP following the second buffer exchange). The observation that ATP, but not ADP, ATP-γ-S, or ADP+ATP-γ-S, promotes mismatch rebinding suggests that ATP hydrolysis and nucleotide exchange are required for MutS mobile clamps to switch back to a state that allows mismatch binding.
Fig. 2.
Impact of nucleotide exchange on MutS revisiting a mismatch on end-blocked DNA. (A–D) Exchanging buffer after mobile clamp formation (second buffer exchange, Fig. 1E) to introduce ADP (B), ATP-γ-S (C), or ADP+ATP-γ-S (D) instead of ATP (A) significantly reduces the MutS mobile clamp population revisiting the mismatch. Color bars and state descriptions are as defined in Fig. 1F. (E and F) Presteady-state ATPase kinetics of free MutS (purple trace), and mixed with unblocked DNA (blue trace) or doubly end-blocked DNA (red trace) shown at 2-s (E) and 20-s (F) time scales (SI Appendix, Supporting Methods). Exponential+linear fits to the data are overlaid on the traces as dashed lines (amplitudes and rate constants listed below the graph; residuals in SI Appendix, Fig. S3). MutS alone shows burst hydrolysis of one ATP/dimer at k = 9.4 ± 0.2 s−1, the second ATP/dimer at k = 0.3 ± 0.02 s−1, and then a linear, steady state kcat = 0.21 ± 0.06 s−1 (purple trace). On unblocked T-bulge DNA, MutS shows a slow burst of two ATP/dimer hydrolyzed at k = 0.15 ± 0.03 s−1, followed by kcat = 0.12 ± 0.02 s−1 (blue trace). On doubly end-blocked T-bulge DNA, MutS again shows a burst of two ATP/dimer hydrolyzed at k = 0.24 ± 0.007 s−1, followed by kcat = 0.06 ± 0.004 s−1 (red trace).
Ensemble Studies Confirm MutS Mobile Clamps on DNA Hydrolyze ATP.
The idea that MutS mobile clamps catalyze ATP hydrolysis without dissociating from DNA conflicts with current dogma in the field that MutS clamps do not hydrolyze ATP while sliding on DNA. To address this issue, we conducted bulk presteady-state ATPase experiments comparing free MutS, MutS with unblocked mismatch-containing DNA, and MutS clamps formed on doubly end-blocked mismatch-containing DNA. We confirmed that >95% of the end-blocked DNA had neutravidin attached to the biotinylated ends and that all of the MutS remained bound to end-blocked DNA in the presence of ATP using gel electrophoresis (SI Appendix, Supporting Methods and Fig. S3A).
In the absence of DNA, we observed a rapid burst of hydrolysis of one ATP/MutS from the high-affinity ATPase site on the dimer at 9.4 s−1, as reported previously (40, 41), followed by slower hydrolysis of a second ATP at 0.3 s−1 from the low-affinity site, and then a linear steady-state rate, kcat, at 0.2 s−1 (Fig. 2E, purple trace, and SI Appendix, Fig. S3 B and C). In the presence of unblocked T-bulge DNA, the ATPase activity of MutS is strongly suppressed, although still detectable, as reported previously (Fig. 2E, blue and red traces, and SI Appendix, Fig. S3C) (41, 42). Upon monitoring this reaction over a longer timescale (20 s) (Fig. 2F) in the presence of unblocked T-bulge DNA, we observe a slow burst of two ATPs hydrolyzed per dimer at 0.15 s−1 (∼60-fold slower than the burst in the absence of DNA), which is followed by steady-state turnover at a kcat of 0.12 s−1 (Fig. 2F, blue trace). Note that this kcat value from experiments using unblocked DNA reflects the ATPase activity of MutS cycling between binding a mismatch, forming a clamp, sliding on DNA, and then off the free DNA ends; hence, it does not resolve whether the clamp continues to hydrolyze ATP while still on DNA.
In the presence of doubly end-blocked T-bulge DNA, we again observed a burst of two ATPs hydrolyzed per MutS dimer at 0.24 s−1, which is followed by steady-state turnover at a kcat of 0.06 s−1 (Fig. 2F, red trace). In this case, the MutS sliding clamp is expected to remain on DNA during the 20-s ATPase experiment given its reported ∼10-min lifetime on end-blocked DNA (24). Notably, the burst of ATP hydrolysis at 0.2 s−1 on both blocked and unblocked T-bulge DNA is comparable to the ∼0.4 s−1 rate at which Taq MutS is known to leave the mismatch site as a sliding clamp (24, 30, 43), suggesting that the clamp can hydrolyze ATP as (or right after) it releases the mismatch. Because ATP hydrolysis is not required for mobile clamp formation (10, 19, 24, 26, 28, 44–46), these observations suggest that the forces that drive mobile clamp formation (such as DNA unbending) (30) also promote ATP hydrolysis when possible.
The observed steady-state ATPase activity of MutS on the end-blocked T-bulge substrate after the burst associated with clamp formation indicates that mobile clamps repeatedly hydrolyze ATP while on DNA. If rebinding to the mismatch requires ATP hydrolysis, as our data suggest, the kcat of 0.06 s−1 would correspond to a 16-s mismatch release–rebind cycling time. This time is somewhat longer than the 4- to 5-s cycling time between rebinding events measured by our smFRET measurements (Fig. 1 and SI Appendix, Fig. S2). The discrepancy likely results from slight differences in bulk and single-molecule experimental conditions, and the averaging of mobile clamp heterogeneity in bulk experiments (e.g., if a subpopulation of mobile clamps dissociates from DNA following ATP-dependent mobile clamp formation, and binds the mismatch again from solution, the cycling time between rebinding events could appear longer in bulk measurements). Importantly, both the smFRET and presteady-state ATPase kinetics indicate that MutS mobile clamps hydrolyze ATP while on DNA, challenging current MMR models that envision long-lived ATP hydrolysis-independent clamps. It should also be noted that ATP hydrolysis by MutS mobile clamps on DNA is indirectly supported by prior reports showing that the clamp lifetime on end-blocked DNA depends on ATP concentration (supplementary figure 7 in ref. 24) and that MutS can promote DNA loop formation only in conditions permitting ATP hydrolysis (38, 39).
Our findings suggest that the function of ATP hydrolysis by MutS in MMR after mismatch recognition is not simply to reset the protein after it dissociates from DNA (10, 17, 24), but also to allow it to rebind the mismatch. Specifically, ATP hydrolysis by the MutS mobile clamp induces conformational changes that create another mobile clamp state that is now capable of rebinding the mismatch. Our studies cannot differentiate whether ATP hydrolysis by MutS mobile clamps promotes their active translocation on DNA or if they continue to diffuse passively but with an altered conformation. The latter idea is supported by previous studies of Taq MutS that demonstrated mobile clamps are diffusive in the presence of ATP (36). Accordingly, we suggest that MutS moves as a diffusive clamp between rebinding events independent of ATPase activity, and that ATP hydrolysis only alters the conformation of the clamp such that it can now rerecognize the mismatch, which is consistent with the two-step kinetic model required to fit the dwell-time distribution of FRET 0 events during rebinding (Fig. 1 I, Lower). Before this ATP hydrolysis-dependent conformational change occurs, the mobile clamp could repeatedly bypass the mismatch without recognizing it. As such, ATP hydrolysis licenses the diffusive mobile clamp to rebind the mismatch. This ATP hydrolysis-dependent clamp conformation may also have other functions yet to be discovered. In this regard, a recent study of yeast MMR revealed that MutSα (Msh2-Msh6) mutants with impaired ATP binding are defective for repair in vivo even though they continue to recruit MutLα (Mlh1-Pms1) to mismatched DNA in vitro (47), suggesting the ATPase properties of MutS mobile clamps may also be important in signaling downstream MMR events.
MutL Traps MutS into an Immobile State.
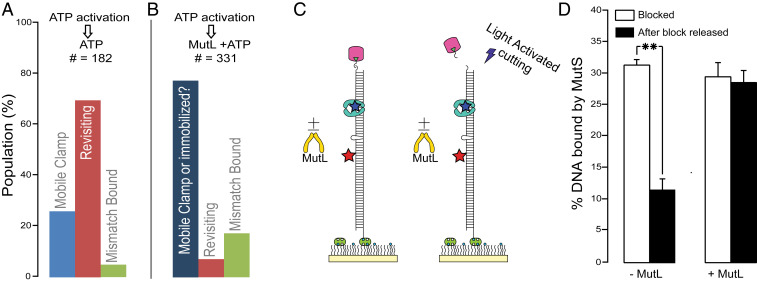
MutL recruitment is the next step in MMR after mismatch- and ATP-induced activation of MutS. We examined the impact of MutL by adding it simultaneously with ATP to preformed MutS mobile clamps (Fig. 1E, second buffer exchange). Strikingly, the fraction of revisiting clamps drops from 69 to 7%, and most of the MutS (76%) remains in a FRET 0 state (Fig. 3 A and B). This MutL-induced FRET 0 state of MutS could be due to MutL locking MutS into a mobile state that bypasses the mismatch or into an immobile state that is held distant (>10 nm) from the mismatch. To differentiate between these two possibilities, we 1) loaded single MutS mobile clamps onto DNA with a photocleavable end-block (Materials and Methods and Figs. 1E and 3C), 2) exchanged to buffer containing ATP or ATP+MutL (Fig. 1E, second buffer exchange), 3) removed the end-block by photocleavage (Fig. 3C and SI Appendix, Figs. S4 and S5), and 4) counted the number of DNAs colocalized with MutS. Releasing the end-block decreases the fraction of MutS-bound DNA from 31 to 12% in the absence of MutL (Fig. 3D and SI Appendix, Fig. S6), as mobile MutS clamps slide off the free DNA end (the clamps have a 2-s lifetime on the unblocked DNA substrate) (28). In the presence of MutL, however, the fraction of MutS-bound DNA remains unchanged (Fig. 3D and SI Appendix, Fig. S6), indicating that MutL traps MutS on DNA in an immobile state away from the mismatch (FRET 0). Again, this finding contrasts with some reports of mobile MutS–MutL clamps (17, 35), but is supported by other reports showing that MutL slows MutS release from DNA (19, 48–50) and that yeast MutSα–MutLα complexes exhibit the same slow dissociation rate on blocked and unblocked mismatched DNA (19).
Fig. 3.
Effect of MutL on MutS revisiting the mismatch. (A and B) Adding MutL with ATP in the second buffer exchange (B) greatly reduces the revisiting MutS population compared to ATP (A) alone. Color bars are as defined in Fig. 1F. In B the darker blue indicates that the FRET zero could arise from a mobile clamp that does not revisit the mismatch or an immobilized MutS–MutL complex far enough from the mismatch to yield no FRET emission from the acceptor. (C) Schematic of experiments with UV-photocleavable DNA end-block. (D) Fraction of Cy5–T-bulge DNAs colocalizing with AF555-MutS before (white bars) and after (black bars) end-block removal, in the absence (Left) and presence (Right) of MutL. This fraction decreases from 31 to 12% when clamps slide off DNA. Error bars are SEM from three experiments; **P ≤ 0.005 for two-sample unpaired t test; two-tailed P value (P = 0.005). The modest increase in mismatch-bound population upon addition of MutL (green; compare B to A) may be because it immobilizes revisiting MutS clamps at the mismatch, consistent with our previous finding that MutL can trap MutS at the mismatch before it forms a mobile clamp (18).
Discussion
Although several different models have been proposed for MMR signaling after mismatch recognition (11, 12, 17, 27, 35, 47, 51–56), a prevailing view envisions formation of an ATP hydrolysis-independent MutS–MutL sliding clamp, and is based on two widely accepted premises: 1) ATP or nonhydrolyzable ATP analogs activate mismatch-bound MutS to form a long-lived mobile clamp that appears to no longer recognize the mismatch (10, 17, 24, 35), and 2) in the E. coli methyl-directed MMR system, MutS–MutL must activate latent MutH endonuclease activity at hemimethylated GATC sites that can be up to 1,000 base pairs away. It is possible that MutS–MutL mobility after mismatch recognition is required for methyl-directed MMR in E. coli; indeed, E. coli MutS–MutL sliding clamps have been observed interacting with MutH, although the mobility of the E. coli MutS–MutL clamps is about a factor of 10 slower than MutS alone (17). In addition, recent studies of E. coli MMR suggest that MutL can be activated by MutS into a mobile MutL signaling clamp that can travel and activate MutH (13). MMR in most other organisms, however, is fundamentally different in that it does not require activation of an endonuclease (MutH) at distant, fixed sites; instead, the mobile processivity clamp (PCNA/β-clamp), which has a long lifetime on DNA (57), can activate MutL endonuclease in a stationary MutS–MutL complex on DNA.
Our findings challenge both underlying foundational ideas of the mobile MutS–MutL clamp model, at least for nonmethyl-directed MMR. First, we show that Taq MutS mobile clamps do hydrolyze ATP while bound to DNA, and this hydrolysis allows frequent revisits to the mismatch. Moreover, we find that Taq MutS–MutL complexes are immobilized on DNA, whether they form at or away from the mismatch (Fig. 3D) (18). Below, we discuss the implications of these findings individually and the resulting, reimagined model of MMR.
Free-Energy Considerations Reveal the Role of ATP Hydrolysis for MutS Mobile Clamp Mismatch Rebinding.
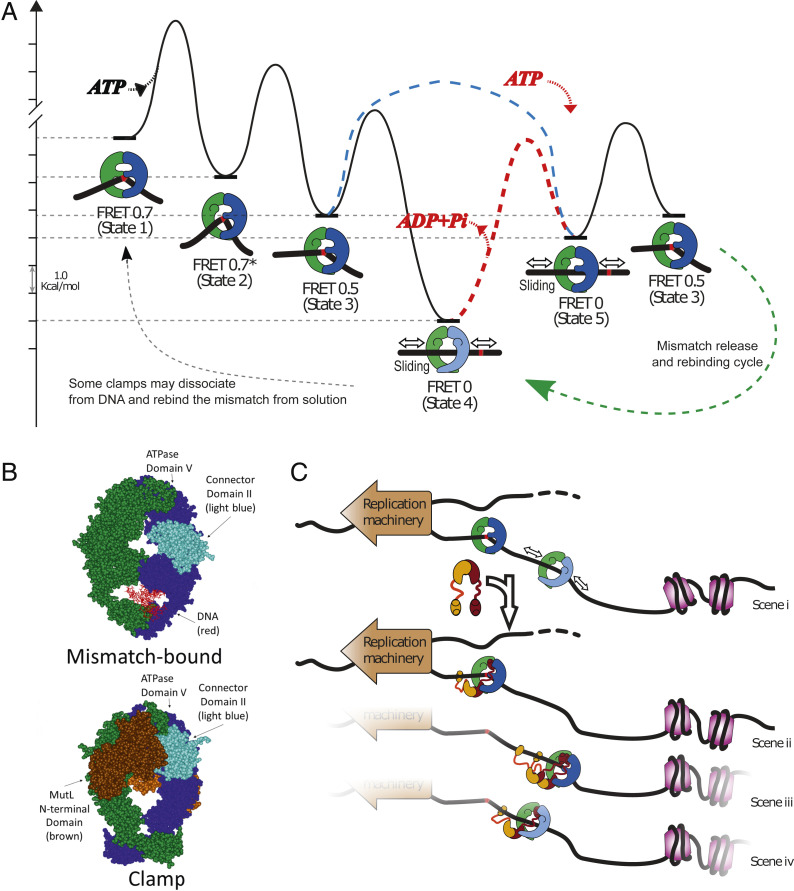
The ATP hydrolysis requirement for MutS mobile clamps to rebind the mismatch is expected based on energetics, because ATP binding alone allows MutS to progress in the opposite direction from a mismatch recognition state to a mobile clamp (10, 19, 24, 26, 28, 44–46). Specifically, ATP-γ-S is sufficient to drive formation of MutS mobile clamps, but it does not support mismatch rebinding by these clamps (Fig. 2). To quantify and visualize these arguments, we generated a free-energy landscape (Fig. 4A) based on our kinetic data (18, 28, 30). It should be noted that the lifetimes of all of the states will likely vary depending on experimental conditions, such as temperature and the model organism. Using Eyring theory (SI Appendix, Supporting Methods), we estimated transition free energies for MutS undergoing each step in the pathway from initial mismatch recognition (Fig. 4A, state 1), through the ATP-dependent conformational changes while bound to the mismatch (Fig. 4A, states 2 and 3) that lead to mobile clamp formation (Fig. 4A, state 4 or 5).
Fig. 4.
Modeling MutS and MutL actions in DNA MMR. (A) Free-energy landscape based on the rates of known steps (left to right) taken by MutS during transition from mismatch recognition (state 1) to mobile clamp (state 4) (SI Appendix, Supporting Methods). There are three known conformational states of the mismatch-bound MutS–DNA complex on the pathway to the mobile clamp (state 4): State 1: mismatch recognition and accompanying DNA bending (FRET 0.7); state 2: conformational change associated with increased DNA bending but no change in protein-DNA FRET (FRET 0.7*); state 3: additional conformational change associated with decreased DNA bending and lower protein-DNA FRET (FRET 0.5) (30). The initial mobile clamp (state 4) can undergo ATP-dependent conformational change to a new mobile clamp state (state 5), which can rebind the mismatch into intermediate state 3. The dashed red line indicates the event requiring ATP hydrolysis/nucleotide exchange (the red ATP binding and ADP+Pi leaving) to enable mismatch rebinding by the MutS clamp (the height of this transition is only illustrative, as it is unknown). The precise timing of ATP hydrolysis and ATP binding as MutS progress from state 4 through state 5 to state 3 remains undetermined. In addition, ATP hydrolysis could occur during the transition from the initial mismatch recognition state (state 1) to the mobile clamp state. Our experiments cannot determine the order in which states 4 and 5 occur upon initial mobile clamp formation with the light blue dashed line connecting state 3 directly to state 5 depicting an additional possibility for the first transition from initial mismatch recognition to mobile clamp. We only determine that once the mobile clamp is formed, ATP hydrolysis appears to be necessary to allow formation of state 5 (red pathway and red ATP binding/ADP+Pi release), which is capable of rebinding the mismatch. The light shading on MutS prior to this transition depicts a mobile clamp state (state 4) that is not capable of rebinding the mismatch. The dashed green line with arrow on the lower right represents the cycle of repetitive mismatch release and rebinding, including state 3, state 4, and state 5. The dashed gray line on the lower left represents stochastic events that could impact the ATPase turnover rate from bulk measurements (e.g., dissociation of a subpopulation of mobile clamps from DNA and rebinding to the mismatch from solution, instead of rebinding the mismatch on DNA). (B) Crystal structures compare E. coli MutS in the mismatch recognition state (1E3M.pdb) (Upper) (65) and the MutS–MutL sliding clamp state (5AKC.pdb) (Lower) (49). The connector domain II (residues 126 to 286) (light blue), ATPase domains (residues 568 to 800), the MutL N-terminal domain (brown), and the DNA (red) are indicated. The second MutL N-terminal domain on the back of the clamp is a lighter shade of brown. (C) MutS alone becomes a mobile clamp following mismatch detection (scene i). Its movement on DNA is bounded by the replication machinery and packaging of newly synthesized DNA (nucleosome assembly, purple), and may help keep DNA flanking the mismatch transiently clear for repair. The limited range of motion also increases the probability of repeated encounters between MutS and the mismatch and facilitates localization of MutS–MutL complexes nearby. (scenes ii to iv) One or more MutL proteins (burgundy and ochre) can stop MutS at the mismatch (scene ii, MutS in state 3) or away from the mismatch in mismatch rebinding (scene iii, MutS dark shading, state 5) or nonbinding (scene iv, MutS light shading, state 4) states.
The long-lived mobile clamp can adopt (at least) two different conformational states depending on ATP binding and hydrolysis. State 4 is the mobile clamp observed in the presence of ATP-γ-S, which is incapable of mismatch rebinding, and state 5 is the mobile clamp observed in the presence of ATP, which is competent to rebind the mismatch into state 3. The existence of two mobile clamp states of which only one is competent to rebind the mismatch, is consistent with the two-step kinetics required to fit the dwell-time distributions of the sliding state (FRET 0) between rebinding events (Fig. 1I). ATP hydrolysis is depicted as occurring between state 4 and state 5 because we observed that ATP is required to revisit the mismatch; however, during the initial mobile clamp formation in conditions that allow hydrolysis, it is possible that state 5 precedes state 4 (Fig. 4A, light blue dashed line). In the absence of hydrolysis, only state 4 is attained, and state 5 is not populated. Finally, on the transition from initial mismatch recognition to the first mobile clamp formation, ATP hydrolysis could occur somewhere between states 1 and 5.
For simplicity, we drew the energy landscape with state 4 preceding state 5 on the initial mismatch recognition pathway. The energy landscape illustrates the downhill path leading to the first mobile clamp (Fig. 4A, state 4), which is much more stable (lower free energy) than the mismatch-bound intermediate (state 3) in the absence of hydrolysis. This difference is calculated from the lifetime of state 3 (∼1 s) (18, 28, 30) and the lifetime of MutS mobile clamp on blocked DNA in the absence of ATP hydrolysis (∼600 s) (24), which suppresses mismatch rebinding by MutS (Fig. 2). ATP hydrolysis raises the energy of this mobile clamp, leading to a second mobile clamp state (state 5) that can rebind the mismatch. This diagram also illustrates why ATP-γ-S or AMPPNP is sufficient to drive formation of MutS mobile clamps (10, 19, 24, 26, 28, 44) because it is an energetically downhill process leading to the most stable state (steps 1 to 4). Subsequently, ATP hydrolysis is necessary to raise the energy level of the mobile clamp (state 4 or 5) and allow mismatch rebinding every 4 to 5 s. It also explains why mobile clamps predominately rebind the mismatch into state 3 (FRET 0.5) (Fig. 1 H and I) and not states 1 or 2 (FRET 0.7), because these latter states have higher free energies than state 3.
Structural Data Suggest ATP Hydrolysis by MutS Mobile Clamps Alters the Conformation of the DNA Binding Channel.
A key finding of our study is that ATP hydrolysis allows MutS clamps to rebind the mismatch. As discussed above, MutS exists in a mobile clamp state on DNA both before and after ATP hydrolysis, but in different conformations. Before the ATP hydrolysis-dependent conformational change, the mobile clamp may repeatedly bypass the mismatch without recognizing it, whereas after ATP hydrolysis the clamp can rebind the mismatch.
Insights into the mechanism whereby nucleotides control this process can be gleaned from a recent crystal structure of an E. coli MutS sliding clamp in complex with the N-terminal domains of MutL (49). In this structure, both nucleotide binding sites of MutS are occupied by the nonhydrolyzable ATP analog, AMPPNP. The DNA binding domains I are not visible, but the adjacent connector domains II are ordered and have moved away from DNA to interact with the ATPase domains V (Fig. 4B). Domains IV have tilted such that the DNA is pushed into a new channel opened by the outward motion of domains I and II. In this configuration, MutS forms a loose ring that is expected to slide freely on the DNA, consistent with solution data. The absence of well-ordered domains I that are essential for mismatch binding and the crossing of domains IV imply that MutS is unlikely to reengage the mismatch in this state. We envision this or a similar MutS conformation to be the nonrebinding mobile clamp state 4 in our model (Fig. 4A). Minimally, for this state to become competent for mismatch binding, domain IV must uncross and domain I of the mismatch-binding subunit must drop into a position that allows contact with the mismatch. Because the connector domain II interacts with ATPase domain V in this structure, the authors suggested that ATP binding stabilizes the connector domain in the open conformation (49). Taking this idea one step further, we suggest that ATP hydrolysis by this MutS mobile clamp (state 4) may release domain II, permitting domain I to interact with DNA again (state 5). Now when it encounters a mismatch while sliding on DNA, MutS is capable of binding it again (state 3). State 3 was previously suggested to have one domain I of the MutS dimer in contact with the mismatch and the other domain I open (30), much like the configuration described here for state 5; thus, state 5 would be primed to rebind a mismatch. This mechanochemical coupling model, based on structural and solution data, explains the need for ATP hydrolysis for a MutS mobile clamp to rebind the mismatch.
Previous studies have shown that MutL can trap MutS in state 3 before it leaves the mismatch (18, 30). Our present study demonstrates that MutL can also stop MutS mobile clamps after they leave the mismatch, indicating it binds MutS in either state 4 or state 5 or both. This interaction is possible since one MutS connector domain is open, exposing the MutL binding site, in both state 4 and state 5 (Fig. 4C). The ability of MutL to interact with multiple states of MutS on DNA, along with the ability of MutS to repetitively bind to the mismatch, likely increases the odds of initiating repair near the mismatch.
Conclusions
A comprehensive model of the actions undertaken by MutS and MutL to begin mismatch repair emerges from the studies discussed above (2, 53). Once MutS recognizes a mismatch, it undergoes an ordered sequence of conformational changes associated with ADP-to-ATP exchange that license its interaction with MutL (16–18, 23, 28, 30, 44, 49). If MutS is bound by MutL before leaving the mismatch as a mobile clamp, the MutS–MutL complex remains at or near the site (with MutS DNA binding domains I in dynamic motion) (18). If MutS leaves the mismatch site prior to MutL binding, ATP hydrolysis allows the clamp to reset to a conformation that can rebind the mismatch (Fig. 4A). MutS can continue repeating this ATPase-dependent cycle of sliding away and returning to the mismatch until it is found and immobilized by MutL. While the mismatch is free, additional MutS mobile clamps could be loaded onto the DNA. During DNA synthesis, the replication fork and assembling nucleosomes may function as barriers to restrict the range of MutS movement and increase the likelihood of repeated encounters with the mismatch and interaction with MutL near the mismatch (Fig. 4C). In both scenarios, MutS–MutL complexes would be localized in the vicinity of the mismatch, which, in addition to marking the DNA region needing repair, might also help keep it clear of proteins, such as nucleosomes, until the PCNA/β-clamp can activate MutL endonuclease. Localization of MutS–MutL complexes at or near the mismatch before initial clamp formation or after (by rebinding) provides a compelling explanation for the clustering of multiple MutL-catalyzed nicks near the mismatch observed in reconstituted human and yeast MMR (14, 58, 59). Constraining the nicks in the neighborhood of the mismatch likely enhances repair efficiency by providing multiple, local entry points for strand excision (59). The number of proteins and the stoichiometric ratio of MutS to MutL in the MutS–MutL repair initiation complexes will depend on protein concentrations as well as the timing of the events in the assembly pathway. For example, we previously found that Taq MutS–MutL complexes trapped at the mismatch contain one to three MutS proteins and one to four MutL proteins (more MutL relative to MutS) (18), similar to other in vitro (19, 58) and in vivo studies (12, 21), including our atomic force microscopy analysis of human MMR proteins (60).
In summary, our findings require a reenvisioning of MMR, at least in organisms that utilize nonmethyl-directed MMR, from a model in which MutS has a passive role after mismatch recognition simply functioning as an ATP hydrolysis-independent mobile platform that helps transport MutL to a distant strand discrimination signal, to a model in which ATP hydrolysis by MutS mobile clamps drives repeated rebinding to the mismatch, localizing formation of immobile MutS–MutL complexes in its vicinity. The next stage of repair would ensue when PCNA/β-clamp interacts with mismatch proximal complexes to activate MutL-catalyzed nicking of the daughter strand. These findings, combined with atomic force microscopy and tethered particle-motion data on human MutSα and MutLα in our companion paper (60), provide a view of MMR initiation, with MutS–MutL complexes assembling to mark and protect the region around a mismatch until the error-containing strand is nicked in preparation for excision and then resynthesis to complete MMR. Beyond MMR, given that DNA-damage induced apoptosis appears to be initiated by recognition of damage by MutS homologs and subsequent recruitment of MutL homologs (9, 61), it will be interesting in the future to examine whether MutS exhibits similar behavior upon interaction with damaged DNA.
Materials and Methods
DNA Substrates and Proteins.
The construction of mismatched, end-blocked DNA substrates has been described in detail elsewhere (18). To make photocleavable DNA, we used a modified oligonucleotide purchased from Integrated DNA Technologies with an internal photocleavable group (called Int PC Spacer;/iSpPC/) between the last 5′ base and the digoxigenin group in the digoxigenin modified primer (5′-/5DigN/iSpPC/GAG TCA GTG AGC GAG GAA GC-3′).
The expression and purification of Taq MutS and MutL proteins has been described previously (18, 28, 41, 43). MutS (M88C or E315C) was labeled with Alexa Fluor 555-maleimide (AF555) with labeling efficiency ranging from 60 to 100%. We have previously verified the mismatch binding affinity and ATPase activity of these MutS mutants is similar to wild-type Taq MutS (18, 30).
Further details about construction of smFRET DNA, about the end-blocked DNA used in the ensemble ATPase studies, and about proteins we used are available in the SI Appendix, Supporting Methods.
smFRET Assay Using End-Blocked DNA.
Flow chambers built between a quartz slide and a coverslip were passivated by Poly (Ethylene Glycol) (PEG), which included a small fraction of biotinylated-PEG (details in SI Appendix, Supporting Methods). The 550-base pair biotin/dig Cy5-T-bulge DNA was added to a passivated flow chamber whose surface was treated with streptavidin to achieve well-spaced immobilized DNA molecules. Antidigoxigenin (anti-dig) antibody (Roche Diagnostics, 11333089001) was then added to the chamber at 20 μg/mL for 15 min to block the free end of the immobilized DNA. To minimize the number of DNA molecules with multiple MutS mobile clamps, the experimental protocol (Fig. 1E) was optimized by first incubating MutS with DNA in buffer without ATP, resulting in MutS bound stably at the mismatch, which in turn blocks additional MutS binding. Specifically, 5 nM (dimer; 10 nM monomer) AF555 Taq MutS was injected into the chamber to bind the T-bulge (bufferA: 20 mM Tris⋅HCl, pH 7.8, 100 mM sodium acetate, and 5 mM MgCl2). After 15 min, the chamber was washed with bufferA containing 2 mM ATP to 1) remove unbound MutS molecules from solution and prevent additional MutS binding to DNA, and 2) activate conformational changes in mismatch-bound MutS for conversion into mobile clamps. This protocol results in few DNAs being loaded with multiple MutS mobile clamps, and about 10% of DNA loaded with a single MutS mobile clamp (the remaining DNA did not have MutS bound) (SI Appendix, Table S1). SI Appendix, Fig. S1 shows that 80% of MutS colocalized with DNA is at the mismatch before addition of ATP, and 5% remains at the mismatch after addition of ATP (nonzero, stable FRET), confirming that ATP efficiently converts mismatch-bound MutS to mobile clamps (SI Appendix, Fig. S1). We have previously established that a mismatch is required for efficient MutS loading onto surface immobilized DNA, as negligible MutS binding is detected in our assay on fully matched duplex DNA regardless of the presence of nucleotides in the buffer (18, 28, 30, 62). After MutS mobile clamps were established, the buffer was exchanged to bufferA containing various nucleotides as indicated in the text and figures (2 mM ATP, 2 mM ADP, 2 mM ATP-γ-S, or 0.1 mM ADP + 0.1 mM ATP-γ-S). In experiments with MutL, the solutions contained bufferA augmented with 2 mM ATP + 200 nM MutL. For final imaging, all experiments also included glucose oxidase, catalase, and 2% (wt/vol) glucose in the buffers for oxygen scavenging and 100 μM cyclooctatetraene for triplet-state quenching (imaging buffer).
The prism-type total internal reflection, single-molecule fluorescence microscope, and our data analysis routines have been described previously (18, 30, 62, 63). FRET efficiency is calculated as E = IA/(IA + ID), where IA and ID are background and leakage corrected acceptor and donor intensities, respectively. Additional details about the microscope and data analysis are available in SI Appendix, Supporting Methods. Note that while most of the revisiting events were transitions between FRET 0 and 0.5, about 2% exhibited transitions from FRET 0 to 0.5 to 0.7, which matches the levels in initial mismatch binding events shown in Fig. 1C (0.7 → 0.5 → 0). For population analyses, single-molecule fluorescence or FRET time traces with donor lifetime shorter than 100 s or donor number over 2 were rejected. Traces with at least one sequence of transitions of zero → nonzero → zero were categorized as FRET-switching molecules. Traces with constant zero FRET were categorized as “FRET zero” molecules while traces with constant nonzero FRET were categorized as “FRET non-zero.” Note that nonspecifically surface-attached MutS that randomly colocalize with a DNA will falsely contribute to the “FRET zero” population.
DNA End-Block Release Assay.
For experiments monitoring MutS clamps on photocleavable end-blocked DNA, 550-base pair Cy5–T-bulge DNA containing biotin on one end and the photocleavable digoxigenin on the other was immobilized on the surface of PEG-passivated, streptavidin-coated chambers and blocked with anti-dig as described above. To remove the anti-dig end-block, the chambers were exposed to 365-nm light (Doctor UV, DRUV-HH-365HP, 7 Watt, at 1-cm distance, 36 W/cm2) for a total of 90 s, smoothly moving the 5-mm diameter illumination spot around in the 5-mm × 20-mm chamber. SI Appendix contains characterization of the efficiency of the photocleavage reaction (SI Appendix, Supporting Methods and Fig. S4) and controls (SI Appendix, Supporting Methods Figs. S4 and S5).
To determine the effects of ATP and MutL on MutS clamps sliding on photocleavable end-blocked DNA (Fig. 3C), AF555-MutS was first injected to chambers prepared with photocleavable, end-blocked Cy5–T-bulge DNA in buffer without any nucleotide for 15 min. Buffer exchange with ATP-containing buffer (no additional MutS) then established single MutS mobile clamps on the end-blocked DNA. Next, imaging buffer containing 2 mM ATP or 2 mM ATP + 200 nM MutL was injected into the chamber. Two chambers were prepared identically in this way, and one was exposed to UV light for 90 s to remove the end-block while the other was left alone for 90 s before measuring MutS occupancy. All buffer wash volumes and incubation times were kept uniform across all four conditions tested in Fig. 3D to minimize any effects from spontaneous MutS dissociation [MutS mobile clamp lifetime on blocked DNA is 10 min (24)]. The fraction of MutS-bound DNAs was calculated as the number of MutS molecules colocalized with DNA over the number of DNA molecules. All experiments were repeated three times with sets of independently prepared chambers. Each repeat included three videos of 40-s length under each condition. No traces were rejected based on a short time to photobleaching.
Further evidence that releasing the end-block permits the MutS mobile clamp to slide off the resulting free end is provided by the decrease in the mismatch revisiting MutS fraction from 46% (end-blocked DNA) to 6% (end-block released) in the absence of MutL (SI Appendix, Fig. S6 A and B). In the presence of MutL, 2% of the MutS clamps revisit the mismatch on end-blocked DNA and this fraction remains unchanged upon releasing the end-block (SI Appendix, Fig. S6B), which also supports the conclusion that MutL stops MutS clamp movement on DNA. Traces with at least one zero → nonzero → zero FRET transition were categorized as revisiting molecules. The fraction of revisiting was calculated as the number of revisiting MutS molecules over the number of MutS colocalized with DNA.
ATPase and Gel Mobility-Shift Assays, Energy Landscape Construction, and Diffusion Calculations.
SI Appendix contains additional methodological details about the ATP hydrolysis and Pi release assay with end-blocked DNA, the gel mobility-shift assay of MutS–DNA complexes, construction of the energy landscape (Fig. 4A), and estimating the number of times diffusive MutS sliding clamps cross the mismatch between rebinding events.
Supplementary Material
Acknowledgments
We thank Paul Modrich, Tom Kunkel, and Hong Wang for critical reading of the manuscript. This research funded by National Institute of General Medical Sciences Grants R01 GM109832 (to D.A.E. and K.R.W.), R01 GM079480 and R35 GM127151 (to D.A.E.), R01 GM132263 (to K.R.W.), R15 GM114743 (to M.M.H.), R35 GM127145 (to T.C.E.), and F32 GM123602 (to S.J.L.); and National Cancer Institute Grant K01 CA218304 (to S.J.L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.T. is a guest editor invited by the Editorial Board.
Data deposition: The data for this paper are available from Dryad, https://doi.org/10.5061/dryad.612jm641d.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918517117/-/DCSupplemental.
Data Availability.
Primary data for this paper are available from Dryad at https://doi.org/10.5061/dryad.612jm641d.
References
- 1.Modrich P., Mechanisms in E. coli and human mismatch repair (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55, 8490–8501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunkel T. A., Erie D. A., Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49, 291–313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boland C. R., Lynch H. T., The history of Lynch syndrome. Fam. Cancer 12, 145–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch H. T., Snyder C. L., Shaw T. G., Heinen C. D., Hitchins M. P., Milestones of Lynch syndrome: 1895-2015. Nat. Rev. Cancer 15, 181–194 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Lebbink J. H., Drost M., de Wind N., DNA mismatch repair: From biophysics to bedside. DNA Repair (Amst.) 38, 1–2 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Heinen C. D., Mismatch repair defects and Lynch syndrome: The role of the basic scientist in the battle against cancer. DNA Repair (Amst.) 38, 127–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martín-López J. V., Fishel R., The mechanism of mismatch repair and the functional analysis of mismatch repair defects in Lynch syndrome. Fam. Cancer 12, 159–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh P., Yamane K., DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 129, 391–407 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta D., Heinen C. D., The mismatch repair-dependent DNA damage response: Mechanisms and implications. DNA Repair (Amst.) 78, 60–69 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Gradia S. et al., hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell 3, 255–261 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L., DNA mismatch repair: Functions and mechanisms. Chem. Rev. 106, 302–323 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hombauer H., Campbell C. S., Smith C. E., Desai A., Kolodner R. D., Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell 147, 1040–1053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mardenborough Y. S. N. et al., The unstructured linker arms of MutL enable GATC site incision beyond roadblocks during initiation of DNA mismatch repair. Nucleic Acids Res. 47, 11667–11680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadyrov F. A., Dzantiev L., Constantin N., Modrich P., Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126, 297–308 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Pluciennik A. et al., PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 107, 16066–16071 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acharya S., Foster P. L., Brooks P., Fishel R., The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell 12, 233–246 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Liu J. et al., Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature 539, 583–587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu R. et al., MutL traps MutS at a DNA mismatch. Proc. Natl. Acad. Sci. U.S.A. 112, 10914–10919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendillo M. L., Mazur D. J., Kolodner R. D., Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J. Biol. Chem. 280, 22245–22257 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Reyes G. X., Schmidt T. T., Kolodner R. D., Hombauer H., New insights into the mechanism of DNA mismatch repair. Chromosoma 124, 443–462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elez M., Radman M., Matic I., Stoichiometry of MutS and MutL at unrepaired mismatches in vivo suggests a mechanism of repair. Nucleic Acids Res. 40, 3929–3938 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elez M. et al., Seeing mutations in living cells. Curr. Biol. 20, 1432–1437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hingorani M. M., Mismatch binding, ADP-ATP exchange and intramolecular signaling during mismatch repair. DNA Repair (Amst.) 38, 24–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong C. et al., MutS switches between two fundamentally distinct clamps during mismatch repair. Nat. Struct. Mol. Biol. 18, 379–385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lebbink J. H. et al., Magnesium coordination controls the molecular switch function of DNA mismatch repair protein MutS. J. Biol. Chem. 285, 13131–13141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazur D. J., Mendillo M. L., Kolodner R. D., Inhibition of Msh6 ATPase activity by mispaired DNA induces a Msh2(ATP)-Msh6(ATP) state capable of hydrolysis-independent movement along DNA. Mol. Cell 22, 39–49 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Jiang J. et al., Detection of high-affinity and sliding clamp modes for MSH2-MSH6 by single-molecule unzipping force analysis. Mol. Cell 20, 771–781 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Qiu R. et al., Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. EMBO J. 31, 2528–2540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yildiz A. et al., Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 (2003). [DOI] [PubMed] [Google Scholar]
- 30.LeBlanc S. J. et al., Coordinated protein and DNA conformational changes govern mismatch repair initiation by MutS. Nucleic Acids Res. 46, 10782–10795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonik M., Felekyan S., Gaiduk A., Seidel C. A. M., Separating structural heterogeneities from stochastic variations in fluorescence resonance energy transfer distributions via photon distribution analysis. J. Phys. Chem. B 110, 6970–6978 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Choi U. B., McCann J. J., Weninger K. R., Bowen M. E., Beyond the random coil: Stochastic conformational switching in intrinsically disordered proteins. Structure 19, 566–576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santoso Y., Torella J. P., Kapanidis A. N., Characterizing single-molecule FRET dynamics with probability distribution analysis. ChemPhysChem 11, 2209–2219 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Nir E. et al., Shot-noise limited single-molecule FRET histograms: Comparison between theory and experiments. J. Phys. Chem. B 110, 22103–22124 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorman J. et al., Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 109, E3074–E3083 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho W.-K. et al., ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure 20, 1264–1274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blackwell L. J., Wang S., Modrich P., DNA chain length dependence of formation and dynamics of hMutSalpha.hMutLalpha.heteroduplex complexes. J. Biol. Chem. 276, 33233–33240 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Allen D. J. et al., MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 16, 4467–4476 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blackwell L. J., Martik D., Bjornson K. P., Bjornson E. S., Modrich P., Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 273, 32055–32062 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Antony E., Hingorani M. M., Mismatch recognition-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry 42, 7682–7693 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antony E., Hingorani M. M., Asymmetric ATP binding and hydrolysis activity of the Thermus aquaticus MutS dimer is key to modulation of its interactions with mismatched DNA. Biochemistry 43, 13115–13128 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs-Palmer E., Hingorani M. M., The effects of nucleotides on MutS-DNA binding kinetics clarify the role of MutS ATPase activity in mismatch repair. J. Mol. Biol. 366, 1087–1098 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma A., Doucette C., Biro F. N., Hingorani M. M., Slow conformational changes in MutS and DNA direct ordered transitions between mismatch search, recognition and signaling of DNA repair. J. Mol. Biol. 425, 4192–4205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monti M. C. et al., Native mass spectrometry provides direct evidence for DNA mismatch-induced regulation of asymmetric nucleotide binding in mismatch repair protein MutS. Nucleic Acids Res. 39, 8052–8064 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Junop M. S., Obmolova G., Rausch K., Hsieh P., Yang W., Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell 7, 1–12 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Selmane T., Schofield M. J., Nayak S., Du C., Hsieh P., Formation of a DNA mismatch repair complex mediated by ATP. J. Mol. Biol. 334, 949–965 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Graham W. J. 5th, Putnam C. D., Kolodner R. D., The properties of Msh2-Msh6 ATP binding mutants suggest a signal amplification mechanism in DNA mismatch repair. J. Biol. Chem. 293, 18055–18070 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schofield M. J., Nayak S., Scott T. H., Du C., Hsieh P., Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J. Biol. Chem. 276, 28291–28299 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Groothuizen F. S. et al., MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. eLife 4, e06744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhairosing-Kok D. et al., Sharp kinking of a coiled-coil in MutS allows DNA binding and release. Nucleic Acids Res. 47, 8888–8898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hura G. L. et al., DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proc. Natl. Acad. Sci. U.S.A. 110, 17308–17313 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiricny J., Postreplicative mismatch repair. Cold Spring Harb. Perspect. Biol. 5, a012633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kunkel T. A., Erie D. A., DNA mismatch repair. Annu. Rev. Biochem. 74, 681–710 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Liu D., Keijzers G., Rasmussen L. J., DNA mismatch repair and its many roles in eukaryotic cells. Mutat. Res. 773, 174–187 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Lenhart J. S., Pillon M. C., Guarné A., Biteen J. S., Simmons L. A., Mismatch repair in Gram-positive bacteria. Res. Microbiol. 167, 4–12 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Li Y., Schroeder J. W., Simmons L. A., Biteen J. S., Visualizing bacterial DNA replication and repair with molecular resolution. Curr. Opin. Microbiol. 43, 38–45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao N. et al., Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells 1, 101–113 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Kadyrov F. A. et al., Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J. Biol. Chem. 282, 37181–37190 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kadyrov F. A. et al., A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 106, 8495–8500 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bradford K. C. et al., Dynamic human MutSα–MutLα complexes compact mismatched DNA. Proc. Natl. Acad. Sci. U.S.A. 117, 16302–16312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiricny J., The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 7, 335–346 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Sass L. E., Lanyi C., Weninger K., Erie D. A., Single-molecule FRET TACKLE reveals highly dynamic mismatched DNA-MutS complexes. Biochemistry 49, 3174–3190 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gauer J. W. et al., Single-molecule FRET to measure conformational dynamics of DNA mismatch repair proteins. Methods Enzymol. 581, 285–315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Obmolova G., Ban C., Hsieh P., Yang W., Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407, 703–710 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Lamers M. H. et al., The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature 407, 711–717 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primary data for this paper are available from Dryad at https://doi.org/10.5061/dryad.612jm641d.