Significance
p53 is the most frequently mutated, well-studied tumor-suppressor gene, yet the switch from p53-induced cell-cycle arrest to apoptosis remains poorly understood. We unexpectedly identify a critical role for the only human pseudo-caspase FLIP(L) in regulating this switch. Genetically or pharmacologically inhibiting expression of FLIP(L), which we identify as a new p53 transcriptional target, promoted apoptosis over cell-cycle arrest in response to p53 activation. Mechanistically, FLIP(L) inhibited p53-induced apoptosis by blocking ligand-independent activation of caspase-8 by the TRAIL-R2/DR5 death receptor. Notably, FLIP(L) also modulated transcription of canonical p53 target genes: increasing expression of cell-cycle regulator p21 and suppressing upregulation of apoptosis-inducing PUMA. Thus, we report unexpected, therapeutically exploitable roles for FLIP(L) in regulating the switch from p53-induced cell-cycle arrest to apoptosis.
Keywords: p53, apoptosis, FLIP, TRAIL-R2, entinostat
Abstract
p53 is the most frequently mutated, well-studied tumor-suppressor gene, yet the molecular basis of the switch from p53-induced cell-cycle arrest to apoptosis remains poorly understood. Using a combination of transcriptomics and functional genomics, we unexpectedly identified a nodal role for the caspase-8 paralog and only human pseudo-caspase, FLIP(L), in regulating this switch. Moreover, we identify FLIP(L) as a direct p53 transcriptional target gene that is rapidly up-regulated in response to Nutlin-3A, an MDM2 inhibitor that potently activates p53. Genetically or pharmacologically inhibiting expression of FLIP(L) using siRNA or entinostat (a clinically relevant class-I HDAC inhibitor) efficiently promoted apoptosis in colorectal cancer cells in response to Nutlin-3A, which otherwise predominantly induced cell-cycle arrest. Enhanced apoptosis was also observed when entinostat was combined with clinically relevant, p53-activating chemotherapy in vitro, and this translated into enhanced in vivo efficacy. Mechanistically, FLIP(L) inhibited p53-induced apoptosis by blocking activation of caspase-8 by the TRAIL-R2/DR5 death receptor; notably, this activation was not dependent on receptor engagement by its ligand, TRAIL. In the absence of caspase-8, another of its paralogs, caspase-10 (also transcriptionally up-regulated by p53), induced apoptosis in Nutlin-3A-treated, FLIP(L)-depleted cells, albeit to a lesser extent than in caspase-8-proficient cells. FLIP(L) depletion also modulated transcription of canonical p53 target genes, suppressing p53-induced expression of the cell-cycle regulator p21 and enhancing p53-induced up-regulation of proapoptotic PUMA. Thus, even in the absence of caspase-8/10, FLIP(L) silencing promoted p53-induced apoptosis by enhancing PUMA expression. Thus, we report unexpected, therapeutically relevant roles for FLIP(L) in determining cell fate following p53 activation.
The mechanisms through which p53 activation differentially regulates cell survival (by initiating cell-cycle arrest and DNA damage repair) and cell death (by promoting apoptosis) remain enigmatic (1). Canonically, phosphorylation of the p53 N terminus blocks its interaction with the E3 ubiquitin ligase Mouse Double Minute 2 (MDM2) (2) reducing ubiquitination of the C terminus and stabilizing p53. Stabilized p53 directly activates target genes to induce cell-cycle arrest mediated by the cyclin-dependent kinase inhibitor p21/CDKN1A and other effectors (3, 4). In the event of sustained stress and/or irreparable DNA damage, sequential posttranslational modification of p53 is thought to direct p53 activity toward proapoptotic target genes, including those encoding BAX, PUMA, and NOXA (5, 6) and the death receptor TRAIL receptor 2 (TRAIL-R2 also known as death receptor-5, DR5) (7). We and others have contributed to progress in identifying direct and indirect p53 target genes using chromatin immunoprecipitation sequencing (ChIP-Seq) (8) (summarized in ref. 9); however, the mechanisms underlying the switch from cell-cycle arrest to cell death induction remain poorly understood, although p53-mediated transcriptional up-regulation of PUMA has been suggested to be crucial in a number of studies (10, 11).
Despite p53/TP53 being the most frequently mutated gene in cancer, ∼50% of all tumors retain wild-type p53 (WT-p53) and typically circumvent or suppress p53’s functions through alternative nonmutational mechanisms (12). Reawakening the latent tumor suppressive functions in cancers retaining WT-p53 therefore is an attractive clinical concept (12). Targeting the p53-MDM2 interaction has received significant attention and has led to the development of the selective small molecule inhibitors of MDM2 (MDM2i) such as Nutlin-3A (13). While highly effective at stabilizing p53 (12), such agents promote variable cellular fates depending on cell type in all but MDM2-amplified cells, in which they induce cell death (14). The observation that potent p53 stabilization elicited by MDM2i is often insufficient to induce cell death suggests the existence of fundamental mechanisms that regulate the cell death-inducing effects of stabilized p53. Strategies targeting such p53-mediated cell death inhibitory mechanisms have potential to not only enhance MDM2i-induced cell death, but also to augment the efficacy of p53-induced cell death induced by radiotherapy and DNA damaging chemotherapies.
In this study, we unexpectedly identify a critical role for the apoptosis regulator and only human pseudo-caspase FLIP(L) (Fas-associated death domain [FADD]-like interleukin-1β-converting enzyme inhibitory protein) in regulating the switch from p53-induced cell-cycle arrest to apoptosis. Importantly, this biology is therapeutically exploitable using class-I HDAC inhibitors, which prevent p53-mediated transcriptional up-regulation of FLIP(L) leading to synergistic inductions of apoptosis when these agents are combined with Nutlin-3A or DNA damaging chemotherapy.
Results
FLIP(L) Inhibits p53-Dependent Apoptosis.
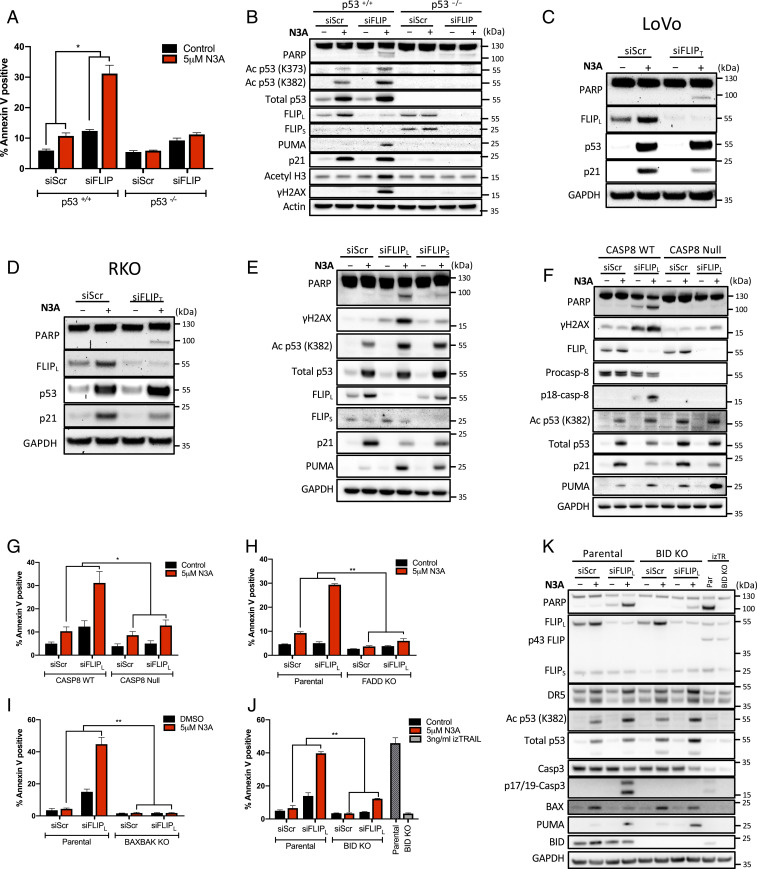
Despite potent effects on p53 stabilization, the specific MDM2 inhibitor Nutlin-3A preferentially induces cell-cycle arrest in p53-WT colorectal cancer (CRC) cells (15). To identify critical regulators of p53-dependent CRC cell death, we evaluated the effect of Nutlin-3A in p53-WT HCT116 cells transfected with siRNA pools targeting key antiapoptotic regulators. Only three of these enhanced the effects of Nutlin-3A: Mcl-1, cIAP1, and FLIP, with the depletion of the latter having the greatest effect (SI Appendix, Fig. S1A). As controls, we used an siRNA pool targeting p53, which reversed growth inhibition in response to Nutlin-3A, and a p53-null HCT116 daughter cell line (SI Appendix, Fig. S1A). Moreover, we also identified sgRNAs targeting CFLAR/FLIP as a sensitizer to Nutlin-3A in gene-level model-based analysis of genome-wide CRISPR/Cas9 knockout (MAGeCK) analysis (16) of a genome-wide CRISPR screen (17) (SI Appendix, Fig. S1B and Dataset S1 A and B). We confirmed the predictions of the siRNA and CRISPR screens by demonstrating quantitatively (Fig. 1A) and qualitatively (using PARP cleavage as a marker; Fig. 1B) that RNAi-mediated depletion of FLIP synergistically enhanced apoptotic cell death induced by Nutlin-3A in a p53-dependent manner (Fig. 1 A and B). These results were further confirmed in LoVo and RKO p53-WT CRC cell line models (Fig. 1 C and D and SI Appendix, Fig. S1 C and D). Moreover, splice form-specific siRNA indicated that selective FLIP(L) depletion was sufficient to promote Nutlin-3A-induced cell death, with depletion of FLIP(S) having only a modest effect on PARP cleavage (Fig. 1E). These results also demonstrate that in the presence of FLIP(L), Nutlin-3A induces very little apoptosis in p53-WT CRC models, with high levels of p21 induction (Fig. 1 B–D) driving cell-cycle arrest in G1 and G2 (18).
Fig. 1.
FLIP(L) inhibits p53-dependent apoptosis. (A) Annexin-V/PI FACS and (B) Western blot assessment of cell death in HCT116 p53 WT and null cells transfected with 10 nM FLIP or scrambled control (siScr) siRNA for 6 h prior to treatment with 5 µM Nutlin-3A (N3A) for a further 24 h. Western blot assessment of cell death in LoVo (C) and RKO (D) p53 WT cells transfected for 6 h with 10 nM siScr or a FLIPT-specific siRNA prior to treatment with 5 μM N3A for a further 24 h. (E) Western blot analysis of HCT116 p53 WT cells transfected with 10 nM FLIPL- or FLIPS-specific siRNAs or control siRNA for 6 h prior to treatment with 5 μM N3A for a further 24 h. (F–K) Western blot and Annexin-V/PI FACS analysis of effects of siRNA-mediated depletion of FLIPL in combination with 5 µM N3A as in A and B in HCT116 isogenic models for caspase-8 (CASP8) (F and G), FADD (H), BAX/BAK DKO (I), and BID (J and K) (treatment with 3 ng/mL isoleucine zipper TRAIL (IZTRAIL) serves as a positive control). Error bars in A and G–J are represented as mean ± SEM of at least three independent experiments. P values *P < 0.05; **P < 0.01; calculated by two-way ANOVA.
The best-described function of FLIP is its role as a regulator of caspase-8-dependent apoptosis. Consistent with this, a caspase-8-deficient HCT116 model (19) was significantly more resistant to the cell death-inducing effects of Nutlin-3A/siFLIP(L) than parental cells (Fig. 1 F and G). FLIP and caspase-8 interact via the adaptor FADD (20), and the cell death induced by Nutlin-3A/siFLIP(L) was also FADD dependent (Fig. 1H and SI Appendix, Fig. S1E). HCT116 cells have been identified as “type-2” in terms of caspase-8-mediated apoptosis (21), meaning that they require amplification of extrinsically derived apoptotic signals via the mitochondria. Consistent with this, the apoptosis induced by Nutlin-3A/siFLIP(L) was completely attenuated in BAX/BAK double knockout (DKO) cells (Fig. 1I and SI Appendix, Fig. S1F) and also partially attenuated in SMAC null cells (SI Appendix, Fig. S1G). In the extrinsic pathway, crosstalk between caspase-8 and the mitochondria is mediated by the BH3 protein BID, which when cleaved by caspase-8 to its truncated form (tBID) translocates to the mitochondria and promotes mitochondrial outer membrane permeabilization (MOMP). In line with this, the mechanism of apoptosis induced by Nutlin-3A/siFLIP(L) was significantly attenuated, although not completely prevented, in BID knockout cells (Fig. 1 J and K). By comparison, the effects of the direct extrinsic apoptosis pathway activator isoleucine zipper-TRAIL (IZ-TRAIL) were completely abrogated by BID deletion, confirming this model as apoptotically type-2 (Fig. 1 J and K).
FLIP(L) Is a Direct, Acetylation-Dependent p53-Target Gene.
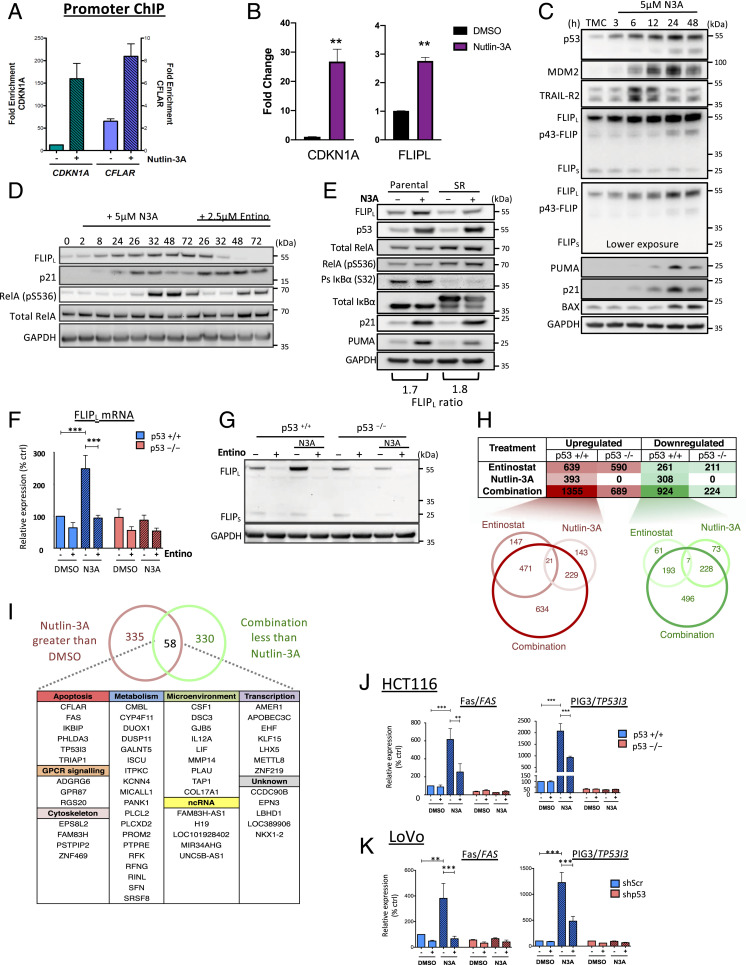
From the Western blot analyses in Fig. 1, it was apparent that Nutlin-3A treatment up-regulated expression FLIP(L) in p53-WT CRC cells. Reanalysis of previous ChIP-Seq data from our group in human foreskin keratinocytes treated with cisplatin (8) or publicly available ChIP-Seq data from p53-WT MCF7 breast cancer cells treated with Nutlin-3A (22) identified direct, treatment-induced binding of p53 to the CFLAR/FLIP promoter (SI Appendix, Fig. S2A), and we confirmed this in HCT116 cells following treatment with Nutlin-3A (Fig. 2A). This coincided with induction of transcription of FLIP(L) as detected by qRT-PCR (Fig. 2B), which was reflected at the protein level for FLIP(L) but not for FLIP(S), which was expressed at much lower levels than the long splice form (Fig. 2C). FLIP is a known NF-κB target (23); however, time course analysis revealed that FLIP(L) protein expression was induced in response to Nutlin-3A prior to activation of the canonical NF-κB pathway as assessed by phospho-RelA (Fig. 2D and SI Appendix, Fig. S2B). In addition, while the absolute levels of FLIP(L) expression was decreased by both the IκB-superrepressor (24) and the IKK inhibitor BAY-117082 (confirming CFLAR/FLIP as an NF-κB target gene), the magnitude of p53-dependent induction of FLIP(L) was maintained (Fig. 2E and SI Appendix, Fig. S2C). Up-regulation of FLIP(L) coincided with TRAIL-R2 up-regulation, both of which preceded up-regulation of the canonical p53 apoptotic targets BAX and PUMA (Fig. 2C).
Fig. 2.
FLIP(L) is a direct, acetylation-dependent p53-target gene. (A) p53 ChIP assay in p53 WT HCT116 cells using primers directed against the FLIP/CFLAR promoter region. Relative promoter occupancy between treatments was calculated by fold enrichment of target region versus a nonspecific region (Cyclin D1/CCND1). (B) Quantitative RT-PCR of p21/CDKN1A and FLIPL/CFLAR mRNA expression following treatment with 5 µM Nutlin-3A (N3A) for 24 h. (C) Western blot assessment of HCT116 parental cells treated with 5 µM N3A for the indicated times (TMC, time matched control). (D) Western blot analysis of p53 WT HCT116 cells treated with 5 µM N3A with or without 2.5 µM Entinostat (Entino) for the indicated times. (E) HCT116 IkB-superrepressor (SR) and matched parental cells treated with 5 μM N3A for 24 h. (F and G) Quantitative RT-PCR and Western blot analysis of FLIP expression in p53 WT and null HCT116 cells treated with 5 µM N3A with or without 2.5 µM Entino for 8 (F) or 24 h (G). (H) Differential gene expression analysis of mRNA-Seq performed in HCT116 p53 WT and null cells from two independent experiments. RNA was extracted 8 h posttreatment with dimethyl sulfoxide (DMSO), 5 μM N3A, 2.5 μM Entino, or their combination. Mapped read counts for transcripts were RPKM (reads per kilobase per million) normalized for relative expression and differentially expressed transcripts between each treatment detected by three-way ANOVA (mean RPKM ≥1.7x, false discovery rate [FDR] 0.05). The total numbers of significantly up/down-regulated genes vs. DMSO for each treatment are tabulated. Venn diagrams illustrating numbers of transcripts significantly up-regulated (red) or down-regulated (green) in response to Entino, N3A, and the combination in p53 WT cells. (I) Venn diagram of numbers of transcripts whose transcriptional up-regulation in response to N3A is significantly attenuated in the combination treatment, and list of those 58 genes subcategorized based on cellular function. (J and K) Quantitative RT-PCR of Fas/FAS and PIG3/TP53I3 in p53 WT and null HCT116 (J) and LoVo (K) cells treated as in F and SI Appendix, Fig. S2D. Data in B, F, J, and K are normalized to RPL24 control for each sample and represented as mean ± SEM of three independent experiments. **P < 0.01; ***P < 0.001 calculated by Student’s t test (B) or two-way ANOVA (F, J, and K). Data in A are represented as mean ± SD of two independent experiments.
Acetylation of a dense cluster of lysine residues in p53’s C terminus has been suggested to modulate transactivation of apoptotic target genes (25, 26), and we previously found that the clinically relevant class-I histone deacetylase (HDAC) inhibitor Entinostat inhibits FLIP expression in several cancer models (27, 28). It was therefore notable that Nutlin-3A-induced FLIP(L) protein and mRNA expression was attenuated by cotreatment with Entinostat in CRC models (Fig. 2 F and G and SI Appendix, Fig. S2 D and E). To identify other acutely induced p53 target genes affected similarly to FLIP by Entinostat, we conducted mRNA-Seq of samples collected after an 8-h treatment, a timepoint which precedes activation of apoptotic cell death and at which p53 was stabilized with significant increases in expression of direct p53 targets p21/CDKN1A, PUMA/BBC3, and TRAIL-R2/TNFRSF10B (SI Appendix, Fig. S2 F and G). As expected mRNA-Seq analysis revealed that Nutlin-3A alone caused a significant increase (≥1.7-fold) in 393 genes enriched for p53 signaling, apoptosis, and direct p53 target genes (Fig. 2H and Datasets S2 A–D and S3A data viewable in HDAC_visualizeR Shiny App). No significantly altered genes were identified in the p53-null model in response to Nutlin-3A, underlining the selectivity of this MDM2 inhibitor. Even at this early timepoint, 308 significantly repressed genes were identified; these were enriched for cell cycle and FOXM1/E2F4 targets, likely mediated through indirect suppression downstream of p21 activation (reviewed in ref. 29) (Dataset S3 A and B). There was significant overlap between the genes induced or repressed by Entinostat in both the p53-WT and -null models, indicating that a large proportion of the effects of single agent Entinostat are not affected by p53 status (Dataset S3C); most notable was p53-independent suppression of NF-κB.
Surprisingly, of the 393 Nutlin-3A-induced genes, 143 were not significantly up-regulated (≥1.7-fold) in combination-treated cells (Fig. 2H and Dataset S2F), of which 58, including FLIP, were significantly reduced (≥1.7-fold) compared to Nutlin-3A treatment alone (Fig. 2I and Dataset S2 A–D and F). Of these 58 genes, there was enrichment for TNF and p53 signaling, including direct p53 targets such as Fas/CD95 death receptor, leukemia inhibitory factor (LIF), PIG3/TP53I3, 14-3-3σ/SFN, as well as FLIP (Fig. 2 H–K, SI Appendix, Fig. S2, and Datasets S2F and S3D), suggesting that inhibition of class-I HDACs antagonizes p53-mediated up-regulation of a discrete subset of its target genes. Notably, only 31 genes were significantly more up-regulated in the combination arm relative to the single agent treatments (Fig. 2H and Datasets S2G and S3E), and only one of these, NOXA, is a canonical direct p53-induced proapoptotic target (SI Appendix, Fig. S2 H and I). Altogether, these results establish FLIP(L) as a direct p53 target that is induced in a manner that is independent of its canonical regulation by NF-κB, but dependent on class-I HDAC activity.
Entinostat Enhances Nutlin-3A- and Chemotherapy-Induced Apoptosis.
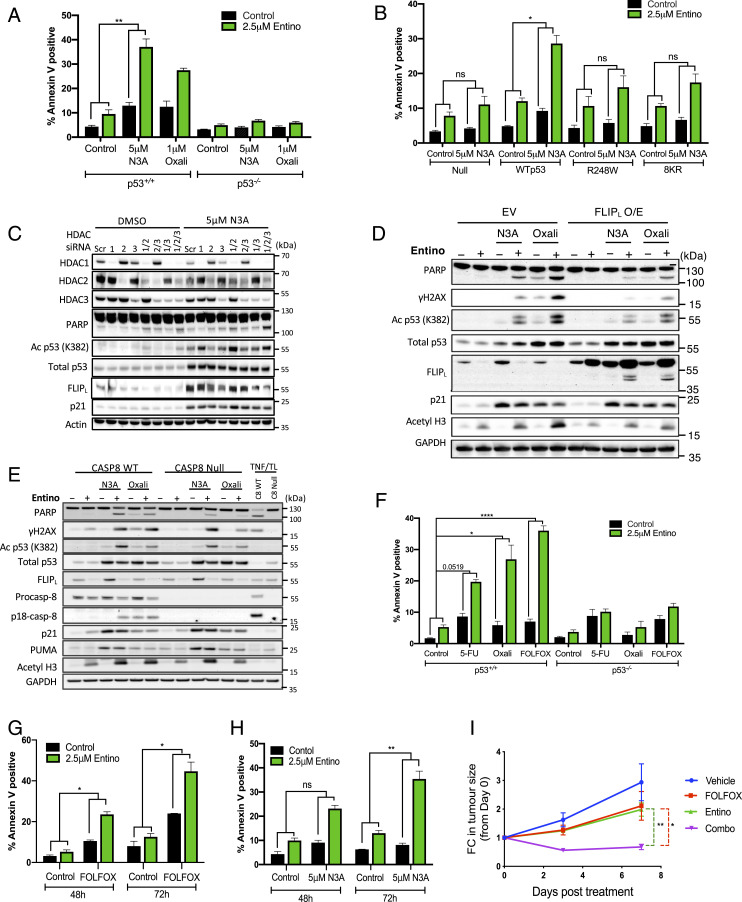
Similar to the effects of siFLIP(L) SI Appendix, (Fig. 1 and SI Appendix, Fig. S1) and consistent with its ability to suppress FLIP(L) expression (SI Appendix, Fig. S2 and Fig. 2), Entinostat enhanced Nutlin-3A-induced apoptosis in p53-WT but not p53-null HCT116 cells (Fig. 3A and SI Appendix, Fig. S3A). Entinostat also enhanced apoptosis induced by the DNA damaging agent oxaliplatin in a p53-dependent manner (SI Appendix, Fig. S3A and Fig. 3A). To assess the importance of p53-mediated transcription for these apoptotic effects, we evaluated the Nutlin-3A/Entinostat combination in HCT116 p53-null cells retrovirally expressing WT-p53, the clinically relevant R248W DNA binding p53 mutant, or an acetylation mutant form of p53 (Lys-Arg [8KR]) (30). Significant cell death was only observed in cells expressing WT-p53 (SI Appendix, Fig. S3B and Fig. 3B), indicating that both DNA binding and acetylation of p53 are required.
Fig. 3.
Entinostat enhances Nutlin-3A- and chemotherapy-induced apoptosis. (A) Annexin-V/PI FACS analysis of p53 WT and null HCT116 cells treated with 5 μM Nutlin-3A (N3A) or 1 μM oxaliplatin (Oxali) for 24 h prior to treatment with 2.5 μM Entino for a further 24 h. (B) Annexin-V/PI FACS analysis of parental p53 null HCT116 cells and daughter cells ectopically expressing wild-type (WTp53), R248W mutant, or 8KR acetylation mutant p53. Cells were treated with 5 μM N3A for 24 h prior to treatment with 2.5 μM Entino for an additional 24 h. (C) Western blot analysis of protein expression in p53 WT HCT116 cells transfected with control (Scr) or HDAC1/2/3-targeted siRNAs for 24 h prior to treatment with 5 μM N3A for a further 48 h. (D) Western blot analysis of parental or stable FLIPL overexpressing HCT116 cells treated with 5 μM N3A or 1 μM Oxali for 24 h prior to treatment with 2.5 μM Entino for a further 24 h. (E) Western blot assessment of Caspase-8 WT and null cells pretreated for 24 h with either 5 µM N3A or 1 µM Oxali prior to treatment of 2.5 µM Entino for a further 24 h. A 24-h treatment with 10 ng/mL TNFα/1 µM Biranapant (Tetralogics-TL) serves as a positive control. (F) Comparison of cell death induced by 2.5 μM 5-FU, 1 μM Oxali alone, or in combination (FOLFOX) for 24 h prior to treatment with Entino for a further 24 h in p53 WT and null HCT116 cells. (G and H) Annexin-V/PI FACS of cell death induced after 48 h and 72 h concurrent exposure of CT26 murine CRC cells to 2.5 µM Entino and either FOLFOX (1.25 μM 5-FU plus 1 μM Oxali) (G) or 5 µM N3A (H). (I) Assessment of fold change (FC) in tumor size of BALB/c mice (six per group at day 3/d3; three per group at day 7/d7), injected with 1 × 106 CT26 murine CRC cells, over 7 d of daily treatment with vehicle (30% cyclodextrin/phosphate buffered saline [PBS]), FOLFOX (5-FU 10 mg/kg, oxaliplatin 1 mg/kg), Entinostat (10 mg/kg), or their combination. Error bars in A, B, F, G, and H are represented as mean ± SEM of at least three independent experiments. *P < 0.05; **P < 0.01, ****P < 0.0001; ns = not significant calculated by two-way ANOVA. Three mice per group were analyzed in I, *P < 0.05; **P < 0.01; ***P < 0.001 calculated by Student’s t test.
siRNA-mediated down-regulation of HDACs 1/2/3 individually or in combination revealed that simultaneous depletion of all three nuclear HDACs is necessary to maximally suppress Nutlin-3A-induced FLIP(L) up-regulation and enhance Nutlin-3A-induced PARP cleavage (Fig. 3C). For both siRNA- and small molecule-mediated targeting of HDAC1/2/3, the cell death induced was coincident with acetylation of p53K373/K382 (Fig. 3C and SI Appendix, Fig. S3 A and B). Similar p53-dependent effects of Entinostat were observed in further p53 isogenic paired CRC models (LoVo and RKO; SI Appendix, Fig. S3 C–F) and with second generation MDM2 inhibitors (SI Appendix, Fig. S3G). Moreover, acute induction of cell death in response to Nutlin-3A/Entinostat translated into enhanced long-term growth inhibition compared to the individual agents (SI Appendix, Fig. S3H).
Consistent with FLIP(L) down-regulation being a key mechanism underpinning Entinostat’s enhancement of p53-induced apoptosis, cells overexpressing FLIP(L) (Fig. 3D) or lacking caspase-8 (Fig. 3E and SI Appendix, Fig. S3I) were more resistant to apoptosis induced by Entinostat/Nutlin-3A. Similar FLIP(L)-dependent (Fig. 3D) and caspase-8-dependent (Fig. 3E) effects were observed for Entinostat/oxaliplatin cotreatments. Moreover, combining Entinostat with the backbone chemotherapeutic agent 5-fluorouracil (5-FU) or the clinically relevant chemotherapy doublet of 5-FU and oxaliplatin (“FOLFOX”) also resulted in p53-dependent enhancement of cell death in HCT116 cells (Fig. 3F). Similar effects were observed for the combination of Entinostat with FOLFOX or Nutlin-3A in the p53 wild-type CT26 murine model of microsatellite stable (MSS), KRAS mutant CRC (Fig. 3 G and H), and importantly, this enhanced efficacy was also observed in vivo (Fig. 3I).
Nutlin-3A Promotes Apoptosis via a Ligand-Independent TRAIL-R2 Complex.
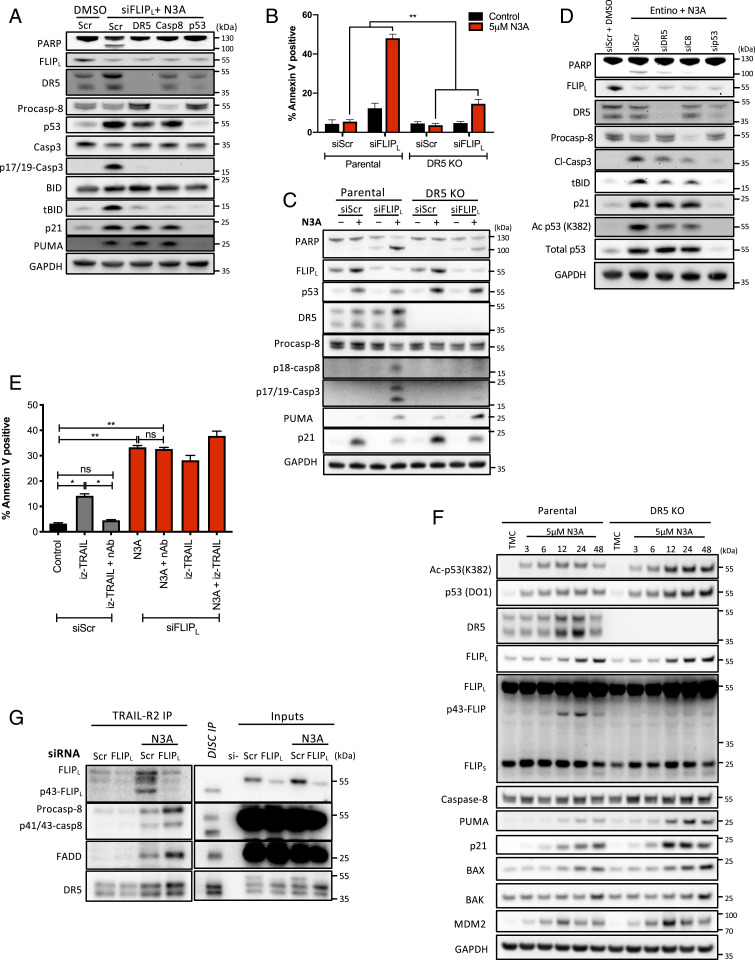
Canonically, FLIP and FADD regulate procaspase-8 processing and activation at death-inducing signaling complexes (DISCs) formed by death receptors and by the cytoplasmic ripoptosome formed by RIPK1 following depletion of cIAP1/2 (cellular inhibitor of apoptosis proteins) (23). To identify the upstream regulator(s) of caspase-8-mediated apoptosis in Nutlin-3A-treated, FLIP(L)-depleted cells, we reexamined the mRNA-Seq data for the effects of Nutlin-3A and Entinostat on death receptor expression. FAS/Fas and TNFRSF10A/DR4/TRAIL-R1 were both up-regulated in response to Nutlin-3A, but this increase was suppressed by cotreatment with Entinostat. TNFRSF10B/DR5/TRAIL-R2, a known direct p53 target (7), was expressed at a higher level than the other two death receptors and was, as expected, significantly up-regulated in response to Nutlin-3A; however in this case, the up-regulation was unaffected by cotreatment with Entinostat in mRNA-Seq and qRT-PCR (SI Appendix, Figs. S2G and S4A). RIPK1 was expressed at a low level that was unaffected transcriptionally by either treatment. Notably, RNAi-mediated down-regulation of TRAIL-R2, but not the other death receptors nor RIPK1 (SI Appendix, Fig. S4B), significantly abrogated the apoptosis induced by Nutlin-3A/siFLIP(L) and did so to a similar extent as silencing caspase-8 or p53 (Fig. 4A). Cleavage and activation of BID was also reduced in TRAIL-R2- and caspase-8-depleted cells (Fig. 4A). These results were confirmed in two independent p53-WT CRC models (SI Appendix, Fig. S4C) and using an HCT116 model in which TRAIL-R2 expression was deleted (Fig. 4 B and C). Silencing TRAIL-R2 or caspase-8 (and as expected p53) in HCT116 cells also partially inhibited apoptosis signaling in response to Nutlin-3A/Entinostat cotreatment (Fig. 4D).
Fig. 4.
Nutlin-3A promotes apoptosis via a ligand-independent TRAIL-R2 complex. (A) Western blot assessment of p53 WT HCT116 cells transfected with the indicated siRNAs (10 nM) for 24 h followed by transfection with siFLIPL for 6 h and treatment with 5 µM Nutlin-3A (N3A) for a further 24 h. (B) Annexin-V/PI FACS and (C) Western blot assessment of cell death in HCT116 DR5/TRAIL-R2 CRISPR KO and matched parental cells transfected for 6 h with 10 nM scrambled (siScr) or a FLIPL siRNA prior to treatment with 5 µM N3A for a further 24 h. (D) Western blot assessment of p53 WT HCT116 cells transfected with the indicated siRNAs (10 nM) for 24 h followed by cotreatment with 5 µM N3A and 2.5 µM Entino for 24 h. (E) Annexin-V/PI FACS analysis of the effect of TRAIL neutralizing antibody (nAb) treatment (100 ng/mL) on cell death in cells transfected for 6 h with 10 nM scrambled (siScr) or FLIPL siRNA prior to treatment with 3 ng/mL izTRAIL and/or 5 µM N3A for a further 24 h. (F) Western blot assessment of HCT116 DR5/TRAIL-R2 CRISPR KO and matched parental cells treated with 5 µM N3A for the indicated times (TMC, time matched control). (G) Postlysis DR5/TRAIL-R2 IP performed in HCT116 parental cells transfected for 6 h with 10 nM scrambled (siScr) or a FLIPL siRNA prior to treatment with 5 µM N3A for a further 24 h. Error bars in B and E are represented as mean ± SEM of at least three independent experiments. *P < 0.05; **P < 0.01; ns = not significant calculated by two-way ANOVA (B) and one-way ANOVA (E).
Canonically, TRAIL-R2-mediated activation of caspase-8 and apoptosis is triggered following binding of its ligand TRAIL, which is normally expressed on the surface of immune effector cells (20); it was therefore highly notable that coincubation with a TRAIL neutralizing antibody failed to rescue cell death induced by Nutlin-3A/siFLIP(L) (Fig. 4E), a result confirmed in both RKO and LoVo models (SI Appendix, Fig. S4 D and E). Moreover, addition of IZ-TRAIL did not significantly enhance cell death induced by Nutlin-3A/siFLIP(L) (Fig. 4E and SI Appendix, Fig. S4F). As expected, FADD-, CASP8/10-, BID- and BAX/BAK knockout models were resistant to IZ-TRAIL; however, the TRAIL-R2 knockout model (which still expresses TRAIL-R1/DR4) retained IZ-TRAIL sensitivity (SI Appendix, Fig. S4 G and H); this suggests that Nutlin-3A/siFLIP(L)-induced apoptosis operates via a mechanism that is distinct from the canonical extrinsic cell surface ligation of TRAIL-R2. Furthermore, in time-course experiments, the up-regulation of TRAIL-R2 and FLIP(L) coincided with the generation of the p43-form of FLIP(L), which is normally only generated at the death-inducing signaling complex (DISC) when it heterodimerizes with caspase-8 (Figs. 2C and 4F) (20). Moreover, the generation of p43-FLIP(L) was almost completely attenuated in TRAIL-R2 knockout cells (Fig. 4F).
Altogether, these results suggested formation of a ligand-independent TRAIL-R2/FLIP(L)/caspase-8 complex in cells treated with Nutlin-3A. To assess this, we isolated (postlysis) TRAIL-R2 complexes formed in response to Nutlin-3A and observed that interactions between TRAIL-R2 and its canonical DISC components FADD, FLIP(L), and caspase-8 were enhanced (Fig. 4G). Moreover, both FLIP(L) and caspase-8 were processed to p43 and p41/43 forms indicative of formation of catalytically active complexes. Importantly, depletion of FLIP(L), enhanced caspase-8 and FADD association with TRAIL-R2 in this ligand-independent complex (Fig. 4G), consistent with the apoptotic phenotype observed in FLIP(L)-depleted, Nutlin-3A-treated cells.
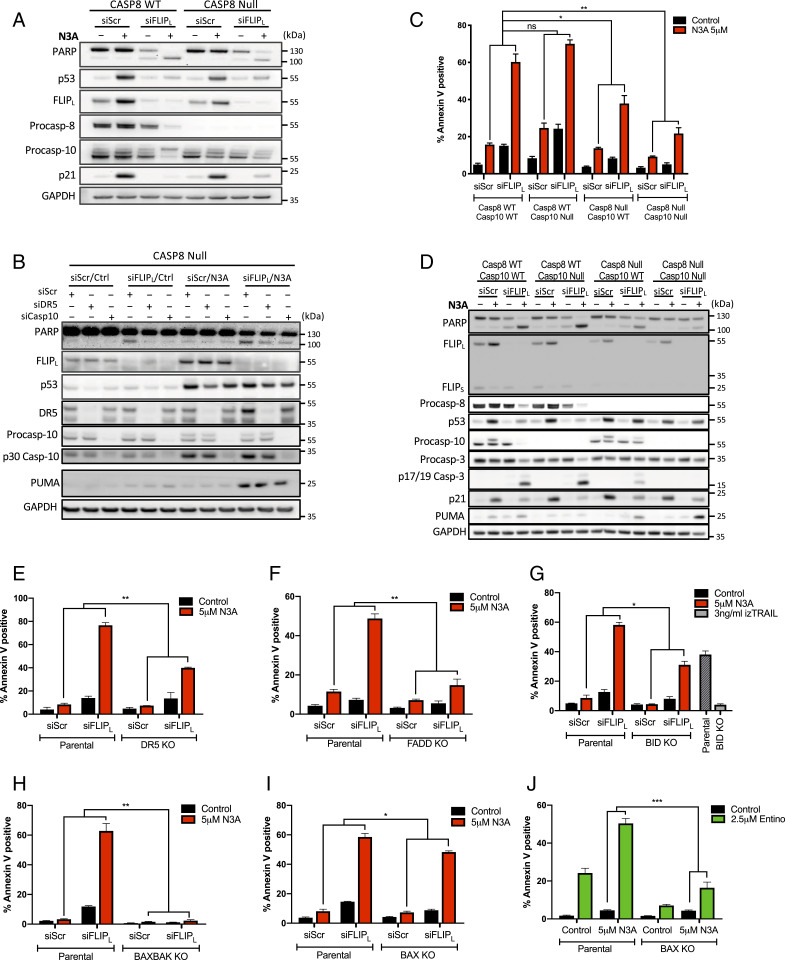
In the Absence of Caspase-8, Caspase-10 Activates Apoptosis in Nutlin-3A-Treated, FLIP(L)-Depleted Cells.
At 48 h, the apoptosis induced by Nutlin-3A/siFLIP(L) was notably less caspase-8 dependent than at 24 h (Fig. 5A). As processing of procaspase-10, a paralog of procaspase-8 and FLIP was observed in both CASP8-WT and -null cells in response to Nutlin-3A/siFLIP(L) (Fig. 5A), we investigated its involvement in mediating the apoptotic phenotype at 48 h. siCASP10 partially rescued PARP cleavage induced by Nutlin-3A/siFLIP(L) in caspase-8-deficient cells and did so to a similar extent as siTRAIL-R2 (Fig. 5B), indicating that this later cell death is also partly TRAIL-R2 dependent. Moreover, CRISPR knockout of caspase-10 in caspase-8-null cells, also further attenuated apoptosis induced by Nutlin-3A/siFLIP(L) at 48 h (Fig. 5 C and D). However, in the presence of caspase-8, loss of caspase-10 did not significantly affect the levels of apoptosis induced by Nutlin-3A/siFLIP(L) (Fig. 5 C and D). Caspase-10 knockout in caspase-8-deficient cells also further inhibited apoptosis induced by Nutlin-3A/Entinostat compared to knockout of caspase-8 alone; however, this was only evident at 24 h, as by 48 h codeletion of caspase-8/10 failed to rescue the cell death induced by the combination (SI Appendix, Fig. S5 A–D). Collectively, these results show that in the absence of caspase-8, caspase-10 can promote apoptosis induced by Nutlin-3A in FLIP(L)-depleted cells, albeit to a lesser extent. In contrast, caspase-10 was unable to compensate for loss of caspase-8 to activate IZ-TRAIL-induced apoptosis (SI Appendix, Fig. S5E).
Fig. 5.
In the absence of Caspase-8, Caspase-10 activates apoptosis in Nutlin-3A-treated, FLIP(L)-depleted cells. (A) Western blot analysis of HCT116 Caspase-8 parental and CRISPR KO cells transfected with 10 nM FLIPL or control siRNA for 6 h prior to treatment with 5 µM Nutlin-3A (N3A) for a further 48 h. (B) Western blot analysis of HCT116 Caspase-8 CRISPR KO cells transfected with siRNA targeting DR5, Caspase-10, or their combination for 24 h prior to subsequent transfection of 10 nM FLIPL-specific siRNA prior to treatment with 5 µM N3A for a further 48 h. (C) Annexin-V/PI FACS and (D) Western blot analysis of effects of FLIPL siRNA/N3A treatment as in A in HCT116 Caspase-8/10 CRISPR single and dual KO cells. (E–I) Annexin-V/PI FACS analysis of cell death in HCT116 isogeneic for DR5/TRAIL-R2 (E), FADD (F), BID (G), BAX/BAK DKO (H), and BAX (I) treated with FLIPL siRNA/N3A as in A, C, and D. A total of 3 ng/mL izTRAIL acts as a positive control for BID KO. (J) Annexin-V/PI FACS analysis of cell death in HCT116 isogenic for BAX treated with 2.5 µM Entino in combination with 5 µM N3A. Error bars in C and E–J are represented as mean ± SEM of at least three independent experiments. *P < 0.05; **P < 0.01; ns = not significant calculated by two-way ANOVA.
Surprisingly, even in the absence of both caspases-8 and -10, the combination of Nutlin-3A and siFLIP(L) still induced significant levels of apoptosis at 48 h compared to Nutlin-3A alone (Fig. 5 C and D). Knockout of TRAIL-R2, FADD or BID also significantly reduced cell death at 48 h, however similar to the caspase-8/10 double knockout model, significant levels of cell death were still observed when compared to Nutlin-3A treatment alone (Fig. 5 E–G and SI Appendix, Fig. S5 F–H). Collectively, this implies that FLIP(L) has functions that are independent of its canonical interactome. The cell death induced by Nutlin-3A/siFLIP(L) at the later timepoint was however completely attenuated in cells lacking both BAX and BAK, indicating intrinsic mitochondrially mediated apoptosis (Fig. 5H and SI Appendix, Fig. S6A). Knockout of both BAX and BAK (and as expected p53) also completely rescued the cell death induced by Nutlin-3A/Entinostat SI Appendix, Fig. S6 B–E. Of note, the single deletion of BAX conferred greater resistance to apoptosis induced by Nutlin-3A/Entinostat than Nutlin-3A/siFLIP(L) (Fig. 5 I and J and SI Appendix, Fig. S6 F and G). These results indicate overlapping but nonidentical apoptotic mechanisms of action for siFLIP(L) and Entinostat in combination with Nutlin-3A.
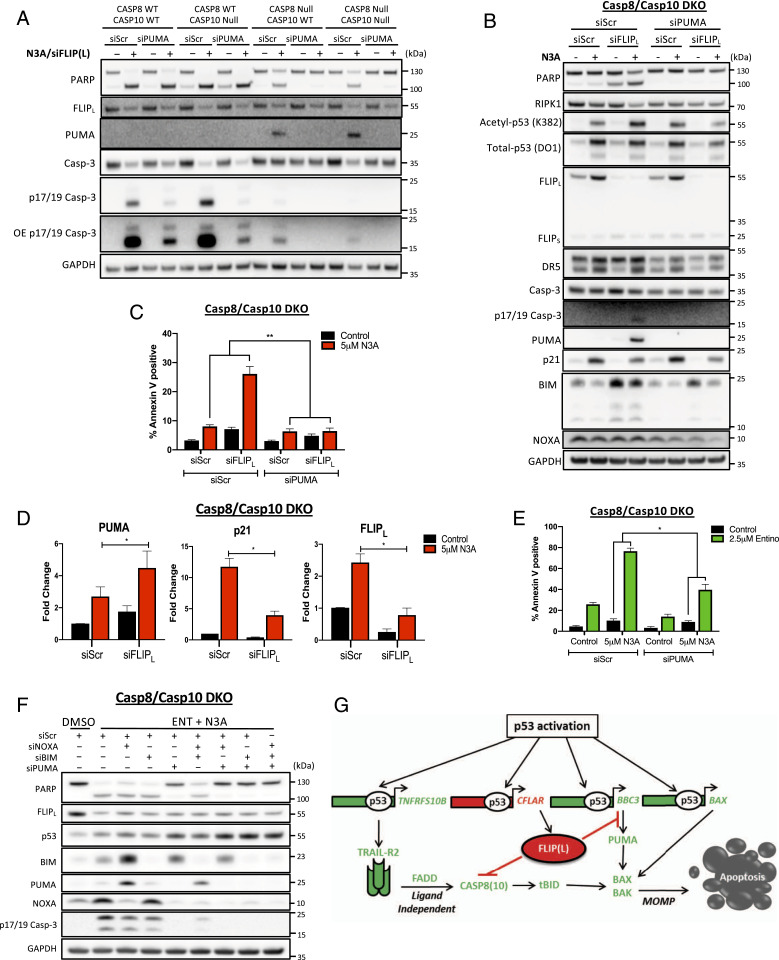
FLIP(L) Inhibits p53-Mediated Up-Regulation of PUMA.
It was notable that Nutlin-3A-induced up-regulation of the canonical p53 target PUMA was consistently enhanced in FLIP(L)-depleted cells; this was most easily detected in cell lines in which apoptosis induction was inhibited (Figs. 1F, 4 C and F, and 5 B and D and SI Appendix, Figs. S1E and S5 G and H). We therefore assessed whether PUMA was responsible for mediating the cell death phenotype induced at later timepoints by Nutlin-3A/siFLIP(L) in cells lacking caspase-8 and/or caspase-10. In caspase-8-deficient models, PUMA was again found to be up-regulated in response to Nutlin-3A/siFLIP(L) (Fig. 6 A and B), and cosilencing of PUMA indeed rescued the residual cell death induced by Nutlin-3A/siFLIP(L) in caspase-8/10-deficient models (Fig. 6 A–C). Of note, silencing PUMA reduced the levels of active caspase-3 detected in caspase-8-proficient cells in response to Nutlin-3A/siFLIP(L), although it failed to inhibit cleavage of the caspase-3 substrate PARP, suggesting that sufficient caspase-3 activation is induced in caspase-8-proficient cells to drive apoptosis independently of PUMA. However, in caspase-8-deficient/caspase-10-proficient cells, silencing PUMA inhibited the generation of both active caspase-3 and cleaved PARP in response to Nutlin-3A/siFLIP(L), indicating that in the absence of caspase-8, cell death induction requires cooperation between caspase-10 and PUMA. Further analysis of mRNA expression indicated that silencing FLIP(L) enhanced Nutlin-3A-induced up-regulation of PUMA mRNA (Fig. 6D), which notably was concomitant with suppression of p21 mRNA induction, consistent with observations made at the protein level for p21 (Figs. 4C, 5 A and D, and 6B).
Fig. 6.
FLIP(L) inhibits p53-mediated up-regulation of PUMA. (A) Western blot assessment of HCT116 Caspase-8/10 CRISPR single and dual KO (DKO) cells transfected for 24 h with 10 nM PUMA or control (Scr) siRNA prior to transfection with Scr/FLIPL siRNA for 6 h and treatment with 5 µM Nutlin-3A (N3A) for a further 48 h. Western blot (B) and Annexin-V/PI FACS (C) assessment of cell death in HCT116 Caspase-8/10 DKO treated as in A. (D) Quantitative RT-PCR of PUMA, p21, and FLIPL mRNA expression following transfection with Scr/FLIPL siRNA for 6 h and treatment with 5 µM N3A for a further 48 h. (E) Annexin-V/PI FACS assessment of cell death in HCT116 Caspase-8/10 DKO cells transfected with 10 nM PUMA or control (Scr) siRNA for 24 h prior to treatment with 5 µM N3A and 2.5 µM Entino for a further 48 h. (F) Western blot assessment of HCT116 Caspase-8/10 DKO cells transfected with 5 nM of the indicated siRNAs for 24 h prior to treatment with 5 µM N3A and 2.5 µM Entino for a further 48 h. (G) Schematic overview of mechanisms through which FLIPL supresses cell death induced by p53 activation. Error bars in C and E are represented as mean ± SEM of at least three independent experiments. Data in D are normalized to RPL24 control for each sample and represented as mean ± SEM of four independent experiments. *P < 0.05; **P < 0.01; calculated by two-way ANOVA (C and E) and Student’s t test (D).
Silencing PUMA also significantly but incompletely rescued the cell death induced by Nutlin-3A/Entinostat in caspase-8/10-deficient cells (Fig. 6 E and F). As has been previously reported in ref. 31, another BH3-only protein BIM was significantly up-regulated in response to Entinostat; moreover, we found that this induction was p53 independent (SI Appendix, Fig. S6H). Notably, cosilencing BIM alongside PUMA completely rescued the cell death induced by Nutlin-3A/Entinostat in caspase-8/10-deficient cells (SI Appendix, Fig. S6I and Fig. 6F), indicating involvement for BIM in mediating this apoptotic phenotype. However, the resistance of p53-null cells to Entinostat/Nutlin-3A (Fig. 3) shows that Entinostat-induced up-regulation of BIM is insufficient to drive apoptosis in the absence of p53-mediated up-regulation of other components of the apoptotic machinery, such as PUMA and BAX.
Discussion
In the 50% of tumors harboring WT p53, its function is typically suppressed; reactivation of p53’s latent tumor suppressive functions in these cancers is a highly attractive therapeutic approach (12). However, despite the development of selective p53 activating agents (best exemplified by MDM2 inhibitors, e.g., Nutlin-3A), none of these molecules have yet been clinically approved (32). This is likely because these agents primarily induce cell-cycle arrest rather than cell death in all but MDM2-amplified cancer cells (14, 33). Indeed, the mechanisms that regulate p53-induced cell death remain surprisingly poorly understood, and strategies that enhance cell death induced by p53 activating agents are highly sought after.
We found that direct transcriptional up-regulation of FLIP(L) in response to Nutlin-3A prevents p53-mediated induction of apoptosis, providing a mechanistic explanation for why p53 activation often fails to activate apoptosis despite robust up-regulation of apoptosis target genes. We speculate that acute concomitant induction of FLIP(L) and TRAIL-R2 in response to p53 stabilization enables epithelial cells to be primed to undergo apoptosis, while preventing induction of apoptosis en masse, which in epithelial cells would result in barrier dysfunction. Rapid commitment to cell death could then occur in response to secondary signaling event(s) (absent in cells treated with selective MDM2 inhibitors), which overcome FLIP(L)’s antiapoptotic functions. The rapid up-regulation of the long FLIP splice form in response to p53 stabilization may not only put the brakes on p53-induced apoptosis, but also inhibit proinflammatory RIPK1-dependent necroptotic cell death as the FLIP(L)-caspase-8 heterodimer [but not the FLIP(S)-caspase-8 heterodimer] can cleave RIPK1, thereby preventing necroptosis (34). More complex signals than MDM2 inhibition such as DNA damaging chemotherapy result in oscillating levels of p53 and its positive and negative regulators. In this context, modifications to p53 not induced by Nutlin-3A (such as acetylation) and modulation of additional signaling pathways may lead to down-regulation of FLIP(L), a short-lived protein, thereby triggering apoptosis. Nonetheless, FLIP is also important in the context of DNA damaging chemotherapy as demonstrated herein (Fig. 3D) and in our previous publications (35, 36), in which we demonstrated that FLIP depletion (by RNAi) synergized with 5-FU and oxaliplatin to induce apoptosis in p53 wild-type HCT116 cells. Importantly, these effects were significantly reduced in p53-deficient models, and, consistent with our findings with Nutlin-3A, the apoptosis-inhibitory effects in p53 wild-type HCT116 cells were predominantly mediated by FLIP(L).
While the caspase-8 inhibitory, antiapoptotic effects of FLIP(S) are well established, whether FLIP(L) is a promoter or inhibitor of caspase-8-mediated apoptosis is controversial (23), although we have recently shed light on this issue (37). Herein, we clearly demonstrate that FLIP(L) up-regulation in response to Nutlin-3A prevents caspase-8-mediated CRC cell death. Consistent with the canonical mechanism of action, the apoptosis induced in response to FLIP(L)-depletion in Nutlin-3A-treated cells is not only caspase-8 dependent, but also dependent on the adaptor protein FADD and the downstream effector BID, the cleavage of which promotes amplification of the cell death signal via the mitochondrial apoptotic pathway by activating BAX and (primarily) BAK (38). The upstream mechanism of action of caspase-8 was notable, since FADD-dependent recruitment of FLIP/caspase-8 to death receptors usually occurs in response to extrinsic ligation of these receptors. However, we found that although TRAIL-R2 was clearly the upstream receptor primarily responsible for activation of caspase-8 in FLIP(L)-depleted Nutlin-3A-treated cells, its ligation by TRAIL was not required. TRAIL-R2-dependent, TRAIL-independent cell death in response to chemotherapy (39) and during endoplasmic reticulum (ER) stress (40) has been described, with the latter recently reported to be mediated by misfolded proteins binding to the TRAIL-R2 ectodomain (41), although other mechanisms such as enhanced receptor density in the ER promoting autoactivation are also possible (42). Indeed, we detected a ligand-independent TRAIL-R2-FLIP(L)-caspase-8 complex formed in response to Nutlin-3A, the recruitment of caspase-8 into which was enhanced by FLIP(L) depletion. Thus, p53 activation promotes formation of a ligand-independent TRAIL-R2 complex containing FADD, FLIP(L), and caspase-8 in colon cancer cells, which in the absence of FLIP(L) drives apoptosis induction.
Although highly caspase-8 dependent in the first 24 h, significant levels of caspase-8-independent cell death were induced in Nutlin-3A/siFLIP(L) cells by 48 h. We initially examined whether this was mediated by the caspase-8 and FLIP(L) paralog caspase-10. Similar to FLIP(L), the role of caspase-10 at the DISC is controversial, with it described as being able to compensate for lack of caspase-8 (43) unable to compensate (44), and acting to inhibit caspase-8-mediated apoptosis (45). Our results shed light on the complex biology of caspase-10: in the presence of caspase-8, deletion of caspase-10 had no significant effect on cell death induced by siFLIP(L)/Nutlin-3A; however, in the absence of caspase-8, codeletion of caspase-10 significantly decreased apoptosis induction. These results indicate that caspase-10 can drive apoptosis in siFLIP(L)/Nutlin-3A-treated cells in the absence of caspase-8 (albeit less efficiently). In contrast, we found no evidence for caspase-10 being able to drive ligand-dependent TRAIL-R2-mediated apoptosis in the absence of caspase-8, highlighting differences between p53-dependent, ligand-independent TRAIL-R2 apoptotic signaling and canonical ligand-dependent receptor signaling.
Even in the absence of caspase-8 and caspase-10, apoptosis was still observed in FLIP(L)-depleted, Nutlin-3A-treated cells. This cell death was also independent of TRAIL-R2, FADD, and BID, but was BAX/BAK dependent, indicative of a hitherto unreported role for FLIP(L) in regulating the intrinsic mitochondrial apoptotic pathway. Further analyses revealed an unexpected role for FLIP(L) in suppressing p53-mediated transcriptional up-regulation of PUMA/BBC3, such that in siFLIP(L)/Nutlin-3A-treated cells, PUMA expression was up-regulated and drove the apoptosis induced in caspase-8/10-deficient cells. Moreover, p53-induced p21 transcription was suppressed in FLIP(L)-depleted cells. Thus, in addition to its role in preventing ligand-independent TRAIL-R2-mediated apoptosis in response to p53 stabilization, these results are consistent with additional prosurvival roles for FLIP(L): suppression of p53-mediated up-regulation of PUMA and promotion of p53-mediated up-regulation of p21.
Class-I HDACs have been shown to regulate acetylation of p53 in its lysine-rich C terminus, the hyperacetylation of which has been correlated with enhancement of apoptosis (46–50). In support of this, we found that the clinically relevant class-I HDAC inhibitor Entinostat enhanced Nutlin-3A-induced apoptosis in p53 wild-type CRC cells and that this effect was lost in cells expressing a nonacetylatable mutant form of p53. We therefore expected Entinostat to enhance Nutlin-3A-induced transcription of proapoptotic p53 target genes. However, mRNA-Seq analyses revealed that Entinostat enhanced Nutlin-3A-induced expression of only 31 genes, and only one canonical p53 proapoptotic target (PMAIP1/NOXA). In fact, Entinostat suppressed induction of 58 Nutlin-3A-induced genes, most notably FLIP. Indeed, the mechanism of cell death induced by the combination of Nutlin-3A and Entinostat was almost identical to that induced by Nutlin-3A/siFLIP(L) with a couple of notable exceptions. As previously reported (31) and confirmed by us, Entinostat significantly up-regulates BIM; this up-regulation has been suggested to be mediated by acetylation of FOXO1 (51). Along with PUMA, BIM was found to be responsible for the apoptosis induced by Nutlin-3A/Entinostat when the extrinsic apoptotic pathway was blocked by caspase-8/10 codeletion. As BIM preferentially activates BAX, and BID preferentially activates BAK (38), this likely explains the differential dependence of the Nutlin-3A/Entinostat and Nutlin-3A/siFLIP(L) combinations on BAX and BAK, respectively (Fig. 5 I and J).
In summary, this work has uncovered clinically relevant biology, in which early p53-mediated up-regulation of FLIP(L) prevents commitment of CRC cells to apoptosis by blocking activation of caspase-8 from a p53-induced, ligand-independent TRAIL-R2 complex and by suppressing p53-mediated induction of PUMA (Fig. 6G). From a cancer therapeutics point of view, we show the potential of class-I HDAC inhibitors to overcome FLIP(L)-mediated resistance to MDM2 inhibitors and DNA damaging chemotherapeutic regimens in p53-WT cancers.
Materials and Methods
Cell Culture.
HCT116 p53+/+, HCT116 p53−/−, and HCT116 p53−/R248W cells and BAX−/− (52) were kind gifts received from Bert Vogelstein, Johns Hopkins University, Baltimore, MD. HCT116 CASP8+/+ and HCT116 CASP8−/− cells (19) were kind gifts received from Galit Lahav, Harvard Medical School, Boston, MA. HCT116 cells stably expressing the NF-κB superrepressor were a kind gift from Aideen Ryan, NUI Galway, Galway, Ireland. HCT116 SMAC KO, DR5 KO, and BAX/BAK DKO cells were a kind gift from Markus Rehm, University of Stuttgart, Stuttgart, Germany. HCT116 BID KO cells were a kind gift from Lin Zhang, University of Pittsburgh, Pittsburgh, PA. LoVo, RKO, and CT26 cells were purchased from American Type Culture Collection (ATCC). HCT116 and RKO cell lines were cultured in McCoy’s 5A medium supplemented with 10% fetal bovine serum (FBS), 50 U/mL penicillin, 0.1 mg/mL streptomycin, 2 mM l-glutamine, and 1 mM sodium pyruvate (all Gibco). LoVo cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 10% FBS, 50 U/mL penicillin, and 0.1 mg/mL streptomycin. CT26 cells were cultured in Roswell Park Memorial Institute (RPMI) medium (Gibco) supplemented with 10% FBS, 50 U/mL penicillin, 0.1 mg/mL streptomycin, 2 mM l-glutamine, and 1 mM sodium pyruvate. All cells were maintained at 37 °C in a 5% CO2 humidified atmosphere and regularly screened for the presence of mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza).
Supplementary Material
Acknowledgments
Work in this manuscript was primarily supported by studentships (principal investigator [PI]: S.S.M.) sponsored by Northern Ireland Department for Education and Department for Employment and Learning, as well as funding from Cancer Research UK (CRUK) Program grants C11884/A24367 (PI: D.B.L. and coinvestigator [CI]: S.S.M.) and C212/A13721 (PI: P.G.J. and CI: D.B.L.), ECMC C36697/A25176, RCUK | Biotechnology and Biological Sciences Research Council (BBSRC) BB/T002824/1 (Co-PI: S.S.M. and D.B.L.). RNA-Seq data generation in this study was supported by the “Queen’s Faculty of Medicine Health and Life Sciences Genomics Core Technology Unit.” We thank Professor Bert Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD) for kindly gifting the HCT116 p53+/+ and p53−/− cell lines; Professor Markus Rehm (University of Stuttgart, Germany) for supplying the HCT116 DR5 KO, BAX/BAK DKO, and SMAC KO cells; and Professor Galit Lahav (Department of Systems Biology, Harvard Medical School, Boston, MA) for providing the HCT116 caspase-8 CRISPR cells. We also thank Dr. Lin Zhang (University of Pittsburgh, Pittsburgh, PA) for supplying the HCT116 BID KO cells.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The raw data and RNA-Seq analyses discussed in this publication have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database and linked sequence read archive files, which are accessible at accession no. GSE113682.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001520117/-/DCSupplemental.
Data Availability.
The raw data and RNA-Seq analyses discussed in this publication have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus and linked sequence read archive files, which are accessible through GEO Series accession no. GSE113682. Resulting analyses can also be visualized and downloaded from our Shiny app (https://functionalgenomicsgroup.shinyapps.io/HDAC_visualiseR/).
References
- 1.Kastenhuber E. R., Lowe S. W., Putting p53 in context. Cell 170, 1062–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J., The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 (1992). [DOI] [PubMed] [Google Scholar]
- 3.el-Deiry W. S. et al., WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J., The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Nakano K., Vousden K. H., PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7, 683–694 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Villunger A. et al., p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302, 1036–1038 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Takimoto R., El-Deiry W. S., Wild-type p53 transactivates the KILLER/DR5 gene through an intronic sequence-specific DNA-binding site. Oncogene 19, 1735–1743 (2000). [DOI] [PubMed] [Google Scholar]
- 8.McDade S. S. et al., Genome-wide characterization reveals complex interplay between TP53 and TP63 in response to genotoxic stress. Nucleic Acids Res. 42, 6270–6285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer M., Grossmann P., Padi M., DeCaprio J. A., Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 44, 6070–6086 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J., Wang Z., Kinzler K. W., Vogelstein B., Zhang L., PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 1931–1936 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeffers J. R. et al., Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4, 321–328 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Brown C. J., Lain S., Verma C. S., Fersht A. R., Lane D. P., Awakening guardian angels: Drugging the p53 pathway. Nat. Rev. Cancer 9, 862–873 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Vassilev L. T. et al., In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Tovar C. et al., Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc. Natl. Acad. Sci. U.S.A. 103, 1888–1893 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrysik Z. et al., Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome Res. 27, 1645–1657 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W. et al., MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalem O. et al., Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen H., Moran D. M., Maki C. G., Transient nutlin-3a treatment promotes endoreduplication and the generation of therapy-resistant tetraploid cells. Cancer Res. 68, 8260–8268 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paek A. L., Liu J. C., Loewer A., Forrester W. C., Lahav G., Cell-to-cell variation in p53 dynamics leads to fractional killing. Cell 165, 631–642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphreys L., Espona-Fiedler M., Longley D. B., FLIP as a therapeutic target in cancer. FEBS J. 285, 4104–4123 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Scaffidi C. et al., Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17, 1675–1687 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikulenkov F. et al., Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 19, 1992–2002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riley J. S., Malik A., Holohan C., Longley D. B., DED or alive: Assembly and regulation of the death effector domain complexes. Cell Death Dis. 6, e1866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo J.-L., Maeda S., Hsu L.-C., Yagita H., Karin M., Inhibition of NF-kappaB in cancer cells converts inflammation- induced tumor growth mediated by TNFalpha to TRAIL-mediated tumor regression. Cancer Cell 6, 297–305 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Marouco D., Garabadgiu A. V., Melino G., Barlev N. A., Lysine-specific modifications of p53: A matter of life and death? Oncotarget 4, 1556–1571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y., Zhao W., Chen Y., Zhao Y., Gu W., Acetylation is indispensable for p53 activation. Cell 133, 612–626 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley J. S. et al., Prognostic and therapeutic relevance of FLIP and procaspase-8 overexpression in non-small cell lung cancer. Cell Death Dis. 4, e951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerr E. et al., Identification of an acetylation-dependant Ku70/FLIP complex that regulates FLIP expression and HDAC inhibitor-induced apoptosis. Cell Death Differ. 19, 1317–1327 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer M., Steiner L., Engeland K., The transcription factor p53: Not a repressor, solely an activator. Cell Cycle 13, 3037–3058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drosten M. et al., Loss of p53 induces cell proliferation via Ras-independent activation of the Raf/Mek/Erk signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 111, 15155–15160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue S., Riley J., Gant T. W., Dyer M. J. S., Cohen G. M., Apoptosis induced by histone deacetylase inhibitors in leukemic cells is mediated by Bim and Noxa. Leukemia 21, 1773–1782 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Khoo K. H., Verma C. S., Lane D. P., Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 13, 217–236 (2014). Erratum in: Nat. Rev. Drug Discov.13, 314 (2014). [DOI] [PubMed] [Google Scholar]
- 33.París R., Henry R. E., Stephens S. J., McBryde M., Espinosa J. M., Multiple p53-independent gene silencing mechanisms define the cellular response to p53 activation. Cell Cycle 7, 2427–2433 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pop C. et al., FLIP(L) induces caspase 8 activity in the absence of interdomain caspase 8 cleavage and alters substrate specificity. Biochem. J. 433, 447–457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longley D. B. et al., c-FLIP inhibits chemotherapy-induced colorectal cancer cell death. Oncogene 25, 838–848 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Wilson T. R. et al., c-FLIP: A key regulator of colorectal cancer cell death. Cancer Res. 67, 5754–5762 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Humphreys L. M. et al., A revised model of TRAIL-R2 DISC assembly explains how FLIP(L) can inhibit or promote apoptosis. EMBO Rep. 21, e49254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarosiek K. A. et al., BID preferentially activates BAK while BIM preferentially activates BAX, affecting chemotherapy response. Mol. Cell 51, 751–765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson T. R. et al., Procaspase 8 overexpression in non-small-cell lung cancer promotes apoptosis induced by FLIP silencing. Cell Death Differ. 16, 1352–1361 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Lu M. et al., Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 345, 98–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam M., Marsters S. A., Ashkenazi A., Walter P., Misfolded proteins bind and activate death receptor 5 to trigger apoptosis during unresolved endoplasmic reticulum stress. eLife 9, e52291 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan G. P. et al., TRAIL receptors serve as stress-associated molecular patterns to promote ER-stress-induced inflammation. Dev. Cell 52, 714–730.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Wang J., Chun H. J., Wong W., Spencer D. M., Lenardo M. J., Caspase-10 is an initiator caspase in death receptor signaling. Proc. Natl. Acad. Sci. U.S.A. 98, 13884–13888 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sprick M. R. et al., Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J. 21, 4520–4530 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horn S. et al., Caspase-10 negatively regulates caspase-8-mediated cell death, switching the response to CD95L in favor of NF-κB activation and cell survival. Cell Rep. 19, 785–797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knights C. D. et al., Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J. Cell Biol. 173, 533–544 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellert H. S. et al., Deacetylation of the DNA-binding domain regulates p53-mediated apoptosis. J. Biol. Chem. 286, 4264–4270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sykes S. M. et al., Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brochier C. et al., Specific acetylation of p53 by HDAC inhibition prevents DNA damage-induced apoptosis in neurons. J. Neurosci. 33, 8621–8632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laptenko O. et al., The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol. Cell 57, 1034–1046 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y. et al., Acetylation of FoxO1 activates Bim expression to induce apoptosis in response to histone deacetylase inhibitor depsipeptide treatment. Neoplasia 11, 313–324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sur S. et al., A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc. Natl. Acad. Sci. U.S.A. 106, 3964–3969 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data and RNA-Seq analyses discussed in this publication have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus and linked sequence read archive files, which are accessible through GEO Series accession no. GSE113682. Resulting analyses can also be visualized and downloaded from our Shiny app (https://functionalgenomicsgroup.shinyapps.io/HDAC_visualiseR/).