Significance
LINE-1 retrotransposons are mobile genetic elements that have significantly contributed to the evolution of our genome. However, unhindered LINE-1 mobility can be detrimental and cause various genetic disorders. Here, we identify the antiviral protein TRIM5α as host factor that controls LINE-1 elements and protects our genome from novel insertions. In addition, we show that TRIM5α recognizes LINE-1 ribonucleoprotein complexes as a molecular pattern and initiates innate immune signaling upon sensing. Thus, our findings confirm the role of TRIM5α as pattern recognition receptor in innate immunity and uncover its important role in safeguarding the genome against endogenous mobile genetic elements.
Keywords: TRIM5α, LINE-1, retrotransposon, restriction factor, innate immune sensing
Abstract
Mobile genetic elements have significantly shaped our genomic landscape. LINE-1 retroelements are the only autonomously active elements left in the human genome. Since new insertions can have detrimental consequences, cells need to efficiently control LINE-1 retrotransposition. Here, we demonstrate that the intrinsic immune factor TRIM5α senses and restricts LINE-1 retroelements. Previously, rhesus TRIM5α has been shown to efficiently block HIV-1 replication, while human TRIM5α was found to be less active. Surprisingly, we found that both human and rhesus TRIM5α efficiently repress human LINE-1 retrotransposition. TRIM5α interacts with LINE-1 ribonucleoprotein complexes in the cytoplasm, which is essential for restriction. In line with its postulated role as pattern recognition receptor, we show that TRIM5α also induces innate immune signaling upon interaction with LINE-1 ribonucleoprotein complexes. The signaling events activate the transcription factors AP-1 and NF-κB, leading to the down-regulation of LINE-1 promoter activity. Together, our findings identify LINE-1 as important target of human TRIM5α, which restricts and senses LINE-1 via two distinct mechanisms. Our results corroborate TRIM5α as pattern recognition receptor and shed light on its previously undescribed activity against mobile genetic elements, such as LINE-1, to protect the integrity of our genome.
TRIM5α is a powerful intracellular restriction factor to retroviral infection (1). It blocks infectivity at an early postentry step and directly targets incoming viral capsid. TRIM5α is a member of the tripartite motif (TRIM) protein family and contains the characteristic amino-terminal RING domain, one B-box, and a coiled-coil (CC) region (2). While the RING domain comprises an E3 ubiquitin ligase activity, the CC region is important for TRIM5α dimerization and the B-box mediates multimerization of the dimers. The TRIM motif connects via a flexible linker region (L2) to the carboxyl-terminal SPRY domain (PRY-SPRY, B30.2), which mediates the interaction with retroviral capsid structures. The exact mode of restriction is still unclear but it has been shown that binding of the SPRY domains to viral capsid structures initiates the higher order assembly of TRIM5α dimers around the capsids (3, 4). The formation of such a hexagonal lattice has been suggested to initiate the destabilization of the incoming capsid. The multimerization of TRIM5α also compensates for the low affinity of the SPRY domain for viral capsid by increasing the avidity of the binding (5). The interaction between SPRY domain and viral capsid is highly species specific. While rhesus TRIM5α potently restricts HIV-1 infection, human TRIM5α is only weakly active against HIV-1 (1). Nevertheless, human TRIM5α has been described to block HIV-1 infection in Langerhans cells (6) and upon activation of the immunoproteasome and therefore contributes to the antiviral state induced by type I IFN (7). Human TRIM5α is also active against other retroviruses, such as N-tropic murine leukemia virus (N-MLV) (8–10). On top of direct capsid destabilization, TRIM5α has been suggested to act as a pattern recognition receptor for retroviral capsids (11). The interaction of TRIM5α with retroviral capsids has been shown to initiate innate immune signaling resulting in the activation of the transcription factors AP-1 and NF-κB (11). Upon TRIM5α overexpression or recognition of viral capsid, TRIM5α induces ubiquitin-dependent signaling via its RING domain by recruiting E2 ubiquitin (Ub)-conjugating enzymes. TRIM5α multimerization on the viral capsid leads to a trivalent RING domain arrangement, which induces the polymerization of K63-linked ubiquitin chains at the amino terminus of TRIM5α (12). The generation of polyubiquitin chains in turn activates the TAK1 kinase complex and, subsequently, the downstream MAP kinase and NF-κB signaling pathways leading to AP-1 and NF-κB activation. However, how these signaling processes are initiated and how they affect retroviral replication is not fully understood.
Typical for an innate immune factor involved in direct pathogen interaction, human TRIM5α has undergone multiple episodes of positive selection. Interestingly, these episodes predate the origin of primate lentiviruses, ruling out lentiviruses as causative agents for the selection process (13, 14). Human TRIM5 is active against N-MLV, a gamma-retrovirus closely related to various human endogenous retroviruses that have previously invaded the genome (9, 15, 16). Thus, it has been speculated that the evolution of TRIM5α may have been influenced by endogenous retroviruses or retroelements (17, 18). The only known autonomously active transposable element (TE) in humans is Long Interspersed Element 1 (LINE-1), which belongs to the class of nonlong terminal repeat (non-LTR) retroelements (19, 20). LINE-1 transcripts consist of a 5′ untranslated region (UTR) containing the promoter, a 3′ UTR encoding a polyA signal, and two major ORFs, ORF1 and ORF2. A third, small, antisense-encoded ORF, ORF0, has recently been identified to promote LINE-1 mobility (21). The protein encoded by ORF1, ORF1p, is a trimeric, RNA binding protein with high affinity for LINE-1 RNA (22, 23). ORF2p harbors the endonuclease and reverse transcriptase activities of LINE-1. Upon translation, ORF1p and ORF2p form ribonucleoprotein complexes (RNPs) in the cytoplasm by binding “in cis” to the RNA molecule they have been translated from (22–25). After relocation of the RNPs to the nucleus, ORF2p cleaves genomic DNA and uses single-stranded genomic DNA to prime reverse transcription of its RNA. This process called target primed reverse transcription (TPRT) leads to the integration of a new copy of LINE-1 into the host genome. Retrotransposition events have been described primarily in the germline during early embryogenesis and have been associated with various genetic disorders, such as hemophilia, or neurofibromatosis (26). More recently, it became evident that somatic retrotransposition of LINE-1 elements can also occur in adults, most prominently in neuronal precursor cells contributing to the genetic mosaicism of neurons (27–29). To maintain genome integrity and avoid genetic disorders it is important to keep LINE-1 elements at bay. Mechanisms to prevent retrotransposition include epigenetic silencing of the promoter, RNA interference, and the activity of host restriction factors, such as APOBEC3 proteins, Mov10, or SAMHD1 (30).
Here, we show that human TRIM5α contributes to safeguarding the genome by restricting the retrotransposition of LINE-1 elements. Upon interaction with LINE-1 RNPs in the cytoplasm, TRIM5α induces innate immune signaling resulting in the up-regulation of the transcription factors AP-1 and NF-κB. LINE-1 promoter studies demonstrate that the TRIM5α-mediated innate immune signaling inhibits the LINE-1 promoter. Together, we show that human TRIM5α senses and restricts LINE-1 mobile genetic elements by blocking its promoter activity in a negative feedback loop and thereby protects the genome from novel LINE-1 integrates.
Results
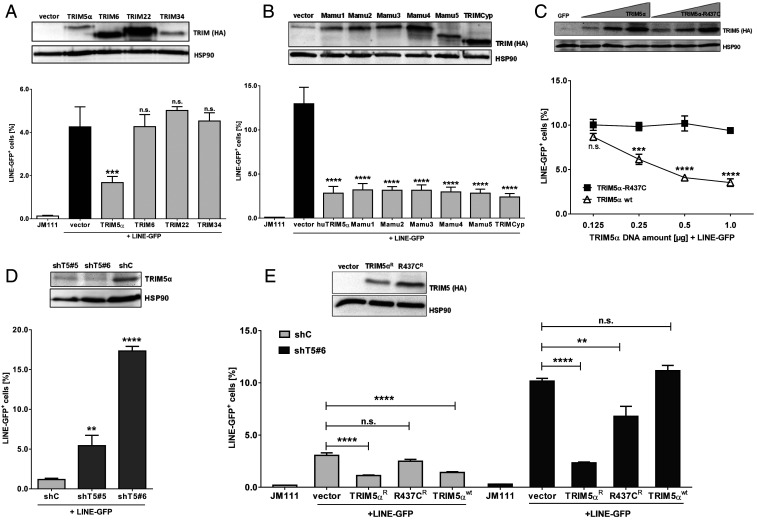
Human and Rhesus TRIM5α Restricts LINE-1 Retrotransposition.
Members of the TRIM protein family are involved in diverse intrinsic antiviral immune mechanisms, such as sensing of RNA viruses (TRIM25), restriction of retroviruses (TRIM5α), or inhibition of transposable elements (TRIM28) (31, 32). Human TRIM5 is part of a cluster of four closely related TRIM genes, TRIM5, TRIM6, TRIM22, and TRIM34 (33). First, we asked whether TRIM5α or one of its close paralogues is involved in restricting transposable elements. We tested the ability of the different TRIM proteins to restrict LINE-1 elements using a well-established reporter assay (34). Briefly, upon transfection of a LINE-GFP reporter plasmid, the intron-containing GFP reporter gene is only expressed when the LINE-1 RNA is spliced, reverse transcribed, and integrated into the host genome. Hence, GFP expression serves as a surrogate for successful retrotransposition. Upon transient transfection of HEK 293T cells, we found that human TRIM5α significantly reduced LINE-GFP activity by ∼3- to 5-fold compared to empty vector-transfected control cells (Fig. 1A and SI Appendix, Fig. S1). The inhibition of LINE-1 proved to be specific for TRIM5α since the other members of the cluster did not block LINE-1 retrotransposition (Fig. 1A). Since human and rhesus TRIM5α differ in their activity against HIV-1, we wondered whether rhesus TRIM5α is also active against LINE-1. Thus, we analyzed six known alleles of rhesus TRIM5α, Mamu1-5 and Mamu7 (TRIMCyp), which can be distinguished by polymorphic variations of the SPRY domain (35, 36). In the case of TRIMCyp, the coding sequence of the prolyl isomerase Cyclophilin A (CypA), which also binds to retroviral capsid structures, has replaced the SPRY domain as a consequence of a LINE-1 retrotransposition event (36, 37). Similar to human TRIM5α, coexpression of all variants of rhesus TRIM5α potently inhibited the activity of LINE-GFP by ∼4-fold, suggesting that the SPRY domain of TRIM5α recognizes LINE-1 differently from exogenous retroviruses (Fig. 1B). Interestingly, TRIMCyp also restricted LINE-1 replication, suggesting that, similar to retroviral capsids, CypA might bind to LINE-1 RNPs and that the TRIMCyp restriction is mediated by a similar interaction. Human TRIM5α bearing the rare polymorphism R437C in the SPRY domain has lost its ability to restrict nonpermissive viruses such as N-MLV (38). To determine whether the R437C substitution also affects the ability of TRIM5α to restrict LINE-1 elements, we transfected LINE-GFP together with increasing amounts of plasmids encoding TRIM5α or R437C. While both proteins where efficiently expressed upon transfection, only wild-type (WT) protein restricted LINE-GFP activity in a dose-dependent manner (Fig. 1C). In contrast, R437C did not inhibit LINE-1, which further underlines the specificity of the TRIM5α-mediated block and the importance of the SPRY domain for restriction (Fig. 1C). To test whether endogenous TRIM5α also restricts LINE-1, we generated TRIM5α knockdown cells by transducing HEK 293T cells with lentiviral particles encoding different shRNAs targeting TRIM5α (shT5) or scrambled control shRNA (shC) (Fig. 1D). Upon selection, we identified two polyclonal cell lines, shT5 #5 and #6, that showed strongly reduced endogenous TRIM5α level compared to shC cells (Fig. 1D). In shT5 #5 cells, LINE-GFP activity was enhanced ∼5-fold, while in shT5 #6 cells, with even lower amounts of endogenous TRIM5α, LINE-GFP retrotransposition was up-regulated 15-fold (Fig. 1D). To verify that the enhanced activity of LINE-GFP was due to the absence of TRIM5α, we reconstituted shC and shT5 #6 cells with shRNA-resistant TRIM5α (TRIM5αR) and analyzed the reconstituted cells in LINE-GFP reporter assays (Fig. 1E). In shC cells, expression of both TRIM5α and TRIM5αR limited LINE-GFP retrotransposition. In contrast, in shT5 cells only the transfection of resistant TRIM5αR but not of shRNA-sensitive TRIM5α or R437CR efficiently reduced LINE-1 activity, confirming that the knockdown of endogenous TRIM5α is responsible for the enhanced LINE-1 retrotransposition in shTRIM5α cells (Fig. 1E and SI Appendix, Fig. S2).
Fig. 1.
Human and rhesus TRIM5α restrict LINE-1 retrotransposition. (A) HEK 293T cells were transfected with LINE-GFP or a retrotransposition-defective plasmid (JM111) and either empty vector or vector encoding human TRIM5α-HA, TRIM6-HA, TRIM22-HA, or TRIM34-HA. Five days posttransfection, GFP-positive cells were quantified by flow cytometry. Expression of the TRIM proteins was analyzed by immunoblot 2 d posttransfection. Membranes were probed with antibodies targeting the HA tag and the housekeeping gene HSP90. (B) HEK 293T cells were transfected with LINE-GFP together with the HA-tagged human TRIM5α, rhesus TRIM5 alleles Mamu1-5, TRIMCyp, or empty vector. Expression of the TRIM proteins was analyzed by immunoblot 2 d posttransfection. Membranes were probed with antibodies targeting the HA tag and the housekeeping gene HSP90. (C) HEK 293T cells were transfected with LINE-GFP and increasing amounts of human TRIM5α-HA or R437C-HA. Expression of the TRIM proteins was analyzed by immunoblot 2 d posttransfection. Membranes were probed with antibodies targeting the HA tag and the housekeeping gene HSP90. (D) HEK 293T cells expressing two different shRNAs targeting TRIM5α (shT5 #5 and shT5 #6) or scrambled shRNA (shC) were transfected with LINE-GFP. The knockdown of TRIM5α was confirmed by immunoblot using a TRIM5α-specific antibody. (E) ShC or shT5 #6 cells were transfected with LINE-GFP and WT TRIM5α or the shRNA-resistant variants TRIM5αR-HA and R437CR-HA. Expression of the TRIM5α proteins was confirmed by immunoblot using HA-specific antibodies. Five days posttransfection, GFP-positive cells were quantified by flow cytometry. The percentage of LINE-GFP-positive cells is shown as mean of triplicate transfections. Error bars represent SD. One out of three independent experiments is shown. (A, B, D, and E) Statistical analysis was done using one-way ANOVA followed by Tukey’s post hoc test or (C) two-way ANOVA followed by Bonferroni post hoc test. **P < 0.01; ***P < 0.001; ****P < 0.0001, n.s., not significant.
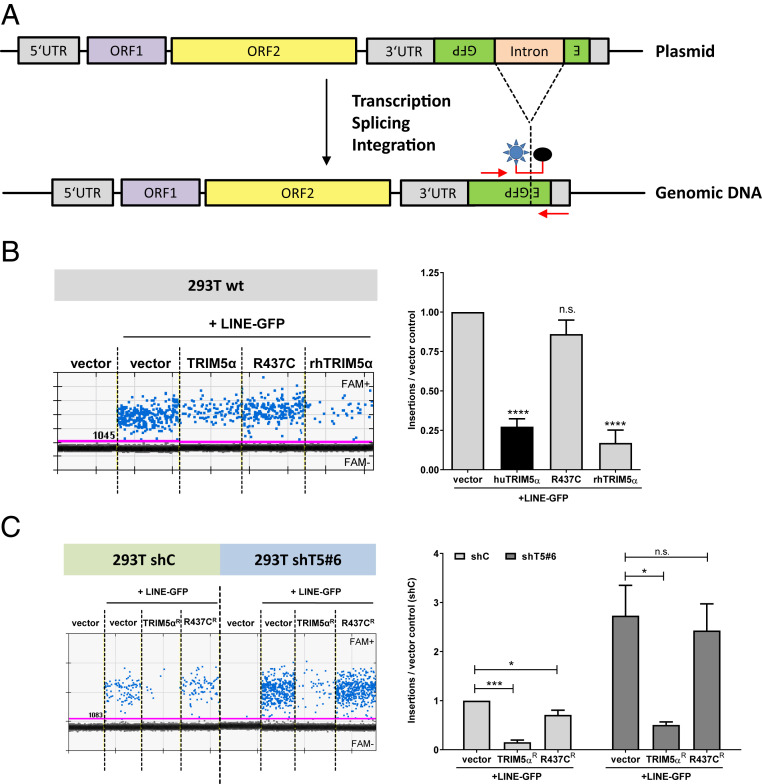
TRIM5α Reduces the Number of Novel LINE-GFP Integrates.
To assess LINE-1 restriction by TRIM5α independently, we developed a digital droplet PCR (ddPCR) approach targeting novel LINE-GFP integrates. For ddPCR, we used GFP-specific oligos to avoid amplification of endogenous elements and an exon-spanning, fluorescent probe recognizing only spliced, and therefore integrated, LINE-GFP elements (Fig. 2A). First, we transfected HEK 293T cells with LINE-GFP reporter plasmid together with rhesus or human TRIM5α, as well as TRIM5α R437C, and isolated genomic DNA 5 d posttransfection. We quantified genomic LINE-GFP insertions, as well as cellular genome copies by targeting the cellular gene RPP30. While the coexpression of human and rhesus TRIM5α reduced the number of LINE-1 integrates by four- to sevenfold, the inactive mutant R437C only slightly affected LINE-1 integration (Fig. 2B). Thus, TRIM5α reduces the number of novel LINE-1 integrates, confirming the anti-LINE-1 activity of TRIM5α independently of reporter gene expression. To determine whether endogenous TRIM5α affects the number of novel LINE-1 integrates, we transfected shTRIM5α and shC cells with LINE-GFP together with shRNA-resistant TRIM5αR and R437CR (Fig. 2C). We found that the number of LINE-GFP integrates per cell was approximately fourfold enhanced in shT5 cells compared to shC cells upon transfection of control vector, suggesting that endogenous TRIM5α reduces LINE-1 integrates (Fig. 2C). In contrast, transfection of TRIM5αR but not of inactive R437CR suppressed LINE-1 integration in shT5 cells, confirming the TRIM5α specificity of the observed phenotype (Fig. 2C).
Fig. 2.
TRIM5α reduces the number of novel LINE-1 integrates. (A) Genomic integration events of LINE-GFP reporter elements were quantified by digital droplet PCR using oligos specific for GFP. The FAM/BHQ1-labeled probe was designed to target the spliced GFP reporter gene, present only upon successful integration of the DNA into the host genome. (B) HEK 293T cells were transfected with LINE-GFP and human (hu) TRIM5α, R437C, or rhesus (rh) TRIM5α. Genomic DNA of transfected cells was isolated 5 d posttransfection and FAM+ integration events were quantified by ddPCR. Insertions per cell are the quotient of LINE-1-positive events and events for the reference gene RPP30 divided by 2. One out of three independent experiments is shown. Results are depicted as the mean of quadruplicate transfection with error bars representing the SD. (C) HEK 293T shC or shT5 #6 cells were transfected with LINE-GFP and shRNA-resistant TRIM5αR and R437CR. LINE-1 insertions were normalized on vector control samples and are shown as mean of triplicate transfections with error bars representing the SD. The average of three biological replicates is shown. Statistical analysis was done using one-way ANOVA followed by Tukey’s post hoc test. *P < 0.05; ***P < 0.001; ****P < 0.0001, n.s., not significant.
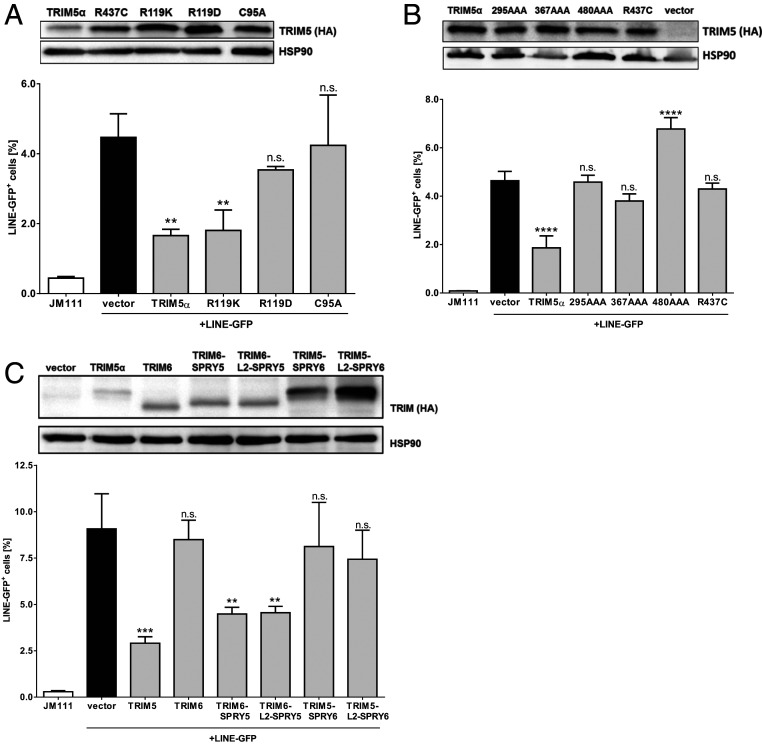
A Functional SPRY Domain and B-Box Are Important for TRIM5α-Mediated Restriction of LINE-1.
TRIM5α interacts with retroviral capsid structures via its SPRY domain and forms multimeric lattices around the capsid, mediated by the CC region and the B-box (4). To determine whether multimerization of TRIM5α is important for LINE-1 restriction, we tested TRIM5α mutants with a functionally defective B-box in LINE-GFP retrotransposition assays (Fig. 3A). The mutation C95A disrupts one of the two zinc-binding sites within the B-box, while charge swapping mutations at residue 119, such as R119D, disturb a hydrophobic patch within the B-box and therefore block proper oligomerization and retroviral restriction (39, 40). In LINE-GFP assays, we found that introducing the mutation R119D resulted in a loss of restriction, whereas a conservative swap preserving the charge, R119K, did not affect LINE-1 restriction. Similarly, TRIM5α C95A lost its ability to restrict LINE-1, indicating that a functional B-box is important for restriction (Fig. 3A). Together, these results suggest that the multimerization of TRIM5α is essential for LINE-1 restriction. The TRIM5α SPRY mutant R437C does not restrict LINE-1 (Fig. 1C). To corroborate this finding, we tested the previously described SPRY mutants 295AAA, 367AAA, and 480AAA, which display triple amino acid changes at different surface patches of the SPRY domain, in retrotransposition reporter assays (41) (Fig. 3B). We found that all tested SPRY mutants failed to block retrotransposition, strongly suggesting that a functional SPRY domain is essential for efficient LINE-1 restriction. In addition, we generated chimeric proteins by exchanging the SPRY domains of TRIM5α and the inactive TRIM6 protein (Fig. 3C). To minimize the possibility that correct folding and positioning of the SPRY domains is impaired, we used two different sets of chimeras, either with or without the linker region (L2) connecting the CC domain and SPRY. In contrast to WT TRIM6, both TRIM6 chimeras harboring the TRIM5α SPRY domain (TRIM6-SPRY5 and TRIM6-L2-SPRY5) restricted LINE-GFP, whereas the SPRY domain of TRIM6 fused to TRIM5α abrogated restriction (Fig. 3C). Together, our results show that the multimerization of TRIM5α via its B-box, as well as the SPRY domain are crucial for LINE-1 restriction, which is reminiscent of the binding and inhibition of exogenous retroviruses.
Fig. 3.
B-Box and SPRY domain are essential for TRIM5α-mediated LINE-1 restriction. (A) HEK 293T cells were transfected with LINE-GFP and TRIM5α-HA or the B-box mutants R119K, R119D, or C95A. (B) HEK 293T cells were transfected with LINE-GFP and TRIM5α-HA, R437C, or the SPRY domain mutants 295AAA, 367AAA, 480AAA. (C) HEK 293T cells were transfected with LINE-GFP and chimeric TRIM5 and TRIM6 proteins, containing the SPRY domain (SPRY) or the SPRY domain together with the linker 2 region (L2-SPRY) of either TRIM5α (SPRY5) or TRIM6 (SPRY6). (A–C) Five days posttransfection, GFP-positive cells were quantified by flow cytometry. The percentage of GFP-positive cells is shown as the mean of triplicate transfections with error bars representing the SD. One of three independent experiments is shown. Cell lysates were analyzed by immunoblot using HA-specific and HSP90-specific antibodies 2 d posttransfection. Statistical analysis was done using one-way ANOVA followed by Tukey’s post hoc test. **P < 0.01; ***P < 0.001; ****P < 0.0001, n.s., not significant.
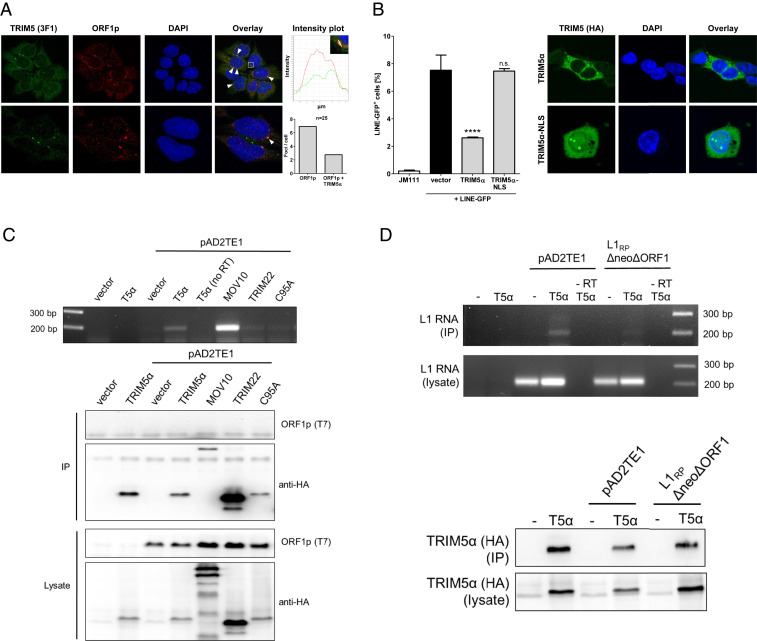
TRIM5α Interacts with LINE-1 in the Cytoplasm.
Since TRIM5α interacts with the retroviral core upon entry, we asked whether TRIM5α might colocalize with LINE-1. Thus, we analyzed endogenous proteins in the embryonal carcinoma cell line 2102EP, which expresses elevated LINE-1 protein levels (42, 43). Using antibodies targeting TRIM5α and ORF1p (44), we identified both proteins to form characteristic aggregates in the cytoplasm, in addition to a more diffuse cytoplasmic distribution (Fig. 4A). While LINE-1 cytoplasmic foci have often been associated with stress granules, TRIM5α cytoplasmic bodies have been described as rather dynamic structures with unknown function (45, 46). We found TRIM5α to be present in about 40% of the cytoplasmic LINE-1 foci (Fig. 4A). Not all TRIM5α and LINE-1 foci colocalize, which might be due to a possible heterogeneity in LINE-1 foci, the dynamic nature of TRIM5α aggregates, or the anti-LINE-1 activity of TRIM5α. In addition, we observed a similar pattern of colocalization in other cell lines such as HEK 293T cells, showing that the colocalization of both proteins is not restricted to 2102EP cells (SI Appendix, Fig. S3). To determine whether the cytoplasmic interaction of TRIM5α and LINE-1 has functional consequences, we introduced a nuclear localization signal (NLS), “PAAKRVKLD,” at amino acid 6 of TRIM5α-HA prior to the RING domain to generate TRIM5α-NLS. Two days upon transfection of HEK 293T cells, TRIM5α localized almost exclusively to the cytoplasm, while TRIM5α-NLS was also found in the nucleus (Fig. 4B). Although TRIM5α-NLS was not exclusively expressed in the nucleus, the characteristic TRIM5α aggregates were only visible within the nucleus, suggesting nuclear localization of a majority of functional TRIM5α-NLS (Fig. 4B). We then compared the anti-LINE-1 activity of WT TRIM5α and TRIM5α-NLS in LINE-GFP reporter assays and found that TRIM5α-NLS lost the ability to restrict LINE-1 retrotransposition (Fig. 4B). These findings suggest that the cytoplasmic localization of TRIM5α is necessary to block retrotransposition, indicating that a direct interaction of TRIM5α with LINE-1 is important for restriction.
Fig. 4.
TRIM5α interacts with LINE-1 RNPs in the cytoplasm. (A) Human embryonal carcinoma cells 2102EP were probed with ORF1p-specific (red) and TRIM5α-specific antibodies (green) an analyzed by confocal microscopy. Cellular DNA was probed with DAPI. Colocalizing signals are colored in yellow, marked (white arrow) and counted in 25 cells under the microscope. Enlargement: Intensity plots illustrate the colocalization of fluorescent signals in defined areas. (B) HEK 293T cells were transfected with TRIM5α-HA or the nuclear localization signal-containing variant TRIM5α-NLS-HA. Five days posttransfection, GFP-positive cells were quantified by flow cytometry. The frequency of GFP-positive cells is shown as mean of triplicate transfections with errors bars indicating the SD. One out of three independent experiments is shown. Statistical analysis was done using one-way ANOVA followed by Tukey’s post hoc test. ****P < 0.0001, n.s., not significant. (C) HEK 293T cells were transfected with LINE-1 expression plasmid encoding T7 tagged ORF1p (pAD2TE1) and empty vector or the indicated HA-tagged protein. After 24 h, cells were lysed and HA-tagged proteins were precipitated using anti-HA antibodies bound to magnetic beads. cDNA was synthesized from RNPs using MLV-RT and a LEAP primer targeting the polyA of bound mRNAs. Resulting cDNA was amplified by PCR using primers targeting the 3′ end of LINE-1 and the LEAP primer sequence. Protein expression in lysates and precipitates was controlled by immunoblot. (D) HEK 293T cells were transfected with WT LINE-1 (pAD2TE1) or a construct lacking ORF1p expression (L1RPΔneoΔORF1) (44) together with empty vector or TRIM5α-HA. After 24 h, cells were lysed and TRIM5α-HA was precipitated using anti-HA antibodies. LEAP reaction and PCR were performed as in C. Due to the lack of ORF1p in cell lysates, LINE-1 RNA input from cell lysates was analyzed. One out of three independent experiments is shown.
Thus, we asked whether TRIM5α directly interacts with LINE-1 RNPs but immunoprecipitation assays analyzing TRIM5α and LINE-1 proteins remained inconclusive due to unspecific binding of TRIM5α to the beads. To test for a possible interaction by different means, we analyzed whether TRIM5α coprecipitates LINE-1 RNA, the central component of RNPs. We transfected HEK 293T cells with expression plasmids encoding LINE-GFP and TRIM5α-HA and precipitated TRIM5α-HA from cell lysates 24 h posttransfection (Fig. 4C). We reverse transcribed precipitated RNA using recombinant reverse transcriptase (MLV-RT) and amplified the resulting cDNA by PCR using LINE-GFP-specific oligonucleotides. LINE-1 was only amplified from samples with TRIM5α or the RNA helicase Mov10 but not from control lysates or lysates containing the inactive TRIM proteins TRIM22 and TRIM5α-C95. This suggests that LINE-1 RNA is specifically precipitated by TRIM5α (Fig. 4C). To determine whether TRIM5α binds directly to LINE-1 RNA or indirectly via interaction with LINE-1 proteins, we analyzed the LINE-1 construct L1RPΔneoΔORF1, which is deficient for ORF1p expression due to a 330-bp deletion in ORF1 (44) (Fig. 4D). Similar to WT LINE-1 RNA, we detected ΔORF1 RNA expression in cell lysates upon transfection. However, in contrast to WT LINE-1, we found very little ΔORF1 RNA in TRIM5α immunoprecipitates (Fig. 4D). This strongly suggests that TRIM5α does not directly bind to LINE-1 RNA but precipitates the RNA by interacting with LINE-1 RNPs, most likely with the main component ORF1p.
TRIM5α Overexpression Reduces Cytosolic LINE-1 Foci.
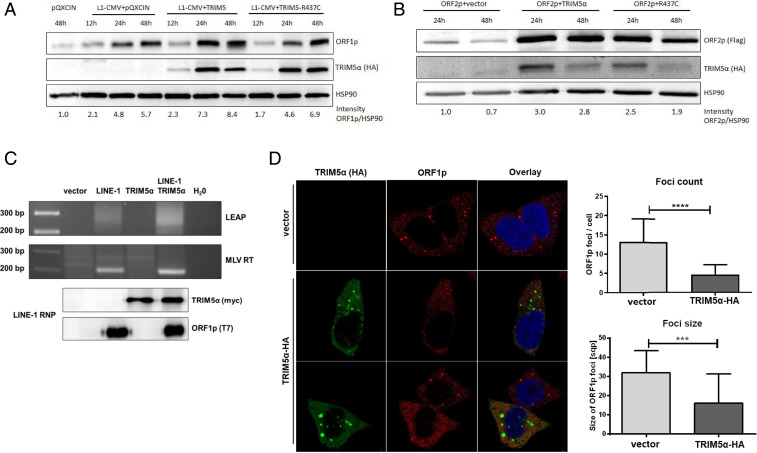
Since TRIM5α induces the degradation of retroviral capsids, we asked whether TRIM5α expression also affects LINE-1 protein abundance. Thus, we overexpressed LINE-1 together with HA-tagged TRIM5α or R437C in HEK 293T cells and analyzed ORF1p protein levels at various time points posttransfection by immunoblot using an ORF1p-specific antibody (Fig. 5A). In LINE-1 transfected cells, we found an increase in ORF1p level over time, which was not affected by the presence of restricting TRIM5α or inactive R437C, suggesting that ORF1p degradation is not contributing to TRIM5α restriction (Fig. 5A and SI Appendix, Fig. S4). Similarly, we did not detect reduced overall amounts of LINE-1 ORF2p upon expression of TRIM5α (Fig. 5B and SI Appendix, Fig. S4). On the contrary, expression of TRIM5α or R437C both led to an increase in ORF2p level, indicating that TRIM5α rather stabilizes ORF2p level. In line with these findings, we also did not detect a decrease in endogenous LINE-1 ORF1p level upon TRIM5α overexpression (SI Appendix, Fig. S5). Together, our data suggest that TRIM5α blocks LINE-1 without inducing the degradation of ORF1p and ORF2p. Next, we asked whether the interaction of TRIM5α with LINE-1 RNPs affects the ability of ORF2p to reverse transcribe LINE-1 RNA (Fig. 5C). Thus, we analyzed the RT activity of ORF2p in vitro using the well-established L1 element amplification protocol (LEAP) (47). We transfected LINE-1 reporter plasmid together with TRIM5α or empty vector and lysed the transfected HEK 293T cells after 48 h. LINE-1 RNPs were purified by ultracentrifugation of the lysates through a sucrose cushion and controlled for ORF1p content by immunoblot. Next, LINE-1 RNA was reverse transcribed by ORF2p present in the purified RNPs or by adding exogenous MLV-RT. Upon amplification of the resulting cDNA using LINE-1-specific oligos, we detected LINE-GFP-specific PCR products in RNPs from LINE-1 transfected cells. Importantly, the presence of TRIM5α did not affect the ability of ORF2p to reverse transcribe its RNA (Fig. 5C). Since TRIM5α has been shown to disrupt retroviral cores, we asked whether TRIM5α mediates the disassembly of functional LINE-1 RNPs. We transfected HEK 293T cells with TRIM5α-HA or control vector and analyzed the formation of LINE-1 cytoplasmic foci by immunofluorescence using antibodies targeting TRIM5α-HA and endogenous ORF1p (44) (Fig. 5D). Strikingly, we found a significant decrease in overall number and size of LINE-1 ORF1p foci in cells expressing TRIM5α. Since LINE-1 RNPs are thought to accumulate in these cytoplasmic foci, the decrease in foci suggests that TRIM5α inhibits formation or mediates the disassembly of LINE-1 RNPs rather than inducing degradation of single LINE-1 proteins.
Fig. 5.
TRIM5α overexpression reduces cytosolic LINE-1 foci. (A) HEK 293T cells were cotransfected with the LINE-1 expression plasmid pAD2TE1 and empty vector, TRIM5α-HA, or R437C-HA. Cells were lysed 12 h, 24 h, and 48 h posttransfection and protein expression was analyzed by immunoblot using ORF1p-specific, HA-specific, or HSP90-specific antibodies. (B) HEK 293T cells were transfected with ORF2-3×-Flag expression plasmid together with empty vector, TRIM5α, or R437C. Cells were lysed 24 h and 48 h posttransfection and ORF2p expression was analyzed by immunoblot using Flag-specific, HA-specific, and HSP90-specific antibodies. (A and B) Band intensities were quantified using an AIDA image analyzer. (C) HEK 293T cells were transfected with LINE-1 expression plasmid (pAD2TE1) together with empty vector or TRIM5α-myc. After 48 h, cells were lysed and RNPs were extracted by ultracentrifugation. RNA was isolated from RNPs and cDNA was synthesized either by MLV-RT or by ORF2p using the LEAP primer targeting RNA polyA. H20 control samples did not contain input RNA (MLV) or RNPs (LEAP). RT products were amplified by PCR with primers targeting the 3′ UTR of LINE-1 and LEAP-specific sequence added by the LEAP prime and visualized on an agarose gel. RNPs were analyzed by immunoblot to control for the presence of ORF1p-T7 and TRIM5α-myc. (D) HEK 293T cells were transfected with pQXCIN or TRIM5α-HA and stained for immunofluorescence 48 hours postinfection (hpi) with antibodies against TRIM5α-HA (HA) and endogenous ORF1p (ORF1p). sqp., square pixel. Exemplary images of one out of three independent experiments are shown. A total of 25 cells per condition were analyzed using ImageJ software. Statistical analysis was done using an unpaired t test with ****P < 0.0001; ***P = 0.0001.
TRIM5α Overexpression Restricts LINE-1 Promoter Activity.
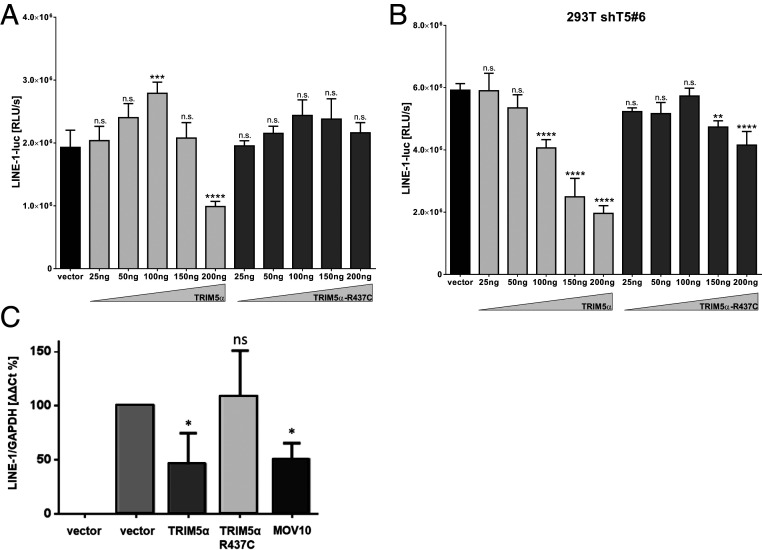
Previous studies report that overexpression of TRIM5α promotes innate immune signaling pathways resulting in the activation of the transcription factors AP-1 and NF-κB (11). We therefore asked whether the innate signaling cascades initiated by TRIM5α might affect transcription from the LINE-1 promoter. Thus, we transfected HEK 293T cells with increasing amounts of TRIM5α or R437C together with a reporter plasmid expressing luciferase under control of the LINE-1 promoter (LINE-1-luc). Two days posttransfection, we found that high amounts of WT TRIM5α but not of inactive R437C strongly reduced LINE-1 promoter activity (Fig. 6A). Of note, overexpression of TRIM5α did not reduce cytomegalovirus (CMV) promoter-driven luciferase expression from a control plasmid, excluding a negative effect of TRIM5α on transcription in general or on luciferase protein stability (SI Appendix, Fig. S6). To exclude an effect of endogenous TRIM5α on the promoter analysis, we repeated the experiment in HEK 293T cells expressing shRNA targeting TRIM5α (Fig. 6B). In the absence of endogenous protein, we found that transiently expressed TRIM5α reduced LINE-1 promoter activity at even lower concentrations, while overexpression of R437C did not affect luciferase activity (Fig. 6B). To confirm the effect of TRIM5α expression on the LINE-1 promoter by independent means, we quantified RNA expression from LINE-1 reporter plasmids by qRT-PCR (Fig. 6C). At 8 h posttransfection, we found that WT TRIM5α significantly reduced LINE-1 RNA level similar to the known inhibitor Mov10. In contrast, R437C transfected cells did not show decreased LINE-1 RNA level compared to empty vector transfected cells. Of note, LINE-1 RNA level did not differ significantly between samples after 24 h, suggesting an early effect of TRIM5α expression on LINE-1 promotor activity. Together, these results suggest that overexpression of TRIM5α but not of inactive R437C inhibits LINE-1 promoter activity, most likely by inducing innate signaling events as described previously (11).
Fig. 6.
TRIM5α reduces LINE-1 promoter activity. (A) HEK 293T cells or (B) shT5 #6 cells were cotransfected with a LINE-1 promoter-driven luciferase expression plasmid (LINE-1-luc) and increasing amounts of TRIM5α, R437C, or empty vector. Two days posttransfection, cells were lysed and luciferase activity was determined. The mean luciferase activity (relative light units [RLUs]) of quadruplicate reactions is depicted with error bars representing the SD. (C) HEK 293T cells were transfected with LINE-GFP reporter plasmids together with TRIM5α-myc, R437C-myc, Mov10-myc, or empty vector at a ratio of 3:1. Cellular mRNA was isolated at 8 h posttransfection and quantified in quadruplicates by qRT-PCR using oligos targeting the intron sequence of the GFP reporter gene. Results were normalized on GAPDH using the ΔΔCt analysis. The average of four biologically independent assays is shown with error bars indicating the SD. Statistical analysis was done using one-way ANOVA followed by Tukey’s post hoc test. *P < 0.05; **P < 0.01; ****P < 0.0001, n.s., not significant. One out of three independent experiments is shown.
TRIM5α Induces Innate Immune Signaling in the Presence of LINE-1.
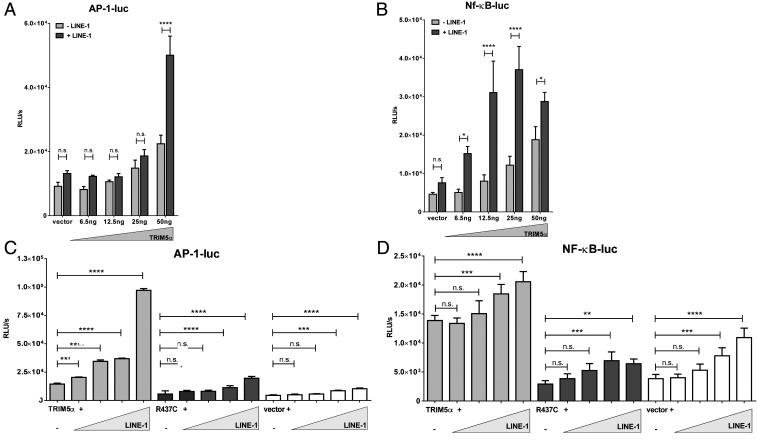
The presence of retroviruses has been shown to amplify immune signaling mediated by TRIM5α, suggesting that TRIM5α acts as pattern recognition receptor scavenging for incoming retroviral capsid lattices (11, 18, 48). To test whether TRIM5α also senses the presence of LINE-1, we set up reporter assays by transiently transfecting HEK 293T cells with plasmids expressing luciferase under the control of an AP-1-dependent (AP-1-luc) or an NF-κB-dependent (NF-κB-luc) promoter (Fig. 7). We transfected low amounts of TRIM5α to avoid induction of immune signaling already due to overexpression of the protein. We found that coexpression of increasing amounts of TRIM5α slightly enhanced the AP-1 and NF-κB-driven luciferase expression compared to empty vector transfected cells (Fig. 7 A and B). Intriguingly, the signaling activity was strongly enhanced in the presence of cotransfected LINE-1 (three- to fourfold), indicating that the presence of LINE-1 amplifies the TRIM5α-mediated induction of innate immune signaling (Fig. 7 A and B). Next, we transfected HEK 293T cells with low amounts of TRIM5α together with increasing amounts of LINE-1 or control vector and the AP-1-luc and NF-κB-luc reporter plasmids. While TRIM5α alone only marginally enhanced luciferase activity, coexpression of LINE-1 resulted in a dose-dependent increase in AP-1-luc and NF-κB-luc expression (Fig. 7 C and D). The up-regulation of luciferase activity was only found in TRIM5α-expressing cells but was absent in R437C or control transfected cells. Similarly, in shT5 #6 cells, we observed a direct correlation between the amount of transfected LINE-1 and luciferase reporter activities upon transfection of TRIM5αR (SI Appendix, Fig. S7 A and B). The RING domain of TRIM5α has been shown to be crucial for innate immune signaling (11). Consistently, we did not observe enhanced NF-κB or AP-1 signaling upon cotransfection of TRIM5αR-ΔRING and increasing amounts of LINE-1 (SI Appendix, Fig. S7 A and B). In addition, we only detected a minor increase in signaling upon transfection of TRIM5α-ΔSPRY (SI Appendix, Fig. S7 A and B). This suggests that the SPRY-mediated interaction with LINE-1 is important for signaling, similar to what has been proposed for retroviruses (11). Next, we transfected increasing amounts of LINE-1 and luciferase reporter plasmid in shC and shT5 #6 cells. Although the effects were less pronounced compared to TRIM5α overexpression, we observed an enhanced luciferase signal in response to LINE-1 in shC compared to shT5 cells, suggesting that also endogenous TRIM5α induces AP-1 and NF-κB signaling in response to LINE-1 (SI Appendix, Fig. S7 C and D).
Fig. 7.
TRIM5α induces innate signaling upon LINE-1 interaction. (A and B) HEK 293T cells were transfected with reporter plasmids expressing luciferase under the control of an (A) AP-1 (AP-1-luc) or (B) NF-κB-dependent promoter (NF-κB-luc), together with LINE-1 plasmid (+ LINE-1) or control vector (− LINE-1), and increasing amounts of TRIM5α expression plasmid. (C and D) HEK 293T cells were transfected with (C) AP-1-luc or (D) NF-κB-luc, together with TRIM5α, R437C, or empty vector, and increasing amounts of LINE-1. Two days posttransfection, luciferase activity of the cell lysates was determined. Mean luciferase activity (RLUs) of quadruplicate transfections is depicted with error bars representing the SD. One out of three independent experiments is shown. (A and B) Statistical analysis was done using two-way ANOVA followed by Bonferroni post hoc test or (C and D) one-way ANOVA followed by Tukey’s test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant.
The TRIM5α-Mediated Immune Signaling in Response to LINE-1 Restricts the Promoter Activity of LINE-1.
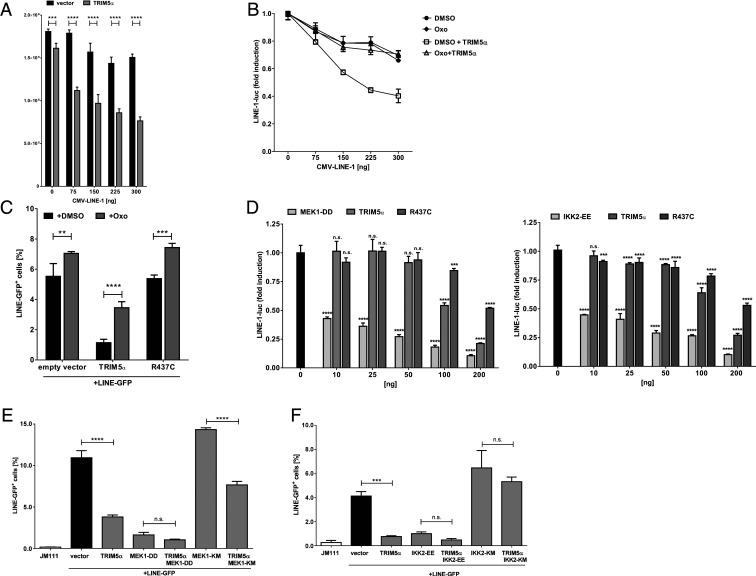
First, we transfected the LINE-1 promoter reporter plasmid, LINE-1-luc, together with increasing amounts of a LINE-1 construct containing an additional CMV promoter (pAD2TE1, CMV-LINE-1) in HEK 293T cells (Fig. 8A). Here, we used LINE-1 under control of an additional CMV promotor to ensure stable expression of LINE-1, since TRIM5α does not affect CMV promoter activity (SI Appendix, Fig. S6). We found that in the absence of exogenous TRIM5α, transfection of increasing amounts of CMV-LINE-1 did only minimally affect LINE-luc expression. In contrast, TRIM5α-transfected cells showed a dose-dependent decrease in LINE-1 promoter activity upon cotransfection with increasing amounts of CMV-LINE-1 (Fig. 8A). Thus, the TRIM5α-mediated immune signaling in response to LINE-1 results in a decreased LINE-1 promoter activity. Several groups established that the multimerization of TRIM5α initiates immune signaling by triggering polyubiquitination of TRIM5α, followed by activation of the TAK1 kinase complex (11, 12, 18, 48). To test whether TRIM5α also signals through TAK1 to block LINE-1 promoter activity, we expressed increasing amounts of CMV-LINE-1 together with LINE-1-luc reporter plasmid and TRIM5α in the presence of the TAK1 inhibitor 5Z-7-oxozeaenol (Oxo) (Fig. 8B). In untreated TRIM5α-expressing cells, the amount of transfected CMV-LINE-1 negatively correlated with LINE-1 promoter activity. However, this dose-dependent reduction was absent from TRIM5α-transfected cells treated with Oxo or in cells lacking TRIM5α (Fig. 8B). This indicates that in response to enhanced LINE-1 levels, TRIM5α induces AP-1 and NF-κB pathways via TAK1 activation to block LINE-1 transcription. In addition, we found that Oxo relieved the TRIM5α-mediated block to LINE-GFP retrotransposition (Fig. 8C), demonstrating that the TRIM5α-dependent activation of the TAK1 complex contributes to restriction. Interestingly, Oxo did not completely abrogate inhibition, indicating that additional mechanisms contribute to restriction, such as the disassembly of LINE-1 RNPs following the direct interaction of TRIM5α with LINE-1 (Fig. 5D). Oxo also slightly increased retrotransposition in R437C and empty vector transfected cells, suggesting a basal activation of TAK1 upon LINE-1 transfection, most likely via endogenous TRIM5α (Fig. 8C).
Fig. 8.
TRIM5α-mediated immune signaling inhibits LINE-1 promoter activity. (A) HEK 293T cells were transfected with a LINE-1 promoter-driven luciferase expression plasmid (LINE-1-luc), TRIM5α or empty vector, and increasing amounts of pAD2TE1, which contains an additional CMV promotor (CMV-LINE-1). Two days posttransfection, cells were lysed and luciferase activity was determined. RLUs are shown as mean of quadruplicate transfections with error bars indicating the SD. (B) HEK 293T cells were transfected with LINE-1-luc, TRIM5α or empty vector, and increasing amounts of CMV-LINE-1. Six hours posttransfection, cells were treated with either DMSO or the TAK1 inhibitor (5Z)-7-oxozeaenol (Oxo). Two days posttransfection, cellular luciferase activity was determined and normalized to lysates lacking CMV-LINE-1 (0 ng). RLUs are shown as mean of quadruplicate transfections with error bars indicating the SD. (C) HEK 293T cells were transfected with LINE-GFP and TRIM5α, R437C, or empty vector. Six hours posttransfection, DMSO or Oxo was added to the culture medium. Five days posttransfection, GFP-positive cells were quantified by flow cytometry. RLUs are shown as mean of triplicate transfections with error bars indicating the SD. (D) HEK 293T cells were transfected with LINE-1-luc and increasing amounts of the constitutively active MAP kinase kinase MEK-1 (MEK1-DD), the constitutively-active NF-κB activator IKK2 (IKK2-EE), TRIM5α, or R437C. Two days posttransfection, cellular luciferase activity was determined and normalized to lysates lacking CMV-LINE-1 (0 ng). RLUs are shown as mean of quadruplicate transfections with error bars indicating the SD. (E) HEK 293T cells were transfected with LINE-GFP and TRIM5α, MEK1-DD, a dominant-negative MEK-1 variant (MEK1-KM), or a combination of TRIM5α with one of the kinases. Five days posttransfection, GFP-positive cells were quantified by flow cytometry and are plotted as mean of triplicate transfections with error bars indicating the SD. One of two experiments is depicted. (F) HEK 293T cells were transfected with LINE-GFP and TRIM5α, IKK2-EE, a dominant-negative IKK2 variant (IKK2-KM), or a combination of TRIM5α with one of the kinases. Five days posttransfection, GFP-positive cells were quantified by flow cytometry and are plotted as mean of triplicate transfections with error bars indicating the SD. If not indicated otherwise, one out of three independent experiments is shown. (A–D) Statistical analysis was done using two-way ANOVA followed by Bonferroni correction or (E and F) one-way ANOVA followed by Tukey’s post hoc test. **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant.
To further validate the inhibitory effect of the TRIM5α-mediated immune signaling, we analyzed the role of the NF-κB pathway and the mitogen-activated protein kinase (MAPK) pathway, which activates AP-1, in LINE-1 restriction. MEK1 is a MAP kinase kinase (MAPKK) and acts as part of the MAP kinase signaling pathway. We found that coexpression of the constitutively active variant of MEK1, MEK1-DD, inhibited the LINE-1 promoter activity similar to overexpression of TRIM5α (Fig. 8D). Likewise, expression of the constitutively active variant of the kinase IKK2, IKK2-EE, which activates NF-κB signaling by phosphorylating the inhibitor IκB, also resulted in a strongly reduced LINE-1 promoter activity (Fig. 8D). In addition to the effect on the LINE-1 promoter, we found that overexpression of the constitutive-active variants MEK1-DD and IKK2-EE also inhibited LINE-1 retrotransposition in GFP reporter assays (Fig. 8 E and F). Thus, activation of both pathways by independent means results in LINE-1 promotor inhibition, confirming LINE-1 promotor inhibition as a mechanism of the TRIM5α-mediated restriction. In contrast, coexpression of the inactive variants IKK2-KM and MEK-KM did not affect LINE-GFP activity and even relieved, at least in part, the TRIM5α-mediated block to LINE-1, suggesting that both NF-κB and MAPK signaling are required for the TRIM5α-mediated inhibition of LINE-1 (Fig. 8 E and F). Fittingly, coexpression of the dominant-negative NF-κB inhibitor DN-IκB also relieved the TRIM5α-mediated restriction of LINE-1 (SI Appendix, Fig. S8) (49). This further corroborates our finding that the TRIM5α-mediated activation of immune signaling pathways contributes to restriction of LINE-1 by inhibiting the LINE-1 promoter in a negative feedback loop.
Discussion
Within this study, we found that TRIM5α inhibits the retrotransposition of LINE-1 elements, the only autonomously active mobile genetic elements in the human genome. In LINE-1 reporter assays and ddPCR-based integration assays, we identified human and rhesus TRIM5α to be active against LINE-1 (Figs. 1B and 2B). This finding stands in stark contrast to the previously described species specificity of TRIM5α towards HIV. Since the specificity of TRIM5α restriction is mediated by its SPRY domain, we asked whether other TRIM proteins harboring a SPRY domain might also block LINE-1. Thus, we analyzed TRIM6, TRIM22, and TRIM34, which cluster together with TRIM5α on chromosome 11, in LINE-GFP retrotransposition assays. However, we found that these TRIM proteins lack anti-LINE-1 activity and that the block is specific for TRIM5α (Fig. 1A). In addition, we analyzed six previously described alleles of rhesus TRIM5α (Mamu1-5 and Mamu7), which differ in their SPRY domain and consequently target different lentiviral strains (35, 50). We found that all alleles tested repressed retrotransposition equally effectively, showing that the antiretroviral specificity of the different alleles does not correlate with LINE-1 restriction (Fig. 1B). Interestingly, Mamu7 or rhesus TRIMCyp, in which the SPRY domain is replaced by Cyclophilin A and which is not active against HIV-1 (36, 50), did also strongly inhibit LINE-1 retrotransposition similarly to human TRIM5α. The sensitivity toward TRIMCyp suggests that prolyl isomerase Cyclophilin A binds to LINE-1 RNPs and that TRIMCyp interacts with RNPs via a similar mechanism. Interestingly, recent work showed that another prolyl isomerase, PIN1, indeed binds to ORF1p and that this interaction is important for function (51). Together, our results suggest that the restriction of endogenous LINE-1 elements is a conserved function for TRIM5α proteins of primate origin.
Although the specificity of LINE-1 restriction differs from the block to lentiviral infection, the requirement of the SPRY domain and the B-box (Fig. 3), as well as the cytoplasmic localization of TRIM5α (Fig. 4B), suggests a similar mechanism of recognition for HIV and LINE-1. Thus, a direct interaction of the SPRY domain with an unknown structure in LINE-1 RNPs seems very likely. Indeed, we found that TRIM5α coprecipitates with LINE-1 RNA (Fig. 4 C and D), which is central to LINE-1 RNP formation (52, 53), strongly suggesting that TRIM5α interacts with LINE-1 RNPs. TRIM5α did not precipitate RNA from a LINE-1 construct lacking functional ORF1, indicating that TRIM5α does not directly bind LINE-1 RNA but via interaction with LINE-1 RNPs, most likely ORF1p (Fig. 4D). Fittingly, we found that TRIM5α colocalizes with ORF1p in distinct foci in the cytoplasm, demonstrating a spatial proximity of LINE-1 and TRIM5α (Fig. 4A). Both proteins do not always colocalize and also cluster in separate foci. This might be due to the reported dynamic nature of TRIM5α (45) or due to the anti-LINE-1 activity of TRIM5α. Supporting the latter, we found that overexpression of TRIM5α significantly reduces the number and the size of ORF1p foci as well as colocalization (Fig. 5D). Mechanistically, it is very well conceivable that TRIM5α mediates the dissolving of the complexes by targeting functional LINE-1 RNPs, thereby interfering with LINE-1 retrotransposition.
Of note, we were not able to confirm direct binding of TRIM5α to single LINE-1 proteins in immunoprecipitation assays. Thus, similar to retroviral capsid recognition (4), TRIM5α might recognize a repetitive epitope with relatively low affinity and only upon multimerization on the target structure avidity would increase and lead to a specific recognition of LINE-1 RNPs by TRIM5α. ORF1p trimers represent the most abundant building blocks of LINE-1 RNPs and package LINE-1 RNA into large assemblies that theoretically allow for the formation of structured hexagonal networks (54), a formation also typical for retroviral capsids. Since TRIM5α has been shown to multimerize and form hexagonal lattices on retroviral capsid surfaces, it is tempting to speculate that similar mechanisms contribute to recognition and restriction of LINE-1.
Upon recognition of viral capsids, TRIM5α induces ubiquitin-dependent signaling pathways resulting in the activation of the transcription factors AP-1 and NF-κB and culminating in the induction of the so-called antiviral state (11). Similarly, we found that TRIM5α induces enhanced AP-1 and NF-κB signaling in the presence of LINE-1, suggesting that TRIM5α acts as a pattern recognition receptor for the intracellular accumulation of LINE-1 RNPs (Fig. 7). Despite the importance of ubiquitination for immune signaling and restriction, the role of the RING domain during viral restriction is still unclear (55–57). Thus, we decided to focus on the important downstream pathways that activate AP-1 and NF-κB through initiation of mitogen-activated protein kinase (MAPK) signaling cascades. Our LINE-1 retrotransposition assays in the presence of a pharmacological TAK1 inhibitor or modified AP-1 and NF-κB signaling intermediates suggest that TRIM5α induces the same signaling pathways in response to LINE-1 as in retroviral recognition (Fig. 8). These results indicate that the pattern recognition receptor TRIM5α recognizes LINE-1 RNP in the cytoplasm and subsequently induces AP-1 and NF-κB activation to down-regulate LINE-1 promoter activity in a negative feedback loop. To decipher the mechanism of the TRIM5α-mediated LINE-1 promoter block, we asked whether AP-1, NF-κB, or both transcription factors are important for inhibition. We found that transfection of LINE-1 together with low levels of TRIM5α led to an up-regulation of both AP-1-dependent and NF-κB-dependent reporter gene expression, emphasizing the importance of both transcription factors for LINE-1 inhibition (Fig. 7). In line with these findings, overexpression of constitutively active members of both the MAP kinase and the NF-κB pathway resulted in efficient inhibition of LINE-1 in retrotransposition assays, while overexpression of inactive mutants did not block LINE-GFP (Fig. 8 E and F). Similarly, expression of dominant-negative IκB, DN-IκB, which has been shown to inhibit NF-κB signaling, did impair the TRIM5α-mediated restriction of LINE-GFP, indicating that the induction of NF-κB is also crucial for the TRIM5α-mediated restriction of LINE-1 retrotransposition (SI Appendix, Fig. S8). Importantly, we found that blocking the TRIM5α-mediated signaling did not completely restore LINE-GFP activity (Fig. 8C), suggesting additional inhibitory effects of TRIM5α on retrotransposition, such as the direct interaction and disassembly of LINE-1 RNPs by TRIM5α with LINE-1 RNPs (Fig. 5 C and D). While the induction of AP-1 and NF-κB suppressed the promoter of LINE-1 (Fig. 8D), we did not find evidence for direct binding of the transcription factors to the promoter sequence. Mutating two putative consensus-binding motifs for each transcription factor within the LINE-1 promoter did not affect the TRIM5α-mediated down-regulation of its activity (SI Appendix, Fig. S9). Although AP-1 and NF-κB might interact with alternative binding sites, our findings do not exclude an indirect effect of TRIM5α signaling on LINE-1. NF-κB and AP-1 regulate the expression of many inflammatory and antiviral genes, one of which might mediate LINE-1 promoter inhibition or act on LINE-1 in general. Next steps will clearly include the identification of the yet unknown AP-1 and NF-κB-activated inhibitors of LINE-1 promoter activity and therefore shed light on the regulation of LINE-1 transcription in general.
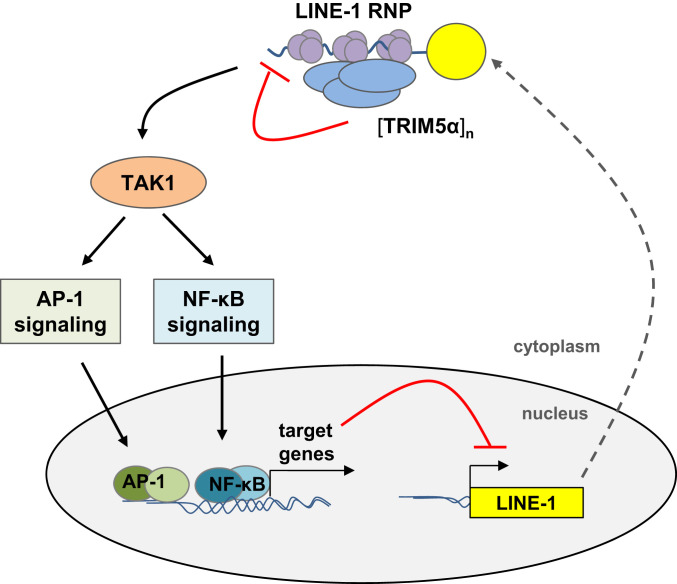
Together, we identified the endogenous retroelement LINE-1 as an “intracellular pathogen” targeted by the human restriction factor TRIM5α. We found that TRIM5α inhibits LINE-1 retrotransposition by two distinct mechanisms (Fig. 9). In addition to direct interaction and interference with LINE-1 RNPs in the cytoplasm, we confirm the role of TRIM5α as pattern recognition receptor and identify LINE-1 RNPs as a potential molecular pattern recognized by TRIM5α. Upon engagement, TRIM5α activates innate immune signaling pathways leading to a down-regulation of the LINE-1 promoter. Thus, TRIM5α joins the ranks of a “guardian of the genome” due to sensing and restriction of excess LINE-1 in the cytoplasm. It will be exciting in future studies to unravel the role of TRIM5α in LINE-1-associated malignancies including monogenetic diseases, tumorigenesis, or autoinflammation.
Fig. 9.
Model of TRIM5α-mediated restriction of LINE-1 retroelements. TRIM5α colocalizes and interacts with LINE-1 RNPs in the cytoplasm via its SPRY domain. The sensing of LINE-1 RNPs initiates the multimerization of TRIM5α and the downstream activation of the TAK1 complex. Subsequently, activated MAP kinase and NF-κB signaling cascades lead to a block of LINE-1 retrotransposition, most likely via expression of AP-1 and NF-κB target genes that down-regulate LINE-1 promoter activity.
Methods
Detailed materials and methods for plasmids and constructs, cell culture, retrotransposition reporter assays, signaling reporter assays, immunoblot and immunofluorescence analysis, RNA-IP, ddPCR assays, and LEAP assays are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Andrea Kirmaier and Welkin Johnson (Boston College) and William Diehl (University of Massachusetts) for sharing rhesus TRIM5α-encoding plasmids, expertise, and methods. We thank Gerald Schumann (Paul Ehrlich Institute), John Goodier (Johns Hopkins University), John Moran (University of Michigan), and Oliver Weichenrieder (Max Planck Institute for Developmental Biology) for sharing reagents and fruitful discussions. T.G., B.V., J.D., J.L., and S.W. were funded by the University Hospital Erlangen Interdisciplinary Center for Clinical Research (IZKF) Grant A67, the IZKF ELAN-Fond (14-08-01-1), and the German Research Foundation (GR-3355/6-1; GRK2504/A4).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922366117/-/DCSupplemental.
Data Availability.
All data necessary for replication are included in the submission.
References
- 1.Stremlau M. et al., The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Nisole S., Stoye J. P., Saïb A., TRIM family proteins: Retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3, 799–808 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Ganser-Pornillos B. K., Pornillos O., Restriction of HIV-1 and other retroviruses by TRIM5. Nat. Rev. Microbiol. 17, 546–556 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganser-Pornillos B. K. et al., Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. U.S.A. 108, 534–539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y. L. et al., Primate TRIM5 proteins form hexagonal nets on HIV-1 capsids. eLife 5, e16269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribeiro C. M. et al., Receptor usage dictates HIV-1 restriction by human TRIM5α in dendritic cell subsets. Nature 540, 448–452 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Jimenez-Guardeño J. M., Apolonia L., Betancor G., Malim M. H., Immunoproteasome activation enables human TRIM5α restriction of HIV-1. Nat. Microbiol. 4, 933–940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatziioannou T., Perez-Caballero D., Yang A., Cowan S., Bieniasz P. D., Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. U.S.A. 101, 10774–10779 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yap M. W., Nisole S., Lynch C., Stoye J. P., Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. U.S.A. 101, 10786–10791 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perron M. J. et al., TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11827–11832 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pertel T. et al., TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher A. J. et al., Trivalent RING assembly on retroviral capsids activates TRIM5 ubiquitination and innate immune signaling. Cell Host Microbe 24, 761–775.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawyer S. L., Wu L. I., Emerman M., Malik H. S., Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U.S.A. 102, 2832–2837 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp P. M., Bailes E., Robertson D. L., Gao F., Hahn B. H., Origins and evolution of AIDS viruses. Biol. Bull. 196, 338–342 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Andersson M. L. et al., Diversity of human endogenous retrovirus class II-like sequences. J. Gen. Virol. 80, 255–260 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Belshaw R. et al., Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. U.S.A. 101, 4894–4899 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tareen S. U., Sawyer S. L., Malik H. S., Emerman M., An expanded clade of rodent Trim5 genes. Virology 385, 473–483 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lascano J., Uchil P. D., Mothes W., Luban J., TRIM5 retroviral restriction activity correlates with the ability to induce innate immune signaling. J. Virol. 90, 308–316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancks D. C., Kazazian H. H. Jr., Active human retrotransposons: Variation and disease. Curr. Opin. Genet. Dev. 22, 191–203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck C. R., Garcia-Perez J. L., Badge R. M., Moran J. V., LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 12, 187–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denli A. M. et al., Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 163, 583–593 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Hohjoh H., Singer M. F., Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15, 630–639 (1996). [PMC free article] [PubMed] [Google Scholar]
- 23.Martin S. L., Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 11, 4804–4807 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei W. et al., Human L1 retrotransposition: Cis preference versus trans complementation. Mol. Cell. Biol. 21, 1429–1439 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esnault C., Maestre J., Heidmann T., Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Kazazian H. H. Jr., Moran J. V., Mobile DNA in health and disease. N. Engl. J. Med. 377, 361–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muotri A. R. et al., Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Coufal N. G. et al., L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Upton K. R. et al., Ubiquitous L1 mosaicism in hippocampal neurons. Cell 161, 228–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodier J. L., Restricting retrotransposons: A review. Mob. DNA 7, 16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martín-Vicente M., Medrano L. M., Resino S., García-Sastre A., Martínez I., TRIM25 in the regulation of the antiviral innate immunity. Front. Immunol. 8, 1187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grassi D. A., Jönsson M. E., Brattås P. L., Jakobsson J., TRIM28 and the control of transposable elements in the brain. Brain Res. 1705, 43–47 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Sawyer S. L., Emerman M., Malik H. S., Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 3, e197 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostertag E. M., Prak E. T., DeBerardinis R. J., Moran J. V., Kazazian H. H. Jr., Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28, 1418–1423 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirmaier A. et al., TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 8, e1000462 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson S. J. et al., Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc. Natl. Acad. Sci. U.S.A. 105, 3557–3562 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman R. M. et al., Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 4, e1000003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X. et al., Inhibitory effect of human TRIM5alpha on HIV-1 production. Microbes Infect. 12, 768–777 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Diaz-Griffero F. et al., A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 83, 10737–10751 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstone D. C. et al., Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc. Natl. Acad. Sci. U.S.A. 111, 9609–9614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sebastian S. et al., An invariant surface patch on the TRIM5alpha PRYSPRY domain is required for retroviral restriction but dispensable for capsid binding. J. Virol. 83, 3365–3373 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Josephson R. et al., Qualification of embryonal carcinoma 2102Ep as a reference for human embryonic stem cell research. Stem Cells 25, 437–446 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Perez J. L. et al., LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 16, 1569–1577 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Raiz J. et al., The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40, 1666–1683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campbell E. M. et al., TRIM5 alpha cytoplasmic bodies are highly dynamic structures. Mol. Biol. Cell 18, 2102–2111 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodier J. L., Zhang L., Vetter M. R., Kazazian H. H. Jr., LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27, 6469–6483 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kulpa D. A., Moran J. V., Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 13, 655–660 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Uchil P. D. et al., TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J. Virol. 87, 257–272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voll R. E. et al., NF-kappa B activation by the pre-T cell receptor serves as a selective survival signal in T lymphocyte development. Immunity 13, 677–689 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Wilson S. J. et al., Rhesus macaque TRIM5 alleles have divergent antiretroviral specificities. J. Virol. 82, 7243–7247 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cook P. R., Jones C. E., Furano A. V., Phosphorylation of ORF1p is required for L1 retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 112, 4298–4303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khazina E. et al., Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 18, 1006–1014 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Basame S. et al., Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition. J. Mol. Biol. 357, 351–357 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Khazina E., Weichenrieder O., Human LINE-1 retrotransposition requires a metastable coiled coil and a positively charged N-terminus in L1ORF1p. eLife 7, e34960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diaz-Griffero F. et al., Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology 349, 300–315 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Fletcher A. J. et al., TRIM5α requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription. EMBO J. 34, 2078–2095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campbell E. M. et al., TRIM5α-Mediated ubiquitin chain conjugation is required for inhibition of HIV-1 reverse transcription and capsid destabilization. J. Virol. 90, 1849–1857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data necessary for replication are included in the submission.