Abstract
Histone acetylation plays important roles in regulating DNA metabolic processes, including many DNA repair pathways. The nucleotide excision repair (NER) pathway is critical for removing bulky, helix-distorting DNA lesions, such as UV light-induced photoproducts, but the activity of this pathway is significantly inhibited when lesions reside in nucleosomes. Recent studies have indicated that histone acetyltransferase (HAT) activity may be induced in response to UV damage, in order to facilitate the repair of UV-induced lesions in chromatin. Budding yeast (Saccharomyces cerevisiae) is an important model system for studying the functional roles of histone acetylation and HATs in NER, due to the ease of genetically altering HAT activity or acetylated lysine residues in histones. Here, we describe protocols for measuring the repair of cyclobutane pyrimidine dimers (CPDs), the major UV-induced photoproduct, in yeast strains deficient in HAT activity, either due to gene deletion or rapid anchor-away depletion of the HAT enzyme. Methods for measuring CPD repair in bulk chromatin, as well as individual chromatin loci, are detailed below.
Keywords: Nucleotide excision repair, Chromatin, Histone, Anchor-away, Histone acetyltransferase, Cyclobutane pyrimidine dimer, UV light
1. Introduction
The packaging of DNA into chromatin not only helps compact eukaryotic DNA in the nucleus, but also represents an important mechanism for regulating the activity of many DNA metabolic processes, including DNA repair [1]. Chromatin is primarily comprised of nucleosomes, in which ~147 bp of DNA is wrapped around an octamer of histone proteins [2]. However, even this lowest level of chromatin organization presents a formidable barrier to many DNA repair pathways [3–6]. To enable DNA repair and other DNA metabolic processes to operate in chromatin, nucleosomes are dynamically rearranged and modified by a number of different classes of chromatin modifying enzymes. An important class of chromatin modifying enzymes is lysine (K) acetyltransferases (KATs), which acetylate lysine residues in histones (and other proteins) to regulate chromatin dynamics and accessibility [7]. Recent studies have implicated histone acetylation in regulating a number of DNA repair pathways, including nucleotide excision repair [8–10].
Nucleotide excision repair (NER) is responsible for removing bulky, helix-distorting DNA lesions induced by ultraviolet (UV) light, cigarette smoke, and other exogenous and endogenous sources [11]. This class of lesions must be efficiently repaired by the NER pathway to avoid mutagenesis or cell death. Indeed, individuals with defects in the NER pathway (e.g., xeroderma pigmentosum patients) have greatly elevated rates of skin cancer due to their inability to repair UV-induced photoproducts [12, 13] such as cyclobutane pyrimidine dimers (CPDs). Regional variations in NER activity across the genome due to differences in chromatin state and DNA accessibility have been shown to correlate with mutation density in skin cancers [14], highlighting the impact of chromatin and chromatin-associated modifications, such as histone acetylation, on NER activity and cancer-associated mutagenesis.
Previous studies have suggested that NER is facilitated in chromatin by the action of histone modifying enzymes, such as histone acetyltransferases (HATs) [8–10]. The functional roles of HATs and acetylated histone residues in NER can be studied in yeast by analyzing the impact of HAT deletions or histone lysine mutants on repair of UV-induced CPD lesions. This can be assessed on a global scale by quantifying the number of CPD lesions in the genome using T4 endonuclease V (T4 endoV) digestion (e.g., [15]). T4 endoV generates single-stranded DNA nicks specifically at sites of CPD lesions in DNA [16], and the number of T4 endoV-induced nicks per kilobase (kb) of genomic DNA can be quantified by denaturing alkaline gel electrophoresis [17, 18]. Removal of CPD lesions by NER during the repair time course is reflected by a concomitant decrease in the number of T4 endoV-induced nicks. Hence, assessing CPD removal using this assay provides an accurate measure of global repair kinetics in yeast, both in wild-type cells or HAT or histone mutant cells. Additionally, the repair of CPD lesions can be quantified at individual genomic loci in different chromatin environments using a related experimental approach, in which the repair of CPD lesions is analyzed by southern blot using a locus-specific probe following T4 endoV digestion and alkaline gel electrophoresis [19–22]. Typically, repair is analyzed at yeast loci representing different chromatin environments, including transcriptionally active chromatin (RPB2 locus), transcriptionally repressed chromatin (GAL10 locus, repressed when yeast are grown in glucose-containing media), and transcriptionally silent heterochromatin (HML locus). These experiments can provide insights into the role of HATs and histone acetylation in different chromatin environments.
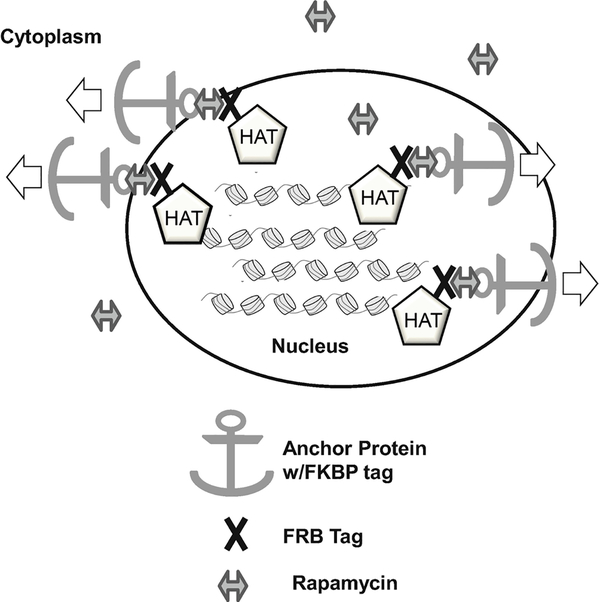
A potential complicating factor in studying yeast deletion mutants is that in some cases, differences in repair can be a secondary effect of changes in repair gene expression due to the deletion mutant (e.g., [23]). This is particularly important for studying HATs and other chromatin modifying enzymes, since these enzymes are known to be important regulators of gene expression. Hence, an important alternative method for studying the roles of HATs in NER is to rapidly deplete the HAT enzyme from the yeast nucleus using the anchor-away approach [24] (Fig. 1). The speed by which this approach depletes HAT activity can be used to mitigate potential indirect effects due to HAT-dependent changes in gene expression. The anchor-away strategy can also be used to study the function of essential chromatin modifying enzymes, for which the deletion mutant is inviable. In this chapter, we describe methods for studying the functional roles of HATs and histone acetylation in NER in yeast.
Fig. 1.
Schematic of the anchor-away system. After addition of the chemical inducer of dimerization, rapamycin, the FRB tagged protein on the histone acetyltransferase (HAT) protein dimerizes with the FKBP tag on the anchor protein (usually a ribosomal protein, such as RPL13A). This leads to mislocalization of the HAT from the nucleus to the cytoplasm of the yeast cell, and spatial separation from the chromatin
2. Materials
2.1. Growth, UV Irradiation, and Repair Incubation of Yeast
Yeast extract peptone dextrose (YPD), synthetic complete (SC) media, or other desired yeast media containing 2% glucose.
UV-C light source.
Rapamycin, resuspended to 50 mg/mL in DMSO (LC labs).
2.2. Yeast Genomic DNA Isolation
Lysis Buffer: 2% Triton-X 100, 1% SDS, 100 mM NaCl, 10 mM Tris–Cl pH 8, 1 mM Na2EDTA.
Phenol-Chloroform-Isoamyl alcohol (25:24:1).
TE pH 8.0.
100% Ethanol.
70% Ethanol.
RNaseA (10 mg/mL).
Acid-washed glass beads.
2.3. T4 Endonuclease V Digestion
10× T4 endonuclease reaction buffer: 40 mM Tris–HCl (pH 7.4), 20 mM EDTA, 250 mM NaCl, 250 μg/mL BSA.
T4 Endonuclease V (see Subheading 3.9 for purification protocol).
2.4. Alkaline Gel Electrophoresis
10× alkaline buffer: 500 mM NaCl, 100 mM EDTA.
Agarose.
HindIII digested λ phage.
6× Alkaline Loading Dye: 15 mg bromocresol green, 25 mg xylene cyanol FF, 120 μL 0.5 M EDTA pH 8, 3 mL 1 M NaOH, 20% glycerol, ddH2O to 10 mL.
Neutralization Buffer: 1 M Tris Base, 1.5 M NaCl, HCl to pH 7.5.
Tris-Boric Acid-EDTA (TBE).
SYBR Gold.
2.5. Gel Scanning and Data Analysis
Typhoon FLA 7000 Capture Software.
ImageQuant.
2.6. Site-Specific Repair Analysis: Restriction Enzyme and T4 Endonuclease V Digestion
Restriction enzymes to excise the fragment of interest and appropriate buffer, for example: EcoRI+EcoRV for GAL10, PvuII+BsaHI for HML, DraI for RPB2.
2.7. Southern Blotting
20× SSC buffer: 175 g NaCl, 90 g Sodium Citrate, ddH2O to 1 L.
Hybridization Solution: 5× SSC Buffer, 5× Dehnhardt’s Solution, 40% formamide, 0.5% SDS.
50× Dehnhardt’s Solution: 1% Ficoll 400, 1% Polyvinylpyrolidine, 1% BSA.
Prime-It II Random Primer Labeling Kit.
α-P32 dATP (or dCTP).
PCR product specific to the region of interest.
Sonicated herring/salmon sperm DNA.
2× SSC + 0.1% SDS.
1× SSC + 0.1% SDS.
0.1× SSC + 0.1% SDS.
Nylon-N+ membrane.
Whatman blot paper.
2.8. Analysis of Southern Blot Data
Typhoon FLA 7000 Capture Software.
ImageQuant.
2.9. Purification of T4 Endonuclease V
T7 express lysγ chemically competent E. coli.
pET16-T4 endoV plasmid.
S.O.C. media: 2% Vegetable Peptone, 0.5% Yeast Extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4.
20 mM Glucose.
LB+ 100 μg/mL Ampicillin agar plates.
LB+ 100 μg/mL Ampicillin.
500 mM Isopropyl ß-D-1-thiogalactopyranoside (IPTG).
2× Laemmli buffer: 65.8 mM Tris–HCl, pH 6.8, 26.3% (w/v) glycerol, 2.1% SDS, 0.01% bromophenol blue.
Extraction buffer: 20 mM NaPO4 pH 6.5, 50 mM Dextrose, 05 M NaCl, 0.5% Triton X-100, and 20 mM Imidizole.
Lysozyme.
SigmaFast EDTA-free protease inhibitors (Sigma-Aldrich).
Whatman paper, Buchner funnel and 125 mL Erlenmeyer flask with vacuum.
60 mL syringe and 0.2um PALL Acrodisc Supor Membrane.
Ni2+ column (5 mL HisTrap FF crude, GE Biosciences).
Ni2+ Column Wash Buffer: 20 mM NaPO4 pH 6.5, 75 mM KCl, and 20 mM Imidizole.
T4 EndoV Elution Buffer: 20 mM NaPO4 pH 6.5, 75 mM KCl, and 250 mM Imidizole.
CM column (HiTrap CM, GE Biosciences).
CM column Low Salt Buffer: 20 mM NaPO4 pH 6.5, and 0.5 mM EDTA.
CM column High Salt Buffer: 20 mM NaPO4 pH 6.5, 0.5 mM EDTA, and 1 M KCl.
Dialysis Cassette (Molecular Weight Cutoff of 7.5 kDa).
Storage Buffer: 50 mM Tris pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1% Triton X-100, and 50% glycerol.
3. Methods
3.1. Growth, UV Irradiation, and Repair Incubation of Yeast
Unless otherwise indicated, Saccharomyces cerevisiae cells are grown at 30 °C with agitation.
3.1.1. Growth and UV Irradiation of Yeast HAT Deletion or Histone Lysine Mutants
Start a 5 mL overnight culture of the yeast strain in desired media.
Subculture into 55 mL of fresh media so the starting OD is ~0.1–0.25, and grow until the cells reach an OD600 of 0.6–1.
Aliquot 10 mL for “No” time point in a 15 mL falcon tube. Spin down in tabletop centrifuge at ~1900 × g (i.e., 3000 rpm in Sorvall Legend RT table top centrifuge) for 2 min, remove media, wrap the tube containing the yeast cell pellet in foil, and freeze at −80 °C.
Spin down 40 mL of cells in a tabletop centrifuge at 1900 × g for 2 min and resuspend them in 40 mL of sterile water.
Expose the cells to 100 J/m2 UV-C light using a pencil box or similar to hold the cells in a thin layer of water during exposure (see Notes 1 and 2).
Aliquot 10 mL of culture into 15 mL falcon tube for 0 h timepoint. Spin down in tabletop centrifuge at 1900 × g for 2 min, remove water, wrap the tube containing the yeast cell pellet in foil, and freeze at −80 °C.
Spin down remaining cells in a 50 mL falcon tube using a tabletop centrifuge at 1900 × g for 2 min, remove water, and resuspend in 30 mL media.
Aliquot 10 mL of culture into a 15 mL falcon tube for each subsequent time point (1, 2, and 3 h), spin down, remove media, wrap the tube containing the yeast cell pellet in foil, and freeze at −80 °C at least 1 h or overnight.
3.1.2. Growth and UV Irradiation of Yeast Anchor-Away Strains
Start a 5 mL overnight culture in desired media.
Subculture into 55 mL of fresh media so the starting OD is ~0.1–0.25, and grow until the cells reach an OD600 of 0.3–0.65 (see Note 3).
Add rapamycin directly to the culture to a final concentration of 1 μg/mL, and continue to incubate with shaking for 3 h (see Note 4).
Aliquot 10 mL of cell culture into a 15 mL falcon tube for “No” timepoint. Spin down in a tabletop centrifuge 1900 × g for 2 min, remove media, wrap the tube containing the yeast cell pellet in foil, and freeze at −80 °C.
Spin down 40 mL of cells in a tabletop centrifuge 1900 × g for 2 min and resuspend them in 40 mL of sterile water.
Expose the cells to 100 J/m2 UV-C light using a pencil box or similar to hold the cells in a thin layer of water during exposure (see Notes 1 and 2).
Aliquot 10 mL for 0 timepoint in a 15 mL falcon tube. Spin down in a tabletop centrifuge 1900 × g for 2 min, remove water, wrap the tube containing the yeast cell pellet in foil, and freeze at −80 °C.
Spin down remaining cells in a 50 mL falcon tube using a tabletop centrifuge at 1900 × g for 2 min, remove water, and resuspend in 30 mL media containing 1 μg/mL rapamycin.
Aliquot 10 mL for each subsequent time point (1, 2, and 3 h) into a 15 mL falcon tube, spin down in a tabletop centrifuge 1900 × g for 2 min, remove media, wrap in foil, and freeze at −80 °C at least 1 h or overnight.
3.2. Yeast Genomic DNA Isolation
Thaw cells at room temperature (RT).
For each timepoint, place ~250 μL acid-washed glass beads into a 1.5 mL Eppendorf tube and add 300 μL Phenol-Chloroform-Isoamyl alcohol (PCI).
Resuspend the thawed cell pellet in 200 μL Lysis Buffer and transfer it to a PCI/bead tube.
Vortex at the highest setting for 4 min (2 min × 2 times).
Add 300 μL TE pH 8.0.
Centrifuge tubes at the highest speed in a microcentrifuge for 10 min.
Transfer the aqueous (top) layer to a fresh 1.5 mL Eppendorf tube containing 900 μL 100% ethanol and mix by inversion.
Incubate at −80 °C for 10–20 min or −20 °C overnight.
Centrifuge for 10 min at the highest speed in a microcentrifuge to pellet DNA.
Pour off the ethanol, and add 500 μL 70% ethanol. Spin for 3–5 min.
Remove the 70% ethanol with a pipette tip, and allow the DNA pellet to dry for 5–10 min.
Dissolve the pellet in 80 μL TE pH 8.0 + 0.2 mg/mL RNase A.
Incubate at 37 °C for at least 15 min.
3.3. T4 Endonuclease V Digestion
Vortex the DNA well and check the amount of DNA present by measuring absorbance of an aliquot at 260 nm or running 1 μL on an agarose gel and staining with SYBR Safe or ethidium bromide.
Determine the volume of genomic DNA to use for digestion (~50 μg).
- Set up the digest:
- 3 μL 10× T4 endonuclease reaction buffer.
- 1 μL T4 endonuclease V (for +) (see Note 5).
- X μL Genomic DNA (50 μg).
- ddH2O to 30 μL.
Incubate at 37 °C for a minimum of 2 h.
3.4. Alkaline Gel Electrophoresis
Prepare a 1.2% agarose gel in water (see Note 6).
Heat until agarose is dissolved and allow to cool before adding 10× alkaline buffer to a final 1× concentration.
Pour into gel casting tray and allow to solidify for ~2 h.
Add 1× Alkaline Buffer to gel apparatus to cover the gel.
Spin down T4 digested DNA samples briefly.
Add 5 μL 6× Alkaline Loading Dye to the DNA samples.
Load samples into the wells (see Note 7).
For a ladder, load 1 μL HindIII digested λ phage that has been diluted in a loadable volume of water and alkaline loading dye to 1× final concentration.
Run the gel at 30 V for 20 h (see Note 6).
Remove the gel from the gel box and rinse the gel briefly with water.
Neutralize the gel with Neutralization Buffer by soaking it for 1 h with gentle rocking.
Rinse briefly with water.
Dilute SYBR Gold in 0.5× TBE at 1:10,000 ratio and stain gel 1 h with gentle rocking covered in foil.
Destain with water for 1 h with the gel covered in foil.
Rinse briefly with water, then destain with water for an additional 1 h covered in foil.
3.5. Gel Scanning and Data Analysis
Scan the gel using fluorescence scanner (i.e., GE Typhoon). An example gel image is shown in Fig. 2.
Using ImageQuant software (Molecular Dynamics) or similar, determine the ensemble average distance (xmed) traveled (mm) in each lane.
Use the ladder (with known size fragments) to determine an equation for the relationship between DNA fragment length and distance traveled using the method described by Southern [25]. This method fits a linear equation relating DNA fragment length to the reciprocal of the average distance traveled.
Use the equation to determine the median DNA length (in nucleotides) of the distribution (Lmed ).
Multiply the median length by 0.6 to yield the number average length (Ln ) of the (presumed) Poisson distribution of fragments in the ensemble (e.g., see [18]).
-
For each imepoint, subtract the reciprocal of the value in step 5 for the −T4 EndoV lane from the reciprocal of the +T4 EndoV lane value:
. This gives then number of CPDs/kb [18].
Determine % repair by normalizing this value to the 0 h timepoint (0%).
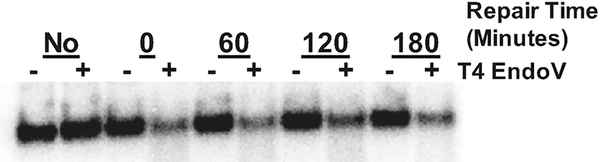
Fig. 2.
Alkaline gel scan representing global CPD damage and repair of a wild-type yeast strain. DNA was isolated from yeast collected at each timepoint before or after UV irradiation, incubated with (+) or without (−) T4 EndoV, resolved by alkaline gel electrophoresis and stained with SYBR Gold. A decrease in the average DNA fragment sizes in the UV irradiated sample (0 min +T4 EndoV) relative to the unirradiated control (No) reflects T4 EndoV cleavage at UV-induced CPDs. The increase in the average DNA fragment sizes in the +T4 EndoV samples at subsequent time points (e.g., 60, 120, 180 min) reflects repair of CPD lesions during the time course
3.6. Site-Specific Repair Analysis: Restriction Enzyme and T4 Endonuclease V Digestion
Grow, treat with rapamycin (if applicable), damage, and harvest yeast cells as for the global assay (Subheading 3.1).
Isolate genomic DNA as for the global assay (Subheading 3.2).
Following RNase digestion, treat the entire DNA sample for a minimum of 2 h with 30–50 U of each restriction enzyme to release an ~2 kb fragment around the region of interest (EcoRI+EcoRV for GAL10, PvuII+BsaHI for HML, DraI for RPB2) in supplied buffer so the total volume of the digest is 300 μL.
Purify DNA by adding 300 μL PCI and briefly vortexing. Centrifuge in a microcentrifuge for 10 min at top speed, and transfer the top layer to a new tube.
Add 2.5 volumes of ethanol and 1/10 volume 3 M sodium acetate and allow DNA to precipitate at least 1 h at −20 °C.
Spin down DNA for 30 min at top speed in a microcentrifuge at 4 °C, and remove ethanol.
Add 500 μL 70% ethanol and spin an additional 10 min at top speed in a microcentrifuge at 4 °C.
Remove the ethanol from the DNA pellet, and allow it to dry.
Resuspend the pellet in 65 μL water.
Determine the volume of DNA required for subsequent T4 endoV digestion so about 30 μg of DNA is loaded in each well of the alkaline gel.
Digest with T4 endonuclease V as for the global assay (Subheading 3.3).
3.7. Site-Specific Repair Analysis: Southern Blotting
Prepare a 1% agarose gel in water.
Heat until agarose is dissolved and allow to cool before adding 10× alkaline buffer to a final 1× concentration.
Allow the gel to solidify ~2 h.
Add 6× alkaline loading dye to the DNA samples to a final 1× concentration and load the samples into the gel.
Run the gel at 30 V for 20 h (see Note 6).
Remove the gel from the gel box, and neutralize the gel with Neutralization Buffer for 1 h.
Soak the gel in 20× SSC buffer for 5 min.
Prepare the transfer apparatus by placing a glass plate over a glass baking dish. Wrap Whatman paper around the glass plate so the Whatman paper goes down into the baking dish. Wet the Whatman paper with 20× SSC buffer and add 20× SSC buffer to the baking dish.
Place the gel on top of the Whatman paper upside down.
Wet a piece of nylon membrane in 20× SSC buffer and place it on top of the gel. Use a plastic 10 mL pipette (or similar) to roll out any bubbles.
Wet a piece of Whatman paper in 20× SSC buffer and place it on top of the membrane. Use a plastic 10 mL pipette (or similar) to roll out any bubbles.
Surround the gel with parafilm, covering the baking dish to avoid evaporation, to maintain capillary action through the gel and not through the blotting paper.
Add a package of bifold paper towels to the top of this that are similar in size to the Whatman paper.
Allow the gel to transfer overnight (12–18 h).
Dry the membrane by placing it on a dry piece of Whatman paper at 55 °C for 2–3 h.
Pre-warm hybridization solution in a 37 °C in a water bath.
To 15 mL hybridization solution, add 30 μL of 10 μg/mL sonicated herring/salmon sperm DNA that has been boiled for 10 min then rapidly chilled on ice.
Place dried membrane in a hybridization tube, then add the hybridization buffer + ssDNA.
Rotate at 42 °C for at least 1 h.
Make radiolabeled probe using a Random Labeling Kit, α-P32 dATP and a PCR product specific to the region of interest.
Add 25 μL labeled probe to the hybridization buffer and allow to rotate for at least 12 h at 42 °C.
- Carry out the following washes of the membrane using buffers that have been pre-warmed to 37 °C.
- Twice for 10 min in 2 × SSC + 0.1% SDS at RT.
- Twice for 15 min in 1 × SSC + 0.1% SDS at 65 °C.
- Twice for 10 min in 0.1 × SSC + 0.1% SDS at 65 °C.
Place the membrane on Whatman paper, wrap in plastic wrap, and expose to phosphor screen.
3.8. Analysis of Southern Blot Data
Scan the phosphor screen using a phosphor imager (i.e., GE Typhoon). An example Southern blot image is shown in Fig. 3.
Using ImageQuant software (or similar) draw a box around the band in each lane and determine the intensity of signal in the box (see Notes 8 and 9).
- Divide the intensity of the +T4 EndoV lane [P0(+T4 endo)] by the −T4 EndoV lane [P0(−T4 endo)] for each timepoint to obtain the fraction of CPDs for any repair time (t) (see Note 10):
- Transform the value from step 3 (Ft) by taking the negative natural log of the value. This gives the average number of lesions per fragment:
- Average number of lesions / frament =-ln Ft
-
To determine the percent repair, normalize to the 0 h timepoint:
, where Ft and F0 indicate fractional inten-sities obtained in step 3 for repair times t and 0.
Fig. 3.
Autoradiogram of a Southern Blot representing CPD damage and repair at an individual yeast locus. DNA was isolated from yeast collected at each timepoint before or after UV irradiation. This DNA was digested with appropriate restriction enzymes to release an ~2 kb fragment before T4 EndoV treatment (+) or control (−). Following alkaline gel electrophoresis, DNA was transferred to a nylon membrane and probed using a radiolabeled probe specific to the region of interest (HML locus). A decrease in the band intensity in the UV irradiated samples is due to CPD cleavage by T4 EndoV
3.9. Purification of T4 Endonuclease V
Transform pET16-T4 endoV (encodes T4 endoV with an N-terminal His-tag) into T7 Express lysγ chemically competent E. coli (NEB #C3010I) by mixing 10 ng of pET16-T4 endoV plasmid with 50 μL of E. coli suspension in a 15 mL culture tube on ice for 30 min.
Transfer tubes to at 42 °C water bath for 45 s and then return to ice for 2 min.
Add 1 mL of S.O.C. media and incubate at 37 °C for 1 h while shaking tubes at 225 rpm.
Remove 50 μL of culture and spread onto an LB plate containing 100 μg/mL ampicillin.
Place plates at 30 °C overnight.
Inoculate 5 mL of liquid LB media containing 100 μg/mL ampicillin with a single E. coli transformant containing pET16-T4 endoV. Grow overnight at 30 °C while shaking at 250 rpm. The following morning, inoculate 2 L of liquid LB broth containing 100 μg/mL ampicillin with 5 mL of the starter culture grown the previous night. Incubate while shaking at 250 rpm at 30 °C until the culture reaches an OD600 of 0.6.
Remove 2 mL of culture and place in a 15 mL culture tube for an un-induced control. Induce T4 endoV expression in the remainder of the culture by adding IPTG to 0.5 mM final concentration. Replace un-induced and induced cultures into the shaking 30 °C incubator for 4 h (see Note 11).
Transfer the un-induced culture and 2 mL of the induced culture to microcentrifuge tubes and collect cells by centrifuging at 6000 × g in a benchtop microcentrifuge. Remove liquid media and add 200 μL of 2× Laemmli buffer and 200 μL of water. Resuspend cell pellets in Laemmli buffer by brief sonication and heat pellets at 95 °C for 2 min. Centrifuge samples at maximum rpm for 5 min in a benchtop microcentrifuge. Monitor induction of T4 endoV by SDS-PAGE.
Collect the induced E. coli cells for the remainder of the 2 L culture by centrifugation at 6000 × g at 4 °C in a Sorvall GSA rotor for 20 min. Remove liquid. Resuspend cell pellet in 40 mL of water and centrifuge again at 10,000 × g in a fixed angle rotor for 5 min. Remove water. Cell pellet can be frozen at −80 °C at this point.
Resuspend pellet on ice in 35 mL extraction buffer supplemented with 40 mg lysozyme, and 1× SigmaFast EDTA-free protease inhibitors. Allow resuspended pellet to sit at room temperature for 15 min, until lysed.
Using a Misonix Sonicator 3000 set to a power of 5, sonicate on ice for 30 s followed by a 30 s rest period on ice. Repeat for 6 cycles (a total of 3 min active sonication). The lysate should no longer be viscous.
Centrifuge in a Sorvall SS-34 rotor for 20 min at 27,000 × g at 4 °C to pellet insoluble debris and fragmented DNA. This should form a compact pellet. If the pellet appears viscous, repeat sonication and high speed centrifugation.
Filter the supernatant through Whatman paper using a Buchner funnel into a 125 mL Erlenmeyer flask with vacuum.
Place the filtered extract into a 60 mL syringe and filter through a 0.2 μm PALL Acrodisc Supor Membrane.
Save 100 μL of the filtered extract for diagnostic analysis of the preparation.
Apply the remainder of the filtered extract a Ni2+ column (5 mL HisTrap FF crude, GE Biosciences) equilibrated in 10 column volumes extraction buffer. Collect the flow through for diagnostic analysis (see Notes 12 and 13).
Once the extract has completely entered the column, wash the column with 10 column volumes of extraction buffer.
Lower the salt concentration of the sample by exchanging the buffer on the column with 10 column volumes of Ni2+ Column Wash Buffer.
Elute the T4 endoV with 25 mL of T4 EndoV Elution Buffer. Begin collecting 2 mL fractions once the elution buffer has begun entering the column.
Determine which fractions contain the T4 endoV by running 20 μL of each fraction on an SDS-PAGE gel along with 20 μL of the reserved filtered extract and the flow through of the nickel column to assess the efficiency of the extraction (see Note 14).
Pool fractions containing T4 endoV. Apply the sample to a 5 mL CM column (HiTrap CM, GE Biosciences) equilibrated in CM Column Low Salt Buffer supplemented with 2% CM Column High Salt Buffer.
Elute with gradient to 100% CM Column High Salt Buffer over 10 column volumes. The His-tagged T4 endoV elutes from the CM column at ~41.4 mS.
Determine which fractions contain the T4 endoV by running 20 μL of each fraction on an SDS-PAGE gel.
Pool fractions containing T4 endoV. Place in a dialysis cassette (Molecular Weight Cutoff of 7.5 kDa). At 4 °C, dialyze in 1 L of storage buffer for 1 h.
Transfer the dialysis cassette containing the T4 endoV to 1 L of fresh storage buffer. Dialyze at 4 °C overnight.
Transfer T4 endoV to microcentrifuge tubes and store at −20 °C.
Acknowledgments
We thank Dr. Michael Smerdon, Dr. Peng Mao, and Dalton Plummer for helpful comments and suggestions. This research was supported by grants from NIEHS (R01ES002614 and R21ES027937).
Footnotes
Following UV irradiation, it is important to keep yeast cells in the dark to limit photolyase activity.
It is important to be consistent among yeast strains in an experiment with the concentration of cells irradiated. High-density cultures experience a lower level of DNA damage than low-density cultures at the same UV dose, probably due to cells shielding one another.
The goal is to achieve an OD600 ~ 0.6–1afterthe3 h incubation with rapamycin. The growth rate may vary based on the mutant and the media used. Ideally, the wild-type and mutant yeast strain should be at approximately the same OD600 at the time of UV irradiation. If necessary, log phase cells may be diluted so a similar cell density is irradiated.
The rate at which factors are exported from the yeast nucleus may vary; therefore, the rapamycin treatment time before irradiation may need to be optimized.
The volume/amount of T4 endonuclease V enzyme added to the reaction should be empirically determined to ensure complete digestion of CPD lesions in genomic DNA.
We use a gel casting tray that is 23 cm × 26 cm and a 400 mL agarose gel (agarose + 360 mL water +40 mL 10× alkaline buffer). Different size gels may be used, but the running conditions must be optimized for that gel size.
It is important to run an empty lane to account for background during analysis.
Southern blots should show a single band. Multiple bands can mean either the probe is insufficiently specific and is recognizing something other than the region of interest, or the DNA is under-digested at the restriction enzyme step.
It is important for Southern Blot quantification that the boxes are of equal size and the intensity is corrected for the background signal obtained from the “No Sample” lane(s).
The value of each band is expressed as the percentage of the total intensity of a lane, and the average CPDs/strand is obtained from the intensity of the intact fragment (i.e., P0), assuming a Poisson distribution of UV-damaged fragments, as described by Bohr and Okumoto [26].
Leaky expression of T4 endoV can be toxic to E. coli, resulting in poor induction. Make sure to use lysozyme-expressing cells and limit the length of the induction time.
Try to conduct all chromatography steps at 4 °C to minimize inactivation of the protein.
Do not use protease inhibitors that contain EDTA or buffers containing DTT for any steps prior to purification over the Ni2+ column. These agents will either chelate or react with the Ni2+, damaging the column.
T4 endoV only has 1 tryptophan and absorbs UV poorly at 280 nm. Thus, it is difficult to determine the location of your protein peak by UV absorbance. You must determine what fractions contain T4 endoV by SDS-PAGE stained with Coomassie Blue. The lack of tryptophan also means the protein is not visible on BioRad’s TGX stain-free gels.
References
- 1.Workman JL, Abmayr SM (2014) Fundamentals of chromatin. Springer, New York [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260 [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez Y, Hinz JM, Smerdon MJ (2015) Accessing DNA damage in chromatin: preparing the chromatin landscape for base excision repair. DNA Repair 32:113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao P, Brown AJ, Malc EP, Mieczkowski PA, Smerdon MJ, Roberts SA et al. (2017) Genome-wide maps of alkylation damage, repair, and mutagenesis in yeast reveal mechanisms of mutational heterogeneity. Genome Res 27:1674–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao P, Smerdon MJ, Roberts SA, Wyrick JJ (2016) Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc Natl Acad Sci U S A 113:9057–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nag R, Smerdon MJ (2009) Altering the chromatin landscape for nucleotide excision repair. Mutat Res 682:13–20 [DOI] [PubMed] [Google Scholar]
- 7.Lee KK, Workman JL (2007) Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol 8:284–295 [DOI] [PubMed] [Google Scholar]
- 8.Mao P, Wyrick JJ (2016) Emerging roles for histone modifications in DNA excision repair. FEMS Yeast Res 16:fow090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.House NC, Koch MR, Freudenreich CH (2014) Chromatin modifications and DNA repair: beyond double-strand breaks. Front Genet 5:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waters R, van Eijk P, Reed S (2015) Histone modification and chromatin remodeling during NER. DNA Repair 36:105–113 [DOI] [PubMed] [Google Scholar]
- 11.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T (2006) DNA repair and mutagenesis, 2nd edn. ASM Press, Washington, DC [Google Scholar]
- 12.Hoeijmakers JH (2009) DNA damage, aging, and cancer. N Engl J Med 361:1475–1485 [DOI] [PubMed] [Google Scholar]
- 13.Spivak G, Hanawalt PC (2015) Photosensitive human syndromes. Mutat Res 776:24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adar S, Hu J, Lieb JD, Sancar A (2016) Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc Natl Acad Sci U S A 113:E2124–E2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodges AJ, Gallegos IJ, Laughery MF, Meas R, Tran L, Wyrick JJ (2015) Histone sprocket arginine residues are important for gene expression, DNA repair, and cell viability in Saccharomyces cerevisiae. Genetics 200: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lloyd RS (1999) The initiation of DNA base excision repair of dipyrimidine photoproducts. Prog Nucleic Acid Res Mol Biol 62:155–175 [DOI] [PubMed] [Google Scholar]
- 17.Sutherland BM, Shih AG (1983) Quantitation of pyrimidine dimer contents of nonradioactive deoxyribonucleic acid by electrophoresis in alkaline agarose gels. Biochemistry 22: 745–749 [DOI] [PubMed] [Google Scholar]
- 18.Bespalov VA, Conconi A, Zhang X, Fahy D, Smerdon MJ (2001) Improved method for measuring the ensemble average of strand breaks in genomic DNA. Environ Mol Mutagen 38:166–174 [DOI] [PubMed] [Google Scholar]
- 19.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC (1985) DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40:359–369 [DOI] [PubMed] [Google Scholar]
- 20.Gong F, Fahy D, Smerdon MJ (2006) Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol 13:902–907 [DOI] [PubMed] [Google Scholar]
- 21.Nag R, Kyriss M, Smerdon JW, Wyrick JJ, Smerdon MJ (2010) A cassette of N-terminal amino acids of histone H2B are required for efficient cell survival, DNA repair and Swi/Snf binding in UV irradiated yeast. Nucleic Acids Res 38:1450–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhuri S, Wyrick JJ, Smerdon MJ (2009) Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae. Nucleic Acids Res 37:1690–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kou H, Zhou Y, Gorospe RM, Wang Z (2008) Mms19 protein functions in nucleotide excision repair by sustaining an adequate cellular concentration of the TFIIH component Rad3. Proc Natl Acad Sci U S A 105:15714–15719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haruki H, Nishikawa J, Laemmli UK (2008) The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31:925–932 [DOI] [PubMed] [Google Scholar]
- 25.Southern EM (1979) Measurement of DNA length by gel electrophoresis. Anal Biochem 100:319–323 [DOI] [PubMed] [Google Scholar]
- 26.Bohr VA, Okumoto DS (1988) Analysis of frequency of pyrimidine dimers in specific genomic sequences In: Friedberg EC, Hanawalt PC (eds) DNA repair: a laboratory manual of research procedures, vol 3 Marcel Dekker Inc., New York, pp 347–366 [Google Scholar]