Abstract
A novel nocardioform strain, CICC 11023T, was isolated from a tissue biopsy of neck lesions of a patient with primary cutaneous nocardiosis and characterized to establish its taxonomic position. The morphological, biochemical, physiological and chemotaxonomic properties of strain CICC 11023T were consistent with classification in the genus Nocardia . Whole-cell hydrolysates were rich in meso-diaminopimelic acid, galactose, arabinose and fructose. Mycolic acids were present. The major polar lipids were diphosphatidylglycerol, phosphatidylethanolamine, one unidentified phospholipid and two unidentified lipids, and the predominant menaquinone was cyclo MK-8 (H4, ω-cyclo). The main fatty acids (>5 %) were C18 : 0 10-methyl (TBSA), C16 : 0, summed feature 4 (C16 : 1 trans 9/C15 : 0 iso 2OH), C15 : 0 and C17 : 0 10-methyl. Phylogenetic analyses based on 16S rRNA gene sequences revealed that the isolate is most closely related (>98 % similarity) to the type strains Nocardia ninae OFN 02.72T, Nocardia iowensis UI 122540T and Nocardia alba YIM 30243T, and phylogenetic analysis of gyrB gene sequences showed similarity (89.1–92.2 %) to Nocardia vulneris NBRC 108936T, Nocardia brasiliensis IFM 0236T and Nocardia exalbida IFM 0803T. DNA–DNA hybridization results for strain CICC 11023T compared to Nocardia type strains ranged from 20.4 to 35.4 %. The genome of strain CICC 11023T was 8.78 Mbp with a G+C content of 67.4 mol% overall. The average nucleotide identity (ANI) values between strain CICC 11023T and N. alba YIM 30243T were low (OrthoANIu=77.47 %), and the ANI values between strain CICC 11023T and N. vulneris NBRC 108936 T were low (OrthoANIu=83.75 %). Consequently, strain CICC 11023T represents a novel Nocardia species on the basis of this polyphasic study, for which the name Nocardia colli sp. nov. is proposed. The type strain is CICC 11023T (=KCTC 39837T).
Keywords: Nocardia colli sp. nov., pathogen, cutaneous nocardiosis
The genus Nocardia , belonging to the suborder Corynebacterineae [1], was established by Trevisan in 1889 [2] and consists of Gram-stain-positive, variably acid-fast, strictly aerobic bacteria that form filamentous, branched cells that fragment into pleomorphic, rod-shaped or coccoid elements [3]. Since Pijper and Pullinger identified Nocardia transvalensis as the pathogenic micro-organism associated with a case of mycetoma in a South African patient in 1927 [4], more and more cases of clinical Nocardia infection have been reported worldwide every year. This increased prevalence is partly due to advances in phylogenetic analyses based on 16S rRNA and partial gyrB gene sequences, allowing for the more rapid identification of nocardial isolates compared to standard phenotypic techniques [5, 6]. More than 40 species within the genus Nocardia have been reported as clinically relevant, and many of these show resistance to several classes of antimicrobials [7]. Nocardia species are widely distributed in the environment and cause a variety of suppurative and granulomatous infections of humans and animals, including cutaneous, subcutaneous, lymphocutaneous, pulmonary, cerebral or disseminated nocardiosis. Treatment of nocardiosis often requires long-term therapy with a combination of drugs [8]. In the present study, a novel strain of Nocardia was isolated from a patient with primary cutaneous nocardiosis and the phenotypic, morphological, chemotaxonomic and molecular characteristics of strain CICC 11023T are presented.
A 36-year-old woman, who is a farmer by occupation, presented to the Department of Dermatology of the Second Affiliated Hospital of Kunming Medical University (Kunming, Yunnan Province, PR China) with a 10 year history of gradually enlarging and infiltrating painless papulo-nodular lesions of the neck and chest [9]. Two strains were isolated from an aerobic culture of the biopsied skin tissue specimens at 25 °C in Sabouraud agar medium after 1 week. One of the strains showed 99.8 % 16S rRNA gene sequence similarity to Staphylococcus epidermidis ATCC 14990T. S. epidermidis is part of the normal human flora, typically the skin flora, and is less commonly found in the mucosal flora [10]. The other strain, KY2-1, was deposited in the China National Research Institute of Food and Fermentation Industries, China Centre of Industrial Culture Collection (CICC) as strain CICC 11023T and the Korean Collection for Type Cultures (KCTC), Biological Resource Centre (BRC), Korea Research Institute of Bioscience and Biotechnology as strain KCTC 39837T.
Characteristic chemotaxonomic properties of the genus Nocardia are based on mycolic acids and fatty acid compositions [11]. To identify the whole-cell fatty acid composition, strain CICC 11023T and reference Nocardia type strains were grown in trypticase soy broth (TSB) with shaking at 150 r.p.m. for 7 days at 28 °C. Extraction and analysis of the cellular fatty acids were based on the standard protocol of the Sherlock Microbial Identification (midi) System, version 6.0 [12], and peaks were identified using the peak-naming table TSBA6 compiled by the China General Microbiological Culture Collection Centre (cgmcc) [13]. Analysis of the acyl cell wall was performed according to a glycolate test by diethyl ether extraction as previously reported [14] . The whole-cell sugars and diaminopimelic acids were determined by thin-layer chromatography using previously described methods [15, 16]. Menaquinones were extracted and purified through the method described by Collins et al. [17] and analysed by high-performance liquid chromatography [18] . Phospholipids were extracted by two-dimensional thin-liquid chromatography [19] and identified by following a previously reported procedure [20]. Analysis of mycolic acids was carried out using a previously described method [19].
In general, a >5 % fatty acid content is considered to present a ‘major fatty acid’ [21]. Analyses of the fatty acids by gas–liquid chromatography revealed that the main fatty acids (>5 %) of strain CICC 11023T were C18 : 0 10-methyl (TBSA, 30.36 %), C16 : 0 (20.52 %), summed feature 4 (C16 : 1 trans 9/C15 : 0 iso 2OH; 14.33 %), C15 : 0 (13.01%) and C17 : 0 10-methyl (5.41%). The fatty acid patterns of the novel strain and the reference strains are presented in Table 1. Comparisons of the fatty acid profiles showed that all seven tested strains contained C10 : 0, C12 : 0, C14 : 0, C16 : 0, C17 : 0, C18 : 0, C18 : 0 10-methyl (TBSA) and C18 : 1ω9c; however, strain CICC 11023T exhibited relatively large amounts of C17 : 0 and C18 : 0 10-methyl (TBSA), and small amounts of C16 : 0 and C18 : 1ω9c. Thus, compared to the six reference strains, strain CICC 11023T showed a distinct major fatty acid pattern. The whole-cell hydrolysates were rich in meso-diaminopimelic acid, galactose, arabinose and fructose. The major polar lipids consisted of diphosphatidylglycerol, phosphatidylethanolamine, one unidentified phospholipid and two unidentified lipids, and the predominant menaquinone was cyclo MK-8(H4, ω-cyclo) (86.5 %). The strain also contained mycolic acid, which is characteristic of the genera Nocardia and Rhodococcus [22]. The chemotaxonomic features of strain CICC 11023T were consistent with those of members of the genus Nocardia [6].
Table 1.
Main fatty acid compositions (>5 %) of strain CICC 11023T and the type strains of related Nocardia species
Strains: 1, CICC 11023T; 2, Nocardia ninae OFN 02.72T; 3, Nocardia iowensis UI 122540T; 4, Nocardia alba YIM 30243T; 5, Nocardia vulneris NBRC 108936T; 6, Nocardia brasiliensis IFM 0236T; 7, Nocardia exalbida IFM 0803T. All data are from this study. Values are percentages (%) of total fatty acids. −, Not detected.
|
Fatty acid |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
|---|---|---|---|---|---|---|---|
|
C15 : 0 |
13.0 |
– |
– |
– |
– |
<5 |
– |
|
C16 : 0 |
20.5 |
35.8 |
31.4 |
24.7 |
32.9 |
40.0 |
25.9 |
|
C17 : 0 10-methyl |
5.4 |
– |
<5 |
<5 |
– |
– |
– |
|
C18 : 0 |
<5 |
<5 |
<5 |
5.0 |
<5 |
7.9 |
5.7 |
|
C18 : 0 10-methyl (TBSA) |
30.3 |
8.1 |
15.7 |
11.7 |
6.2 |
12.9 |
8.2 |
|
C18 : 1ω9c |
<5 |
24.0 |
11.4 |
19.5 |
18.0 |
24.0 |
17.1 |
|
Summed feature 3* |
– |
13.4 |
30.8 |
18.3 |
16.0 |
– |
12.8 |
|
Summed feature 4* |
14.3 |
– |
– |
– |
– |
– |
– |
*Summed features represent groups of two or three fatty acids that could not be separated by GLC using the midi system. Summed feature 3 contained C16 : 1ω7c and/or C16 : 1ω6c; summed feature 4 contained C16 : 1 trans 9 and/or C15 : 0 iso 2OH.
Pigmentation, production of aerial hyphae, and morphological characteristics were observed under a light microscope (Olympus CX41) and a scanning fiber-optic electron microscope (FEI Quanta). Strain CICC 11023T was grown separately on Gause 1, ISP 2, ISP 3, ISP 4 and ISP 5 at 30 °C for 5 days, and then examined for colour determination using colour chips from the ISCC–NBS colour charts (standard sample no. 2106). Growth at 21, 28, 37 and 45 °C was measured on ISP 2 for 5 days. The pH range for growth using the buffer system described in [23] (pH 4–10 at intervals of 0.5 pH units) and the requirement for NaCl (1, 4, 7 and 10 %) were determined in ISP 2 broth. Phenotypic characteristics such as Gram-staining, catalase and oxidase activity, and hydrolysis of casein, Tweens 20 and 80, egg yolk and starch were examined using the methods described by Smibert and Krieg [24]. Utilization of various substrates as sole carbon sources was tested at the CICC using the GN2 MicroPlate Gram-negative identification test panel (Biolog), and the result was determined after incubation at 30 °C for 24 h. Physiological and biochemical properties were further determined with API 20NE, API 20E and API ZYM strips (bioMérieux). Tests were generally performed according to the manufacturer’s instructions. The API 20NE tests were read after 24–48 h at 28 °C, the API 20E tests were read after 18–24 h at 36 °C, and the API ZYM tests were read after 4 h of incubation at 37 °C [13].
Morphological characteristics of strain CICC 11023T presented typical properties of the genus Nocardia . Strain CICC 11023T was aerobic, Gram-stain-positive, non-motile, with modified acid alcohol-positive actinomycetes forming extensively branched grey-white substrate mycelium and aerial mycelium with fragments that appeared as short coli-like bodies under scanning electronic microscopy (0.5×0.7–0.9 µm in diameter). When grown on Gause 1, ISP 3 and ISP 5 media at 30 °C for 5 days, the surface of colonies appeared as a velvet powder, with a grey aerial mycelium and substrate mycelium, and a grey spore heap. When grown on ISP 2 at 30 °C for 5 days, the colonies had a corrugated surface, with a white aerial mycelium, light brown substrate mycelium and a white spore heap. Culture inserts on ISP 2 at 30 °C for 5 days showed formation of short spore chains, a spore chain flex and mycelium breaking into a rod-like curved body after 8 days. Growth was weak on ISP 4 at 30 °C for 5 days. No soluble pigments were found on any medium. Strain CICC 11023T grew at 21, 28 and 37 °C, but not at 45 °C. Positive reactions were observed for milk peptonization, catalase, aesculin, urease and cellulose; negative reactions were observed for milk coagulation, nitrate reduction, gelatin liquefaction, tyrosinase, hydrolysis of starch, casein, xanthine, hypoxanthine and production of H2S. Strain CICC 11023T utilized l-arabinose, rhamnose, d-fructose, salicin, d-xylose, inositol, lactose, melibiose, d-glucose, raffinose, sucrose, d-mannitol, maltose, trehalose and arabinose as sole carbon sources. The main differential characteristics between strain CICC 11023T and closely related Nocardia species are presented in Table 2.
Table 2.
Differential phenotypic characteristics between strain CICC 11023T and closely related Nocardia species
Strains: 1, CICC 11023T; 2, Nocardia ninae OFN 02.72T; 3, Nocardia iowensis UI 122540T; 4, Nocardia alba YIM 30243T; 5, N ocardia vulneris NBRC 108936T; 6, Nocardia brasiliensis IFM 0236T; 7, Nocardia exalbida IFM 0803T. All data were obtained in this study unless indicated otherwise. +, positive; −, negative; w, weak.
|
Characteristic |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
|---|---|---|---|---|---|---|---|
|
Growth at 37 °C |
+ |
+ |
+ |
− |
+ |
+ |
+ |
|
Growth at 45 °C |
− |
− |
+ |
− |
− |
− |
− |
|
Milk coagulation |
− |
− |
− |
− |
− |
− |
− |
|
Milk peptonization |
+ |
− |
− |
− |
− |
− |
− |
|
Carbon utilization: |
|
|
|
|
|
|
|
|
Glucose |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Mannitol |
+ |
− |
− |
+ |
+ |
w |
+ |
|
Inositol |
+ |
− |
− |
+ |
− |
− |
− |
|
Arabinose |
+ |
+ |
− |
− |
− |
− |
− |
|
Maltose |
+ |
w |
+ |
+ |
− |
+ |
+ |
|
Galactose |
+ |
+ |
− |
− |
+ |
+ |
w |
|
Raffinose |
+ |
− |
− |
− |
− |
− |
− |
|
Rhamnose |
w |
− |
− |
+ |
− |
− |
− |
|
Sorbitol |
w |
− |
− |
− |
− |
− |
− |
|
Decomposition: |
|
|
|
|
|
|
|
|
Adenine |
+ |
+ |
− |
− |
− |
− |
− |
|
Casein |
− |
− |
− |
− |
+ |
− |
− |
|
Tyrosine |
− |
− |
+ |
− |
+ |
+ |
+ |
|
Xanthine |
− |
− |
+ |
− |
− |
− |
+ |
|
Hypoxanthine |
− |
+ |
+ |
− |
+ |
+ |
+ |
|
Uric acid |
+ |
+ |
+ |
− |
+ |
− |
− |
|
Aesculin |
+ |
− |
− |
+ |
− |
− |
− |
|
Polar lipids* |
DPG, PE, uPL, uL1, uL2 |
DPG, PE, PI, PIM |
DPG, PE, PI, PIM |
DPG, PE, PI, PIM, GL |
DPG, PE, PI, PIM |
DPG, PE, PI, PIM |
DPG, PE, PI, PIM |
|
DNA G+C content (mol%) |
65.6 |
67.6 |
70.5 |
72 |
68.4 |
69.6 |
68 |
*DPG, diphosphatidylglycerol; GL, glycolipid; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PIM, phosphatidylinositolmannoside; uL, unidentifiedlipid; uPLunidentifiedphospholipid.
Antibiotic sensitivity analysis of strain CICC 11023T was performed using Etest (bioMérieux) to determine the minimal inhibitory concentration values for some antibiotics according to the manufacturer instructions. Sulfonamides have been the mainstay of antimicrobial therapy for human nocardiosis [25]. Thus, the patient was treated with oral Co-SMZ (containing 0.4 g sulfamethoxazole and 0.08 g trimethoprim; two tablets/time, three times/day, twice the first dose) for 8 weeks and achieved very good improvement with this treatment: the nocardiosis resolved 6 months after the administration of Co-SMZ [9]. No recurrence of the infection was observed for approximately 3 years. Although the majority of these infections can be treated with sulfonamides, there are in vitro differences noted in the antimicrobial susceptibility among different cases of Nocardia . The drug susceptibility testing showed that strain CICC 11023T was susceptible to levofloxacin, ofloxacin, tobramycin and clindamycin, and was resistant to fosfomycin, imipenem, vancomycin and erythromycin.
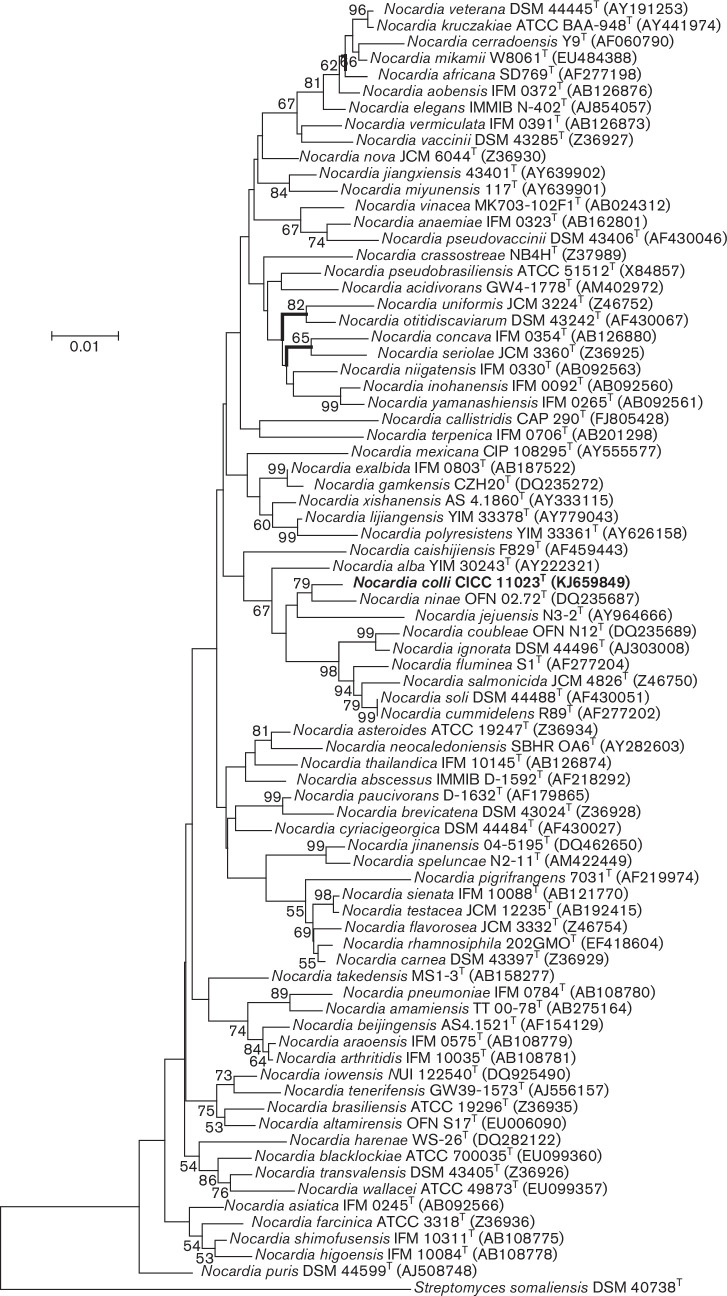
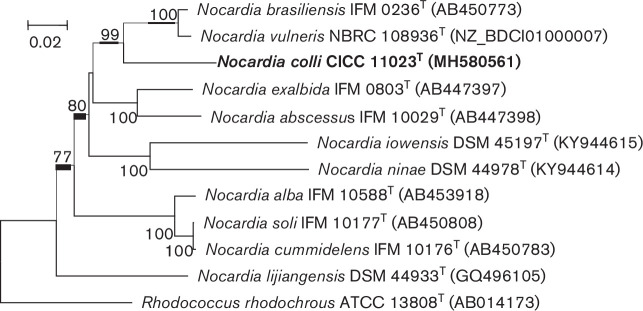
Extraction of genomic DNA, PCR amplification and sequencing of the 16S rRNA gene from strain CICC 11023T were performed as described previously [26] . The program clustal x was used to conduct multiple alignments with sequences of the most closely related Actinobacteria strains and for calculations of sequence similarity [27]. Phylogenetic trees were reconstructed using the neighbour-joining [28], maximum-parsimony [29] and maximum-likelihood [30] algorithms in mega version 4.0 [31]. The stability of the clades in the trees was appraised using a bootstrap value with 1000 replications [32] . The 16S rRNA gene sequence (1506 bp) of strain CICC 11023T was determined. Phylogenetic analysis showed that strain CICC 11023T was most closely related to members of the genus Nocardia , and sequence similarity calculations obtained by pairwise comparisons indicated that the closest relatives of strain CICC 11023T were Nocardia ninae OFN 02.72T (98.4 %), Nocardia iowensis UI 122540T (98.3 %) and Nocardia alba YIM 30243T (98.1 %; Fig. 1). The gyrB gene sequence for strain CICC 11023T (1094 bp) was also determined and analysed according to the methods reported by Takeda et al. [5]. The closest phylogenetic neighbours were Nocardia vulneris NBRC 108936T (92.2 %), Nocardia brasiliensis IFM 0236T (91.8 %) and Nocardia exalbida IFM 0803T (89.1 %; Fig. 2). The Nocardia type species were clustered based on gyrB sequence similarity values of 93.5 % and above [5]. Therefore, N. ninae OFN 02.72T, N. iowensis UI 122540T, N. alba YIM 30243T, N. vulneris NBRC 108936T, N. brasiliensis IFM 0236T and N. exalbida IFM 0803T as reference strains were used for phenotypic comparisons and DNA–DNA hybridization (DDH) tests.
Fig. 1.
Neighbour-joining tree based on 16S rRNA gene sequences showing relationships between Nocardia colli CICC 11023T and closely related type strains of the genus Nocardia . Streptomyces somaliensis DSM 40738T was used as outgroup. Bootstrap values were expressed as percentages of 1000 replications. The branching is supported by the results from the three algorithms used. Bar, 0.01 substitutions per nucleotide position. GenBank accession numbers are given in parentheses.
Fig. 2.
Phylogenetic trees derived from the gyrB gene sequences showing relationships between Nocardia colli CICC 11023T and closely related type strains of the genus Nocardia . The trees were created using the neighbour-joining method. Rhodococcus rhodochrous ATCC 13808T was used as outgroup. Bootstrap values were expressed as percentages of 1000 replications. Bar, 0.02 substitutions per nucleotide position. GenBank accession numbers are given in parentheses. The numbers on the tree represent bootstrap values for the branch points. Bootstrap values greater than 50 % significance are indicated.
The G+C content was determined using the method of Mesbah et al. [33] and was found to be 65.6 mol%. DDH experiments were carried out at the CGMCC using dot-blot hybridization and a simple fluorimetric method based on thermal denaturation temperatures [34] to evaluate the DNA–DNA relatedness between strain CICC 11023T and its most closely related species: N. ninae OFN 02.72T (35.4 %), N. iowensis UI 122540T (20.4 %), N. alba YIM 30243T (25 %), N. vulneris NBRC 108936T (21.6 %), N. brasiliensis IFM 0236T (22.2 %) and N. exalbida IFM 0803T (23.3 %). In accordance with the recommended threshold value of 70 % DNA–DNA relatedness for species delineation [35], strain CICC 11023T represents a species distinct from N. ninae OFN 02.72T, N. iowensis UI 122540T, N. alba YIM 30243T, N. vulneris NBRC 108936T, N. brasiliensis IFM 0236T and N. exalbida IFM 0803T.
The extracted genomic DNA of strain CICC 11023T was sequenced by combining Illumina HiSeq at the CGMCC. An Illumina library with an insert size of about 400 bp was prepared from 500 ng of DNA using the TruSeq DNA Sample Prep Kit according to the manufacturer's instructions. Genes were predicted within the completed genomic sequence using Glimmer software 3.02 [36]. tRNA genes were predicted using tRNAscan-SE 1.3.1 [37], and rRNA genes were identified using RNAmmer 1.2 [38]. The protein sequence of the predicted gene was blastp-aligned with the Nr, Swiss-prot, string and GO databases, respectively (blast 2.2.28+), thereby obtaining annotation information for the predicted gene. Konstantinidis and Tiedje [39] proposed that the 70 % DDH standard seen as a pragmatic cut-off value for the delineation of species corresponds to 94 % average nucleotide identity (ANI) value in the definition of prokaryotic species. The orthologous ANI algorithm used the usearch program [40]. The final genome of strain CICC 11023T comprised 48 scaffolds with a total size of 8.78 Mb and a G+C content of 67.4 mol% overall, 68.05 mol% for the gene regions and 62.84 mol% for the intergenetic regions. The assembled contigs were annotated with the ncbi Prokaryotic Genome Annotation Pipeline pipeline [41], yielding a total of 9563 coding genes. General features of the genome of strain CICC 11023T are shown in Table S3. The ANI values of strain CICC 11023T were calculated between N. alba YIM 30243T and N. vulneris NBRC 108936T, respectively. The OrthoANIu value between CICC 11023T and N. alba YIM 30243T was 77.47 %, the DDH value was 25 %, and the OrthoANIu value between CICC 11023T and N. vulneris NBRC 108936T was 83.75 %), the DDH value was 21.6 %.
In conclusion, the morphological and chemotaxonomical characteristics and the results of phylogenetic analyses support that strain CICC 11023T had characteristics typical of a member of the genus Nocardia . The differential characteristics shown in Table 2 indicate that strain CICC 11023T has several different phenotypic properties that allow discrimination from the closest related species of the genus Nocardia , including utilization of raffinose, sorbitol, milk peptonization, and decomposition of aesculin. In addition, the cellular fatty acid analysis clearly suggested that CICC 11023T contained relatively large amounts of C17 : 0 and C18 : 0 10-methyl (TBSA), and small amounts of C16 : 0 and C18 : 1ω9c. The unique 16S rRNA and gyrB gene sequences and low level of DDH support that strain CICC 11023T represents a new species of the genus Nocardia with low ANI values (<94 %). The name Nocardia colli sp. nov. is proposed.
Description of Nocardia colli sp. nov.
Nocardia colli (col′li. L. neut. gen. n. colli of the neck).
Strain CICC 11023T is an aerobic, Gram stain-positive, non-motile, modified acid alcohol-positive actinomycetes bacterium, which forms an extensively branched grey-white substrate mycelium and aerial mycelium with fragments forming short coli-like bodies under scanning electronic microscopy (0.5×0.7–0.9 µm in diameter). The growth temperature range of strain CICC 11023T is 21–37 °C with an optimum growth temperature of 28 °C. The salt tolerance is in the range of 1–4% and the optimum growth salinity is 1 %. The strain shows positive reactions for milk peptonization, catalase, aesculin, urease and cellulose; negative reactions for milk coagulation, nitrate reduction, gelatin liquefaction, tyrosinase, hydrolysis of starch, casein, xanthine, hypoxanthine and production of H2S. Strain CICC 11023T can utilize l-arabinose, rhamnose, d-fructose, salicin, d-xylose, inositol, lactose, melibiose, d-glucose, raffinose, sucrose, d-mannitol, maltose, trehalose and arabinose as sole carbon sources. No soluble pigments are produced. The main fatty acids (>5 %) are C18 : 0 10-methyl (TBSA), C16 : 0, summed feature 4 (C16 : 1 trans 9/C15 : 0 iso 2OH), C15 : 0 and C17 : 0 10-methyl. The main menaquinone is cyclo MK-8(H4, ω-cyclo). The phospholipids are diphosphatidylglycerol, phosphatidylethanolamine, one unidentified phospholipid and two unidentified lipids. The type strain is susceptible to levofloxacin, ofloxacin, tobramycin and clindamycin; resistant to fosfomycin, imipenem, vancomycin and erythromycin. The organism is a pathogen of cutaneous infection in normal immunocompetent patients. The DNA G+C content of the type strain is 65.6 mol%.
The type strain, CICC 11023T (=KCTC 39837T), was isolated from aerobic culture of a biopsied skin tissue specimen from a 36-year-old female patient with primary cutaneous nocardiosis in Yunnan Province, south-west China.
Supplementary Data
Funding information
This work was supported by the Special Coordination Funds of Science and Technology Department of Yunnan Province and Kunming Medical University (2010CD174).
Acknowledgements
We are grateful to Professor Lihua Xu (Yunnan Institute of Microbiology, Yunnan University, China) for technical assistance.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ANI, average nucleotide identity; DDH, DNA–DNA hybridization; TSB, trypticase soy broth.
The nucleotide sequence of the 16S rRNA gene of strain CICC 11023T that we determined has been submitted to GenBank under the accession number KJ659849. The gyrB gene sequence of strain CICC 11023T determined in this study has been deposited in GenBank under the accession number MH580561. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession VXLC00000000. The version described in this paper is version VXLC01000000.
Two supplementary tables are available with the online version of this article.
References
- 1.Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new Hierarchic classification system, actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. doi: 10.1099/00207713-47-2-479. [DOI] [Google Scholar]
- 2.Trevisan VB. I Generi E Le specie delle Batteriacee. L. Zanaboni E Gabuzzi. 1889 http://scholar.google.com/scholar?cluster=12213764842208350008&hl=en&oi=scholarr
- 3.Beaman BL, Beaman L. Nocardia species: host-parasite relationships. Clin Microbiol Rev. 1994;7:213–264. doi: 10.1128/CMR.7.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conville PS, Brown JM, Steigerwalt AG, Brown-Elliott BA, Witebsky FG. Nocardia wallacei sp. nov. and Nocardia blacklockiae sp. nov., Human Pathogens and Members of the "Nocardia transvalensis Complex". J Clin Microbiol. 2008;46:1178–1184. doi: 10.1128/JCM.02011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeda K, Kang Y, Yazawa K, Gonoi T, Mikami Y. Phylogenetic studies of Nocardia species based on gyrB gene analyses. J Med Microbiol. 2010;59:165–171. doi: 10.1099/jmm.0.011346-0. [DOI] [PubMed] [Google Scholar]
- 6.Tindall BJ, Busse H-J, Ludwig W, Rosselló-Móra R, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 7.Conville PS, Nocardia WFG, Rhodococcus G. In: Manual of Clinical Microbiology. 10th ed. American Society of Microbiology;; 2011. Actinomadura, Streptomyces, and other aerobic Actinomycetes. [Google Scholar]
- 8.Welsh O, Vera-Cabrera L, Salinas-Carmona MC. Current treatment for Nocardia infections. Expert Opin Pharmacother. 2013;14:2387–2398. doi: 10.1517/14656566.2013.842553. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Zhou T, Deng D, Guo Y. A case of cutaneous nocardiosis with involvement of the trachea, anterior mediastinum and sternum. Case Rep Dermatol. 2010;2:177–182. doi: 10.1159/000321635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fey PD, Olson ME. Current concepts in biofilm formation of Staphylococcus epidermidis . Future Microbiol. 2010;5:917–933. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li WJ, Jiang Y, Kroppenstedt RM, Xu LH, Jiang C-L. Nocardia alba sp.nov., a novel actinomycete strain isolated from soil in China. Syst Appl Microbiol. 2004;27:308–312. doi: 10.1078/0723-2020-00270. [DOI] [PubMed] [Google Scholar]
- 12.Athalye M, Noble WC, Minnikin DE. Analysis of cellular fatty acids by gas chromatography as a tool in the identification of medically important coryneform bacteria. J Appl Bacteriol. 1985;58:507–512. doi: 10.1111/j.1365-2672.1985.tb01491.x. [DOI] [PubMed] [Google Scholar]
- 13.Kong BH, Li YH, Liu M, Liu Y, Li CL, et al. Massilia namucuonensis sp. nov., isolated from a soil sample. Int J Syst Evol Microbiol. 2013;63:352–357. doi: 10.1099/ijs.0.039255-0. [DOI] [PubMed] [Google Scholar]
- 14.Uchida K, Kudo T, Suzuki K-ichiro, Nakase T. A new rapid method of glycolate test by diethyl ether extraction, which is applicable to a small amount of bacterial cells of less than one milligram. J Gen Appl Microbiol. 1999;45:49–56. doi: 10.2323/jgam.45.49. [DOI] [PubMed] [Google Scholar]
- 15.Lechevalier MP, Lechevalier H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol. 1970;20:435–443. doi: 10.1099/00207713-20-4-435. [DOI] [Google Scholar]
- 16.Becker B, Lechevalier MP, Gordon RE, Lechevalier HA. Rapid differentiation between Nocardia and Streptomyces by paper chromatography of whole-cell hydrolysates. Appl Microbiol. 1964;12:421–423. doi: 10.1128/am.12.5.421-423.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins MD, Pirouz T, Goodfellow M, Minnikin DE. Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol. 1977;100:221–230. doi: 10.1099/00221287-100-2-221. [DOI] [PubMed] [Google Scholar]
- 18.Groth I, Schumann P, Rainey FA, Martin K, Schuetze B, et al. Demetria terragena gen. nov., sp. nov., a new genus of actinomycetes isolated from compost soil. Int J Syst Bacteriol. 1997;47:1129–1133. doi: 10.1099/00207713-47-4-1129. [DOI] [PubMed] [Google Scholar]
- 19.Minnikin DE, Hutchinson IG, Caldicott AB, Goodfellow M. Thin-Layer chromatography of methanolysates of mycolic acid-containing bacteria. J Chromatogr A. 1980;188:221–233. doi: 10.1016/S0021-9673(00)88433-2. [DOI] [Google Scholar]
- 20.Collins MD, Jones D. Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4-diaminobutyric acid. J Appl Bacteriol. 1980;48:459–470. doi: 10.1111/j.1365-2672.1980.tb01036.x. [DOI] [Google Scholar]
- 21.Wang Y, Jiang Y. Chemotaxonomy of Actinobacteria. In: Dhanasekaran D, editor. Actinobacteria - Basics and Biotechnological Applications. InTech; 2016. editor. [Google Scholar]
- 22.Kageyama A, Torikoe K, Iwamoto M, Masuyama J-I, Shibuya Y, et al. Nocardia arthritidis sp. nov., a new pathogen isolated from a patient with rheumatoid arthritis in Japan. J Clin Microbiol. 2004;42:2366–2371. doi: 10.1128/JCM.42.6.2366-2371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nie G-X, Ming H, Li S, Zhou E-M, Cheng J, et al. Amycolatopsis dongchuanensis sp. nov., an actinobacterium isolated from soil. Int J Syst Evol Microbiol. 2012;62:2650–2656. doi: 10.1099/ijs.0.038125-0. [DOI] [PubMed] [Google Scholar]
- 24.Smibert RM, Krieg NR. Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR, editors. Methods for General and Molecular Bacteriology. Washington, DC: American Society of Microbiology; 1994. [Google Scholar]
- 25.Hornef MW, Gandorfer A, Heesemann J, Roggenkamp A. Humoral response in a patient with cutaneous nocardiosis. Dermatology. 2000;200:78–80. doi: 10.1159/000018325. [DOI] [PubMed] [Google Scholar]
- 26.Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R, et al. Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia . Int J Syst Evol Microbiol. 2007;57:1424–1428. doi: 10.1099/ijs.0.64749-0. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol. 1971;20:406–416. doi: 10.1093/sysbio/20.4.406. [DOI] [Google Scholar]
- 30.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 32.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 33.Mesbah M, Premachandran U, Whitman WB. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol. 1989;39:159–167. doi: 10.1099/00207713-39-2-159. [DOI] [Google Scholar]
- 34.Gillis M, Ley JD, Cleene MD. The determination of molecular weight of bacterial genome DNA from renaturation rates. Eur J Biochem. 1970;12:143–153. doi: 10.1111/j.1432-1033.1970.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 35.Wayne LG. International Committee on systematic bacteriology announcement of the report of the ad hoc Committee on reconciliation of approaches to bacterial Systematics. J Appl Bacteriol. 1988;64:283–284. doi: 10.1111/j.1365-2672.1988.tb01872.x. [DOI] [PubMed] [Google Scholar]
- 36.Delcher AL. Glimmer Release Notes Version 3.02. 2006. [Google Scholar]
- 37.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. Search and clustering orders of magnitude faster than blast. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 41.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. Ncbi prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.