Abstract
Background and Aims:
Cirrhosis is characterized by extensive fibrosis of the liver and is a major cause of liver-related mortality. Cirrhosis is partially heritable but genetic contributions to cirrhosis have not been systemically explored. Here, we carry out association analyses with cirrhosis in two large biobanks and determine the effects of cirrhosis associated variants on multiple human disease/traits.
Methods:
We carried out a genome-wide association analysis of cirrhosis as a diagnosis in UK BioBank (UKBB; 1088 cases vs. 407 873 controls) and then tested top-associating loci for replication with cirrhosis in a hospital-based cohort from the Michigan Genomics Initiative (MGI; 875 cases of cirrhosis vs. 30 346 controls). For replicating variants or variants previously associated with cirrhosis that also affected cirrhosis in UKBB or MGI, we determined single nucleotide polymorphism effects on all other diagnoses in UKBB (PheWAS), common metabolic traits/diseases and serum/plasma metabolites.
Results:
Unbiased genome-wide association study identified variants in/near PNPLA3 and HFE, and candidate variant analysis identified variants in/near TM6SF2, MBOAT7, SERPINA1, HSD17B13, STAT4 and IFNL4 that reproducibly affected cirrhosis. Most affected liver enzyme concentrations and/or aspartate transaminase-to-platelet ratio index. PheWAS, metabolic trait and serum/plasma metabolite association analyses revealed effects of these variants on lipid, inflammatory and other processes including new effects on many human diseases and traits.
Conclusions:
We identified eight loci that reproducibly associate with population-based cirrhosis and define their diverse effects on human diseases and traits.
Keywords: fibrosis, genetics, phenotype, SNP
1 ∣. INTRODUCTION
Cirrhosis is characterized by extensive scarring of the liver and can develop after many liver diseases. Cirrhosis is associated with medical complications including ascites, variceal bleeding and hepatocellular carcinoma.1 Cirrhosis accounts for nearly $10 billion in healthcare costs in the United States alone and over one million deaths world-wide in 2015.1,2 While the most common causes of cirrhosis – viral hepatitis, non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease – are environmentally influenced, there is significant variation in whether people develop cirrhosis from these diseases.3 Genetics contributes to development of cirrhosis with the heritability of liver fibrosis and cirrhosis estimated to be about 50%,4 but the genetic determinants of disease have not been systematically explored.
The genetics of cirrhosis has so far been studied within small liver disease-specific populations. PNPLA3 and TM6SF2 variants associate with increased fibrosis in individuals with NAFLD, alcoholic liver disease and hepatitis C virus infection.5-7 Variants near MBOAT7 associate with alcoholic cirrhosis.6 Variants in or near GCKR, LYPLAL1 and HSD17B13 associate with fibrosis in NAFLD patients.8-11 In hepatitis C virus-infected patients, rs910049 (in the major histocompatibility complex region) and single nucleotide polymorphisms (SNPs) in MERTK and TULP1 associated with liver fibrosis progression,12,13 as does a SNP near PCSK7 in individuals with hereditary hemochromatosis.14 Finally, two monogenic liver diseases are caused by relatively common variants: hereditary hemochromatosis, which in 80%-90% of cases is caused by rs1800562-A (HFE C282Y), and alpha-1 antitrypsin deficiency, which is caused by mutations in SERPINA1, the most common severely deleterious of which is rs28929474-T (E342K).15,16 However, penetrance is incomplete for all these variants, including those in HFE and SERPINA1, and whether these variants lead to cirrhosis in the general population remains uncertain.
Here, we aim to evaluate the genetics of all-cause cirrhosis in the population. We performed a genome-wide association study (GWAS) for all cause cirrhosis (1088 cases vs. 407 873 controls) in approximately 410 000 individuals of British-Caucasian descent from the UK BioBank (UKBB).17 We test top-associating variants and previously reported cirrhosis variants for replication with cirrhosis (875 cases vs. 30 346 controls) in the Michigan Genomics Initiative (MGI).18 We determined the effects of all replicating variants on human disease/traits, on metabolic diseases from publically available GWAS analyses and on serum metabolites.
2 ∣. METHODS
2.1 ∣. Ethics statement
All research in this study was approved by the Institutional Review Board of the University of Michigan. UKBB protocols were approved by the National Research Ethics Service Committee and all participants provided written informed consent. Analyses in this project were conducted under UKBB Resource Project 18120. All MGI participants provided written informed consent approved by the University of Michigan Institutional Review Board.
2.2 ∣. Cohorts
The UKBB includes genotypic, clinical and demographic information of over 400 000 individuals aged 40-69 at time of recruitment. Genotyping and data collection were previously described.19 In brief, participants were genotyped on one of two custom arrays: UK BiLEVE Axiom Array (n = 50 520) or UKBB Axiom Array (n = 438 692) with >95% overlap. SNPs were imputed using the Haplotype Reference Consortium. For imputed SNPs, only SNPs with an imputation quality cut-off of 0.85 were used in analyses. SNPs with minor allele count <20 were excluded. After quality control, 18 530 078 SNPs in 408 961 white-British individuals were included in analysis.
Michigan Genomics Initiative is a prospective cohort with ongoing enrolment and at time of analysis included 35 888 subjects. All patients undergoing elective surgery at Michigan Medicine (Ann Arbor, MI) are potentially eligible for enrolment in this cohort. Participants underwent genotyping of peripheral blood on the Illumina HumanCoreExome v.12.1 array, a GWAS and exome array consisting of >500 000 SNPs.18 In addition, full laboratory information and diagnosis codes are available. Imputation was performed as previously described.20 In brief, samples were imputed to Haplotype Reference Consortium (release 1 for chromosomes 1-22 and 1.1 for X); SNPs were excluded if imputation quality was low (r2 < 0.3) or minor allele count was <4. After quality control, 30 751 457 imputed SNPs were available.
2.3 ∣. GWAS
This study had three stages (Figure 1). We limited analyses to individuals of European ancestry as there were not enough individuals from other ancestries to carry out powered analyses. Stage 1 was a GWAS for cirrhosis as defined by International Classification of Diseases (ICD)-10 code K70.2-4, K71.7, or K74.X in UKBB. SNPs within 500 kilobases of a lead SNP with lower p value were removed to identify independent signals. In stage 2, independent SNPs with P < 5 × 10−6 in UKBB were tested for association with cirrhosis in MGI as defined by the presence of an ICD-9 code for cirrhosis (571.5, 571.2 and 571.6), ICD-10 code for cirrhosis (K74.X, K70.2-4 and K71.7), or for cirrhosis being present in pathology or radiology reports using text searching. Text search of radiology and pathology reports was performed for the character ‘cirrho’, and subjects with that character were included as having cirrhosis with the following exceptions: (a) if the word ‘without’ or ‘no’ appeared in the same sentence as ‘cirrho’, subjects were considered controls; (b) if the words ‘primary biliary cirrhosis’ appeared in a sentence, that sentence was ignored for text search purposes to avoid falsely identifying individuals with primary biliary cholangitis as having cirrhosis and (c) if the words ‘evaluate’, ‘assess’ or ‘rule out’ appeared in a sentence with ‘cirrho’, that sentence was ignored for text search purposes. All subjects without ICD-9/-10 diagnosis code or positive text search were included as controls. A gastroenterologist (VLC) manually reviewed 200 randomly selected text strings and identified no false-positive cirrhosis diagnoses. Only SNPs with a minor allele count >6 in MGI for cirrhosis were tested for replication as the others were underpowered to see an effect. Only lead SNPs were included for replication.
FIGURE 1.
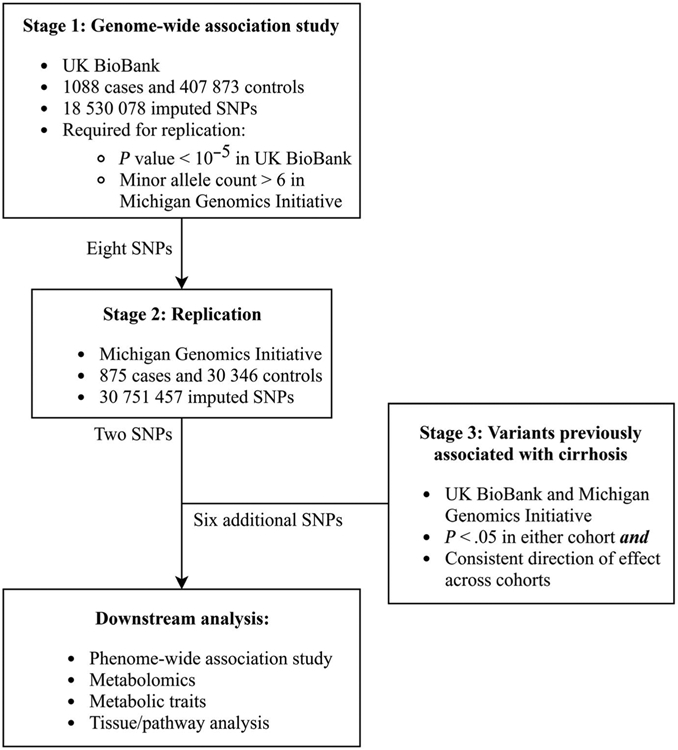
Study design
Association analyses in stage 1 and 2 were performed using Scalable and Accurate Implementation of GEneralized mixed model, which substantially reduces p value inflation otherwise seen in GWAS for rare traits,21 with a logistic mixed model and the saddlepoint approximation. Covariates included in this analysis were age, age,2 sex and principal components 1-10 to account for ethnic background. We used only a minimal number of covariates to maximize power and to avoid adjusting for potential mediators such as body mass index or diabetes mellitus.
Stage 3 analysis included replicating SNPs as well as SNPs that have been previously reported to associate with cirrhosis in subpopulations, as described in the introduction (Table S1).5,8,9,12-16,22-32 As above, we included only SNPs with minor allele count >6 in the European populations of UKBB and MGI, which corresponded to minor allele frequency >0.006. For stage 3 SNPs, a cut-off of P < .05 in either UKBB or MGI was used because there was prior knowledge of their role in liver disease and fibrosis.
All SNP coordinates were reported based on GRCh38.p12 or, if updated during the most recent patch, p13.
C statistics for prediction of cirrhosis were calculated first using a minimal model that included only age, age,2 sex and principal components 1-10, then adding cirrhosis-increasing SNPs. These were computed separately in MGI and UKBB.
2.4 ∣. Phenome-wide association study (PheWAS)
To assess the effects of SNPs on other human diagnoses and diseases, we carried out PheWAS on all SNPs identified in stage 1-3 analysis based on published data on 778 traits (http://geneatlas.roslin.ed.ac.uk/phewas/.17 For statistical significance of individual traits, a Bonferroni-corrected P value cut-off of 0.05 corrected for eight SNPs and 778 traits was used (P = 9.2 × 10−6; Z = 4.46). Significant diagnoses with associated ICD-10 codes or blood cell traits are shown. Hierarchical clustering on SNPs (but not traits) was performed using the gplot package using default settings. We conducted sensitivity analysis where we adjusted for cirrhosis status the associations between each significant SNP-trait pair replicating the models of the original PheWAS.17 We conducted among unrelated Caucasian individuals in UKBB logistic regression (for binary traits) or linear regression (for quantitative traits) with each trait as the primary dependent variable and each SNP as the primary independent variable. Covariates were age, age,2 sex, principal components 1-20, genotyping batch and assessment centre, with or without adjustment for cirrhosis status. A Z score change of >1.96 (corresponding to P < .05 for a difference between the associations) between the cirrhosis-adjusted and -unadjusted analyses was used as a cut-off for significant effect modification by cirrhosis.
We also tested the effect of all stage 1-3 SNPs on mean alanine aminotransferase (ALT), alkaline phosphatase (AP) and AST to platelet ratio index (APRI) in MGI. ALT, AP and APRI were inverse-normally transformed and linear regression was performed for SNP effects, adjusted for age, age,2 sex and principal components 1-10. As above, we used a minimal set of covariates to avoid including mediators and to maximize statistical power.
2.5 ∣. Metabolomic analysis
All SNPs identified in stage 1-3 analysis were tested for association with serum metabolites from previously published data.33 In brief, the authors performed genotyping and high-throughput nuclear magnetic resonance serum/plasma metabolomics on 14 datasets from 10 European cohorts. Data on 123 metabolites in up to 24 925 individuals were included. Because the traits were highly cross-correlated (Figure S1), data are only shown for certain representative metabolites. A Bonferroni-corrected significance cut-off of 0.05 corrected for eight SNPs and 123 metabolites was used (P = 2.5 × 10−5; Z = 4.05).
2.6 ∣. Cross-trait analyses
All SNPs identified in stage 1-3 analysis were tested for association with metabolic traits using publicly available data from nine GWAS. These studies were as follows: CARDIoGRAM-CAD34 (coronary artery disease; http://www.cardiogramplusc4d.org/data-downloads), DIAGRAM35 (type 2 diabetes; http://diagram-consortium.org/downloads.html), GCLC36 (high- and low-density lipoproteins, total cholesterol, triglycerides; http://csg.sph.umich.edu/abecasis/public/lipids2013/), GIANT37 (body mass index, height, waist-to-hip ratio, waist-to-hip ratio adjusted for body mass index; https://www.broad/institute.org/collaboration/giant/index.php/GIANT_consortium_data_files), MAGIC38 (fasting insulin and glucose levels; http://www.magicinvestigators.org/downloads/), a leptin GWAS39 and a body fat percentage GWAS40 (https://walker05.u.hpc.mssm.edu for the last two). There is no adequately powered GWAS for blood pressure with full publicly available data, so GWAS for systolic and diastolic blood pressure were performed on individuals of white-British ancestry in UKBB using Scalable and Accurate Implementation of GEneralized mixed model and a linear mixed model.21 In addition, data from the GOLD consortium were used to determine the effect of these SNPs on hepatic steatosis, as measured by lower liver attenuation on computed tomography relative to a ‘phantom’ control.9 Heat maps and clustering were performed using Z scores of SNP-trait associations. Only traits that were associated with at least one SNP at nominal significance (P < .05) were included in the heat map, so only seven GWAS were included. For statistical significance, a Bonferroni-adjusted p value cut-off of 0.05 divided by seven SNPs (one of the identified SNPs and its proxies were not available in any of the public GWAS data) and 17 traits (P = 4.2 × 10−4; Z = 3.52) was used.
2.7 ∣. Tissue and pathway analyses
Analysis of pathway and tissue enrichment was performed using FUMA-GWAS as previously reported.41 The background gene set used was ‘all’. Tissue enrichment was based on Genome-Tissue Expression Project version 6 and pathway enrichment on Reactome and Gene Ontology biological process terms.
3 ∣. RESULTS
3.1 ∣. Genome-wide association study
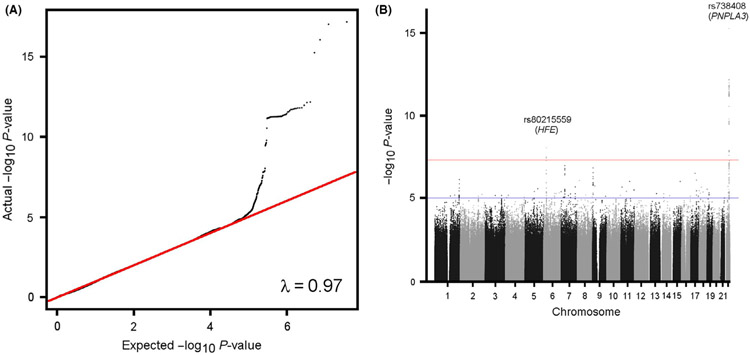
We carried out genome-wide association analyses for ~70 million imputed SNPs with cirrhosis (n = 1088 cases identified using ICD-10 codes vs. 407 873 controls without this diagnosis) in UKBB controlling for age, gender, and the first 10 principal components using a Scalable and Accurate Implementation of GEneralized logistic mixed model. There was no significant inflation of the association statistic with a lambda of 0.97 (Figure 2A). Two SNPs associated with cirrhosis at genome-wide significance levels (P < 5 × 10−8) and an additional 12 had a suggestive association with P < 5 × 10−6 (Figure 2B, Table 1, Table S2). We tested SNPs with suggestive association with cirrhosis (P < 5 × 10−6) in UKBB with cirrhosis in MGI (n = 875 cases based on ICD-10 codes and text search, vs. 31 221 controls). Only eight SNPs were common enough to have enough power to be evaluated for association with cirrhosis in MGI (Table 1, Table S2). Of these, two were significantly associated with increased cirrhosis prevalence (P < .05) in MGI: rs738408-T (PNPLA3 exon) and rs80215559-C (SLC17A2 intron) (Table 1). Of note, rs80215559 is in complete link-age with rs1800562 (r2 = 1.0 in CEU/GBR), which is also associated with cirrhosis at genome-wide significance levels (P = 3.3 × 10−8) and corresponds to the HFE C282Y mutation, the primary cause of hereditary hemochromatosis.16 Conditional analysis of the effect of rs80215559 on cirrhosis conditional on rs1800562 eliminated its association with cirrhosis (odds ratio 1.61 [95% CI 1.33-1.84] to 1.00 [95% CI 1.00-1.00]), suggesting that its effect is because of the HFE C282Y mutation.
FIGURE 2.
Quantile-quantile and Manhattan plots for GWAS of cirrhosis in UK BioBank. A, Quantile-quantile plot. B, Manhattan plot. GWAS, genome-wide association study
TABLE 1.
Loci that associate with cirrhosis in UK BioBank or Michigan Genomics Initiative
| CHR:POS | Variant | Gene | Ref/Alt | UK BioBank |
Michigan Genomics Initiative |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | Beta | SE | P value | EAF | Beta | SE | P value | ||||
| 22:43928850 | rs738408 | PNPLA3 (m) | C/T | 0.216 | 0.472 | 0.055 | 6.73E-18 | 0.233 | 0.269 | 0.058 | 3.54E-06 |
| 6:25917997 | rs80215559 | SLC17A2 (i), HFE (b) | T/C | 0.077 | 0.477 | 0.083 | 9.13E-09 | 0.059 | 0.280 | 0.106 | 8.39E-03 |
| 22:43928847 | rs738409 | PNPLA3 (m) | C/G | 0.216 | 0.470 | 0.055 | 9.58E-18 | 0.233 | 0.269 | 0.058 | 3.56E-06 |
| 6:26092913 | rs1800562 | HFE (m) | G/A | 0.078 | 0.454 | 0.082 | 3.30E-08 | 0.062 | 0.300 | 0.102 | 3.09E-03 |
| 19:19268740 | rs58542926 | TM6SF2 (m) | C/T | 0.076 | 0.378 | 0.084 | 6.01E-06 | 0.075 | 0.086 | 0.093 | 3.54E-01 |
| 14:94378610 | rs28929474 | SERPINA1 (m) | C/T | 0.020 | 0.727 | 0.162 | 7.35E-06 | 0.018 | 0.456 | 0.180 | 1.11E-02 |
| 4:87310241 | rs72613567 | HSD17B13 (s) | -/A | 0.721 | 0.178 | 0.048 | 2.30E-04 | NA | |||
| 19:54173307 | rs626283 | MBOAT7 (d) | G/C | 0.440 | 0.121 | 0.044 | 5.48E-03 | 0.433 | 0.031 | 0.050 | 5.31E-01 |
| 2:191099907 | rs7574865 | STAT4 (i) | T/G | 0.224 | 0.117 | 0.052 | 2.38E-02 | 0.224 | 0.077 | 0.059 | 1.87E-01 |
| 19:39248147 | rs12979860 | IFNL4 (i) | T/C | 0.299 | 0.081 | 0.047 | 8.40E-02 | 0.317 | 0.115 | 0.053 | 3.05E-02 |
Note: UK BioBank had 1088 cases and 407 874 controls, and Michigan Genomics Initiative had 875 cases and 30 346 controls. Additional variants are shown in Table S1.
Abbreviations: (b), biologically relevant gene; (i), intron; (m), missense mutation; (s), splice variant; Alt, alternate allele; CHR: POS, chromosome: position; EAF, effect allele frequency; NA Not available; Ref, reference allele; SE, standard error.
Bold value indicates that the association is statistically significant.
3.2 ∣. Candidate variant analysis
We also examined the effects of SNPs that have previously been associated with cirrhosis owing to any cause of liver disease (Table S1). Rs738409-G (PNPLA3), rs1800562-G (HFE), rs28929474-C (SERPINA1), rs58542926-C (TM6SF2), rs6834314-A and rs72613567-(no insertion) (HSD17B13), rs641738-T and rs626283-G (MBOAT7), and rs7574865-T (STAT4) associated with cirrhosis in UKBB, and rs738409-G (PNPLA3), rs1800562-G (HFE), rs28929474-C (SERPINA1), rs6834314-A (HSD17B13), and rs12979860-T (IFNL4) associated with cirrhosis in MGI (Table 1). The allele for each gene with the lowest p value in UKBB was included in subsequent analysis, except in the case of rs738409, which we chose in lieu of rs738408 as rs738409 is in high LD (R2 = 1) and corresponds to the causal variant.42
The C statistics for a minimal model and a model including all eight cirrhosis-increasing SNPs were, respectively, 0.63 and 0.67 in UKBB and 0.61 and 0.63 in MGI.
3.3 ∣. Effects on liver enzymes
We investigated whether these SNPs had effects on markers of liver injury such as serum ALT, a marker of hepatocellular injury; AP, a marker of cholestasis/infiltrative disease; and APRI, a non-invasive marker of liver fibrosis.43 Rs6834314 was used as a proxy for rs72613567 (R2 = 0.94), as the latter was not available in MGI. Both rs738409-G (PNPLA3) and rs8021559-C (HFE) associated with increased APRI, consistent with a role in worsening fibrosis (P = 1.75 × 10−17 and 7.41 × 10−3, respectively) (Table 2). In addition, rs738409-G (PNPLA3) associated with increased ALT (P = 1.42 × 10−14), but not AP, while rs80215559-C (HFE) associated with increased AP (P = 8.46 × 10−3) but not ALT (Table 2). PNPLA3, HFE, SERPINA1, MBOAT7 and HSD17B13 variants increase AP while TM6SF2 decreased AP (P < .05 for all). PNPLA3, HFE, TM6SF2, HSD17B13 all increased ALT (P < .05 for all). PNPLA3, HFE, SERPINA1 and HSD17B13 all associated with increased APRI (P < .05 for all). STAT4 and IFNL4 did not associate with abnormal liver enzymes in MGI.
TABLE 2.
Association between liver enzymes and cirrhosis-affecting variants in Michigan Genomics Initiative
| Variant | Gene | Alanine aminotransferase |
Alkaline phosphatase |
AST-to-platelet ratio index |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P value | Beta | SE | P value | Beta | SE | P value | ||
| rs738409 | PNPLA3 (m) | 0.091 | 0.012 | 1.42E-14 | 0.009 | 0.012 | 4.39E-01 | 0.102 | 0.012 | 1.75E-17 |
| rs80215559 | SLC17A2 (i), HFE (b) | 0.020 | 0.022 | 3.55E-01 | 0.058 | 0.022 | 8.46E-03 | 0.059 | 0.022 | 7.41E-03 |
| rs1800562 | HFE (m) | 0.268 | 0.037 | 3.93E-13 | 0.059 | 0.021 | 4.60E-03 | 0.063 | 0.021 | 2.60E-03 |
| rs28929474 | SERPINA1 (m) | 0.268 | 0.037 | 3.93E-13 | 0.264 | 0.037 | 9.67E-13 | 0.214 | 0.037 | 8.82E-09 |
| rs58542926 | TM6SF2 (m) | 0.085 | 0.019 | 8.00E-06 | −0.103 | 0.019 | 6.28E-08 | 0.027 | 0.019 | 1.54E-01 |
| rs626283 | MBOAT7 (d) | 0.009 | 0.010 | 3.92E-01 | 0.026 | 0.010 | 1.27E-02 | 0.004 | 0.010 | 6.70E-01 |
| rs6834314 | HSD17B13(u) | 0.039 | 0.011 | 1.11E-02 | −0.014 | 0.011 | 2.15E-01 | 0.036 | 0.011 | 1.26E-03 |
| rs7574865 | STAT4 (i) | 0.013 | 0.012 | 2.86E-01 | −0.002 | 0.012 | 8.96E-01 | 0.019 | 0.012 | 1.03E-01 |
| rs12979860 | IFNL4 (i) | 0.004 | 0.011 | 6.85E-01 | 0.009 | 0.011 | 4.15E-01 | 0.001 | 0.011 | 9.14E-01 |
Note: N = 19 598 for alanine aminotransferase, 19 493 for alkaline phosphatase, and 19 269 for aspartate aminotransferase-to-platelet ratio index. Abbreviations: (b), biologically relevant gene; (d), downstream gene; (i), intron; (m), missense mutation.
Bold value indicates that the association is statistically significant.
3.4 ∣. Tissue/pathway analyses and PheWAS
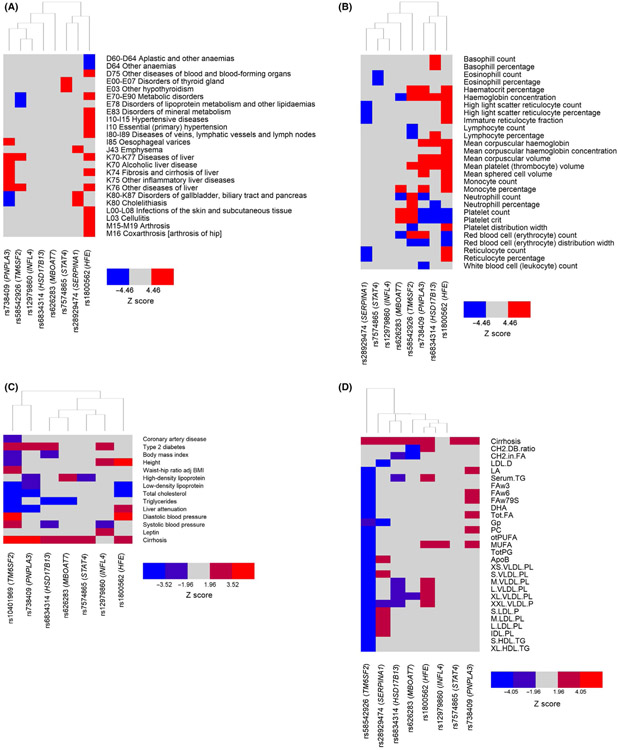
The genes implicated by the variants were upregulated in liver (Figure S2A).41 Gene Ontology analysis showed that these same genes were enriched for effects on pathways related to inflammation, lipid synthesis and protein complex synthesis (Figure S2B). However, these analyses were based on previously annotated pathways, and we sought to use a more unbiased approach to determine whether cirrhosis-associated SNPs had effects on other human diseases and traits using PheWAS analysis of diagnoses/traits in UKBB (Table S3). Nearly all traits were either ICD-10 diagnoses (Figure 3A) or blood cell traits (Figure 3B). We replicated known disease processes: rs1800562-A (HFE) associated with numerous red blood cell-related traits and increased arthroses and skin infections and rs2892947-T (SERPINA1) associated with increased emphysema. On PheWAS, PNPLA3 and TM6SF2 variants clustered together owing to effect on various liver diseases as well as, unexpectedly, decreased neutrophil count, haemoglobin traits and platelet traits. TM6SF2 and SERPINA1 variants had distinct effects on anthropometric traits: the former decreased peripheral fat while the latter increased it (Table S3). Multiple variants associated with blood cell traits; some of the unique associations included HSD17B13 and basophil count/percentage, STAT4 with eosinophil count/percentage, TM6SF2 with neutrophil count and PNPLA3 with leukopenia (Figure 3B).
FIGURE 3.
Representative pleiotropic effects of cirrhosis-increasing alleles. (A and B) Heat plot of the effect of cirrhosis-increasing alleles on phenome-wide association study in UK BioBank. Diagnoses are shown in (A) and blood cell traits in (B). (C) Heat plot of the effect of cirrhosis-increasing alleles on metabolic traits from publicly available GWAS. (D) Heat plot of the effect of cirrhosis-increasing alleles on serum/plasma metabolites. ‘Cirrhosis’ refers to cirrhosis diagnosis in UK BioBank (Methods). *IFNL4 trended towards a significant association with cirrhosis in UK BioBank (Z = 1.73, P = .08). Full lists of PheWAS analyses and metabolite labels are shown in Tables S3-S5. ApoB, ApoB; CH2.DB.ratio, CH2 groups in fatty acids; CH2.in.FA, CH2 groups to double bonds ratio; DHA, 22:6 docosahexaenoic acid; FAw3, Omega-3 fatty acids; FAw6, Omega-6 fatty acids; Gp, Glycoprotein acetyls mainly a1-acid glycoprotein; GWAS, genome-wide association study; IDL. PL, Phospholipids in intermediate-density lipoprotein; L.LDL.PL, Phospholipids in large low-density lipoproteins; L.VLDL.PL, Phospholipids in large very low-density lipoproteins; LDL.D, LDL diameter; M.LDL.PL, Phospholipids in medium low-density lipoproteins; M.VLDL.PL, Phospholipids in medium very low-density lipoproteins; MUFA, Mono-unsaturated fatty acids; otPUFA, Other polyunsaturated fatty acids than 18:2; PC, Phosphatidylcholine and other cholines; S.HDL.TG, Triglycerides in small high-density lipoproteins; S.LDL.P, Concentration of small low-density lipoprotein particles; S.VLDL.PL, Phospholipids in small very low-density lipoproteins; Serum.TG, Serum total triglycerides; Tot.FA, Total fatty acids; TotPG, Total phosphoglycerides; XL.HDL.TG, Triglycerides in very large high-density lipoproteins; XL.VLDL.PL, Phospholipids in very large very low-density lipoproteins; XS.VLDL.PL, Phospholipids in very small very low-density lipoproteins; XXL. VLDL.P, Concentration of chylomicrons and extremely large very low-density lipoproteins particles
Cirrhosis is itself associated with numerous physiological alterations including in lipid profiles, platelet count and red blood cell count. Thus, as sensitivity analysis, we performed a PheWAS in UKBB adjusted for cirrhosis status. For continuous traits, none of the Z scores changed significantly (Z score change >1.96) after adjustment for cirrhosis (Table S4). For binary traits, associations between the SNPs and several liver-related traits (including oesophageal varices and liver fibrosis/cirrhosis itself) changed in significance. However, no associations between SNPs and non-liver binary traits changed in significance; indeed, other than liver-related traits, no Z scores changed by >1 (Table S4). This analysis indicates that the associations between cirrhosis-increasing SNPs and non-liver diseases/traits are not due solely to the effect of these SNPs on cirrhosis.
3.5 ∣. Metabolic traits and metabolomics
We also characterized whether cirrhosis-associated variants associated with any of 19 different metabolic phenotypes (Methods; Table S5; Figure 3C). Owing to missing data, we used rs10401969 as a proxy for rs58542926 (TM6SF2; R2 = 0.95) and rs6834314 for rs72613567 (HSD17B13; R2 = 0.94); there were no acceptable link-age disequilibrium proxies (R2 > 0.5) for rs28929474 (SERPINA1) in European populations with available data in the GWASs, so this variant was not included in this analysis. Several variants with known metabolic effects clustered together: rs738409-G (PNPLA3) and rs5854292-T (TM6SF2) associated with decreased liver attenuation (increased liver fat) and altered lipid profiles. We also identified a separate cluster of variants which had not been thought to mediate liver disease through metabolic alterations, with effect on numerous metabolic traits. For example, rs1800562-A (HFE) decreased low-density lipoprotein and total cholesterol, and increased height and blood pressure. Rs626283-C (MBOAT7) decreased serum triglycerides.
We finally determined the effect of cirrhosis-associated variants on 123 metabolites based on previously published data (Figure 3D, Table S6).33 Because the metabolites were highly cross-correlated (Figure S1), we performed hierarchical clustering and with metabolite clusters shown in Table S6. Consistent with our previous report, the cirrhosis promoting allele rs58542926-T (TM6SF2) associated with decreased concentration of very low-density, intermediate- and high-density lipoproteins.44 The cirrhosis promoting allele rs2892474-T (SERPINA1) also associated with decreased glycoproteins with several other suggestive associations. MBOAT7 associated with fewer CH2 groups in fatty acids.
4 ∣. DISCUSSION
In this study, we identified common and low-frequency genetic variants that associate with population-based cirrhosis. PheWAS and metabolomic analysis identified distinct metabolic effects of cirrhosis associated variants suggesting that liver disease caused by many mechanisms can lead to cirrhosis.
Previously, rare diseases such as cirrhosis, which has an estimated prevalence of 1%, have been difficult to study at a population level.2 With the use of large cohorts of densely genotyped and well-characterized individuals, here we show that even previously considered rare liver diseases such as alpha-1 anti-trypsin deficiency and hereditary hemochromatosis contribute to population-based cirrhosis. Furthermore, using biobank PheWAS analysis, we verified known genetic pleiotropisms associated with cirrhosis promoting variants including dyslipidaemia, diabetes and body fat composition for six identified variants.45-47 Others have reported associations between HFE C282Y and arthritis, diabetes, chronic pain and brain iron accumulation,48-50 and SERPINA1 variants and hepatitis and fibrosis.51-54 Another recent study identified a SNP in MARC1 associated with cirrhosis, which was also associated with cirrhosis in UKBB (P = .047); differences in strengths of associations are likely due to differences in statistical power and definitions of cirrhosis.55 However, we also identified previously unreported associations such as increased body fat with SERPINA1, increased basophil count with HSD17B13, increased neutrophil count despite decreased overall leukocyte count with PNPLA3, and multiple sclerosis with HFE that might give new insights into disease pathophysiology caused by variants at these loci.
Most cirrhosis-increasing variants increased AP, suggesting a common pathway towards liver cholestasis with cirrhosis. TM6SF2 was a strong notable exception where the cirrhosis-promoting allele decreased AP. The mechanism of that is not clear but one intriguing possibility is that cholesterol (which cannot be excreted in very low-density lipoprotein in individuals with TM6SF2 variants56) is shunted to bile synthesis which increases farsenoid × receptor agonism and lowers AP.57 Notably, PNPLA3 and HSD17B13 variants did not associate with AP but did increase ALT, suggesting hepatocellular toxicity and potentially distinct disease mechanisms. Some variants (in PNPLA3, HFE, SERPINA1 and HSD17B13) also associated with AST-to-platelet ratio index, suggesting that we may be able to use non-invasive scores to find more variants that associate with cirrhosis in populations where we do not have diagnoses or direct imaging or pathology verification of the disease.
Genetic variants implicated genes that were enriched for expression in liver which suggests likely tissue autonomous effect of these variants. HFE and SERPINA1 were part of acute phase and inflammatory response pathways but these genes as well as all the other ones (PNPLA3, TM6SF2, HSD17B13, and MBOAT7) either were part of lipid metabolism affecting pathways or affected serum lipids in our metabolomics analyses. While cirrhosis itself alters lipid profiles,58 there was a low prevalence of cirrhosis in both cohorts, so these lipid changes are unlikely to be driven by cirrhosis alone. Another interesting possibility is that disruptions in lipid signalling represent a final common pathway to cirrhosis. PNPLA3 and HSD17B13, for example, may alter metabolism of vitamin A to retinoic acid to effect cirrhosis and is known to promote stellate cell activation and liver fibrosis.8,59 Whether the effect of other variants will ultimately converge on this as a mechanism to all fibrosis remains to be determined. The biochemical effects of the variants that promote liver damage, however, are diverse and thus it is not likely that one solution to eliminating liver damage will be possible even if the final common fibrotic pathway is curbed.
Clustering of genes differs based on whether previously annotated or agnostic methods are used. When previously annotated pathways such as Reactome are used, PNPLA3, TM6SF2, HSD17B13 and MBOAT7 cluster because they affect lipid traits, which is distinct from HFE and SERPINA1 which more affect acute phase and inflammatory responses. Using more agnostic approaches, we show here that MBOAT7 also affects inflammation-related traits including neutrophil count and (at nominal significance) ankylosing spondylitis, peritonitis and appendicitis. The effect of MBOAT7 on the immune system has been suggested before – it is expressed at high levels in immune cells and MBOAT7 variants are associated with increased inflammation with chronic hepatitis C infection60 – but here we demonstrate this at the population level. Conversely, HFE and SERPINA1 affect serum lipids and SERPINA1 also affects body fat composition and weight.
Cirrhosis-promoting variants protect against development of other human disease suggesting that simply reversing their effects may not be universally beneficial. For example, cirrhosis-causing variants decrease coronary artery disease/ischaemic heart disease (TM6SF2, SERPINA1),61,62 diabetes (TM6SF2), total cholesterol (PNPLA3, HFE, TM6SF2) and triglycerides (MBOAT7, HSD17B13, TM6SF2). These findings imply that medications may need to be personalized based on individual risk factors and environmental exposures to be able to reverse fibrosis while maintaining an adequate cardiovascular safety profile.
Non-alcoholic fatty liver disease is a major cause of cirrhosis in the population. Notably, though, genes that predispose to hepatic steatosis do not necessarily increase risk of fibrosis. We previously showed that variants in/near GCKR and LYPLAL1 increase non-alcoholic steatohepatitis/fibrosis,9 but in this study we did not find that they increased cirrhosis in the population. This discrepancy could be in part because they had a weaker effect on fibrosis than variants in TM6SF2, PNPLA3 or HSD17B13. Also, GCKR and LYPLAL1 variants may primarily affect steatosis or inflammation and not fibrosis: in our previous study, most individuals with non-alcoholic steatohepatitis also had fibrosis, so we were notable to distinguish between effects on inflammation alone vs. fibrosis. It will be critical to distinguish between steatohepatitis and fibrosis in future studies investigating the effect of genetics on outcomes in NAFLD.
Limitations of our study include that it included only individuals of European ancestry and may not have identified cirrhosis-increasing alleles that may be prevalent in only other ancestry groups. Interestingly, though, we replicated variants previously associated with cirrhosis in Asian populations, suggesting that many variants will have effects across populations. Furthermore, the UKBB analyses rely on ICD codes, and while the codes we used for cirrhosis are specific, they are not sensitive, which may reduce overall sensitivity for identifying cirrhosis-altering variants.63 This limitation may be why only two SNPs were identified on GWAS, whereas the rest were identified based on candidate variant analysis. Finally, rare diseases such as primary biliary cholangitis are under-represented in the general population, so genetic effects that require particular exposures to develop cirrhosis only in some but not all populations may not be detected.64 On a related note, we lacked adequate power to sub-classify patients with cirrhosis based on disease aetiology. Strengths include that this is the largest GWAS for cirrhosis to date.
We demonstrate that eight loci promote cirrhosis in population-based data and define their effects on human diseases and traits including many novel pleiotropic effects. We identified previously unrecognized metabolic and immune effects of several of these SNPs and defined novel gene clustering. This work gives new insights into the pathophysiology of cirrhosis.
Supplementary Material
Key points.
Some genes cause cirrhosis, which is scarring of the liver that occurs after prolonged injury, but the genetics of cirrhosis have not been previously studied in an unbiased manner.
We identified two genetic mutations that cause cirrhosis at genome-wide significance and validated six other mutations previously suggested to affect cirrhosis in small studies now in large biobanks.
We show that these variants affect multiple human disease and traits which give us new insights into the diverse causes of cirrhosis.
ACKNOWLEDGEMENTS
VLC was supported in part by a University of Michigan Training in Basic and Translational Digestive Sciences T32 grant (NIDDK 5T32DK094775) and The University of Michigan Department of Internal Medicine. EKS, SKH, XD and YC are supported in part by R01DK106621 (to EKS), R01DK107904 (to EKS) and The University of Michigan Department of Internal Medicine. The authors acknowledge the Michigan Genomics Initiative participants, Precision Health at the University of Michigan, the University of Michigan Medical School Central Biorepository, and the University of Michigan Advanced Genomics Core for providing data and specimen storage, management, processing, and distribution services, and the Center for Statistical Genetics in the Department of Biostatistics at the School of Public Health for genotype data curation, imputation, and management in support of the research reported in this publication. Analyses in the UK BioBank were done under approved project 18120 (EKS).
Funding information
University of Michigan Training in Basic and Translational Digestive Sciences, Grant/Award Number: NIDDK 5T32DK094775; The University of Michigan Department of Internal Medicine, Grant/Award Number: R01DK106621 and R01DK107904
Abbreviations:
- ALT
alanine aminotransferase
- AP
alkaline phosphatase
- APRI
AST-to-platelet ratio index
- GWAS
genome-wide association study
- ICD
International Classification of Diseases
- MGI
Michigan Genomics Initiative
- NAFLD
non-alcoholic fatty liver disease
- PheWAS
phenome-wide association study
- SNP
single nucleotide polymorphism
Footnotes
CONFLICTS OF INTEREST
Vincent Chen, Yanhua Chen, Xiaomeng Du, Samuel Handelman, Elizabeth Speliotes: no financial conflicts of interest to disclose.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- 1.Mokdad AA, Lopez AD, Shahraz S, et al. Liver cirrhosis mortality in 187 countries between 1980 and 2010: a systematic analysis. BMC Med. 2014;12(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mellinger JL, Shedden K, Winder GS, et al. The high burden of alcoholic cirrhosis in privately insured persons in the United States. Hepatology. 2018;68(3):872–882. [DOI] [PubMed] [Google Scholar]
- 3.Wong RJ, Aguilar M, Cheung R, et al. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 2015;148(3):547–555. [DOI] [PubMed] [Google Scholar]
- 4.Loomba R, Schork N, Chen C-H, et al. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology. 2015;149(7):1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Trépo E, Nahon P, et al. PNPLA3 and TM6SF2 variants as risk factors of hepatocellular carcinoma across various etiologies and severity of underlying liver diseases. Int J Cancer. 2019;144(3):533–544. [DOI] [PubMed] [Google Scholar]
- 6.Buch S, Stickel F, Trépo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47(12):1443–1448. [DOI] [PubMed] [Google Scholar]
- 7.Hernaez R, McLean J, Lazo M, et al. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third National Health and Nutrition Examination Survey. Clin Gastroenterol Hepatol. 2013;11(9):1183–1190.e1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y, Belyaeva OV, Brown PM, et al. HSD17B13 is a hepatic retinol dehydrogenase associated with histological features of non-alcoholic fatty liver disease. Hepatology. 2019;69(4):1504–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7(3):e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirola CJ, Garaycoechea M, Flichman D, et al. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J Lipid Res. 2019;60(1):176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abul-Husn NS, Cheng X, Li AH, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378(12):1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patin E, Kutalik Z, Guergnon J, et al. Genome-wide association study identifies variants associated with progression of liver fibrosis from HCV infection. Gastroenterology. 2012;143(5):1244–1252.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rüeger S, Bochud P-Y, Dufour J-F, et al. Impact of common risk factors of fibrosis progression in chronic hepatitis C. Gut. 2015;64(10):1605–1615. [DOI] [PubMed] [Google Scholar]
- 14.Stickel F, Buch S, Zoller H, et al. Evaluation of genome-wide loci of iron metabolism in hereditary hemochromatosis identifies PCSK7 as a host risk factor of liver cirrhosis. Hum Mol Genet. 2014;23(14):3883–3890. [DOI] [PubMed] [Google Scholar]
- 15.Greene CM, Marciniak SJ, Teckman J, et al. alpha1-Antitrypsin deficiency. Nat Rev Dis Primers. 2016;2:16051. [DOI] [PubMed] [Google Scholar]
- 16.Radford-Smith DE, Powell EE, Powell LW. Haemochromatosis: a clinical update for the practising physician. Intern Med J. 2018;48(5):509–516. [DOI] [PubMed] [Google Scholar]
- 17.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dey R, Schmidt EM, Abecasis GR, Lee S. A fast and accurate algorithm to test for binary phenotypes and its application to PheWAS. Am J Hum Genet. 2017;101(1):37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguire LH, Handelman SK, Du X, Chen Y, Pers TH, Speliotes EK. Genome-wide association analyses identify 39 new susceptibility loci for diverticular disease. Nat Genet. 2018;50(10):1359–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W, Nielsen JB, Fritsche LG, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50(9):1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang D-K, Ma X-P, Wu X, et al. Genetic variations in STAT4, C2, HLA-DRB1 and HLA-DQ associated with risk of hepatitis B virus-related liver cirrhosis. Sci Rep. 2015;5:16278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attallah AM, Omran D, Marie MS, et al. IL-28B rs12979860 polymorphism affect the course of chronic hepatitis and the development of HCC in Egyptian patients with hepatitis C type 4. Br J Biomed Sci. 2018;75(4):157–162. [DOI] [PubMed] [Google Scholar]
- 24.Besheer T, Arafa M, El-Maksoud MA, et al. Diagnosis of cirrhosis in patients with chronic hepatitis C genotype 4: Role of ABCB11 genotype polymorphism and plasma bile acid levels. Turk J Gastroenterol. 2018;29(3):299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medrano LM, Rallón N, Berenguer J, et al. Relationship of TRIM5 and TRIM22 polymorphisms with liver disease and HCV clearance after antiviral therapy in HIV/HCV coinfected patients. J Transl Med. 2016;14:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ezzikouri S, Elfihry R, Chihab H, et al. Effect of MBOAT7 variant on hepatitis B and C infections in Moroccan patients. Sci Rep. 2018;8(1):12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang L, Mo Z, Li S, Qin X. The association between PIN1 genetic polymorphisms and the risk of chronic hepatitis B and hepatitis B virus-related liver cirrhosis: a case-control study. Medicine (Baltimore). 2018;97(35):e12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Sharkawy R, Thabet K, Lampertico P, et al. A STAT4 variant increases liver fibrosis risk in Caucasian patients with chronic hepatitis B. Aliment Pharmacol Ther. 2018;48(5):564–573. [DOI] [PubMed] [Google Scholar]
- 29.Dai Z-J, Liu X-H, Wang M, et al. IL-18 polymorphisms contribute to hepatitis B virus-related cirrhosis and hepatocellular carcinoma susceptibility in Chinese population: a case-control study. Oncotarget. 2017;8(46):81350–81360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang XQ, Hong XJ, Bai XJ. Susceptibility to active decompensated cirrhosis is associated with polymorphisms of intercellular adhesion molecule-1 (ICAM-1) in chronic HBV carriers. J Viral Hepatitis. 2008;15(3):173–178. [DOI] [PubMed] [Google Scholar]
- 31.Migita K, Maeda Y, Abiru S, et al. Polymorphisms of interleukin-1beta in Japanese patients with hepatitis B virus infection. J Hepatol. 2007;46(3):381–386. [DOI] [PubMed] [Google Scholar]
- 32.Song Q-L, He X-X, Yang H, et al. Association of a TANK gene polymorphism with outcomes of hepatitis B virus infection in a Chinese Han population. Viral Immunol. 2012;25(1):73–78. [DOI] [PubMed] [Google Scholar]
- 33.Kettunen J, Demirkan A, Würtz P, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The CDC. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The DGR, Meta-analysis C. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Global Lipids Genetics C, Wilier CJ, Schmidt EM, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shungin D, Winkler TW, Croteau-Chonka DC, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kilpeläinen TO, Carli JFM, Skowronski AA, et al. Genome-wide meta-analysis uncovers novel loci influencing circulating leptin levels. Nat Commun. 2016;7:10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y, Day FR, Gustafsson S, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat Commun. 2016;7:10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe K, Taskesen E, vanBochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li JZ, Huang Y, Karaman R, et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest. 2012;122(11):4130–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wai CT, Greenson JK, Fontana RJ, et al. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003;38(2):518–526. [DOI] [PubMed] [Google Scholar]
- 44.Kim DS, Jackson AU, Li YK, et al. Novel association of TM6SF2 rs58542926 genotype with increased serum tyrosine levels and decreased apoB-100 particles in Finns. J Lipid Res. 2017;58(7):1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hyysalo J, Gopalacharyulu P, Bian H, et al. Circulating triacylglycerol signatures in nonalcoholic fatty liver disease associated with the I148M variant in PNPLA3 and with obesity. Diabetes. 2014;63:312–322. [DOI] [PubMed] [Google Scholar]
- 46.Palmer CNA, Maglio C, Pirazzi C, et al. Paradoxical lower serum triglyceride levels and higher type 2 diabetes mellitus susceptibility in obese individuals with the PNPLA3 148M variant. PLoS ONE. 2012;7(6):e39362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Speliotes EK, Butler JL, Palmer CD, et al. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52(3):904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pilling LC, Tamosauskaite J, Jones G, et al. Common conditions associated with hereditary haemochromatosis genetic variants: cohort study in UK Biobank. BMJ. 2019;364:k5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elliott LT, Sharp K, Alfaro-Almagro F, et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562(7726):210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamosauskaite J, Atkins JL, Pilling LC, et al. Hereditary hemochromatosis associations with frailty, sarcopenia and chronic pain: evidence from 200,975 older UK biobank participants. J Gerontol A Biol Sci Med Sci. 2019;74(3):337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark VC, Marek G, Liu C, et al. Clinical and histologic features of adults with alpha-1 antitrypsin deficiency in a non-cirrhotic cohort. J Hepatol. 2018;69(6):1357–1364. [DOI] [PubMed] [Google Scholar]
- 52.Kim RG, Nguyen P, Bettencourt R, et al. Magnetic resonance elastography identifies fibrosis in adults with alpha-1 antitrypsin deficiency liver disease: a prospective study. Aliment Pharmacol Ther. 2016;44(3):287–299. [DOI] [PubMed] [Google Scholar]
- 53.Hamesch K, Mandorfer M, Pereira VM, et al. Liver fibrosis and metabolic alterations in adults with alpha-1-antitrypsin deficiency caused by the Pi*ZZ mutation. Gastroenterology. 2019;157(3):705–719.e18. [DOI] [PubMed] [Google Scholar]
- 54.Tanash HA, Piitulainen E. Liver disease in adults with severe alpha-1-antitrypsin deficiency. J Gastroenterol. 2019;54(6):541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emdin CA, Haas M, Khera AV, et al. A missense variant in mitochondrial amidoxime reducing component 1 gene and protection against liver disease. bioRxiv;2019:594523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahdessian H, Taxiarchis A, Popov S, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci USA. 2014;111(24): 8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trauner M, Gulamhusein A, Hameed B, et al. The nonsteroidal FXR agonist cilofexor (GS-9674) improves markers of cholestasis and liver injury in patients with PSC. Hepatology. 2019;70:788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Privitera G, Spadaro L, Marchisello S, Fede G, Purrello F. Abnormalities of lipoprotein levels in liver cirrhosis: clinical relevance. Dig Dis Sci. 2018;63(1):16–26. [DOI] [PubMed] [Google Scholar]
- 59.Trepo E, Romeo S, Zucman-Rossi J, Nahon P. PNPLA3 gene in liver diseases. J Hepatol. 2016;65(2):399–412. [DOI] [PubMed] [Google Scholar]
- 60.Thabet K, Asimakopoulos A, Shojaei M, et al. MBOAT7 rs641738 increases risk of liver inflammation and transition to fibrosis in chronic hepatitis C. Nat Commun. 2016;7:12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holmen OL, Zhang HE, Fan Y, et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet. 2014;46(4):345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pirola CJ, Sookoian S. The dual and opposite role of the TM6SF2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: a meta-analysis. Hepatology. 2015;62(6):1742–1756. [DOI] [PubMed] [Google Scholar]
- 63.Nehra MS, Ma Y, Clark C, Amarasingham R, Rockey DC, Singal AG. Use of administrative claims data for identifying patients with cirrhosis. J Clin Gastroenterol. 2013;47(5):e50–e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou WC, Zhang QB, Qiao L. Pathogenesis of liver cirrhosis. World J Gastroenterol. 2014;20(23):7312–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.