Abstract
Cell dedifferentiation is the process by which cells grow reversely from a partially or terminally differentiated stage to a less differentiated stage within their own lineage. This extraordinary phenomenon, observed in many physiological processes, inspires the possibility of developing new therapeutic approaches to regenerate damaged tissue and organs. Meanwhile, studies also indicate that dedifferentiation can cause pathological changes. In this review, we compile the literature describing recent advances in research on dedifferentiation, with an emphasis on tissue-specific findings, cellular mechanisms, and potential therapeutic applications from an engineering perspective. A critical understanding of such knowledge may provide fresh insights for designing new therapeutic strategies for regenerative medicine based on the principle of cell dedifferentiation.
Subject terms: Tissue engineering, Regenerative medicine
Introduction
Regeneration has always been a dream of humans, as shown in various fairy tales. Some invertebrates and amphibians have extraordinary capacity for regeneration. Zebrafish are able to fully regenerate their hearts, even after 20% of the ventricle is removed1. Earthworms readily reform two new duplicates after being cut in half2. Some amphibians, such as newts, create new limbs, tails and jaws with identical structures or functions to the lost body parts after amputation3, which is regarded as complete regeneration. However, at the organismal level, humans (like many other mammals) have a limited capacity for regeneration. The structure or functions of a damaged organ may be repaired to a compromised extent compared to the native status, and such “incomplete” regeneration usually occurs within this specific organ4. Typical cases include scarring in the skin that restores coverage without perspiration, as well as fibrocartilage that provides a structure different from articulate cartilage (and therefore inferior functions). Thus, these complete regenerative phenomena are prompting researchers to determine the underlying mechanisms and transform this mythological image into reality for humans. Dedifferentiation is regarded as one of the mechanisms involved in regeneration, as it enables cells, especially those without proliferative potential, to proliferate again and redifferentiate, leading to the replacement of the lost cells.
There are a variety of cell types throughout multicellular organisms. All of these cells originate from a simple zygote through a series of processes such as cell division and differentiation5. During differentiation, the fundamental basis of development of an organism, a less specialized cell type gradually transforms into a more specialized cell type, which is constrained to a stable morphology, structure and function. Moreover, the process of differentiation reduces the self-renewal ability and pluripotency of the cell. Differentiation continues during adulthood to maintain homeostasis. Dedifferentiation is a cellular process by which cells grow in reverse, from a partially or terminally differentiated stage to a less differentiated stage within their own lineage. In general, the phenomenon is manifested by a change in the shape, gene expression pattern, protein expression pattern and function. For example, dedifferentiated chondrocytes experience a change in phenotype after repeated culture in monolayers. The phenotype of spherical chondrocytes changes to spindle-shaped fibroblastic-like cells. Meanwhile, dedifferentiation results in a shift in the expression of the type II collagen (Col II) gene to type I collagen (Col I) and thus a corresponding change in the production of Col II protein to Col I protein in the extracellular matrix (ECM)6. In addition, these changes also lead to a switch from hyaline cartilage to fibrous cartilage, which would considerably change the function of articular cartilage7.
In recent decades, an increasing number of researchers have proposed that the induction of dedifferentiation shows promise to repair injured tissue in the clinic4,8. Terminally differentiated cells obtained in a noninvasive manner are potentially transformed to pluripotent stem cells through dedifferentiation. Then, these cells would be propagated in vitro, differentiated into objective cells and transplanted in vivo to achieve the regeneration of the damaged organ. Based on accumulating evidence, signaling pathways play a critical role in the process of dedifferentiation. Activation of the Wnt/β-catenin signaling pathway induces the dedifferentiation of epidermal cells9, articular chondrocytes10, or endothelial cells11 for regeneration. In addition, some specific biomolecules trigger the dedifferentiation of vascular smooth muscle cells12 or chondrocytes13 via the mitogen-activated protein kinase (MAPK) pathways. Also, innovative biomaterials are designing and creating to regulate dedifferentiation14,15. As our understanding of the molecular mechanisms of dedifferentiation improves, therapeutic methods using dedifferentiation present tremendous potential.
In this review, we compile the literature on recent advances in research of dedifferentiation, with an emphasis on the current knowledge of dedifferentiation processes, possible mechanisms and the therapeutic applications with an engineering perspective. A comprehensive understanding of such knowledge may provide fresh insights into the potential use of dedifferentiation in translational science.
Dedifferentiation in tissue regeneration (as a physiological mechanism)
The phenomena of dedifferentiation exists in various tissues and organs from plants, amphibians and animals, manifesting mainly as re-entry into the cell cycle, the acquisition of a stem cell-like phenotype, the expression of stem cell markers and redifferentiation to regenerate damaged tissues16. Here, the dedifferentiation of mammalian cardiomyocytes and neurons, which have been studied extensively, will be discussed in detail.
Dedifferentiation of cardiomyocytes
Mammalian cardiomyocytes stop multiplying soon after birth, which leads to the inability to replace the heart muscle cells that are damaged by myocardial infarction and other illnesses17. However, Porrello et al.18 observed a transient regenerative potential of the neonatal murine heart induced by the dedifferentiation of cardiomyocytes. Moreover, cardiomyocyte remodeling, which includes the restructuring of the normally existing structures and rearrangement of the cytoskeleton, is vital for the normal function of heart19. A recently proposed hypothesis states that cardiomyocyte remodeling is closely related to dedifferentiation, which is characterized by a switch in the expression pattern of mature cardiomyocytes to a more immature state, enabling cardiomyocytes to survive under unfavorable conditions, enter the cell cycle to proliferate and recover their functions20,21. MacLellan’s group demonstrated in mouse model that dedifferentiation may be an essential prerequisite for cardiomyocyte proliferation22. Both early genes, such as α-smooth muscle actin (α-SMA) and GATA4, and characteristic markers that are generally expressed in stem cells, such as Runx1 and Dab2, are re-expressed in dedifferentiated cardiomyocytes, which might facilitate proliferation and improve the multipotent differentiation capacity22,23. The dedifferentiation of cardiomyocytes may be an alternative pathway to induce regeneration, as observed in the newt, whose heart is regenerated through the dedifferentiation of mature cardiomyocytes and subsequent proliferation (Fig. 1). The corresponding mechanisms have not been completely elucidated. Numerous studies have been performed to provide insights into the underlying mechanisms.
Fig. 1. Schematic showing an overview of cardiomyocyte dedifferentiation during cardiac regeneration.
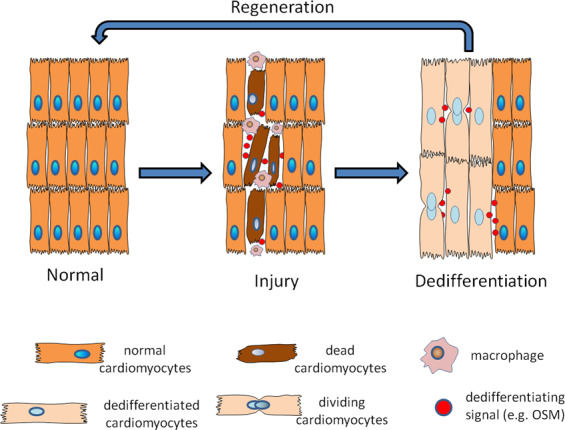
Macrophages infiltrate the damaged heart to remove debris and release OSM to induce the dedifferentiation of surviving cardiomyocytes. Then, cardiomyocytes switch from the expression pattern of mature cardiomyocytes to a more immature state, enabling the cells to survive under unfavorable conditions, enter the cell cycle to proliferate, re-establish cell–cell contacts and recover their functions.
An in vitro model in which adult rabbit cardiomyocytes were cocultured with cardiac fibroblasts was established24. The model exhibits similar morphological changes to dedifferentiating cardiomyocytes in vivo, and thus may be a useful tool to investigate the factors and the pathways involved in dedifferentiation in vivo. Recently, Kubin et al.25 reported a key role for the inflammatory cytokine oncostatin M (OSM), a member of the interleukin-6 family, in the mechanism regulating cardiomyocyte dedifferentiation. OSM triggers the Ras/MEK/Erk cascade to induce dedifferentiation of in vitro rat cardiomyocyte via OSM receptor β. A positive feedback loop mediates murine cardiomyocyte dedifferentiation; it comprises Yes-associated protein (YAP), transcriptional enhancer factor (TEAD1) and OSM—where the YAP-TEAD1 pathway stimulates OSM expression, OSM activates YAP-TEAD1, and OSM also upregulates OSM receptors through the YAP-TEAD1 pathway26. The size of OSM-treated cardiomyocytes also increases, which may allow them to re-establish cellular contacts to promote cellular survival27.
In addition, during the process of limb regeneration in urodele amphibians, the retinoblastoma (RB) protein has a very important role in facilitating the re-entry of differentiated cells into the cell cycle. During this process, mature cells dedifferentiate through the inactivation of RB and then enter the cell cycle28. The findings from numerous studies on the role of RB are consistent with this finding. The inactivation of RB, which is attributed to phosphorylation by increased numbers of cyclin D1/cdk4 complexes, is related to cardiomyocyte hypertrophy in congestive heart failure29. According to MacLellan et al.30, mice lacking RB and RB-like 2 have a larger heart and exhibit increased cardiomyocytes proliferation. Zaglia et al.31 were the first researchers to report that GATA4, a zinc-finger transcription factor, is expressed by fibroblasts in the adult rat heart, but not in fibroblasts from the gut, skin and skeletal muscle. In addition, the authors established an in vitro coculture experiment with cardiac fibroblasts and cardiomyocytes. Cardiac fibroblasts enhance the dedifferentiation of adult cardiomyocytes, a change that is strongly correlated with cell cycle reprogramming, as evidenced by the expression of DNA synthesis markers. Stimulation of GATA4 by overexpression of GRP78 (glucose-regulated protein of 78 kDa) can promote cardiomyocyte growth in mice32. However, further studies are needed to investigate the underlying mechanisms by which GATA4 affects cell division.
Dedifferentiation of neurons
Following nerve injury, Schwann cells regenerate. The mature myelinating Schwann cells dedifferentiate, regain the expression pattern associated with immature Schwann cells, proliferate and subsequently redifferentiate to induce nerve repair33. Schwann cells remove myelin and axonal debris after dedifferentiation by themselves or by recruiting circulating macrophages34. Upon dedifferentiation, Schwann cells downregulate myelin-associated genes that are essential for the myelination process, such as myelin protein zero and Krox20/Egr-235, and re-express molecules associated with immature states, such as p75 neurotrophin receptor (p75NTR), neural cell adhesion molecule (NCAM) and L136. Moreover, Schwann cells produce neurotrophic factors to support axon regeneration37 (Fig. 2).
Fig. 2. Schematic showing an overview of nerve regeneration after injury induced by the dedifferentiation of Schwann cells.
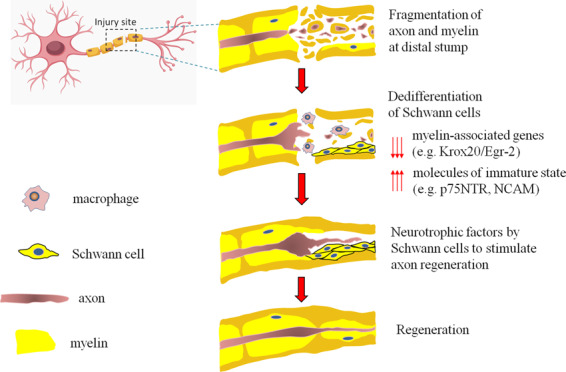
The fragmentation of axons and myelin occurs at the injury site. The mature myelinating Schwann cells dedifferentiate, regain the expression pattern associated with immature Schwann cells and proliferate. Then, Schwann cells remove myelin and axonal debris after dedifferentiation by themselves or by recruiting circulating macrophages and produce neurotrophic factors to support axon regeneration.
Based on accumulating evidence, several intracellular signaling pathways are involved in Schwann cell dedifferentiation. Notch signaling negatively regulates myelination, accelerates the dedifferentiation of mature Schwann cells and promotes the proliferation of immature Schwann cells38. A series of studies has focused on mitogen-activated protein kinase (MAPK) pathways in Schwann cell dedifferentiation. Although extracellular signal-regulated kinase (ERK) signaling is implicated in Schwann cell dedifferentiation39, increased ERK phosphorylation alone is not sufficient to cause dedifferentiation without colony-stimulating factor (CSF)-1-activated macrophage reactions40. The transcription factor c-Jun, which is the terminal component of the Jun-N-terminal kinase (JNK) pathway, is a negative regulator of myelination41. Parkinson et al.42 genetically depleted c-Jun from murine Schwann cells and observed a delay in demyelination after injury and the suppression of dedifferentiation. Recently, the transient reactivation of mTOR complex 1 was shown to be necessary to increase c-Jun translation and Schwann cell dedifferentiation after mice nerve injury43. Additionally, c-Jun contributes to the production of neurotrophic factors, including glial cell line-derived neurotrophic factor (GDNF) and Artemin, which is beneficial for axonal regeneration44,45. The activation of p38 MAPK induces c-Jun expression and drives rat Schwann cell dedifferentiation46. Park and colleagues47 reported an important role for the Rac1 GTPase (Rac) in Schwann cell dedifferentiation. Rac controls the demyelination of murine Schwann cells in Schmidt-Lantermann incisures and induces c-Jun expression in injured sciatic nerves. Rac also regulates the expression of GDNF and p75NTR, which are related to regeneration. Furthermore, the neuregulin-Rac-MKK7-JNK/c-Jun pathway is required for Schwann cell dedifferentiation following rat nerve injury48. As shown in the study by Jung’s group, extracellular adenosine triphosphate inhibits rat Schwann cell proliferation and Schwann cell dedifferentiation by blocking the lysosomal activation49. Likewise, increased cAMP levels induce and maintain the differentiation of Schwann cells, while the removal of cAMP rapidly induces the dedifferentiation and proliferation of Schwann cells42,50. Moreover, according to Viader et al.51, miRNAs expressed in Schwann cells play a key role in modulating murine Schwann cell response to nerve injury. The authors identified interactions of miR-34a with Notch1 and Ccnd1 to promote the dedifferentiation and proliferation of Schwann cells, and miR-140 influenced the remyelination.
In addition, astrocytes, which are widely distributed throughout the central nervous system (CNS), dedifferentiate into neural stem/progenitor cells (NSPCs) upon exposure to appropriate stimuli based on compelling evidence52,53. Conditioned medium from scratch-insulted astrocytes (ACM) induces astrocytes to regain NSPCs potentials and these NSPCs of astrocytic origin redifferentiated into neuronal and glial cells54,55. Overexpression of Ezh2 increases the expression of neural stem cell genes and downregulates the expression of astrocytic genes, but these changes were insufficient to induce a complete dedifferentiation56. A series of studies has been conducted by Yang et al.57 to investigate the underlying mechanisms involved in the astrocyte dedifferentiation process. Using an in vitro scratch-wound model, the authors revealed that ACM treatment stimulated the rat astrocytes to dedifferentiate into radial glial progenitor cells with the characteristic radial glial-like morphology, re-expression of nestin, RC2 and paired homeobox proteins, and increased multipotent differentiation capability. Concomitantly, the progressive increase in the level of ErbB2 protein level suggested that the activation of the ErbB2 signaling pathways may lead to all these phenotypes in astrocytes. Furthermore, fibroblast growth factor 4 (FGF4), which is actively involved in neurogenesis58, is required for the astrocyte dedifferentiation because it activates the PI3K/Akt/p21 signaling pathway59. Astrocytes lacking FGF4 do not undergo dedifferentiation, while the application of FGF4 alone to astrocyte cultures fails to elicit the reprogramming of astrocytes towards NSPCs. In addition, in vitro experiments confirmed that the increase in sonic hedgehog (Shh) production after invasive injury plays a key role in rat astrocyte dedifferentiation60. The authors observed a significant inhibition of astrocyte dedifferentiation into NSPCs using a neutralizing antibody to inhibit Shh. Moreover, the addition of Shh alone had little effect on the dedifferentiation process and reversion to NSPCs. Based on these results, Shh is necessary but not sufficient to induce astrocyte dedifferentiation. However, in a subsequent study, Sirko drew a distinct conclusion that Shh signaling is necessary and sufficient to promote the proliferation of astrocytes in a mouse model and trigger their ability to form neurospheres in vitro61. More recent studies further demonstrate that Shh signaling effectively promotes conversion of murine astrocytes to a pluripotent state62,63. The treatment with two chemically distinct agonists of the Shh signaling promoted the elongation and proliferation of murine astrocytes64. The mechanism was related to the crosstalk between Sox2/Shh-targeted downstream signals and PI3K/Cdk2/Smurf2 signaling65.
Dedifferentiation in diseases (as a pathological factor)
As mentioned above, the induction of dedifferentiation might be beneficial for tissue recovery in various organs. However, it might also cause some pathological events in certain tissues. In this case, cartilage is one of the classic tissue models and will be discussed in details.
Chondrocyte dedifferentiation has some similarities to osteoarthritis, such as a significant decrease in the expressions of aggrecan and Col II66. Chondrocyte dedifferentiation induces substantial changes in the levels of cell surface markers and cytoskeletal proteins, as well as the appearance of nonspecific ECM, which would ultimately lead to osteoarthritis. In addition to changes in the aggrecan to versican and Col II to Col I ratios and the deposition of collagen type X67, which are markers of the chondrocyte dedifferentiation status, the ratio of the cell surface markers CD14 to CD90 at the mRNA and protein levels has been corroborated as a new cell membrane-based differentiation index68. In addition, CD166 is an indicator of chondrocyte redifferentiation69. As shown in the study by Hong and Reddi, microRNA-221 and microRNA-222 are upregulated during dedifferentiation, while microRNA-140, microRNA-143 and microRNA-145 are downregulated70. Moreover, chondrocyte dedifferentiation controls the mechanical properties of the cell, such as increasing cell stiffness by enhancing membrane-actin adhesion71. However, chondrocyte dedifferentiation is not related to cell division72.
An increasing number of studies have been conducted to explore the cytokines and signaling pathways that are involved in chondrocyte dedifferentiation. MAPK signaling, consisting of ERK, JNK and p38, has very important roles in chondrocyte phenotype and metabolism. Rosenzweig et al.73 investigated the role of MAPK signaling in the dedifferentiation of in vitro primary bovine chondrocyte using inhibitors to block ERK, JNK, and p38. The inhibition of ERK and JNK by their respective inhibitors PD98059 and SP600125 significantly increased the expression of both chondrogenic marker genes and fibrotic genes, while blockade of p38 with SB203580 significantly upregulated Col II expression but inhibited Col I expression. Thus, p38 has a substantial contribution to the dedifferentiation of primary chondrocytes under normal monolayer culture conditions. Interestingly, Kim and colleagues74 reported that cytokine-induced apoptosis inhibitor-1 (CIAPIN-1) induced the dedifferentiation of in vitro cultured rabbit articular chondrocytes by activating ERK-1/2 and inactivating p38. The inhibition of ERK-1/2 by PD98059 appears to suppress the CIAPIN-1-induced dedifferentiation, while the addition of SB203580 to inhibit p38 promotes dedifferentiation. This inconsistency is possibly because—(i) other signaling molecules activated and (ii) the extent of p38 inhibition. The latter could be more likely because this group further observed that p38 inhibition under both CIAPIN-1 promoter and inhibitor SB203580 enhanced dedifferentiation. Two more subsequent findings by this group75,76 are similar, both involving a pre-inducing factor. Kim and colleagues also performed a series of in vitro studies in this area. They investigated the effects of thymoquinone (TQ) and PEP-1-SIRT2 on the dedifferentiation and inflammation of rabbit chondrocytes13,77. TQ or PEP-1-SIRT2 induce the expression of cyclooxygenase-2 (COX-2), causing the dedifferentiation of rabbit articular chondrocytes78 and subsequently leading to the dedifferentiation of chondrocytes through the ERK pathways. TQ-induced inflammation is mediated by the phosphoinositide 3-kinase (PI3K) and p38 pathways, while PEP-1-SIRT2-induced inflammation is mediated by the p38 and ERK pathways. The authors also reported that salinomycin (SAL) causes the dedifferentiation of rabbit articular chondrocytes via the ERK pathway, as determined by increased ERK activity without alterations in JNK and p38 activity following treatment with SAL79. Recently, the authors revealed that PEP-1-glutaredoxin-1 increased endoplasmic reticulum-stress and then stimulated the dedifferentiation of rabbit chondrocytes via the ERK-1/2 pathway75. Another Korean lab clarified the molecular mechanisms by which interleukin (IL)-1β, a proinflammatory cytokine associated with various cellular activities, induced the dedifferentiation of in vitro cultured primary rabbit chondrocytes80. IL-1βinduces the expression of nicotinamide phosphoribosyltransferase (NAMPT) and then activates SIRT1, which subsequently induces the activation of ERK and p38. In turn, ERK and p38 activate SIRT1, forming a positive feedback loop81. Integrin αvβ5 was reported to activate ERK signaling and to subsequently enhance the dedifferentiation of in vitro cultured primary human articular chondrocytes82. In addition, the Notch pathway is involved in the regulation of matrix metalloproteinase 13 (MMP-13) activity and the differentiation of human articular chondrocytes in vitro83. In rabbit articular chondrocytes in vitro, 2-deoxy-D-glucose (2DG) induces dedifferentiation via the β-catenin pathway84. A recent study revealed a feed-forward loop driven by USP14 (a deubiquitinating enzyme) and the NF-kappa B pathway that enhances the effect of IL-1β on the dedifferentiation of human chondrocytes in vitro85.
The architecture of the actin cytoskeleton modulates the phenotype of chondrocytes during dedifferentiation86. The disruption of the actin cytoskeleton inhibits the dedifferentiation of in vitro cultured rabbit chondrocytes by blocking PKCα and –ζ signaling87. The integrity of the actin cytoskeleton is related to the differentiated phenotype of chondrocytes through the PI3K/Akt and p38 pathways88. In addition, the Rho family GTPase ROCK plays a key role in organizing the actin cytoskeleton89. Treatment with a ROCK inhibitor alters human chondrocyte morphology in vitro and activates SOX9 and chondrogenic gene expression, suggesting the suppression of dedifferentiation90. Berberine, a compound widely present in many plants, reorganizes the actin cytoskeleton and subsequently induces dedifferentiation via PI3K/Akt and p38 pathways91. Focal adhesion kinase (FAK) is involved in cell adhesion, migration and cell proliferation. The role of FAK in the chondrocytic dedifferentiation process was investigated by Shin et al.92 An increasing number of passages in monolayer culture resulted in higher levels of focal adhesion complexes. In addition, FAK knockdown facilitates the restoration of the expression of cartilage-specific genes, including Col II, aggrecan and SOX9, in rat dedifferentiated chondrocytes in vitro. In another study, FAK knockdown inhibited porcine chondrocyte proliferation in vitro, and FAK was required to modulate the expression of Col II93. Ding and colleagues94 further explored the effects of spreading areas and aspect ratios of single chondrocytes undergoing dedifferentiation using a micropatterning technique in vitro. Rat chondrocytes incubated under conditions of larger sizes and higher aspect ratios preferentially undergo dedifferentiation.
Implications from recent studies of dedifferentiation
The information above provides interesting implications. First, dedifferentiation may have either positive or negative consequences to tissue repair in different tissue/organs. For example, after injury, dedifferentiated cardiomyocytes were preferentially protected from apoptosis; they can survive and re-enter cell cycle for tissue repair16. Meanwhile, dedifferentiated chondrocytes gradually lose the ability of proliferation and undergo hypertrophy and eventually apoptosis95. Therefore, understanding the impact of cell dedifferentiation in specific tissue is essential for devising further therapeutic strategies—such as induction versus suppression (and to what extent). To date, much remains unknown about the causes and consequences of dedifferentiation under specific physiological or pathological circumstances, which requires more mechanistic studies at different levels.
Second, it should be noted that the findings from non-human models provide valuable evidence for assessing the application potential of dedifferentiation for future trials and translations in humans. Many studies show that a main reason for the huge difference of regenerative capacity between amphibians and mammals is the induction of dedifferentiation, which does not often occur after injury in mammals8. Inspired by the studies on dedifferentiation of amphibians such as newt, researchers found that murine myotubes could dedifferentiate to a certain extent when stimulated by an extract obtained from newt regenerating limbs96. Also, partial dedifferentiation of Schwann cells can be induced by local factors such as hormones and neurotrophic factors97. These results suggest that such extrinsic signals can play an important role in dedifferentiation. Although current interpretations remain on a rather speculative level and await further verification, in vitro investigations on dedifferentiation provide valuable insights into the molecular mechanisms and the pathways associated with cell dedifferentiation, with extensive in vivo studies in demand.
Third, by exploring the regenerative phenomena in invertebrates and amphibians, researchers have uncovered the cellular and molecular mechanisms to develop regenerative strategies for humans. Dedifferentiation and trans-differentiation, part of the physiological response to injury in numerous organs, are mechanistically associated with natural regeneration98. Dedifferentiation refers to a reversion of a fully differentiated cell within its own lineage, which allows the cell to proliferate again to replace those damaged cells in many cases99. Trans-differentiation, first reported in the newt for regeneration of lens over 100 years ago, involves dedifferentiation in the first step and subsequent differentiation into another cell type. It may also be observed during experimental induction of trans-differentiation that cells can directly trans-differentiate to a distinct lineage through an “unnatural” intermediate phase. Thus, a strategy for regenerative medicine would be either (i) to induce dedifferentiation and then proliferation of target cells or (ii) to stimulate trans-differentiation of cells into the desired cell types. In either case, a clear dissection of the changes and impacts of the genes involved in dedifferentiation is fundamental.
Engineering approaches to regulate dedifferentiation for therapeutic purposes
Although mesenchymal stem cells (MSCs) are promising “seed” cells in tissue regeneration owing to their excellent characteristics such as self-renewal capacity100, immunoregulatory function101, and multilineage differentiation potential102, MSC-based therapies for tissue regeneration have not yet been accepted clinically103. Current efforts focus on the development of bioreactor system, which could combine scaffolds, biochemical factors, and mechanical stimuli to simulate the in vivo microenvironment so that MSCs-based cellular products can meet the requirements for clinical use104. Recent studies have provided evidence that dedifferentiation is a promising approach for tissue regeneration4. An increasing number of trials have used engineering approaches to regulate dedifferentiation.
Creation of tissue microenvironment
Scaffold materials, which can provide a three-dimensional culture condition that more closely resembles the in vivo environment, play a critical role in the biological characteristics and proliferation rate of seeding cells, subsequently determining the fate and function of cells105. Recently, researchers have focused on designing and preparing innovative biomaterials to induce dedifferentiation. Yahalom-Ronen et al.15 established an in vitro culture system for murine P1 neonatal cardiomyocytes. They constructed softer substrates to stimulate dedifferentiation and the cell cycle re-entry of cardiomyocytes, suggesting that the development of soft scaffolds might facilitate the regeneration of the adult human heart. A keratin hydrogel was created and used to repair a 1-cm defect in rat sciatic nerve injury models106. Compared with Matrigel, the keratin hydrogel group induced more rapid dedifferentiation of Schwann cells, leading to the faster activation of macrophages and more efficient clearance of myelin debris and ECM remodeling. Thus, the positive regulation of Schwann cell dedifferentiation encourages effective nerve regeneration.
On the other hand, many studies of biomaterials have focused on preventing the dedifferentiation of chondrocytes, as the dedifferentiation of chondrocytes severely compromises the therapeutic outcome of cartilage repair. Dedifferentiation is induced when chondrocytes are exposed to degradative enzymes during passaging or incubated in a standard culture flask with a flat and rigid culture surface. Through a series of studies, Quinn and colleagues107 developed a “continuous expansion” (CE) in vitro culture technique that avoids repeated passaging and maintains the chondrocytic phenotype. Using this new method, bovine chondrocytes were cultured on high-extension silicone rubber (HESR) dishes with continuously expanding and elastic properties. Furthermore, the authors functionalized the HESR dishes with a cartilage ECM extract from a cow108. HESR dishes functionalized with a cartilage extract were more effective at inhibiting chondrocytic dedifferentiation than control dishes. Another group further developed biomimetic materials using decellularized ECM derived from human articular chondrocytes14. These biomaterials were more effective at maintaining the phenotype and enhancing the proliferation of chondrocytes than standard culture plates. Moreover, similar results were obtained using MSC-derived ECM109,110, indicating that a cell type-specific ECM is more similar to the native microenvironment and reduces the dedifferentiation of chondrocytes. Gelatin methacryloyl/hyaluronic acid methacrylate hydrogels displayed their positive effects on down-regulation of dedifferentiation gene markers111. In addition, a biomimetic hydrogel was prepared based on alginate and tuned by altering the concentration of chlorosulfonic acid112. The sulfation of alginate, which mimics proteoglycans in cartilage, promotes in vitro bovine chondrocyte proliferation, increase cartilage matrix deposition, and inhibits the expression of dedifferentiation markers. A blended gel composed of alginate and collagen also provided a preferable gel matrix environment for the suppression of rat chondrocyte dedifferentiation in vitro113. More interestingly, Ding and colleagues114 concentrated on the effects of external nanoscale features on the dedifferentiation of in vitro cultured primary rat chondrocytes. Poly(ethylene glycol) (PEG) hydrogels were modified with nanopatterned arginine–glycine–aspartate (RGD) and used to culture chondrocytes (Fig. 3). The nanoscale distribution of RGD revealed a crucial and independent role for this material in regulating cell dedifferentiation. Overall, all of these aspects should be specifically considered when designing and optimizing advanced biomaterials for regenerative medicine.
Fig. 3. Effects of a nanoscale spatial arrangement of RGD on dedifferentiation of chondrocytes.
a Schematic illustrating the experimental design. PEG hydrogel surfaces were nanopatterned with RGD peptides and then cultured with chondrocytes to explore the effect of material techniques of nanopatterning on the dedifferentiation of chondrocytes. b Schematic illustrating the effects of RGD nanopatterns on the dedifferentiation of chondrocytes. RGD with a nanospacing greater than 70 nm is beneficial for maintaining the chondrocyte phenotype. Reprinted (adapted) with permission from Li et al.114, copyright 2015, American Chemistry Society.
Biochemical regulation
Numerous biomolecules have shown the potential in modulating dedifferentiation and have been applied in regenerative medicine. The methods used to deliver biomolecules mainly include—(i) direct addition in the form of recombinant proteins and (ii) gene delivery of their complementary DNAs (cDNAs) using viral or nonviral vectors.
MMPs, which are capable of degrading ECM proteins, regulate Schwann cell signaling and mitosis. As shown in a study by the Shubayev group, MMP-9 inhibits Schwann cell mitosis during nerve regeneration, while the MMP inhibitor, GM6001, counteracts the effect of MMP-9 and further increases the division of murine Schwann cells115. Based on these findings, the group subsequently performed in vivo experiments; GM6001 was injected into rats immediately after sciatic nerve crush to stimulate Schwann cell division, reduce myelin protein expression and subsequently support axonal regeneration116. In another interesting study, leukemia inhibitory factor (LIF), a promyelination factor, was encapsulated in nanoparticle carriers and delivered to enhance remyelination and promote nerve regeneration117, highlighting the feasibility of biochemically regulating tissue development with engineering approaches. Similarly, recombinant human midkine was added to the growth medium of rat chondrocytes for in vitro monolayer expansion and not only promoted cell proliferation but also suppressed dedifferentiation, avoiding a shift in the chondrocyte to fibroblast phenotype and decreasing the expression of chondrocytic genes118. In addition, andrographolide, the main active component of the traditional Chinese medicine Andrographis paniculata, also prevents the dedifferentiation of rabbit articular chondrocytes in vitro, and 3 μM was reported to be the optimal dose119. Although positive effects have been observed by directly supplementing culture media with biomolecules, this method will result in instability in vivo, limiting further clinical applications. Therefore, gene-transfer strategies appear to be a promising alternative.
Two main types of gene delivery vectors have bee identified: viral vectors, such as adenoviral vectors and lentiviral vectors, and nonviral vectors, such as polymers and liposomes. Viral vectors have better transfection efficiencies while nonviral vectors have advantages, such as ease of synthesis and low immunogenicity. Endothelial nitric oxide (eNO) plays a pivotal role in regulating Schwann cell dedifferentiation. NO synthesized by eNO synthase decelerates axon regeneration after crush of the XII nerve120. Therefore, researchers have constructed adenoviral vectors to express a truncated mutant form of eNO synthase followed by an intraneural injection of the adenovirus121. Rat Schwann cell dedifferentiation was induced, as evidenced by the overexpression of dedifferentiated Schwann cell marker growth-associated protein 43 and an increase in the bands of Bungner, the cellular substrate for guiding growing axons. On the other hand, various biomolecules are being developed to inhibit the dedifferentiation of chondrocytes. Based on the results of the previous studies showing that IL-1β induces the dedifferentiation of chondrocytes and is inhibited by cytokine response modifier A (CrmA)81,122,123, hyaluronic acid-chitosan microspheres were designed for the controlled release of CrmA124. Rat chondrocytes treated with these microspheres in vitro maintain the chondrocytic phenotype and production of cartilage-specific proteins even after cocultured with IL-1β. Additionally, chondrocytes expressing wild-type p53-inducible phosphatase (Wip1) following microporation generally retain a chondrocyte phenotype for a longer time during in vitro expansion125.
A “Tet-Off” transgenic mouse model was established by Tzahor and colleagues126 to modulate the expression of a constitutively active (ca) form of ErbB2 in murine cardiomyocytes following the administration of the tetracycline analog doxycycline (DOX). Notably, caErbB2 promotes the dedifferentiation and subsequently induces cell division, highlighting the proliferative and regenerative potential of adult cardiomyocytes.
Molecular/cellular modulation
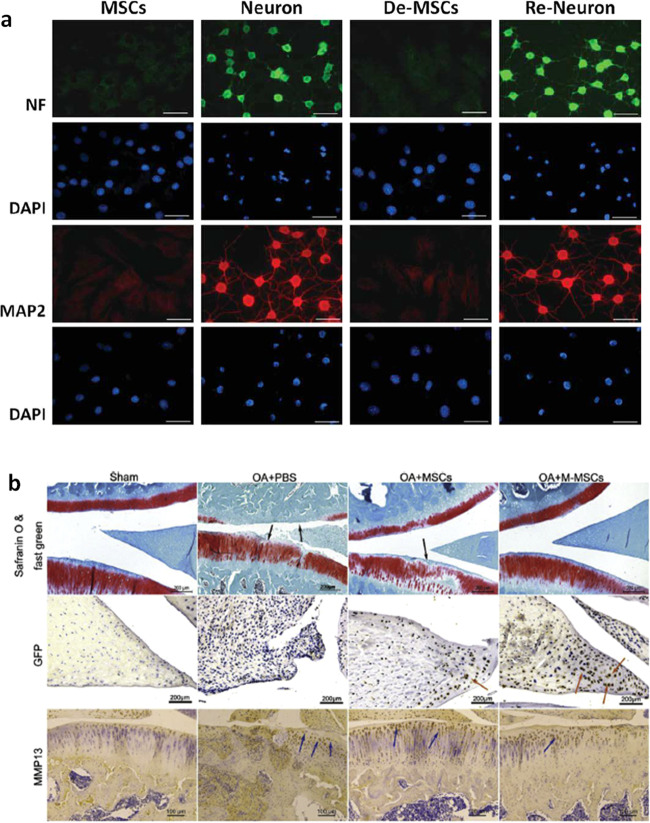
Studies have shown that MSC-derived neurons dedifferentiate into MSCs after the removal of extrinsic stimuli. Liu et al.127 explored the therapeutic potential of the dedifferentiated MSCs (De-MSCs) in neuron repair (Fig. 4a). In vitro monoclonal rat MSCs were harvested and induced to differentiate into neurons with modified neuronal medium for 24–48 h. Then, the De-MSCs were obtained after incubation in complete MSC medium for another 48 h. The researchers were the first to show that De-MSCs tended to undergo neuronal differentiation, as evidenced by genetic and functional assays, compared to uncommitted MSCs. Moreover, in vitro and in vivo, De-MSCs show increased cell survival, greater resistance to a hostile environment and a higher efficacy of neuronal differentiation, suggesting a better therapeutic effect. In addition to being derived from the same lineage, dedifferentiation appeared to be a prerequisite for bone marrow-derived neurons to differentiate into a different lineage128. Moreover, direct translineage differentiation from neuronal cells to epithelial was not observed under appropriate defined conditions. Interestingly, the dedifferentiation-reprogramming method was also effective in chondrogenesis. According to Li et al.129, rat MSCs in vitro that are incubated in chondrogenic induction medium undergo chondrogenesis and then are allowed to grow and subculture in growth medium to expand and dedifferentiate (Fig. 4b). Based on these findings, chondrogenically manipulated MSCs (M-MSCs) harvested in this manner display higher viability and chondrogenic potential.
Fig. 4. Dedifferentiation-reprogrammed mesenchymal stem cells with promising therapeutic potential for cartilage and nerves.
a Immunofluorescence staining revealed the preferential neuronal differentiation of De-MSCs, as evidenced by the higher expression of neuronal markers, including neurofilament-M (NF-M) and microtubule-associated protein 2 (MAP2) (used with permission from John Wiley and Sons/Hsiao et al.127). b Histology and immunohistochemistry revealed that the M-MSC group exhibited a greater therapeutic effect on OA, as evidenced by the increased ECM deposition, increased number of GFP-positive cells, and reduction in MMP-13 expression129 (Copyright 2017, with permission from Elsevier).
Other factors exerting an inhibitory effect on dedifferentiation are also studied. A study by Li et al. reveals that DNA methylation is an important regulator of chondrocyte dedifferentiation. The authors applied the DNA methylation inhibitor 5-azacytidine (5-AzaC) to human chondrocytes in vitro, resulting in an increase in the DNA methylation level of the Col I-A1 promoter, which slowed chondrocyte dedifferentiation130. Low-dose γ-radiation also effectively inhibits the IL-1β-induced dedifferentiation of in vitro cultured primary rabbit articular chondrocytes131. Other researchers have also delivered biomolecules132,133 or optimized culture conditions, such as materials134,135, oxygen tension136,137, and mechanical stimulation138, to drive efficient redifferentiation of dedifferentiated chondrocytes for use in regenerative medicine.
Conclusions and future perspectives
Discovering the interesting phenomenon of cell dedifferentiation, followed by uncovering its underlying mechanisms, opens new avenues for controlling cell fate for regenerative applications. It is possible that in different tissue/organs, dedifferentiation occurs in various frequencies and to different extents; and perhaps much remains unknown about the causes and consequences of this phenomenon under specific physiological or pathological circumstances. However, there must be a reason that the organisms develop the capability of dedifferentiation through the long period of evolution—to adapt, to survive, to heal, to evade from dangers, and to evolve. As such, recapitulating dedifferentiation in regenerative medicine may provide a new, rational, evolution-inspired strategy to bypass inherent limitations with therapeutic cells (such as inability to proliferate) and initiate tissue development under better control.
Future studies can be directed in three aspects. First, more mechanistic studies at different levels are required to understand the initiation of cell dedifferentiation. Genetic, epigenetic, transcriptional, metabolic, and mechanobiological controls of the cell fate decision towards dedifferentiation are all worth investigation, which will offer comprehensive clues for designing engineering tools. Second, tissue regeneration involves multiple cell types of different lineages and characteristics, especially in the case of a complex organ such as the liver, kidney, and heart. It is very likely that the change in the behavior of one cell type affects the function of another. Hence, the consequence of engineered dedifferentiation of a group of target cells on other cells in the tissue niche should be carefully examined. Any potential adverse effects—either short-term (e.g., cell death, acute inflammation, and off-target effect) or long-term (e.g., carcinogenicity caused by uncontrolled cell proliferation after induced dedifferentiation) risks—need to be avoided. Third, for any practice in which cell dedifferentiation demonstrates preliminary signs of safety and effectiveness, extensive work is demanded to standardize the protocol. Major questions include: how far should the cells be reversed along the way of dedifferentiation, how many percentages of the cells are effectively induced to dedifferentiate, and when is the best timing to apply the dedifferentiated cells into tissue repair. As such, exciting future work lies ahead from biological, clinical, and engineering perspectives for translating the discovery of cell dedifferentiation into tissue regeneration.
Acknowledgements
This work was supported by Science and Technology Program of Guangzhou, China (grant number 201804010479), Key Program for Scientific Research Project of Guangzhou Colleges and Universities (grant number 1201610097), the Science and Technology Development Fund, Macau SAR (File no. 0018/2019/AFJ and 0097/2019/A2) and University of Macau (File no. MYRG2019-00080-ICMS).
Author contributions
Both authors had substantial contribution to this review. Y.Y. reviewed the literature, drafted the manuscript, and created figures. C.W. supervised the development of this work, revised the manuscript, and approved the submission.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, et al. Fifteen new earthworm mitogenomes shed new light on phylogeny within the Pheretima complex. Sci. Rep. 2016;6:20096. doi: 10.1038/srep20096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarig R, Tzahor E. The cancer paradigms of mammalian regeneration: can mammals regenerate as amphibians? Carcinogenesis. 2017;38:359–366. doi: 10.1093/carcin/bgw103. [DOI] [PubMed] [Google Scholar]
- 4.Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 5.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlier E, et al. Chondrocyte dedifferentiation and osteoarthritis (OA) Biochem. Pharmacol. 2019;165:49–65. doi: 10.1016/j.bcp.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi VJ, et al. Redifferentiated chondrocytes in fibrin gel for the repair of articular cartilage lesions. Am. J. Sports Med. 2019;47:2348–2359. doi: 10.1177/0363546519857571. [DOI] [PubMed] [Google Scholar]
- 8.Pesaresi M, Sebastian-Perez R, Cosma MP. Dedifferentiation, transdifferentiation and cell fusion: invivo reprogramming strategies for regenerative medicine. FEBS J. 2018 doi: 10.1111/febs.14633. [DOI] [PubMed] [Google Scholar]
- 9.Zhang CP, et al. Wnt/ss-catenin signaling is critical for dedifferentiation of aged epidermal cells in vivo and in vitro. Aging Cell. 2012;11:14–23. doi: 10.1111/j.1474-9726.2011.00753.x. [DOI] [PubMed] [Google Scholar]
- 10.Laadhar L, et al. Wnt signaling is involved in human articular chondrocyte dedifferentiation in vitro. Osteoporos. Int. 2014;25:S419–S420. [Google Scholar]
- 11.Kohler EE, et al. Low-dose 6-bromoindirubin-3‘-oxime induces partial dedifferentiation of endothelial cells to promote increased neovascularization. Stem Cells. 2014;32:1538–1552. doi: 10.1002/stem.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan Y, et al. Macrophage migration inhibitory factor triggers vascular smoothmuscle cell dedifferentiation by a p68-serum response factor axis. Cardiovascular Res. 2017;113:519–530. doi: 10.1093/cvr/cvx025. [DOI] [PubMed] [Google Scholar]
- 13.Eo SH, Kim DW, Choi SY, Kim HA, Kim SJ. PEP-1-SIRT2 causes dedifferentiation and COX-2 expression via the MAPK pathways in rabbit articular chondrocytes. Exp. Cell Res. 2015;339:351–359. doi: 10.1016/j.yexcr.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Mao Y, et al. Extracellular matrix derived from chondrocytes promotes rapid expansion of human primary chondrocytes in vitro with reduced dedifferentiation. Acta Biomater. 2019;85:75–83. doi: 10.1016/j.actbio.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Yahalom-Ronen, Y., Rajchman, D., Sarig, R., Geiger, B. & Tzahor, E. Reduced matrix rigidity promotes neonatal cardiomyocyte dedifferentiation, proliferation and clonal expansion. Elife4, e07455 (2015). [DOI] [PMC free article] [PubMed]
- 16.Szibor M, Poling J, Warnecke H, Kubin T, Braun T. Remodeling and dedifferentiation of adult cardiomyocytes during disease and regeneration. Cell. Mol. Life Sci. 2014;71:1907–1916. doi: 10.1007/s00018-013-1535-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eguizabal C, Montserrat N, Veiga A, Belmonte JCI. Dedifferentiation, transdifferentiation, and reprogramming: future directions in regenerative medicine. Semin. Reprod. Med. 2013;31:82–94. doi: 10.1055/s-0032-1331802. [DOI] [PubMed] [Google Scholar]
- 18.Porrello ER, et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad. Sci. USA. 2013;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali H, Braga L, Giacca M. Cardiac regeneration and remodelling of the cardiomyocyte cytoarchitecture. FEBS J. 2020;287:417–438. doi: 10.1111/febs.15146. [DOI] [PubMed] [Google Scholar]
- 20.Wang CY, et al. Cardiomyocyte dedifferentiation and remodeling in 3D scaffolds to generate the cellular diversity of engineering cardiac tissues. Biomater. Sci. 2019;7:4636–4650. doi: 10.1039/c9bm01003c. [DOI] [PubMed] [Google Scholar]
- 21.Taegtmeyer H, Sen S, Vela D. Return to the fetal gene program: a suggested metabolic link to gene expression in the heart. Ann. N. Y. Acad. Sci. 2010;1188:191–198. doi: 10.1111/j.1749-6632.2009.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, Y. Q. et al. Single-cell imaging and transcriptomic analyses of endogenous cardiomyocyte dedifferentiation and cycling. Cell Discov.5, 30 (2019). [DOI] [PMC free article] [PubMed]
- 23.Poling J, et al. The Janus face of OSM-mediated cardiomyocyte dedifferentiation during cardiac repair and disease. Cell Cycle. 2012;11:439–445. doi: 10.4161/cc.11.3.19024. [DOI] [PubMed] [Google Scholar]
- 24.Dispersyn GD, Geuens E, Ver Donck L, Ramaekers FC, Borgers M. Adult rabbit cardiomyocytes undergo hibernation-like dedifferentiation when co-cultured with cardiac fibroblasts. Cardiovasc Res. 2001;51:230–240. doi: 10.1016/s0008-6363(01)00326-1. [DOI] [PubMed] [Google Scholar]
- 25.Kubin T, et al. Oncostatin M is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell. 2011;9:420–432. doi: 10.1016/j.stem.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda S, et al. Hippo deficiency leads to cardiac dysfunction accompanied by cardiomyocyte dedifferentiation during pressure overload. Circulation Res. 2019;124:292–305. doi: 10.1161/CIRCRESAHA.118.314048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruoslahti E. Stretching is good for a cell. Science. 1997;276:1345–1346. doi: 10.1126/science.276.5317.1345. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka EM, Gann AA, Gates PB, Brockes JP. Newt myotubes reenter the cell cycle by phosphorylation of the retinoblastoma protein. J. Cell Biol. 1997;136:155–165. doi: 10.1083/jcb.136.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wohlschlaeger J, et al. Reversible regulation of the, retinoblastoma protein/E2F-1 pathway during “reverse cardiac remodelling” after ventricular unloading. J. Heart Lung Transplant. 2010;29:117–124. doi: 10.1016/j.healun.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 30.MacLellan WR, et al. Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol. Cell Biol. 2005;25:2486–2497. doi: 10.1128/MCB.25.6.2486-2497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaglia T, et al. Cardiac interstitial cells express GATA4 and control dedifferentiation and cell cycle re-entry of adult cardiomyocytes. J. Mol. Cell Cardiol. 2009;46:653–662. doi: 10.1016/j.yjmcc.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Zhang GY, et al. GRP78 (glucose-regulated protein of 78 kDa) promotes cardiomyocyte growth through activation of GATA4 (GATA-binding protein 4) Hypertension. 2019;73:390–398. doi: 10.1161/HYPERTENSIONAHA.118.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carr MJ, Johnston APW. Schwann cells as drivers of tissue repair and regeneration. Curr. Opin. Neurobiol. 2017;47:52–57. doi: 10.1016/j.conb.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Webber C, Zochodne D. The nerve regenerative microenvironment: early behavior and partnership of axons and Schwann cells. Exp. Neurol. 2010;223:51–59. doi: 10.1016/j.expneurol.2009.05.037. [DOI] [PubMed] [Google Scholar]
- 35.Warner LE, et al. Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat. Genet. 1998;18:382–384. doi: 10.1038/ng0498-382. [DOI] [PubMed] [Google Scholar]
- 36.Jessen KR, Mirsky R. Negative regulation of myelination: relevance for development, injury, and demyelinating disease. Glia. 2008;56:1552–1565. doi: 10.1002/glia.20761. [DOI] [PubMed] [Google Scholar]
- 37.Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat. Rev. Neurosci. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- 38.Woodhoo A, et al. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat. Neurosci. 2009;12:839–847. doi: 10.1038/nn.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Napoli I, et al. A central role for the ERK-signaling pathway in controlling Schwann cell plasticity and peripheral nerve regeneration in vivo. Neuron. 2012;73:729–742. doi: 10.1016/j.neuron.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 40.Groh J, et al. CSF-1-activated macrophages are target-directed and essential mediators of Schwann cell dedifferentiation and dysfunction in Cx32-deficient mice. Glia. 2015;63:977–986. doi: 10.1002/glia.22796. [DOI] [PubMed] [Google Scholar]
- 41.Mirsky R, et al. Novel signals controlling embryonic Schwann cell development, myelination and dedifferentiation. J. Peripheral Nerv. Syst.: JPNS. 2008;13:122–135. doi: 10.1111/j.1529-8027.2008.00168.x. [DOI] [PubMed] [Google Scholar]
- 42.Parkinson DB, et al. c-Jun is a negative regulator of myelination. J. Cell Biol. 2008;181:625–637. doi: 10.1083/jcb.200803013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norrmen C, et al. mIORC1 is transiently reactivated in injured nerves to promote c-Jun elevation and Schwann cell dedifferentiation. J. Neurosci. 2018;38:4811–4828. doi: 10.1523/JNEUROSCI.3619-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fontana X, et al. c-Jun in Schwann cells promotes axonal regeneration and motoneuron survival via paracrine signaling. J. Cell Biol. 2012;198:127–141. doi: 10.1083/jcb.201205025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arthur-Farraj PJ, et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 2012;75:633–647. doi: 10.1016/j.neuron.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang DP, et al. p38 MAPK activation promotes denervated Schwann cell phenotype and functions as a negative regulator of Schwann cell differentiation and myelination. J. Neurosci. 2012;32:7158–7168. doi: 10.1523/JNEUROSCI.5812-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung J, et al. Actin polymerization is essential for myelin sheath fragmentation during Wallerian degeneration. J. Neurosci. 2011;31:2009–2015. doi: 10.1523/JNEUROSCI.4537-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin YK, et al. The Neuregulin-Rac-MKK7 pathway regulates antagonistic c-jun/Krox20 expression in Schwann cell dedifferentiation. Glia. 2013;61:892–904. doi: 10.1002/glia.22482. [DOI] [PubMed] [Google Scholar]
- 49.Shin YH, Lee SJ, Jung J. Extracellular ATP inhibits Schwann cell dedifferentiation and proliferation in an ex vivo model of Wallerian degeneration. Biochem. Biophys. Res. Commun. 2013;430:852–857. doi: 10.1016/j.bbrc.2012.11.057. [DOI] [PubMed] [Google Scholar]
- 50.Monje PV, Soto J, Bacallao K, Wood PM. Schwann cell dedifferentiation is independent of mitogenic signaling and uncoupled to proliferation: role of cAMP and JNK in the maintenance of the differentiated state. J. Biol. Chem. 2010;285:31024–31036. doi: 10.1074/jbc.M110.116970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viader A, Chang LW, Fahrner T, Nagarajan R, Milbrandt J. MicroRNAs modulate Schwann cell response to nerve injury by reinforcing transcriptional silencing of dedifferentiation-related genes. J. Neurosci. 2011;31:17358–17369. doi: 10.1523/JNEUROSCI.3931-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corti S, et al. Direct reprogramming of human astrocytes into neural stem cells and neurons. Exp. Cell Res. 2012;318:1528–1541. doi: 10.1016/j.yexcr.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moon JH, et al. Nanog-induced dedifferentiation of p53-deficient mouse astrocytes into brain cancer stem-like cells. Biochem. Biophys. Res. Commun. 2011;412:175–181. doi: 10.1016/j.bbrc.2011.07.070. [DOI] [PubMed] [Google Scholar]
- 54.Yang H, Cheng XP, Li JW, Yao Q, Ju G. De-differentiation response of cultured astrocytes to injury induced by scratch or conditioned culture medium of scratch-insulted astrocytes. Cell Mol. Neurobiol. 2009;29:455–473. doi: 10.1007/s10571-008-9337-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H, et al. Evidence for heterogeneity of astrocyte de-differentiation in vitro: astrocytes transform into intermediate precursor cells following induction of ACM from scratch-insulted astrocytes. Cell Mol. Neurobiol. 2010;30:483–491. doi: 10.1007/s10571-009-9474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sher F, Boddeke E, Copray S. Ezh2 expression in astrocytes induces their dedifferentiation toward neural stem cells. Cell Reprogram. 2011;13:1–6. doi: 10.1089/cell.2010.0052. [DOI] [PubMed] [Google Scholar]
- 57.Yang H, et al. ErbB2 activation contributes to de-differentiation of astrocytes into radial glial cells following induction of scratch-insulted astrocyte conditioned medium. Neurochem. Int. 2011;59:1010–1018. doi: 10.1016/j.neuint.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 58.Kosaka N, et al. FGF-4 regulates neural progenitor cell proliferation and neuronal differentiation. FASEB J. 2006;20:1484–1485. doi: 10.1096/fj.05-5293fje. [DOI] [PubMed] [Google Scholar]
- 59.Feng GD, et al. Fibroblast growth factor 4 is required but not sufficient for the astrocyte dedifferentiation. Mol. Neurobiol. 2014;50:997–1012. doi: 10.1007/s12035-014-8649-1. [DOI] [PubMed] [Google Scholar]
- 60.Yang H, et al. Sonic hedgehog released from scratch-injured astrocytes is a key signal necessary but not sufficient for the astrocyte de-differentiation. Stem Cell Res. 2012;9:156–166. doi: 10.1016/j.scr.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 61.Sirko S, et al. Reactive glia in the injured brain acquire stem cell properties in response to sonic hedgehog. [corrected] Cell Stem Cell. 2013;12:426–439. doi: 10.1016/j.stem.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 62.Hill, S. A. et al. Sonic hedgehog signaling in astrocytes mediates cell type-specific synaptic organization. Elife8, e45545 (2019). [DOI] [PMC free article] [PubMed]
- 63.Sanchez MA, Sullivan GM, Armstrong RC. Genetic detection of Sonic hedgehog (Shh) expression and cellular response in the progression of acute through chronic demyelination and L updates amok for remyelination. Neurobiol. Dis. 2018;115:145–156. doi: 10.1016/j.nbd.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 64.Ugbode CI, Smith I, Whalley BJ, Hirst WD, Rattray M. Sonic hedgehog signalling mediates astrocyte crosstalk with neurons to confer neuroprotection. J. Neurochemis. 2017;142:429–443. doi: 10.1111/jnc.14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang H, et al. Sonic hedgehog effectively improves Oct4-mediated reprogramming of astrocytes into neural stem cells. Mol. Ther. 2019;27:1467–1482. doi: 10.1016/j.ymthe.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall, A. C. The role of chondrocyte morphology and volume in controlling phenotypeimplications for osteoarthritis, cartilage repair, and cartilage engineering. Curr. Rheumatol. Rep.21, 38 (2019). [DOI] [PMC free article] [PubMed]
- 67.Wu L, et al. Extracellular matrix domain formation as an indicator of chondrocyte dedifferentiation and hypertrophy. Tissue Eng. Part C. Methods. 2014;20:160–168. doi: 10.1089/ten.tec.2013.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diaz-Romero J, Nesic D, Grogan SP, Heini P, Mainil-Varlet P. Immunophenotypic changes of human articular chondrocytes during monolayer culture reflect bona fide dedifferentiation rather than amplification of progenitor cells. J. Cell Physiol. 2008;214:75–83. doi: 10.1002/jcp.21161. [DOI] [PubMed] [Google Scholar]
- 69.Kruger M, Kruger JP, Kinne RW, Kaps C, Endres M. Are surface antigens suited to verify the redifferentiation potential and culture purity of human chondrocytes in cell-based implants. Tissue Cell. 2015;47:489–497. doi: 10.1016/j.tice.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 70.Hong E, Reddi AH. Dedifferentiation and redifferentiation of articular chondrocytes from surface and middle zones: changes in microRNAs-221/-222, -140, and -143/145 expression. Tissue Eng. Part A. 2013;19:1015–1022. doi: 10.1089/ten.TEA.2012.0055. [DOI] [PubMed] [Google Scholar]
- 71.Sliogeryte K, Botto L, Lee DA, Knight MM. Chondrocyte dedifferentiation increases cell stiffness by strengthening membrane-actin adhesion. Osteoarthr. Cartil. 2016;24:912–920. doi: 10.1016/j.joca.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 72.Minegishi Y, Hosokawa K, Tsumaki N. Time-lapse observation of the dedifferentiation process in mouse chondrocytes using chondrocyte-specific reporters. Osteoarthr. Cartil. 2013;21:1968–1975. doi: 10.1016/j.joca.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 73.Rosenzweig DH, Ou SJ, Quinn TM. P38 mitogen-activated protein kinase promotes dedifferentiation of primary articular chondrocytes in monolayer culture. J. Cell Mol. Med. 2013;17:508–517. doi: 10.1111/jcmm.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu SM, Yeo HJ, Choi SY, Kim SJ. Cytokine-induced apoptosis inhibitor-1 causes dedifferentiation of rabbit articular chondrocytes via the ERK-1/2 and p38 kinase pathways. Int. J. Biochem. Cell Biol. 2016;80:10–18. doi: 10.1016/j.biocel.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 75.Yu SM, Choi YJ, Kim SJ. PEP-1-glutaredoxin-1 induces dedifferentiation of rabbit articular chondrocytes by the endoplasmic reticulum stress-dependent ERK-1/2 pathway and the endoplasmic reticulum stress-independent p38 kinase and PI-3 kinase pathways. Int. J. Biol. Macromolecules. 2018;111:1059–1066. doi: 10.1016/j.ijbiomac.2018.01.127. [DOI] [PubMed] [Google Scholar]
- 76.Han Y, Kim SJ. Simvastatin induces differentiation of rabbit articular chondrocytes via the ERK-1/2 and p38 kinase pathways. Exp. Cell Res. 2016;346:198–205. doi: 10.1016/j.yexcr.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 77.Yu SM, Kim SJ. The thymoquinone-induced production of reactive oxygen species promotes dedifferentiation through the ERK pathway and inflammation through the p38 and PI3K pathways in rabbit articular chondrocytes. Int. J. Mol. Med. 2015;35:325–332. doi: 10.3892/ijmm.2014.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee WK, Yu SM, Cheong SW, Sonn JK, Kim SJ. Ectopic expression of cyclooxygenase-2-induced dedifferentiation in articular chondrocytes. Exp. Mol. Med. 2008;40:721–727. doi: 10.3858/emm.2008.40.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu SM, Kim SJ. Salinomycin causes dedifferentiation via the extracellular signal-regulated kinase (ERK) pathway in rabbit articular chondrocytes. J. Pharm. Sci. 2015;127:196–202. doi: 10.1016/j.jphs.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 80.Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117:561–574. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 81.Hong EH, et al. Nicotinamide phosphoribosyltransferase is essential for interleukin-1beta-mediated dedifferentiation of articular chondrocytes via SIRT1 and extracellular signal-regulated kinase (ERK) complex signaling. J. Biol. Chem. 2011;286:28619–28631. doi: 10.1074/jbc.M111.219832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fukui N, et al. alphavbeta5 integrin promotes dedifferentiation of monolayer-cultured articular chondrocytes. Arthritis Rheum. 2011;63:1938–1949. doi: 10.1002/art.30351. [DOI] [PubMed] [Google Scholar]
- 83.Sassi N, et al. Notch signaling is involved in human articular chondrocytes de-differentiation during osteoarthritis. J. Receptors Signal. Transduct. 2014;34:48–57. doi: 10.3109/10799893.2013.856920. [DOI] [PubMed] [Google Scholar]
- 84.Yu SM, Kim HA, Kim SJ. 2-Deoxy-D-glucose regulates dedifferentiation through beta-catenin pathway in rabbit articular chondrocytes. Exp. Mol. Med. 2010;42:503–513. doi: 10.3858/emm.2010.42.7.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li MG, Zhao JQ, Jia LG. USP14-mediated I kappa B alpha degradation exacerbates NF-kappa B activation and IL-1 beta-stimulated chondrocyte dedifferentiation. Life Sci. 2019;218:147–152. doi: 10.1016/j.lfs.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 86.Parreno J, et al. Interplay between cytoskeletal polymerization and the chondrogenic phenotype in chondrocytes passaged in monolayer culture. J. Anat. 2017;230:234–248. doi: 10.1111/joa.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim SJ, Hwang SG, Kim IC, Chun JS. Actin cytoskeletal architecture regulates nitric oxide-induced apoptosis, dedifferentiation, and cyclooxygenase-2 expression in articular chondrocytes via mitogen-activated protein kinase and protein kinase C pathways. J. Biol. Chem. 2003;278:42448–42456. doi: 10.1074/jbc.M304887200. [DOI] [PubMed] [Google Scholar]
- 88.Park EH, et al. Integrity of the cortical actin ring is required for activation of the PI3K/Akt and p38 MAPK signaling pathways in redifferentiation of chondrocytes on chitosan. Cell Biol. Int. 2008;32:1272–1278. doi: 10.1016/j.cellbi.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 89.Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 90.Matsumoto E, Furumatsu T, Kanazawa T, Tamura M, Ozaki T. ROCK inhibitor prevents the dedifferentiation of human articular chondrocytes. Biochem. Biophys. Res. Commun. 2012;420:124–129. doi: 10.1016/j.bbrc.2012.02.127. [DOI] [PubMed] [Google Scholar]
- 91.Yu SM, et al. Berberine induces dedifferentiation by actin cytoskeleton reorganization via phosphoinositide 3-kinase/Akt and p38 kinase pathways in rabbit articular chondrocytes. Exp. Biol. Med. (Maywood) 2016;241:800–807. doi: 10.1177/1535370216631028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shin H, et al. Focal adhesion assembly induces phenotypic changes and dedifferentiation in chondrocytes. J. Cell Physiol. 2016;231:1822–1831. doi: 10.1002/jcp.25290. [DOI] [PubMed] [Google Scholar]
- 93.Kim YH, Lee JW. Targeting of focal adhesion kinase by small interfering RNAs reduces chondrocyte redifferentiation capacity in alginate beads culture with type II collagen. J. Cell Physiol. 2009;218:623–630. doi: 10.1002/jcp.21637. [DOI] [PubMed] [Google Scholar]
- 94.Cao B, Peng R, Li Z, Ding J. Effects of spreading areas and aspect ratios of single cells on dedifferentiation of chondrocytes. Biomaterials. 2014;35:6871–6881. doi: 10.1016/j.biomaterials.2014.04.107. [DOI] [PubMed] [Google Scholar]
- 95.Hwang HS, Kim HA. Chondrocyte apoptosis in the pathogenesis of osteoarthritis. Int. J. Mol. Sci. 2015;16:26035–26054. doi: 10.3390/ijms161125943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McGann CJ, Odelberg SJ, Keating MT. Mammalian myotube dedifferentiation induced by newt regeneration extract. Proc. Natl Acad. Sci. USA. 2001;98:13699–13704. doi: 10.1073/pnas.221297398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen ZL, Yu WM, Strickland S. Peripheral regeneration. Annu. Rev. Neurosci. 2007;30:209–233. doi: 10.1146/annurev.neuro.30.051606.094337. [DOI] [PubMed] [Google Scholar]
- 98.Merrell AJ, Stanger B. Adult cell plasticity in vivo: de-differentiation and transdifferentiation are back in style. Nat. Rev. Mol. Cell Bio. 2016;17:413–425. doi: 10.1038/nrm.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat. Rev. Genet. 2010;11:710–722. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mendez-Ferrer S, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–U859. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tan JM, et al. Induction therapy with autologous mesenchymal stem cells in living-related kidney transplants a randomized controlled trial. JAMA. 2012;307:1169–1177. doi: 10.1001/jama.2012.316. [DOI] [PubMed] [Google Scholar]
- 102.Sensebe, L., Gadelorge, M. & Fleury-Cappellesso, S. Production of mesenchymal stromal/stem cells according to good manufacturing practices: a review. Stem Cell Res. Ther.4, 66 (2013). [DOI] [PMC free article] [PubMed]
- 103.Presen, D. M., Traweger, A., Gimona, M. & Redl, H. Mesenchymal stromal cell-based bone regeneration therapies: from cell transplantation and tissue engineering to therapeutic secretomes and extracellular vesicles. Front. Bioeng. Biotechnol.7, 352 (2019). [DOI] [PMC free article] [PubMed]
- 104.Confalonieri D, Schwab A, Walles H, Ehlicke F. Advanced therapy medicinal products: a guide for bone marrow-derived MSC application in bone and cartilage tissue engineering. Tissue Eng. Part B-Rev. 2018;24:155–169. doi: 10.1089/ten.TEB.2017.0305. [DOI] [PubMed] [Google Scholar]
- 105.Khademhosseini A, Langer R. A decade of progress in tissue engineering. Nat. Protoc. 2016;11:1775–1781. doi: 10.1038/nprot.2016.123. [DOI] [PubMed] [Google Scholar]
- 106.Pace LA, Plate JF, Smith TL, Van Dyke ME. The effect of human hair keratin hydrogel on early cellular response to sciatic nerve injury in a rat model. Biomaterials. 2013;34:5907–5914. doi: 10.1016/j.biomaterials.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 107.Rosenzweig DH, et al. Culture of primary bovine chondrocytes on a continuously expanding surface inhibits dedifferentiation. Tissue Eng. Part A. 2012;18:2466–2476. doi: 10.1089/ten.tea.2012.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rosenzweig DH, Solar-Cafaggi S, Quinn TM. Functionalization of dynamic culture surfaces with a cartilage extracellular matrix extract enhances chondrocyte phenotype against dedifferentiation. Acta Biomater. 2012;8:3333–3341. doi: 10.1016/j.actbio.2012.05.032. [DOI] [PubMed] [Google Scholar]
- 109.Pei M, He F. Extracellular matrix deposited by synovium-derived stem cells delays replicative senescent chondrocyte dedifferentiation and enhances redifferentiation. J. Cell Physiol. 2012;227:2163–2174. doi: 10.1002/jcp.22950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang YH, et al. Mesenchymal stem cell-derived extracellular matrix enhances chondrogenic phenotype of and cartilage formation by encapsulated chondrocytes in vitro and in vivo. Acta Biomater. 2018;69:71–82. doi: 10.1016/j.actbio.2017.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pahoff S, et al. Effect of gelatin source and photoinitiator type on chondrocyte redifferentiation in gelatin methacryloyl-based tissue-engineered cartilage constructs. J. Mater. Chem. B. 2019;7:1761–1772. doi: 10.1039/c8tb02607f. [DOI] [PubMed] [Google Scholar]
- 112.Ozturk E, et al. Sulfated hydrogel matrices direct mitogenicity and maintenance of chondrocyte phenotype through activation of FGF signaling. Adv. Funct. Mater. 2016;26:3649–3662. doi: 10.1002/adfm.201600092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jin, G. Z. & Kim, H. W. Efficacy of collagen and alginate hydrogels for the prevention of rat chondrocyte dedifferentiation. J. Tissue Eng.9, 2041731418802438 (2018). [DOI] [PMC free article] [PubMed]
- 114.Li SY, et al. Effects of nanoscale spatial arrangement of arginine-glycine-aspartate peptides on dedifferentiation of chondrocytes. Nano Lett. 2015;15:7755–7765. doi: 10.1021/acs.nanolett.5b04043. [DOI] [PubMed] [Google Scholar]
- 115.Chattopadhyay S, Shubayev VI. MMP-9 controls Schwann cell proliferation and phenotypic remodeling via IGF-1 and ErbB receptor-mediated activation of MEK/ERK pathway. Glia. 2009;57:1316–1325. doi: 10.1002/glia.20851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu H, et al. Matrix metalloproteinase inhibition enhances the rate of nerve regeneration in vivo by promoting dedifferentiation and mitosis of supporting schwann cells. J. Neuropathol. Exp. Neurol. 2010;69:386–395. doi: 10.1097/NEN.0b013e3181d68d12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rittchen S, et al. Myelin repair in vivo is increased by targeting oligodendrocyte precursor cells with nanoparticles encapsulating leukaemia inhibitory factor (LIF) Biomaterials. 2015;56:78–85. doi: 10.1016/j.biomaterials.2015.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu C, et al. Recombinant human midkine stimulates proliferation and decreases dedifferentiation of auricular chondrocytes in vitro. Exp. Biol. Med. 2011;236:1254–1262. doi: 10.1258/ebm.2011.011022. [DOI] [PubMed] [Google Scholar]
- 119.Luo LK, Wei QJ, Liu L, Zheng L, Zhao JM. Andrographolide enhances proliferation and prevents dedifferentiation of rabbit articular chondrocytes: an in vitro study. Evid. Based Complement Alternat. Med. 2015;2015:984850. doi: 10.1155/2015/984850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sunico CR, Portillo F, Gonzalez-Forero D, Kasparov S, Moreno-Lopez B. Evidence for a detrimental role of nitric oxide synthesized by endothelial nitric oxide synthase after peripheral nerve injury. Neuroscience. 2008;157:40–51. doi: 10.1016/j.neuroscience.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 121.Sunico CR, Moreno-Lopez B. Evidence for endothelial nitric oxide as a negative regulator of Schwann cell dedifferentiation after peripheral nerve injury. Neurosci. Lett. 2010;471:119–124. doi: 10.1016/j.neulet.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 122.Santangelo KS, Nuovo GJ, Bertone AL. In vivo reduction or blockade of interleukin-1beta in primary osteoarthritis influences expression of mediators implicated in pathogenesis. Osteoarthr. Cartil. 2012;20:1610–1618. doi: 10.1016/j.joca.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fujino M, et al. CrmA gene expression protects mice against concanavalin-A-induced hepatitis by inhibiting IL-18 secretion and hepatocyte apoptosis. Gene Ther. 2003;10:1781–1790. doi: 10.1038/sj.gt.3302067. [DOI] [PubMed] [Google Scholar]
- 124.Ma BL, et al. Inhibition of interleukin-1beta-stimulated dedifferentiation of chondrocytes via controlled release of CrmA from hyaluronic acid-chitosan microspheres. BMC Musculoskelet. Disord. 2015;16:61. doi: 10.1186/s12891-015-0521-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cha BH, Lee JS, Kim SW, Cha HJ, Lee SH. The modulation of the oxidative stress response in chondrocytes by Wip1 and its effect on senescence and dedifferentiation during in vitro expansion. Biomaterials. 2013;34:2380–2388. doi: 10.1016/j.biomaterials.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 126.D’uva G, et al. ERBB2 triggers mammalian heart regeneration by promoting cardiorlyocyte dedifferentiation and proliferation. Nat. Cell Biol. 2015;17:627–U207. doi: 10.1038/ncb3149. [DOI] [PubMed] [Google Scholar]
- 127.Liu Y, et al. Dedifferentiation-reprogrammed mesenchymal stem cells with improved therapeutic potential. Stem Cells. 2011;29:2077–2089. doi: 10.1002/stem.764. [DOI] [PubMed] [Google Scholar]
- 128.Liu Y, et al. Switching from bone marrow-derived neurons to epithelial cells through dedifferentiation and translineage redifferentiation. Cell Biol. Int. 2010;34:1075–1083. doi: 10.1042/CBI20100516. [DOI] [PubMed] [Google Scholar]
- 129.Lin S, et al. Stepwise preconditioning enhances mesenchymal stem cell-based cartilage regeneration through epigenetic modification. Osteoarthr. Cartil. 2017;25:1541–1550. doi: 10.1016/j.joca.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 130.Duan L, et al. DNA methylation profiling in chondrocyte dedifferentiation in vitro. J. Cell Physiol. 2017;232:1708–1716. doi: 10.1002/jcp.25486. [DOI] [PubMed] [Google Scholar]
- 131.Hong EH, et al. Low-dose gamma-radiation inhibits IL-1beta-induced dedifferentiation and inflammation of articular chondrocytes via blockage of catenin signaling. IUBMB Life. 2014;66:128–137. doi: 10.1002/iub.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jimenez G, et al. Activin A/BMP2 chimera AB235 drives efficient redifferentiation of long term cultured autologous chondrocytes. Sci. Rep. 2015;5:16400. doi: 10.1038/srep16400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yao Y, et al. In vitro study of chondrocyte redifferentiation with lentiviral vector-mediated transgenic TGF-beta3 and shRNA suppressing type I collagen in three-dimensional culture. J. Tissue Eng. Regen. Med. 2011;5:e219–e227. doi: 10.1002/term.425. [DOI] [PubMed] [Google Scholar]
- 134.Schrobback K, et al. Adult human articular chondrocytes in a microcarrier-based culture system: expansion and redifferentiation. J. Orthop. Res. 2011;29:539–546. doi: 10.1002/jor.21264. [DOI] [PubMed] [Google Scholar]
- 135.Zeng L, et al. Redifferentiation of dedifferentiated chondrocytes in a novel three-dimensional microcavitary hydrogel. J. Biomed. Mater. Res. A. 2015;103:1693–1702. doi: 10.1002/jbm.a.35309. [DOI] [PubMed] [Google Scholar]
- 136.Schrobback K, et al. Effects of oxygen and culture system on in vitro propagation and redifferentiation of osteoarthritic human articular chondrocytes. Cell Tissue Res. 2012;347:649–663. doi: 10.1007/s00441-011-1193-7. [DOI] [PubMed] [Google Scholar]
- 137.Babur BK, et al. The interplay between chondrocyte redifferentiation pellet size and oxygen concentration. PLoS ONE. 2013;8:e58865. doi: 10.1371/journal.pone.0058865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Levett PA, et al. Chondrocyte redifferentiation and construct mechanical property development in single-component photocrosslinkable hydrogels. J. Biomed. Mater. Res. A. 2014;102:2544–2553. doi: 10.1002/jbm.a.34924. [DOI] [PubMed] [Google Scholar]