Abstract
Sex differences in 116 local gray matter volumes (GMVOL) were assessed in 444 males and 444 females without correcting for total intracranial volume (TIV) or after adjusting the data with the scaling, proportions, power-corrected proportions (PCP), and residuals methods. The results confirmed that only the residuals and PCP methods completely eliminate TIV-variation and result in sex-differences that are “small” (∣d∣ < 0.3). Moreover, as assessed using a totally independent sample, sex differences in PCP and residuals adjusted-data showed higher replicability ( 93%) than scaling and proportions adjusted-data 68%) or raw data ( 45%). The replicated effects were meta-analyzed together and confirmed that, when TIV-variation is adequately controlled, volumetric sex differences become “small” (∣d∣ < 0.3 in all cases). Finally, we assessed the utility of TIV-corrected/ TIV-uncorrected GMVOL features in predicting individuals’ sex with 12 different machine learning classifiers. Sex could be reliably predicted (> 80%) when using raw local GMVOL, but also when using scaling or proportions adjusted-data or TIV as a single predictor. Conversely, after properly controlling TIV variation with the PCP and residuals’ methods, prediction accuracy dropped to 60%. It is concluded that gross morphological differences account for most of the univariate and multivariate sex differences in GMVOL
Subject terms: Sexual dimorphism, Brain
Introduction
The study of neuroanatomical sex differences in the brain is a subject of considerable scientific importance1–3 that also arouses great interest in the popular press and lay audiences4,5. However, precisely quantifying sex differences in the volumes of specific brain regions is a challenging task that is severely complicated by the existing sex differences in the overall body and head size6–8. Thus, although there is an increasing consensus about the need to parse out the quantitative contribution of “direct” or “specific” effects of sex on regional brain volumes from those derived from gross morphology differences between females and males9–18, there is far less agreement about how to do this. Thus, several adjusting variables (for a review, see O’Brien, 2006) and statistical methods are currently used to adjust gross morphology variation7,8,15,16.
Within this context, we recently conducted a broad systematic study to compare how five different TIV-adjustment methods (scaling as implemented by the non-linear modulation option of the VBM8 toolbox, proportions, power-corrected-proportions, covariate regression, or the residuals methods) affected the number, size, and direction of sex differences in 116 local gray matter volumes (GMVOL) in the so-called “UJI-sample”19. Our results confirmed and extended those of other previous studies by showing that: (1) males have larger TIV-uncorrected (raw) GMVOL in all brain areas, but these differences are largely due to TIV-variation9,10,13,14,17; (2) different TIV-adjustment methods end up producing different patterns of sex differences that are not equally valid15,16. Regarding the latter, we observed that the scaling and proportions adjustment methods inverted, but did not eliminate, the preexisting relationships between TIV and local GMVOL, thus resulting in larger adjusted volumes in females than in males and promoting sex differences that were very distinct in number, size, and direction from those observed in a subgroup of females and males matched on their TIV19. Conversely, data adjusted with the three other methods had no influence of TIV-variation and resulted in fewer, smaller, and bi-directional sex differences that closely resembled those observed in the sample of TIV-matched males and females19.
The first aim of the present study was to confirm these results by directly replicating them in a larger sample (hereinafter referred as the “HCP sample”) composed of 444 females and 444 males. Replication of findings is a cornerstone of scientific progress because it makes it possible to increase the precision of effect size estimates and provide information about whether an earlier published effect should be considered a true effect, a false positive, or the result of an interaction with a contextual moderator20,21. Replication should not be assessed based on coincidence analyses of “significant/non-significant effects”22–24, but on other metrics specifically developed to compare the effects found in different samples from a single population (i.e. prediction intervals23,25). Therefore, in the present study, prediction intervals were calculated to assess the extent to which the direction and size (Cohen’s d values) of the sex differences in GMVOL obtained in the present study replicated the ones we previously observed in the “UJI-sample”19.
This replication assessment also allowed us to address a largely unexplored question, namely, the extent to which the replicability of sex differences in GMVOL is affected by the method employed to adjust TIV-variation. In this regard, although TIV-adjustment is known to increase measurement error and reduce the reliability of local GMVOL measurements26,27, it actually improves the detection of between-group differences in GMVOL27. Moreover, random measurement error in TIV values has been found to increase variability and reduce between-groups mean differences in proportions-adjusted data, but not in residuals-adjusted data28. Therefore, it can be tentatively hypothesized that at least some TIV-adjustment methods could increase the replicability of sex differences in GMVOL, especially when considering the replicability of effect sizes based on means and standard deviations, such as Cohen’s d. However, to our knowledge, this proposal has not been empirically tested.
As a third and final objective, in the present study we also explored the effect of different TIV-adjustment methods when assessing multivariate sex differences. Assessing multivariate sex differences is important because a series of small univariate differences might (or might not) aggregate into a larger overall difference, and because multivariate statistics provide non-redundant, distinct information from what univariate measures convey29–31. Multivariate sex differences can be assessed through effect size indexes such as Mahalanobis’ D (the multivariate equivalent of Cohen’s d; see29,30). However, D and other related effect sizes are more meaningful when summarizing a “coherent, theoretically justified set of variables”31 p. 11) than when comparing whole-brain averages of local effects running in disparate directions (for a detailed discussion, see31). Alternatively, multivariate differences can be investigated through classification/prediction statistical techniques collectively referred to as machine learning or statistical learning32–34. These techniques are increasingly being used in the study of brain sex differences31,35–41 because they make it possible to estimate the degree of statistical distinctiveness or separateness of the brains of females and males at the multivariate level, with the added conceptual appeal of focusing on individual scores instead of on score summaries such as means. However, to our knowledge, no previous study has specifically analyzed to what extent this multivariate distinctiveness is affected by TIV-adjustment.
Therefore, in the present study, we assessed how four currently used TIV-adjustment methods affect the collective utility of 116 local GMVOL when inputed as features of 12 different machine learning algorithms constructed to differentiate the brains of females and males and predict individuals’ sex. Following current recommendations, classification algorithms were fitted, tested, and validated in separate groups of participants33,34. More specifically, each algorithm was initially fitted in a randomly selected training subsample that comprised 311 females and 311 males (70% of total) from the HCP-sample. The obtained classifiers were internally validated34 in the testing subsample, which was composed of the hold-out participants from the HCP sample (133 females and 133 males). Finally, the classification algorithms were externally validated34 in a completely independent sample (the so-called external validation subsample), composed of 171 females and 171 males of the UJ- sample).
Results and discussion
Univariate sex differences in the HCP sample
Raw data
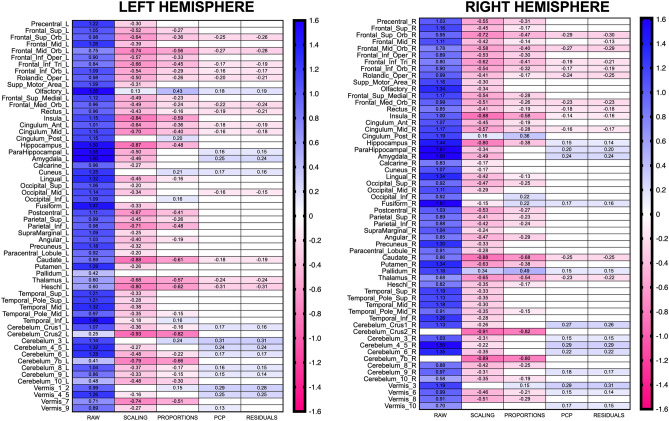
Males had larger TIVs than females (Difference in means: 212.80 ml, 95% CI [197.73, 227.86]; d = 1.86, p = 2.65–122). Males also exhibited larger raw GMvol than females in the 116 anatomical regions considered in the present study. In 114 cases (98.28% of the total), the confidence intervals for these differences did not include the zero value, making it possible to reject a null hypothesis of no difference between means (uncorrected p-values range: 0.0002—2.26–98). As Fig. 1 depicts, in these 114 VOIs, Cohen’s d values ranged from 0.25 (#93, Cerebellum_Crus2_L; overlap = 90.0%, PS = 57.0%) to 1.61 (#40, Parahippocampal_R; overlap = 42.1%, PS = 87.3%); average d = 1.06, 95%CI [0.99, 1.10]. For more detailed information about the sex differences observed in this data set, see Supplementary Table 1A.
Figure 1.
Size and location of sex differences in each dataset of the HCP sample. Panels left and right present odd- and even-numbered brain anatomical regions of the AAL atlas, which (with the exception of the lobules of the cerebellar vermis) are located in the left and right hemisphere, respectively. Heatmaps display the Cohen’s d values for statistically significant sex differences (a more detailed description of all effects is provided in Supplementary Tables 1A–5A). Blue colored cells and positive d values correspond to M > F effects, whereas red colored cells and negative d values correspond to F > M effects.
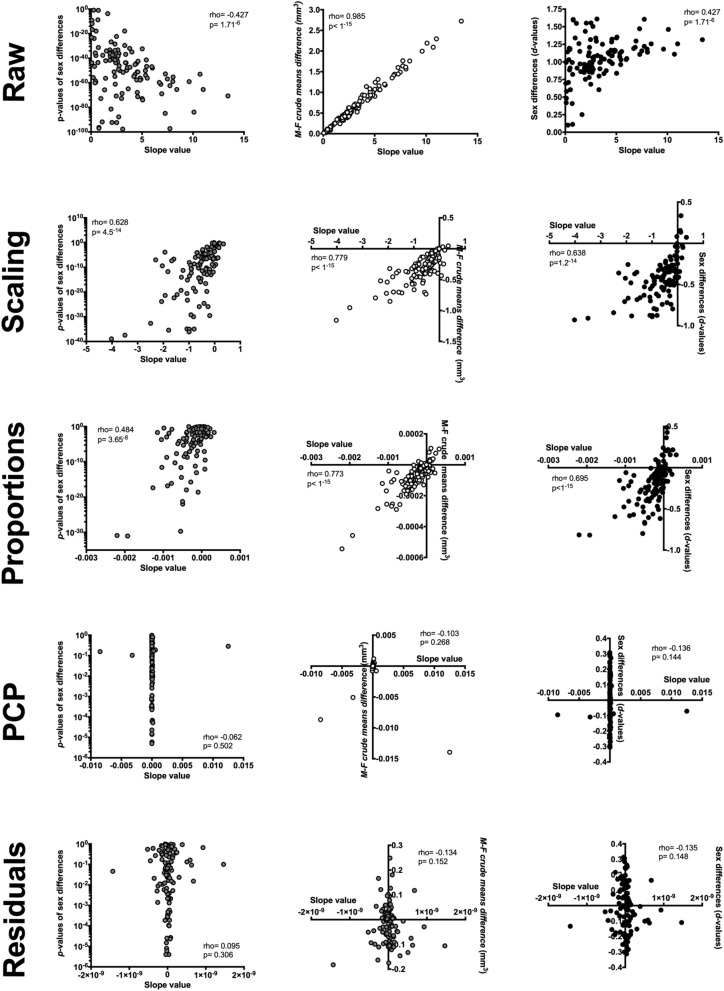
TIV variation was directly related to GMvol variation in the 116 VOIraw considered in the present study (p-values < 1.81–5 in all cases; Supplementary Table 1B). The percent of variance accounted for by TIV differed at each VOI, ranging between 2.05% (#94, Cerebellum_Crus2_R) and 73.59% (#56, Fusiform_gyrus_R). As Fig. 2 shows, the slope values of these TIV-VOIraw linear regressions were correlated with the p-values (rho = -0.43, p < 1.7–6) and the size (unstandardized mean difference, rho = 0.99, p < 1–15; d-values, rho = 0.42, p < 1.7–6) of the sex differences observed in raw GMvol. These results confirm that the significance levels, size, and direction of the sex differences in raw GMvol are largely dependent on TIV variation.
Figure 2.
Correlation between TIV-VOI slopes and observed sex differences in each data set. Ordinal correlations (Spearman’s rho) were calculated between the slope values of the TIV-VOI regression lines (provided in Supplementary Tables 1B–5B) and the p-values (left column), unstandardized means difference (central column), and Cohen’s d values (right column) of the sex differences obtained in the raw, scaling, proportions, PCP, and residuals datasets. Note that the scales and labels’ positioning are customized in each figure to better show the very distinct patterns of correlations observed in each dataset.
Scaled data
On the one hand, females exhibited larger scaled GMvol (GMscaling) than males in most of the VOIs. In 107 cases (92.24% of total), the 95% CI of these differences did not include the zero value, allowing to reject the null hypothesis of no differences between means (uncorrected p-values range: 0.046 to 1.21–39). As Fig. 1 shows, F > M differences were observed in 104 of these 107 VOIs, with d values ranging from -0.15 (#56, Fusiform_R; overlap = 93.9%; PS = 54.3%) to -0.93 (#93, Cerebellum_Crus2_L; overlap = 64.2%; PS = 74.5%); average d = − 0.47, CI [− 0.43, − 0.50]. On the other hand, males had larger GMscaling in the Olfactory_L (#21; d = 0.13, overlap = 94.6%; PS = 53.8%), Cingulum_post_R (#36; d = 0.15, overlap = 93.8%; PS = 54.4%), and Pallidum_R (#76; d = 0.34, Overlap = 86.7%; PS = 59.4%). For more detailed information about the sex differences observed in this data set, see Supplementary Table 2A.
The scaling method reduced and, in most cases, inverted the direction, but it did not eliminate the effects of TIV on GMvol variation (Supplementary Table 2B). Thus, in 90 cases, TIV-VOIscaling linear regressions had slope values that were statistically different from 0 (p-values ranging from 0.047 to < 1–15), with TIV explaining percentages of variance ranging between 0.45% (#53, Occipital_inf_L) and 22.44% (#94, Cerebellum_Crus2_R). As Fig. 2 reveals, the slope values of the 116 regression TIV-VOIscaling lines were correlated with the p-values (rho = 0.63, p = 4.5–14) and the size (unstandardized mean difference, rho = 0.78, p < 1–15; d values, rho = 0.64, p = 1.2–14) of the observed sex differences in GMscaling.
Proportions-adjusted data
As in the case of GMscaling, proportions-adjusted GMvol (GMprop) were larger in females than in males. In 76 cases (65.51% of total), the 95%CI of these differences did not include the zero value and allowed to reject the null hypothesis of no difference between means (uncorrected p-values range: 0.045—9.6–32). In 64 of these 76 cases (84.2%), females exhibited larger GMprop than males, with Cohen’s d values ranging from − 0.13 (#48, Lingual_R) to − 0.82 (#94, Cerebellum_Crus2_R); average d = − 0.35, CI [− 0.39, − 0.30]. These d values (depicted in Fig. 1) translated into degrees of overlap ranging between 68.2% and 94.7%, and PS ranging between 53.8% and 71.9%. In the 12 cases where males had larger GMprop than females, differences ranged from d = 0.15 (#109, vermis_1_2; overlap = 94.1%; PS = 54.2%) to d = 0.49 (#76, pallidum_R; overlap = 80.8%, PS = 63.5%). For more detailed information about the sex differences observed in this data set, see Supplementary Table 3A.
The proportions method reduced and, in most cases, inverted but did not eliminate the effects of TIV on GMVOL variation (Supplementary Table 3B). In 77 anatomical regions, the slope values for linear TIV-VOIprop were significantly different from zero (p-values range: 0.03 to 1–15) and, at these VOIs, TIV explained between 0.5% (#85, Temporal_Mid_L) and 24.76% (#94, Cerebellum_Crus2_R) of the observed variance. As Fig. 2 shows, the slope values of the 116 TIV-VOIprop linear relationships were correlated with p-values (rho = 0.482, p = 3.65–8) and the size (unstandardized mean difference, rho = 0.77, p < 1–15; d values, rho = 0.65, p < 1–15) of the sex differences observed in GMprop (Fig. 2).
PCP-adjusted data
Sex differences in PCP-adjusted GMvol (GMPCP) showed a clearly bidirectional pattern. In 50 VOIs, the 95% CI of the between-means difference did not include the zero value and allowed to reject the null hypothesis (uncorrected p-values range: 0.047 to 5.0–6). Within this subset (depicted in Fig. 1), there were 26 M > F differences, with d values ranging between 0.11 (#115, Vermis_9; overlap = 94.7%, PS = 53.8%) and 0.31 (#95, Cerebellum_3_L; overlap = 87.8%; PS = 58.6%); average d = 0.20, CI [0.18, 0.23]. In addition, F > M differences were observed in 24 brain anatomical regions (Fig. 1; Supplementary Table 4A). In these 24 cases, d values ranged between − 0.14 (#30, Insula_R; overlap = 94.3%, PS = 54.0%) and − 0.31 (#79, Heschl_L; overlap = 87.9%; PS: 58.4%); average d = − 0.21, CI [− 0.19, − 0.23]. For more detailed information about the sex differences observed in this dataset, see Supplementary Table 4A.
Sex differences in GMPCP were devoid of any influence of TIV. Linear regression analyses indicated that TIV did not account for any of the variance observed in the 116 VOIs in this dataset (r2 values ranged between 2.19–4 and 5.57–12; p-values > 0.66 in all cases). All the slope values were also virtually zero (absolute values ranging from 0.01 to 2.79–9; Supplementary Table 4B), and, consequently, they were uncorrelated with the p-values (rho = − 0.06, p = 0.502) and the size (unstandardized mean difference, rho = − 0.10, p = 0.268; d values, rho = − 0.14, p = 0.144) of the sex differences observed in GMPCP (Fig. 2).
Residuals-adjusted data
Sex differences in the residuals-adjusted GMvol (GMres) were very similar to those observed in the GMPCP. In 50 cases, the CIs for these differences did not include the zero value and allowed us to reject the null hypothesis of no differences between means (uncorrected p-values ranging from 0.046 to 4–6). On the one hand, females had larger local GMres in 25 VOIs, with d values ranging from − 0.13 (#8, Frontal_Mid_R: overlap = 94.7%; PS = 53.8%) to -0.32 (#79, Heschl_L; overlap = 87.6% PS = 58.8%); average d = − 0.22, CI [− 0.20, − 0.24]. On the other hand, in the 25 anatomical regions in which males had larger GMres, d values ranged from to 0.14 (#105 Cerebellum_9_L; overlap = 94.5%; PS = 53.9%) to 0.31 (#95, Cerebellum_3_L; overlap = 87.6%; PS = 58.8%); average d = 0.20, CI [0.18, 0.23]. These results are depicted in Fig. 1 and described in detail in Supplementary Table 5A.
As in the case of GMPCP, sex differences in GMres were devoid of any influence of TIV variation. In this dataset, TIV-VOI linear regression analyses yielded r2 values ranging from 6.67–20 to 1.98–28 (uncorrected p-values > 0.99 in all cases; Supplementary Table 5B). All slope values were also virtually 0 (absolute values ranging from 1.47–9 to 6.88–15), and as Fig. 2 shows, they were uncorrelated with the p-values (rho = 0.09, p = 0.308) and the size (unstandardized mean difference, rho = − 0.13, p = 0.152; d values, rho = − 0.14, p = 0.148) of the sex differences observed in GMres.
Summary
The results obtained make it possible to draw three main conclusions: First, as previously described8,10,13,14,18,19, raw GMVOL conflate sex and TIV variation effects, resulting in large differences that invariably favor the sex with larger TIV (males). Second, as previously shown8,10,14–16,19,42, not all the currently used methods are equally effective in statistically removing TIV effects because sex differences calculated using scaling and proportions-adjusted data are still partially due to TIV variation. Accordingly, these two methods are increasingly viewed as suboptimal TIV-adjustment methods14,16,19,42,43. Third, when TIV variation is properly controlled, sex differences appear to be bidirectional, and their size is very much reduced, approaching zero in many cases. This last observation also agrees with those of previous studies10,13,16,18,19,44.
Replication of univariate sex differences
Despite being similar to those of other studies, the results described in the previous section do not provide direct information about which sex differences in GMVOL are replicated, or to what extent the replicability of sex differences in GMVOL is affected by different TIV-adjustment methods. In order to answer these two questions, we estimated the 95% prediction intervals for the d values of the sex differences in GMVOL previously observed in the UJI sample (Supplementary Table 6A), and we calculated the replication rates observed in each dataset.
As summarized in Table 1, the number of replicated effects greatly differed for raw (52; 44.83%), scaling (72; 62.06%), proportions- (86; 74.14%), PCP- (110; 94.82%) and residuals- (106; 91.38%) adjusted data (χ2 = 102.77, df = 4, p-value < 2.2–16, see pairwise comparisons in Table 1 and Supplementary Table 6B). Based on these results, it is clear that replication rates were higher for those datasets in which TIV variation had been properly controlled than for those in which it had not. Moreover, this effect was observed even though the sex differences in TIV obtained in the HCP sample fell within the prediction interval of the difference observed in the UJI sample (see Supplementary Table 6A). Therefore, it might be tentatively concluded that by controlling the effects of TIV (which can vary in different samples), the PCP and residuals methods provide not only TIV-independent but also more replicable estimates of sex differences in GMVOL. However, because this is the first time such an effect has been described, this conclusion requires verification by future independent studies.
Table 1.
Number and description of replicated effects. The first row of the table provides the number of replicated effects (those Cohen’s d values of the HCP sample falling within the prediction interval of their counterparts in the UJI sample). Superscripted letters (A to E) denote a statistically significant different proportion of replicated effects from what was observed in the raw, scaling, proportions, PCP, or residuals datasets, respectively. Effects were designated as “differences” if the 95% CI of their mean difference did not include the zero value and as “no-differences” if they did. Brain areas showing the largest and smallest sex differences are reported along with M > F and F > M averaged Cohen’s d values and their corresponding percent of overlap (o) and superiority (PS). More detailed information about these comparisons and outcomes is provided in Supplementary Table 6.
| Raw (A) | Scaling (B) | Proportions (C) | PCP (D) | Residuals (E) | |
|---|---|---|---|---|---|
| Number of replicated effects |
52 B, C, D, E |
72 A, D, E |
86 A, D, E |
110 A, B, C |
106 A, B, C |
| Differences | 52 | 63 | 49 | 47 | 51 |
| No-differences | 0 | 9 | 37 | 63 | 55 |
| Differences M > F | 52 | 2 | 6 | 22 | 22 |
| Differences F > M | 0 | 56 | 43 | 25 | 29 |
|
Differences M > F d maximum |
1.51 Amygdala R |
0.16 Cingulum post_R |
0.30 Cingulum post_R |
0.31 Pallidum R |
0.26 Cerebellum 4_5_R |
|
Differences M > F d minimum |
0.40 Cerebellum 10_L |
0.13 Olfactory R |
0.13 Occipital inf_R |
0.12 Hippocampus R |
0.12 Vermis 9 |
|
Differences M > F d average |
0.96 O: 63.09% PS: 75.16% |
0.14 O: 94.42% PS: 53.94% |
0.20 O: 92.03% PS: 55.62% |
0.18 O: 92.83% PS: 55.06% |
0.18 O: 92.83% PS: 55.06% |
|
Differences F > M d maximum |
– |
− 0.80 Hippocampus R |
− 0.66 Thalamus L |
− 0.30 Frontal_sup Orb_R |
− 0.30 Frontal_sup Orb_R |
|
Differences F > M d minimum |
– |
− 0.15 Fusiform R |
− 0.13 Rolandic Oper_R |
− 0.12 Frontal Sup_R |
− 0.11 Precentral R |
|
Differences F > M d average |
– |
− 0.39 O: 84.54% PS: 60.86% |
− 0.31 O: 87.64% PS: 58.70% |
− 0.18 O: 92.83% PS: 55.06% |
− 0.17 O: 93.15% PS: 55.84% |
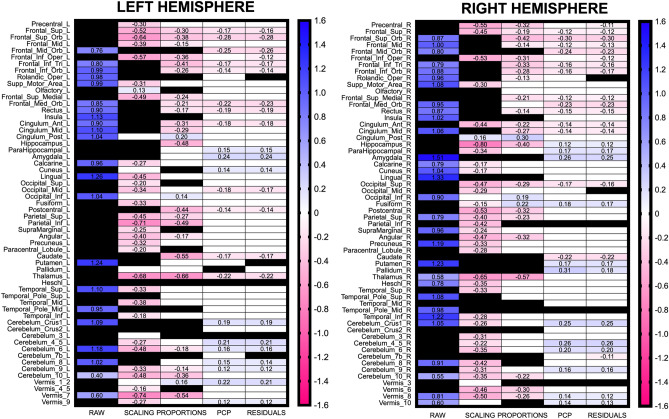
The anatomical locations of replicated and non-replicated effects are depicted in Fig. 3. This figure displays the averaged d values for each replicated effect whose 95% confidence interval did not include the zero value (referred to as “sex differences” in Table 1). In the same figure, replicated effects whose CIs included the zero value are depicted as white cells (and referred to as “no-differences” in Table 1), but their values and corresponding CIs can be found in Supplementary Table 6C. Non-replicated effects are depicted as black colored cells. For all replicated effects, new prediction intervals estimating the range of d values that could be expected in future replication studies assessing sex differences in GM in these anatomical regions were also calculated (Supplementary Table 6D).
Figure 3.
Averaged Cohen’s d values for replicated sex differences. As in Fig. 1, panels left and right present odd- and even-numbered brain anatomical regions of the AAL atlas. Heatmaps display the averaged Cohen’s d values for replicated sex differences in each dataset, with red colored cells and negative d values corresponding to F > M effects, and blue colored cells and positive d values corresponding to M > F effects, respectively. Replicated “no-differences” and non-replicated effects are depicted as cells colored in white or black, respectively. An effect was considered to be satisfactorily replicated if its d value in the HCP sample fell within the 95% prediction interval of the same effect in the UJI sample (see Replication of univariate sex differences in the Materials and Methods section, and Supplementary Table 6 for further details).
From the averaged d values calculated for replicated effects, it can again be concluded that in properly TIV-adjusted data, sex effects in local GMVOL are bidirectional and “small” (average |d| 0.12; average Overlap 95%; average PS 53.5%). Conversely, when TIV variation is not adequately controlled, sex differences appear much larger in size, and their direction is skewed, either in favor of males (raw data) or females (scaling and proportions datasets).
Multivariate classification
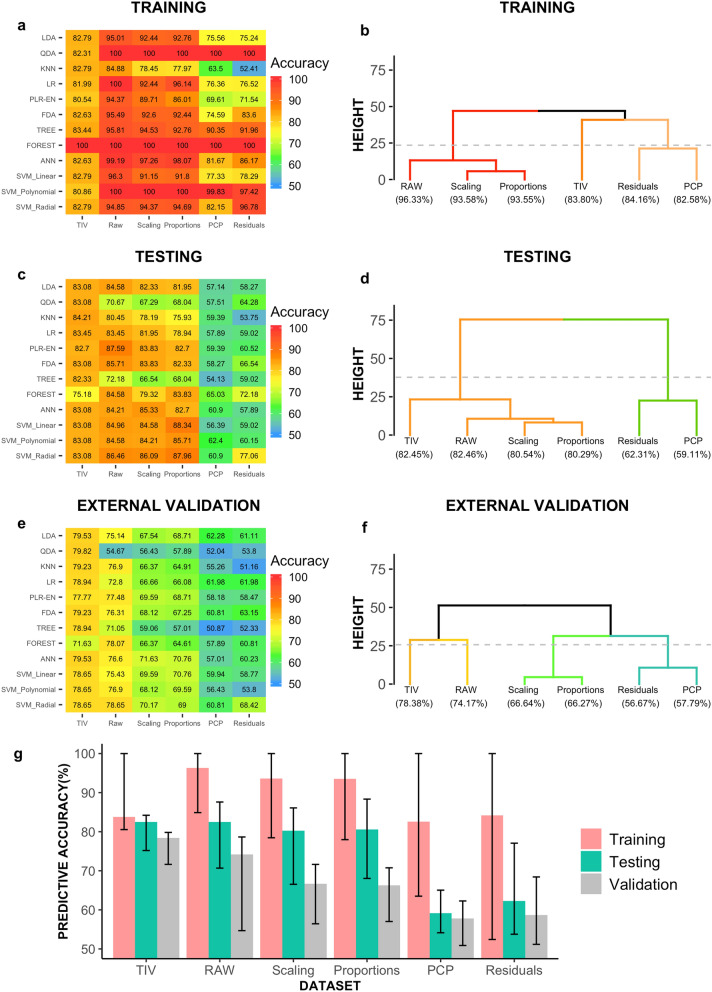
Figure 4 depicts the classification accuracy achieved by each classifier in the training, testing, and external validation subsamples of the raw, scaling, proportions, PCP, and residuals datasets (for more detailed output, see Supplementary Table 7). As panels A and B of the same figure show, average classification accuracy rates were high (> 80%) in the training subsamples of all the datasets. However, a finer-grained evaluation with the PAM clustering algorithm revealed three distinguishable patterns of results (see Supplementary Table 8). The first cluster was composed of the raw, scaling, and proportions datasets, which exhibited the highest accuracy levels and a high degree of homogeneity, with almost all the classifiers yielding a perfect or nearly perfect (> 90%) classification of females and males. The second cluster consisted solely of the TIV dataset, and it exhibited a large degree of homogeneity, but slightly lower accuracy levels ( 84%). Finally, the third cluster was composed of the PCP and the residuals datasets, which exhibited an average accuracy similar to what was observed in the second cluster, but with a much larger variation in the classifiers (range: 52.41–100%).
Figure 4.
Sex prediction accuracy. The heatmaps depicted in panels a, c, and e show the accuracy rate (percent of correctly predicted cases) for sex prediction obtained by each classifier (rows) in each dataset (columns) in the training, testing, and external validation subsamples, respectively (see Multivariate classification in the Materials and Methods section, and Supplementary Table 7 for further details). Note that, in these panels, the order of the rows and columns is constant but arbitrary. The dendrograms depicted in panels b, d, and f display the hierarchical relationships (average linkage based on Euclidean distances) between the prediction results obtained in the TIV, raw, scaling, proportions, PCP, and residuals datasets and their aggregation into performance-based clusters in the training, testing, and external validation subsamples, respectively (see Multivariate classification in the Materials and Methods section, and Supplementary Table 8 for further details). In these panels, average performance is reported as bracketed numbers under the dataset labels, and the dashed horizontal line indicates 50% of the maximum height of each dendrogram. Panel g summarizes all the previous results by showing the average (bars) and the maximum and minimum (“whiskers”) of the prediction accuracy observed in each dataset in each subsample.
As could be expected, accuracy rates in the testing subsample were lower than in the training subsample (Fig. 4, panel C), thus revealing that the very high accuracy rates observed in the training subsamples were partly due to overfitting. This decrease was more pronounced when using the local GMVOL of the raw (− 13.87%), scaling (− 13.28%), proportions (− 13.01%), PCP (− 23.46%), and residuals (− 21.85%) datasets as multivariate predictors than when using TIV as a single predictor (− 1.34%). Consequently, the results observed in the testing subsamples were ordered differently from in the training subsamples (entanglement: 25.9%; Supplementary Table 8), now showing only two very clearly separated clusters (see Fig. 4, panel D). The first cluster was composed of all the datasets that incorporated variation due to gross morphology differences between males and females (TIV, raw, scaling, and proportions), and it was characterized by high (> 80%) classification accuracy rates. Conversely, the second cluster, composed of those datasets that were free of any influence of TIV-variation (PCP and residuals), showed much lower ( 60%) accuracy rates. It should be noted that accuracy rates showed a wide variation among the classifiers in both clusters. Nevertheless, differences between clusters were evident, even when considering the results obtained with each classifier separately.
As could be expected34, accuracy rates were slightly lower in the external validation than in the testing subsample (Fig. 4, panel E). This decrease was larger in the proportions (− 14.02%), scaling (− 13.9%) and raw (− 8.29%) datasets than in the TIV (− 4.07%), residuals (− 3.46%), or PCP (− 1.32%) datasets. This differential reduction in the predictive accuracy did not substantially change the datasets’ ordering (entanglement: 5.75%, see tanglegram in Supplementary Table 8), but it divided the homogeneous cluster 1 from the testing subsample into 3 clusters, while leaving cluster 2 unaffected. Thus, in the external validation subsample, 4 clusters were observed: TIV > Raw > scaling proportions > Residuals PCP (Fig. 4, panel F). In the first cluster (TIV dataset), average accuracy was 78.38%, and all the classifiers yielded very similar prediction accuracy rates (range 71.63–79.82%). In the raw dataset (cluster 2), most of the methods performed slightly worse than in cluster 1, but the poor accuracy exhibited by the QDA classifier (54.67%) was mainly responsible for its slightly lower average accuracy (74.17%). The average accuracy of the third cluster (scaling and proportions datasets) was around 66%, with the QDA and simple classification trees clearly performing below this average. Finally, the fourth cluster was composed of the PCP and residuals datasets, which once again exhibited the lowest average accuracy rates (around 58%), with the QDA, KNN, and simple classification trees showing almost chance-level performance.
These results (summarized in Fig. 4, panel G) confirm and extend those of other previously published studies. In this regard, Chekroud and collaborators 45 obtained 92% (CI: 88.9–94.5%) accuracy when predicting sex in a large cohort of young females and males (training subsample, n = 1,156, testing subsample = 400) through an elastic net -penalized logistic regression (PLR-EN) that incorporated TIV-uncorrected subcortical GM volumes and cortical thickness measurements as predictors. Similarly, using PLR-EN and a support vector machine with a radial kernel (SVMradial) as classifiers and a complex array of predictors (raw GM density estimates, scaled GM volumes, age, and intellectual quotient), Anderson and co-workers37 found classification accuracy rates above 90% when predicting sex in large cohorts of incarcerated (training subsample, n = 930; testing subsample, n = 370) and non-incarcerated (training subsample, n = 922; testing subsample, n = 526) individuals. These reported accuracy rates are similar but slightly higher than those observed with the same classifiers in the raw dataset of the present study, a fact that is probably related to the use of larger training samples33,46 and other procedural discrepancies (e.g. different predictors). However, when considered together, the results of these two preceding studies and our results confirm that sex might be very accurately predicted by TIV-uncorrected GM features.
We also observed that sex prediction accuracy becomes much lower when TIV-variation on local GMVOL is appropriately controlled, but this is not as true when non-appropriate TIV-adjustment methods are employed (Fig. 4). To our knowledge, no previous study had been specifically designed to address this question. However, some reports had provided scattered evidence suggesting that the accuracy of sex prediction is reduced when using appropriate TIV-adjustment methods. Thus, Joel and co-workers reported that an anomaly detector algorithm discriminated between the brains of females and males better when brain features were not corrected for TIV-variation than when the same features were corrected with the PCP method36. Similarly, in their previously mentioned study45, Chekroud and collaborators observed that the sex prediction accuracy of their PLR-EN dropped from 92 to 70% (CI: 65.0–74.2%) when TIV-variation was “regressed out”. On the other hand, two other reports have provided estimates of sex prediction accuracy when using as predictors the same 116 scaled GMVOL that were employed in the present study. Thus, when re-analyzing some previously published data47, Rosenblatt found that a SVMlinear correctly predicted sex in about 80% of the cases35. With the same data, DelGiudice et al. (2015) found that principal component analysis combined with LDA properly predicted the individuals’ sex in about 70% of the cases. All these results are again very similar to those obtained in the present study, and they confirm that the method chosen to control TIV-variation has a major impact on sex predictability.
In summary, based on the results of this and other preceding studies, it can be concluded that sex can be appropriately predicted from raw GM local brain volumes. However, as also occurs when considering univariate sex differences in GMVOL in isolation, the distinctiveness of the brains of females and males at the multivariate level is very much dependent on their gross morphological differences (operationalized here in terms of TIV variation). Indeed, when using TIV as a single predictor, sex might be inferred with approximately the same accuracy and slightly less variance as when 116 raw GM local volumes are used. Conversely, when TIV variation is completely ruled out, the multivariate distinctiveness of the brains of females and males is very much reduced, and high misclassification rates are observed.
Limitations
The present study has some limitations that should be considered. Some of these limitations are related to the samples used. First, it should be noted that the present study used two convenience samples, rather than random samples obtained through optimized epidemiological procedures. Moreover, the samples used covered a relatively narrow age range. Thus, although these limitations are common in non-clinical brain volumetric studies, the representativeness of conveniences samples is not fully guaranteed, and the results obtained may have limited generalizability, especially for much younger or older populations. Second, although these samples could be considered “large” and ensured the necessary sensitivity to evaluate univariate sex differences, they resulted in case/variable ratios that might be suboptimal for some of the multivariate analyses included in the present study.
In addition, one of the objectives of the present study was to provide a direct replication of the results obtained in a previous study by our team on univariate sex differences in GMVOL19. This goal constrained some methodological decisions, including the use of a VOI-based approach and, particularly, the AAL atlas. The use of predefined VOIs has several advantages (e.g. avoids circularity, reduces the number of between-group comparisons,…), and it contributes to more accurate estimation of effect sizes48,49. However, the use of any predefined template (and that of the AAL atlas in particular) reduces anatomical precision and introduces other limitations and challenges50,51 that without compromising the validity of the present results, impede a direct comparison with estimates of GMVOL sex differences obtained with voxel-wise approaches.
Finally, the present study explored the use of the local GMVOL in predicting sex as a feasible approach to assess the degree of multivariate distinctiveness of male and female brains. In this attempt, the performance of 12 classification algorithms with distinct statistical assumptions and intrinsic operations was evaluated. However, even this ample exploration does not exhaust all the possible methods, and different results could be obtained with other classification algorithms, other predictors, or different parameters of the classifiers tested in the present study. This limitation does not reduce the validity of our results and conclusions about the effects of different TIV-adjustment methods on sex prediction. However, this limitation suggests that additional caution is needed when using the prediction accuracy rates obtained in this study as estimates of the multivariate morphological distinctiveness of the brains of females and males.
Conclusion
Our results show that univariate and multivariate sex differences in GMVOL are largely dependent on male–female differences in TIV, and that when this source of variation is parsed out univariate and multivariate sex differences are very much reduced. Our results also show that not all currently used TIV-adjustment methods are equally effective to remove TIV variation, and that which method is finally used has a major impact on the size (and, in the case of univariate differences, also the direction and, probably, the replicability) of the estimated sex differences. Consequently, choosing an appropriate TIV-adjustment method becomes a critical methodological decision that should be carefully considered and explicitly reported when designing new studies or when summarizing/ meta-analyzing preceding results.
Materials and methods
Participants
This study was conducted using data from two samples. The “HCP-sample” was composed of 444 females an 444 males included in the 1,200 Subject Release of the Human Connectome Project (HCP) 52, who did not differ in age (Meanfemales = 28.76, SD = 3.59; Meanmales = 28.52, SD = 3.40). On the other hand, the “UJI-sample” 19 was composed of 171 females and 185 males with similar ages (Meanfemales = 22.39, SD: 3.04; Meanmales = 21.64, SD: 4.90) See Supplementary Table 9 for further details.
Imaging data and TIV-adjustment
MRI acquisition
The MRI acquisition details of the HCP-sample might be found at the reference manual of the S1200 release of the HCP (https://www.humanconnectome.org/storage/app/media/documentation/s1200/HCP_S1200_Release_Reference_Manual.pdf). The details of the MRI data for the UJI subsample can be found in19.
Image pre-processing
All images were preprocessed with the VBM8 toolbox (version r445) implemented in the “New Segment” toolbox of the SPM8 (https://www.fil.ion.ucl.ac.uk/spm/software/spm8/) software (version 6316). This protocol includes four main steps: (1) segmentation of the images into gray matter, white matter, and cerebrospinal fluid; (2) registration to a standard template provided by the International Consortium of Brain Mapping (ICBM); (3) a high-dimensional DARTEL normalization of the gray matter segments to the MNI template; and (4) a data quality check (in which no outliers or incorrectly aligned cases were detected). After applying this procedure, which does not include any correction for overall head size, voxels were mapped into 116 regions according to the Automated Anatomical Labeling atlas (AAL, 50) by calculating the total gray matter volume for each region of interest (VOI) and participant via a MATLAB script (https://www0.cs.ucl.ac.uk/staff/g.ridgway/vbm/get_totals.m).
On this initial dataset (referred to as “raw”) sex differences unadjusted for TIV-variation were evaluated. Moreover, all the TIV adjustment methods (except the “scaling” method) were applied a posteriori to this initial output to generate TIV-adjusted datasets. On the other hand, TIV was estimated using native-space tissue maps obtained in the VBM8 segmentation step. Briefly, TIV was calculated as the sum of GM, WM and CSF total values multiplied by voxel size and divided by 1,000 to obtain a milliliter (ml) measurement. Although automated TIV estimation is less precise than that obtainable by manual segmentation43, this possible bias is not a major concern in the present study that used the same TIV estimation procedure when comparing different TIV-adjustment methods in a large sample of participants.
TIV-adjustment methods
Briefly, the four TIV-adjustment methods compared in the present study were:
–Scaling: Scaling is a normalization-related option provided in several image processing software packages that intends to remove the effects of head size (TIV) variation in local volumes using a two-step procedure. First, all brains are deformed as to make them to have exactly the same size. Second, the obtained normalized GM segments are multiplied by the non-linear determinants of the normalization deformation matrix. In this way, all GM segments are scaled to have the same size while local differences in volume are preserved. In the present study, the scaling method was implemented by using the non-linear modulation option included in the VBM8 toolbox53.
–Proportions adjustment method (proportions): This method attempts to provide adjusted VOIs by simply dividing each individual’s unadjusted VOI value by the value of its TIV8.
–The power-corrected proportions method (PCP): This method was recently proposed14 as a way to improve the proportions approach by introducing an exponential correcting parameter (VOIadj = VOI/TIVb) in the denominator. This parameter (b) corresponds to the slope value of the LOG(VOI) ~ LOG(TIV) regression line.
–The residuals method (residuals): This method was originally described by27 and it aims to remove TIV-VOI relationships through the formula VOIadj = VOI − b(TIV − ), where b is the slope value of the TIV-VOI regression line, and denotes the mean of the TIV values for all the participants.
Statistical analyses
Univariate sex differences in the HCP sample
Following current recommendations54,55, the statistical analyses focused on estimating effect sizes and 95% confidence intervals (CI) rather than on testing statistical significance.
Standardized effect sizes for between-mean differences (Cohen’s d) and their 95% CIs were calculated for each VOI in the raw, scaling, proportions, PCP, and residuals datasets of the HCP-sample. In the present study, positive Cohen d values indicate larger GMVOL in males than in females (M > F), whereas negative Cohen d values denote larger GMVOL in females than in males (F > M). To facilitate interpretation29, d values were transformed into the Weitzman’s (also known as percent of overlap and ORL-1) and the percent of superiority (PS). The percent of overlap denotes the proportion of scores that overlap in two normal distributions whose means differ in some magnitude. PS denotes the probability that a randomly sampled member of population a will have a score that is higher than the score attained by a randomly sampled member of population b29.
Following current recommendations56, unstandardized effect sizes for sex differences in GMVOL were also calculated. The 95% CIs of these differences were used to identify statistically significant sex differences (e.g. a 95% CI for the difference between two means that includes the zero value makes it possible to reject a nil null hypothesis at p < 0.0557). Exact p-values were obtained through separate Student’s t tests for independent groups. No corrections for multiple comparisons were introduced initially, but FWER and FDR adjusted p-values using the Benjamin-Hochberg 58 and Bonferroni-Dunn 59 methods, respectively, were also calculated (see Supplementary Tables 1A–5A).
Previous studies have shown that raw GMVOL are directly related to TIV12,14,16,42, and that the strength of these relationships (slope values of linear TIV-VOIraw regressions) is ordinally correlated (Spearman’s rho) with the size and p-values of the sex differences found in these VOIraw19. Conversely, VOIs adjusted (VOIadj) with appropriate methods no longer show a linear relationship with TIV, and the size and p-values of the sex differences in GMVOL are uncorrelated with the TIV-VOIadj slope values19. Therefore, in the present study, we employed the same regression-based approach to assess the efficacy of each TIV-adjustment method in eliminating the effects of TIV variation.
Replication of univariate sex differences
Following current recommendations23,25, effects’ replication was assessed by calculating Prediction Intervals (PIs). More specifically, appropriate PIs were calculated to assess to what extent the d values obtained in each dataset of the HCP sample replicated those previously observed in the same datasets of the UJI sample19. PIs estimate the range of values within which a parameter (e.g., Cohen’s d value) would fall in future replication studies if differences among studies were solely due to sampling error23,25. Thus, when a replication result falls outside the prediction interval, the results of the original study are not properly replicated, and it can be concluded that factors other than sampling error were operating to produce distinct results in each study.
PIs for the sex differences in GMVOL observed in the UJI sample were calculated with the predictionInterval package for R60. A second step was to identify whether each of these PIs captured the corresponding d value in the HCP sample (see25 for details). From these data, the percent of successfully replicated effects (replication rates) in each dataset was estimated and compared to the others. These comparisons were conducted by means of the χ2 test for independence, followed by appropriate dyadic comparisons using the pairwise tests of independence for nominal data from the rcompanion package for R61. All replicated effects were meta-analyzed with the metafor package for R62, hence obtaining weighted average d values and their corresponding CIs. From these new estimates, 95% PIs estimating the range of expected values of d at each VOI in possible future replication studies were also calculated (Supplementary Table 6).
Multivariate classification
To assess the effects of TIV-adjustment on the utility of the 116 VOIs defined by the AAL atlas in predicting sex categorically defined as male or female, we tested 12 supervised classification algorithms (see below) in the raw, scaling, proportions, PCP- and residuals-adjusted datasets. Moreover, to provide a reference point for judging the results obtained, the same analyses were repeated using TIV as a single predictor of sex. Before being used as predictors, all these variables were transformed into z-scores to avoid distortions due to their different ranges33,63.
Following current recommendations33,34, classification algorithms were fitted, tested, and validated in separate groups of participants with the same number of females and males (hence avoiding classification distortions due to between-class imbalance64,65). Thus, each algorithm was initially fitted in a randomly selected training subsample (311 females and 311 males) from the HCP-sample, internally validated34 in the testing subsample (the 133 females and 133 males hold-out participants from the HCP sample) and externally validated34 in the so-called external validation subsample (171 males and 171 females randomly extracted from the UJI-sample). The classifiers’ performance was primarily evaluated in terms of overall accuracy (percent of correctly classified cases and its 95% CI), although a standardized measure of the concordance between the predicted and actual sex of the participants in each sample (Cohen’s Kappa and its 95% CI) is also provided in Supplementary Table 7.
Instead of relying on the estimates provided by a single classifier, we opted to calculate, report, and compare the prediction accuracy rates obtained with 12 classification methods. It was important to test several methods because the predictive accuracy achieved by a particular classifier is very much dependent on whether or not the data characteristics satisfy the assumptions (e.g. normality, linearity…) under which the classifier operates33,66, and these data characteristics are likely to differ across the datasets compared in the present study or across samples from different studies. Described briefly, the classifiers tested were:
Linear discriminant analysis (LDA)
LDA has traditionally been the parametric method of reference for classification studies. LDA assumes normality and equality of variances/covariances33,67. In the present study, LDA was implemented using the default options of the rda function of the MASS package for R68.
Quadratic discriminant analysis (QDA)
QDA is a similar classification method to LDA, but (1) QDA does not assume a common covariance matrix; (2) QDA classification is based on quadratic decision boundaries; (3) QDA is more sensitive to small sample size (or n/ predictor ratios), and it presents greater variance but less bias than LDA33,69. In the present study, QDA was implemented using the default options of the qda function of the MASS package for R68.
K-nearest neighbors (KNN)
KNN is a simple but often powerful classifier that does not make any assumptions about the data distribution70. When K must be kept constant in order to compare several sets of predictors, it is customary to fix K as the square root of the number of subjects included in the training sample71. Therefore, in the present study, the K value was pre-established as K = 25 ( = 24.93), and the KNN classifier was implemented through the knn function of the class package for R68.
Logistic regression (LR)
LR was implemented using the glm function of the stats package for R. LR is a linear classification method similar to LDA, but it does not assume normality, and it is less sensitive to outlier effects, hence outperforming LDA when the normality assumption is severely violated72..
Penalized logistic regression with an elastic net (PLR-EN)
PLR-EN was implemented using the glmnet function of the glmnet package for R73. PLR-EN is a form of logistic regression that reduces the number of variables in the regression model by penalizing the coefficients of the variables that contribute less to the prediction, using an “elastic” criterion that sets some of these coefficients to exactly zero while merely shrinking other coefficients toward zero74. Compared to traditional LR procedures, PLR-EN often (but not always) exhibits reduced bias and increased predictive performance75.
Flexible discriminant analysis (FDA)
FDA can briefly be described as performing LDA in an enlarged feature space, usually showing much higher predictive accuracy than LDA33,76. In the present study, non-penalized FDA was implemented using the fda function of the mda package77, employing the adaptive additive-spline regression function of the BRUTO subroutine of this R package.
Tree-based classifiers
Classification trees do not make any strong assumptions about the data and they operate by segmenting the feature space into a number of non-overlapping regions through a recursive binary splitting process33,78. At the risk of overfitting, accuracy might be enhanced by aggregating a large number of decision trees into a single random forest, each of them using a limited subset of predictors (ordinarily, ). In the present study. a simple classification tree and a complex random forest (500 trees with 10 randomly selected predictors each) were implemented using the tree package for R79.
Artificial neuronal networks (ANN)
ANNs are very powerful but opaque learning algorithms that extract linear combinations of inputs as derived features, which in turn are used to non-linearly model the classification problem33,80. In the present study, a simple ANN was constructed by using the default specifications of the neuralnet package for R81.
Support-vector machines (SVMs)
SVMs is a generic name for a series of very flexible procedures that produce nonlinear classification boundaries by constructing linear boundaries into an enlarged feature space using all or just a fraction of the cases33,82. In the present study, the tune function (tenfold cross-validation) was used to automatically select the optimal values for the regularization (C; tested range: from 1–3 to 13) and kernel-width (γ; tested range: 0.0001, 0.001, 0.01, 0.1, 0,5, 1, 2, 3, 4, 5) parameters when building the SVMs with linear, radial, and polynomial (degree = 3) kernels, using the svm function of the e1071 package for R83.
To identify which datasets exhibited similar predictive performance across methods in the training, testing, and external validation subsamples, a robust outlier clustering method (the partitioning around medoids algorithm; PAM) was applied84. Thus, for each subsample, the PAM algorithm of the cluster package for R 85 was run four times, each time setting the number of clusters (K) to 2, 3, 4, or 5, respectively. The K value that maximized the average silhouette was considered the optimal number of clusters in each subsample (see 84 for further details). To provide a graphical representation of the clusters’ composition and the between-cluster dissimilarities in each subsample, three separate dendrograms were constructed with the dendextend package for R86 by subjecting the accuracy rates obtained in each dataset to a hierarchical cluster analysis (average linkage based on Euclidean distances) and then cutting them at appropriate heights to illustrate the clusters previously identified by the PAM algorithm. Of note, in all cases, between-cluster separation was at least fivefold larger than the average within-cluster dissimilarity, and all the obtained clusters only merged at above 50% of the maximum height of their dendrograms (see Supplementary Table 8). These observations indicate that the identified clusters are not a product of random variation, but rather they correspond to specific/ meaningful predictive performance profiles.
Ethics approval and consent to participate
This study was carried out in accordance with the recommendations of the ethical standards of the American Psychological Association. The protocol was approved by the Ethics Standards Committees of the Universitat Jaume I. In accordance with the Declaration of Helsinki, all subjects of the HCP and UJI samples gave written informed consent prior to participating.
Supplementary information
Acknowledgements
Data were provided [in part] by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. The authors also thank Dr. César Avila from Universitat Jaume I for kindly providing a second set of scan images used in this study (UJI sample). This research was supported by a grant (UJI B2017-05) awarded to CS-S. This funding source did not play any role in designing the study or in the collection, analysis, and interpretation of the data.
Abbreviations
- AAL
Automated anatomical labeling atlas
- ANN
Artificial neuronal net
- CI
Confidence interval
- F
Females
- FDA
Flexible discriminant analysis
- FDR
False discovery rate
- FWER
Family wise error rate
- GM
Gray matter
- GMPCP
Gray matter volumes adjusted with the power-corrected-proportions’ method
- GMprop
Gray matter volumes adjusted with the proportions’ method
- GMraw
Gray matter volumes of the raw dataset
- GMres
Gray matter volumes adjusted with the residuals’ method
- GMVOL
Gray matter volume
- HCP
Human connectome project
- KNN
K-nearest neighbors
- LDA
Linear discriminant analysis
- LR
Logistic regression
- M
Males
- MRI
Magnetic resonance imaging
- PAM
Partitioning around medoids algorithm.
- PCP
Power-corrected proportions
- PI
Prediction interval
- PLR-EN
Penalized logistic regression (elastic net)
- PS
Percent of superiority
- QDA
Quadratic discriminant analysis
- SVM
Support vector machine
- TIV
Total intracranial volume
- VBM
Voxel based morphometry
- VOI
Volume of interest
- VOIadj
Adjusted VOI
Author contributions
C.S.-S. and C.F. designed the study. N.A. and A.J.C.-G. processed the scan images on which C.S.-S. and M.V.I.-G. conducted the statistical analyses. C.S.-S. wrote the first draft of the manuscript. C.F. and M.V.I.-G. contributed to write several sections of the final version of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Data availability
This study was primarily conducted using data from the open source 1,200 Subject Release (S1200) of the Human Connectome Project (HCP). The access to this sample should be directly requested to the Washington University—University of Minnesota Consortium of the Human Connectome Project (WU-Minn HCP). The second sample used in this study (UJI sample) was kindly provided by Dr. César Ávila of Universitat Jaume I. Requests for accessing this second sample should be directly addressed to, and authorized by, Dr. César Ávila.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/29/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Supplementary information
is available for this paper at 10.1038/s41598-020-69361-9.
References
- 1.Hirtz D, et al. How common are the ‘common’ neurologic disorders? Neurology. 2007 doi: 10.1212/01.wnl.0000252807.38124.a3. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy MM. Incorporating sex as a variable in preclinical neuropsychiatric research. Schizophr. Bull. 2015 doi: 10.1093/schbul/sbv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zagni E, Simoni L, Colombo D. Sex and gender differences in central nervous system-related disorders. Neurosci. J. 2016 doi: 10.1155/2016/2827090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maney DL. Just like a circus: The public consumption of sex differences. Curr. Top. Behav. Neurosci. 2015 doi: 10.1007/7854_2014_339. [DOI] [PubMed] [Google Scholar]
- 5.O’Connor C, Joffe H. Gender on the brain: A case study of science communication in the new media environment. PLoS ONE. 2014 doi: 10.1371/journal.pone.0110830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters M, et al. Unsolved problems in comparing brain sizes in Homo sapiens. Brain Cogn. 1998 doi: 10.1006/brcg.1998.0983. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien LM, et al. Adjustment for whole brain and cranial size in volumetric brain studies: A review of common adjustment factors and statistical methods. Harvard Rev. Psychiatry. 2006 doi: 10.1080/10673220600784119. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien LM, et al. Statistical adjustments for brain size in volumetric neuroimaging studies: Some practical implications in methods. Psychiatry Res. Neuroimaging. 2011 doi: 10.1016/j.pscychresns.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonard CM, et al. Size matters: Cerebral volume influences sex differences in neuroanatomy. Cereb. Cortex. 2008 doi: 10.1093/cercor/bhn052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jäncke L, Mérillat S, Liem F, Hänggi J. Brain size, sex, and the aging brain. Hum. Brain Mapp. 2015 doi: 10.1002/hbm.22619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ardekani BA, Figarsky K, Sidtis JJ. Sexual dimorphism in the human corpus callosum: An MRI study using the OASIS brain database. Cereb. Cortex. 2013 doi: 10.1093/cercor/bhs253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes J, et al. Head size, age and gender adjustment in MRI studies: A necessary nuisance? Neuroimage. 2010 doi: 10.1016/j.neuroimage.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Ritchie SJ, et al. Sex differences in the adult human brain: Evidence from 5216 UK Biobank participants. Cereb. Cortex. 2018 doi: 10.1093/cercor/bhy109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu D, Johnson HJ, Long JD, Magnotta VA, Paulsen JS. The power-proportion method for intracranial volume correction in volumetric imaging analysis. Front. Neurosci. 2014 doi: 10.3389/fnins.2014.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordenskjöld R, et al. Intracranial volume normalization methods: Considerations when investigating gender differences in regional brain volume. Psychiatry Res. Neuroimaging. 2015 doi: 10.1016/j.pscychresns.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Pintzka CWS, Hansen TI, Evensmoen HR, Håberg AK. Marked effects of intracranial volume correction methods on sex differences in neuroanatomical structures: A HUNT MRI study. Front. Neurosci. 2015 doi: 10.3389/fnins.2015.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lüders E, Steinmetz H, Jäncke L. Brain size and grey matter volume in the healthy human brain. NeuroReport. 2002 doi: 10.1097/00001756-200212030-00040. [DOI] [PubMed] [Google Scholar]
- 18.Jäncke L, Staiger JF, Schlaug G, Huang Y, Steinmetz H. The relationship between corpus callosum size and forebrain volume. Cereb. Cortex. 1997 doi: 10.1093/cercor/7.1.48. [DOI] [PubMed] [Google Scholar]
- 19.Sanchis-Segura C, et al. Sex differences in gray matter volume: How many and how large are they really? Biol. Sex Differ. 2019 doi: 10.1186/s13293-019-0245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwaan RA, Etz A, Lucas RE, Donnellan MB. Making replication main stream. Behav. Brain Sci. 2017 doi: 10.1017/S0140525X17001972. [DOI] [PubMed] [Google Scholar]
- 21.Simons DJ. The value of direct replication. Perspect. Psychol. Sci. 2014 doi: 10.1177/1745691613514755. [DOI] [PubMed] [Google Scholar]
- 22.Asendorpf JB, et al. Recommendations for increasing replicability in psychology. Eur. J. Pers. 2013 doi: 10.1002/per.1919. [DOI] [Google Scholar]
- 23.Patil P, Peng RD, Leek JT. What should researchers expect when they replicate studies? A statistical view of replicability in psychological science. Perspect. Psychol. Sci. 2016 doi: 10.1177/1745691616646366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cumming G. Replication and p intervals: P values predict the future only vaguely, but confidence intervals do much better. Perspect. Psychol. Sci. 2008 doi: 10.1111/j.1745-6924.2008.00079.x. [DOI] [PubMed] [Google Scholar]
- 25.Spence JR, Stanley DJ. Prediction interval: What to expect when you’re expecting … A replication. PLoS ONE. 2016 doi: 10.1371/journal.pone.0162874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arndt S, Cohen G, Alliger RJ, Swayze VW, Andreasen NC. Problems with ratio and proportion measures of imaged cerebral structures. Psychiatry Res. Neuroimaging. 1991 doi: 10.1016/0925-4927(91)90031-K. [DOI] [PubMed] [Google Scholar]
- 27.Mathalon DH, Sullivan EV, Rawles JM, Pfefferbaum A. Correction for head size in brain-imaging measurements. Psychiatry Res. Neuroimaging. 1993 doi: 10.1016/0925-4927(93)90016-B. [DOI] [PubMed] [Google Scholar]
- 28.Sanfilipo, M. P., Benedict, R. H. B., Zivadinov, R. & Bakshi, R. Correction for intracranial volume in analysis of whole brain atrophy in multiple sclerosis: The proportion vs. residual method. Neuroimage, 10.1016/j.neuroimage.2004.03.037 (2004). [DOI] [PubMed]
- 29.Grissom, R. J. & Kim, J. J. Effect Sizes for Research: Univariate and Multivariate Applications, 2nd Edn (Routledge, Multivariate application tests, 2012). 10.4324/9780203803233
- 30.Del Giudice, M. Multivariate misgivings: Is D a valid measure of group and sex differences? Evolut. Psychol. (2013). [PubMed]
- 31.Del Giudice, M. Measuring sex differences and similarities. In Gender and Sexuality Development: Contemporary Theory and Research. (ed. VanderLaan, D.P., Wong, W. I.) (2019).
- 32.Kotsiantis SB, Zaharakis ID, Pintelas PE. Machine learning: A review of classification and combining techniques. Artif. Intell. Rev. 2006 doi: 10.1007/s10462-007-9052-3. [DOI] [Google Scholar]
- 33.Hastie, T., Tibshirani, R., Friedman, J. The Elements of Statistical Learning The Elements of Statistical LearningData Mining, Inference, and Prediction, 2nd Edn. Springer Series in Statistics (2009). 10.1007/978-0-387-84858-7
- 34.Bzdok D, Ioannidis JPA. Exploration, inference, and prediction in neuroscience and biomedicine. Trends Neurosci. 2019 doi: 10.1016/j.tins.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Rosenblatt J. Multivariate revisit to ‘sex beyond the genitalia’. Proc. Natl. Acad. Sci. USA. 2016 doi: 10.1073/pnas.1523961113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joel D, et al. Analysis of human brain structure reveals that the brain “types” typical of males are also typical of females, and vice versa. Front. Hum. Neurosci. 2018 doi: 10.3389/fnhum.2018.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson NE, et al. Machine learning of brain gray matter differentiates sex in a large forensic sample. Hum. Brain Mapp. 2018 doi: 10.1002/hbm.24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weis S, et al. Sex classification by resting state brain connectivity. Cereb. Cortex. 2019 doi: 10.1093/cercor/bhz129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Putten MJAM, Olbrich S, Arns M. Predicting sex from brain rhythms with deep learning. Sci. Rep. 2018 doi: 10.1038/s41598-018-21495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wachinger C, Golland P, Kremen W, Fischl B, Reuter M. BrainPrint: A discriminative characterization of brain morphology. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Del Giudice, M., Lippa, R. A., Puts, P. D. A. & Bailey, Drew H J. Bailey, Michael P. Schmitt, D. Mosaic Brains? A Methodological Critique of Joel et al. (2015). Online document. (2015). 10.13140/RG.2.1.1038.8566.
- 42.Voevodskaya, O. et al. The effects of intracranial volume adjustment approaches on multiple regional MRI volumes in healthy aging and Alzheimer’s disease. Front. Aging Neurosci.6 (2014). [DOI] [PMC free article] [PubMed]
- 43.Malone IB, et al. Accurate automatic estimation of total intracranial volume: A nuisance variable with less nuisance. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fjell AM, et al. Minute effects of sex on the aging brain: A multisample magnetic resonance imaging study of healthy aging and Alzheimer’s disease. J. Neurosci. 2009 doi: 10.1523/JNEUROSCI.0115-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chekroud AM, Ward EJ, Rosenberg MD, Holmes AJ. Patterns in the human brain mosaic discriminate males from females. Proc. Natl. Acad. Sci. U.S.A. 2016 doi: 10.1073/pnas.1523888113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foody GM. Sample size determination for image classification accuracy assessment and comparison. Int. J. Remote Sens. 2009 doi: 10.1080/01431160903130937. [DOI] [Google Scholar]
- 47.Joel D, et al. Sex beyond the genitalia: The human brain mosaic. Proc. Natl. Acad. Sci. 2015 doi: 10.1073/pnas.1509654112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poldrack RA, et al. Scanning the horizon: Towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci. 2017 doi: 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kriegeskorte N, Lindquist MA, Nichols TE, Poldrack RA, Vul E. Everything you never wanted to know about circular analysis, but were afraid to ask. J. Cereb. Blood Flow Metab. 2010 doi: 10.1038/jcbfm.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tzourio-Mazoyer N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002 doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 51.Evans AC, Janke AL, Collins DL, Baillet S. Brain templates and atlases. NeuroImage. 2012 doi: 10.1016/j.neuroimage.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 52.Van Essen DC, et al. The WU-Minn human connectome project: An overview. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurth, F., Luders, E. & Gaser, C. VBM8—toolbox manual. Funct. Imaging (2010).
- 54.Wasserstein RL, Lazar NA. The ASA’s statement on p-values: Context, process, and purpose. Am. Stat. 2016;70:129–133. doi: 10.1080/00031305.2016.1154108. [DOI] [Google Scholar]
- 55.American Psychological Association APA sixth edition. Am. Psychol. Assoc. 2010 doi: 10.1006/mgme.2001.3260. [DOI] [Google Scholar]
- 56.Baguley T. Standardized or simple effect size: What should be reported? Br. J. Psychol. 2009 doi: 10.1348/000712608X377117. [DOI] [PubMed] [Google Scholar]
- 57.Cumming G. The new statistics: Why and how. Psychol. Sci. 2014 doi: 10.1177/0956797613504966. [DOI] [PubMed] [Google Scholar]
- 58.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 2018;57:289–300. [Google Scholar]
- 59.Dunn OJ. Multiple comparisons among means. J. Am. Stat. Assoc. 1961 doi: 10.1080/01621459.1961.10482090. [DOI] [Google Scholar]
- 60.Stanley, D. predictionInterval: Prediction Interval Functions for Assessing Replication Study Results. R package version 1.0.0. (2016).
- 61.Mangiafico, S. rcompanion: Functions to Support Extension Education Program Evaluation. R package version 2.2.2. (2019).
- 62.Viechtbauer, W. Conducting meta-analyses in R with the metafor. J. Stat. Softw. (2010).
- 63.Ali, S. & Smith-Miles, K. A. Improved support vector machine generalization using normalized input space. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) (2006). 10.1007/11941439-40.
- 64.Ali, A., Shamsuddin, S. M. & Ralescu, A. L. Classification with class imbalance problem: A review. Int. J. Adv. Soft Comput. Appl. (2015).
- 65.García, V., Sánchez, J. S., Mollineda, R. A. & Sotoca, R. A. J. M. The class imbalance problem in pattern classification and learning. Data Eng. (2007).
- 66.Kiang MY. A comparative assessment of classification methods. Decis. Support Syst. 2003 doi: 10.1016/S0167-9236(02)00110-0. [DOI] [Google Scholar]
- 67.Moore, B. A. & McLachlan, G. J. Discriminant analysis and statistical pattern recognition. J. R. Stat. Soc. Ser. A (Statistics Soc.) (1994). 10.2307/2983518
- 68.Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S 4th Edn(World, 2002). 10.2307/2685660
- 69.Bose S, Pal A, Saharay R, Nayak J. Generalized quadratic discriminant analysis. Pattern Recognit. 2015 doi: 10.1016/j.patcog.2015.02.016. [DOI] [Google Scholar]
- 70.Hu Q, Yu D, Xie Z. Neighborhood classifiers. Expert Syst. Appl. 2008 doi: 10.1016/j.eswa.2006.10.043. [DOI] [Google Scholar]
- 71.Lantz, B. Machine Learning with R 2nd Edn. Machine Learning with R (2015). 10.1007/978-981-10-6808-9
- 72.Holden JE, Finch WH, Kelley K. A comparison of two-group classification methods. Educ. Psychol. Meas. 2011 doi: 10.1177/0013164411398357. [DOI] [Google Scholar]
- 73.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010 doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zou, H. & Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. (2005). 10.1111/j.1467-9868.2005.00503.x
- 75.De Mol C, De Vito E, Rosasco L. Elastic-net regularization in learning theory. J. Complex. 2009 doi: 10.1016/j.jco.2009.01.002. [DOI] [Google Scholar]
- 76.Hastie T, Tibshirani R, Buja A. Flexible discriminant analysis by optimal scoring. J. Am. Stat. Assoc. 1994 doi: 10.1080/01621459.1994.10476866. [DOI] [Google Scholar]
- 77.Friedrich Leisch, K. H. and & Ripley., B. D. mda: Mixture and Flexible Discriminant Analysis. R package version 0.4-10. (2017).
- 78.Breiman, L., Friedman, J. H., Olshen, R. A. & Stone, C. J. Classification and Regression Trees (Routledge, Multivariate application tests, 2017). 10.1201/9781315139470
- 79.Ripley., B. D. tree: Classification and Regression Trees. R package version 1.0-40. (2019).
- 80.Tavanaei A, Ghodrati M, Kheradpisheh SR, Masquelier T, Maida A. Deep learning in spiking neural networks. Neural Networks. 2019 doi: 10.1016/j.neunet.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 81.Fritsch, S., Guenther, F. & Wright, M. N. neuralnet: Training of Neural Networks. R package version 1.44.2. (2019).
- 82.Scholkopf, B. & Smola, A. J. Learning with Kernels: Support Vector Machines, Regularization. Optimization, and Beyond. (2001). 10.1198/jasa.2003.s269
- 83.Meyer, D. et al. e1071: Miscelaneous Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071). (2019).
- 84.Kaufman, L. & Rousseeuw, P. J. Finding Groups in Data: An Introduction to Cluster Analysis (Wiley Series in Probability and Statistics). Eepe.Ethz.Ch (1990). 10.1007/s13398-014-0173-7.2
- 85.Maechler, M., Rousseeuw, P., Struyf, A., Hubert, M., Hornik, K. cluster: Cluster Analysis Basics and Extensions. R package version 2.1.0. (2019).
- 86.Galili T. dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study was primarily conducted using data from the open source 1,200 Subject Release (S1200) of the Human Connectome Project (HCP). The access to this sample should be directly requested to the Washington University—University of Minnesota Consortium of the Human Connectome Project (WU-Minn HCP). The second sample used in this study (UJI sample) was kindly provided by Dr. César Ávila of Universitat Jaume I. Requests for accessing this second sample should be directly addressed to, and authorized by, Dr. César Ávila.