Abstract
The term machine learning refers to a collection of tools used for identifying patterns in data. As opposed to traditional methods of pattern identification, machine learning tools relies on artificial intelligence to map out patters from large amounts of data, can self-improve as and when new data becomes available and is quicker in accomplishing these tasks. This review describes various techniques of machine learning that have been used in the past in the prediction, detection and management of infectious diseases, and how these tools are being brought into the battle against COVID-19. In addition, we also discuss their applications in various stages of the pandemic, the advantages, disadvantages and possible pit falls.
Keywords: COVID 19, Artificial intelligence, Machine learning, Outcome prediction
Introduction
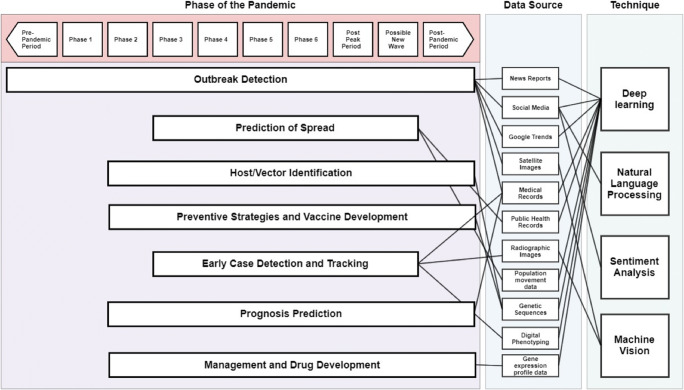
As the global population grows, even though we are still battling some of the pathogens that have been with us since the advent of known human history such as tuberculosis, we are also witnessing a trend in increasing emergence of novel pathogens from non-human hosts, and this is posing a major threat to the public health [1]. Within the last ten years, we witnessed the emergence of new viruses that could potentially spread across international borders and wreak global havoc, the latest of this being the novel coronavirus (COVID-19). The recent development of machine learning-based tools for healthcare providers allows novel ways to combat such global pandemics. The term machine learning encompasses the collection of tools and techniques for identifying patterns in data [2]. In traditional methods of identifying patterns from data, we approach the system with our presumptions as to which components of the data (age, sex, pre-existing conditions) affect the outcome of interest (patient survival). However, in machine learning, we provide data and the machine identifies trends and patterns, enabling us to formulate a model to predict the outcome of patients. The authors will attempt to provide a narrative review of such tools, how they are useful in healthcare, and how they are being utilized in the prediction, prevention, and management of COVID-19. We will also include discussions on such tools used in past infectious diseases such as the SARS-CoV-1 and MERS-CoV viruses and how they may be translatable to COVID-19. The discussion is structured under the techniques for outbreak detection, prediction models for spread, prevention and vaccine development, early case detection and tracking, prognosis prediction of affected patients, and drug development (Fig. 1).
Fig. 1.
Machine learning tools, data sources and the interventions that are helpful in different stages of a pandemic. Phase 1: Animal influenza virus has not been known to create diseases in the humans. Phase 2: An animal influenza virus has been known to cause disease in humans and is hence a potential pandemic threat. Phase 3: An animal influenza virus has been known to cause solitary or a cluster of diseases in humans but has not created human-to-human transmission. Phase 4: Human to human transmission, sufficient to maintain community level outbreak has been identified. Phase 5: The virus has created sustained community level outbreak in two countries within the same WHO region. Phase 6: In addition to Phase 5, the virus has created sustained community level outbreak in at least one country in another WHO region. Post Peak Period: Level of pandemic in most countries have dropped below peak levels. Possible New Wave: Level of pandemic in most countries is rising again. Post-Pandemic Period: Level of pandemic has returned to seasonal influenza level in most countries
Outbreak detection
Biosurveillance is the science of early detection and prevention of a disease outbreak in the community [3]. Analytics, machine learning, and natural language processing (NLP) are being increasingly used in biosurveillance [4]. Scanning social media, news reports, and other online data can be used to detect localized disease outbreaks before they even reach the level of pandemics [5]. The Canadian company Blue Dot successfully used machine learning algorithms to detect early outbreaks of COVID-19 in Wuhan, China by the end of December 2019 [6, 7]. Big data analysis of medical records, as well as satellite imaging (eg: cars crowding around a hospital), are some other ways big data analysis has been used in the past to detect localized outbreaks [8, 9]. Google Trends has been used in the past to detect the outbreak of Zika virus infections in populations, using dynamic forecasting models [10]. Sentiment analysis is the technique of using natural language processing in social media to understand the positive and negative emotions of the population [11]. Unsupervised sentiment analysis has been proposed as a method for the early detection of infectious diseases in the population [12]. Also, sentiment analysis could be a valuable tool to understand the public’s reactions or overreactions thereof, towards disease outbreaks, and can provide valuable insights to the government in directing efforts towards public education [13–15]. These techniques of sentinel biosurveillance would help detect pandemics before they become one and can provide valuable time for the health system to prepare for prevention and management.
Prediction of spread
Various statistical, mathematical and dynamic predictive modeling has been used to successfully predict the extent and spread of infectious diseases through the population [16–21]. As opposed to traditional epidemiological predictive models, big-data-driven models have the added advantage of adaptive learning, trend-based recalibration, flexibility and scope to improve based on a newer understanding of the disease process, as well as estimation of the impact of the interventions, such as social distancing, in curbing its spread [22]. The most common is the Susceptible-Exposed-Infectious-Recovered (SEIR) modeling method which is now being used to predict the areas and extent of COVID-19 spread [23, 24]. These techniques can also be used to determine other parameters of the epidemic, such as under-reporting of cases, the effectiveness of interventions, and the accuracy of testing methods [25, 26]. For example, a modeling algorithm attempted to simulate the conditions in which Ebola could spread in the Chinese society, and the effectiveness of the four levels of governmental interventions was evaluated in such conditions [27]. Similar models have also attempted to predict the outbreak and expansion of the Zika virus in real-time in the Americas and were determined to have close to 85% accuracy in quantitative evaluations [28]. An attempt at validating different machine learning algorithms determined that backward propagation neural network (BPNN) demonstrated highest predictive accuracy is modelling Zika virus transmission [29].
Scientists at the Johns Hopkins University developed a COVID-19 prediction modeling based on a previously published stochastic metapopulation epidemic model [30]. A comparison of the prediction of this modeling with real-life data elucidated the lacunae in the understanding of the virus’s dynamics and the model’s limitations [31, 32].
However, a predictive model is only as good as the data it is based on, and in the event of a global pandemic, data sharing across communities is of paramount importance. This was one of the major obstacles in learning about and modeling the 2013–2016 Ebola virus outbreak [33]. The World Health Organization (WHO) has proposed a consensus on expedited data sharing on the COVID-19 outbreak to promote inter-community learning and analytics in this area.
Preventive strategies and vaccine development
Artificial Neural Networks (ANN) were used to predict antigenic regions with a high density of binders (antigenic hotspots) in the viral membrane protein of Severe Acute Respiratory Syndrome Coronavirus (SARS–CoV) [34, 35]. This information is critical to the development of vaccines. Using machine learning for this purpose allows for rapid scanning of the entire viral proteome, allowing faster and cheaper vaccine development. Reverse vaccinology and machine learning were successfully employed to identify six potential vaccine target proteins in the SARS-CoV-2 proteome [36]. Machine learning has also been used in the past to predict the strains of influenza virus that are more likely to cause infection in a population in an upcoming year, and in turn, should constitute the year’s seasonal influenza vaccine. Successful prediction of the future expansion of small subtrees of hemagglutinins (HA) part of the viral antigenic set was possible from training H3N2 and testing on H1N1, using reconstructed timed phylogenetic tree [37]. Machine learning can also be used to predict the hosts of newly discovered viruses based on analysis of nucleoprotein gene sequences and spike gene sequences, and can be a useful additional tool for tracing back viral origins, especially when the data set is large and comparative analysis is difficult or time-consuming [38].
Early case detection and tracking
Early case identification, quarantining, and preventing exposure to the communities are crucial pillars in managing an epidemic such as COVID-19. Mobile phone-based surveys can be useful in early identification of cases, especially in quarantined populations [39, 40]. Such methods have shown success in Italy in identifying influenza patients through a web-based survey [41]. As opposed to traditional methods of survey and analysis, the use of artificial intelligence tools can be used to collect and analyze large amounts of data, identify trends, stratify patients based on risk, and propose solutions to population instead of the individual. Digital phenotyping is the novel concept of collecting smartphone-based active (surveys) and passive (text, voice, location, screen use) data to produce an individual phenotype [42, 43]. This technique can be used to obtain multiple data points and allow stratifying individuals based on their risk. The government of India recently launched a mobile application called “Aarogya Setu” which tracks its users’ exposure to potentially COVID-19 infected patients, using the Bluetooth functionality to scan the surrounding area for other smartphone users. If a patient is tested positive, then the data from the mobile application can be used to track down every app-user who the patient encountered, within the last 30 days [44]. Such techniques of digital phenotyping can be performed even on entry-level smartphones and would be especially useful in low and middle-income countries as a cost-effective method of risk stratification, due to the ubiquitous smartphone availability [45].
The close physical and economic proximity with China should have resulted in high morbidity and mortality due to COVID-19 in Taiwan. However, with the help of machine learning, they were able to bring the number of infected patients to far lower than what was initially predicted. They identified the threat early, mobilized their national health insurance database, and customs and immigration database to generate big data for analytics. Machine learning on this big data helped them stratify their population into lower risk or higher risk based on several factors, including travel history. Persons with higher risk were quarantined at home and were tracked through their mobile phones to ensure that they remained in quarantine. This application of big data, in addition to active case finding efforts, ensured that their case numbers were far fewer than what was initially anticipated [46].
Deep learning algorithms have also been used to identify patterns of infectious disease involvement in imaging results such as CT and MRI. With CT scanning showing high correlations to PCR-positive COVID patients, such algorithms have shown great promise in their ability to detect findings consistent with COVID-19 in CT images of patients [47–49].
Prognosis prediction
Machine learning algorithms have been used previously to predict prognosis in patients affected by the MERS Co-V infection [50]. The patient’s age, disease severity on presentation to the healthcare facility, whether the patient was a healthcare worker, and the presence of pre-existing co-morbidities were the four factors that were identified to be the major predictors in the patient’s recovery. These findings are consistent with the currently observed trends in the COVID-19 disease [51, 52]. Using the data visualization tool Mirador, a mobile application Ebola CARE (Computational Assignment of Risk Estimates) was developed to predict a patient’s outcome after being infected with Ebola [53, 54]. The tool identified 24 clinical and laboratory parameters that possibly affect a patient’s prognosis. There is a need for adaptation of these algorithms to assist physicians in their decision-making process while managing COVID-19. Recovery prediction tools help determine resource allocation, triage, treatment determination, as well as health system preparedness.
Treatment development
Machine learning tools have been used in drug development, drug testing, as well as drug repurposing. They enable us to interpret large gene expression profile data sets to suggest new uses for currently available medications. Deep generation models, also known as AI imagination, can design novel therapeutic agents with possible desired activity [55]. These tools help reduce the cost and time of developing drugs, help in developing novel therapeutic agents, as well as predict possible off-label uses for some therapeutic agents [56]. Bayesian Machine Learning tools have been used to develop drugs against Ebola in in-vitro settings and the findings translated well to in-vivo settings as well [57].
Conclusion
Machine learning provides an exciting array of tools that are flexible enough to allow their deployment in any stage of the pandemic. With the large amount of data that is being generated while studying a disease process, machine learning allows for analysis and rapid identification of patterns that traditional mathematical and statistical tools would take a long time to derive. The flexibility, ability to adapt based on a new understanding of the disease process, self-improvement as and when new data becomes available, and the lack of human prejudice in the approach of analysis makes machine learning a highly versatile novel tool for managing novel infections. However, with such an enhanced ability to derive meaning from large amounts of data, there is a greater demand for higher quality control during the collection, storage, and processing of the data. Besides, standardization of data structures across populations would allow these systems to adapt and learn from data across the globe, which wasn’t possible in the past, and is even more important in learning and managing a global pandemic like COVID-19. In addition, with a new pandemic, there tends to be a lot more “noise” in the data, and hence blindly feeding this immature data, which is ridden with outliers into an AI algorithm should always be approached with caution.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animal performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Mobile & Wireless Health
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Agam Bansal and Rana Prathap Padappayil contributed equally to this manuscript.
Contributor Information
Agam Bansal, Email: agambansal7@gmail.com.
Rana Prathap Padappayil, Email: ranaprathappadappayil@gmail.com.
Chandan Garg, Email: olf.chandan@gmail.com.
Anjali Singal, Email: anjali_singal@rediffmail.com.
Mohak Gupta, Email: xmohakgupta@gmail.com.
Allan Klein, Email: kleina@ccf.org.
References
- 1.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–3. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiens J, Shenoy ES. Machine Learning for Healthcare: On the verge of a major shift in healthcare epidemiology. Clin. Infect Dis. Off Publ. Infect Dis. Soc. Am. 2018;66(1):149–53. doi: 10.1093/cid/cix731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen KC. Chapter 13 - Applications: Biosurveillance, biodefense, and biotechnology. In: Horney JA, editor. Disaster epidemiology [Internet]. Academic Press; 2018 [cited 2020 Mar 29]. p. 143–51. Available from: http://www.sciencedirect.com/science/article/pii/B9780128093184000198
- 4.Jalal A. Analytics, Machine Learning & NLP – use in BioSurveillance and Public Health practice. Online J Public Health Inform [Internet]. 2015 May 7 [cited 2020 Mar 29];7(1). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4512490/
- 5.Al-Garadi MA, Khan MS, Varathan KD, Mujtaba G, Al-Kabsi AM. Using online social networks to track a pandemic: A systematic review. J. Biomed. Inform. 2016;62:1–11. doi: 10.1016/j.jbi.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 6.McCall B. COVID-19 and artificial intelligence: protecting health-care workers and curbing the spread. Lancet Digit Health. 2020;2(4):e166–7. doi: 10.1016/S2589-7500(20)30054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogoch II, Watts A, Thomas-Bachli A, Huber C, Kraemer MUG, Khan K. Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J. Travel. Med. [Internet]. 2020 Mar 13 [cited 2020 Apr 26];27(2). Available from: https://academic.oup.com/jtm/article/27/2/taaa008/5704418 [DOI] [PMC free article] [PubMed]
- 8.Funk S, Camacho A, Kucharski AJ, Eggo RM, Edmunds WJ. Real-time forecasting of infectious disease dynamics with a stochastic semi-mechanistic model. Epidemics. 2018;22:56–61. doi: 10.1016/j.epidem.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson MA, Apfeldorf KM, Dobson S, Devita J, Buczak AL, Baugher B, et al. An open challenge to advance probabilistic forecasting for dengue epidemics. Proc. Natl. Acad. Sci. 2019;116(48):24268–74. doi: 10.1073/pnas.1909865116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teng Y, Bi D, Xie G, Jin Y, Huang Y, Lin B, et al. Dynamic Forecasting of Zika Epidemics Using Google Trends. PLoS One. 2017;12(1):e0165085. doi: 10.1371/journal.pone.0165085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gohil S, Vuik S, Darzi A. Sentiment analysis of health care tweets: review of the methods used. JMIR Public Health Surveill [Internet]. 2018 Apr 23 [cited 2020 Mar 29];4(2). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5938573/ [DOI] [PMC free article] [PubMed]
- 12.Lim S, Tucker CS, Kumara S. An unsupervised machine learning model for discovering latent infectious diseases using social media data. J. Biomed. Inform. 2017;66:82–94. doi: 10.1016/j.jbi.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Choi S, Lee J, Kang M-G, Min H, Chang Y-S, Yoon S. Large-scale machine learning of media outlets for understanding public reactions to nation-wide viral infection outbreaks. Methods San Diego Calif. 2017;01(129):50–9. doi: 10.1016/j.ymeth.2017.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wirz CD, Xenos MA, Brossard D, Scheufele D, Chung JH, Massarani L. Rethinking social amplification of risk: Social media and Zika in three languages. Risk Anal. Off Publ. Soc. Risk. Anal. 2018;38(12):2599–624. doi: 10.1111/risa.13228. [DOI] [PubMed] [Google Scholar]
- 15.Gui X, Wang Y, Kou Y, Reynolds TL, Chen Y, Mei Q, et al. Understanding the Patterns of Health Information Dissemination on Social Media during the Zika Outbreak. AMIA Annu. Symp. Proc. AMIA Symp. 2017;2017:820–9. [PMC free article] [PubMed] [Google Scholar]
- 16.Chae S, Kwon S, Lee D. Predicting Infectious disease using deep learning and big data. Int. J. Environ. Res. Public Health [Internet]. 2018 Aug [cited 2020 Mar 28];15(8). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6121625/ [DOI] [PMC free article] [PubMed]
- 17.Venna SR, Tavanaei A, Gottumukkala RN, Raghavan VV, Maida AS, Nichols S. A Novel Data-Driven Model for Real-Time Influenza Forecasting. IEEE Access. 2019;7:7691–701. [Google Scholar]
- 18.Riad MH, Sekamatte M, Ocom F, Makumbi I, Scoglio CM. Risk assessment of Ebola virus disease spreading in Uganda using a two-layer temporal network. Sci. Rep. 2019;9(1):1–17. doi: 10.1038/s41598-019-52501-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kucharski AJ, Russell TW, Diamond C, Liu Y, Edmunds J, Funk S, et al. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect. Dis. [Internet]. 2020 Mar 11 [cited 2020 Mar 28];0(0). Available from: https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(20)30144-4/abstract [DOI] [PMC free article] [PubMed]
- 20.Gambhir M, Bozio C, O’Hagan JJ, Uzicanin A, Johnson LE, Biggerstaff M, et al. Infectious disease modeling methods as tools for informing response to novel influenza viruses of unknown pandemic potential. Clin Infect Dis Off Publ Infect Dis Soc Am. 2015;60(Suppl 1):S11–19. doi: 10.1093/cid/civ083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen SA, Redd SC. Using results from infectious disease modeling to improve the response to a potential H7N9 influenza pandemic. Clin. Infect Dis. Off Publ. Infect. Dis. Soc. Am. 2015;60(Suppl 1):S9–10. doi: 10.1093/cid/civ090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koo JR, Cook AR, Park M, Sun Y, Sun H, Lim JT, et al. Interventions to mitigate early spread of SARS-CoV-2 in Singapore: a modelling study. Lancet Infect. Dis. 2020 Mar 23; [DOI] [PMC free article] [PubMed]
- 23.Binti Hamzah F, Lau C, Nizri H, Ligot D, Lee G, Tan C, et al. CoronaTracker: Worldwide COVID-19 Outbreak Data Analysis and Prediction. Bull. World Health Organ. 2020 Mar 19;
- 24.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–97. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao S, Musa SS, Lin Q, Ran J, Yang G, Wang W, et al. Estimating the Unreported Number of Novel Coronavirus (2019-nCoV) Cases in China in the First Half of January 2020: A Data-Driven Modelling Analysis of the Early Outbreak. J. Clin. Med. 2020;9(2):388. doi: 10.3390/jcm9020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karako K, Song P, Chen Y, Tang W. Analysis of COVID-19 infection spread in Japan based on stochastic transition model. Biosci. Trends. 2020;advpub. [DOI] [PubMed]
- 27.Zhang P, Chen B, Ma L, Li Z, Song Z, Duan W, et al. The large scale machine learning in an artificial society: Prediction of the ebola outbreak in beijing [Internet]. Vol. 2015, Computational Intelligence and Neuroscience. Hindawi; 2015 [cited 2020 Mar 29]. p. e531650. Available from: https://www.hindawi.com/journals/cin/2015/531650/ [DOI] [PMC free article] [PubMed]
- 28.Akhtar M, Kraemer MUG, Gardner LM. A dynamic neural network model for predicting risk of Zika in real time. BMC Med. 2019;17(1):171. doi: 10.1186/s12916-019-1389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang D, Hao M, Ding F, Fu J, Li M. Mapping the transmission risk of Zika virus using machine learning models. Acta Trop. 2018;185:391–9. doi: 10.1016/j.actatropica.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 30.Zlojutro A, Rey D, Gardner L. A decision-support framework to optimize border control for global outbreak mitigation. Sci. Rep. 2019;9(1):1–14. doi: 10.1038/s41598-019-38665-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. [Internet]. 2020 Feb 19 [cited 2020 Mar 29];0(0). Available from: https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(20)30120-1/abstract [DOI] [PMC free article] [PubMed]
- 32.ncov-model-2 – CSSE [Internet]. [cited 2020 Mar 29]. Available from: https://systems.jhu.edu/research/public-health/ncov-model-2/
- 33.Modjarrad K, Moorthy VS, Millett P, Gsell P-S, Roth C, Kieny M-P. Developing Global Norms for Sharing Data and Results during Public Health Emergencies. PLoS Med. 2016;13(1):e1001935. doi: 10.1371/journal.pmed.1001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang GL, Khan AM, Srinivasan KN, August JT, Brusic V. Neural models for predicting viral vaccine targets. J. Bioinforma. Comput. Biol. 2005;03(05):1207–25. doi: 10.1142/s0219720005001466. [DOI] [PubMed] [Google Scholar]
- 35.Soam SS, Bhasker B, Mishra BN. Improved prediction of MHC class I binders/non-binders peptides through artificial neural network using variable learning rate: SARS corona virus, a case study. Adv. Exp. Med. Biol. 2011;696:223–9. doi: 10.1007/978-1-4419-7046-6_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ong E, Wong MU, Huffman A, He Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. bioRxiv. 2020 Mar 21;2020.03.20.000141. [DOI] [PMC free article] [PubMed]
- 37.Hayati M, Biller P, Colijn C. Predicting the short-term success of human influenza virus variants with machine learning. Proc. Biol. Sci. 2020;287(1924):20200319. doi: 10.1098/rspb.2020.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Sun F. Comparative studies of alignment, alignment-free and SVM based approaches for predicting the hosts of viruses based on viral sequences. Sci. Rep. 2018;8(1):1–9. doi: 10.1038/s41598-018-28308-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao ASRS, Vazquez JA. Identification of COVID-19 Can be quicker through artificial intelligence framework using a mobile phone-based survey in the populations when cities/towns are under quarantine. Infect. Control Hosp. Epidemiol. 2020;3:1–18. doi: 10.1017/ice.2020.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toninelli D, Toninelli D, Pinter R, Pedraza P de, Pinter R, Pedraza P de. Mobile research methods. Ubiquity press [Internet]. 2015 Sep 25 [cited 2020 Mar 29]; Available from: https://www.ubiquitypress.com/site/chapters/10.5334/bar.c/
- 41.Paolotti D, Carnahan A, Colizza V, Eames K, Edmunds J, Gomes G, et al. Web-based participatory surveillance of infectious diseases: the Influenzanet participatory surveillance experience. Clin. Microbiol Infect Off Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2014;20(1):17–21. doi: 10.1111/1469-0691.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Insel TR. Digital phenotyping: Technology for a new science of behavior. JAMA. 2017;318(13):1215–6. doi: 10.1001/jama.2017.11295. [DOI] [PubMed] [Google Scholar]
- 43.Onnela J-P, Rauch SL. Harnessing smartphone-based digital phenotyping to enhance behavioral and mental health. Neuropsychopharmacology. 2016;41(7):1691–6. doi: 10.1038/npp.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aarogya Setu - Apps on Google Play [Internet]. [cited 2020 Apr 10]. Available from: https://play.google.com/store/apps/details?id=nic.goi.aarogyasetu&hl=en_US
- 45.Bastawrous A, Armstrong MJ. Mobile health use in low- and high-income countries: an overview of the peer-reviewed literature: J. R. Soc. Med. [Internet]. 2013 Apr 1 [cited 2020 Mar 29]; Available from: https://journals.sagepub.com/doi/10.1177/0141076812472620 [DOI] [PMC free article] [PubMed]
- 46.Wang CJ, Ng CY, Brook RH. Response to COVID-19 in Taiwan: Big data analytics, new technology, and proactive testing. JAMA [Internet]. 2020 Mar 3 [cited 2020 Mar 29]; Available from: https://jamanetwork.com/journals/jama/fullarticle/2762689 [DOI] [PubMed]
- 47.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). medRxiv. 2020 Mar 11;2020.02.14.20023028. [DOI] [PMC free article] [PubMed]
- 48.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of chest CT and RT-PCR Testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 Cases. Radiology. 2020;26:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, et al. Rapid ai development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning CT image analysis. ArXiv200305037 Cs [Internet]. 2020 Mar 24 [cited 2020 Mar 29]; Available from: http://arxiv.org/abs/2003.05037
- 50.John M, Shaiba H. Main factors influencing recovery in MERS Co-V patients using machine learning. J. Infect. Public Health. 2019;12(5):700–4. doi: 10.1016/j.jiph.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goh KJ, Choong MC, Cheong EH, Kalimuddin S, Duu Wen S, Phua GC, et al. Rapid progression to acute respiratory distress syndrome: Review of current understanding of critical illness from COVID-19 Infection. Ann. Acad. Med. Singap. 2020;49(1):1–9. [PubMed] [Google Scholar]
- 52.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet [Internet]. 2020 Mar 11 [cited 2020 Mar 25];0(0). Available from: https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30566-3/abstract [DOI] [PMC free article] [PubMed]
- 53.Mirador [Internet]. [cited 2020 Mar 29]. Available from: http://andrescolubri.net/projects/mirador
- 54.Colubri A, Silver T, Fradet T, Retzepi K, Fry B, Sabeti P. Transforming clinical data into actionable prognosis models: Machine-learning framework and field-deployable app to predict outcome of ebola patients. PLoS Negl. Trop. Dis. [Internet]. 2016 Mar 18 [cited 2020 Mar 29];10(3). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4798608/ [DOI] [PMC free article] [PubMed]
- 55.Zhavoronkov A. Artificial intelligence for drug discovery, biomarker development, and generation of novel chemistry. Mol. Pharm. [Internet]. 2018 Oct 1 [cited 2020 Apr 11]; Available from: https://pubs.acs.org/doi/full/10.1021/acs.molpharmaceut.8b00930 [DOI] [PubMed]
- 56.Réda C, Kaufmann E, Delahaye-Duriez A. Machine learning applications in drug development. Comput Struct Biotechnol J. 2020;18:241–52. doi: 10.1016/j.csbj.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anantpadma M, Lane T, Zorn KM, Lingerfelt MA, Clark AM, Freundlich JS, et al. Ebola Virus Bayesian Machine Learning Models Enable New in Vitro Leads. ACS Omega. 2019;4(1):2353–61. doi: 10.1021/acsomega.8b02948. [DOI] [PMC free article] [PubMed] [Google Scholar]