Abstract
Japanese encephalitis virus (JEV), a major cause of Japanese encephalitisis, is an arbovirus that belongs to the genus Flavivirus of the family Flaviviridae. Currently, there is no effective drugs available for the treatment of JEV infection. Therefore, it is important to establish efficient antiviral screening system for the development of antiviral drugs. In this study, we constructed a full-length infectious clone of eGFP-JEV reporter virus by inserting the eGFP gene into the capsid-coding region of the viral genome. The reporter virus RNA transfected-BHK-21 cells generated robust eGFP fluorescence signals that were correlated well with viral replication. The reporter virus displayed growth kinetics similar to wild type (WT) virus although replicated a little slower. Using a known JEV inhibitor, NITD008, we demonstrated that the reporter virus could be used to identify inhibitors against JEV. Furthermore, an eGFP-JEV-based high throughput screening (HTS) assay was established in a 96-well format and used for screening of 1443 FDA-approved drugs. Sixteen hit drugs were identified to be active against JEV. Among them, five compounds which are lonafarnib, cetylpyridinium chlorid, cetrimonium bromide, nitroxoline and hexachlorophene, are newly discovered inhibitors of JEV, providing potential new therapies for treatment of JEV infection.
Keywords: Japanese encephalitis virus, Reporter virus, High-throughput system, Antiviral drugs
Highlights
-
•
The eGFP-JEV reporter virus was established.
-
•
The eGFP-JEV -based high throughput screening (HTS) assay was developed with a calculated Z′ factor of >0.5.
-
•
A library of 1443 FDA-approved drugs was screened to determine their ability to inhibit JEV infection using this HTS assay.
-
•
5 novel hit drugs were identified to be active against JEV infection.
1. Introduction
Japanese encephalitis virus (JEV) is a mosquito-borne virus that belongs to the genus Flavivirus in the family Flaviviridae, which includes many other important human pathogens such as yellow fever virus (YFV), West Nile virus (WNV), tick-borne encephalitis virus (TBEV) and dengue virus (DENV) (Buescher et al., 1959; Ellis et al., 2000). JEV is mainly epidemic in the Asia Pacific region, putting more than 3 billion people at the risk of JEV infection (Turtle and Driver, 2018). It is estimated that more than 67,900 Japanese encephalitis cases caused by JEV occur worldwide each year and the mortality rate is as high as about 20%–30% (Campbell et al., 2011; Griffiths et al., 2014; Hills et al., 2010). Although the use of live-attenuated vaccine has greatly reduced the incidence of JEV infection, there is theoretically a risk of possible reversion toinfectivity. So far, no clinically approved antiviral drugs or therapies are available for treatment of JEV infection. Therefore, it is important to develop antiviral agents to control the virus infection.
JEV genome is a single-stranded plus-sense RNA that is approximately 11-kb in length, containing a single open reading frame (ORF) flanked by untranslated regions (UTRs) at the 5′ and 3′ ends. The ORF encodes a polyprotein which is cleaved by viral and host proteases into three structural proteins (capsid [C], precursor membrane or membrane [preM/M] and envelope [E]) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Sampath and Padmanabhan, 2009). The 5′ and 3′ UTRs form various highly conserved secondary structures involved in the replication, translation and packaging of viral genome. The structural proteins assemble into viral particles (Li et al., 2008; Roby et al., 2015), while the non-structural proteins play essential roles in genome replication, viral particle assemble and immune escape (Brinton, 2013; Morrison et al., 2012; Murray et al., 2008; Nikonov et al., 2013; Shi, 2014).
There are usually two strategies for high-throughput screening (HTS) of antiviral drugs. The first one is to screen active molecules targeting specific viral proteins through in vitro functional assays. For flaviviruses, the non-structural proteins NS3, NS5 and the structural protein E are the main targets. Some candidate agents targeting the flavivirus E-domain III(Zu et al., 2014), NS3-helicase (Fang et al., 2016), NS2B-NS3 interaction (Li et al., 2017b) and NS5-methyltransferase (Brecher et al., 2015; Han and Lee, 2017) have been successfully identified. The second strategy is based on in vivo cellular antiviral assays. In this case, the replicon-containing cell line (Li et al., 2018; Zhang et al., 2017), virus-like particle (VLP) (Wang et al., 2017), and the reporter virus (Li et al., 2017a) are the main screening tools. In contrast to VLP and replicon-containing cell line, the propagation of reporter virus contains the entire viral life cycle as wild type (WT) virus, it can be used to screen antiviral drugs targeting the complete infection steps. Currently, several reporter flaviviruses have been developed for different research purposes, including Rluc-JEV (Li et al., 2017a), Rluc-DENV(Zou et al., 2011) and eGFP-DENV(Schoggins et al., 2012).
In this study, we constructed and characterized a JEV reporter virus with an eGFP gene (eGFP-JEV). An eGFP-JEV-based HTS assay was established in a 96-well format and used for screening of 1443 compounds from an FDA-approved drug library. Using this system, 16 hit drugs inhibiting JEV infection were identified, and five of them were firstly reported to have inhibitory effect on flavivirus replication, offering potential new therapies for the treatment of JEV infection.
2. Materials and methods
2.1. Cell lines, viruses, antibodies and reagents
Aedes albopictus mosquito C6/36 cells were cultured in RPMI-1640 medium (Invitrogen, Darmstadt, Germany) with 10% fetal bovine serum (FBS) and 100 U/ml penicillin–streptomycin (PS) at 28 °C. All other cells were grown at 37 °C with 5% CO2. Baby hamster kidney fibroblast (BHK-21) cells, and human hepatoma (HuH-7) cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Darmstadt, Germany) with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin. JEV virus was derived from the infectious cDNA clone of pACYC-JEV-SA14 (Li et al., 2014b). The 4G2 antibody against the E protein of Flavivirus was kindly provided by Dr. Qin, Cheng-Feng (Beijing Institute of Microbiology and Epidemiology, China), and is cross-reactive with the JEV E protein. Texas Red-conjugated goat anti-mouse IgG was purchased from Protein Tech Group. Nucleoside analogue inhibitor NITD008 was synthesized as reported previously (Yin et al., 2009). A library of FDA-approved drugs was purchased from Selleck Chemicals.
2.2. Plasmid construction
The reporter JEV genome carrying eGFP (green fluorescent protein) was constructed with pACYC-JEV-SA14 (Li et al., 2014a) as a backbone. The “KpnI-T7 promoter-5′ UTR-Capsid-38 amino acids-AscI” and “PacI-C-prM-E″ fragments were amplified using pACYC-JEV-SA14 as a template. The “AscI-eGFP-2A-PacI” fragment was amplified using EV71-eGFP-2A as a template (Shang et al., 2013). The three fragments were fused together by overlapping PCR to obtain a fragment “KpnI-T7 promoter-5’ UTR-Capsid-38 amino acids-AscI-eGFP-2A-PacI-C-prM-E″ as shown in Fig. 1 . This fragment was engineered into pACYC-JEV-SA14 by KpnI and BsrGI sites to generate an eGFP-JEV cDNA clone. The complete sequence of the cDNA clone of eGFP-JEV was validated by DNA sequencing analysis before the subsequent experiments.
Fig. 1.
Construction of the infectious clone of eGFP-JEV reporter virus. Using the infectious clone pACYC-JEV-SA14 as a backbone, the “KpnI-T7 promoter-5′UTR-capsid38”, “C-prM-E″ and “eGFP-2A” fragment were fused together by overlapping PCR to obtain a fragment “KpnI-T7 promoter-5′UTR-capsid38-eGFP-2A-C-prM-E”. This fragment was ligated into pACYC-JEV-SA14 by KpnI and BsrGI restriction enzyme sites to generate an eGFP-JEV cDNA clone (pACYC-eGFP-JEV-SA14). The pirmer pair JEV-F and JEV-R which was used in the RT-PCR assay to test the stability of eGFP-JEV was marked in red and placed according to their location in the genome.
2.3. In vitro transcription, RNA transfection
The JEV infectious clone and the reporter cDNA plasmids were linearized with XhoI and purified by extraction with phenol/chloroform. The linearized cDNA was transcribed using mMESSENGER mMACHINE T7 Kit (Ambion, Austin, TX, USA). All procedures were performed according to the manufacturer's protocols. RNA was dissolved in RNase-free water and stored at −80 °C. The RNA was transfected into cells with DMRIE-C reagent (Invitrogen) following the protocol described previously (Deng et al., 2016).
2.4. Immunofluorescence assay (IFA)
The eGFP-JEV genomic RNA was transfected into BHK-21 cells. At 24, 48, and 72 h post-transfection (hpt), the cells on the coverslips were fixed in 4% paraformaldehyde for 10 min at room temperature. The fixed cells were washed three times with PBS and incubated with 4G2 antibody (1:500 dilution in PBS) for 1 h at room temperature. After washing three times with PBS, the cells were incubated with Texas Red-conjugated goat anti-mouse IgG antibody for 40 min in the dark. After washing three times with PBS, the cells were mounted on a glass slide with 95% glycerol and cell images were captured under a fluorescence microscope.
2.5. Reverse transcription PCR (RT-PCR)
To examine the genetic stability of the eGFP gene of eGFP-JEV, total RNAs were extracted from cells transfected with eGFP-JEV or cells infected with each passaged viruses using Trizol reagent (Takara). The fragment from 5′UTR to prM which covers the eGFP gene was amplified by one-step RT-PCR using a PrimeScript RT-PCR kit (TAKARA) with the primers JEV-F (5′-AGAAGTTTATCTGTGTGAACT-3′) and JEV-R (5′-TAGACTTCTTGGTTGTCACAC-3′). The RT-PCR products were analyzed by electrophoresis on 1% agarose gel.
2.6. Real-time RT-PCR
To clarify whether the compounds specifically inhibit viral replication or generally suppress cellular RNA transcription, the viral RNA and β-actin RNA were quantified by real-time PCR after the cells were infected by virus and treated with different concentrations of compounds. Real-time PCR was performed using a one-step PrimeScript RT-PCR kit (TAKARA). For JEV genome quantification, the primer pair (Forwad: 5′-TACAACATGATGGGAAAGCGAGAGAAAAA-3′ and reverse: 5′-GTGTCCCAGCCGGCGGTGTCATCAGC-3′) were used, and the number of genomic RNA copies was determined with a standard curve of the in vitro-transcribed JEV RNA. The primer pair (Forwad: 5′-CCACACTGTGCCCATCTACG-3′ and reverse: 5′-AGGATCTTCATGAGGTAGTCAGTCAG-3′) were used to detect the β-actin RNA level.
2.7. Viral growth kinetics
BHK-21 cells were seeded in 6-well plates (2 × 105 cells per well) one day before infection. The cells were infected with eGFP-JEV or WT-JEV virus (at a multiplicity of infection of 0.1). The supernatants were collected at 2, 24, 36, 48, 60, 72, 84 h post-infection (hpi), and viral titers were quantified using the single-layer plaque assay on BHK-21 cells.
2.8. Single-layer plaque assay
The viral titer and morphology were determined by single-layer plaque assay. The virus was serially diluted 10-fold, and 100 μL of each dilution was added into a single well of a 24-well plate containing BHK-21 cells (1 × 105 cells per well). The plates were incubated at 37 °C with 5% CO2 for 1 h before adding DMEM-2% FBS containing 1% methylcellulose. After 3 days of incubation at 37 °C with 5% CO2, these cells were fixed and stained with 3.7% formaldehyde and 1% crystal violet in water. The morphology and number of plaques were recorded after washing with water.
2.9. Antiviral assay of eGFP-JEV and WT-JEV
BHK-21 cells was seeded in 12-well plates (1 × 105 cells per well) were infected with WT-JEV or eGFP-JEV at an MOI of 0.1. Huh7 was seeded on 12-well plates (2 × 105 cells per well) were infected with WT-JEV at an MOI of 0.5. Thereafter, the infected cells were incubated with various concentrations of NITD008, lonafarnib, cetylpyridinium chlorid, cetrimonium bromide, nitroxoline and hexachlorophene respectively. The supernatants were collected at 48 hpi or 72 hpi, and viral titers were determined by the single-layer plaque assay. For eGFP-JEV based antiviral assay, the expression of eGFP gene was recorded at 72 hpi. The antiviral activity of the compounds was expressed as 50% effective concentration (EC50) which meant the drug concentration required to achieve 50% of viral titer reduction and was calculated by nonlinear regression using GraphPad Prism 5.0 software.
2.10. HTS assay
The eGFP-JEV based HTS assay was developed in a 96-well plate format using NITD008 as a positive control and 0.5% DMSO as a negative control. The compounds of FDA-approved drug library were all dissolved in DMSO at a stock concentration of 1 mM. We diluted each of the compound to 5 μM with DMEM-2% FBS as the working concentration. Huh7 cells were seeded into 96-well plates at the cell density of 1.0 × 104 per well and cultured for 24 h at 37 °C. Then the cells were infected with eGFP-JEV (MOI = 0.5). At the same time, the compound or DMSO was added to the culture medium. At 48 hpi, the number of the eGFP-positive cells was read by a PerkinElmer high content screening system. To evaluate the performance of the HTS assay, the Z′ factor values were calculated. The Z’ factor between 0.5 and 1 indicates an excellent assay with good separation between controls (Zhang et al., 1999).
2.11. Time-of-addition assay
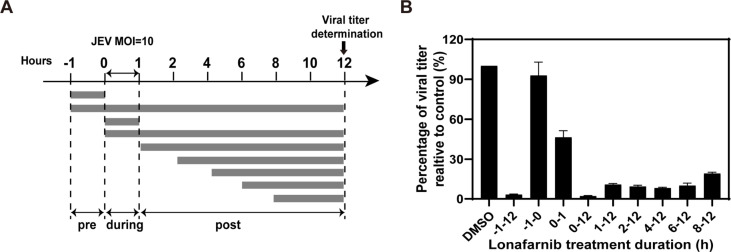
To determine the step inhibited by lonafarnib during viral life cycle, a time-of-addition assay was performed. Huh7 cells were infected with WT-JEV at an MOI of 10 for 1 h at 37 °C, and lonafarnib (5 μM) was added to the infected cells at the following time points: pre-infection (−1 to 0 h), during infection (0–1 h), and post-infection (1, 2, 4, 6, 8 h). Meanwhile, 0.5% DMSO was added as control. Supernatants were harvested at 12 hpi, and viral titers were determined by plaque assay. Inhibition rates are calculated as the percentage of viral titer realtive to control.
2.12. Cytotoxicity assays
Huh7 cells were seeded in 96-well plates (1 × 104 cells per well) and allowed to grow for 24 h before treatments. Afterward, compounds at concentrations in a 2-fold dilution series were added to the cell. At 48 h, the cells were incubated with 10 μL CCK8 reagent (cell counting kit-8, Bimake) for 1 h at 37 °C. The absorbance at 450 nm was read by a Microplate Reader (Varioskan Flash, Thermo Fisher). Cell viability was expressed as a percentage of the treated cells to the control (untreated) cells. For each compound concentration, six wells were performed in parallel, mean values of the cell viability were calculated. The CC50 was calculated by nonlinear regression using GraphPad Prism 5.0 software to determine the cytotoxic concentration at which 50% of the cells are viable.
3. Results
3.1. Construction of eGFP-JEV reporter virus
Using the infectious clone pACYC-JEV-SA14 as a backbone (Li et al., 2014b), we constructed a cDNA clone of JEV encoding an eGFP reporter gene. As shown in Fig. 1, three fragments were inserted between the 5′UTR and the capsid gene, which include the N-terminal 38-amino acid sequence of the C protein, a green fluorescent protein gene, and the FMDV-2A (Foot-and-mouth disease virus protease 2A) sequence. The N-terminal 38 residues sequence of capsid contains important cis-acting RNA elements required for genomic cyclization. The FMDV-2A autoproteolytic cleavage sequence is inserted to facilitate the processing and cleavage of eGFP from the viral protein.
3.2. Characterization of eGFP-JEV reporter virus
Viral RNAs transcribed from the eGFP-JEV infectious clone were transfected into BHK-21 cells to generate viral stocks. The eGFP gene expression was observed and the viral E protein expression was detected by IFA using anti-E antibody 4G2 at different time points after transfection. Fluorescence microscopy analysis of the transfected cells revealed that almost all cells expressing the E protein were also eGFP-positive (Fig. 2 A). The supernatant was collected and subjected to plaque assay to determine the plaque morphology. We found that the plaque size of eGFP-JEV was smaller than that of WT-JEV (Fig. 2B).
Fig. 2.
Characterization of eGFP-JEV reporter virus. (A) (the first column from left) Immunostaining of viral E protein in the eGFP-JEV RNA transfected BHK-21 cells by IFA using 4G2 antibody; (the second column from left) eGFP expression detection under the fluorescence microscope; (the second column from right) DAPI-stained nuclei detection. The merged images were shown on the right. (B) Comparison of plaque morphology between eGFP-JEV and WT-JEV; (C–D) Comparison of growth kinetic between eGFP-JEV and WT-JEV by IFA and plaque assay. BHK-21 cells were seeded in 6-well plates (2 × 105 cells per well). The cells were infected with eGFP-JEV and WT-JEV virus (at an MOI of 0.1), respectively. The infected cells were analyzed by IFA (detecting viral E protein expression) on 48 hpi and 72 hpi. At the same time, culture media were collected at 2, 24, 36, 48, 60, 72, 84 hpi, respectively, and viral titers were quantified using the plaque assay on BHK-21 cells. Error bars indicate the standard deviations from three independent experiments.
To characterize the growth properties of the reporter virus, BHK-21 cells were infected with WT-JEV and eGFP-JEV viruses at the same MOI. The results of IFA (detecting viral E protein) showed that in both the WT-JEV and eGFP-JEV infected cells, the number of E-positive cells increased from day 1 to day 3 (Fig. 2C). However, eGFP-JEV virus-infected cells produced fewer IFA positive cells than that of WT-JEV at each time point. Consistent with the IFA results, the titers of eGFP-JEV virus gradually increased from day 1 to day 3, but were lower than that of WT-JEV at each time point (Fig. 2D). These results showed that the growth trend of eGFP-JEV was similar with WT-JEV and the replication of eGFP-JEV reporter virus could be easily tracked by eGFP signals.
3.3. Stability of eGFP-JEV reporter virus
To test the stability of eGFP-JEV, we serially passaged the virus in C6/36 cells. The virus harvested from viral RNA-transfected C6/36 cells was defined as the P0 virus, and was used for blind passage in C6/36 cells. The viruses from each passage were defined as P1 to P5 passages. For each passage, the expression of eGFP gene in the infected cells was detected under a fluorescence microscope, the RNAs were extracted and subjected to RT-PCR, and the viral morphology was determined by plaque assay. As shown in Fig. 3 A, nearly 90% eGFP-positive cells were exhibited in the P0–P4 viruses infected cells, whereas only 50% eGFP-positive cells were detected in the P5 virus infected cells when the CPE was apparently observed. RT-PCR assay was performed using the primers spanning the 5′UTR and M gene which include the eGFP gene. The expected products for eGFP-JEV and WT-JEV are about 1.5 kb and 0.7 kb respectively. The P1–P4 viruses infected cells exhibited a dominant 1.5 kb band and a very weak 0.7 kb band consistently, whereas the P5 displayed a main 0.7 kb band (Fig. 3B). In addition, the P1–P4 viruses exhibited homogeneously small plaque morphology as P0 virus, but the plaque size of P5 virus turned as large as WT-JEV (Fig. 3C). These results demonstrated that the eGFP gene of the eGFP-JEV reporter virus could be stably maintained within four rounds of passage and was significantly lost at P5. Therefore, we chose the P1 virus for the following antiviral study.
Fig. 3.
Stability of eGFP-JEV reporter virus. (A) Fluorescence detection of the expression of eGFP during virus passage in C6/36 cell. The eGFP-JEV was serially passaged in C6/36 cells for five rounds. Viruses from each passage (P1–P5) were used to infect C6/36 cells, the expression of eGFP was detected under a fluorescent microscope at 96–120h after infection. (B) Detection of the eGFP gene expression during virus passage using RT-PCR. Viral RNAs were extracted from the cells infected with each passage of virus. The fragment covering the reporter gene region was amplified using RT-PCR with the primer pair JEV-F and JEV-R (see the primer sequences in Materials and Methods). (C) Plaque morphology of each passage of eGFP-JEV.
3.4. Inhibitory effects of NITD008 on eGFP-JEV reporter virus
NITD008, a well-known anti-flavivirus compound, has shown to be able to inhibit JEV replication (Li et al., 2017a). To determine the availability of the reporter virus for antiviral drug screening, we evaluated the antiviral activity of NITD008 using the eGFP-JEV reporter virus. Two-fold serial dilutions of NITD008 were used in this study with a maximum concentration of 3 μM. NITD008 showed consistently dose-dependent inhibitory effects on both eGFP expression (Fig. 4 A) and viral titers (Fig. 4B) of eGFP-JEV. In addition, the EC50 of NITD008 for eGFP-JEV was 0.22 μM, which was similar to that of WT-JEV (EC50 = 0.27 μM) (Fig. 4C). The results not only confirmed that the expression level of eGFP can represent the replication of WT-JEV, but also indicated that the eGFP-JEV reporter virus system is suitable for screening potential anti-JEV compounds.
Fig. 4.
Inhibitory effects of NITD008 on eGFP-JEV reporter virus. (A) Effect of different concentrations of NITD008 on the expression of eGFP in eGFP-JEV infected cells. BHK-21 cells were seeded at a density of 105 cells/well in the 12-well plate. At 24 h after inoculation, the cells were infected with eGFP-JEV at an MOI of 0.1 and incubated with the indicated concentrations of NITD008, the eGFP expression was detected under a fluorescent microscope at 72 hpi. Effects of different concentrations of NITD008 on the viral titers of eGFP-JEV (B) and WT-JEV (C). BHK-21 cells were infected with eGFP-JEV or WT-JEV at an MOI of 0.1, and treated with different concentrations of NITD008. The viral titers in supernatants were determined by plaque assay at 72 hpi. Error bars indicate the standard deviations from three independent experiments.
3.5. Optimization of the eGFP-JEV-based HTS assay
We chose a human-derived cell line Huh7 to establish an eGFP-JEV-based HTS assay. The HTS conditions including the cell density, the multiplicity of infection and the time of observation were optimized. Different numbers of Huh7 cells (7,500, 10,000 and 20,000) were seeded into each well of a 96-well plate, followed by infection with different MOIs of eGFP-JEV (0.1, 0.2 and 0.5). The number of eGFP-positive cells was quantified at 36, 48, and 60 hpi respectively by a high-content screening system. As shown in Fig. 5 B, 10,000 Huh7 cells infected with 0.5 MOI of eGFP-JEV showed a better eGFP signal at 48 hpi. Therefore, we chose this condition for the following experiments. As the compound is usually dissolved in DMSO, we tested the effect of different concentrations of DMSO on eGFP-JEV replication in a 96-well plate to determine the optimal solvent concentration for the screen assay. As shown in Fig. 5D, DMSO had little effect on the expression of eGFP at ≤ 0.5% (v/v) concentration, but 1% DMSO significantly reduced the percentage of eGFP-positive cells to 30%. Therefore, we chose 0.5% DMSO as the work concentration to dissolve the compounds. We used the known inhibitor NITD008 to verify the feasibility of these identified conditions. Under the above experimental conditions, the calculated Z′ value was 0.83 at 0.3 μM of NITD008 (HTS assays with Z' ≥ 0.5 are considered robust) (Fig. 5E and F). These results demonstrate that eGFP-JEV reporter virus could be used in a high throughput assay for screening inhibitors of JEV.
Fig. 5.
Optimization of eGFP-JEV-based HTS assay. (A–C) Effects of different seeding density, multiplicity of infection and time course of infection on the expression of eGFP in infected cells. (D) Effect of different concentrations of DMSO on the expression of eGFP in infected cells. (E) Effect of different concentrations of NITD008 on the expression of eGFP in infected cells. (F) 96-well plate uniformity assessment of the eGFP-JEV infection for HTS assay. Error bars indicate the standard deviations from three independent experiments.
3.6. Screening of an FDA-approved drug library for inhibitors of JEV infection
We screened 1443 compounds in an FDA-approved drug library using the eGFP-JEV based HTS to select the potential antiviral drugs. A schematic diagram of eGFP-JEV-based HTS method was shown in Fig. 6 A. All drugs were used at a concentration of 5 μM, the drugs with an inhibition rate greater than 90% were selected as candidates. The Z′ values of each screening were between 0.5 and 1, with an average value of 0.7 (data not shown), indicating that our results were reliable. Twenty-six hits were identified though the HTS assay (Fig. 6B). To exclude false positives caused by drug cytotoxicity or nonspecific fluorescent interference, eGFP-JEV was incubated with different concentrations of those 26 compounds at 96-well plates, drugs showing apparent cytotoxicity and those with no inhibitory effect on eGFP expression were excluded (data not shown). Finally, 16 hit drugs displaying high inhibitory activity against eGFP-JEV and little cytotoxicity were identified. Among them, 11 drugs have been previously reported to be able to inhibit the infection of JEV (5 drugs listed in Group I) or other flaviviruses (6 drugs listed in Group II), confirming the reliability of the eGFP-JEV based HTS system. Interestingly, 5 drugs (listed in Group III), which are lonafarnib, cetylpyridinium chlorid, cetrimonium bromide, nitroxoline and hexachlorophene, are newly discovered inhibitors of JEV in our study (Fig. 6C).
Fig. 6.
Screening of an FDA-approved compound library for inhibitors of JEV using eGFP-JEV reporter virus. (A) Flow chart of the eGFP-JEV-based HTS assay. (B) Scatter plot of primary screening data of 1443 compounds in the FDA-approved drug library. Hit durgs having greater than 90% antiviral activity (represented by horizontal black dot line) were selected for secondary screening. (C) Classification of the anti-JEV compounds screened from the FDA-approved drug library. Group I includes the compounds previously reported to possess anti-JEV activity; Group II are the compounds previously reported to be capable to inhibit the replication of other flaviviruses; Group III is the newly discovered anti-JEV compounds in this study.
3.7. Antiviral activity of the newly discovered inhibitors on WT-JEV
We next confirmed the anti-JEV effects of these newly discovered compounds using the authentic WT-JEV. Huh7 cells were infected with WT-JEV at an MOI of 0.5 with the maintenance of different concentrations of the five compounds. The supernatants were collected at 48 hpi and subjected to plaque assay to determine viral titer. The EC50 values were analyzed to determine the drug concentrations causing 50% reduction of viral titer. As shown in Fig. 7 A, all of the five compounds showed inhibitory effect on virus propagation. The EC50 were 0.982 μM for lonafarnib, 0.455 μM for cetylpyridinium chlorid, 2.232 μM for cetrimonium bromide, 2.482 μM for nitroxoline and 0.421 μM for hexachlorophene, respectively. The effects of the cytotoxicity of the compounds on Huh7 cells were detected with a CCK-8 assay to determine the CC50 values (Fig. 7B). The CC50 were 11.36 (lonafarnib), 7.195 μM (cetylpyridinium chlorid), 7.878 μM (cetrimonium bromide), 9.294 μM (nitroxoline) and 10.04 μM (hexachlorophene) respectively. We next quantified the RNA levels of viral genome and cellular β-actin in the cells which were infected with WT-JEV and treated with various dosages of lonafarnib, cetylpyridinium chlorid, cetrimonium bromide and nitroxoline respectively by qRT-PCR. Consistent with viral titer reduction, the viral RNA copies decreased in a dose-dependent manner of the four tested drugs (Fig. 7C). None of the drugs had any effect on the mRNA level of cellular actin (Fig. 7D), suggesting that the inhibitory effect of these newly discovered compounds on virus replication was due to their antiviral activity rather than generally inhibiting cellular RNA transcription.
Fig. 7.
Evaluation of anti-JEV abilities and toxicity of the newly discovered compounds. (A) Dose-response curves of 5 new hits for inhibition of JEV infection. Huh7 cells were infected at an MOI of 0.5 with WT-JEV and treated with different concentrations of compounds. The viral titers in supernatants were determined by plaque assay at 48 hpi. (B) Cytotoxicity test of 5 new hits. (C–D) Quantification of the cellular β-actin RNA and viral-RNA levels of the cells which were infected with WT-JEV and treated with different compounds. Huh7 cells were infected at an MOI of 0.5 with WT-JEV and treated with the indicated concentrations of compounds. Total RNAs were extracted from parallel wells at 48hpi. Cellular β-actin RNA and viral RNA levels were detected by qRT-PCR. (E) EC50, CC50, and SI values of 5 new hit compounds selected from the reconfirmation screen. Error bars indicate the standard deviations from three independent experiments.
3.8. Time of addition assay for lonafarnib
The selective index (SI) of each newly discovered compound was calculated as the ratio of CC50 to EC50 (Fig. 7E). Lonafarnib, hexachlorophene and cetylpyridinium chlorid possessed good selective index (>10). Since lonafarnib is an orally administered drug and showed lower cytotoxicity, we chose lonafarnib to perform a time-of-addition assay to explore at which stage the drug blocked during viral life cycle. Huh7 cells were infected with WT-JEV at an MOI of 10 for 1h, and washed three times with PBS subsequently to remove the unattached viruses. Lonafarnib was added to the cells either pre-, during, or post infection at a concentration of 5 μM (Fig. 8 A). The supernatants were collected at 12 hpi and subjected to plaque assay. As shown in Fig. 8B, viral titers were significantly reduced when the compound was added at different time points post infection, suggesting the inhibition of virus replication. In addition, treatment with the compound during viral infection (0–1) resulted 60% reduction in viral titer, implying that lonafarnib also affected virus entry stage. However, there was no effect on virus propagation when lonafarnib was added pre-infection, indicating that a cellular response may be not required for lonafarnib to exert its antiviral activity. These results primarily demonstrated that lonafarnib might block viral propagation mainly through significant suppression of virus replication and modest inhibition of virus entry process during viral life cycle.
Fig. 8.
Time-of-addition experiment of lonafenib. (A)Schematic illustration of the time-of-addition experiment. Huh7 cells were infected with WT-JEV at an MOI of 10, and treated with 5 μM lonafarnib pre (-1-0 h), during (0–1 h) and post (1, 2, 4, 6, 8 h) infection, 0.5% DMSO was added at the same time as control. (B) Percentage of viral titer relative to control. At 12 hpi, the supernatants were collected to determine the viral titers, the effects of lonafarnib added at different time points were expressed as the percentage of viral titer to that of DMSO. Error bars indicate the standard deviations from three independent experiments.
4. Discussion
Currently, vaccine is the only approach to prevent JEV infection, and no antiviral therapeutics are available. Thus, development of HTS assay is important for JEV drug discovery. In this study, we constructed an eGFP-JEV reporter virus containing eGFP gene, and developed the eGFP-JEV-based HTS assay for antiviral screening. For the construction of eGFP-JEV reporter virus, the eGFP-2A fragment was inserted between the 5′UTR and C protein of JEV genome (Fig. 1). Upon the cleavage by the FMDV 2A peptide, the eGFP protein was released and visible under a fluorescence microscope. As shown in Fig. 2A, C, and D, the expression level of eGFP correlated well with the level of JEV replication. In addition, a dose-dependent reduction in eGFP fluorescence signals was observed in eGFP-JEV-infected cells with NITD008 (a well-known anti-flavivirus inhibitor) treatment, implying that this reporter virus could be used as a convenient and effective HTS system for screening anti-JEV compounds (Fig. 4). As the stability of the reporter virus is crucial for the reproducibility and reliability of HTS assay, eGFP-JEV was serially passaged in C6/36 cells to investigate its genetic stability. It was found that the eGFP reporter gene was stably retained in P4 virus, indicating that the reporter virus was stable at least within four serial passages. It can meet the measurement requirements of the eGFP-JEV based HTS system.
Under the optimized HTS condition (Figs. 5), 26 hit drugs were firstly identified out of the screened 1443 FDA approved drugs. Considering the possible interference of the unspecific fluorescence and drug cytotoxicity, the first set of compounds were subjected to the dose-dependent inhibition assay using eGFP-JEV. At this stage, 16 drugs with significant inhibitory activity on eGFP expression and low cytotoxicity were screened out (Fig. 6). Among them, 5 drugs (mycophenolic acid, cyclosporin A, manidipine, pimecrolimus and lacidipine) had been reported to be effective in inhibiting JEV infection (Sebastian et al., 2011; Tu et al., 2012; Wang et al., 2017). Having these five drugs independently identified as hits in our screening further confirmed the reliability of the eGFP-JEV based HTS system in identifying potential anti-JEV inhibitors. In addition, it was found that some drugs that had been reported to inhibit other flaviviruses (Balasubramanian et al., 2017; Bullard-Feibelman et al., 2017; de Freitas et al., 2019; Dong et al., 2019; Fryk et al., 2016, 2017 bib_Fryk_et_al_2016 bib_Fryk_et_al_2017; Gan et al., 2018; Kuivanen et al., 2017; Papin et al., 2005; Shahen et al., 2018; Simanjuntak et al., 2015) could also inhibit JEV. For instance, sofosbuvir, an initially approved nucleotide polymerase inhibitor for the distantly related hepatitis C virus (HCV), were active against a variety of flaviviruses including ZIKV (Bullard-Feibelman et al., 2017), DENV (Gan et al., 2018), YFV (de Freitas et al., 2019) as well as JEV determined here. Mefloquine showing antiviral activities against ZIKV and DENV (Balasubramanian et al., 2017) was also identified as a potent inhibitor of JEV here. We reasoned that such broad-spectrum anti-flavivirus activities of this group of hits may mainly attribute to the common features of virion composition and viral protein structures shared by flaviviruses. Nevertheless, further in-depth investigations are still needed to explore the anti-JEV mechanism of these hits in the future.
Notably, 5 new hit drugs were identified for the first time to possess anti-JEV activity using this HTS system. It included three approved antiseptics: cetylpyridinium chloride, cetrimonium bromide and hexachlorophene; and two orally administered drugs: lonafarnib and nitroxoline. Nitroxoline is a quinoline antibiotic for the treatment or prophylaxis of acute urinary tract infections with potential antitumor activity (Veschi et al., 2018). To our knowledge, this was also the first time for nitroxoline associated with the interference with viral infection. Lonafarnib is an orally active inhibitor of farnesyltransferase that has entered clinical trials for treatment hepatitis delta virus (HDV) infection (Koh et al., 2015). Lonafarnib abrogates HDV production by inhibiting prenylation of the large delta hepatitis antigen to prevent its interaction with hepatitis B surface antigen to form secreted particles (Koh et al., 2015). Here, using a time-of-addition assay, we primarily demonstrated that lonafarnib dramatically inhibit virus replication, and also partially affected virus entry process during the viral life cycle of JEV. A recent study had identified that lonafarnib can combine with the RNA-dependent RNA polymerase of SARS-CoV and SARS-CoV-2 by virtual screening (Ruan et al., 2020). Therefore, the possible anti-JEV mechanisms of lonafarnib might be the inhibition of prenylation modification of the viral proteins, or the binding with the RdRp to inhibit viral replication, or other unknown factors, which needs further investigation. In the future studies, we are going to investigate the anti-JEV mechanisms of these newly discovered hit drugs, especially those two oral drugs.
5. Conclusions
In summary, we have developed a novel eGFP-JEV based HTS system. The use of eGFP-JEV for HTS assay provided direct visibility of the suppression of viral infection by JEV inhibitors, expediting greatly the process of antiviral development. Using this system, sixteen drugs that effectively inhibited JEV were identified from the FDA-approved drug library, and five of them are newly discovered compounds which have antiviral activity against JEV and flavivirus, providing potential new therapies for the treatment of JEV infection.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (grant number 2016YFD0500400), National Natural Science Foundation of China (grant number 81702005), and China Postdoctoral Science Foundation (grant number 2018M632945). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- Balasubramanian A., Teramoto T., Kulkarni A.A., Bhattacharjee A.K., Padmanabhan R. Antiviral activities of selected antimalarials against dengue virus type 2 and Zika virus. Antivir. Res. 2017;137:141–150. doi: 10.1016/j.antiviral.2016.11.015. [DOI] [PubMed] [Google Scholar]
- Brecher M., Chen H., Li Z., Banavali N.K., Jones S.A., Zhang J., Kramer L.D., Li H. Identification and characterization of novel broad-spectrum inhibitors of the flavivirus methyltransferase. ACS Infect. Dis. 2015;1:340–349. doi: 10.1021/acsinfecdis.5b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton M.A. Replication cycle and molecular biology of the West Nile virus. Viruses. 2013;6:13–53. doi: 10.3390/v6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher E.L., Scherer W.F., Rosenberg M.Z., Gresser I., Hardy J.L., Bullock H.R. Ecologic studies of Japanese encephalitis virus in Japan. II. Mosquito infection. Am. J. Trop. Med. Hyg. 1959;8:651–664. doi: 10.4269/ajtmh.1959.8.651. [DOI] [PubMed] [Google Scholar]
- Bullard-Feibelman K.M., Govero J., Zhu Z., Salazar V., Veselinovic M., Diamond M.S., Geiss B.J. The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antivir. Res. 2017;137:134–140. doi: 10.1016/j.antiviral.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G.L., Hills S.L., Fischer M., Jacobson J.A., Hoke C.H., Hombach J.M., Marfin A.A., Solomon T., Tsai T.F., Tsu V.D., Ginsburg A.S. Estimated global incidence of Japanese encephalitis: a systematic review. Bull. World Health Organ. 2011;89:766–774. doi: 10.2471/BLT.10.085233. 774A-774E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Freitas C.S., Higa L.M., Sacramento C.Q., Ferreira A.C., Reis P.A., Delvecchio R., Monteiro F.L., Barbosa-Lima G., James Westgarth H., Vieira Y.R., Mattos M., Rocha N., Hoelz L.V.B., Leme R.P.P., Bastos M.M., Rodrigues G.O.L., Lopes C.E.M., Queiroz-Junior C.M., Lima C.X., Costa V.V., Teixeira M.M., Bozza F.A., Bozza P.T., Boechat N., Tanuri A., Souza T.M.L. Yellow fever virus is susceptible to sofosbuvir both in vitro and in vivo. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C.L., Liu S.Q., Zhou D.G., Xu L.L., Li X.D., Zhang P.T., Li P.H., Ye H.Q., Wei H.P., Yuan Z.M., Qin C.F., Zhang B. Development of neutralization assay using an eGFP chikungunya virus. Viruses. 2016;8 doi: 10.3390/v8070181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Kang S., Dimopoulos G. Identification of anti-flaviviral drugs with mosquitocidal and anti-Zika virus activity in Aedes aegypti. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P.M., Daniels P.W., Banks D.J. Japanese encephalitis. Vet. Clin. N. Am. Equine Pract. 2000;16:565–578. doi: 10.1016/s0749-0739(17)30096-2. x-xi. [DOI] [PubMed] [Google Scholar]
- Fang J., Li H., Kong D., Cao S., Peng G., Zhou R., Chen H., Song Y. Structure-based discovery of two antiviral inhibitors targeting the NS3 helicase of Japanese encephalitis virus. Sci. Rep. 2016;6:34550. doi: 10.1038/srep34550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryk J.J., Marks D.C., Hobson-Peters J., Prow N.A., Watterson D., Hall R.A., Young P.R., Reichenberg S., Sumian C., Faddy H.M. Dengue and chikungunya viruses in plasma are effectively inactivated after treatment with methylene blue and visible light. Transfusion. 2016;56:2278–2285. doi: 10.1111/trf.13729. [DOI] [PubMed] [Google Scholar]
- Fryk J.J., Marks D.C., Hobson-Peters J., Watterson D., Hall R.A., Young P.R., Reichenberg S., Tolksdorf F., Sumian C., Gravemann U., Seltsam A., Faddy H.M. Reduction of Zika virus infectivity in platelet concentrates after treatment with ultraviolet C light and in plasma after treatment with methylene blue and visible light. Transfusion. 2017;57:2677–2682. doi: 10.1111/trf.14256. [DOI] [PubMed] [Google Scholar]
- Gan C.S., Lim S.K., Chee C.F., Yusof R., Heh C.H. Sofosbuvir as treatment against dengue? Chem. Biol. Drug Des. 2018;91:448–455. doi: 10.1111/cbdd.13091. [DOI] [PubMed] [Google Scholar]
- Griffiths M.J., Turtle L., Solomon T. Japanese encephalitis virus infection. Handb. Clin. Neurol. 2014;123:561–576. doi: 10.1016/B978-0-444-53488-0.00026-2. [DOI] [PubMed] [Google Scholar]
- Han S.R., Lee S.W. Inhibition of Japanese encephalitis virus (JEV) replication by specific RNA aptamer against JEV methyltransferase. Biochem. Biophys. Res. Commun. 2017;483:687–693. doi: 10.1016/j.bbrc.2016.12.081. [DOI] [PubMed] [Google Scholar]
- Hills S.L., Griggs A.C., Fischer M. Japanese encephalitis in travelers from non-endemic countries, 1973-2008. Am. J. Trop. Med. Hyg. 2010;82:930–936. doi: 10.4269/ajtmh.2010.09-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh C., Canini L., Dahari H., Zhao X., Uprichard S.L., Haynes-Williams V., Winters M.A., Subramanya G., Cooper S.L., Pinto P., Wolff E.F., Bishop R., Ai Thanda Han M., Cotler S.J., Kleiner D.E., Keskin O., Idilman R., Yurdaydin C., Glenn J.S., Heller T. Oral prenylation inhibition with lonafarnib in chronic hepatitis D infection: a proof-of-concept randomised, double-blind, placebo-controlled phase 2A trial. Lancet Infect. Dis. 2015;15:1167–1174. doi: 10.1016/S1473-3099(15)00074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen S., Bespalov M.M., Nandania J., Ianevski A., Velagapudi V., De Brabander J.K., Kainov D.E., Vapalahti O. Obatoclax, saliphenylhalamide and gemcitabine inhibit Zika virus infection in vitro and differentially affect cellular signaling, transcription and metabolism. Antivir. Res. 2017;139:117–128. doi: 10.1016/j.antiviral.2016.12.022. [DOI] [PubMed] [Google Scholar]
- Li J.Q., Deng C.L., Gu D., Li X., Shi L., He J., Zhang Q.Y., Zhang B., Ye H.Q. Development of a replicon cell line-based high throughput antiviral assay for screening inhibitors of Zika virus. Antivir. Res. 2018;150:148–154. doi: 10.1016/j.antiviral.2017.12.017. [DOI] [PubMed] [Google Scholar]
- Li L., Lok S.M., Yu I.M., Zhang Y., Kuhn R.J., Chen J., Rossmann M.G. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science (New York, N.Y.) 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- Li X.D., Li X.F., Ye H.Q., Deng C.L., Ye Q., Shan C., Shang B.D., Xu L.L., Li S.H., Cao S.B., Yuan Z.M., Shi P.Y., Qin C.F., Zhang B. Recovery of a chemically synthesized Japanese encephalitis virus reveals two critical adaptive mutations in NS2B and NS4A. J. Gen. Virol. 2014;95:806–815. doi: 10.1099/vir.0.061838-0. [DOI] [PubMed] [Google Scholar]
- Li X.D., Li X.F., Ye H.Q., Deng C.L., Ye Q., Shan C., Shang B.D., Xu L.L., Li S.H., Cao S.B., Yuan Z.M., Shi P.Y., Qin C.F., Zhang B. Recovery of a chemically synthesized Japanese encephalitis virus reveals two critical adaptive mutations in NS2B and NS4A. J. Gen. Virol. 2014;95:806–815. doi: 10.1099/vir.0.061838-0. [DOI] [PubMed] [Google Scholar]
- Li X.F., Li X.D., Deng C.L., Dong H.L., Zhang Q.Y., Ye Q., Ye H.Q., Huang X.Y., Deng Y.Q., Zhang B., Qin C.F. Visualization of a neurotropic flavivirus infection in mouse reveals unique viscerotropism controlled by host type I interferon signaling. Theranostics. 2017;7:912–925. doi: 10.7150/thno.16615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Brecher M., Deng Y.Q., Zhang J., Sakamuru S., Liu B., Huang R., Koetzner C.A., Allen C.A., Jones S.A., Chen H., Zhang N.N., Tian M., Gao F., Lin Q., Banavali N., Zhou J., Boles N., Xia M., Kramer L.D., Qin C.F., Li H. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res. 2017;27:1046–1064. doi: 10.1038/cr.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison J., Aguirre S., Fernandez-Sesma A. Innate immunity evasion by Dengue virus. Viruses. 2012;4:397–413. doi: 10.3390/v4030397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C.L., Jones C.T., Rice C.M. Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat. Rev. Microbiol. 2008;6:699–708. doi: 10.1038/nrmicro1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonov A., Molder T., Sikut R., Kiiver K., Mannik A., Toots U., Lulla A., Lulla V., Utt A., Merits A., Ustav M. RIG-I and MDA-5 detection of viral RNA-dependent RNA polymerase activity restricts positive-strand RNA virus replication. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin J.F., Floyd R.A., Dittmer D.P. Methylene blue photoinactivation abolishes West Nile virus infectivity in vivo. Antivir. Res. 2005;68:84–87. doi: 10.1016/j.antiviral.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Roby J.A., Setoh Y.X., Hall R.A., Khromykh A.A. Post-translational regulation and modifications of flavivirus structural proteins. J. Gen. Virol. 2015;96:1551–1569. doi: 10.1099/vir.0.000097. [DOI] [PubMed] [Google Scholar]
- Ruan Z., Liu C., Guo Y., He Z., Huang X., Jia X., Yang T. SARS-CoV-2 and SARS-CoV: virtual Screening of Potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12) J. Med. Virol. 2020 doi: 10.1002/jmv.26222. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath A., Padmanabhan R. Molecular targets for flavivirus drug discovery. Antivir. Res. 2009;81:6–15. doi: 10.1016/j.antiviral.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Dorner M., Feulner M., Imanaka N., Murphy M.Y., Ploss A., Rice C.M. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proc. Natl. Acad. Sci. U. S. A. 2012;109:14610–14615. doi: 10.1073/pnas.1212379109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian L., Madhusudana S.N., Ravi V., Desai A. Mycophenolic acid inhibits replication of Japanese encephalitis virus. Chemotherapy. 2011;57:56–61. doi: 10.1159/000321483. [DOI] [PubMed] [Google Scholar]
- Shahen M., Guo Z., Shar A.H., Ebaid R., Tao Q., Zhang W., Wu Z., Bai Y., Fu Y., Zheng C., Wang H., Shar P.A., Liu J., Wang Z., Xiao W., Wang Y. Dengue virus causes changes of MicroRNA-genes regulatory network revealing potential targets for antiviral drugs. BMC Syst. Biol. 2018;12:2. doi: 10.1186/s12918-017-0518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang B., Deng C., Ye H., Xu W., Yuan Z., Shi P.Y., Zhang B. Development and characterization of a stable eGFP enterovirus 71 for antiviral screening. Antivir. Res. 2013;97:198–205. doi: 10.1016/j.antiviral.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Shi P.Y. Structural biology. Unraveling a flavivirus enigma. Science (New York, N.Y.) 2014;343:849–850. doi: 10.1126/science.1251249. [DOI] [PubMed] [Google Scholar]
- Simanjuntak Y., Liang J.J., Lee Y.L., Lin Y.L. Repurposing of prochlorperazine for use against dengue virus infection. J. Infect. Dis. 2015;211:394–404. doi: 10.1093/infdis/jiu377. [DOI] [PubMed] [Google Scholar]
- Tu Y.C., Yu C.Y., Liang J.J., Lin E., Liao C.L., Lin Y.L. Blocking double-stranded RNA-activated protein kinase PKR by Japanese encephalitis virus nonstructural protein 2A. J. Virol. 2012;86:10347–10358. doi: 10.1128/JVI.00525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtle L., Driver C. Risk assessment for Japanese encephalitis vaccination. Hum. Vaccines Immunother. 2018;14:213–217. doi: 10.1080/21645515.2017.1380756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veschi S., De Lellis L., Florio R., Lanuti P., Massucci A., Tinari N., De Tursi M., di Sebastiano P., Marchisio M., Natoli C., Cama A. Effects of repurposed drug candidates nitroxoline and nelfinavir as single agents or in combination with erlotinib in pancreatic cancer cells. J. Exp. Clin. Canc. Res. 2018;37:236. doi: 10.1186/s13046-018-0904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Liu Y., Guo J., Wang P., Zhang L., Xiao G., Wang W. Screening of FDA-approved drugs for inhibitors of Japanese encephalitis virus infection. J. Virol. 2017;91 doi: 10.1128/JVI.01055-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Chen Y.L., Schul W., Wang Q.Y., Gu F., Duraiswamy J., Kondreddi R.R., Niyomrattanakit P., Lakshminarayana S.B., Goh A., Xu H.Y., Liu W., Liu B., Lim J.Y., Ng C.Y., Qing M., Lim C.C., Yip A., Wang G., Chan W.L., Tan H.P., Lin K., Zhang B., Zou G., Bernard K.A., Garrett C., Beltz K., Dong M., Weaver M., He H., Pichota A., Dartois V., Keller T.H., Shi P.Y. An adenosine nucleoside inhibitor of dengue virus. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20435–20439. doi: 10.1073/pnas.0907010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhang Q.Y., Li X.D., Liu S.Q., Deng C.L., Zhang B., Ye H.Q. Development of a stable Japanese encephalitis virus replicon cell line for antiviral screening. Arch. Virol. 2017;162:3417–3423. doi: 10.1007/s00705-017-3508-9. [DOI] [PubMed] [Google Scholar]
- Zou G., Xu H.Y., Qing M., Wang Q.Y., Shi P.Y. Development and characterization of a stable luciferase dengue virus for high-throughput screening. Antivir. Res. 2011;91:11–19. doi: 10.1016/j.antiviral.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Zu X., Liu Y., Wang S., Jin R., Zhou Z., Liu H., Gong R., Xiao G., Wang W. Peptide inhibitor of Japanese encephalitis virus infection targeting envelope protein domain III. Antivir. Res. 2014;104:7–14. doi: 10.1016/j.antiviral.2014.01.011. [DOI] [PubMed] [Google Scholar]