Abstract
Breast cancer initiation and progression are often observed as the result of dysregulation of normal developmental processes and pathways. Studies focused on normal mammary stem/progenitor cell activity have led to an understanding of how breast cancer cells acquire stemness-associated properties including tumor initiation, survival and multi-lineage differentiation into heterogeneous tumors that become difficult to target therapeutically. Importantly, more recent investigations have provided valuable insight into how key developmental regulators can impact multiple phases of metastasis, where they are repurposed to not only promote metastatic phenotypes such as migration, invasion and EMT at the primary site, but also to regulate the survival, initiation and maintenance of metastatic lesions at secondary organs. Herein, we discuss findings that have led to a better understanding of how embryonic and pluripotency factors contribute not only to normal mammary development, but also to metastatic progression. We further examine the therapeutic potential of targeting these developmental pathways, and discuss how a better understanding of compensatory mechanisms, crosstalk between pathways, and novel experimental models could provide critical insight into how we might exploit embryonic and pluripotency regulators to inhibit tumor progression and metastasis.
Introduction
Embryonic development involves the intricate and dynamic coordination of cellular and molecular processes under strict spatial and temporal control. For example, a unicellular zygote gives rise to progeny cells that not only multiply in number, but also migrate and differentiate at specific times and places in the embryo. This ultimately results in a mature, multicellular organism at the time of birth. The mammary gland is one of a few tissues, along with the uterus, ovary, brain and testis, that develops predominately after birth. A diverse set of cell types drive the formation of this highly complex organ through interaction with, and regulation by, local and systemic hormones and growth factors. Because of the highly structured order of events and substantial availability of mouse models to study mammary gland development, we have gained vital knowledge of the molecular underpinnings that regulate this process.
Mammary gland development in mice can be described in three distinct stages: embryonic, pubertal and adult [1]. During the embryonic stage, there are three key events: establishment of the bilateral milk lines (ventral epidermal ridges from which the mammary gland and nipples originate), placode development, primitive mammary bud formation and branching [2]. Birth to the onset of puberty is considered a relatively quiescent state where the growth of the ductal tree within the mammary gland is isometric with body growth [1]. With the onset of puberty, ovarian, and pituitary-derived hormones (primarily) mediate branching morphogenesis, resulting in the ductal tree filling the mammary fat pad. In the case of mice, lobules do not appear until the onset of pregnancy. Of note, although there is significant expansion of the ductal tree network into the fat pad, space remains in order to support additional branching during diestrus and/or pregnancy [1]. The mammary gland continues to respond to secretion of ovarian hormones during each estrous cycle, which allows for the formation of alveolar and lateral buds. Pregnancy ushers in a dynamic and extensive remodeling of the mammary gland mainly involving proliferation and differentiation of the alveolar buds into cells capable of producing and secreting milk [1].
Human mammary gland development shares many common features with mouse mammogenesis, although there are also some clear differences. For instance, humans only have one pair of mammary placodes, versus the five pairs in mice, and have multiple ductal trees in each mammary gland that concentrate at the nipple, versus a single ductal tree connected to the teat as seen in mice [2]. Another major difference is that lobules are present in the human mammary gland prior to pregnancy, whereas the formation of lobules for most laboratory strains of mice does not occur until pregnancy [1].
Two main cell lineages make up the hierarchal organization of the mammary gland, where the major function of the basal population is contraction, whereas the luminal population is critical for milk production [3]. Over the years, many studies have identified a critical population of mammary stem cells (MaSCs) that sits atop of this hierarchy and can give rise to the progenitors and differentiated cells of both lineages. The first evidence of the existence of MaSCs was obtained through transplantation studies, where small segments of mammary epithelium (~0.5–1 mm in length) implanted into the epithelium-free “cleared” mammary fat pads of recipient mice were able to reconstitute the entire mammary gland [4–6]. These data were an early indication that parts of the mammary gland had regenerative capacity, and begged the question of whether a specialized population of cells within the mammary gland are responsible for its regenerative capacity [4]. Subsequent studies further supported the theory of a specialized population of MaSCs by using flow cytometry to isolate rare populations of cells based largely on surface marker expression of CD24, CD29 and CD49f, which could repopulate an entire, functional, mammary gland. For instance, Visvader and colleagues found that CD24med/CD49fhigh expressing cells represented adult MaSCs, referred to as mammary repopulating units (MRUs), that could reliably regenerate entire functioning mammary glands via repeated transplantations [7]. Around the same time, Eaves and colleagues used cell sorting and limiting dilution transplant assays to purify a single population of adult mouse mammary cells that had the ability to regenerate an entire mammary gland [8]. Importantly, these cells, identified as mammary stem cells, were shown to give rise to, and be distinct from, mammary epithelial progenitor cells, with gene expression patterns similar to basal cells [8]. Taken together, these two studies established that a single cell had the capacity to regenerate a complete mammary ductal tree. Since these seminal studies, controversies have arisen as to the origin, timing and designations of multipotent MaSCs [9–11]. Nonetheless, along with subsequent studies discussed in this review, these data support the supposition that rare populations exist within the mammary gland that are critical for proper mammary gland development.
More recent studies using lineage-tracing and single-cell RNA sequencing analyses have been used to better understand the hierarchal organization of mammary stem cells within the mammary gland. For example, lineage-tracing and single-cell RNA-seq studies have presented evidence of a hierarchy that places the basal population as the progenitors for luminal cell lineages [12,13]. However, it is important to note that although single-cell RNA-seq has significantly increased the ability to dissect the heterogeneity of precursors and progenitors involved in mammary gland development, the identification of a distinct stem cell population in the normal mouse or human mammary gland using this technology remains a challenge. Nevertheless, these studies did reinforce previous reports by Visvader and colleagues that suggested the regenerative capacity of the mammary gland originates from the basal population [7,12]. Importantly, during this time other lineage-tracing studies challenged this model by proposing that rather than one bipotent basal cell population, there are unipotent basal and luminal cells that are responsible for the maintenance of the mammary gland [14]. Specifically, multipotent, MaSCs (associated with embryonic mammary development) are mainly present during embryonic development where they are critical for the expansion of basal and luminal progenitor cells ([14] and references therein). After birth, these more differentiated progenitor cells (also known as lineage-restricted stem cells [15–17]) serve to produce all of the necessary cell types needed throughout postnatal development, puberty, pregnancy, lactation and involution [14]. Building on this work, reports from Blanpain and colleagues combined lineage-tracing with scRNAseq analyses to study the dynamics of differentiation within the embryonic multipotent progenitor (EMP) population during mammary gland development [15]. Initially, they found that lineage commitment of EMPs occurs early on in mammary development, with basal cells becoming completely unipotent by 1 day postnatal [15]. Additionally, flow cytometry and scRNAseq analyses demonstrated that EMPs express both basal and luminal-associated gene signatures, suggesting that multipotency is associated with a hybrid basal-luminal cell state [15]. Further, Scheele and Lloyd-Lewis and colleagues used neutral, low-density lineage tracing (to reduce the chances of clonal convergence) to better understand the contributions of proliferative stem/progenitor cells during mammary gland development [16,17]. Similar to Blanpain and colleagues, these reports suggest that lineage commitment of stem/progenitor cells occurs in the postnatal mammary gland, with a significant level of redundancy and heterogeneity within the stem/progenitor pool [16,17]. Together, these reports provide critical insights into the temporal contributions of lineage-restriction and the molecular profiles of stem/progenitor populations during mammary gland development. Moreover, these findings support a more complex hierarchal model where instead of one MaSC population that gives rise to all cell types in the ducts, multipotent MaSCs can differentiate into more specialized progenitor cell types (basal and/or luminal) that function to properly develop and maintain the mammary epithelium.
Intriguingly, the molecular mechanisms and signaling pathways that regulate the functions of the embryonic MaSCs and more differentiated, unipotent progenitor populations are almost universally shared with malignant cells. Like normal MaSCs, tumor-initiating cells (TICs) from the breast are defined by their unique ability to self-renew and give rise to more differentiated tumor cell progeny. One of the first studies to identify a TIC population isolated CD24−/CD44+ cells from human breast tumors and demonstrated that this cell population could initiate tumors with heterogeneous progeny over multiple transplantations in vivo [18]. Such studies demonstrated that a rare population of cells within a tumor could hijack normal stem cell functions to promote tumor initiation. Additionally, mouse models of spontaneous mammary carcinoma, such as the MMTV-PyMT model, have been used to show that during hyperplasia, the mouse MaSC population (isolated by Lin−CD24+CD29hi and/or Lin−CD24+CD49fhi expression [7,19]) is expanded and, when isolated, has increased tumorigenicity in vivo [20]. Other studies have also identified additional markers to isolate and study functions of breast TICs that are similar to those used to identify normal mammary stem cells, including ALDH, WNT (active signaling and target genes) and STAT3 [21–27]. More recently, studies have also established that populations outside of the embryonic MaSC population have the ability to induce tumorigenesis. For example, Nebreda and colleagues found that the protein kinase p38α regulates the transcription factor Runx1 to control the luminal progenitor cell fate in mice [28]. Intriguingly, downregulation of p38α in a mouse model of breast cancer derived from differentiated luminal cells decreases TICs and overall tumor burden [28]. Another example of additional cell populations that contribute to the initiation of tumorigenesis came from Visvader and colleagues, who reported that constitutive Notch signaling regulates the conversion of a MaSC-enriched population, based on flow cytometry analysis, down a luminal lineage. Interestingly, sustained Notch signaling in this context resulted in expansion of luminal progenitor cells and increased tumorigenesis [29]. Lastly, a recent report by Li and colleagues used mouse models to demonstrate that subsets of precancerous cells can give rise to distinct cancer phenotypes, with keratine 6a+ (stem) and WAP+ (more differentiated) cells leading to adenocarcinoma and metaplastic carcinoma, respectively [30]. These data suggest that a MaSC-enriched population, along with progenitor and differentiated populations, could serve as drivers for tumor initiation and determine tumor subtypes during oncogenic transformation.
It has recently been proposed that disseminated tumor cells (DTCs), in addition to circulating tumor cells (CTCs), contain a unique population of metastasis-initiating cells (MICs) that possess the ability to generate detectable metastases at secondary sites [14]. Importantly, recent studies have suggested that the MIC population specifically regulates stemness and differentiation properties that allow for better survival and establishment of newly arriving tumor cells at secondary sites, and thus share many properties with TICs [14]. In addition, given the role of tissue-specific cell-fate regulators in mediating breast cancer metastasis [22,31], recent studies have begun to examine whether more global embryonic cell-fate regulators, such as pluripotency factors, function in the MIC population to promote metastatic outgrowth [32–34]. Although it remains difficult to isolate, study and characterize MICs [14], recent advances in genomic sequencing, imaging and animal modeling have begun to provide insights into the molecular mechanisms that regulate MICs. Through these studies, MICs have been shown to display a diverse set of properties including the ability to evade the immune system, to exit from dormancy, to regulate secondary niches (both pre and post arrival at the secondary site), and to survive the harsh conditions they encounter as they first inhabit the microenvironment at the metastatic site [14]. It is proposed that central to all of these abilities is the fact that MICs have an extraordinary level of plasticity, allowing them to adjust to a plethora of different stimuli and microenvironments [14]. Thus, current studies examining MICs have mainly focused on the role of pluripotency and altered differentiation states as a way to better understand how we might target this population to inhibit metastasis. In this review, we discuss the impact of HH, WNT and NOTCH developmental pathways, which are among a handful of highly conserved signaling nodes that control numerous developmental processes such as patterning, morphogenesis and cell-fate determination, on normal, multipotent MaSCs during mammary gland development. Importantly, we will also examine how their roles in mammary gland development have provided critical insight into how stemness properties can influence metastasis. Additionally, we will summarize the emerging roles of pluripotency factors in metastatic progression and colonization. Lastly, we discuss how this knowledge has led to the identification of potential novel therapeutic approaches to combat breast cancer progression.
Developmental Signaling Pathways in Mammary Stem Cells and Breast Cancer Metastasis
The Hedgehog (HH) Pathway
The HH Pathway in Mammary Stem Cells
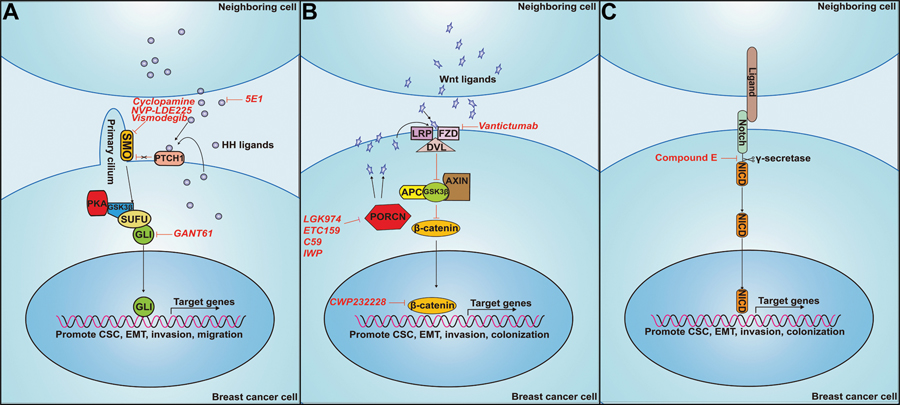
Initiation of the canonical Hedgehog (HH) signaling cascade relies on the presence of the HH ligands. In mammals, the ligands include Sonic Hedgehog (SHH), Desert Hedgehog (DHH) and Indian Hedgehog (IHH) [35,36]. In the absence of HH ligands, the Patched family of transmembrane ligand-binding receptors [PATCHED-1 (PTCH1), and to a lesser extent PATCHED-2 (PTCH2)] inhibits the G-protein-coupled receptor SMOOTHENED (SMO) by keeping it sequestered in vesicle membranes. Sequestration of SMO to vesicles results in phosphorylation [via Protein Kinase A (PKA), Glycogen Synthase Kinase 3 Beta (GSK-3β) or Casein Kinase 1 (CK1)] of members of the GLI transcription factor family (GLI1, GLI2 and GLI3). These phosphorylation events lead to the production of proteolytically cleaved truncated forms of GLI2 and/or GLI3 that translocate to the nucleus and act as transcriptional repressors of HH target genes to suppress the pathway. GLI1 lacks the cleavage site and thus functions exclusively as a transcriptional activator as a full-length protein (Fig. 1A).
Figure 1: Means to inhibit canonical HH, WNT and NOTCH pathways.
A. HH Pathway. Binding of HH ligands (SHH, DHH or IHH) to PTCH receptors relieves PTCH-mediated inhibition of SMO. SMO inhibits PKA, GSK3β, and SUFU from binding to GLI, allowing for stabilization of the activator confirmation, translocation to the nucleus and activation of HH target genes. The monoclonal antibody, 5E1, binds to SHH and prevents its binding to PTCH receptors. Cyclopamine binds to, and prevents, SMO-mediated activation of GLI transcription factors. GANT61 reduces GLI transcription factor DNA binding to block GLI-mediated HH target gene expression. B. WNT pathway. Binding of WNT ligands to the receptor FZD results in translocation of DVL to the membrane. DVL binds to the APC/GSK3β/AXIN complex, leading to its dissociation from β-CATENIN. β-CATENIN is released and stabilized, allowing for translocation to the nucleus, where it activates transcription of target genes. The antibody Vantictumab binds to FZD and prevents WNT ligand binding. LGK974, ETC159, C59 and IWP inhibit PORCN-mediated processing and secretion of WNT ligands that activate the canonical pathway. C. Notch pathway. During canonical signaling, NOTCH receptors on one cell bind to ligand receptors on another cell, after which the NOTCH receptor is cleaved by the protease complex, γ-SECRETASE, leading to release of the NOTCH Intracellular Domain (NOTCH-ICD). NOTCH-ICD translocates to the nucleus to activate transcription of target genes. Compound E functions to block the cleavage of both γ-secretase and NOTCH-ICD, preventing NOTCH-ICD translocation to the nucleus.
When HH ligand is present, it binds to PTCH receptors, resulting in a conformational change to PTCH that results in the release of SMO from vesicles to primary cilia. Cilia associated SMO inhibits the phosphorylation (and cleavage in the case of GLI2 and GLI3) of one or more of the GLI proteins, allowing GLI family members to translocate away from the cilium and accumulate in the nucleus, where they act as transcription factors to activate Hh target genes that are important for cellular survival, proliferation, migration, invasion and for the induction of stem cell properties [35] (Fig. 1A).
Studies have suggested that certain components of the Hh pathway could regulate stemness in the mammary gland [35,37], although they remain controversial. For example, CD24+CD29hi mammary stem cell enriched populations isolated from heterozygous Ptch1 mice showed decreased long-term label retention compared to the same cells isolated from their wildtype counterparts [38], suggestive of role for Hh signaling in mammary stemness. However, conclusions regarding stemness in this study were drawn primarily from cell surface marker expression, and not from functional experiments. Others have attempted to focus on functional studies of Hh signaling, albeit with somewhat confounding results, to determine whether pathway activity is important for mammary stem/progenitor cell populations. For example, limiting dilution transplantation experiments using primary mammary epithelial cells isolated from dissected mammary glands from an activated human SMO mouse mammary gland model (MMTV-SmoM2) demonstrated that SMO activation decreased stem cell frequency [39]. Despite this observed decrease in stemness in vivo, SMO activation increased the ability of these cells to form mammospheres in vitro [39]. These data suggest that while SMO can promote increases in progenitor populations, it does so at the expense of loss of regenerative stem cells. However, it is important to note that the MMTV-SmoM2 model results in constitutively active Hh signaling, and therefore represents artificial activation of the pathway. Nevertheless, in line with this finding, DiRenzo and colleagues demonstrated that Ptch1+/− mice have an increased Lin−/CD24+/CD29low mammary progenitor cell population [38]. These mice exhibit constitutively active Hh signaling in MaSC-enriched populations based on flow cytometry analysis, releasing them from quiescence and leading to an expansion of the mammary progenitor pool [38]. Still, caveats must be considered for these data as well, as the models consist of constitutively active components of the pathway, and thus artificially represent Hh pathway signaling.
Similarly, Wicha and colleagues observed increased expression of several HH signaling components including SMO, PTCH1, GLI1 and GLI2 by comparing gene expression of mammosphere-derived cells grown in suspension to enrich for mammary stem/progenitor cells versus mammosphere-derived cells cultured on a collagen substratum to induce differentiation [40]. Moreover, treatment with SHH increased primary and secondary mammosphere formation. This effect was shown to be dependent on SMO because treatment with cyclopamine, a direct antagonist of SMO, blocked SHH-mediated increases in mammosphere formation [40]. Nevertheless, although these studies suggest HH signaling proteins may promote progenitor-associated properties in the normal mammary gland, they do so with artificial, rather than endogenous, signaling activation. Given these data, it is not surprising that in vivo transplantation of Shh or Ihh deleted mammary epithelium into cleared mammary fat pads resulted in completely normal mammary gland development [37]. Similarly, whole body homozygous Gli1 KO mice (which are viable) display no overt mammary gland phenotypes, and normal mammary development was also observed when embryonic mammary glands derived from whole body Gli2 KO mice (which are perinatal lethal) were transplanted into cleared fat pads. However, the lack of mammary phenotypes does not rule out a role for Hh signaling in mammary stem/progenitor cells, as functional redundancy between the protein family members may mask effects. Furthermore, the lesser studied ligand Dhh and receptor Ptch2, were not examined in this context, and it is thus possible that the pathway could potentially play key roles in mammary stem cells.
Studies attempting to identify the underlying mechanisms by which the downstream effectors of the Hh pathway, the Gli transcription factors, regulate mammary gland development remain difficult, likely in part due to perinatal lethality of Gli2 and Gli3, and partly due to issues of potential redundancy (similar to the ligands) [41]. Joyner and colleagues provided evidence for functional redundancy of Gli transcription factors by replacing Gli2 with Gli1 in Gli2-null KO mice, which was sufficient to restore activation of the Hh signaling pathway and organismal viability in the Gli2 KO context [42]. Further, although Gli1 KO mice are viable with no mammary phenotypes detected, heterozygous KO of Gli2 in conjunction with homozygous Gli1 KO results in severe developmental defects that are lethal perinatally, suggesting dose-dependent compensation of the Gli transcription factors [42]. Interestingly, Cowin and colleagues showed that despite Gli2 and Gli3 being expressed during embryonic mammary gland development, Hh signaling could not be detected in this context when using Gli1 or Ptch-lacZ reporters [43]. However, mice carrying an intragenic deletion in Gli3, resulting in loss of Gli3 expression (null alleles), displayed abnormal early mammary marker expression and loss of mammary bud pairs. Importantly, this effect was attributed to Gli3 loss of function in the somite underlying the mammary gland. Thus, further studies are needed to shed light on potential roles for Gli3 in the mammary epithelium. In further support of the presence of active Hh signaling in the mammary gland, it was shown that Gli3 loss results in an increase in mammary mesenchyme Gli1 expression, suggesting that role of Gli3 is to repress otherwise active Hedgehog/Gli1-mediated activity during embryonic mammary gland development. Studies from Veltmaat and colleagues support this model, as they observed, through characterization of Gli3-null mice, that Gli3-mediated activation of Fgf10 expression was necessary to induce mammary cell fate in the ectoderm [44]. Taken together, these data suggest that Gli3, rather than Gli1 or Gli2, is the predominant mediator of embryonic mammary gland development. Still, as outlined above, some studies remain contradictory and thus further investigations are warranted. Because most studies focus on the study of Hh pathway components in isolation, there remain unanswered questions around redundancy. Therefore, studies focusing on compensatory mechanisms of Gli transcription factors and Hh ligands may shed more light on their functional roles in mammary stem cell activity and development overall.
Role of HH Pathway During Breast Cancer Metastasis
While the role of Hh signaling during normal mammary gland development and in MaSC-associated populations remains debated, a key role for the HH signaling network in transformation and primary breast tumor growth has been clearly elucidated [37,45]. In this review, we focus our attention on how active HH signaling influences breast cancer stemness and metastasis. A study by Wei and colleagues recently linked GLI signaling to stemness and metastatic properties. Using an inducible Gli1 knockdown (KD) model, the authors demonstrated that Gli1 is critical downstream of estrogen stimulation to promote both the expansion of the TIC population and the enhancement of metastatic phenotypes such as invasion and EMT in vitro, suggesting a link between stemness properties and metastatic capabilities [46].
However, it should be noted that the role of Hh/Gli signaling in tumor progression is complex, and does not only involve autocrine functions even within the tumors. We recently demonstrated that breast tumor cells can be induced to activate GLI-mediated transcription in response to paracrine signals emanating from breast tumor cells that had undergone an epithelial to mesenchymal transition (EMT) [47]. Activation of GLI in the more epithelial breast cancer cells results in increased migration, invasion, and anoikis resistance in vitro [47]. Intriguingly, treatment of carcinoma cells with the GLI inhibitor, GANT61, inhibits paracrine mediated, EMT-induced metastasis-associated phenotypes in vitro and paracrine-mediated metastasis of the epithelial breast cancer cells in vivo. Of importance, the paracrine mechanism of GLI activation in heterogenous populations containing EMT and non-EMT cells is not always driven by HH ligands, and instead non-canonical paracrine activation of the pathways is often observed. In line with this finding, PDX tumor growth could be inhibited more effectively with GANT61, than with a SMO inhibitor, suggesting that GLI can act in a SMO-independent manner to promote breast tumor progression [47].
While the study above demonstrates a role for Hh/Gli mediated cross talk between tumor cells in heterogeneous breast tumors, Hh/Gli signaling also functions to mediate crosstalk with peritumoral cells within the tumor microenvironment. For instance, using several breast cancer xenograft models, it was shown that GLI1 activity in tumor cells can promote the expression of critical osteolytic factors, Rank ligand (RANKL) and osteopontin (OPN), in neighboring osteoblasts to facilitate aberrant osteoblast differentiation. This signaling event leads to overstimulation of osteoblast differentiation and subsequent increases in osteoclast differentiation [48], and ultimately culminates in enhanced osteolysis, releasing growth and survival factors that provide a more favorable environment for metastatic outgrowth in bone tissue. In addition, Bianco and colleagues reported that activated HH signaling in TNBC cells leads to Smo-dependent secretion of the pro-angiogenic factor VEGF-A, leading to enhanced tumor vascularization [49]. Importantly, treatment with the selective SMO antagonist NVP-LDE225 could significantly inhibit endothelial cell organization and vascularization in vivo. Similarly, Shevde and colleagues demonstrated that SHH/GLI1 upregulation in tumor cells promotes secretion of cysteine-rich angiogenic inducer 61 (CYR61) into the tumor microenvironment to promote vascularization and easier access to the blood stream for metastasis [50]. Collectively, these data suggest that HH signaling can promote metastasis by multiple mechanisms.
Overall, these studies and others support the hypothesis that Hh/Gli signaling is important in mediating metastatic progression, and that blockade of critical components of the pathway may be able to block multiple aspects of metastatic disease [51–55]. However, despite some clinical success with the use of Hh-targeted therapies (SMO inhibtors: vismodegib, sonidegib and glasdegib) in multiple tumor types such as basal cell carcinoma (BCC), acute myeloid leukemia (AML) and medulloblastoma, results from Hh pathway inhibitor treatment in breast cancer have been largely disappointing [56]. For example, clinical trials using the SMO antagonist, Vismodegib, in combination with the NOTCH pathway inhibitor, RO4929097 for treatment of advanced breast cancer (defined as metastatic or unresectable) have been conducted. The rationale for this therapeutic strategy came from preclinical studies demonstrating that both the HH and NOTCH pathways play critical roles in cancer stemness (trial ID NCT01071564). However, the clinical trial had to be terminated due to life-threatening complications arising from arrythmia (Available from: http://clinicaltrials.gov/show/NCT01071564). Additional clinical trials using SMO inhibitors in combination with other targeted therapies or chemotherapy are ongoing or have been completed, with too small numbers to report conclusive results [56]. Moreover, recently it has also been shown that BCC patients can develop resistance to SMO inhibitors and relapse through acquired mutations in SMO [57]. This has led to a shift towards therapeutics that target downstream mediators of the pathway, such as GLI1 and GLI2, as means to bypass SMO inhibitor resistance and address non-canonical mechanisms of pathway activation. Nonetheless, because of the various roles of HH signaling in the crosstalk between tumor cells and the surrounding microenvironment, therapeutic strategies should be tailored to a specific patient. The tailoring of therapy will be critical, as recent reports suggest stromal-derived HH signaling can both promote and suppress tumor growth and metastasis, and that this is tumor type and stage dependent [36]. Also, since it is thought that only a small number of cells within a tumor respond to and/or utilize Hh signaling, bulk tumor size may not be the most useful way to evaluate efficacy. Further, as with all of the signaling pathways discussed in this review, tumor heterogeneity and subtype must be considered when determining the role activated signaling plays in tumor progression, and whether targeting of the pathway will be helpful to the patient. Therefore, continued investigation into the roles of Hh signaling both in cancer cells, as well as in the microenvironment, are needed to better understand and more effectively inhibit its tumor promotional functions.
The WNT Pathway
Role of WNT Signaling in Mammary Stem cells
WNT signaling plays a fundamental role in embryonic development. Genetic mouse models examining multiple Wnt signaling components such as Wnt ligands, and downstream transcriptional complexes made up of Cadherin-associated protein beta 1 (β-catenin), Lymphoid enhancer-binding factor (Lef1) and T-cell factor (Tcf), have demonstrated that the Wnt pathway is critical for the morphogenesis of multiple organ systems such as the intestine, bone, lungs, kidneys, hematopoietic system and mammary gland [58]. From a molecular standpoint, the WNT pathway can be further divided into two distinct intracellular pathways: canonical and non-canonical [58,59]. The canonical WNT pathway is defined by the involvement of β-CATENIN as the main mediator of downstream pathway activation. In the context of canonical signaling, absence of WNT ligands results in β-CATENIN being readily bound by the destruction complex (DC) mainly consisting of Adenomatous Polyposis Coli (APC), Axis inhibitor (AXIN), and Glycogen Synthase Kinase 3 Beta (GSK3β). This complex formation results in β-CATENIN phosphorylation and subsequent proteasome-mediated degradation. Upon binding of WNT ligands to the FZD receptor, DISHEVELLED (DVL) is recruited to the membrane, where it binds AXIN and GSK3β, leading to phosphorylation of Transmembrane Low-density Lipoprotein Receptor-related Protein 5/6 (LRP5/6) and stabilization of β-CATENIN. β-CATENIN is then free to translocate to the nucleus, where it forms complexes with members of the TCF/LEF family to activate transcription of target genes involved in self-renewal, EMT and cell proliferation [58,59] (Fig. 1B).
Non-canonical WNT signaling, which can be further subdivided into planar cell polarity and calcium WNT pathways, functions independent of β-CATENIN [60,61]. Although both non-canonical pathways still largely involve WNT ligand-mediated activation of FZD, the downstream signaling events are distinct. The non-canonical planar cell polarity pathway involves activation of RHO family GTPases such as RHO, RAC and CDC42 and subsequent phosphorylation and activation of downstream effectors by JUN N-terminal kinase (JNK) or RHO kinase [58,59]. This pathway ultimately results in downstream target gene activation that promotes cytoskeleton reorganization and polarized cell movements [58,59]. Activation of the non-canonical calcium WNT pathway results in a different cascade of protein activity, with a significant dependence on heterotrimeric G proteins [58,59]. Briefly, interaction of FZD with trimeric G-proteins leads to activation of either Phospholipase C (PLC) or Phosphodiesterase (PDE) [58,59]. PLC activation results in calcium release from the endoplasmic reticulum, whereas PDE inhibits calcium release. The release of intracellular calcium leads to calcineurin- and CaMKII-mediated activation of the transcription factor NFAT, and expression of genes involved in various cellular processes such as migration and adhesion [58,59].
The WNT pathway influences stemness in multiple tissues, including the mammary gland, through both autocrine and paracrine mechanisms [62,63]. Mouse models of multiple Wnt pathway components such as Lrp5, β-catenin and Wnt4 have demonstrated that the pathway is essential for mammary stem cell maintenance and function during mammogenesis [64]. A relatively novel Wnt target gene, protein C receptor (Procr) has been used as a marker for multipotent mouse MaSCs [22]. Importantly, Procr+ cells isolated from the basal population (the population presumed to be enriched for MaSCs) possess higher colony forming abilities in vitro and mammary gland reconstitution capacity in vivo compared to either Procr- or the total basal cell populations [22]. Further evidence for an association of Wnt and mammary stemness comes from expression studies, which demonstrate that Limb Bud and Heart Development Homolog (LBH), a transcriptional co-factor in the WNT pathway, is associated with Lin−CD29highCD24+ basal MaSC population [65]. In addition, loss of Lbh in purified basal MaSCs leads to increased differentiation into luminal epithelial cells (as compared to control cells) after growth in differentiation medium. Overall, the data support a role for the Wnt-pathway as an essential signaling cascade to promote mammary stem cell maintenance and activity.
In addition to autocrine Wnt actions, paracrine roles for the molecule have been described in mammary stem cell biology. For example, IHC and gene expression studies of the mouse estrous cycle suggest that progesterone-expressing luminal cells can drive expansion of adult MaSCs from the basal population during the luteal diestrus phase, through a mechanism that relies on Wnt-mediated paracrine signaling [66]. In particular, gene expression analysis from isolated luminal and basal cells from mouse mammary glands showed that luminal-derived Wnt4 ligand downstream of progesterone signaling was met with corresponding upregulation of Lrp5 on basal cells and increased stem cell activity [66]. These data suggest that Wnt4 could act as a critical downstream paracrine effector of progesterone-induced adult MaSC expansion. However, more functional studies are needed to confirm these observations. Additionally, future studies examining Wnt singaling in the context of mammary stem/progenitor cell function should consider the diversity of ligands and their interactions during signal transduction. For example, while Zeng and Nusse showed that Wnt3a is sufficient to promote long-term culture of adult MaSCs [62], Werb and colleagues demonstrated that although Wnt5a promotes the growth of mammary stem/progenitor cells, its close ligand relative Wnt5b is capable of inhibiting mammary stem/progenitor cell growth capacity [67]. Despite these opposing phenotypes within the same pathway, a focus on the spatial and temporal heterogeneity, signal potency and cell-context dependent signaling could help decipher the effects of the pathway on mammary stem/progenitor function overall. Thus, continued development of technologies such as live-cell imaging, multiplex staining and 3D organoid culture models will become even more critical in order address these complex questions.
Role of WNT Signaling During Breast Cancer Metastasis
Altered activity of the Wnt pathway is implicated in numerous cancer types such as colon, melanoma, hepatocellular and of course, breast [68]. In the context of breast cancer, canonical WNT signaling is activated (as measured by β-CATENIN nuclear accumulation) in approximately 50% of all breast cancers and correlates with poor survival [69]. Of note, only a small percentage of breast tumors contain somatic mutations of WNT signaling components [70]. Instead, many studies have identified aberrant expression of pathway components as a prominent mechanism of oncogenic function [63]. Additionally, WNT signaling components such as CTNNB1, LEF1 and AXIN2 are often amplified or more highly expressed in breast cancer cell lines and breast tumors when compared to their normal counterparts, and play a critical role in multiple aspects of tumor progression and metastasis.
Many studies have uncovered roles for the critical WNT downstream regulator, β-CATENIN, in cancer stemness and EMT, and these effects of β-CATENIN impinge on both tumor initiation and metastasis [68]. Here, we focus on the role of β-CATENIN in regulating stemness to promote metastatic progression and its potential as a therapeutic target for inhibiting MIC-mediated metastasis [71].
Some years ago, Nam and colleagues observed that β-CATENIN activity was significantly increased in breast cancer stem cells compared to the bulk tumor, suggesting that, consistent with its association and role within normal mammary stem cells, WNT signaling could also play a role within the breast cancer stem cell, or tumor-initiating, population [72]. Indeed, this group demonstrated that Wnt/β-catenin signaling was not only necessary and sufficient for expansion of the breast cancer stem cell population, but that it also promoted the aggressive, migratory nature of breast cancer stem cells, which was important for increased metastasis in vivo [71]. Moreover, using a highly metastatic mouse mammary carcinoma cell model (4T1), they demonstrated that both specific KD of Wnt1 and treatment with the Wnt/β-Catenin signaling inhibitor, FH535, led to suppression of tumorsphere formation and decreased the breast CD44+/CD24- CSC population. Intriguingly, using both experimental and spontaneous in vivo models of metastasis, their data suggested that decreases in tumor growth and metastasis were, in part, due to a dramatic reduction in the breast CSC population, as measured by a marked reduction in ALDH1 expression, which is a critical factor required for maintaining the pluripotency and tumor initiation properties of breast CSCs in vivo [73]. Overall, these studies indicate that Wnt signaling is involved in multiple aspects of cancer stem cell function and activity that drive tumor progression. Thus, continued research into the mechanisms of the pathway in this context could be crucial for how we might target tumor progression through inhibition of cancer stemness-associated phenotypes.
The same group also demonstrated that treatment of breast cancer cells with the small molecular inhibitor, CWP232228, which prevents β-catenin from binding to Tcf in the nucleus, inhibited the growth of breast cancer stem cells and reduced breast cancer stem cell activity, as measured by tumorsphere formation [72]. Consequently, treatment with CWP232228 after tail vein injection of 4T1 cells led to decreased metastatic burden and increased overall survival [72]. These data suggest that therapies targeting β-catenin could be effective at inhibiting metastasis in patients where Wnt signaling is active [71], in part through inhibiting the ability of β-catenin to activate stem cell programs as well as other pro-metastatic genes. However, since the WNT signaling axis plays such a prominent role in normal tissue homeostasis, toxicity may limit the efficacy of pathway inhibitors. For example, orally administered Tankyrase inhibitors, which target APC-mutated tumors (80% of which are colorectal cancers), cause significant gastrointestinal toxicity [74,75]. Notably, Phase I clinical trials involving the Frizzled inhibitory antibodies vantictumab and ipafricept had to be temporarily halted due to bone loss in treated patients [74]. Still, this effect could be alleviated by limiting doses and mitigating side effects (i.e. simultaneous administration of an anti-resportive, such as alendronate, to reduce bone loss). Nevertheless, reports have shown that WNT pathway inhibitors including those targeting Porcupine, which prevent palmitoylation and transport of WNT ligands, as well as antibodies targeting Frizzled, have resulted in inhibition of cancer cell proliferation with reduced toxicity (intestine and skin) in mouse models [75–78].
While many studies have established the WNT pathway as a critical mediator of tumor progression in terms of stemness, survival and EMT [69,70], more recent studies have identified microenvironmental roles for WNT signaling specifically at the secondary site, where aberrant activation has been implicated in the acquisition of stemness properties by tumor cells to enable the survival of metastatic cells and initiation of metastatic outgrowth. For example, Wang and Ouyang demonstrated that the ECM protein, Periostin, produced by stromal fibroblasts, promotes Wnt signaling in the lung microenvironment to mediate crosstalk with newly arriving tumor cells [79]. This pathway results in high concentrations of Wnt ligands within the metastatic niche, thereby increasing survival and proliferation of MIC/TICs [79]. Similarly, Massagué and colleagues demonstrated that the cancer cell-derived lung metastasis associated protein, Tenascin-C (TNC), participates in an autocrine feedback loop that is critical for newly arriving tumor cells to initially survive and outgrow within the lung parenchyma. Mechanistically, TNC leads to enhanced WNT and NOTCH signaling in breast cancer cells, increasing their stemness associated properties and fitness to survive the harsh lung microenvironment. Interestingly, their data also suggest that stroma-derived TNC, specifically from infiltrating myofibroblasts and other stromal sources, becomes critical for the continued growth of micrometastases to develop large metastatic nodules [80]. Lastly, Clarke and colleagues demonstrated that bone marrow-derived IL1β promotes colonization of the bone through activation of NFkB and CREB signaling within tumor cells, leading to tumor cell Wnt signaling and CSC colony formation [81]. Importantly then, advances in the modeling of non-cell autonomous (in this case originating from stromal sources) interactions will be critical for developing therapeutics to effectively inhibit Wnt-mediated tumor progression, specifically in the case of metastatic initiation and maintenance.
The Notch Pathway
Role of NOTCH Signaling in Mammary Stem Cells
The NOTCH signaling pathway is involved in the development of multiple organ systems including heart, pancreas, blood vessels, nervous system and the mammary gland [82]. In general, the pathway consists of NOTCH receptors (NOTCH1–4) interacting with surface bound or secreted ligands (known collectively as DSL ligands: Delta, Serrate and Lag-2). In mammals, the two main sets of Notch ligands are Delta-Like (DELTA-LIKE-1, DELTA-LIKE-2, DELTA-LIKE-3 and DELTA-LIKE-4) and Serrate-Like (JAGGED1 and JAGGED2) [83]. Activation of NOTCH signaling occurs when NOTCH receptors on one cell bind to ligand receptors on another cell. Following binding, the NOTCH receptor is cleaved by the protease complex, γ-SECRETASE, leading to release of the NOTCH Intracellular Domain (NOTCH-ICD). NOTCH-ICD is then translocated to the nucleus where it interacts with cofactors such as Recombination Signal Binding Protein for Immunoglobulin Kappa J Region (RBP-JK) and E1A binding protein p300 (P300) to activate transcription of target genes involved in numerous cellular processes including proliferation, differentiation and self-renewal [82] (Fig 1C).
NOTCH receptors have been identified as critical mediators of stemness properties in mammary epithelial cells, where Notch1 and 4 have been the most extensively studied. The NOTCH1 receptor specifically regulates asymmetric cell division in human breast epithelial stem cells [84]. Additionally, the Notch1 receptor marks the ER- luminal progenitor population [84]. Interestingly, although these cells are unipotent in mice, they become highly plastic when conducting transplantation experiments, suggesting that Notch1 receptor expression, likely in conjunction with transplantation itself, is associated with plasticity in mammary epithelial cells in vivo [84]. Further, using Notch1-GFP based lineage tracing to track Notch1 receptor-expressing cells and their progeny during embryonic mammary gland development in vivo, the authors demonstrated that Notch1-GFP-marked progeny were represented in all mammary lineages, suggesting that Notch1 expression could be linked to a pool of embryonic mammary stem cells [84]. A more recent study used two mouse models, one containing Confetti reporters to mark Notch receptor+ cells, and another that contained a gain-of-function Notch1 receptor expressed specifically in mammary epithelial cells, to demonstrate that Notch1 was critical for inhibiting the transdifferentiation of multipotent mammary stem cells to basal progenitors [85]. Further, late in luminal differentiation, Notch1 receptors also inhibit conversion of ER- cells to ER+ cells. Similarly, Notch4 receptor overexpression is sufficient to inhibit differentiation of murine mammary epithelial cells in vitro [86]. Lastly, Egan and colleagues demonstrated through conditional mouse models that loss of the N-acetylglucosamine transferase, Lfng, which prevents ligand-mediated activation of Notch receptors, results in a significant increase in mammary stem cell associated CD24+CD49fhi population. Intriguingly, further analysis of the bulk population revealed that the majority of the cells within this population were CD61+Sca1−, a profile associated with pubertal MaSCs and luminal progenitors [87,88]. Lfng loss also resulted in some decreases in the number of CD24hiC49f+ CD61+ luminal progenitor cells. These data suggested that posttranslational modifications of Notch receptors by Lfng in the mouse mammary gland promotes expansion of the pubertal-associated MaSCs and bipotent progenitor cells, at the cost of luminal progenitor cells.
Similarly, Notch ligands have been implicated in the function and maintenance of mammary stem cell populations. The expression of Notch pathway ligands is diverse; mammary stem cell-enriched populations mainly express Delta1, luminal cell populations mainly express Jagged1, and all subsets express varying levels of Jagged2, Delta3 or Delta4 [29]. This complex expression pattern largely defines the ligand functions throughout mammary gland development, where the ligands can both promote and inhibit various aspects of mammary stem cell function. For example, a recent study by Kang and colleagues showed that Delta-Like-1 (Dll1) is enriched in mammary stem cells and is required for mammary stem cell survival and ductal morphogenesis [89]. In addition, the authors found that bipotent MaSC-derived Dll1 is a critical mediator of crosstalk between mammary epithelial cells and macrophages in the stem cell niche to promote stem cell activity [89]. Further association with Notch ligand and stemness was identified by Wicha and colleagues who demonstrated that activated Notch signaling results in mammary stem cell self-renewal and branching morphogenesis [86]. These effects could be reversed through treatment with antibodies targeting Notch4 receptor or inhibitors against y-Secretase activity. Intriguingly, NOTCH pathway ligands are also involved in inhibiting the expansion of mammary stem cells through non-cell autonomous means, underscoring a complex role for Notch signaling in mammary stem cell function. For example, Ostrowski and colleagues demonstrated that stromal-derived JAGGED1 is critical for NOTCH3 receptor-mediated inhibition of stem cell proliferation and activity, and that this effect was downstream of PTEN activity [90]. Collectively, these data emphasize the importance of stromal-epithelial cell crosstalk for bipotent MaSC function, where Notch signaling plays a key role in ensuring the proper control of the mammary stem cell population.
Role of NOTCH Signaling in Breast Cancer Metastasis
Numerous reports have associated NOTCH-ICD activity with stemness, self-renewal and differentiation during development, with subsequent research providing critical insight into how NOTCH signaling is dysregulated to promote transformation and primary tumor growth [91–93]. For the purposes of this review, we will summarize some of the seminal findings related to NOTCH signaling and its roles in promoting stemness and metastasis. Notch receptor activity within tumor cells has been shown to promote EMT, which is linked to stem cell characteristics and metastasis. Specifically, studies have identified the EMT-inducing transcription factor, Snail Family Transcriptional Repressor 2 (SLUG), as a direct transcriptional target of NOTCH1, leading to downregulation of the epithelial cell-associated protein E-cadherin and initiation of EMT [94,95]. Similarly, a signaling axis involving STAT5, FYN and NOTCH2, where STAT5 and FYN form a positive feedback loop to increase DLL4/JAGGED1 expression and NOTCH2-mediated pathway activation, has been shown to be critical for maintaining the mesenchymal phenotype in basal type breast cancer cells. This mesenchymal phenotype is dependent on NOTCH2, as knockdown of NOTCH2 results in a decrease in multiple markers associated with EMT including TWIST, SNAI1, SNAI2 (SLUG), VIM and ZEB1 [96]. Additional studies demonstrate that NOTCH4 promotes tumor progression through regulating TIC activity in ER+ breast cancer cells leading to resistance to endocrine therapies [97]. However, the role of NOTCH3 in cancer stemness and tumor progression is more complicated, as reports have demonstrated both tumor suppressive and tumor promotional activities of the protein. For example, exome sequencing of human tumors have identified nonsense and missense mutations in NOTCH3 in multiple cancers, including breast cancer [98]. Further, reintroduction of NOTCH3 through ectopic overexpression in TNBC cells results in inhibition of EMT, decreased tumor growth and cellular senescence, suggesting that NOTCH3 acts as an inhibitor of tumor progression [99,100]. In contrast, more recent studies have shown that NOTCH3 expression is associated with metastatic colonization, where it promotes invasive properties, cell seeding and secondary site growth through increased self-renewal and inhibition of differentiation in metastatic ER+ and TNBC cells [101]. These data suggest, similar to the pathways discussed above, that context and timing will be of important consideration when developing targeted therapies to modulate NOTCH signaling and inhibit tumor progression and metastasis.
NOTCH ligands and receptors also function within the primary and metastatic niches to regulate stemness phenotypes and promote metastasis. For example, JAGGED1-expressing astrocytes promote NOTCH signaling in metastatic breast cancer cells in the brain to promote the self-renewal of TICs and metastatic colonization in the brain [102]. Crucially, treatment with a blood-brain barrier permeable Notch inhibitor, Compound E, has been shown to be effective at inhibiting brain metastasis in vivo [102]. In addition, within the hypoxic niche, Jagged-2 is upregulated in bone marrow stroma to promote self-renewal and EMT in neighboring breast TICs [103]. Further, NOTCH3 has been shown to regulate the secretion of IL-6 from osteoblasts to promote outgrowth of tumor cells in the bone [97], demonstrating another critical niche function for NOTCH signaling in metastasis.
Given these data, it is not surprising that overexpression of multiple NOTCH signaling components are found to be associated with poor prognosis. For example, several studies have shown that high levels of JAGGED1 and NOTCH1 ligands correlate with poorer prognosis in breast cancer patients, and JAGGED1 is also associated with a more aggressive, basal breast cancer phenotype and with increased recurrence [104,105]. However, because different NOTCH signaling proteins play roles in multiple aspects of tumor progression and metastasis, targeting one component of the Notch pathway therapeutically will likely not be effective. Because Notch receptors/transcriptional regulators have high sequence homology and can be activated by the same sets of ligands, compensation for loss of other family members may frequently occur. In addition, crosstalk between Notch signaling and other developmental pathways can allow for activation of the Notch pathway independent of Notch ligands [74]. Thus, therapeutic strategies should consider inhibition of multiple developmental pathways that crosstalk with Notch signaling to allow for more effective suppression of metastatic phenotypes.
Pluripotency Factors in Mammary Stem Cells and Breast Cancer Metastasis
Pluripotency Factors as Regulators of Mammary Stem Cells
The above sections underscore the role of critical developmental stem cell pathways in metastasis and suggest that key pluripotency factors could also be important contributors to these processes, due to their roles as master regulators of stemness and plasticity during normal development. The transcription factors SRY-box 2 (SOX2), POU Class 5 Homeobox 1 (OCT4), Nanog Homeobox (NANOG), Kruppel Like Factor 4 (KLF4) and cellular MYC Proto-Oncogene (cMYC) are known as master regulators of pluripotency and stemness due to their critical roles in regulating target genes that are important for self-renewal, differentiation and survival [106]. Similar to embryonic development, where these core factors are essential for the function and maintenance of embryonic stem cells, studies have also identified important roles for pluripotency factors in regulating mammary stem cells over the course of mammary gland development and differentiation, with an especially strong association with the expansion of breast tissue during pregnancy and lactation [107]. For instance, multiple master pluripotency factors including SOX2, NANOG, OCT4 and KLF4 are expressed in both luminal progenitors and bipotent MaSCs, are associated with the expansion of the MaSC-enriched populations during pregnancy and lactation, and decrease in expression as cells begin to differentiate [108,109] (Fig. 2). Studies from Hartmann and colleagues examined the expression of pluripotency factors during lactation and found that SOX2, NANOG, OCT4 and KLF4 were highly expressed in isolated breast milk-derived stem cells when compared to non-lactating, non-pregnant breast cells [110]. Interestingly, these specialized cells were able to self-renew in anchorage-independent conditions, had multiple lineage potential and were also capable of differentiating into all three germ layers. Additionally, these cells could transport through the bloodstream to different organs, where they were shown to integrate and differentiate into functional cells of multiple target organs [111].
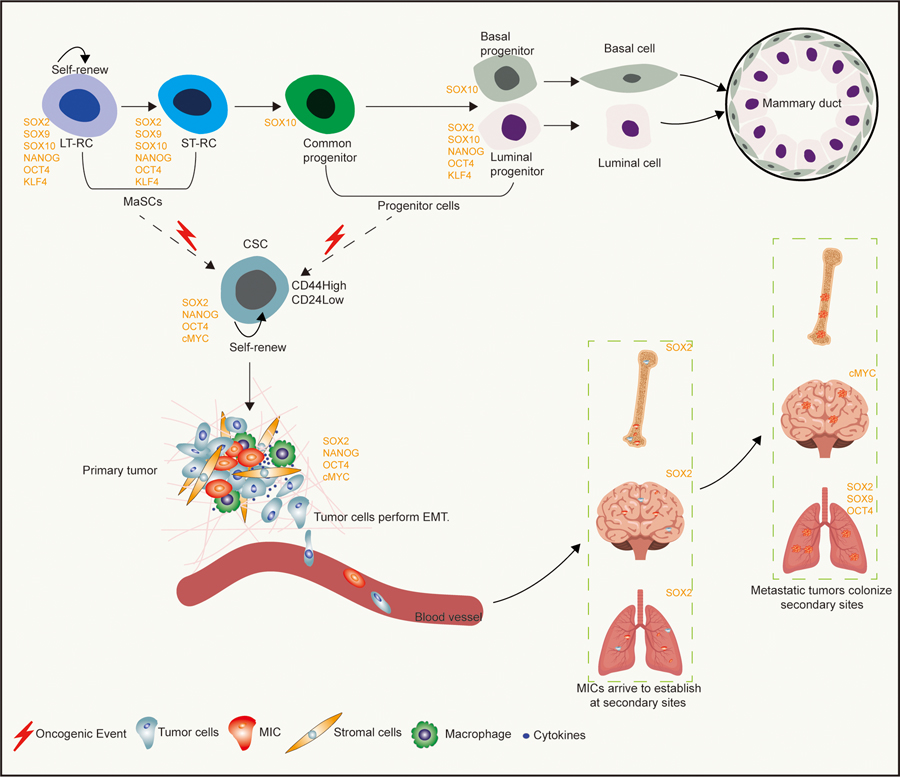
Figure 2: The role of pluripotency factors during normal mammary gland development, tumor initiation and breast cancer metastasis.
Expression of pluripotency factors including SOX2, NANOG, OCT4 and KLF4, as well as cell-fate regulators such as SOX9 and SOX10 are associated with, and promote the maintenance of, putative multipotent MaSC populations. As mammary gland development progresses, multipotent MaSCs further differentiate into bipotent progenitors that can give rise to more specialized unipotent basal and/or luminal progenitors that function to properly develop and maintain the mammary epithelium. Studies suggest that breast cancer stem cells (CSCs) can arise from either multipotent MaSCs or progenitor populations. Often breast CSCs express similar pluripotency and cell-fate regulators as normal mammary multipotent stem and progenitor populations. These critical developmental factors remain important as primary breast tumors become metastatic, where it has been shown that pluripotency factors such as cMYC, SOX2, OCT4 and NANOG promote dedifferentiation (EMT), escape from the primary site and establishment/colonization of secondary sites. Long-Term Repopulating Stem Cells, LT-RCs. Short-Term Repopulating Cells, ST-RCs. The red lightning symbol represents oncogenic events that lead to the transformation of normal stem/progenitor populations to CSCs.
Additional functional studies have indicated that pluripotency factors play a key role in mammary stem cell biology. For example, Liu and colleagues demonstrated that Sox2 is critical downstream of Lgr to activate Wnt signaling and promote mammary stem cell activity [112]. Importantly, they identified Sox2 as a transcriptional target of the Lgr4/Wnt/β-catenin/Lef1 pathway and demonstrated that Sox2 overexpression could rescue the decrease in mammary stem cell repopulation capacity found in Lgr−/− mice [112]. Another pluripotency factor, cMyc, has been shown to be essential for mammary stem cell activity (Fig. 2). Faraldo and colleagues showed, using limiting-dilution and serial transplantation assays, that deletion of cMyc from mammary basal epithelial cells decreases self-renewal. Interestingly, stimulation with Estrogen and Progesterone downstream of cMyc loss could partially rescue repopulation defects in vivo, in part due to compensation by N-Myc. However, when cMyc-KO glands were transplanted a second time, the resulting mammary epithelium completely lacked stem and progenitor cells and failed to repopulate cleared fat pads, suggesting that cMyc regulates both the maintenance and self-renewal of mammary stem cells, and that N-Myc does not compensate for all activities of cMyc [113]. A report from Wahl and colleagues identified Sox10, which has been shown to be important for the pluripotency of neural crest cells [114–116], as not only being highly expressed in fetal MaSCs (fMaSCs), but also being functionally critical for fMaSCs function and self-renewal [117]. Specifically, the authors showed that Cre-mediated deletion of Sox10 in fMaSCs results in a decrease in 3D mammary organoid formation, with the structures that did form being smaller and failing to develop multi-lineage organoids compared to their wildtype counterparts [117]. Further, Sox10-null fMaSCs failed to outgrow after transplantation into cleared mammary fat pads, whereas Sox10 wildtype fMaSCs were successful in reconstituting the mammary gland after transplantation. Together, these data indicate that Sox10 is a critical mediator of fMaSC activity and is required for proper stem/progenitor cell function (Fig. 2). Overall, these reports have provided critical insight into the molecular mechanisms that underlie normal mammary stem cell functions and have been instructive into how this specialized population becomes susceptible to transformation (Fig. 2).
Pluripotency Factors as Regulators of Breast Cancer Metastasis
Although pluripotency factors have a well-established role in the activity and function of breast cancer stemness including transformation in vitro and tumor initiation in vivo [118–121], more recent studies have identified critical roles during tumor progression and metastasis, where they have been shown to regulate several critical properties of metastatic tumor cells including stemness and plasticity. For example, a study by Weinberg and colleagues demonstrated that Sox9, a stem cell-associated transcription factor shown to be important for cell-fate in multiple different tissues [122], cooperates with the stemness and EMT-inducing transcription factor Slug, to induce and maintain MaSC-enriched basal cells [31]. Importantly, Sox9 and Slug expression in MCF7ras cells (which normally can only form micrometastases), enabled the formation of macrometastases [31]. Additionally, knockdown of either Sox9 or Slug in highly metastatic MDA-MB-231 cells resulted in decreased macrometastasis formation after tail vein injection [31].
Numerous additional studies have connected pluripotency factors with metastasis (Fig. 3). For example, a recent study by Chun and colleagues demonstrated that degradation of OCT4 was downregulated in TICs and that restoration of the E3 ubiquitin ligase Carboxy Terminus of HSP70-Interacting Protein (CHIP), which is normally downregulated in breast TICs and targets Oct4, was sufficient to decrease TIC function. Importantly, tail-vein injection of MDA-MB-231 cells with overexpression of CHIP or overexpression of both CHIP and OCT4 rarely resulted in lung metastasis, as measured by node number. In comparison, mice injected with MDA-MB-231 cells possessing stable KD of CHIP or overexpression of OCT4 alone had a significant increase in lung metastases [123]. These data suggest that OCT4 protein stabilization is critical for metastasis of TNBC cells in vivo. Similarly, Xu and colleagues have shown in a transgenic mouse model that inducible overexpression of Nanog in mammary epithelial cells, in combination with Wnt-1 overexpression, is sufficient to induce mammary tumorigenesis and metastasis [124]. Further, studies have shown that pluripotency factors can work in concert to promote metastatic phenotypes. For example, Liu and colleagues showed that simultaneous OE of Oct4 and Nanog increased, whereas combined KD of Oct4 and Nanog decreased, EMT and invasion of breast cancer cells [125]. These data suggest that pluripotency factors do not necessarily function in isolation to influence metastatic phenotypes.
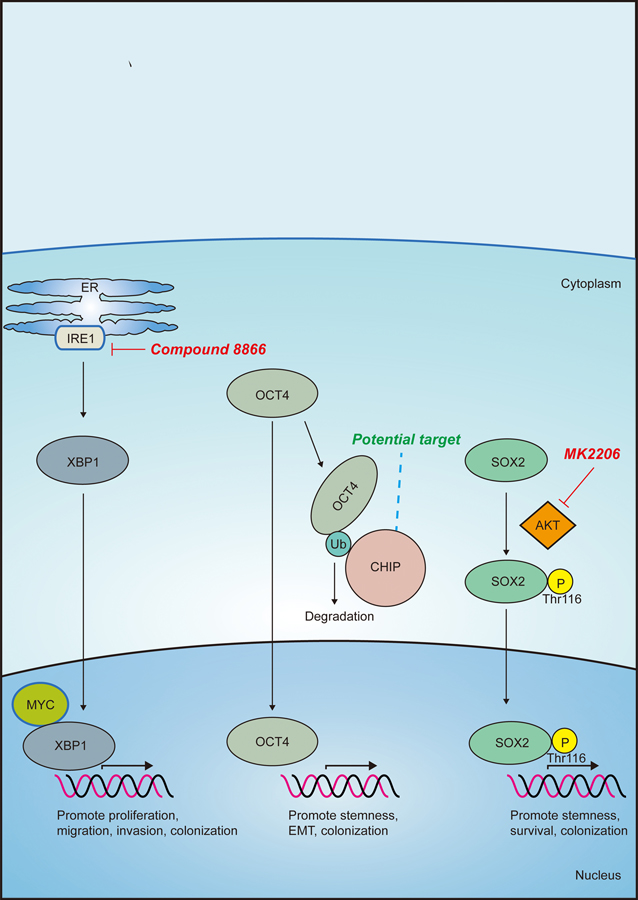
Figure 3: Targeting pluripotency factors to inhibit breast cancer progression.
The small molecule inhibitor 8866 inhibits MYC-driven human breast tumors via blockade of IRE1-mediated activation of the cMYC/XBP1 transcriptional complex. Targeting activation of E3 ubiquitin ligase Carboxy Terminus of HSP70-Interacting Protein (CHIP) could lead to increased degradation of the breast CSC-associated factor OCT4. Inhibiting phosphorylation of threonine 116 on SOX2 could prevent SOX2-mediated transcription and promotion of CSC-related genes. The AKT inhibitor, MK2206 prevents phosphorylation-mediated translocation of SOX2 to the nucleus to activate target genes involved in breast cancer proliferation and stemness.
In addition to a well-established role as an oncogene and cell cycle regulator, cMYC, another pluripotency factor, is also a core regulator of metastatic progression in breast cancer, where it is required for migration, invasion and angiogenesis [126,127]. In the brain, cMYC is critical for metastatic colonization of breast cancer due to its ability to regulate macrophage infiltration, GAP junction formation between tumor cells and astrocytes, and invasion [128]. A recent report from Chen and colleagues showed that in vivo growth of MYC-driven human breast tumors could be restrained through pharmacological inhibition of the RNase activity of inositol-requiring enzyme 1 (IRE1) with the small molecule inhibitor 8866 [129] (Fig. 3). However, given that more recent studies have also identified metastasis inhibitory roles for cMyc [130], any clinical strategy to modulate its function must consider how context, timing and tissue type may influence drug responses.
Like cMYC, SOX2 is a key regulator of breast cancer metastasis. Studies have identified roles for SOX2 across multiple aspects of the metastatic cascade including EMT, migration, invasion, anoikis resistance and angiogenesis [131–134] (Fig. 2). However, its most prominent role in tumor progression involves promotion of stemness and inhibition of differentiation. This is especially evident in breast cancer, where SOX2 has been widely used to distinguish breast cancer stem cells from the bulk tumor population [135], and where its overexpression is associated with more aggressive subtypes of breast cancer [136]. Consistent with these data, our group demonstrated that the developmental transcription factor, Six2, regulates Sox2 to promote a breast cancer stem cell program that enables metastatic colonization [136]. Specifically, in several triple-negative breast cancer (TNBC) models, Six2 enhances the expression of genes associated with embryonic stem cell programs. Six2 directly binds the Sox2 Srr2 enhancer, promoting Sox2 expression and downstream expression of Nanog, which are both key pluripotency factors [136]. Regulation of Sox2 by Six2 enhances cancer stem cell properties and increases metastatic colonization. Six2 and Sox2 expression correlate highly in breast cancers including TNBC, where a Six2 expression signature was shown to be predictive of metastatic burden and poor clinical outcome. Our findings demonstrate that a SIX2/SOX2 axis is required for efficient metastatic colonization, underscoring a key role for stemness factors in outgrowth at secondary sites [136]. Intriguingly, a report published shortly after this study demonstrated that Six1 regulates Sox2 in the context of glioma, suggesting that both family members can regulate this key pluripotency factor [137]. Taken together, these data indicate that, similar to their established roles in stem/progenitor phenotypes during kidney development, SIX family members also function to regulate cancer stemness to promote metastasis in the context of breast and other cancers.
Because of its role as a master pluripotency factor during embryonic development, SOX2 is controlled at the transcriptional, translational and post-translational levels to ensure proper expression and function. Since these mechanisms of SOX2 regulation are also used by tumor cells to promote metastasis, it provides multiple avenues to target and inhibit SOX2 function to inhibit tumor progression. For example, recent examination of human breast cancer patient data has led to the hypothesis that targeting the kinase that is responsible for phosphorylation of the threonine 116 phosphorylation (T116) site of SOX2 may be an attractive means to inhibit SOX2 function [138] (Fig. 3). Specifically, Lai and colleagues showed that not only was increased phosphorylated-Sox2T116 associated with increased tumorigenicity in multiple breast cancer cell lines, but that it was also predominantly found within breast cancer stem cells, suggesting that this phosphorylation state of SOX2 is important for breast cancer stem cell identity [138]. Consequently, when examining a breast cancer cohort, they found that phosphorylated-Sox2T116 levels correlated with increased histological tumor grade and vascular invasion [138]. Interestingly, a recent report has shown that phosphorylation of threonine 116 by AKT stabilizes SOX2 in esopheageal cancer cells [139]. Further, treatment with the AKT inhibitor, MK2206, resulted in decreased SOX2 protein, cell proliferation and stemness properties, as measured by tumorsphere formation. These data suggest that targeting AKT could provide a means to inhibit SOX2-positive CSCs (Fig. 3).
Many studies have examined how increased levels of pluripotency factors influence prognostic indicators such as disease-free survival, resistance and overall survival [140]. Studies examining either expression of OCT4, SOX2, NANOG, KLF4, or cMYC singly, or combined expression of OCT4 and NANOG, showed that higher levels of mRNA expression in breast cancer patient samples across multiple subtypes is significantly correlated with decreased overall survival [140]. Additionally, higher levels of SOX2 mRNA are particularly predictive of decreased resistance- and disease-free survival [140]. Intriguingly, in addition to being associated with multiple prognostic markers including decreased overall survival, cMYC is also more highly expressed in patients that present with metastatic disease at diagnosis [141]. However, several studies have also shown that pluripotency factors can be associated with improved prognosis [140]. These confounding data could be due to tumor heterogeneity, as TIC-associated factors such as Sox2 may exist in a small minority of cells within the bulk tumor, and therefore their level of expression could be masked. Thus, the use of single-cell sequencing technologies may provide significant insight into our understanding of how small populations of cells, like TICs/MICS, function to influence the metastatic outcomes. When used in combination with continued refinement of appropriate markers to identify the TIC population, substantial progress can be made towards understanding the molecular underpinnings of developmental pathway networks and how they function to ensure proper mammary gland development, as well as how their dysregulation promotes tumorigenesis and metastasis. Because modulation of these programs is associated with drug response and clinical prognosis, it is imperative that we continue to examine the underlying biology behind the relationship between stemness, pluripotency and metastasis to develop more effective therapeutic strategies to improve patient outcomes.
Concluding Remarks
Advances in imaging technologies, genomic sequencing, mouse models of metastasis and, more recently, single-cell RNA-seq analysis have greatly improved our understanding of how metastatic tumor cells colonize secondary sites. Such advances have led to substantial progress in the development of novel therapeutic strategies, such as Mechanistic Target of Rapamycin Kinase (mTOR) and Cyclin Dependent Kinase (CDK4/6) inhibitors, to specifically treat advanced/metastatic breast cancer [142]. However, survival of patients with metastatic disease remains poor. The poor prognosis associated with metastasis is largely due to established metastases being inherently more resistant to current treatments compared to primary tumor cells. Given the clinical implications of metastatic outgrowth, it is unfortunate that most current studies mainly focus on preventing the earlier steps of metastasis, such as migration and invasion. It is well known that cancer cells enter the circulation long before a patient is diagnosed, and that many patients relapse at distant sites long after the primary tumor is eradicated. It is also becoming clear that tumor cell behavior and signaling at the primary site can differ dramatically from tumor cell behavior and signaling at different metastatic sites [143]. Therefore, a better understanding of the molecular underpinnings of metastatic establishment, maintenance, outgrowth at the metastatic site are needed to develop more effective therapeutics to target metastasis specifically. As discussed in this review, continued insight into the regulation and function of the TIC/MIC population may lead to major breakthroughs in our understanding of how tumor cells can successfully colonize secondary sites. Indeed, TICs/MICs appear to acquire their core properties of plasticity, stemness and survival, which are needed to complete the metastatic process, by hijacking developmental pathways. Future studies focused on crosstalk between multiple developmental pathways could lead to more effective therapies to target plasticity and stemness in MICs. Additionally, combining MIC-targeted therapies with existing treatment may not only target residual disease, but may also inhibit the development of new metastatic lesions, thereby increasing patient survival.
Acknowledgements
This work was supported by National Institutes of Health grants R01CA221282 (HLF), R01CA224867 (HLF and MTL), 5F99CA223023-02/4K00CA223023-03 (MUJO), and 1F99CA234940-01 (HZ).
Footnotes
The authors’ competing financial interests are disclosed below.
MTL is a founder and limited partner in StemMed Ltd. and a founder and manager in StemMed Holdings, its general partner. MTL is also a founder of, and equity holder in, Tvardi Therapeutics Inc.
References
- 1.McNally S, Stein T. Overview of mammary gland development: A comparison of mouse and human. In: Methods in Molecular Biology [Internet]. 2017. [cited 2019 Jan 19]. p. 1–17. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27796946 [DOI] [PubMed]
- 2.Howard BA, Veltmaat JM. Embryonic Mammary Gland Development; a Domain of Fundamental Research with High Relevance for Breast Cancer Research. J Mammary Gland Biol Neoplasia [Internet]. 2013. June 18 [cited 2018 Jun 29];18(2):89–91. Available from: http://link.springer.com/10.1007/s10911-013-9296-2 [DOI] [PubMed] [Google Scholar]
- 3.Celià-Terrassa T Mammary Stem Cells and Breast Cancer Stem Cells: Molecular Connections and Clinical Implications. Biomedicines [Internet]. 2018. May 4 [cited 2019 Feb 14];6(2). Available from: http://www.ncbi.nlm.nih.gov/pubmed/29734696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DEOME KB, FAULKIN LJ, BERN HA, BLAIR PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res [Internet]. 1959. June [cited 2019 Feb 14];19(5):515–20. Available from: http://www.ncbi.nlm.nih.gov/pubmed/13663040 [PubMed] [Google Scholar]
- 5.Daniel CW, DeOme KB. Growth of mouse mammary glands in vivo after monolayer culture. Science (80-). 1965; [DOI] [PubMed] [Google Scholar]
- 6.Faulkin LJ, Deome KB. Regulation of growth and spacing of gland elements in the mammary fat pad of the c3h mouse. J Natl Cancer Inst 1960; [PubMed]
- 7.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat M-L, et al. Generation of a functional mammary gland from a single stem cell. Nature [Internet]. 2006. January 5 [cited 2015 Oct 14];439(7072):84–8. Available from: http://www.nature.com/doifinder/10.1038/nature04372 [DOI] [PubMed] [Google Scholar]
- 8.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature [Internet]. 2006. February 23 [cited 2015 Dec 6];439(7079):993–7. Available from: 10.1038/nature04496 [DOI] [PubMed] [Google Scholar]
- 9.Lu P, Zhou T, Xu C, Lu Y. Mammary stem cells, where art thou? Wiley Interdisciplinary Reviews: Developmental Biology. 2019. [DOI] [PubMed]
- 10.Makarem M, Spike BT, Dravis C, Kannan N, Wahl GM, Eaves CJ. Stem Cells and the Developing Mammary Gland. J Mammary Gland Biol Neoplasia [Internet]. 2013. June 27 [cited 2019 Jan 19];18(2):209–19. Available from: http://link.springer.com/10.1007/s10911-013-9284-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Wang H, Jiao B. Mammary gland stem cells and their application in breast cancer. Oncotarget. 2017. [DOI] [PMC free article] [PubMed]
- 12.Pal B, Chen Y, Vaillant F, Jamieson P, Gordon L, Rios AC, et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat Commun [Internet]. 2017. December 20 [cited 2019 Jan 2];8(1):1627 Available from: http://www.nature.com/articles/s41467-017-01560-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen QH, Pervolarakis N, Blake K, Ma D, Davis RT, James N, et al. Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat Commun 2018; [DOI] [PMC free article] [PubMed]
- 14.Macias H, Hinck L. Mammary gland development [Internet]. Wiley Interdisciplinary Reviews: Developmental Biology NIH Public Access; 2012. p. 533–57. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22844349 [DOI] [PMC free article] [PubMed]
- 15.Wuidart A, Sifrim A, Fioramonti M, Matsumura S, Brisebarre A, Brown D, et al. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat Cell Biol [Internet]. 2018. June 21 [cited 2019 Jan 19];20(6):666–76. Available from: http://www.nature.com/articles/s41556-018-0095-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lloyd-Lewis B, Davis FM, Harris OB, Hitchcock JR, Watson CJ. Neutral lineage tracing of proliferative embryonic and adultmammary stem/progenitor cells. Dev 2018; [DOI] [PMC free article] [PubMed]
- 17.Scheele CLGJ, Hannezo E, Muraro MJ, Zomer A, Langedijk NSM, Van Oudenaarden A, et al. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature. 2017; [DOI] [PMC free article] [PubMed]
- 18.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci [Internet]. 2003. April 1 [cited 2017 Mar 20];100(7):3983–8. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature [Internet]. 2006. February 4 [cited 2019 Feb 14];439(7079):993–7. Available from: http://www.nature.com/articles/nature04496 [DOI] [PubMed] [Google Scholar]
- 20.Wan L, Lu X, Yuan S, Wei Y, Guo F, Shen M, et al. MTDH-SND1 Interaction Is Crucial for Expansion and Activity of Tumor-Initiating Cells in Diverse Oncogene- and Carcinogen-Induced Mammary Tumors. Cancer Cell. 2014; [DOI] [PMC free article] [PubMed]
- 21.Chakrabarti R, Wei Y, Hwang J, Hang X, Andres Blanco M, Choudhury A, et al. Δnp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat Cell Biol 2014; [DOI] [PMC free article] [PubMed]
- 22.Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L, et al. Identification of multipotent mammary stemcells by protein C receptor expression. Nature. 2015; [DOI] [PubMed]
- 23.Yin P, Wang W, Zhang Z, Bai Y, Gao J, Zhao C. Wnt signaling in human and mouse breast cancer: Focusing on Wnt ligands, receptors and antagonists. Cancer Science. 2018. [DOI] [PMC free article] [PubMed]
- 24.Staniszewska AD, Pensa S, Caffarel MM, Anderson LH, Poli V, Watson CJ. Stat3 Is Required to Maintain the Full Differentiation Potential of Mammary Stem Cells and the Proliferative Potential of Mammary Luminal Progenitors. PLoS One. 2012; [DOI] [PMC free article] [PubMed]
- 25.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell [Internet]. 2007. November [cited 2017 Mar 20];1(5):555–67. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1934590907001336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. WNT/β-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci [Internet]. 2007. January 9 [cited 2019 Feb 3];104(2):618–23. Available from: https://www.pnas.org/content/104/2/618.long?utm_source=TrendMD&utm_medium=cpc&utm_campaign=Proc_Natl_Acad_Sci_U_S_A_TrendMD_0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei W, Tweardy DJ, Zhang M, Zhang X, Landua J, Petrovic I, et al. STAT3 signaling is activated preferentially in tumor-initiating cells in claudin-low models of human breast cancer. Stem Cells. 2014; [DOI] [PubMed]
- 28.del Barco Barrantes I, Stephan-Otto Attolini C, Slobodnyuk K, Igea A, Gregorio S, Gawrzak S, et al. Regulation of Mammary Luminal Cell Fate and Tumorigenesis by p38α. Stem Cell Reports. 2018; [DOI] [PMC free article] [PubMed]
- 29.Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat M-LL, Oakes SR, et al. Notch Signaling Regulates Mammary Stem Cell Function and Luminal Cell-Fate Commitment. Cell Stem Cell [Internet]. 2008. October 9 [cited 2019 Jan 19];3(4):429–41. Available from: https://www.sciencedirect.com/science/article/pii/S1934590908003998#bib5 [DOI] [PubMed] [Google Scholar]
- 30.Bu W, Liu Z, Jiang W, Nagi C, Huang S, Edwards DP, et al. Mammary precancerous stem and non-stem cells evolve into cancers of distinct subtypes. Cancer Res 2019; [DOI] [PMC free article] [PubMed]
- 31.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 Cooperatively Determine the Mammary Stem Cell State. Cell [Internet]. 2012. March 2 [cited 2019 Feb 24];148(5):1015–28. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22385965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Celià-Terrassa T, Kang Y. Distinctive properties of metastasis-initiating cells. Genes Dev [Internet]. 2016. April 15 [cited 2017 Jan 6];30(8):892–908. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27083997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malladi S, MacAlinao DG, Jin X, He L, Basnet H, Zou Y, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell. 2016; [DOI] [PMC free article] [PubMed]
- 34.Gao H, Chakraborty G, Zhang Z, Akalay I, Gadiya M, Gao Y, et al. Multi-organ Site Metastatic Reactivation Mediated by Non-canonical Discoidin Domain Receptor 1 Signaling. Cell. 2016; [DOI] [PMC free article] [PubMed]
- 35.Visbal AP, Lewis MT. Hedgehog signaling in the normal and neoplastic mammary gland. Curr Drug Targets [Internet]. 2010. September [cited 2019 Jan 19];11(9):1103–11. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20545610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu F, Zhang Y, Sun B, McMahon AP, Wang Y. Hedgehog Signaling: From Basic Biology to Cancer Therapy. Cell Chem Biol [Internet]. 2017. March 16 [cited 2019 Jan 19];24(3):252–80. Available from: https://www.sciencedirect.com/science/article/pii/S2451945617300582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monkkonen T, Lewis MT. New paradigms for the Hedgehog signaling network in mammary gland development and breast Cancer [Internet]. Vol. 1868, Biochimica et Biophysica Acta - Reviews on Cancer. Elsevier; 2017. [cited 2019 Jan 19]. p. 315–32. Available from: https://www.sciencedirect.com/science/article/pii/S0304419X17301087#bb0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li N, Singh S, Cherukuri P, Li H, Yuan Z, Ellisen LW, et al. Reciprocal Intraepithelial Interactions Between TP63 and Hedgehog Signaling Regulate Quiescence and Activation of Progenitor Elaboration by Mammary Stem Cells. Stem Cells [Internet]. 2008. May [cited 2019 Feb 2];26(5):1253–64. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18292212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moraes RC, Zhang X, Harrington N, Fung JY, Wu M-F, Hilsenbeck SG, et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development [Internet]. 2007. March 15 [cited 2019 Feb 2];134(6):1231–42. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17287253 [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Dontu G, Mantle ID, Patel S, Ahn N, Jackson KW, et al. Hedgehog Signaling and Bmi-1 Regulate Self-renewal of Normal and Malignant Human Mammary Stem Cells. Cancer Res [Internet]. 2006. June 15 [cited 2019 Jan 21];66(12):6063–71. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16778178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis MT, Ross S, Strickland PA, Sugnet CW, Jimenez E, Hui C, et al. The Gli2 Transcription Factor Is Required for Normal Mouse Mammary Gland Development. Dev Biol [Internet]. 2001. October 1 [cited 2019 Jan 19];238(1):133–44. Available from: https://www.sciencedirect.com/science/article/pii/S0012160601904105 [DOI] [PubMed] [Google Scholar]
- 42.Bai CB, Joyner AL. Gli1 can rescue the in vivo function of Gli2. Development [Internet]. 2001. December [cited 2019 Jan 21];128(24):5161–72. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11748151 [DOI] [PubMed] [Google Scholar]
- 43.Hatsell SJ, Cowin P. Gli3-mediated repression of Hedgehog targets is required for normal mammary development. Development [Internet]. 2006. September 15 [cited 2019 Jan 21];133(18):3661–70. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16914490 [DOI] [PubMed] [Google Scholar]
- 44.Veltmaat JM, Relaix F, Le LT, Kratochwil K, Sala FG, van Veelen W, et al. Gli3-mediated somitic Fgf10 expression gradients are required for the induction and patterning of mammary epithelium along the embryonic axes. Development. 2006; [DOI] [PubMed]
- 45.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature Reviews Molecular Cell Biology. 2013. [DOI] [PubMed]
- 46.Sun Y, Wang Y, Fan C, Gao P, Wang X, Wei G, et al. Estrogen promotes stemness and invasiveness of ER-positive breast cancer cells through Gli1 activation. Mol Cancer. 2014; [DOI] [PMC free article] [PubMed]
- 47.Neelakantan D, Zhou H, Oliphant MUJ, Zhang X, Simon LM, Henke DM, et al. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun [Internet]. 2017. June 12 [cited 2019 Jan 19];8:15773 Available from: http://www.nature.com/doifinder/10.1038/ncomms15773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das S, Tucker JA, Khullar S, Samant RS, Shevde LA. Hedgehog signaling in tumor cells facilitates osteoblast-enhanced osteolytic metastases. PLoS One. 2012; [DOI] [PMC free article] [PubMed]
- 49.Di Mauro C, Rosa R, D’Amato V, Ciciola P, Servetto A, Marciano R, et al. Hedgehog signalling pathway orchestrates angiogenesis in triple-negative breast cancers. Br J Cancer [Internet]. 2017. May 23 [cited 2019 Jan 19];116(11):1425–35. Available from: http://www.nature.com/doifinder/10.1038/bjc.2017.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris LG, Pannell LK, Singh S, Samant RS, Shevde LA. Increased vascularity and spontaneous metastasis of breast cancer by hedgehog signaling mediated upregulation of cyr61. Oncogene. 2012; [DOI] [PMC free article] [PubMed]
- 51.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008; [DOI] [PubMed]
- 52.Theunissen JW, De Sauvage FJ. Paracrine hedgehog signaling in cancer. Cancer Research. 2009. [DOI] [PubMed]
- 53.Heller E, Hurchla MA, Xiang J, Su X, Chen S, Schneider J, et al. Hedgehog signaling inhibition blocks growth of resistant tumors through effects on tumor microenvironment. Cancer Res 2012; [DOI] [PMC free article] [PubMed]
- 54.Habib JG, O’Shaughnessy JA. The hedgehog pathway in triple-negative breast cancer. Cancer Medicine. 2016. [DOI] [PMC free article] [PubMed]
- 55.Carpenter RL, Lo H-W. Hedgehog pathway and GLI1 isoforms in human cancer. Discov Med 2012; [PMC free article] [PubMed]
- 56.Riobo-Del Galdo N, Lara Montero Á, Wertheimer E. Role of Hedgehog Signaling in Breast Cancer: Pathogenesis and Therapeutics. Cells. 2019; [DOI] [PMC free article] [PubMed]
- 57.Sharpe HJ, Pau G, Dijkgraaf GJ, Basset-Seguin N, Modrusan Z, Januario T, et al. Genomic Analysis of Smoothened Inhibitor Resistance in Basal Cell Carcinoma. Cancer Cell. 2015; [DOI] [PMC free article] [PubMed]
- 58.Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities [Internet]. Vol. 169, Cell. Cell Press; 2017. [cited 2019 Jan 19]. p. 985–99. Available from: https://www.sciencedirect.com/science/article/pii/S0092867417305470#bib5 [DOI] [PubMed] [Google Scholar]
- 59.Gómez-Orte E, Sáenz-Narciso B, Moreno S, Cabello J. Multiple functions of the noncanonical Wnt pathway. Trends in Genetics. 2013. [DOI] [PubMed]
- 60.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nature Reviews Molecular Cell Biology. 2009. [DOI] [PubMed]
- 61.Xiao Q, Chen Z, Jin X, Mao R, Chen Z. The many postures of noncanonical Wnt signaling in development and diseases. Biomedicine and Pharmacotherapy. 2017. [DOI] [PubMed]
- 62.Zeng YA, Nusse R. Wnt Proteins Are Self-Renewal Factors for Mammary Stem Cells and Promote Their Long-Term Expansion in Culture. Cell Stem Cell [Internet]. 2010. June 4 [cited 2019 Jan 22];6(6):568–77. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20569694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu QC, Verheyen EM, Zeng YA. Mammary Development and Breast Cancer: A Wnt Perspective. Cancers (Basel) [Internet]. 2016. July 13 [cited 2019 Jan 19];8(7). Available from: http://www.ncbi.nlm.nih.gov/pubmed/27420097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Incassati A, Chandramouli A, Eelkema R, Cowin P. Key signaling nodes in mammary gland development and cancer: β-catenin. Breast Cancer Res [Internet]. 2010. [cited 2019 Jan 19];12(6):213 Available from: http://www.ncbi.nlm.nih.gov/pubmed/21067528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindley LE, Curtis KM, Sanchez-Mejias A, Rieger ME, Robbins DJ, Briegel KJ. The WNT-controlled transcriptional regulator LBH is required for mammary stem cell expansion and maintenance of the basal lineage. Development. 2015; [DOI] [PMC free article] [PubMed]
- 66.Joshi PA, Jackson HW, Beristain AG, Di Grappa MA, Mote PA, Clarke CL, et al. Progesterone induces adult mammary stem cell expansion. Nature [Internet]. 2010. June 10 [cited 2016 Nov 18];465(7299):803–7. Available from: http://www.nature.com/doifinder/10.1038/nature09091 [DOI] [PubMed] [Google Scholar]
- 67.Kessenbrock K, Smith P, Steenbeek SC, Pervolarakis N, Kumar R, Minami Y, et al. Diverse regulation of mammary epithelial growth and branching morphogenesis through noncanonical Wnt signaling. Proc Natl Acad Sci U S A. 2017; [DOI] [PMC free article] [PubMed]
- 68.Blagodatski A, Poteryaev D, Katanaev VL. Targeting the Wnt pathways for therapies. Mol Cell Ther [Internet]. 2014. [cited 2019 Jan 19];2:28 Available from: http://www.ncbi.nlm.nih.gov/pubmed/26056595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene [Internet]. 2017. [cited 2019 Jan 19];36(11):1461–73. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27617575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howe LR, Brown AMC. Wnt signaling and breast cancer. Cancer Biology and Therapy. 2004. [DOI] [PubMed]
- 71.Jang G-B, Kim J-Y, Cho S-D, Park K-S, Jung J-Y, Lee H-Y, et al. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Nat Publ Gr [Internet]. 2015. [cited 2017 Jul 7]; Available from: https://www.nature.com/articles/srep12465.pdf [DOI] [PMC free article] [PubMed]
- 72.Jang GB, Hong IS, Kim RJ, Lee SY, Park SJ, Lee ES, et al. Wnt/β-catenin small-molecule inhibitor CWP232228 preferentially inhibits the growth of breast cancer stem-like cells. Cancer Res 2015; [DOI] [PubMed]
- 73.Charafe-Jauffret E, Ginestier C, Bertucci F, Cabaud O, Wicinski J, Finetti P, et al. ALDH1-positive cancer stem cells predict engraftment of primary breast tumors and are governed by a common stem cell program. Cancer Res 2013; [DOI] [PubMed]
- 74.Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treatment Reviews. 2018. [DOI] [PMC free article] [PubMed]
- 75.Madan B, Virshup DM. Targeting Wnts at the Source--New Mechanisms, New Biomarkers, New Drugs. Mol Cancer Ther 2015; [DOI] [PubMed]
- 76.Proffitt KD, Madan B, Ke Z, Pendharkar V, Ding L, Lee MA, et al. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res 2013; [DOI] [PubMed]
- 77.Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci U S A. 2013; [DOI] [PMC free article] [PubMed]
- 78.Kabiri Z, Greicius G, Madan B, Biechele S, Zhong Z, Zaribafzadeh H, et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Dev 2014; [DOI] [PubMed]
- 79.Wang Z, Ouyang G. Periostin: a bridge between cancer stem cells and their metastatic niche. Cell Stem Cell [Internet]. 2012. February 3 [cited 2015 Nov 3];10(2):111–2. Available from: http://www.sciencedirect.com/science/article/pii/S1934590912000045 [DOI] [PubMed] [Google Scholar]
- 80.Oskarsson T, Acharyya S, Zhang XH-F, Vanharanta S, Tavazoie SF, Morris PG, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med [Internet]. 2011. July 26 [cited 2015 Nov 26];17(7):867–74. Available from: 10.1038/nm.2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eyre R, Alférez DG, Santiago-Gómez A, Spence K, McConnell JC, Hart C, et al. Microenvironmental IL1β promotes breast cancer metastatic colonisation in the bone via activation of Wnt signalling. Nat Commun 2019; [DOI] [PMC free article] [PubMed]
- 82.Bolós V, Grego-Bessa J, de la Pompa JL. Notch Signaling in Development and Cancer. Endocr Rev [Internet]. 2007. May 1 [cited 2019 Jan 19];28(3):339–63. Available from: https://academic.oup.com/edrv/article-lookup/doi/10.1210/er.2006-0046 [DOI] [PubMed] [Google Scholar]
- 83.Callahan R, Raafat A. Notch Signaling in Mammary Gland Tumorigenesis [Internet]. Vol. 6, Journal of Mammary Gland Biology and Neoplasia. 2001. [cited 2019 Jan 22]. Available from: https://link.springer.com/content/pdf/10.1023%2FA%3A1009512414430.pdf [DOI] [PubMed] [Google Scholar]
- 84.Rodilla V, Dasti A, Huyghe M, Lafkas D, Laurent C, Reyal F, et al. Luminal Progenitors Restrict Their Lineage Potential during Mammary Gland Development. PLoS Biol 2015; [DOI] [PMC free article] [PubMed]
- 85.Lilja AM, Rodilla V, Huyghe M, Hannezo E, Landragin C, Renaud O, et al. Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat Cell Biol [Internet]. 2018. June 21 [cited 2019 Jan 19];20(6):677–87. Available from: http://www.nature.com/articles/s41556-018-0108-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res [Internet]. 2004. [cited 2019 Jan 19];6(6):R605–15. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15535842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu K, Usary J, Kousis PC, Prat A, Wang DY, Adams JR, et al. Lunatic Fringe Deficiency Cooperates with the Met/Caveolin Gene Amplicon to Induce Basal-like Breast Cancer. Cancer Cell. 2012; [DOI] [PMC free article] [PubMed]
- 88.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev [Internet]. 2009. November 15 [cited 2016 Oct 26];23(22):2563–77. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19933147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chakrabarti R, Celià-Terrassa T, Kumar S, Hang X, Wei Y, Choudhury A, et al. Notch ligand Dll1 mediates cross-talk between mammary stem cells and the macrophageal niche. Science (80-) [Internet]. 2018. June 29 [cited 2019 Jan 22];360(6396):eaan4153 Available from: http://www.ncbi.nlm.nih.gov/pubmed/29773667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sizemore GM, Balakrishnan S, Hammer AM, Thies KA, Trimboli AJ, Wallace JA, et al. Stromal PTEN inhibits the expansion of mammary epithelial stem cells through Jagged-1. Oncogene [Internet]. 2017. [cited 2019 Jan 22];36(16):2297–308. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27797378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mollen EWJ, Ient J, Tjan-Heijnen VCG, Boersma LJ, Miele L, Smidt ML, et al. Moving breast cancer therapy up a notch. Frontiers in Oncology. 2018. [DOI] [PMC free article] [PubMed]
- 92.Politi K, Feirt N, Kitajewski J. Notch in mammary gland development and breast cancer. Semin Cancer Biol [Internet]. 2004. October 1 [cited 2019 Jan 19];14(5):341–7. Available from: https://www.sciencedirect.com/science/article/pii/S1044579X04000306?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 93.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: A little bit of everything but not all the time. Nature Reviews Cancer. 2011. [DOI] [PubMed]
- 94.Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med 2007; [DOI] [PMC free article] [PubMed]
- 95.Shao S, Zhao X, Zhang X, Luo M, Zuo X, Huang S, et al. Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol Cancer [Internet]. 2015. February 3 [cited 2019 Jan 19];14(1):28 Available from: http://www.ncbi.nlm.nih.gov/pubmed/25645291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee GH, Yoo KC, An Y, Lee HJ, Lee M, Uddin N, et al. FYN promotes mesenchymal phenotypes of basal type breast cancer cells through STAT5/NOTCH2 signaling node. Oncogene. 2018; [DOI] [PubMed]
- 97.Zhang Y, Deng H. Notch and breast cancer metastasis. 2018;(September):1–15. Available from: www.preprints.org
- 98.Cui H, Kong Y, Xu M, Zhang H. Notch3 functions as a tumor suppressor by controlling cellular senescence. Cancer Res 2013; [DOI] [PMC free article] [PubMed]
- 99.Zhang X, Liu X, Luo J, Xiao W, Ye X, Chen M, et al. Notch3 inhibits epithelial-mesenchymal transition by activating Kibra-mediated Hippo/YAP signaling in breast cancer epithelial cells. Oncogenesis. 2016; [DOI] [PMC free article] [PubMed]
- 100.Diluvio G, Del Gaudio F, Giuli MV, Franciosa G, Giuliani E, Palermo R, et al. NOTCH3 inactivation increases triple negative breast cancer sensitivity to gefitinib by promoting EGFR tyrosine dephosphorylation and its intracellular arrest. Oncogenesis. 2018; [DOI] [PMC free article] [PubMed]
- 101.Leontovich AA, Jalalirad M, Salisbury JL, Mills L, Haddox C, Schroeder M, et al. NOTCH3 expression is linked to breast cancer seeding and distant metastasis. Breast Cancer Res 2018; [DOI] [PMC free article] [PubMed]
- 102.Xing F, Kobayashi A, Okuda H, Watabe M, Pai SK, Pandey PR, et al. Reactive astrocytes promote the metastatic growth of breast cancer stem-like cells by activating Notch signalling in brain. EMBO Mol Med 2013; [DOI] [PMC free article] [PubMed]
- 103.Espinoza I, Pochampally R, Xing F, Watabe K, Miele L. Notch signaling: Targeting cancer stem cells and epithelial-to-mesenchymal transition. OncoTargets and Therapy. 2013. [DOI] [PMC free article] [PubMed]
- 104.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res 2005; [DOI] [PubMed]
- 105.Reedijk M, Pinnaduwage D, Dickson BC, Mulligan AM, Zhang H, Bull SB, et al. JAG1 expression is associated with a basal phenotype and recurrence in lymph node-negative breast cancer. Breast Cancer Res Treat 2008; [DOI] [PubMed]
- 106.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126(4):663–76. [DOI] [PubMed] [Google Scholar]
- 107.Seymour T, Twigger A-JJ, Kakulas F, Au A-JT. Pluripotency genes and their functions in the normal and aberrant breast and brain [Internet]. International Journal of Molecular Sciences 2015. Available from: www.mdpi.com/journal/ijms [DOI] [PMC free article] [PubMed]
- 108.Rangel MC, Bertolette D, Castro NP, Klauzinska M, Cuttitta F, Salomon DS. Developmental signaling pathways regulating mammary stem cells and contributing to the etiology of triple-negative breast cancer. Breast Cancer Research and Treatment. 2016. [DOI] [PMC free article] [PubMed]
- 109.Hassiotou F, Hepworth AR, Beltran AS, Mathews MM, Stuebe AM, Hartmann PE, et al. Expression of the Pluripotency Transcription Factor OCT4 in the Normal and Aberrant Mammary Gland. Front Oncol [Internet]. 2013. [cited 2019 Jan 2];3:79 Available from: http://www.ncbi.nlm.nih.gov/pubmed/23596564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hassiotou F, Beltran A, Chetwynd E, Stuebe AM, Twigger AJ, Metzger P, et al. Breastmilk is a novel source of stem cells with multilineage differentiation potential. Stem Cells. 2012; [DOI] [PMC free article] [PubMed]
- 111.Hassiotou F, Heath B, Ocal O, Filgueira L, Geddes D, Hartmann P, et al. Breastmilk stem cell transfer from mother to neonatal organs (216.4). FASEB J 2014;
- 112.Wang Y, Dong J, Li D, Lai L, Siwko S, Li Y, et al. Lgr4 regulates mammary gland development and stem cell activity through the pluripotency transcription factor Sox2. Stem Cells [Internet]. 2013. September [cited 2019 Jan 2];31(9):1921–31. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23712846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moumen M, Chiche A, Deugnier M-A, Petit V, Gandarillas A, Glukhova MA, et al. The Proto-Oncogene Myc Is Essential for Mammary Stem Cell Function. Stem Cells [Internet]. 2012. June [cited 2019 Feb 4];30(6):1246–54. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22438054 [DOI] [PubMed] [Google Scholar]
- 114.Wegner M, Stolt CC. From stem cells to neurons and glia: A Soxist’s view of neural development. Trends in Neurosciences. 2005. [DOI] [PubMed]
- 115.Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron 2003; [DOI] [PubMed]
- 116.Finzsch M, Schreiner S, Kichko T, Reeh P, Tamm ER, Bösl MR, et al. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J Cell Biol 2010; [DOI] [PMC free article] [PubMed]
- 117.Dravis C, Spike BT, Harrell JC, Johns C, Trejo CL, Southard-Smith EM, et al. Sox10 Regulates Stem/Progenitor and Mesenchymal Cell States in Mammary Epithelial Cells. Cell Rep [Internet]. 2015. September [cited 2016 Jun 22];12(12):2035–48. Available from: http://linkinghub.elsevier.com/retrieve/pii/S2211124715009250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iglesias JM, Gumuzio J, Martin AG. Linking Pluripotency Reprogramming and Cancer. Stem Cells Transl Med [Internet]. 2017. February [cited 2019 Feb 4];6(2):335–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28191771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nishi M, Sakai Y, Akutsu H, Nagashima Y, Quinn G, Masui S, et al. Induction of cells with cancer stem cell properties from nontumorigenic human mammary epithelial cells by defined reprogramming factors. Oncogene [Internet]. 2014. January 14 [cited 2019 Feb 4];33(5):643–52. Available from: http://www.nature.com/articles/onc2012614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Y-J, Herlyn M. The emerging roles of Oct4 in tumor-initiating cells. Am J Physiol Physiol [Internet]. 2015. December [cited 2019 Feb 4];309(11):C709–18. Available from: http://www.physiology.org/doi/10.1152/ajpcell.00212.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene [Internet]. 2012. March 8 [cited 2019 Feb 4];31(11):1354–65. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21822303 [DOI] [PubMed] [Google Scholar]
- 122.Jo A, Denduluri S, Zhang B, Wang Z, Yin L, Yan Z, et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes and Diseases. 2014. [DOI] [PMC free article] [PubMed]
- 123.Cho Y, Kang HG, Kim S-J, Lee S, Jee S, Ahn SG, et al. Post-translational modification of OCT4 in breast cancer tumorigenesis. Cell Death Differ [Internet]. 2018. October 6 [cited 2019 Feb 4];25(10):1781–95. Available from: http://www.nature.com/articles/s41418-018-0079-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lu X, Mazur SJ, Lin T, Appella E, Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene [Internet]. 2014. May 17 [cited 2019 Feb 4];33(20):2655–64. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23770853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang D, Lu P, Zhang H, Luo M, Zhang X, Wei X, et al. Oct-4 and Nanog promote the epithelial-mesenchymal transition of breast cancer stem cells and are associated with poor prognosis in breast cancer patients. Oncotarget [Internet]. 2014. November 15 [cited 2019 Feb 4];5(21):10803–15. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25301732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen Y, Olopade OI. MYC in breast tumor progression. Expert Rev Anticancer Ther [Internet]. 2008. October [cited 2019 Feb 5];8(10):1689–98. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18925859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu J, Chen Y, Olopade OI. MYC and Breast Cancer. Genes Cancer [Internet]. 2010. June [cited 2019 Feb 5];1(6):629–40. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21779462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee HY, Cha J, Kim SK, Park JH, Song KH, Kim P, et al. c-MYC Drives Breast Cancer Metastasis to the Brain, but Promotes Synthetic Lethality with TRAIL. Mol Cancer Res [Internet]. 2019. February [cited 2019 Feb 5];17(2):544–54. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30266755 [DOI] [PubMed] [Google Scholar]
- 129.Zhao N, Cao J, Xu L, Tang Q, Dobrolecki LE, Lv X, et al. Pharmacological targeting of MYC-regulated IRE1/XBP1 pathway suppresses MYC-driven breast cancer. J Clin Invest 2018; [DOI] [PMC free article] [PubMed]
- 130.Liu H, Radisky DC, Yang D, Xu R, Radisky ES, Bissell MJ, et al. MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat Cell Biol [Internet]. 2012. May 13 [cited 2019 Feb 26];14(6):567–74. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22581054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weina K, Utikal J. SOX2 and cancer: current research and its implications in the clinic. Clin Transl Med [Internet]. 2014. [cited 2017 Feb 25];3:19 Available from: http://www.ncbi.nlm.nih.gov/pubmed/25114775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wuebben EL, Rizzino A. The dark side of SOX2: cancer - a comprehensive overview. Oncotarget [Internet]. 2017. July 4 [cited 2019 Feb 5];8(27):44917–43. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28388544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim M, Jang K, Miller P, Picon-Ruiz M, Yeasky TM, El-Ashry D, et al. VEGFA links self-renewal and metastasis by inducing Sox2 to repress miR-452, driving Slug. Oncogene [Internet]. 2017. May 15 [cited 2017 May 17]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/28504716 [DOI] [PMC free article] [PubMed]
- 134.Wang H, Xie J. The role of SOX2 in angiogenesis in breast cancer [Internet]. Vol. 11, Int J Clin Exp Pathol 2018. [cited 2019 Feb 5]. Available from: www.ijcep.com/ [PMC free article] [PubMed] [Google Scholar]
- 135.Liu K, Lin B, Zhao M, Yang X, Chen M, Gao A, et al. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal [Internet]. 2013. May [cited 2017 Sep 11];25(5):1264–71. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23416461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Oliphant MUJ, Vincent MY, Galbraith MD, Pandey A, Zaberezhnyy V, Rudra P, et al. Six2 mediates late-stage metastasis via direct regulation of Sox2 and induction of a cancer stem cell program. Cancer Res [Internet]. 2019. January 1;canres.1791.2018. Available from: http://cancerres.aacrjournals.org/content/early/2019/01/03/0008-5472.CAN-18-1791.abstract [DOI] [PMC free article] [PubMed]
- 137.De Lope C, Martín-Alonso S, Auzmendi-Iriarte J, Escudero C, Mulet I, Larrasa-Alonso J, et al. SIX1 represses senescence and promotes SOX2-mediated cellular plasticity during tumorigenesis. Sci Rep [Internet]. 2019. December 5 [cited 2019 Feb 16];9(1):1412 Available from: http://www.nature.com/articles/s41598-018-38176-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gupta N, Gopal K, Wu C, Alshareef A, Chow A, Wu F, et al. Phosphorylation of Sox2 at Threonine 116 is a Potential Marker to Identify a Subset of Breast Cancer Cells with High Tumorigenecity and Stem-Like Features. Cancers (Basel) [Internet]. 2018. February 3 [cited 2019 Feb 26];10(2). Available from: http://www.ncbi.nlm.nih.gov/pubmed/29401647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Z, Kang L, Zhang H, Huang Y, Fang L, Li M, et al. AKT drives SOX2 overexpression and cancer cell stemness in esophageal cancer by protecting SOX2 from UBR5-mediated degradation. Oncogene 2019; [DOI] [PubMed]
- 140.Voutsadakis I The network of pluripotency, epithelial-mesenchymal transition, and prognosis of breast cancer. Breast Cancer Targets Ther [Internet]. 2015. September 7 [cited 2018 Feb 17];7:303–19. Available from: https://www.dovepress.com/the-network-of-pluripotency-epithelialndashmesenchymal-transition-and--peer-reviewed-article-BCTT [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pietiläinen T, Lipponen P, Aaltomaa S, Eskelinen M, Kosma VM, Syrjänen K. Expression of c-myc proteins in breast cancer as related to established prognostic factors and survival. Anticancer Res [Internet]. [cited 2019 Feb 26];15(3):959–64. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7645986 [PubMed] [Google Scholar]
- 142.Ballinger TJ, Meier JB, Jansen VM. Current Landscape of Targeted Therapies for Hormone-Receptor Positive, HER2 Negative Metastatic Breast Cancer. Front Oncol [Internet]. 2018. August 10 [cited 2019 Feb 18];8:308 Available from: https://www.frontiersin.org/article/10.3389/fonc.2018.00308/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kim SJ, Garcia-Recio S, Creighton CJ, Perou CM, Rosen JM. Alterations in Wnt- and/or STAT3 signaling pathways and the immune microenvironment during metastatic progression. Oncogene. 2019; [DOI] [PMC free article] [PubMed]