Abstract
Background
Within the complex wheat flour proteome, the gluten proteins have attracted most of the attention because of their importance in determining the functional properties of wheat flour doughs and their roles in human health conditions such as celiac disease and food allergies. However, certain non-gluten proteins also trigger immunological responses but may be present in flour in low amounts or obscured by the more abundant gluten proteins in two-dimensional gels of total protein preparations.
Methods
Non-gluten proteins were preferentially extracted from the flour with a dilute salt solution and separated by two-dimensional gel electrophoresis. Proteins in 173 gel spots were identified by tandem mass spectrometry after cleavage with trypsin or chymotrypsin. Transgenic wheat lines in which specific groups of gluten proteins were suppressed by RNA interference were used to estimate the amount of carry-over of gluten proteins in the salt-soluble protein fraction.
Results
Fifty-seven different types of non-gluten proteins were identified, including 14 types that are known or suspected immunogenic proteins. The predominant proteins in 18 gel spots were gluten proteins. Some of these also contained non-gluten proteins. Analysis of the salt-soluble proteins from a transgenic line in which omega-1,2 gliadins were eliminated by RNA interference indicated that certain omega-1,2 gliadins were present in large amounts in the salt-soluble fraction and obscured relatively small amounts of beta-amylase and protein disulfide isomerase. In comparison, analysis of a transgenic line in which alpha gliadins were absent revealed that glyceraldehyde-3 phosphate dehydrogenase was a moderately abundant protein that co-migrated with several alpha gliadins.
Conclusions
In this study, we constructed a proteomic map of the non-gluten protein fraction of wheat flour from the US wheat Butte 86 that complements a proteomic map of the total flour proteins developed previously for the same cultivar. Knowing the identities of low abundance proteins in the flour as well as proteins that are hidden by some of the major gluten proteins on two-dimensional gels is critical for studies aimed at assessing the immunogenic potential of wheat flour and determining which wheat proteins that should be targeted in future gene editing experiments to reduce the immunogenic potential of wheat flour.
Keywords: Albumins/globulins, Allergens, Celiac disease, Non-celiac wheat sensitivity, Wheat flour proteome, Mass spectrometry
Background
Wheat flour proteins can be separated on the basis of solubility into the water/salt-insoluble gliadins and glutenins (gluten proteins) and the water/salt-soluble albumins and globulins (non-gluten proteins). The gluten proteins, comprising ~ 80% of the total protein, are a collection of ~ 60–80 different proteins that serve as the primary storage proteins of the grain while the non-gluten proteins are a diverse collection of different types of proteins that have metabolic, structural or defense functions. Much of the past research has focused on the gluten proteins because of their important role in determining the unique viscoelastic properties that give wheat flour its commercial value. These proteins are also of considerable interest because they are a major trigger of human health conditions, including celiac disease (CD), a chronic inflammatory disease of the small intestine that occurs in genetically susceptible individuals, and food allergies [1, 2]. The gliadins are primarily monomeric proteins that are divided into four groups termed alpha, gamma, omega and delta gliadins while the glutenins are polymeric proteins that consist of high molecular weight glutenin subunits (HMW-GS) and low molecular weight glutenin subunits (LMW-GS). Epitopes that stimulate T-cells in CD patients have been identified in proteins within most major classes of gluten proteins, although epitopes found in certain alpha gliadins and a subclass of omega gliadins, the omega-1,2 gliadins, have been found to be immunodominant [3, 4]. Gluten proteins in all major classes also react with IgE from patients with wheat allergy [5]. In particular, the omega-5 gliadins, another subclass of omega gliadins, have been found to trigger the serious food allergy wheat-dependent exercise-induced anaphylaxis that occurs in sensitized individuals when the ingestion of wheat is followed by physical exercise [6].
Non-gluten proteins also have been found to be immunogenic. In studies that examined cohorts of 28 and 60 patients with confirmed cases of wheat allergy, Battais et al. [5, 7] found that IgE from 67 and 72% of patients, respectively, reacted with proteins in the albumin/globulin fraction. Further studies using 1-D and 2-D immunoblots of albumin/globulin fractions demonstrated that a wide range of proteins often react with IgE from single patients and that the reactive proteins vary from individual to individual [8–11]. Non-gluten proteins likely to be allergenic are listed in the Allergome database (http://www.allergome.org/) and include beta amylase (Tri a 17), thioredoxin (Tri a 25), serpin (Tri a 33), triosephosphate isomerase (Tri a 31), peroxidase (Tri a peroxidase), chitinase (Tri a chitinase), peroxiredoxin (Tri a 32), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Tri a 34), xylanase inhibitor (Tri a XI), dehydrin (Tri a 35), 9 kDa lipid transfer protein (Tri a 14) and a number of alpha amylase/trypsin inhibitors (AAI) (Tri a 28, Tri a 29, Tri a 30, Tri a 40). In addition, certain non-gluten proteins have been shown to be reactive with IgG and IgA antibodies from CD patients, suggesting a role for serpins, globulins, purinins, farinins and AAI in CD [12]. AAI also have been implicated in non-celiac wheat sensitivity (NCWS), a response to wheat ingestion that is distinct from CD and food allergies [13], while a globulin from wheat flour has been associated with the development of type 1 diabetes [14].
Because sensitivities to wheat are increasing, there is a need to identify clinically relevant proteins in wheat flour, associate specific flour proteins with particular types of allergies and sensitivities, and determine the frequencies of immunogenic responses to particular proteins. Two-dimensional immunoblot analysis is a powerful method for assessing the reactivity of patient sera to specific proteins in the flour [8, 10–12, 15–17]. The approach combines high resolution two-dimensional gel electrophoresis (2-DE), a robust, reproducible method for the both the separation and quantification of hundreds of proteins, with the identification of individual proteins by tandem mass spectrometry (MS/MS). However, 2-DE analyses of total protein preparations from wheat flour can be problematic because the more abundant gluten proteins overlap with low-abundance non-gluten proteins in some areas of the gels [18]. In a comprehensive analysis of total flour proteins, Dupont et al. [19] found MS/MS evidence for non-gluten proteins in 22% of 2-DE spots containing either LMW-GS or gliadins as the predominant proteins. In their study, LMW-GS overlapped with globulins, peroxidase, serpin and xylanase inhibitor; alpha gliadins overlapped with fructose bisphosphate aldolase, alcohol dehydrogenase and formate dehydrogenase; gamma gliadins overlapped with triticin and aspartic proteinase; and omega gliadins overlapped with beta-amylase. For this reason, salt-soluble protein fractions should also be considered for studies of immunogenic proteins. In this paper, we extracted proteins from flour from the US wheat Butte 86 with a dilute salt solution, separated the proteins by 2-DE, and identified proteins in 173 2-DE spots by MS/MS. The resulting proteomic map should make it possible to use 2-D immunoblots to rapidly identify and prioritize non-gluten proteins to be targeted in future gene editing experiments aimed at reducing the immunogenic potential of wheat flour.
Methods
Proteins were extracted from wheat flour from the US spring wheat cultivar Butte 86 with a buffer containing 100 mM KCl as described in Hurkman and Tanaka [18], quantified by Lowry, and analyzed by 2-DE as described in detail in Dupont et al. [19]. In short, 50 mg flour was suspended in 200 μl of 50 mM Tris-HCl pH 7.8, 100 mM KCl, 5 mM EDTA and incubated for 5 min with intermittent vortex mixing. Samples were centrifuged at 4 °C at 14,500 x g for 15 min. The supernatant was collected and proteins were precipitated by the addition of 4 volumes of cold (− 20 °C) acetone. The pellet was rinsed with cold acetone, air-dried, and stored at -20 °C. After quantification, proteins were solubilized in urea buffer (9 M urea, 4% NP-40, 1% DTT, 2% ampholytes) for isoelectric focusing using capillary tube gels with a pI range of 3–10. NuPAGE 4–12% Bis-Tris protein gels (Life Technologies, Carlsbad, CA, USA) were used for the second dimension separation.
LC-MS analysis
Protein spots were excised from replicate 2-D gels and placed in 96-well plates where they were reduced, alkylated and digested separately with trypsin (Sequencing Grade Modified Trypsin, Promega Corp., Madison, WI, USA) or chymotrypsin (Sequencing Grade Chymotrypsin, Promega Corp., Madison, WI, USA) using a DigestPro according to the directions of the manufacturer (Intavis, Koeln, Germany).
Plates containing peptides from digested gel spots were placed in the autosampler of a NanoLC 425 (Sciex, Framinham, MA, USA) that was interfaced by a nano-electrospray source to an Orbitrap Elite mass spectrometer (Thermo Scientific, San Jose, CA, USA). Ten μl fractions were loaded by the autosampler onto one of three PicoChip reverse phase columns (PicoChip with ReproSil-PUR C18, 10.5 cm, ID 75 μm, 3 μm, 120 Å, ReproSil-PUR-C18-AQ, New Objective Inc., Woburn, MA, USA). The positions of the columns were switched in front of the mass spectrometer with the PicoSlide system for Thermo LTQ/Orbi (New Objective Inc., Woburn, MA, USA). With this configuration, Column A is eluting, Column B is loading and Column C is washing, thereby saving time and reducing carryover between samples.
The loaded sample was washed with solvent A to remove salts and the column position was switched in front of the Orbitrap Elite mass spectrometer and eluted with a gradient of acetonitrile into the mass spectrometer. Solvent A was 5% in acetonitrile and Solvent B was 80% in acetonitrile, both solvents were 0.05% in formic acid. Gradient elution was at a flow rate of 250 nl per minute from 100% Solvent A to 35% Solvent B in 45 min. Peptides were detected in the Orbitrap set to scan a range from 200 to 2000 m/z at a resolution of 60,000. The 10 most intense peaks were subject to collision-induced dissociation (CID). The minimal signal threshold was set to 10,000. Dynamic exclusion with a repeat count of 2 was enabled for duration of 10 s. Normalized collision energy was set to 30%.
Data analysis
The vendor specific .raw files were converted into .mgf files using MSConvert (ProteoWizard, Palo Alto, CA, USA). Spectral data was searched against a database containing all Triticeae sequences from NCBI downloaded on 02/07/2018, full-length Butte 86 sequences from Dupont et al. [19], full length Chinese Spring gluten protein sequences reported by Huo et al. [20, 21], and common MS contaminant sequences contained in the common Repository of Adventitious Proteins (cRAP) database (ftp://ftp.thegpm.org/fasta/cRAP/crap.fasta). Two search engines were used for analysis, Mascot (www.matrixscience.com) and XTandem! (https://www.thegpm.org/TANDEM/).
Results were loaded into Scaffold 4 (Scaffold_4.9.0, Proteome Software, Inc., Portland, OR, USA). The mass accuracy was set to 30 ppm on precursor level and 0.6 Da on MS/MS level. Scaffold summarized the Mascot and X!Tandem peptide hit results from the two different enzyme digestions (trypsin, chymotrypsin) with the MudPit analysis and compress data summarizing option.
Twenty-one spots that contained evidence of gluten proteins in this analysis were excised from replicate gels, digested with thermolysin (Promega Corp., Madison, WI, USA), and analyzed by MS/MS as above. These included spots 26, 27, 45, 54, 55, 60, 61, 80, 82, 83, 84, 87, 91, 101, 102, 104, 105, 106, 189, 190, and 191. Data from all three enzyme digestions were then combined in Scaffold. The predominant protein in each spot was deemed to be the protein for which the greatest number of unique peptides were identified and is reported in Table 1. All proteins identified in each spot are reported in Additional file 1. MS data for predominant proteins and proteins that contained at least half the number of unique peptides found in the predominant proteins are shown in Additional file 2. The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [23] with the dataset identifier PXD017260 and 10.6019/PXD017260.
Table 1.
Predominant proteins identified by MS/MS in 2-DE protein spots from the KCl-soluble fraction of Butte 86 flour. All proteins had identification scores greater than 99% probability with at least 4 peptides as determined by Scaffold. Positions of spots in 2-DE are shown in Fig. 2. Identities of all proteins found in each spot are reported in Additional file 1
| Spot Number | Predominant Protein | Accession #a | Source | # Unique peptides | # Spectra | % Coverage |
|---|---|---|---|---|---|---|
| 1 | pyruvate phosphate dikinase | XP_0201883631 | Ae. tauschii | 19 | 50 | 24 |
| 2 | pyruvate phosphate dikinase | XP_0201883631 | Ae. tauschii | 19 | 83 | 21 |
| 3 | pyruvate phosphate dikinase | XP_0201883631 | Ae. tauschii | 20 | 101 | 26 |
| 4 | aconitate hydratase | XP_020176302 | Ae. tauschii | 13 | 31 | 15 |
| 5 | no ID | |||||
| 6 | heat shock protein 101 | AAD22629 | T.aestivum | 26 | 67 | 31 |
| 7 | elongation factor 2 | EMS59408 | T. urartu | 20 | 52 | 25 |
| 8 | elongation factor 2 | EMS59408 | T. urartu | 11 | 25 | 13 |
| 9 | embryonic protein DC-8-like | XP_020196294 | Ae. tauschii | 14 | 50 | 24 |
| 10 | embryonic protein DC-8-like | XP_020196294 | Ae. tauschii | 20 | 59 | 29 |
| 11 | embryonic protein DC-8-like | XP_020196294 | Ae. tauschii | 17 | 65 | 31 |
| 12b | embryonic protein DC-8-like | XP_020196294 | Ae. tauschii | 30 | 85 | 38 |
| 13 | embryonic protein DC-8-like | XP_020196294 | Ae. tauschii | 42 | 131 | 50 |
| 14 | heat shock protein 90 | ABG57075 | T.aestivum | 19 | 64 | 26 |
| 15 | heat shock protein 70 | XP_020151532 | Ae. tauschii | 43 | 209 | 48 |
| 16 | heat shock protein 70 | EMS51416 | T. urartu | 26 | 85 | 35 |
| 17 | no ID | |||||
| 18 | beta-D-glucan exohydrolase | AAM13694 | T.aestivum | 25 | 84 | 34 |
| 19 | beta-D-glucan exohydrolase | AAM13694 | T.aestivum | 24 | 60 | 33 |
| 20 | beta-D-xylosidase | XP_020155854 | Ae. tauschii | 17 | 44 | 23 |
| 21c | glucose and ribitol dehydrogenase | pir T06212 | Hordeum vulgare | 7 | 19 | 28 |
| 22 | no ID | |||||
| 23 | glucose-6-P isomerase | ABE41790 | T.aestivum | 22 | 67 | 37 |
| 24 | glucose-6-P isomerase | ABE41790 | T.aestivum | 21 | 59 | 38 |
| 25 | globulin 3A | AFM30909 | T.aestivum | 12 | 38 | 24 |
| 26b,c | beta-amylase | XP_020197275 | Ae. tauschii | 64 | 397 | 70 |
| 27c,d | beta-amylase | XP_020197275 | Ae. tauschii | 46 | 287 | 58 |
| 28 | beta-amylase | EMS68884 | T. urartu | 21 | 83 | 37 |
| 29 | beta-amylase | EMS68884 | T. urartu | 22 | 88 | 40 |
| 30 | ketol-acid reductoisomerase | XP_020174215 | Ae. tauschii | 38 | 111 | 47 |
| 31 | leucine aminopeptidase 2 | EMS53149 | T. urartu | 26 | 61 | 47 |
| 32 | atp1 | ACA62607 | T.aestivum | 20 | 67 | 38 |
| 33 | aldehyde dehydrogenase | AKE36953 | T.aestivum | 6 | 17 | 16 |
| 34 | omega gliadin | AKB95614 | T. urartu | 18 | 34 | 51 |
| 35 | enolase | XP_020163593 | Ae. tauschii | 33 | 135 | 78 |
| 36 | enolase | XP_020163593 | Ae. tauschii | 55 | 164 | 89 |
| 37 | enolase | BAJ85279 | Hordeum vulgare | 45 | 167 | 83 |
| 38c | enolase | BAJ85279 | Hordeum vulgare | 19 | 62 | 63 |
| 39 | alanine aminotransferase | XP_020147857 | Ae. tauschii | 38 | 104 | 66 |
| 40 | leghemoglobin reductase | XP_020185250 | Ae. tauschii | 35 | 139 | 69 |
| 41 | cupincin | XP_020147625 | Ae. tauschii | 16 | 42 | 33 |
| 42 | cupincin | XP_020147625 | Ae. tauschii | 30 | 90 | 47 |
| 43 | cupincin | XP_020147625 | Ae. tauschii | 35 | 105 | 51 |
| 44 | cupincin | XP_020147625 | Ae. tauschii | 33 | 151 | 47 |
| 45d | cupincin | XP_020147625 | Ae. tauschii | 35 | 192 | 53 |
| 46 | cupincin | XP_020147625 | Ae. tauschii | 38 | 112 | 53 |
| 47 | globulin-3A | AFM30909 | T.aestivum | 34 | 136 | 45 |
| 48 | globulin-3A | AFM30909 | T.aestivum | 35 | 107 | 38 |
| 49 | globulin-3A | AFM30909 | T.aestivum | 43 | 147 | 46 |
| 50 | globulin-3A | AFM30909 | T.aestivum | 25 | 77 | 28 |
| 51 | translation elongation factor 1 alpha | AAA34306 | T.aestivum | 25 | 109 | 44 |
| 52 | globulin 3 | ACJ65514 | T.aestivum | 5 | 13 | 8 |
| 53 | globulin-3A | AFM30909 | T.aestivum | 25 | 83 | 28 |
| 54d,e | globulin-3A | AFM30909 | T.aestivum | 23 | 107 | 35 |
| 55d,f | LMW-GS | ACA63869 | T.aestivum | 27 | 84 | 53 |
| 56 | globulin-3A | AFM30909 | T.aestivum | 7 | 23 | 14 |
| 57 | LMW-GS | ABC84366 | T.aestivum | 28 | 54 | 60 |
| 58 | LMW-GS | ABC84366 | T.aestivum | 9 | 14 | 29 |
| 59 | ERBB-3 binding protein | XP_020191236 | Ae. tauschii | 11 | 53 | 29 |
| 60d | gamma gliadin | BU-gamma-5 | T.aestivum | 15 | 38 | 41 |
| 61d | gamma gliadin | BU-gamma5 | T.aestivum | 15 | 52 | 41 |
| 62 | triticin | ACB41345 | T.aestivum | 15 | 49 | 26 |
| 63 | isocitrate dehydrogenase | AMP82030 | T.aestivum | 11 | 41 | 27 |
| 64 | serpin | EMS54555 | T. uratu | 34 | 102 | 68 |
| 65 | serpin | CAB52709 | T.aestivum | 51 | 213 | 78 |
| 66 | serpin | ACN59485 | T.aestivum | 35 | 109 | 66 |
| 67 | serpin | ACN59485 | T.aestivum | 90 | 358 | 85 |
| 68 | serpin | ACN59485 | T.aestivum | 55 | 162 | 78 |
| 69 | serpin | ACN59484 | T.aestivum | 16 | 50 | 40 |
| 70 | serpin | ACN59484 | T.aestivum | 25 | 75 | 48 |
| 71 | serpin | AFC89429 | T.aestivum | 30 | 67 | 63 |
| 72 | serpin | AFC89429 | T.aestivum | 87 | 268 | 86 |
| 73 | serpin | AFC89429 | T.aestivum | 65 | 207 | 82 |
| 74 | serpin | AFC89429 | T.aestivum | 11 | 36 | 32 |
| 75 | glycosyltransferase 75 | ADK56176 | T.aestivum | 7 | 18 | 22 |
| 76 | serpin | CAA90071 | T.aestivum | 31 | 123 | 61 |
| 77 | serpin | CAA90071 | T.aestivum | 31 | 61 | 61 |
| 78 | serpin | EMS46390 | T. uratu | 39 | 115 | 67 |
| 79 | alpha gliadin | BAM08450 | T.aestivum | 10 | 23 | 34 |
| 80b,d,f | alpha gliadin | CAY54134 | T.aestivum | 25 | 104 | 45 |
| 81b | ER molecular chaperone | AGN94840 | T.aestivum | 30 | 149 | 34 |
| 82d,f | gamma gliadin | BU-gamma-1 | T.aestivum | 29 | 164 | 50 |
| 83d,f,g | alpha gliadin | BU-alpha-23 | T.aestivum | 38 | 107 | 63 |
| 84d | LMW-GS | ABC84361 | T.aestivum | 17 | 48 | 33 |
| 85 | no ID | |||||
| 86f | LMW-GS | ACJ76984 | T. dicoccoides | 7 | 25 | 16 |
| 87d,f | gamma gliadin | AEA30015 | T.aestivum | 24 | 78 | 53 |
| 88 | alpha gliadin | AAA96522 | T.aestivum | 11 | 24 | 42 |
| 89 | peroxidase | XP_020147267 | Ae. tauschii | 38 | 173 | 53 |
| 90 | peroxidase | AAM88383 | T.aestivum | 47 | 125 | 57 |
| 91b,d,e | peroxidase | AAM88383 | T.aestivum | 62 | 197 | 60 |
| 92 | aldose 1 epimerase | XP_020180146 | Ae. tauschii | 9 | 24 | 22 |
| 93 | globulin 3A | AFM30909 | T.aestivum | 19 | 57 | 25 |
| 94 | 63 kd globulin | XP-020163670 | Ae. tauschii | 24 | 88 | 27 |
| 95 | farinin | XP_020147251 (BU-farinin-1) | Ae. tauschii | 9 | 49 | 31 |
| 96 | 63 kd globulin | XP-020163670 | Ae. tauschii | 24 | 106 | 28 |
| 97 | aspartic proteinase | XP_020157026 | Ae. tauschii | 9 | 27 | 19 |
| 98g | aspartic proteinase | XP_020157026 | Ae. tauschii | 5 | 13 | 13 |
| 99 | cupincin | XP_020147625 | Ae. tauschii | 8 | 31 | 18 |
| 100 | peroxidase | AAM88383 | T.aestivum | 13 | 36 | 34 |
| 101b,d,h | glyceraldehyde 3-P dehydrogenase | ALE18234 | T.aestivum | 49 | 164 | 65 |
| 102b,d | glyceraldehyde 3-P dehydrogenase | ANW11922 | T.aestivum | 79 | 230 | 92 |
| 103b,d | glyceraldehyde 3-P dehydrogenase | ALE18234 | T.aestivum | 37 | 85 | 68 |
| 104b,d | fructose bisphosphate aldolase | CDM85265 | T.aestivum | 60 | 197 | 73 |
| 105b,d | glyceraldehyde 3-P dehydrogenase | ABQ81648 | T.aestivum | 56 | 208 | 73 |
| 106b,d,f | alpha gliadin | CS-alpha-A1 | T.aestivum | 70 | 215 | 77 |
| 107 | malate dehydrogenase | XP_020196362 | Ae. tauschii | 6 | 17 | 17 |
| 108 | guanine nucleotide binding protein | CDM83867 | T.aestivum | 11 | 23 | 40 |
| 109 | malate dehydrogenase | XP_020157252 | Ae. tauschii | 44 | 149 | 73 |
| 110 | malate dehydrogenase | AAT64932 | T.aestivum | 21 | 53 | 54 |
| 111 | desiccation-related protein | XP_020169835 | Ae. tauschii | 16 | 37 | 35 |
| 112 | desiccation-related protein | XP_020169835 | Ae. tauschii | 11 | 34 | 31 |
| 113 | lactoylglutathione lyase | XP_020185375 | Ae. tauschii | 13 | 38 | 41 |
| 114 | lactoylglutathione lyase | XP_020185375 | Ae. tauschii | 51 | 149 | 79 |
| 115 | lactoylglutathione lyase | XP_020185375 | Ae. tauschii | 16 | 54 | 44 |
| 116 | lactoylglutathione lyase | EMS56223 | T. uratu | 7 | 19 | 21 |
| 117 | glucose and ribitol dehydrogenase | XP_020188730 | Ae. tauschii | 16 | 41 | 46 |
| 118 | glucose and ribitol dehydrogenase | XP_020188730 | Ae. tauschii | 28 | 107 | 61 |
| 119 | glucose and ribitol dehydrogenase | XP_020188730 | Ae. tauschii | 31 | 127 | 67 |
| 120 | glucose and ribitol dehydrogenase | XP_020188730 | Ae. tauschii | 29 | 101 | 65 |
| 121 | proteosome subunit alpha type 1 | BAJ85935 | Hordeum vulgare | 10 | 28 | 39 |
| 122 | glucose and ribitol dehydrogenase | BAJ85825 | Hordeum vulgare | 15 | 64 | 39 |
| 123 | dehydrin | AOM63239 | T.aestivum | 12 | 29 | 61 |
| 124 | purinin | ADA62372 (BU-purinin-3) | T.aestivum | 14 | 105 | 40 |
| 125 | 19 kd globulin | XP_020162707 | Ae. tauschii | 12 | 65 | 61 |
| 126 | oil body associated protein | XP_020196637 | Ae. tauschii | 24 | 74 | 81 |
| 127 | 19 kd globulin | XP_020162707 | Ae. tauschii | 9 | 62 | 50 |
| 128 | xylanase inhibitor | AGN71004 | T.aestivum | 34 | 127 | 64 |
| 129 | globulin-1 | ABG68034 | T.aestivum | 12 | 55 | 46 |
| 130 | chitinase | XP_020181917 | Ae. tauschii | 19 | 108 | 41 |
| 131 | globulin | AAR95703 | T. turgidum | 11 | 65 | 50 |
| 132 | globulin | AAR95703 | T. turgidum | 10 | 72 | 50 |
| 133 | tritin | BAA02948 | T.aestivum | 53 | 316 | 75 |
| 134 | farinin | XP_020147251 (BU-farinin-1) | Ae. tauschii | 7 | 35 | 21 |
| 135 | secretory protein | XP_020161391 | Ae. tauschii | 9 | 23 | 38 |
| 136 | secretory protein | XP_020161391 | Ae. tauschii | 5 | 14 | 29 |
| 137 | cyclophilin A | AAK49426 | T.aestivum | 14 | 84 | 57 |
| 138 | chitinase | XP_020181917 | Ae. tauschii | 15 | 28 | 32 |
| 139 | chitinase | AAX83263 | T.aestivum | 20 | 73 | 48 |
| 140 | AAI | prf 1207200A (WASI BU-1) | T.aestivum | 39 | 234 | 82 |
| 141 | AAI | prf 1207200A (WASI BU-1) | T.aestivum | 14 | 72 | 51 |
| 142 | AAI | prf 1207200A (WASI BU-1) | T.aestivum | 31 | 112 | 71 |
| 143 | peroxiredoxin | ACE82290 | T.aestivum | 21 | 91 | 67 |
| 144 | 1-cys peroxiredoxin | AAQ74769 | T.aestivum | 21 | 73 | 65 |
| 145 | purinin | EMS62569 (BU-purinin-1) | T. urartu | 22 | 98 | 68 |
| 146 | purinin | EMS62569 (BU-purinin-1) | T. urartu | 9 | 37 | 52 |
| 147 | triosephosphate isomerase | CDM81250 | T.aestivum | 31 | 102 | 70 |
| 148 | oil body associated protein | XP_020147075 | Ae. tauschii | 30 | 103 | 84 |
| 149 | late embryogenesis abundant protein | XP_020193122 | Ae. tauschii | 10 | 22 | 55 |
| 150 | late embryogenesis abundant protein | XP_020193122 | Ae. tauschii | 14 | 39 | 62 |
| 151 | late embryogenesis abundant protein | BAK04960 | Hordeum vulgare | 6 | 16 | 28 |
| 152 | glutathionine-S-transferase | XP_020196644 | Ae. tauschii | 15 | 57 | 49 |
| 153 | glutathionine-S-transferase | XP_020196644 | Ae. tauschii | 51 | 214 | 60 |
| 154 | glutathionine-S-transferase | XP_020196644 | Ae. tauschii | 21 | 65 | 53 |
| 155 | lysosomal thiol reductase | XP_020155412 | Ae. tauschii | 15 | 73 | 52 |
| 156 | nucleoside diphosphate kinase | EMS63724 | T. urartu | 5 | 16 | 41 |
| 157 | AAI | AAR10959 (WTAI-CM3 BU-1) | T.aestivum | 37 | 388 | 74 |
| 158 | no ID | |||||
| 159 | AAI | CAI84642 (WDAI BU-1) | T.aestivum | 8 | 24 | 66 |
| 160 | no ID | |||||
| 161 | no ID | |||||
| 162 | no ID | |||||
| 163 | ubiquitin | AAP50253 | T.aestivum | 10 | 56 | 83 |
| 164 | AAI | AAV39514 (WDAI BU-1) | T.aestivum | 26 | 244 | 100 |
| 165 | AAI | AAV39517 (WDAI BU-1) | T.aestivum | 22 | 101 | 88 |
| 166 | AAI | sp P16851 (WTAI-CM2 BU-1) | T.aestivum | 24 | 107 | 74 |
| 167 | superoxide dismutase | AFF27607 | T.aestivum | 18 | 61 | 66 |
| 168 | globulin-1 | EMS62417 | T. urartu | 12 | 53 | 15 |
| 169 | AAI | ABO45967 (WMAI BU-1) | T.aestivum | 25 | 135 | 99 |
| 170 | AAI | ABI54484 (WDAI BU-3) | T. dicoccoides | 14 | 58 | 96 |
| 171 | AAI | ABO45967 (WMAI BU-1) | T.aestivum | 19 | 116 | 98 |
| 172 | AAI | AAV91972 (WDAI BU-4) | T.aestivum | 37 | 198 | 100 |
| 173 | AAI | ACM41418 (WTAI-CM16 BU-1) | T. macha | 34 | 228 | 80 |
| 174 | AAI | CAA42453 (WTAI-CM-17 BU-1) | T.aestivum | 25 | 133 | 77 |
| 175 | AAI | CAA42453 (WTAI-CM-17 BU-1) | T.aestivum | 13 | 59 | 50 |
| 176 | AAI | ABO45967 (WMAI BU-1) | T.aestivum | 4 | 17 | 36 |
| 177 | AAI | sp P16851 (WTAI-CM2 BU-1) | T.aestivum | 7 | 25 | 44 |
| 178 | homocysteine methyltransferase | EMS51950 | T. urartu | 20 | 76 | 27 |
| 189d,f | alpha gliadin | BU-alpha-3 | T.aestivum | 15 | 44 | 35 |
| 190d,e | alpha gliadin | BAM08458 | T.aestivum | 23 | 73 | 50 |
| 191b,d | alpha gliadin | BU-alpha-4 | T.aestivum | 19 | 53 | 46 |
a Accession numbers beginning with BU are from Butte 86 and CS are from Chinese Spring. All other accession numbers are from NCBI. The Butte 86 accession is shown in parentheses in cases where a sequence from NCBI is identical to a sequence from Butte 86. Gliadin, purinin and farinin sequences from Butte 86 are from [19]. AAI sequences from Butte 86 are from [22]
b a gluten protein with less than half the number of unique peptides found in the predominant protein was also identified in spot
c also contains an omega gliadin
d replicate sample was digested with thermolysin and analyzed by MS/MS
e also contains a LMW-GS
f also contains a non-gluten protein
g also contains a gamma gliadin
h also contains an alpha gliadin
Flour from transgenic lines missing specific gluten proteins was used to further evaluate spots containing both gluten proteins and non-gluten proteins. Transgenic lines were produced previously using RNA interference. Transgenic line SA-30-118a-5 is missing all omega-1,2 gliadins [16] while transgenic line SA-35a-124j is missing all alpha gliadins [17].
Results
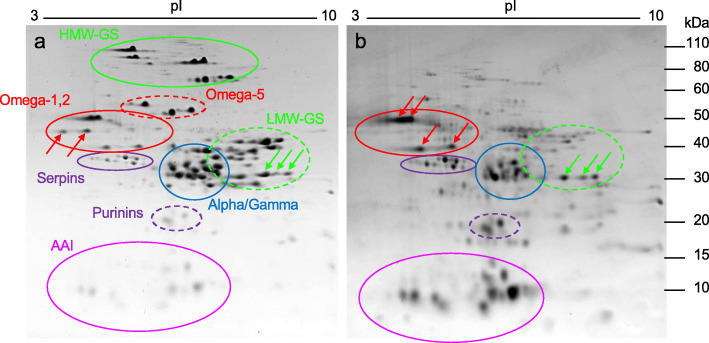
KCl-soluble proteins from Butte 86 flour were separated by 2-DE and the pattern was compared to that of a total SDS protein extract (Fig. 1). The most abundant proteins in the total protein extract, representing 125 of 233 2-DE spots identified previously by MS/MS [19], were the gluten proteins; the HMW-GS and LMW-GS (shown in green), the alpha and gamma gliadins (shown in blue) and the omega gliadins (shown in red) (Fig. 1a). Several groups of non-gluten proteins, including serpins, purinins and alpha-amylase/trypsin inhibitors (AAI), were well-separated from the major gluten proteins but of considerably lower abundance. In comparison, the most abundant proteins in the KCl-soluble fraction (Fig. 1b) were located in the AAI, omega-1,2 gliadin and alpha and gamma gliadin regions of the gel. Additionally, proteins in the serpin and purinin regions were notably more abundant in the KCl-soluble fraction than in the total protein fraction.
Fig. 1.
2-DE separation of total SDS-extracted proteins (a) and KCl-soluble proteins (b) from Butte 86 wheat flour. The positions of major protein groups based on the MS identifications of proteins in Dupont et al. [19] are indicated in red (omega-1,2 gliadins), dashed red (omega-5 gliadins), blue (alpha and gamma gliadins), green (HMW-GS), dashed green (LMW-GS), purple (serpins), dashed purple (purinins), or magenta (AAI) ovals. Red arrows in panel a point to omega-1,2 gliadins that are found in the total protein fraction but not in the KCl-soluble fraction while red arrows in panel b point to proteins likely to be omega-1,2 gliadins that partition into the KCl-soluble fraction. Green arrows point to spots in which the predominant proteins were identified as LMW-GS in the total protein fraction, but peroxidases in the KCl-soluble fraction. The position of size markers in kDa are shown on the right
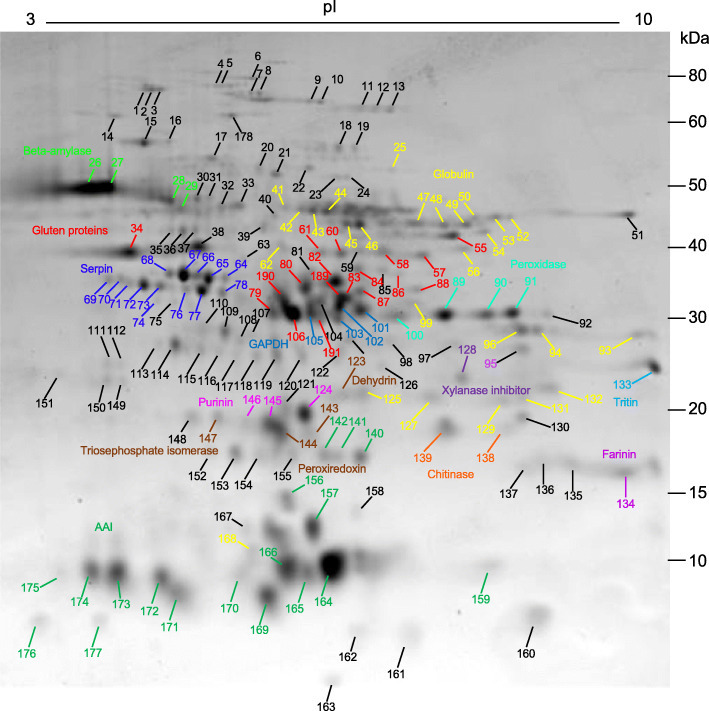
One-hundred-eighty-one spots were excised from 2-D gels of the KCl-soluble proteins and analyzed by MS/MS (Fig. 2). One hundred seventy-three of the spots yielded valid identifications. MS/MS coverage of the proteins ranged from 8 to 100% with an average of 49%. Sixty-one different protein types and 111 sequences were identified as the predominant proteins in these spots (Table 1). The predominant proteins in 26 spots were globulins (Fig. 2). Nine different globulin sequences were identified. Seventeen spots were identified as AAI and 11 different sequences were found. Fourteen spots were serpins and seven different sequences were identified. In addition, the predominant proteins in multiple spots were identified as glucose and ribitol dehydrogenase (6), embryonic protein DC-8 (5), beta-amylase (4), enolase (4), GAPDH (4), lactoylglutathione lyase (4), peroxidase (4), chitinase (3), glutathione-S-transferase (3), late embryogenesis abundant protein (3), malate dehydrogenase (3), pyruvate phosphate dikinase (3), and purinin (3). Forty other non-gluten protein types were identified as the predominant proteins in either one or two 2-DE spots (Table 1). Only eight of these were identified previously in the total protein fraction [19].
Fig. 2.
Identification of proteins in a KCl-soluble fraction of wheat flour. Spots in which the predominant proteins were gluten proteins are labeled in red. Spots in which the predominant proteins were non-gluten proteins that are either known allergens or likely to be immunogenic are labeled in bright green (beta-amylase), dark blue (serpins), medium blue (glyceraldehyde-3-phosphate dehydrogenase (GAPDH), yellow (globulins), teal (peroxidase), magenta (purinins), orange (chitinase), purple (xylanase inhibitor), light blue (tritin), light purple (farinin), brown (dehydrin, peroxiredoxin, triosephosphate isomerase), or green (AAI). Protein identifications can be found in Table 1 and Additional file 1
In some cases, gluten proteins were separated effectively from non-gluten proteins on the basis of solubility. For example, peroxidase was the predominant protein in three moderately abundant spots (89, 90, 91) in the LMW-GS region of the gel from the KCl-soluble fraction (green arrows in Figs. 1b and 2). In the total protein fraction of Dupont et al. [19], the predominant proteins in these three spots were LMW-GS (indicated with green arrows in Fig. 1a). The absence of most LMW-GS in the KCl-soluble fraction also made it easier to visualize many of the minor globulins that are found in this region of the gel (labeled in yellow in Fig. 2).
Despite the fact that most gluten proteins are not readily soluble in salt solutions, gluten proteins were the predominant proteins identified in 18 spots in the KCl fraction (Fig. 2). Eight spots were alpha gliadins, four were gamma gliadins, one was an omega gliadin and five were LMW-GS (Table 1). Fourteen of these spots also contained non-gluten proteins (34, 55, 60, 61, 80, 82, 83, 84, 86, 87, 106, 189, 190, 191). Gluten proteins also were detected as minor components of 14 spots (Table 1, Additional file 1).
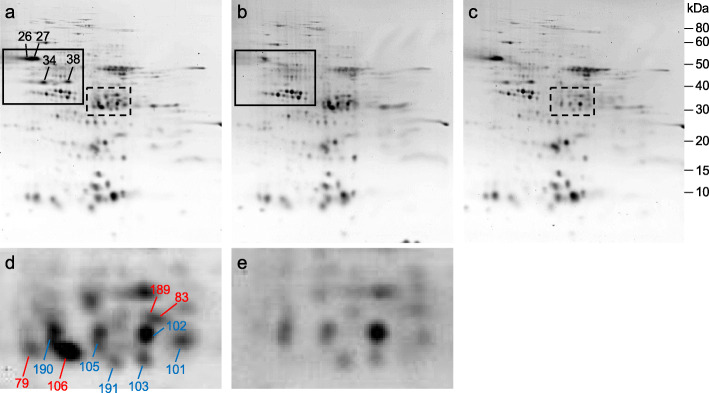
Four major spots (26, 27, 34, 38) in the omega-1,2 gliadin region of the gel were confounding (indicated with red arrows in Fig. 1b, Fig. 2). The predominant proteins identified in spots 26 and 27 were beta-amylase (Table 1). However, protein disulfide isomerase and omega-1,2 gliadin BAN29067 also were identified in these spots (Additional file 1). Spot 34 was identified as omega-1,2 gliadin AKB95614 whereas spot 38 contained both enolase and omega-1,2 gliadin AKB95614. Interestingly, two other spots previously identified as omega-1,2 gliadins (shown with red arrows in Fig. 1a) did not appear in the KCl-soluble fraction even though their sequences were very similar to the omega-1,2 gliadins found in spots 26 and 27 [16].
A number of spots in the region of the gel containing alpha and gamma gliadins also were problematic. Three minor spots in this region contained only alpha gliadins (79, 88, 191) while two minor spots contained only gamma gliadins (60, 61). However, five moderately abundant spots contained both an alpha gliadin as the predominant protein and GAPDH (83, 106, 189, 190, 191). Additionally, the predominant proteins in four other spots were GADPH, but MS/MS evidence also pointed to the presence of alpha gliadins in these spots (101, 102, 103, 105) (Fig. 2, Additional file 1).
To estimate the amounts of different gluten proteins that were carried over into the KCl-soluble protein fraction, we examined the profiles of flour proteins from transgenic lines in which the expression of either the omega-1,2 gliadins or the alpha gliadins was suppressed by RNA interference [16, 17]. Detailed proteomic analyses of total protein fractions from flour of the transgenic lines revealed that all omega-1,2 gliadins were absent in line SA-30-118a-5 [16] while all alpha gliadins were absent in line SA-35a-124j [17]. In 2-D gels of the KCl-soluble protein fraction, spots 26, 27, 34, 38 containing omega-1,2 gliadins were major spots in the non-transgenic flour (Fig. 3a). Spot 26 was significantly reduced while other spots were not apparent in transgenic line SA-30-118a-5 (Fig. 3b), suggesting that significant amounts of certain omega-1,2 gliadins partition into the KCl-soluble fractions and that beta-amylase and protein disulfide isomerase are low abundance proteins. Within the alpha gliadin region of the gel (Fig. 3a, c), several spots in which either alpha gliadins (79) or alpha gliadins and GAPDH (83, 106, 189) were identified in the KCl fraction from Butte 86 were not apparent in transgenic line SA-35a-124j, suggesting that alpha gliadins comprise the bulk of the protein in these spots. Several other spots containing both alpha gliadin and GAPDH were present in similar amounts in Butte 86 and the transgenic line (101, 102, 103, 105, 190, 191), suggesting that most of the protein in these spots is GAPDH and that this protein is fairly abundant in the KCl fraction. Interestingly, GADPH was not identified in the total protein fraction in the Dupont et al. (2011) study [19].
Fig. 3.
KCl-soluble flour proteins in Butte 86 (a) and transgenic lines SA-30-118a-5 (b) and SA-35a-124j (c). The solid box highlights regions of the gels containing the omega-1,2 gliadins while the dashed box highlights regions of gels containing the alpha gliadins. The alpha gliadin regions from Butte 86 and SA-35a-124j were enlarged in panels (d) and (e), respectively. Spots labeled in red in panel d were significantly reduced or absent in SA-35a-124j and are predominantly alpha gliadins that partition into the KCl fraction. Spots labeled in blue in panel (d) were also found in the KCl fraction of SA-35a-124j and contain mostly GAPDH
Discussion
A number of non-gluten proteins that are abundant in the KCl-soluble protein fraction are well-separated from the major gluten proteins and can be identified unambiguously by MS/MS in a total SDS protein extract of Butte 86 flour [19]. These include the serpins, purinins and AAI. However, other non-gluten proteins are overshadowed by the more abundant gluten proteins. In 2-DE gels of total protein extracts, beta-amylases are hidden by omega-1,2 gliadins, GAPDHs are masked by alpha gliadins, and peroxidases and some globulins are obscured by LMW-GS. In this study, non-gluten proteins were preferentially extracted from wheat flour using KCl and 57 different types of non-gluten proteins were identified, including 14 types that are known or suspected immunogenic proteins. Although most gluten proteins are insoluble in KCl, we observed some carry-over of the abundant gluten proteins in the KCl fraction. This was not unexpected and, in contrast to other studies [8–11], was likely noted because protein spots were digested separately with trypsin and chymotrypsin before analysis by MS/MS, thereby facilitating the identification of both non-gluten proteins that are readily digested with trypsin and gluten proteins that have few tryptic peptide cleavage sites. For example, 276 tryptic peptides were identified for beta-amylase in spot 27 and 62 tryptic peptides were identified for enolase in spot 38. The omega-1,2 gliadins in these two spots did not yield any tryptic peptides, but were identified on the basis of 81 and 41 chymotryptic peptides, respectively. Likewise, 72 tryptic peptides were identified for GAPDH in spot 101. Although the alpha gliadin in the same spot did not yield any tryptic peptides, it was identified by 24 chymotryptic peptides (Additional file 2). In general, protein spots that contained either gamma gliadins or LMW-GS were relatively minor while spots containing certain omega-1,2 gliadins or alpha gliadins were quite prominent in the KCl-soluble fraction. Interestingly, two spots found in the omega-1,2 gliadin region of the total protein fraction but not in the KCl-soluble fraction were identified previously as omega-1,2 gliadins that contain single cysteine residues [16]. These cysteine residues likely enable the proteins to be incorporated into glutenin polymers, rendering them insoluble in aqueous solutions even though their sequences are otherwise quite similar to omega-1,2 gliadins that partition into the KCl fraction. Indeed, in past experiments, these omega-1,2 gliadins were accumulated preferentially in the polymer fractions of Butte 86 wheat flour [24].
The availability of transgenic lines missing either the omega-1,2 gliadins or the alpha gliadins made it possible to estimate the abundance of specific non-gluten proteins that overlap with gluten proteins in the KCl-soluble fraction. These analyses demonstrated that beta-amylase and protein disulfide isomerase are low abundance proteins while GAPDH is a protein of moderate abundance.
Di Francesco et al. [25] recently used a shotgun approach to characterize the KCl-soluble proteins from old and modern durum wheats. In their study, proteins were prepared from mature kernels rather than milled flour and the protein extracts were digested only with trypsin. Analysis of the resulting spectral data used a database of protein sequences from Triticum, Oryza, Hordeum, Avena, Secale, Maize and Brachypodium downloaded from the UniProt database. Interestingly, only 21% of identified proteins were from Triticum, 10.8% from Hordeum and 1% from Secale. In the current study, proteins were digested individually with either trypsin or chymotrypsin and spectra were searched against a database containing all Triticeae sequences from NCBI along with a selection of sequences from Chinese Spring and Butte 86. Not surprisingly, many of the same types of proteins, including likely allergens, were identified in both studies. However, it is curious that Di Francesco et al. identified only one globulin in their study while nine different globulin sequences were identified in 26 2-DE spots in the current study. Likewise, Di Francesco et al. identified only three different serpins, while seven different sequences in 14 spots were found in the current study. They did, however, report a number of alpha and gamma gliadins as well as HMW-GS and LMW-GS in the KCl fraction, confirming the carryover of gluten proteins into this fraction that was also observed in our analysis. It is likely that they would have identified more gluten proteins in this fraction if proteins had been digested with both trypsin and chymotrypsin prior to MS analysis.
Conclusions
The proteomic map of KCl-soluble flour proteins generated in this study complements the map of total flour proteins from Butte 86 described by Dupont et al. [19] and serves as a reference map that, combined with 2-D immunoblots using sera from well-characterized patients, should provide new insight into non-gluten proteins that are responsible for immunogenic responses to wheat flour and lead to a better understanding of the complement of proteins associated with different types of allergies. Transgenic lines missing specific groups of gluten proteins should be of value in confirming whether reactivity with patient antibodies is due to non-gluten proteins or to gluten proteins that are carried over into the KCl-soluble fraction. This information is critical for future efforts to develop wheat with reduced immunogenic potential.
Supplementary information
Additional file 1. Proteins identified by MS/MS in 2-DE protein spots from the KCl-soluble fraction of Butte 86 flour. The positions of the spots are shown in Figure 2. For each spot, the predominant protein and all proteins that contained at least half the number of unique peptides as the predominant protein are highlighted in yellow. Proteins that contained less than half the number of unique peptides found in the predominant protein are highlighted in blue. Gluten proteins that contained less than half the number of unique peptides found in the predominant protein are highlighted in green.
Additional file 2. MS/MS data for individual protein spots shown in Figure 2. Scaffold data for all proteins for which the number of unique peptides was equal to or greater than half the number assigned to the predominant protein are shown.
Acknowledgements
The authors thank Leslie Harden and Melissa Erickson-Beltran for expert advice and assistance with mass spectrometry work.
Abbreviations
- 2-DE
Two-dimensional gel electrophoresis
- AAI
Alpha amylase/trypsin inhibitors
- CD
Celiac disease
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- HMW-GS
High molecular weight glutenin subunit
- LMW-GS
Low molecular glutenin subunit
- MS/MS
Tandem mass spectrometry
- NCWS
Non-celiac wheat sensitivity
Authors’ contributions
SA designed experiments, analyzed the data and wrote the manuscript, H-CC was responsible for 2-DE analyses and AS-B did the MS/MS work. All authors read and approved the manuscript.
Funding
Research was funded by USDA Agricultural Research Service CRIS Project 2030–21430-014-00D. USDA is an equal opportunity provider and employer. Mention of a specific product name by the United States Department of Agriculture does not constitute an endorsement and does not imply a recommendation over other suitable products.
Availability of data and materials
The dataset generated in this study has been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12953-020-00164-6.
References
- 1.Scherf KA. Immunoreactive cereal proteins in wheat allergy, non-celiac gluten/wheat sensitivity (NCGS) and celiac disease. Curr Opin Food Sci. 2019;25:35–41. doi: 10.1016/j.cofs.2019.02.003. [DOI] [Google Scholar]
- 2.Scherf KA, Koehler P, Wieser H. Gluten and wheat sensitivities – an overview. J Cereal Sci. 2016;67:2–11. doi: 10.1016/j.jcs.2015.07.008. [DOI] [Google Scholar]
- 3.Sollid LM, Qiao S-W, Anderson RP, Gianfrani C, Konig F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics. 2012;64:455–460. doi: 10.1007/s00251-012-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tye-Din JA, Stewart JA, Dromey JA, Beissbarth T, van Heel DA, Tatham A, et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci Transl Med. 2010;4:41ra51. doi: 10.1126/scitranslmed.3001012. [DOI] [PubMed] [Google Scholar]
- 5.Battais F, Courcoux P, Popineau Y, Kanny G, Moneret-Vautrin DA, Denery-Papini S. Food allergy to wheat: differences in immunoglobulin E-binding proteins as a function of age or symptoms. J Cereal Sci. 2005;42:109–117. doi: 10.1016/j.jcs.2005.01.004. [DOI] [Google Scholar]
- 6.Palosuo K, Alenius H, Varjonen E, Koivuluhta M, Mikkola J, Keskinen H, et al. A novel wheat gliadin as a cause of exercise-induced anaphylaxis. J Allergy Clin Immunol. 1999;103:912–917. doi: 10.1016/S0091-6749(99)70438-0. [DOI] [PubMed] [Google Scholar]
- 7.Battais F, Pineau F, Popineau Y, Aparicio C, Kanny G, Guerin L, Moneret-Vautrin DA, Denery-Papini S. Food allergy to wheat: identification of immunogloglin E and immunoglobulin G-binding proteins with sequential extracts and purified proteins from wheat flour. Clin Exp Allergy. 2003;33:962–970. doi: 10.1046/j.1365-2222.2003.01592.x. [DOI] [PubMed] [Google Scholar]
- 8.Akagawa M, Handoyo T, Ishii T, Kumazawa S, Morita N, Suyama K. Proteomic analysis of wheat flour allergens. J Agric Food Chem. 2007;55:6863–6870. doi: 10.1021/jf070843a. [DOI] [PubMed] [Google Scholar]
- 9.Pastorello EA, Farioli L, Conti A, Pravettoni V, Bonomi S, Iametti S, et al. Wheat IgE-mediated food allergy in European patients: α-amylase inhibitors, lipid transfer proteins and low-molecular-weight glutenins. Int Arch Allergy Immunol. 2007;144:10–22. doi: 10.1159/000102609. [DOI] [PubMed] [Google Scholar]
- 10.Sotkovsky P, Hubalek M, Hernychova L, Novak P, Havranova M, Setinova I, et al. Proteomic analysis of wheat proteins recognized by IgE antibodies of allergic patients. Proteomics. 2008;9:1677–1691. doi: 10.1002/pmic.200700347. [DOI] [PubMed] [Google Scholar]
- 11.Sotkovsky P, Sklenar J, Halada P, Cinova J, Setinova I, Kainarova A, et al. A new approach to the isolation and characterization of wheat flour. Clin Exp Allergy. 2011;41:1031–1043. doi: 10.1111/j.1365-2222.2011.03766.x. [DOI] [PubMed] [Google Scholar]
- 12.Huebener S, Tanaka CK, Uhde M, Zone JJ, Vensel WH, Kasarda DD, et al. Specific nongluten proteins of wheat are novel target antigens in celiac disease humoral response. J Proteome Res. 2015;14:503–511. doi: 10.1021/pr500809b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Junker Y, Zeissig S, Kim S-J, Barisani D, Wieser H, Leffler DA, et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J Exp Med. 2012;209:2395–2408. doi: 10.1084/jem.20102660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacFarlane AJ, Burghardt KM, Kelly J, Simell T, Simell O, Altosaar I, et al. A type 1 diabetes-related protein from wheat (Triticum aestivum) J Biol Chem. 2003;278:54–63. doi: 10.1074/jbc.M210636200. [DOI] [PubMed] [Google Scholar]
- 15.Altenbach SB, Tanaka CK, Pineau F, Lupi R, Drouet M, Beaudouin E, Morisset M, Denery-Papini S. Assessment of the allergenic potential of transgenic wheat (Triticum aestivum) with reduced levels of ω-5 gliadins, the major sensitizing allergen in wheat-dependent exercise-induced anaphylaxis. J Agric Food Chem. 2015;63:9323–9332. doi: 10.1021/acs.jafc.5b03557. [DOI] [PubMed] [Google Scholar]
- 16.Altenbach SB, Chang H-C, Yu XB, Seabourn BW, Green PH, Alaedini A. Elimination of omega-1,2 gliadins from bread wheat (Triticum aestivum) flour: effects on immunogenic potential and end-use quality. Front Plant Sci. 2019;10:580. doi: 10.3389/fpls.2019.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altenbach SB, Chang H-C, Rowe MH, Yu XB, Simon-Buss A, Seabourn BW, Green PH, Alaedini A. Reducing the immunogenic potential of wheat flour: silencing of alpha gliadin genes in a U.S. wheat cultivar. Front Plant Sci. 2020;11:20. doi: 10.3389/fpls.2020.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurkman WJ, Tanaka CK. Improved methods for separation of wheat endosperm proteins and analysis by two-dimensional gel electrophoresis. J Cereal Sci. 2004;40:295–299. doi: 10.1016/j.jcs.2004.09.001. [DOI] [Google Scholar]
- 19.Dupont FM, Vensel WH, Tanaka CK, Hurkman WJ, Altenbach SB. Deciphering the complexities of the wheat flour proteome using quantitative two-dimensional electrophoresis, three proteases and tandem mass spectrometry. Proteome Sci. 2011;9:10. doi: 10.1186/1477-5956-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huo N, Zhang S, Zhu T, Dong L, Mohr T, Hu T, et al. Gene duplication and evolution dynamics in the homeologous regions harboring multiple prolamin and resistance gene families in hexaploid wheat. Front Plant Sci. 2018;9:673. doi: 10.3389/fpls.2018.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huo N, Zhu T, Altenbach S, Dong L, Wang Y, Mohr T, et al. Dynamic evolution of α-gliadin prolamin gene family in homeologous genomes of hexaploid wheat. Sci Rep. 2018;8:5181. doi: 10.1038/s41598-018-23570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altenbach SB, Vensel WH, Dupont FM. The spectrum of low molecular weight alpha-amylase/protease inhibitor genes expressed in the US bread wheat cultivar Butte 86. BMC Research Notes. 2011;4:242. [DOI] [PMC free article] [PubMed]
- 23.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vensel WH, Tanaka CK, Altenbach SB. Protein composition of wheat gluten polymer fractions determined by quantitative two-dimensional gel electrophoresis and tandem mass spectrometry. Proteome Sci. 2014;12:8. doi: 10.1186/1477-5956-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Francesco A, Saletti R, Cunsolo V, Svensson B, Muccilli V, De Vita P, Foti S. Qualitative proteomic comparison of metabolic and CM-like protein fractions in old and modern wheat Italian genotypes by a shotgun approach. J Proteome. 2020;211:103530. doi: 10.1016/j.jprot.2019.103530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Proteins identified by MS/MS in 2-DE protein spots from the KCl-soluble fraction of Butte 86 flour. The positions of the spots are shown in Figure 2. For each spot, the predominant protein and all proteins that contained at least half the number of unique peptides as the predominant protein are highlighted in yellow. Proteins that contained less than half the number of unique peptides found in the predominant protein are highlighted in blue. Gluten proteins that contained less than half the number of unique peptides found in the predominant protein are highlighted in green.
Additional file 2. MS/MS data for individual protein spots shown in Figure 2. Scaffold data for all proteins for which the number of unique peptides was equal to or greater than half the number assigned to the predominant protein are shown.
Data Availability Statement
The dataset generated in this study has been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository.