Abstract
People living with HIV (PLWH) have expressed concerns with the life-long burden and stigma associated with daily pill taking and can experience medication fatigue that can lead to suboptimal treatment adherence and the emergence of drug-resistant viral variants thereby limiting future treatment options1–3. As such, there is strong interest in long-acting antiretroviral (ARV) agents that can be administered less frequently4. Herein, we report GS-CA1 as a novel archetypal small-molecule HIV capsid inhibitor with exceptional potency against HIV-2 and all major HIV-1 types, including viral variants resistant to ARVs currently in clinical use. Mechanism of action studies indicate that GS-CA1 binds directly to HIV-1 capsid and interferes with capsid-mediated nuclear import of viral DNA, HIV particle production and ordered capsid assembly. GS-CA1 selects in vitro for unfit GS-CA1 resistant capsid variants that remain fully susceptible to other classes of ARVs. Its high metabolic stability and low solubility enabled a sustained drug release in mice following a single subcutaneous dosing. GS-CA1 showed high antiviral efficacy as a long-acting injectable monotherapy in a humanized mouse model of HIV-1 infection, outperforming long-acting rilpivirine. Collectively, these studies demonstrate the potential of ultrapotent capsid inhibitors as novel long-acting agents for the treatment of HIV-1 infection.
Current ARVs span six mechanistic classes that each inhibit one of four steps in the HIV replication cycle: viral entry, reverse transcription, viral DNA integration, or proteolysis of viral polyproteins5. While most current ARV therapies are effective and well-tolerated, there remains a pressing medical need to discover and develop new classes of HIV inhibitors that can suppress viral variants resistant to currently used ARVs and to identify long-acting drug products that can be administered less frequently to improve treatment adherence and to provide additional treatment options to PLWH4.
The HIV-1 capsid protein (CA, p24) plays essential roles at multiple steps in the viral replication cycle, making it a mechanistically attractive target for therapeutic interventions6,7. In the late stages of viral replication, CA is expressed as part of the Gag/Gag-Pol precursor polyproteins and provides key protein-protein interactions necessary for virion assembly and release8. Following Gag/Gag-Pol cleavage by the viral protease, mature CA is released and spontaneously assembles into a conical shell comprised of ~1,500 CA monomers assembled into 250 CA hexamers and exactly 12 CA pentamers9. Proper viral capsid formation and integrity is essential for virus infectivity8. After an infectious virion fuses with a naïve target cell, the viral capsid enters the cytoplasm and undergoes controlled disassembly as the single-stranded RNA genome is reverse-transcribed into double-stranded DNA and subsequently transported into the nucleus for integration into the host genome. These early stages of viral replication are affected by mutations in CA that alter capsid stability10 or its interactions with host factors that either restrict or enable the infection11.
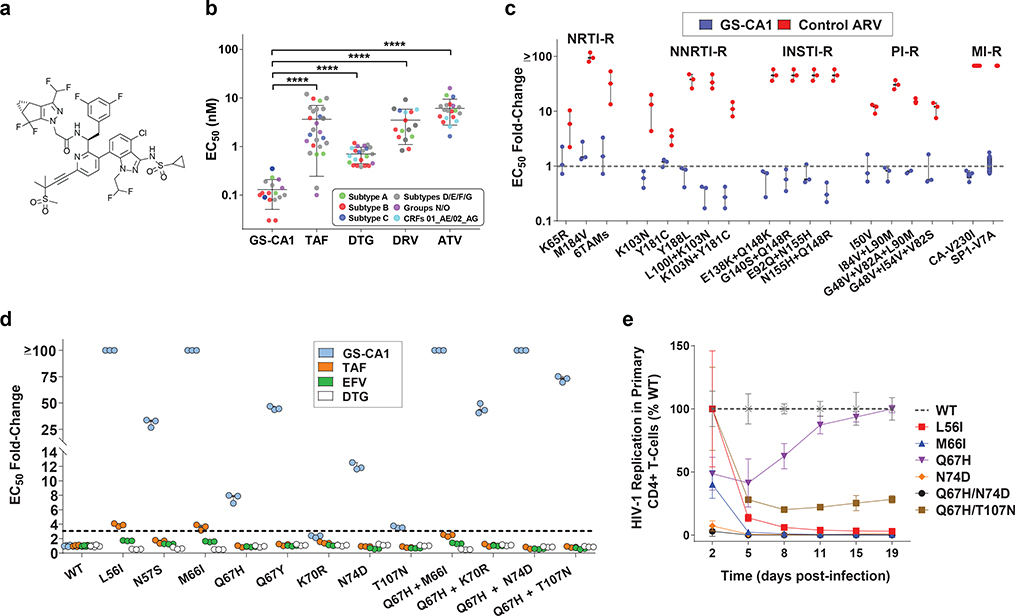
To investigate the utility of inhibiting capsid function, we performed high-throughput screening for small-molecule CA binders and optimized the most promising leads for antiviral activity and drug-like properties. This research led to the discovery of GS-CA1, a novel small-molecule HIV-1 capsid inhibitor with potent antiviral properties (Fig. 1a). In MT-4 cells acutely infected with HIV-1IIIB, GS-CA1 showed high antiviral potency, with a mean 50% effective concentration (EC50) of 240 ± 40 picomolar (pM, n = 97), low cytotoxicity (mean 50% cytotoxicity concentration, CC50 > 50 μM, n = 13), and a selectivity index (CC50/EC50 ratio) of >208,300. In comparison, the previously identified capsid inhibitor PF-3450074 (PF74)12 showed >5,000-fold weaker antiviral potency (mean EC50 = 1,239 ± 257 nanomolar, nM, n = 43), higher cytotoxicity (CC50 = 32.2 ± 9.3 micromolar, μM, n = 39) and a selectivity index of 26. GS-CA1 exhibited higher potency and selectivity in primary human CD4+ T-cells and macrophages acutely infected with HIV-1BaL, with mean EC50 values of 60 ± 10 pM and 100 ± 70 pM, respectively, and mean CC50 values > 50 μM in each cell type (Supplementary Table 1). These properties make GS-CA1 more potent and >10-fold more selective than the nonnucleoside reverse transcriptase inhibitor (NNRTI) efavirenz (EFV), the integrase strand transfer inhibitor (INSTI) dolutegravir (DTG), and the protease inhibitor (PI) atazanavir (ATV). Importantly, GS-CA1 also inhibited a broad range of clinical HIV-1 isolates in human peripheral blood mononuclear cells (PBMCs) with a mean EC50 value of 130 ± 80 pM, representing markedly higher potency than any other ARV tested (Fig. 1b). GS-CA1 also inhibited the in vitro replication of HIV-2 and SIV isolates, albeit with somewhat reduced picomolar potency relative to that against HIV-1 that may be due, in part, to sequence variations within the GS-CA1 binding site of these viruses (Extended Data Fig. 1). GS-CA1 retained full antiviral potency against HIV-1 variants resistant to nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs), NNRTIs, INSTIs, PIs, or the maturation inhibitor (MI) bevirimat (Fig. 1c). Thus, GS-CA1 is a highly potent and selective inhibitor of both wild-type (WT) and drug-resistant HIV variants.
Fig. 1: Antiviral properties of GS-CA1.
a, Chemical structure of GS-CA1. b, Inhibition of HIV-1 clinical isolates in human PBMCs. Symbols represent individual isolates assayed in triplicate cell cultures. Center line and error bars represent mean ± s.d. values across the virus panel. P values relative to GS-CA1 (n = 17) were determined by unpaired two-tailed Mann-Whitney U tests for tenofovir alafenamide (TAF, n = 26, ****P = 3.9 × 10−11), dolutegravir (DTG, n = 22, ****P = 1.7 × 10−9), darunavir (DRV, n = 16, ****P = 1.3 × 10−10), and atazanavir (ATV, n = 20, ****P = 3.3 × 10−9). c, Inhibition of HIV-1 mutants resistant to nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs), nonnucleoside RT inhibitors (NNRTIs), integrase strand transfer inhibitors (INSTIs), protease inhibitors (PIs) or maturation inhibitors (MIs). Symbols represent EC50 fold-change values against each mutant virus relative to its matched wild-type virus, as determined from triplicate cell cultures across at least 3 independent experiments. Mean EC50 fold-change (FC) values per agent against each virus (n = 7 for SP1-V7A, n = 6 for CA-V230I, and n = 3 for all other mutants) across the mutant panel for GS-CA1 (FC = 0.06, n = 18) vs. control ARV (FC = 34.4, n = 18) were significantly different by unpaired two-tailed Mann-Whitney U tests (P = 2.2 × 10−10). SP1, the spacer peptide downstream of immature CA. d, Phenotypic analysis of resistance mutations selected in vitro by GS-CA1. Symbols represent mean ± s.d. EC50 values for each agent against each single-cycle reporter HIV-1 CA mutant relative to the WT virus obtained from triplicate cell cultures in each of 3 independent experiments. Dotted line indicates FC cut-off values used to account for assay variation and defines drug resistance. e, Outgrowth kinetics for replication-competent reporter HIV-1 encoding the most commonly observed GS-CA1-selected variants in primary human CD4+ T-cells. Symbols represent mean ± s.d. luminescence values expressed relative to the WT virus from 6 replicate cell cultures in each of 2 independent experiments.
To evaluate in vitro resistance to GS-CA1, we cultured MT-2 cells infected with HIV-1IIIB and human PBMCs independently infected with 5 HIV-1 isolates in the presence of multiple fixed concentrations of GS-CA1 (1.6 – 12.6 nM). Cultures were monitored for viral outgrowth over a 5-week period and emergent variants characterized by population sequencing. Analysis of breakthrough variants identified a total of seven substituted amino acid positions within CA (L56, N57, M66, Q67, K70, N74 and T107) (Supplementary Table 2). No other genetic changes within Gag-Pol were observed, including the viral protease, reverse transcriptase, integrase or the SP1 spacer peptide. When introduced, alone and in combination, into a single-cycle reporter virus based on HIV-1NL4.3, these CA mutants showed variable levels of fold resistance to GS-CA1 (2.3- to >100-fold) in MT-2 cells while remaining fully susceptible to representative inhibitors from other ARV classes (Fig. 1d). The K70R and T107N variants were secondary resistance-associated mutations (RAMs) as each alone did not confer significant resistance to GS-CA1 until independently combined with Q67H. The minor loss of sensitivity to the NRTI tenofovir alafenamide (TAF) observed with the L56I and M66I mutants (3.8-fold and 3.5-fold, respectively) is likely an artifact of the high multiplicity of infection (MOI) required with these two poorly infectious mutants (each <10% of WT infectivity). In the context of a single-cycle HIV-1NL4.3 infection, all but one of the GS-CA1 RAMs (Q67H) showed markedly reduced infectivity in MT-2 cells relative to the wild-type virus (2% – 58% of WT), suggesting that GS-CA1 resistance comes with a profound fitness cost (Extended Data Fig. 2). The most commonly observed in vitro GS-CA1-selected RAMs were also introduced into a replication-competent reporter HIV-1 and evaluated for their ability to support a spreading infection in primary human CD4+ T-lymphocytes. The results were consistent with the single-cycle data and indicate that the replication capacity of all of the most commonly observed GS-CA1-selected RAMs except Q67H are severely compromised in primary CD4+ T-cells (Fig. 1e).
GS-CA1 RAMs were mapped to the X-ray crystal structure determined for the cross-linked CA hexamer13 (Extended Data Fig. 3a–c). The localized nature of these resistance-associated residues suggests that GS-CA1 binds at the interface formed by two adjacent CA monomers. This binding site is shared by capsid-binding host nuclear import factors that participate in HIV-1 replication, including nucleoporin 153 (Nup153) and cleavage and polyadenylation specificity factor 6 (CPSF6)14–17 (Extended Data Fig. 3b). This inhibitor/host factor binding site is highly conserved (Extended Data Fig. 3c), with all seven amino acid positions associated with GS-CA1 resistance exhibiting a sequence conservation between 94% and 100% across 8 major HIV-1 subtypes (L56 [100%], N57 [99% - 100%], M66 [100%], Q67 [99% - 100%], K70 [100%], N74 [100%], and T107 [94–99%]).
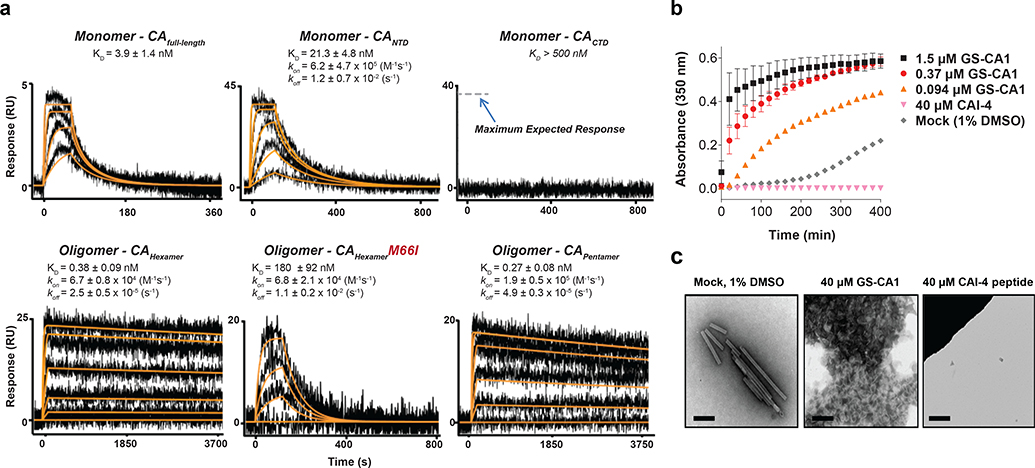
GS-CA1 binding to different forms of HIV-1 CA was evaluated in biosensor experiments with surface plasmon resonance detection (Fig. 2a). Sensorgrams for GS-CA1 showed dose-dependent and saturable binding to all CA surfaces except the isolated C-terminal domain (CTD) of CA. Consistent with the crystallographic mapping of GS-CA1-selected RAMs, GS-CA1 exhibited >10-fold higher affinity binding to, and markedly slower disassociation kinetics from, oligomeric forms of CA (hexamers and pentamers) relative to the CA monomer. Furthermore, the M66I RAM reduced GS-CA1 binding affinity to the CA hexamer nearly 500-fold, suggesting that the crystallographically defined pocket of the CA hexamer is the functional target of GS-CA1 and that this RAM acts directly by reducing its binding affinity.
Fig. 2: GS-CA1 binding to recombinant HIV-1 capsid.
a, Representative binding sensorgrams depicting the interaction of GS-CA1 with different HIV-1NL4.3 CA forms assessed by surface plasmon resonance (SPR) detection. Binding data (black lines) were fit (orange lines) to a simple kinetic model with a mass transport added when necessary. Mean ± s.d. equilibrium binding dissociation constant (KD) and forward and reverse rate constant (kon and koff) values were determined from three independent experiments with similar results. If not shown, kon and koff values could not be precisely determined. b, Mean ± s.d. light scattering (A350) responses showing the rate and extent of spontaneous in vitro CA (20 μM) assembly in 2M NaCl in the absence (mock) or presence of GS-CA1 or CAI-4 from 3 independent experiments. c, Representative electron micrographs showing the effect of GS-CA1 on the morphology of CA assembly products. Six images were obtained from each of 3 independent experiments with similar results. Image showing loss of CA tube formation in the presence of the CAI-4 peptide assembly inhibitor is included as a control. Scale bar = 500 nm.
Recombinant CA can spontaneously oligomerize under high ionic strength conditions to form open-ended, helical tubes comprised of repeating CA hexamers18–21. To investigate the impact of GS-CA1 on CA tube assembly, we monitored the rate and extent of in vitro CA (20 μM) assembly in 2M NaCl by monitoring for changes in light absorbance over time in the presence and absence of GS-CA1 (Fig. 2b). Unlike the control CAI-4 peptide22, which inhibited CA assembly relative to the mock (1% DMSO) control treatment, GS-CA1 showed a dose-dependent increase in the rate and extent of in vitro CA assembly. Ultrastructural analysis of assembly products showed that GS-CA1 induced the formation of short, misshaped and heterogeneous CA polymers that differed from the uniformly long and well-organized CA tubes formed in the absence of GS-CA1 (Fig. 2c). These data indicate that GS-CA1 binding has the potential to interfere with capsid assembly and/or disassembly.
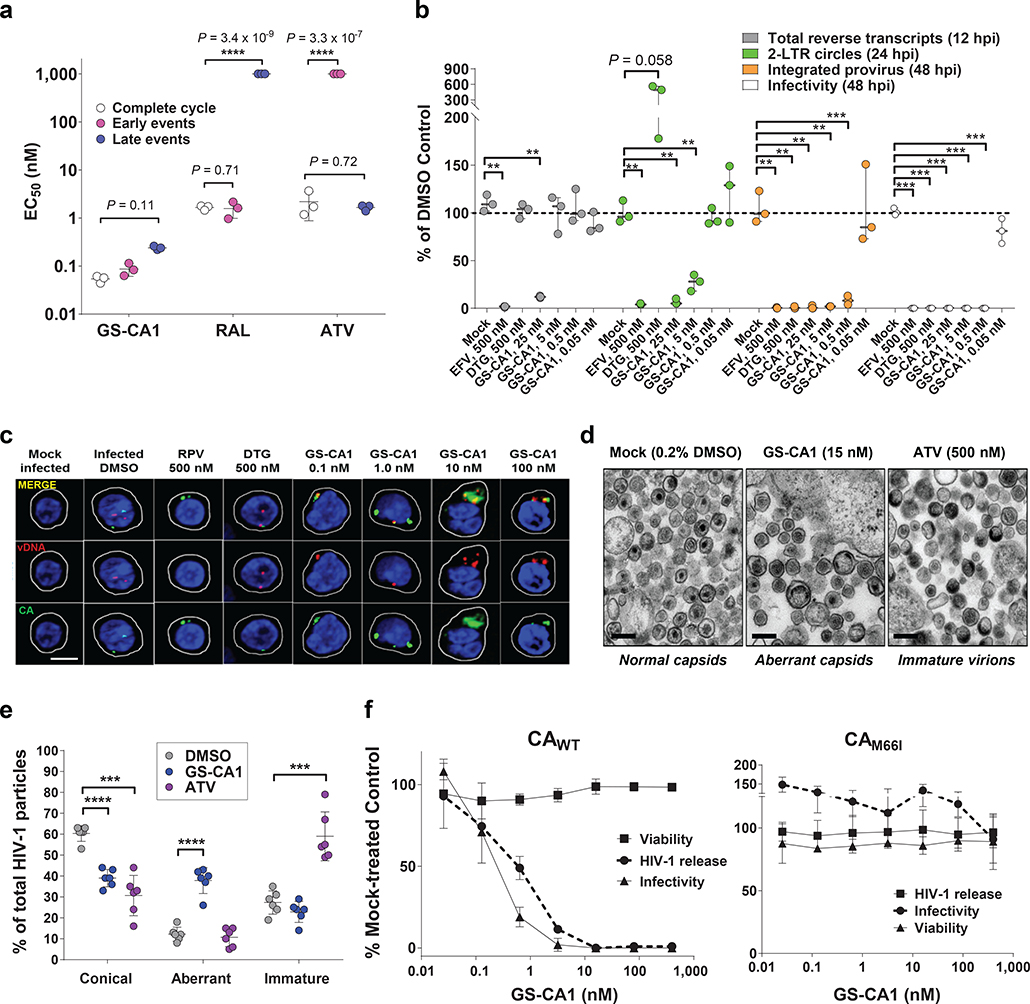
To identify the stages of the viral replication cycle affected by GS-CA1, we utilized a two-part assay system23 to independently measure antiviral potency in cultures of target MT-2 cells (early phase) and virus producer HEK293T cells (late phase)(Fig. 3a). Unlike the control agents raltegravir (RAL, an INSTI that is active only in target cells) and ATV (a PI that is active only in producer cells), GS-CA1 inhibited HIV-1 functions both in target cells (EC50 = 87 pM) and in producer cells (EC50 = 240 pM). GS-CA1 potencies were similar to those observed in the full-cycle assay (EC50 = 53 pM), indicating that GS-CA1 is a picomolar inhibitor of both early and late stages of HIV-1 replication.
Fig. 3: GS-CA1 mechanisms of action.
a, Antiviral activity of GS-CA1, RAL and ATV when present selectively at early stage, late stage, or during a full HIV-1 infection cycle. Center line and error bars represent mean ± s.d. EC50 values from triplicate cell cultures in each of 3 independent experiments. Significant P values relative to matched complete cycle EC50 values by unpaired two-tailed t tests with Welch’s correction are highlighted with asterisks. b, HIV-1 DNA abundance by quantitative PCR. The effect of compounds on particle infectivity and abundance of total HIV-1 cDNA, 2-LTR circles, and integrated provirus in MT-2 cells is shown. Significant P values relative to matched DMSO controls (n = 3 independent cell culture samples per group) by unpaired two-tailed t tests with Welch’s correction from left to right were: (cDNA) **P = 0.0026, **P = 0.0036; (2-LTR) **P = 0.0047, **P = 0.0033, **P = 0.0013; (provirus) **P = 0.0011, **P = 0.0011, **P = 0.001, **P = 0.0012, ***P = 0.0007; (infectivity) ***P = 0.0008, ***P = 0.0008, ***P = 0.0008, and ***P = 0.0008. c, Representative confocal microscopy images of primary human CD4+ T-cells that were either mock- or HIV-1-infected for 12 hours in conjunction with the indicated treatments. Cell membranes (white), nuclei (blue), CA foci (green), vDNA foci (red) and merged (yellow) images from 2 independent experiments, each from a single donor producing similar results. The total number of cell images analyzed were: n = 118 (mock and DMSO), n = 26 (RPV), n = 49 (DTG), n = 55 (0.1 nM GS-CA1), n = 44 (1 nM GS-CA1), n = 43 (10 nM GS-CA1), n = 57 (100 nM GS-CA1). Scale bar = 5 μm. d, Representative thin section electron micrograph images of HIV-1 produced in the presence of the indicated treatment. Six images for each condition from a single experiment were analyzed and produced similar results. Scale bars = 200 nm. e, Quantitation of capsid morphology frequencies within 6 representative images for HIV-1 produced in the presence of 0.2% DMSO (n = 223), 15 nM GS-CA1 (n = 214), or 500 nM ATV (n = 243). Center line and error bars represent mean ± s.d.values. P values relative to matched DMSO controls by unpaired two-tailed t tests with Welch’s correction from left to right were: ****P = 3.1 × 10−6, ***P = 0.0003, ****P = 2.4 × 10−5, ***P = 0.0005. f, Effect of GS-CA1 on HEK293T producer cell viability and single-cycle reporter HIV-1 particle production and infectivity. Collected supernatants were tested for released HIV-1 virion levels by p24 ELISA and for infectivity in MT-2 cells following sample dilution. Symbols represent mean ± s.d. values obtained from triplicate cell cultures collected from each of 3 independent experiments.
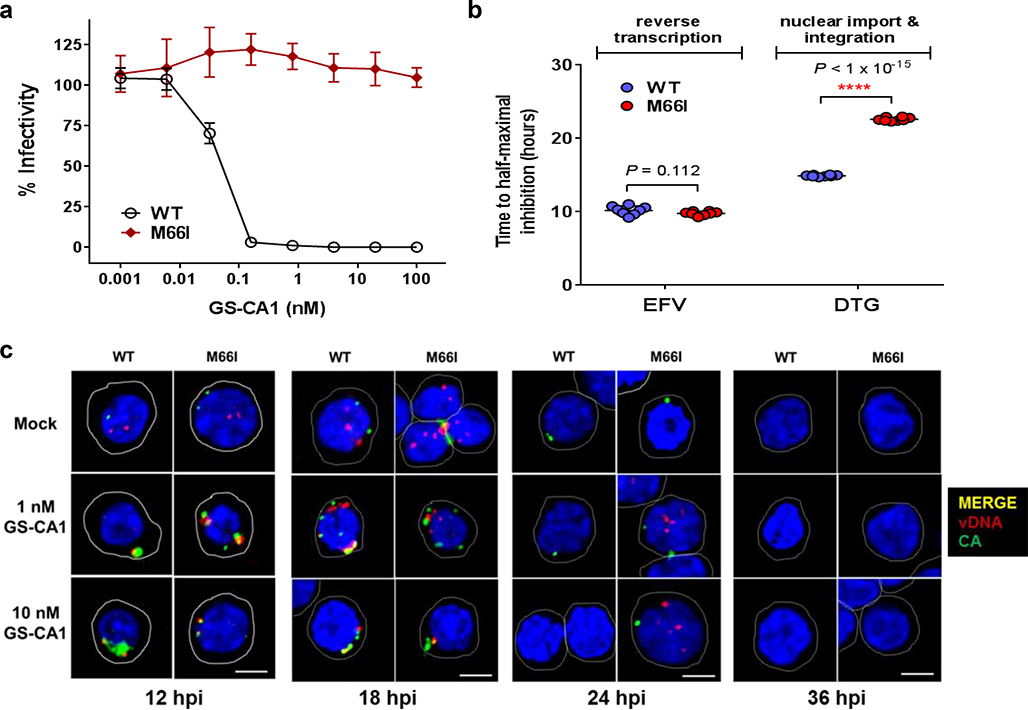
To dissect the steps at which GS-CA1 acts during the early stage of HIV-1 infection, we performed virus entry and time-of-addition studies to demonstrate that GS-CA1 targets one or more post-entry steps occurring after reverse transcription and concomitant with or close to integration (Extended Data Fig. 4; Supplementary Fig. 1). To further dissect these GS-CA1 activities, we infected MT-2 cells with single-cycle reporter HIV-1 at an MOI of 1 and assayed by quantitative PCR for the dose-dependent inhibition of reverse transcription, 2-LTR circle formation (a nuclear episomal form of the viral cDNA that can accumulate as a byproduct of blocked integration), proviral integration, and infectivity (Fig. 3b). Control inhibitors behaved as expected: the RT inhibitor EFV blocked formation of all three replication products, whereas the integrase inhibitor DTG allowed reverse transcription, blocked proviral integration, and increased 2-LTR circle accumulation. Interestingly, GS-CA1 exhibited three concentration-dependent profiles. At high concentrations (25 nM; 500-fold EC50), GS-CA1 resembled EFV, demonstrating that GS-CA1 can interfere with reverse transcription under conditions of high drug exposure. Intermediate GS-CA1 concentrations (5 nM; 100-fold over EC50) did not alter total HIV cDNA synthesis but significantly reduced the formation of integrated proviruses and 2-LTR circles, suggesting that GS-CA1 can prevent nuclear import of viral cDNA. At the lowest effective drug concentration tested (0.5 nM; 10-fold over EC50), GS-CA1 reduced proviral integration without affecting viral cDNA or 2-LTR circle abundance. The block to integration in the absence of a concomitant increase in 2-LTR circles suggests that low GS-CA1 concentrations may simultaneously interfere with both nuclear entry and integration. These effects are consistent with emerging reports that the HIV-1 capsid (or a rearranged remnant) is involved in post-nuclear entry steps preceding integration24–28.
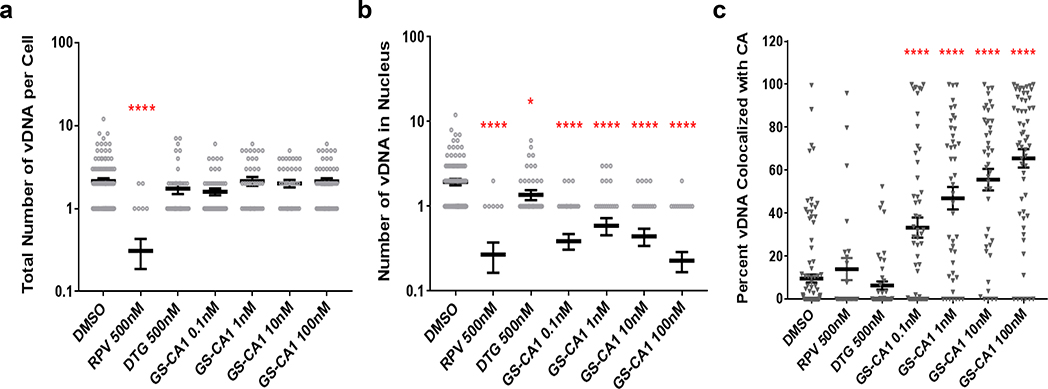
To directly examine how GS-CA1 affects the cellular localization of different viral DNA forms, we incubated infected (MOI = 70) and mock-infected primary human CD4+ T-cells in the presence of different test treatments and then performed ViewHIV analysis (Fig. 3c, Extended Data Fig. 5). This methodology combines branch-chain DNA technology, sandwich hybridization and capsid-specific immunolabeling to enable visualization of early HIV-1 infection events by confocal microscopy29. At 12 hours post-infection (hpi), individual HIV-1-infected cells treated with DMSO vehicle showed both capsid (1.5 ± 0.1 foci/cell) and vDNA (2.1 ± 0.2 foci/cell), with the capsid foci localized to the cell cytoplasm, nucleus or both compartments depending on the number of foci in a given cell. Across the entire collection of HIV-1-infected cells treated with DMSO vehicle (n = 118), capsid foci were localized throughout the cell (43% nuclear), whereas vDNA foci localized predominantly in the nucleus (92%), and the two signals rarely colocalized (10%). Both fluorescent signals were highly specific as neither was observed in mock-infected samples. Control treatments with the RT inhibitor rilpivirine (RPV) dramatically reduced vDNA (to 14% of control) without affecting capsid levels (100% of control), whereas treatment with the integrase inhibitor DTG did not alter either fluorescent signal. In contrast, infected cells treated with GS-CA1 (0.1–100 nM) showed an accumulation of vDNA and capsid signals in the cytoplasm, and the two signals often colocalized in large fluorescent aggregates at or near the nuclear membrane (Fig. 3c). The absence of any change in the accumulation of reverse transcripts in the ViewHIV assay at GS-CA1 concentrations up to 100 nM versus the reduced reverse transcript levels observed with 25 nM GS-CA1 in quantitative PCR studies is likely due to MOI differences (70-fold) between these two assays since, as observed with other classes of ARVs, the in vitro antiviral activity of GS-CA1 decreased with increased input of HIV-1 inoculum in cell cultures (Supplementary Table 3). Although GS-CA1 also induced cytoplasmic accumulation of vDNA from a drug-resistant M66I mutant strain at 12 hpi, the vDNA subsequently entered the nucleus by 24 hpi and was integrated by 36 hpi, suggesting that GS-CA1 cannot durably suppress nuclear entry of vDNA from this GS-CA1 resistant CA variant (Extended Data Fig. 6). For the WT HIV-1 infections, quantitative image analyses confirmed that while GS-CA1 treatment did not change the total number of vDNA foci per cell (Extended Data Fig. 5a), it did significantly reduce the amount of HIV-1 vDNA detected in the nucleus (Extended Data Fig. 5b, 9-fold maximal effect) and increased the colocalization of capsid and vDNA signals in the cytoplasm of infected cells (Extended Data Fig. 5c, 7-fold maximal effect). These data suggest that GS-CA1 inhibits the early phase of viral infection by blocking capsid-dependent translocation of HIV-1 cDNA into the nucleus.
To investigate the effect of GS-CA1 on the late stages of viral replication, we initially produced HIV-1 in the presence and absence of test compounds and analyzed the capsid ultrastructure in released virions by electron microscopy (Fig. 3d,e). HIV-1 virions produced in the presence of 0.2% DMSO (vehicle control) showed conical capsids, with encased viral RNA evident within the wide end of the cones. In contrast, HIV-1 virions produced in the presence of GS-CA1 contained predominantly irregularly shaped capsids in which the viral RNA was either regionally or diffusely confined. Notably, these capsid morphologies were distinct from those observed in virions produced in the presence of the protease inhibitor ATV, which predominantly contained immature Gag/Gag-Pol lattices. Thus, GS-CA1 interferes with the proper assembly of CA into regular conical capsids within released HIV-1 virions.
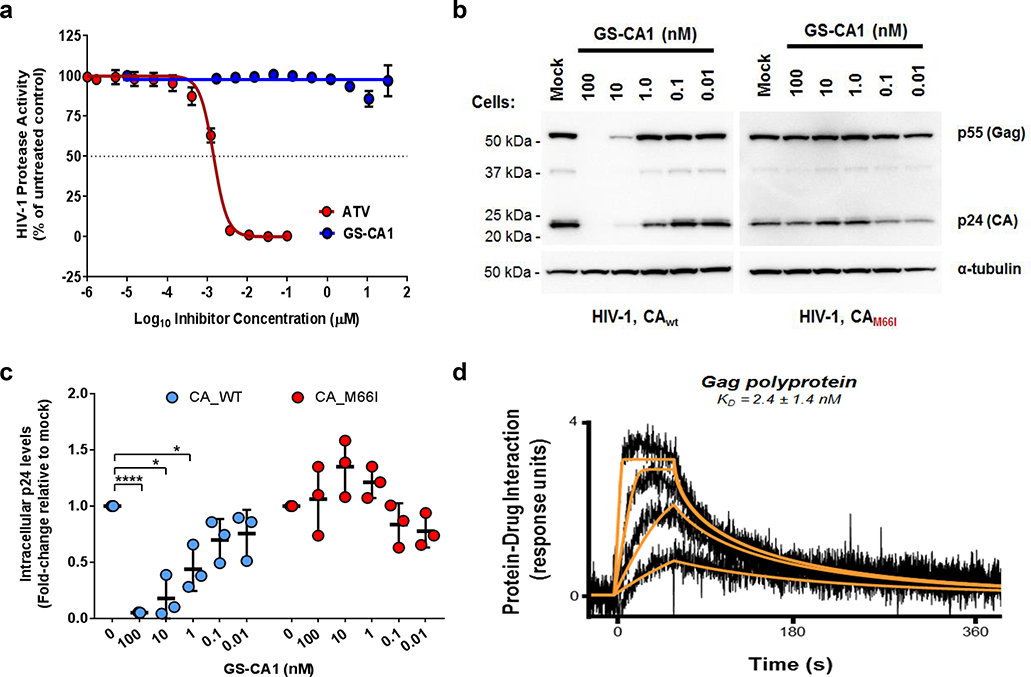
To test whether GS-CA1 had any additional late-stage effects on the viral replication cycle, we measured virus production in HEK293T cells transfected with single-cycle reporter HIV-1 plasmid in the presence and absence of GS-CA1 (Fig. 3f). GS-CA1 showed no cytotoxicity in the producer cells but induced dose-dependent reductions in mature HIV-1 (p24) released into the media, with a mean EC50 of 501 ± 99 pM. Reduced p24 levels were not attributable to reduced Gag/Gag-Pol proteolysis as GS-CA1 did not inhibit HIV-1 protease in vitro up to the highest concentration tested (30 μM) (Extended Data Fig. 7a). Furthermore, the titers of wild-type virus were significantly reduced with GS-CA1 relative to untreated controls, with a late-stage antiviral EC50 of 240 pM, whereas GS-CA1 did not inhibit the release or infectivity of the drug-resistant M66I mutant strain. Interestingly, GS-CA1 reduced intracellular levels of both Gag and processed CA proteins in the producer cells, and this effect was again CA-specific because it was abrogated by the M66I mutation (Extended Data Fig. 7b,c). While the mechanism underlying loss of Gag and CA is unknown, we note that GS-CA1 also binds with high affinity to recombinant Gag (Extended Data Fig. 7d), suggesting that GS-CA1 binding to Gag/Gag-Pol precursor proteins may limit Gag stability, trafficking, and/or Gag assembly. Thus, GS-CA1 effectively limits the production and proper assembly of infectious virus particles via CA-specific mechanisms. Collectively, these findings demonstrate that GS-CA1 interferes with multiple distinct CA-dependent steps during both the early and late stages of HIV-1 infection. This multi-stage mechanism of action profile is unique among all currently approved classes of ARVs.
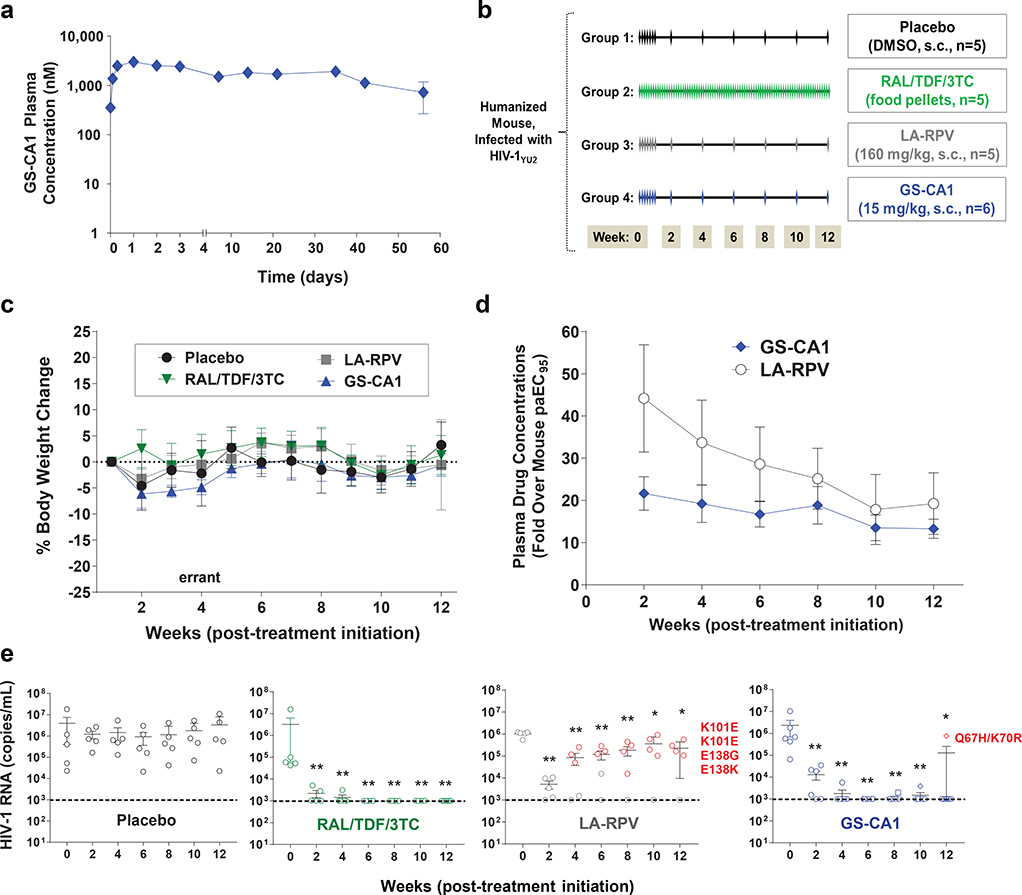
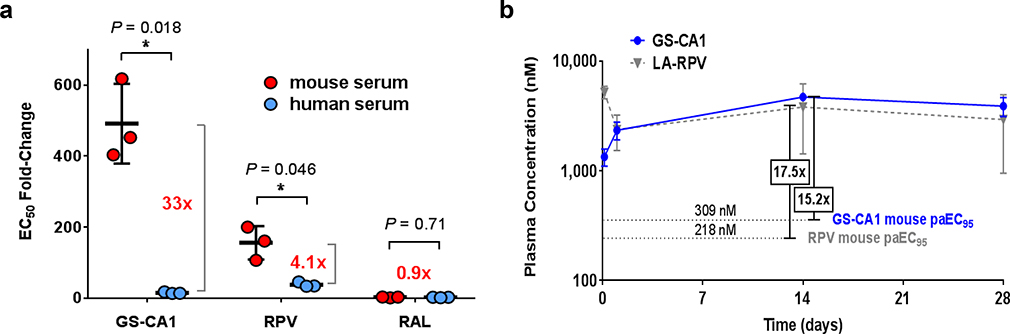
By evaluating the microsome stability and identifying the metabolites of numerous chemical analogs, GS-CA1 was optimized for high metabolic stability, with a predicted hepatic extraction of < 7.13% in mice. This combination of high potency, high metabolic stability, and low solubility of GS-CA1 make it an ideal candidate for a low-dose injectable administration suitable for sustained in vivo drug release. We therefore evaluated the pharmacokinetics of GS-CA1 in mice after its subcutaneous administration. GS-CA1 clearance was exceptionally low in C57Bl/6 mice, with plasma concentrations remaining well above its EC95 value (630 pM) for at least 56 days (length of study) after a single subcutaneous dose of 15 mg/kg, underscoring its potential for long-acting administrations (Fig. 4a). Unfortunately, antiviral assays performed in the presence and absence of different sera indicated very high murine serum protein binding for GS-CA1 relative to human serum (Extended Data Fig. 8a). Thus, in order to reach sufficient free drug exposure levels in mice and to identify conditions that would allow direct comparison of GS-CA1 to a control long-acting agent, we tested various dosing regimens and determined that administration of GS-CA1 at 15 mg/kg daily for 7 days or of long-acting rilpivirine (LA-RPV, 160 mg/kg daily for 7 days) resulted in plasma GS-CA1 and RPV levels that were 15.2-fold and 17.2-fold above their respective mouse serum protein binding-adjusted EC95 (Extended Data Fig. 8b).
Fig. 4: Preclinical PK/PD evaluations of GS-CA1.
a, Plasma drug concentrations over time in male C57Bl/6 mice following a single subcutaneous administration of 15 mg/kg GS-CA1. Symbols represent mean ± s.d. values obtained from 3 mice per time point in a single experiment. b, Efficacy study design. HIV-1YU2-infected humanized female NOG mice were mock-treated (placebo, n = 5) or treated with antiretroviral therapy with daily oral RAL/TDF/3TC (n = 5) or with long-acting RPV (n = 5) or GS-CA1 (n = 6) monotherapy administered by subcutaneous injection. Monotherapy treatments were initiated daily for 7 consecutive days to quickly achieve steady-state drug levels and then switched to every other week administrations, using either a RPV suspension (160 mg/kg) or a GS-CA1 solution (15 mg/kg) formulation. c, Mouse body weights over time in each experimental group as a general indicator of treatment tolerability. Symbols represent mean ± s.d. values as a percent change in body weight relative to week 0 (start of treatment). d, Plasma GS-CA1 and RPV drug concentrations over time in efficacy study. Symbols represent the mean ± s.d. fold over the mouse serum protein binding-adjusted EC95 (paEC95) values for each compound. e, Viral loads in HIV-1YU2-infected humanized mice subjected to the indicated treatment regimens. Center line and error bars represent mean ± s.e.m. values at indicated time points. Colored circles indicate mice that failed treatment, with associated viral resistance mutations in RT (for RPV) or CA (for GS-CA1) noted. Dashed lines represent the limit of detection (103 HIV-1 RNA copies/mL). Significant P values relative to week 0 by unpaired two-tailed Mann-Whitney U tests from left to right were: (RAL/TDF/3TC) **P = 0.0079 (weeks 2–12); (LA-RPV) **P = 0.0079 (weeks 2–6), *P = 0.032, *P = 0.016; (GS-CA1) **P = 0.0022 (weeks 2–10), *P = 0.022.
To directly compare the in vivo efficacy of long-acting monotherapies of GS-CA1 and RPV, we used a humanized mouse model of HIV-1 infection. Humanized mice were infected with HIV-1YU2 and 5-weeks later, the animals were distributed among four experimental groups (5–6 mice/group) to normalize mean baseline HIV-1 RNA levels at 106.4 ± 100.3 copies/mL across each group (Fig. 4b). Control animals in Group 1 received DMSO vehicle subcutaneously (10 μL daily for 7 days, then once every two weeks) and group 2 animals received daily oral doses of RAL/TDF/3TC (873/107/109 mg/kg/day). Group 3 and 4 animals were treated daily for 7 days, then once every two weeks, with the long-acting subcutaneous monotherapies of RPV (160 mg/kg) and GS-CA1 (15 mg/kg), respectively.
Each regimen was well tolerated as no animals either died or required ethical sacrifice due to signs of distress or significant (>25%) body weight loss over the 12-week treatment period (Fig. 4c). Relative to their respective mouse serum protein binding-adjusted EC95 values, plasma RPV levels were similar or higher than GS-CA1 over the 12-week treatment study (Fig. 4d). As expected, mean viral loads remained unchanged in 5/5 vehicle-treated animals, whereas the viral load in 5/5 animals receiving daily oral RAL/TDF/3TC therapy rapidly declined below the detection limit (103 copies/mL) and remained undetectable for the duration of the study. Both long-acting treatments showed similar initial viral load declines in the first 2 weeks of therapy, but the treatment responses diverged dramatically thereafter (Fig. 4e). In the RPV treatment group, mean HIV-1 RNA levels started increasing after 4 weeks of treatment and continued increasing thereafter for the remainder of the 12-week treatment study, with 4/5 animals (80%) returning to baseline viremia by week 10. All 4 mice with virological failures showed RT mutations associated with RPV resistance (E138K in 1/4 animals, E138G in 1/4 animals, and K101E in 2/4 animals). In contrast, HIV-1 RNA levels continued to decline in the GS-CA1 treated animals and remained suppressed for the first 10 weeks of treatment, with 6/6 animals showing HIV-1 RNA levels at or near the lower detection limit. One of these animals showed a return to baseline viremia by week 12 and virologic failure in this animal coincided with emergence of CA mutations Q67H+K70R. This CA variant was also selected in vitro with GS-CA1 (Supplementary Table 1) and conferred 44-fold resistance to GS-CA1 (Fig. 1d). Thus, intermittent subcutaneous low-dose GS-CA1 monotherapy (15 mg/kg every 2 weeks) provided a strong antiviral response and sustained viral suppression in 83% of treated animals, with only one animal showing viral breakthrough at the 12th week of treatment. In contrast, RPV, a licensed NNRTI currently in clinical trials as a long-acting agent30, administered at significantly higher doses (160 mg/kg every 2 weeks), was unable to maintain the same level of durable suppression and resulted in rapid emergence of drug resistance in 80% of treated animals.
The development of an ultra-potent small-molecule HIV capsid inhibitor capable of sustained delivery has implications for the future of antiretroviral therapy. Despite strong interest in long-acting ARVs that can be administered less frequently4, some long-acting drugs in development rely on large dose intramuscular injections that can cause discomfort and require frequent clinic visits for their administration based on their less favorable potency and metabolic stability. The identification of high potency, high metabolic stability ARVs suitable for sustained delivery offers the possibility of infrequent low-dose subcutaneous self-administration. In addition, the development of agents with a novel mechanism of action provides valuable new treatment options for PLWH who have developed resistance to currently approved ARV agents.
Recently, GS-6207 has been reported as the first-in-class capsid inhibitor to enter clinical trials31,32. GS-6207 has been optimized for human administration and is in the same structural class as GS-CA1 with a similar pharmacological profile, including mechanism of action, resistance profile, high potency and metabolic stability. GS-6207 is currently undergoing Phase 1a/1b proof-of-concept studies to determine the optimal dose, safety profile and frequency of subcutaneous administration in PLWH (ClinicalTrials.gov identifier = NCT03739866). The encouraging preclinical data with the HIV-1 capsid inhibitor GS-CA1 reported here, together with emerging Phase 1 study results using the related analog GS-6207, warrant rapid development of this novel class of antiretroviral agents.
Methods
Compounds
GS-CA1, PF74, emtricitabine (FTC), tenofovir (TFV), tenofovir alafenamide (TAF), elvitegravir (EVG), darunavir (DRV), atazanavir (ATV) and bevirimat (BVM) were synthesized at Gilead Sciences (Foster City, CA). Efavirenz (EFV), rilpivirine (RPV), dolutegravir (DTG) and raltegravir (RAL) were purchased from Toronto Research Chemicals, Curragh Chemistries, Porton Shanghai R&D Center and Expicor, respectively.
Cells
Human MT-2, MT-4, and SupT1 T-cell lines were obtained from the NIH AIDS Reagent Program and maintained in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 units/mL penicillin, and 10 μg/mL streptomycin (complete RPMI). MT-2 cells chronically infected with HIV-1IIIB (MT-2/IIIB) were cultured in complete RPMI. The CEM-NKR CCR5+Luc+ infectivity indicator T-cell line33 was obtained from the NIH AIDS Reagent Program cultured in complete RPMI supplemented with 0.8 mg/mL Geneticin. These cell lines were passaged twice weekly and maintained at densities below 800,000 cells/mL. HEK293T cells were obtained from the Gladstone Institute for Virology and Immunology and maintained at densities below 80% confluency in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 10% FBS, 100 units/mL penicillin, and 10 μg/mL streptomycin (complete DMEM). Human peripheral blood mononuclear cells (PBMCs) were collected from consenting healthy volunteers under informed consent and approved by an Institutional Review Board at AllCells. Human PBMCs, CD4+ T-lymphocytes, and monocyte-derived macrophage cultures were prepared as previously described34. Prior to infection, PBMCs and CD4+ T-cells were activated for 48 hours at 37°C by addition of 1 μg/mL phytohemagglutinin (PHA, Sigma-Aldrich) and 50 IU/mL recombinant human interleukin-2 (IL-2, Roche Diagnostics).
Plasmids
The HIV-1 plasmid pKS13ΔEnv encoding firefly luciferase and the expression plasmid pHCMV-G encoding the vesicular stomatitis virus G (VSV-G) envelope were previously described23. To generate plasmid pNL4–3-JRFL-secNLuc encoding replication competent reporter HIV-1, the NL4–3 gp120 ectodomain was replaced with the JRFL ectodomain by BstE2-BstB1 ligation. A synthetic gene encoding secreted NanoLuc luciferase (secNLuc), an internal ribosomal entry site element35 and nef was purchased from GenScript and inserted into pNL4.3-JRFL via XhoI-BamHI ligation. Capsid genes containing resistance-associated mutation(s) were synthesized at GenScript and inserted into pKS13ΔEnv, pNL4–3-JRFL-secNLuc and pJexpress-CA-4Mu22 plasmids via BssHII-ApaI ligation.
HIV-1
HIV-1IIIB was obtained from the NIH AIDS Reagent Program. Clinical HIV-1 isolates from the Southern Research virus collection included 92UG031, 92UG037, 89BZ_167, 90US_873, YU-2, 91US001, 91US004, 92BR025, 98US_MSC5016, 92UG001, 98UG57128, CMU02, 93BR020, JV1083, 90TH_CM235, and 91DJ263 strains. HIV-1 recombinant strains encoding mutation(s) conferring resistance to NRTIs, NNRTIs, INSTIs, PIs or MIs have been previously described34,36.
Reporter HIV-1
Single-cycle HIV-1 encoding firefly luciferase was made by co-transfecting HEK293T cells with pKS13ΔEnv and pHCMV-G plasmids. Replication-competent reporter HIV-1 encoding secreted NanoLuc was made by transfecting HEK293T cells with pNL4.3-JRFL-secNLuc plasmids. Cell-free viral supernatant was collected 3 days post-transfection, clarified using a 0.45 μm syringe filter and stored at −80°C. The amount of HIV in each sample was quantified by p24 antigen ELISA (Perkin Elmer) and a reverse transcriptase (RT) activity assay (Southern Research).
Anti-HIV and cytotoxicity assays
The 5-day cytoprotection antiviral assays using MT-2 and MT-4 T-cell lines have been previously described34. Data analysis was performed using GraphPad Prism 7.0 to calculate EC50 values. A 7-day RT endpoint antiviral assay using fresh human PBMCs independently infected with a panel of clinical HIV-1 isolates was performed by Southern Research as a contracted research study.
For single-cycle antiviral assays, MT-2 cells in complete RPMI ± single-cycle reporter HIV-1 was mixed with 4-fold serially-diluted test compound on 96-well plates (5 × 104 cells/well). Assay plates were kept in a humidified 37°C incubator and developed 3 days later by addition of ONE-Glo Luciferase Assay reagent (Promega). Luminescence data were collected and analyzed using EnVision Manager 1.13.3009 and GraphPad Prism 7.0 softwares, respectively. In some cases, single-cycle assays were set up such that compound was present selectively at early stage, late stage, or during a full course of infection as previously described23.
For cytotoxicity assessment in each assayed cell type, the protocol was identical to that of the respective antiviral assay, including assay duration, except that no virus was added to the plates. The effect of test compounds on cell viability was measured using CellTiter-Glo. Data analysis was performed using GraphPad Prism 7.0 to calculate CC50 values.
HIV-1 replication kinetics in primary human CD4+ T-cells
Mitogen-activated CD4+ T-cells were suspended in complete RPMI, added to 96-well plates (2 × 105 cells/well), and infected with RT-normalized inputs (~2 × 105 cpm equivalents per million cells) of NL4–3-JRFL-secNLuc reporter HIV-1. Each virus, plus RPMI alone as uninfected controls, was added to 6 replicate wells. Final assay wells contained 200 μL complete RPMI supplemented with 10 μg/mL DEAE-dextran and 50 IU/mL IL-2. Assay plates were incubated for 19 days in a humidified 37°C incubator. At indicated time points, a fixed volume of cell-free supernatant (2 μL) was collected from each well and analyzed by addition of Nano-Glo Luciferase Assay reagent (Promega).
Effect of GS-CA1 on HIV-1 particle production
HEK293T cells were co-transfected with pKS13ΔEnv and pHCMV-G plasmids in T-75 flasks. Sixteen hours post-transfection, cells were replated in triplicate 96-well plates (5 × 104 cells/well) in DMEM containing serially-diluted GS-CA1. Two days post-transfection, cell-free virus supernatants were harvested and the amount of HIV in each sample quantified by p24 antigen ELISA. CellTiter-Glo reagent was added to the assay plates to measure viability of transfected cells. The infectivity of each supernatant was analyzed by diluting 10,000-fold and incubating with MT-2 cells for 48 hours prior to assay development with ONE-Glo reagent.
Preparation of recombinant CA proteins
Recombinant HIV-1LAI CA protein was prepared as previously described22. Soluble cross-linked HIV-1NL4.3 CA hexamers (WT and the M66I variant) and CA pentamers were prepared as previously described13,22,37. CA-NTD1–151 and CA-CTD146–231 proteins, each containing a C-terminal GS linker and Avitag sequence, were expressed in BL21-DE3 cells following a 4 hour induction with 0.5 mM isopropyl β-D-1-thiogalactopyranoside and 50 μM D-biotin. To purify these two proteins, bacterial pellets were harvested and the biotinylated CA proteins purified using Pierce Ultralink Monomeric Avidin Resin. Biotinylated CA-NTD1–151 and CA-CTD146–231 proteins were eluted using buffer containing 2 mM Biotin and the samples further purified by fractionation over a Shodex KW2003 column equilibrated in 25 mM Tris-HCl, pH 7.0, 5 mM DTT. Samples were pooled based on SDS-PAGE analysis, flash-frozen and stored at −80oC.
In vitro CA binding
Surface plasmon resonance biosensor binding experiments were performed using the ProteOn XPR36 platform (Bio-Rad) or the Biacore T100 platform (used to study Monomer-CAfull length and Gag proteins). For ProteOn experiments, a GLM sensor chip was first conditioned using 0.5% SDS, 50 mM NaOH, 100 mM HCl, and 10% DMSO. A similar protocol using 100 mM HCl, 50 mM NaOH and 0.5% SDS was used to precondition a CM5 chip for experiments performed on the Biacore T100. Standard primary amine coupling chemistry was used on both platforms to immobilize NeutrAvidin (Thermo Fisher Scientific) at a concentration of 50 μg/mL. Residual activated carboxyls were blocked with ethanolamine. The neutravidin surfaces were stabilized with 1 M NaCl and 50 mM NaOH. Proteins were captured via their avitag, or by first minimally biotinylating proteins using a 1:1 molar ratio of EZ-link Sulfo-NHS-LC-LC-biotin (Thermo Fisher Scientific). GS-CA1 was prepared by first serially diluting compound in 100% DMSO and then added to running buffer (50 mM sodium phosphate pH 7.5, 150 mM NaCl, 0.01 % Polysorbate 20) at a final DMSO concentration of 1%. For each interaction, background binding and drift were subtracted using a neutravidin reference surface and a 1% DMSO solution lacking compound, respectively. Data were analyzed using ProteOn Manager 3.1.0 (Bio-Rad) or Scrubber 2.0 and fit with a simple kinetic model with a term for mass transport added when necessary.
In vitro HIV-1 CA assembly assay
In vitro assembly of HIV CA protein in the presence and absence of GS-CA1 or CAI-4 inhibitory peptide (ITFEDLLDYYK, synthesized at Anaspec Inc.) was monitored by measuring changes in sample absorbance over time at 350 nm. Assembly reactions were performed in 2 M NaCl and visualized by electron microscopy as previously described22.
Ultrastructural analysis of HIV-1 by electron microscopy
MT-2/IIIB cells were washed and cultured at 37°C in complete RPMI containing 0.25% dimethyl sulfoxide (DMSO), 15 nM GS-CA1 or 500 nM ATV. After a 4 day incubation, samples were pelleted, fixed in ice-cold 2% glutaraldehyde, 1% paraformaldehyde in 0.1 M sodium cacodylate buffer pH 7.4, post-fixed in 2% osmium tetroxide in the same buffer, en block stained in 2% aqueous uranyl acetate, dehydrated in acetone, and embedded in LX-112 resin. Samples were ultrathin sectioned and counterstained with 0.8% lead citrate. Grids were examined on a JEOL JEM-1230 transmission electron microscope and photographed with the Gatan Ultrascan 1000 digital camera.
Quantitation of HIV-1 DNA
HEK293T cells were co-transfected with pKS13ΔEnv and pHCMV-G plasmids. Three days later, clarified virus supernatants were treated with DNase (30 U/mL) to eliminate plasmid DNA in samples. MT-2 cells (2 × 106 cells/mL) were infected with equal volumes (0.5 mL) virus (corresponding to an MOI of 1) for 3 hours at 37°C, washed, added to 24-well plates in drug media and transferred to a humidified 37°C incubator. To account for signal from residual plasmid, a few DNase-treated DMSO virus samples were heat-inactivated (1 hour, 65°C), and infections set up in parallel to facilitate background correction during data analysis. For each condition, cells from each of 3 replicate wells were harvested 12 hpi for late RT product quantification, 24 hpi for 2-LTR circle quantitation, and 48 hpi for Alu-LTR product quantification. Viral DNA was isolated from cell pellets using a QIAamp DNA mini kit (Qiagen) and quantified using the TaqMan real-time PCR and ABI Prism 7900HT sequence detection system (Applied Biosystems) or the QX200 Droplet Digital PCR (ddPCR) System (Bio-Rad). For the quantitation of late RT products, primers MH531 (5’-TGTGTGCCCGTCTGTTGTGT-3’) and MH532 (5’-GAGTCCTG CGTCGAGAGAGC-3’) were used in combination with the Late-RT probe 5’-(FAM)-CAGTGGCGCCCGAACAGGGA-(TAMRA)-3’. For the quantitation of 2-LTR circles, primers FP9600 (5’-GCTTAAGCCTCAATAAAGCTTGCCT-3’) and RP71 (5’-GCCTTGTGTGTGGTAGATCC-3’) were used in conjunction with the 2-LTR probe (5’-(FAM)-TCTGAGGGATCTCTAGTTACCAGAGT-(TAMRA)-3’). The control DNA product for the endogenous C-C chemokine receptor type 5 (CCR5) gene was quantified using primers CCR5-Fw (5’-ATGATTCCTGGGAGAGACGC-3’) and CCR5-Rev (5’-AGCCAGGACGGTCACCTT-3’) together with probe CCR5-P (5’-(FAM)-AACACAGCCACCACCCAAGTGATCA- 3’-(BHQ1)). All primers and probes were synthesized by Integrated DNA Technologies, Inc. For each experiment, a standard curve of each amplicon and endogenous CCR5 control gene was measured from 3.5 × 103 to 3.5 × 106 linearized copies of the pKS13ΔEnv vector plus a no-template control, all diluted into 500 ng sheared salmon sperm DNA (Life Technologies). Each amplicon from each sample was measured in triplicate. Quantitative real-time PCR mixtures (25 μL) contained 1× Roche LightCycler 480 probe master mix kit (Roche Diagnostics), 500 nM forward and reverse primers, 200 nM probe, and 500 ng DNA. After initial incubations at 50°C for 2 minutes and 95°C for 10 minutes, samples were subjected to 40 cycles of amplification (95°C for 15 seconds, 60°C for 1 minute). Copy numbers were normalized by CCR5 gene levels and represented as a percent total copy number of the no-drug infected control (set to 100%) for each amplicon measured. Integration junctions were quantified by ddPCR using primers that recognize HIV-1 LTR sequences Alu-F (5’-AACTAGGGAACCCACTGCTTAAG-3’) and cellular Alu-repeat sequences Alu-R (5’-TGCTGGGATTACAGGCGTGAG-3’) together with the Alu-LTR probe Alu-P Zen (5’-(FAM)-ACACTACTT-(ZEN)-GAAGCACTCAAGGCAAGCTTT-3’-(BHQ1)). Reaction mixtures (25 μL) contained 900 nM forward and reverse primers, 250 nM probes, and 50 ng DNA. After droplet generation, ddPCR reactions were incubated at 95°C for 10 minutes, subjected to 40 cycles of amplification (94°C for 30 seconds, 60°C for 1 minute, 72°C for 4 minutes) and quantified on a QX 200 Droplet Reader (BioRad). Copy numbers are represented as a percent total copy number of the no-drug infected control (set to 100%) and were normalized in duplex by measuring endogenous ribonuclease P protein subunit 30 (RPP30) levels that were quantified using the RPP30 PCR primers RPP30-F (5’-GATTTGGACCTGCCAGCG-3’) and RPP30-R (5’-GCGGCTGTCTCCACAAGT-3’) together with the probe RPP30-P (5’-(HEX)- CTGACCTGAAGGCTCT-3’-(BHQ-1)).
ViewHIV analysis in primary human CD4+ T-cells
Activated primary human CD4+ T-cells were mixed with viral supernatants on ice for 40 minutes and then moved to 37°C for 4 hours. The multiplicity of infection (MOI) in these studies was 70. The use of such a high MOI relative to other assays described herein (MOI ≤ 1) was essential to maximize assay sensitivity, ensuring reliable detection of CA and vDNA signals in primary CD4+ T-cells using this imaging method. Infected cultures were washed once with complete RPMI and maintained in a humidified 37°C incubator for 8, 14, 20, or 32 hours, for 12, 18, 24, and 36 hour infections. Mock- and HIV-1-infected cultures were supplemented with 0.01% DMSO, 500 nM RPV, 500 nM DTG, or 0.1–100 nM GS-CA1 as indicated. All samples were then fixed and stained using an anti-p24 monoclonal antibody (AG3.0) from the NIH AIDS Reagent Program (Catalog #4121, Lot 120227, 1:50 dilution) and the images analyzed as previously described29.
Metabolic stability of GS-CA1 in liver microsomes
Test compounds (0.5 μM) were incubated in liver microsomal fractions diluted to 1 mg/mL protein concentration in a buffer containing 0.1 mM potassium phosphate buffer at pH 7.4 and 25 μg alamethicin per mg microsomal protein. The metabolic reaction mixture was pre-warmed to 37°C for 3 minutes and initiated by the addition of a cofactor mixture consisting of 3.3 mM MgCl2, 3.3 mM glucose 6 phosphate, 0.4 U/mL glucose 6 phosphate dehydrogenase, 5 mM uridine diphosphate glucuronic acid and 1.55 mM nicotinamide adenine dinucleotide phosphate at 37°C. The amount of parent drug remaining after different incubation time periods (2, 12, 25, 45 and 65 minutes) was determined by LC-MS/MS, the peak area ratios graphed and the drug half-life (T1/2) and percent predicted hepatic extraction (Eh%) extrapolated.
Pharmacokinetics of GS-CA1 in C57Bl/6 mice
All animal studies conducted in this report were done in strict compliance with all relevant ethical regulations. Nonclinical pharmacokinetic studies were conducted at Covance Labs in a Laboratory Animal Care-accredited facility, and all study protocols were reviewed and approved by their Institutional Animal Care and Use Committee. GS-CA1 was formulated in 100% DMSO at 30 mg/mL and administered via a single subcutaneous injection at 15 mg/kg into 15 male 6- to 10-week old C57BL/6 mice (Envigo RMS, Inc.). Blood was collected from each animal at designed time points and the plasma drug concentrations determined by LC-MS/MS.
Antiviral activity of GS-CA1 and RPV in humanized mice
In vivo efficacy studies were conducted at TransCurebioServices in a Laboratory Animal Care-accredited facility, and the study protocol was reviewed and approved by the local ethic committee (01_TransCurebioServices-AB-01). Female immune-deficient NOD/Shi-scid/IL-2Rγnull (NOG) mice from Taconic Biosciences, Inc. were humanized with CD34+ hematopoietic stem cells isolated from human cord blood. Following cell engraftment, the degree of humanization was evaluated in each animal by measuring the ratio of circulating human CD45+ cells per total leukocytes. Twenty-week old mice with a humanization rate of ≥ 25% were inoculated with 25 ng (p24) of HIV-1 (YU2, Clade B, CCR5 tropic) by intra-peritoneal injection. Plasma viremia was determined 5 weeks post-infection by qRT-PCR. The HIV-infected hu-mice were then distributed among 4 experimental groups (5–6 mice/group) to normalize HIV viral loads across all test arms and treated as follows: Group 1 (Placebo, 10 μL DMSO); Group 2 (Triple-therapy, daily dosing of 2.35 mg tenofovir disoproxil (TDF), 2.4 mg lamivudine (3TC) and 9.2 mg RAL, via drug-food pellet consumption, equal to 107 mg TDF, 109 mg 3TC, and 873 mg RAL per kg per day); Group 3 (160 mg/kg RPV, 20 μL 160 mg/mL RPV suspension formulated in water with 20 mg/mL Poloxamer 338, micronized using a bead mill); Group 4 (15 mg/kg GS-CA1, 10 μL 30 mg/mL GS-CA1/DMSO solution). Groups 1, 3 and 4 were each dosed subcutaneously daily for 7 days, then once every other week. Every two weeks, blood was collected from each animal and the HIV plasma viral load determined by qRT-PCR. Animal weight was monitored prior to HIV infection and then once weekly until sacrifice.
Statistical analysis
GraphPad Prism 7.0 was used for statistical analysis. An unpaired two-tailed Mann-Whitney U test was performed for nonparametric analysis of two groups. An unpaired two-tailed t test, with or without Welch’s correction, was performed for parametric analysis of two groups. A P value ≤ 0.05 was considered statistically significant.
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Data Availability Statement
All data to understand and assess the conclusions of this research are available in the main text and [dummy_]supplementary materials. Raw datasets supporting the findings of this study are available from the corresponding author on reasonable request. The availability of GS-CA1 is subject to a material transfer agreement which can be requested through the corresponding author. Full uncropped Western blots from Extended Data Fig. 7b are available as Source data.
Extended Data
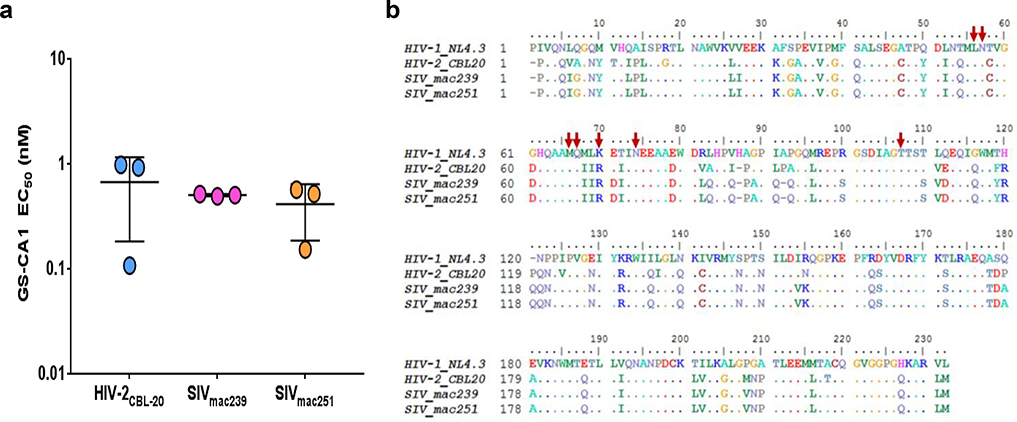
Extended Data Fig. 1: GS-CA1 antiviral activity against HIV-2 and SIV.
a, Antiviral activity of GS-CA1 against one HIV-2 and two SIV isolates replicating in human PBMCs. Center line and error bars represent mean ± s.d. values determined 7 days post-infection using a reverse transcriptase (RT) activity readout. Experiment was performed once using triplicate cell cultures. b, Alignment of capsid amino acid sequences. Dots represent invariant residues. Red arrows highlight HIV-1 residues associated with GS-CA1 resistance.
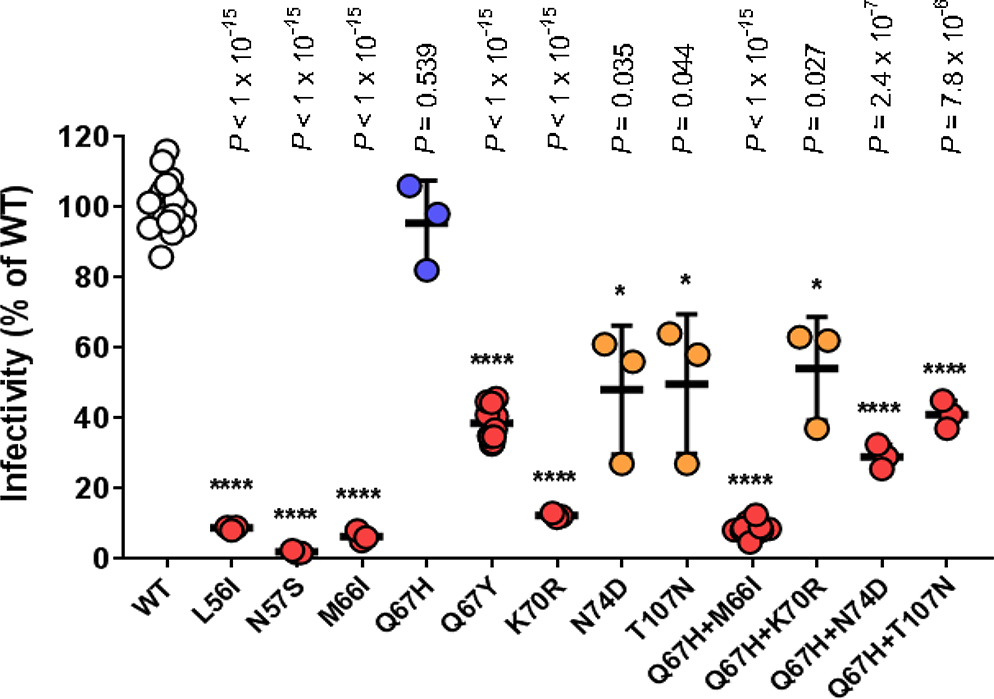
Extended Data Fig. 2: Infectivity of GS-CA1-selected resistance-associated mutants in MT-2 cells.
WT and mutant single-cycle reporter HIV-1NL4.3were produced in parallel in HEK293T cells by transient transfection and the HIV content for each determined by p24 ELISA using a single serial dilution of each sample and quantified across 3 samples within the linear range of the assay. MT-2 cells were infected in duplicate with serially-diluted, p24-normalized WT and mutant viruses and developed 3 days later by One-Glo addition. Center line and error bars represent mean ± s.d. infectivity values, expressed as a percentage of the WT virus, obtained from 3 independent experiments. P values for each mutant (n = 12 replicate cell cultures for Q67Y and Q67H+M66I mutants, 3 replicate cell cultures for all others) relative to WT (n = 18 replicate cell cultures) by unpaired two-tailed t tests with Welch’s correction are indicated.
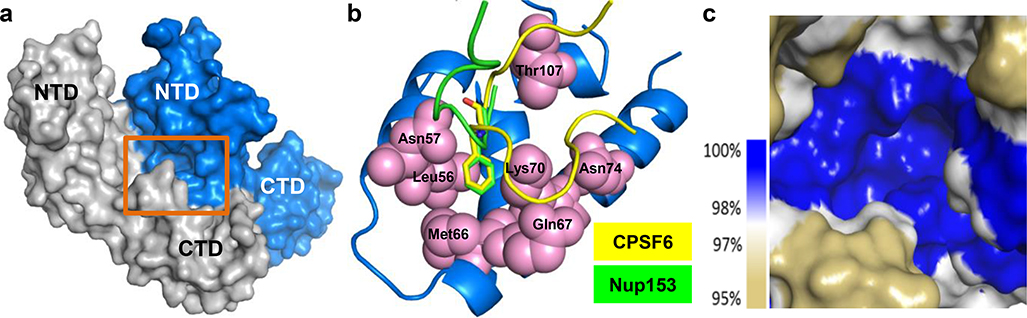
Extended Data Fig. 3: GS-CA1 resistance mutations map to a conserved CA subunit interface.
a, Location of the resistance-defined GS-CA1 binding site (highlighted by orange rectangle) within the HIV-1 CA hexamer structure (ref 13). The resistance-defined GS-CA1 binding subunit is shown in blue and the adjacent CA subunit in silver. b, Location of CA residues associated with GS-CA1 resistance (pink spheres), with CA-binding peptides from cleavage and polyadenylation specificity factor 6 (CPSF6313–327) in yellow and nucleoporin 153 (Nup1531410–1423) in green (ref 17). CPSF6 Phe321 and Nup153 Phe1417 side chains highlighted in stick mode. c, GS-CA1 binding site conservation. The percent CA conservation is depicted as a heat-map based off >4,400 HIV-1 subtype B sequences.
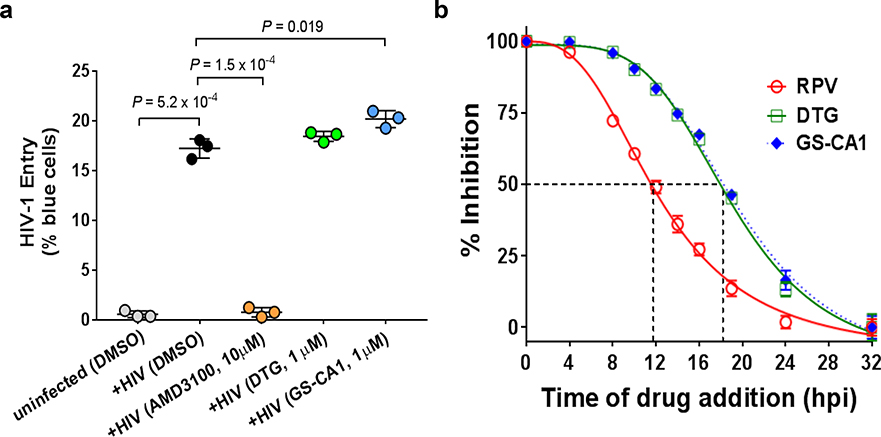
Extended Data Fig. 4: GS-CA1 inhibits HIV-1 replication after virus entry and reverse transcription.
a, Quantitative BlaM-Vpr reporter assay for HIV-1 entry. PBMCs were infected with BlaM-Vpr/HIV-1 (NL4–3 strain) in the presence of GS-CA1 or the designated control compounds, loaded with CCF2 substrate dye, and CD3+ CD4+ CD8− T-cells containing virus were quantified by flow cytometry to detect CCF2 dye cleavage (indicative of virus entry) after 16 hours of incubation. Center line and error bars represent mean ± s.d. values obtained from duplicate cell cultures in each of 3 independent PBMC donors from a single experiment (n=3 per group). Significant P values relative to mock-treated HIV-infected samples by unpaired two-tailed t tests with Welch’s correction are indicated. b, Representative time of addition study. MT-2 cells were infected with HIV-1 reporter virus and drugs RPV (93 nM, RT inhibitor), DTG (193 nM, IN inhibitor) and GS-CA1 (30 nM) were added at indicated time points. Infectivity was measured using a luciferase readout 48 hpi and normalized to mock-treated (DMSO) control. Center line and error bars represent mean ± s.d. values obtained from 8 replicate cell cultures per time point and condition from 2 independent experiments with similar results.
Extended Data Fig. 5: Quantitative analysis of ViewHIV study.
a, Quantitation of total vDNA foci/cell observed in microscopy images. Center line and error bars represent mean ± s.e.m. numbers of vDNA foci/cell 12 hpi. Data in panels a-c obtained from 2 independent transduction experiments, each using an independent CD4+ T-cell donor. The total number of images analyzed for each condition in each panel is from left to right: n = 118, n = 26, n = 49, n = 55, n = 44, n = 43, n = 57. Significant P values relative to DMSO by unpaired two-tailed Mann-Whitney U tests are indicated by asterisks. ****P = 6.1 × 10−12. b, Quantitation of nuclear vDNA foci/cell. Center line and error bars represent mean ± s.e.m. numbers of nuclear vDNA foci/cell 12 hpi. Significant P values relative to DMSO by unpaired two-tailed Mann-Whitney U tests from left to right were: ****P = 1.7 × 10−11, *P = 0.026, ****P = 6 × 10−14, ****P = 4.8 × 10−9, ****P = 2.8 × 10−12, ****P < 1 × 10−15. c, Percent co-localization of vDNA and CA foci. Center line and error bars represent mean ± s.e.m. percentage of all vDNA foci that co-localized with CA foci 12 hpi. Significant P values relative to DMSO by unpaired two-tailed Mann-Whitney U tests from left to right were: ****P = 12 × 10−5 ****P = 1 × 10−8 ****P = 95 × 10−12 ****p < 1 × 10−15.
Extended Data Fig. 6: GS-CA1 resistant M66I mutant delays HIV integration.
a, Antiviral activity of GS-CA1 against WT and CA M66I mutant in MT-2 cells infected with single-cycle reporter HIV-1. Symbols represent mean ± s.d. values obtained from triplicate cell cultures in each of 3 independent experiments. b, Representative time of addition study. MT-2 cells were infected with WT or CA M66I mutant reporter HIV-1 and the drugs efavirenz (EFV, 150 nM, RT inhibitor) and dolutegravir (DTG, 193 nM, IN inhibitor) were added at the indicated times post-infection. Infectivity was measured using a luciferase readout 48 hpi and normalized to mock-treated (DMSO) control. Center line and error bars represent mean ± s.d. values (n = 8 replicate cell cultures per condition) obtained from 2 independent experiments. Significant P values relative to WT virus by unpaired two-tailed t test with Welch’s correction are indicated with an asterisk. c, Representative confocal microscopy images of primary human CD4+ T-cells infected with WT or CA M66I mutant HIV-1 for 12, 18, 24 or 36 hours in conjunction with the indicated treatments. Nuclei are stained in blue (DAPI) and cells are outlined in white. CA (green), vDNA (red) and merged (yellow) representative images are shown for each condition from 2 independent donors from a single experiment. Total number of images analyzed for each row from left to right were: (mock) n = 118, n = 92, n = 18, n = 10, n = 19, n = 17, n = 9, n = 6; (1 nM GS-CA1) n = 44, n = 45, n = 16, n = 16, n = 7, n = 12, n = 3, n = 8; (10 nM GS-CA1) n = 43, n = 56, n = 8, n = 5, n = 4, n = 5, n = 6, n = 13. Scale bar = 5 μm.
Extended Data Fig. 7: GS-CA1 reduces intracellular CA precursor polyprotein levels and particle production.
a, Quantitative HIV-1 protease substrate cleavage assay. The effect of GS-CA1 and atazanavir (ATV, protease inhibitor) on the in vitro cleavage activity of recombinant HIV-1 protease was measured against a fluorogenic HIV-1 protease substrate. Symbols represent mean ± s.d. percent substrate cleavage values obtained from 5 independent experiments performed in duplicate. b, Effect of GS-CA1 on intracellular capsid precursor polyprotein levels. Representative Western blots from 3 independent transfection experiments showing the effect of GS-CA1 on intracellular CA/Gag protein levels. α-Tubulin served as a loading control. HEK293T cells producing WT or M66I HIV-1 were incubated ± GS-CA1 for 48 hours, cell lysates were prepared and normalized inputs were analyzed by anti-CA and anti-tubulin Western blotting. Full uncropped blots are available as Source data. c, Quantitation of total intracellular p24 levels observed in Western blot images, after normalizing to a-tubulin levels. Center line and error bars represent mean ± s.d. normalized p24 values. For each condition, a single cell lysate was prepared and analyzed from each of 3 independent experiments, with similar results. Significant P values relative to matched mock-treated samples by unpaired two-tailed t tests with Welch’s correction from left to right were: ****P = 2.2 × 10−6, *P = 0.016, *P = 0.038. d, Representative sensorgrams showing binding of GS-CA1 to immobilized recombinant HIV-1 Gag polyprotein. Binding data (black lines) were globally fit (orange lines) to a simple kinetic model and used to calculate mean ± s.d. equilibrium dissociation constant (KD) from 3 independent experiments. Due to the fast kinetics of this interaction, values for kon and koff could not be determined with sufficient precision.
Extended Data Fig. 8: GS-CA1 is highly sequestered in mouse but not in human serum.
a, Effect of including 50% serum (mouse or human) in the cell culture media on the antiviral potency of GS-CA1, RPV and RAL. Center line and error bars represent mean ± s.d. EC50 fold-change values obtained ± the indicate serum. Data obtained from 3 independent experiments, each performed with 3 replicate cell cultures per condition. Brackets and numbers highlight potency shift differences between the two sera. Significance P values by unpaired two-tailed t tests with Welch’s correction are indicated with an asterisk. b, Plasma drug concentrations over time in male C57Bl/6 mice (n = 3 per time point) following 7 consecutive daily subcutaneous administrations of either 15 mg/kg GS-CA1 or 160 mg/kg LA-RPV. Symbols represent mean ± s.d. mouse serum protein binding-adjusted EC95 values for each compound, with mean fold mouse paEC95 values indicated in boxes according to day 14 drug levels.
Supplementary Material
Acknowledgments
We are grateful to Alivelu Irrinki for assisting with the FACS portion of the entry studies and Kirsten Stray for supplying the pLAI-RenLuc vector. We appreciate the expertise provided by Jinny Wong at the Gladstone Institute Electron Microscopy Core facility, Carol Lackman-Smith at Southern Research, and Catherine Verhaeghe at TransCure bioServices as part of the work each performed under contracted research agreements. This study was funded by Gilead Sciences, Inc. The anti-HIV-1 p24 AG3.0 monoclonal antibody was obtained from Dr. Jonathan Allan through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH.
Competing Interests Statement: All authors are current or previous employees of Gilead Sciences (except W.I.S., W.M.M., J.M.P., and A.L.B.) and received salary and stock ownership as compensation for their employment. J.O.L., S.D.S., W.C.T., and J.R.Z. are inventors on granted US Patent No. 10,071,985 covering GS-CA1 composition of matter and methods of use.
References
- 1.Claborn KR, Meier E, Miller MB & Leffingwell TR A Systematic Review Of Treatment Fatigue Among HIV-Infected Patients Prescribed Antiretroviral Therapy. Psychology, health & medicine 20, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boretzki J, et al. Highly Specific Reasons for Nonadherence to Antiretroviral Therapy: Results from the German Adherence Study. Patient preference and adherence 11, 1897–1906 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corneli A, et al. Participants’ Explanations for Nonadherence in the FEM-PrEP Clinical Trial. J Acquir Immune Defic Syndr 71, 452–461 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Nyaku AN, Kelly SG & Taiwo BO Long-Acting Antiretrovirals: Where Are We now? Current HIV/AIDS reports 14, 63–71 (2017). [DOI] [PubMed] [Google Scholar]
- 5.De Clercq E Antiretroviral drugs. Curr Opin Pharmacol 10, 507–515 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Thenin-Houssier S & Valente ST HIV-1 Capsid Inhibitors as Antiretroviral Agents. Curr HIV Res 14, 270–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnes SK, Sheehan JH & Aiken C Inhibitors of the HIV-1 capsid, a target of opportunity. Curr Opin HIV AIDS 13, 359–365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed EO HIV-1 Assembly, Release and Maturation. Nat Rev Microbiol 13, 484–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganser BK, Li S, Klishko VY, Finch JT & Sundquist WI Assembly and Analysis of Conical Models for the HIV-1 Core. Science 283, 80–83 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Forshey BM, von Schwedler U, Sundquist WI & Aiken C Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76, 5667–5677 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita M & Engelman AN Capsid-Dependent Host Factors in HIV-1 Infection. Trends Microbiol 25, 741–755 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blair WS, et al. HIV Capsid is a Tractable Target for Small Molecule Therapeutic Intervention. PLoS pathogens 6, 1–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pornillos O, et al. X-ray Structures of the Hexameric Building Block of the HIV Capsid. Cell 137, 1–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharya A, et al. Structural Basis of HIV-1 Capsid Recognition by PF74 and CPSF6. Proc Natl Acad Sci U S A 111, 18625–18630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matreyek KA, Yucel SS, Li X & Engelman A Nucleoporin NUP153 Phenylalanine-Glycine Motifs Engage a Common Binding Pocket Within the HIV-1 Capsid Protein to Mediate Lentiviral Infectivity. PLoS pathogens 9, 1–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AJ, et al. CPSF6 Defines a Conserved Capsid Interface that Modulates HIV-1 Replication. PLoS pathogens 8, 1–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price AJ, et al. Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly. PLoS pathogens 10, 1–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrlich LS, Agresta BE & Carter CA Assembly of Recombinant Human Immunodeficiency Virus Type 1 Capsid Protein In Vitro. J Virol 66, 4874–4883 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganser-Pornillos BK, Cheng A & Yeager M Structure of full-length HIV-1 CA: A Model for the Mature Capsid Lattice. Cell 131, 70–79 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C & Sundquist WI Assembly Properties of the Human Immunodeficiency Virus Type 1 CA Protein. J Virol 78, 2545–2552 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Hill CP, Sundquist WI & Finch JT Image Reconstructions of Helical Assemblies of the HIV-1 CA Protein. Nature 407, 409–413 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Hung M, et al. Large-Scale Functional Purification of Recombinant HIV-1 Capsid. PLoS ONE 8, 1–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balakrishnan M, et al. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PLoS ONE 8, e74163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou L, et al. Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS pathogens 7, e1002194 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen NY, et al. HIV-1 capsid is involved in post-nuclear entry steps. Retrovirology 13, 28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balasubramaniam M, et al. PF74 Inhibits HIV-1 Integration by Altering The Composition of the Preintegration Complex. J Virol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng K, et al. Quantitative Microscopy of Functional HIV Post-Entry Complexes Reveals Association of Replication with the Viral Capsid. eLife 3, e04114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hulme AE, Kelley Z, Foley D & Hope TJ Complementary Assays Reveal a Low Level of CA Associated with Viral Complexes in the Nuclei of HIV-1-Infected Cells. J Virol 89, 5350–5361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chin CR, et al. Direct Visualization of HIV-1 Replication Intermediates Shows that Capsid and CPSF6 Modulate HIV-1 Intra-nuclear Invasion and Integration. Cell reports 13, 1717–1731 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferretti F & Boffito M Rilpivirine long-acting for the prevention and treatment of HIV infection. Curr Opin HIV AIDS 13, 300–307 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Sager JE, et al. Safety and PK of Subcutaneous GS-6207, a Novel HIV-1 Capsid Inhibitor [Abstract]. in Conference on Retroviruses and Opportunistic Infections (CROI) (Seattle, WA, 2019). [Google Scholar]
- 32.Daar ES, et al. Safety and Antiviral Activity Over 10 Days Following A Single Dose of Subcutaneous GS-6207, A First-in-Class, Long-Acting HIV Capsid Inhibitor for People Living with HIV [Abstract]. in 2019 IAS (Mexico City, Mexico, 2019). [Google Scholar]
- 33.Spenlehauer C, Gordon CA, Trkola A & Moore JP A luciferase-reporter gene-expressing T-cell line facilitates neutralization and drug-sensitivity assays that use either R5 or X4 strains of human immunodeficiency virus type 1. Virology 280, 292–300 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Tsiang M, et al. Antiviral Activity of Bictegravir (GS-9883), a Novel Potent HIV-1 Integrase Strand Transfer Inhibitor with an Improved Resistance Profile. Antimicrob Agents Chemother 60, 7086–7097 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alberti MO, et al. Optimized Replicating Renilla Luciferase Reporter HIV-1 Utilizing Novel Internal Ribosome Entry Site Elements for Native Nef Expression and Function. AIDS Res Hum Retroviruses 31, 1278–1296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margot NA, Gibbs CS & Miller MD Phenotypic susceptibility to bevirimat in isolates from HIV-1-infected patients without prior exposure to bevirimat. Antimicrob Agents Chemother 54, 2345–2353 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pornillos O, Ganser-Pornillos BK & Yeager M Atomic-Level Modelling of the HIV Capsid. Nature 469, 424–427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data to understand and assess the conclusions of this research are available in the main text and [dummy_]supplementary materials. Raw datasets supporting the findings of this study are available from the corresponding author on reasonable request. The availability of GS-CA1 is subject to a material transfer agreement which can be requested through the corresponding author. Full uncropped Western blots from Extended Data Fig. 7b are available as Source data.