Highlights
-
•
Simultaneous measurement of IgG to several SARS-CoV-2 antigens.
-
•
Sensitive and specific assay for Trimeric spike, RBD and Nucleocapsid antigen.
-
•
IgG values correlates well with pseudoneutralization measured on the same platform.
-
•
No pseudoneutralisation from cross reactive seasonal coronoavirus antibodies.
Keywords: SARS-CoV-2, Covid19, Immunology, Immunoassays
Abstract
Background
The emergence of SARS-CoV-2 has led to the development of serological assays that could aid in an understanding of the burden of COVID-19 disease. Many available tests lack rigorous evaluation and therefore results may be misleading.
Objectives
The aim of this study was to assess the performance of a novel multiplexed immunoassay for the simultaneous detection of antibodies against SARS-CoV-2 trimeric spike (S), spike receptor binding domain (RBD), spike N terminal domain and nucleocapsid antigen and a novel pseudo-neutralisation assay.
Methods
A multiplexed solid-phase chemiluminescence assay (Meso Scale Discovery) was evaluated for the simultaneous detection of IgG binding to four SARS-CoV-2 antigens and the quantification of antibody-induced ACE-2 binding inhibition (pseudo-neutralisation assay). Sensitivity was evaluated with a total of 196 COVID-19 serum samples (169 confirmed PCR positive and 27 anti-nucleocapsid IgG positive) from individuals with mild symptomatic or asymptomatic disease. Specificity was evaluated with 194 control serum samples collected from adults prior to December 2019.
Results
The specificity and sensitivity of the binding IgG assay was highest for S protein with a specificity of 97.4 % and sensitivity of 96.2 % for samples taken 14 days and 97.9 % for samples taken 21 days following the onset of symptoms. IgG concentration to S and RBD correlated strongly with percentage inhibition measured by the pseudo-neutralisation assay.
Conclusion
Excellent sensitivity for IgG detection was obtained over 14 days since onset of symptoms for three SARS-CoV-2 antigens (S, RBD and N) in this multiplexed assay which can also measure antibody functionality.
1. Introduction
Severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) was first recognised in January 2020 and rapidly spread world-wide [1]. Tests designed to measure antibodies to SARS-CoV-2 antigens were rapidly developed and are important for diagnostics and seroprevalence studies. The latter could help inform disease burden estimates, studies of transmission dynamics and modelling of the epidemic. Antibody tests are particularly important in the context of mild or asymptomatic disease where a swab reverse transcriptase polymerase chain reaction (RT-PCR) test may be negative. For this reason, an understanding of the sensitivity and specificity of the tests being used is critical.
The trimeric spike (S) protein of SARS-CoV-2 is present on the viral surface and in most cases is cleaved by host proteases into the S1 and S2 subunits, responsible for receptor recognition and membrane fusion respectively. S1 uses a region of the molecule, known as the receptor binding domain (RBD) to bind to host ACE-2 receptor and thereby gain entry to the cell [2]. The N terminal domain (NTD) of the spike protein does not interact with the receptor but contains the functional elements required for membrane fusion of the virion. The nucleocapsid (N) protein plays an important role in transcription enhancement and viral assembly [3]. Specific immunoglobulin-G (IgG) and IgM antibody responses to SARS-CoV-2 S, N and RBD of the spike protein develop between 6–15 days following disease-onset [4].
Despite a rapid increase in the number and availability of SARS-CoV-2 serologic assays, most have undergone minimal external evaluation and validation [5]. A recent large scale Spanish seroprevalence study used a point of care IgG test with a stated sensitivity of 97.2 % but on verification found it to have a sensitivity of either 82.1 %, 89.7 %, 99.6 % or 100 % depending on the sample sets used for evaluation [6]. All assays currently suffer from the absence of a defined standard serum so results are reported as positive or negative or as optical density readouts complicating the comparison between assays and studies and for many binding assays the relationship between antibody concentration and function is unclear.
We have evaluated a novel assay designed to simultaneously measure IgG to four SARS-CoV-2 antigens; full-length trimeric S, RBD and NTD of spike as well as N protein. The assay, based on Meso Scale Discovery (MSD) technology, utilises a 96-well based solid-phase antigen printed plate and an electrochemiluminescent detection system. In addition this assay can measure the ability of serum to inhibit the interaction between spike protein components and soluble ACE-2, also called a pseudo-neutralisation assay [7]. To evaluate the sensitivity and specificity of the MSD assay, we were able to utilise a relatively large number of samples obtained from SARS-CoV-2 RT-PCR positive health care workers or patients as well as antibody positive health care staff enrolling in a large SARS-CoV-2 cohort study.
2. Materials and methods
2.1. Serum samples
Sera were obtained from Great Ormond Street Children’s Hospital NHS Foundation Trust (GOSH) and came from; (i) Symptomatic RT-PCR + healthcare workers (ii) staff enrolling in a prospective longitudinal cohort study of SARS-CoV-Serology (COSTARS, IRAS 282713, ClinicalTrials.gov Identifier: NCT04380896) who tested positive for anti-Nucleocapsid IgG (Epitope Diagnostics Inc, San Diego, USA) (iii) Sera from RT-PCR + hospitalised children (n = 10).
Sera for specificity pre-dated 2019 and derived from anonymised samples from healthy adults enrolled in previous studies.
Pooled serum from two individuals with high convalescent antibody levels were used as an interim standard serum calibrated against research reagents NIBSC 20/130 and NIBSC 20/124 (National Institute for Standards and Biological Control, Potters Bar, UK, https://www.nibsc.org/) obtained from COVID-19 recovered patients.
2.2. Serological assays
Samples were screened for IgG to SARS-CoV-2 N protein using a commercially available kit (Epitope Diagnostics Inc, San Diego, USA) as previously described [8].
2.3. Meso scale discovery coronavirus panel for COVID-19 serology
A multiplexed MSD immunoassay (MSD, Rockville, MD) was used to measure the responses to SARS-CoV-2 and other respiratory pathogens. A MULTI-SPOT® 96-well, 10 Spot Plate was coated with four SARS CoV-2 antigens (S, RBD, NTD and N), SARS-CoV-1 and MERS spike trimers, spike proteins from seasonal coronaviruses OCV43S and HKU1, influenza A antigen derived from H3/HongKong and Bovine Serum Antigen. Antigens were spotted at 200−400 μg/mL in a proprietary buffer, washed, dried and packaged for further use (MSD® Coronavirus Plate 1). Proteins were expressed in a mammalian cell expression system (Expi 293 F), purified by ion exchange chromatography, affinity purification, and size exclusion chromatography; the spike proteins were produced as trimers in the pre-fusion form. These assays were developed by MSD in collaboration with the Vaccine Research Center at NIAID (A. McDermott).
To measure IgG antibodies, plates were blocked with MSD Blocker A following which reference standard, controls and samples diluted 1:500 in diluent buffer were added. After incubation, detection antibody was added (MSD SULFO-TAG™ Anti-Human IgG Antibody) and then MSD GOLD™ Read Buffer B was added and plates read using a MESO® SECTOR S 600 Reader.
2.4. Meso scale discovery pseudo-neutralisation assay
Plates were blocked and washed as above, assay calibrator (COVID-19 neutralising antibody; monoclonal antibody against S protein; 200 μg/mL), control sera and test sera samples diluted 1 in 10 in assay diluent were added to the plates. Following incubation Plates an 0.25 μg/mL solution of MSD SULFO-TAG™ conjugated ACE-2 was added after which plates were read as above. Percentage inhibition was calculated relative to the assay calibrator (maximum 100 % inhibition).
2.5. Statistical analysis
Statistical analysis was performed using MSD Discovery Workbench and GraphPad Prism version 8.0 (GraphPad, San Diego, CA). Antibody concentration in arbitrary units (AU) was interpolated from the ECL signal of the internal standard sample using a 4-parameter logistic curve fit. ROC curves showing the sensitivity and specificity (plotted as 100 %-specificity %) calculated using each value in the data as a cut-off were plotted for each antigen. A cut-off antibody concentration was chosen based on the lowest value leading to a positive likelihood ratio (LR) of >10, in order to maximise sensitivity while providing strong evidence to rule-in infection [9]. For S antigen binding, all LR’s were above 10, therefore the LLOD was used as the cut-off for this antigen. Comparisons between groups were performed by Kruskal-Wallis one-way ANOVA with Dunn’s correction for multiple comparisons. Correlation analysis was performed using Spearman correlation. P values of <0.05 were considered as significant. Latent class models with two classes were fitted with the binary antibody responses as outcome variables, using the poLCA package in the R statistical environment. The code used for the latent class analysis is available on request.
3. Results
3.1. Participants and samples
SARS-CoV-2 positive samples (COVID-19 cohort) comprised 169 PCR positive and 27 anti-N IgG positive serum samples from mild symptomatic or asymptomatic cases (total n = 196, 138 females, 56 males [2 missing], median age 37 years). Time between symptom onset and sampling ranged from 4 to 63 days for 168 subjects with verified onset date. Control serum samples comprised 194 anonymised legacy samples obtained from healthy adults, aged predominantly over 50 years.
3.2. Standard serum assignment
An internal standard serum (ISS) was assigned values for S, RBD and N by calibration against the NIBSC control sera. NIBSC 20/130 was used to assign arbitrary unit (AU) values for S and RBD and NIBSC 20/124 for N (Supplementary Figure S1). No endpoint titre corresponding to NTD antigen was available for ISS assignment. The interim values assigned were S 2154 AU, RBD 1837 AU and N 3549 AU and 1000 AU for all other antigens.
3.3. Evaluation of the coronavirus panel for COVID-19 serology
The lower limit of detection (LLOD) was assigned as 1% of the standard value in AU, and upper limit of detection (ULOD) was assigned for NTD and RBD only as the S and N antigen did not reach an upper limit (Table 1 ). For statistical purposes, ULOD was assigned the highest calculated concentration plus 20 % and LLOD as 0.5 %.
Table 1.
The lower limit of detection (LLOD), upper limit of detection (ULOD), quality control (QC) sample range in arbitrary units (AU) and positive/negative cut-off for each SARS-CoV-2 antigen analysed.
| Antigen | LLOD (max.) (AU) | ULOD (min.) (AU) | QC sample range (AU) | Positive/ negative cut-off |
|---|---|---|---|---|
| CoV-2 S | 21.54 | NA | 1092−1478 | 21.5 |
| CoV-2 RBD | 18.37 | 125477 | 2176−2944 | 201.7 |
| CoV-2 N | 35.49 | NA | 3627−4907 | 185.4 |
| CoV-2 NTD | 10.00 | 19452 | 1004−1359 | 1924 |
The mean coefficient of variation (CV) between duplicates was <15 % for all except NTD (17.4 %, data not shown). The mean intra-assay CV was 6.2 % and inter-assay variation <15 % across all SARS-CoV-2 antigens except NTD (19.0 %) on one of four samples (Supplementary Table 1). A QC sample was run on each plate (average CV 10.3 %) and an acceptable performance range was set as within 3 SD of the mean.
3.4. Assay sensitivity and specificity
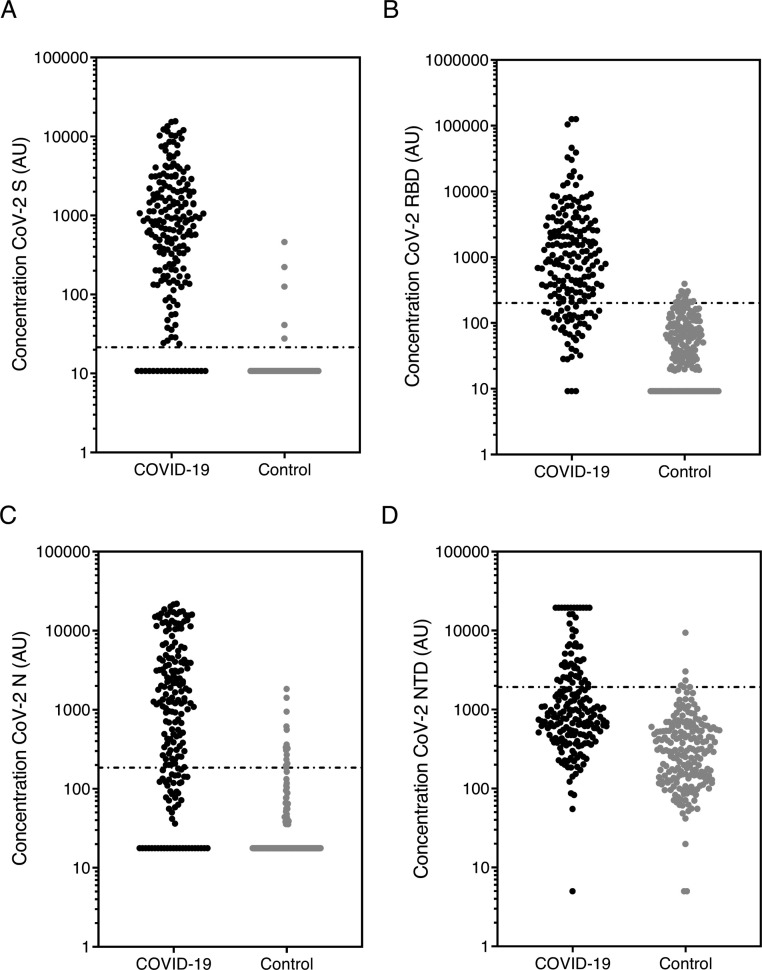
Fig. 1 A-D shows the concentration of IgG to each SARS-CoV-2 antigen.
Fig. 1.
Anti-SARS-CoV-2 IgG concentration.
The concentration of SARS-CoV-2 antibody against (a) spike (S), (b) receptor binding domain (RBD), (c) nucleocapsid (N) and (d) N terminal domain (NTD) was measured using the MSD coronavirus panel. Graphs show data in arbitrary units (AU) (based on the calibrated internal standard serum) in the COVID-19 cohort (n = 196) and controls (n = 194, pre-December 2019). Line shows positive/negative discrimination cut-off.
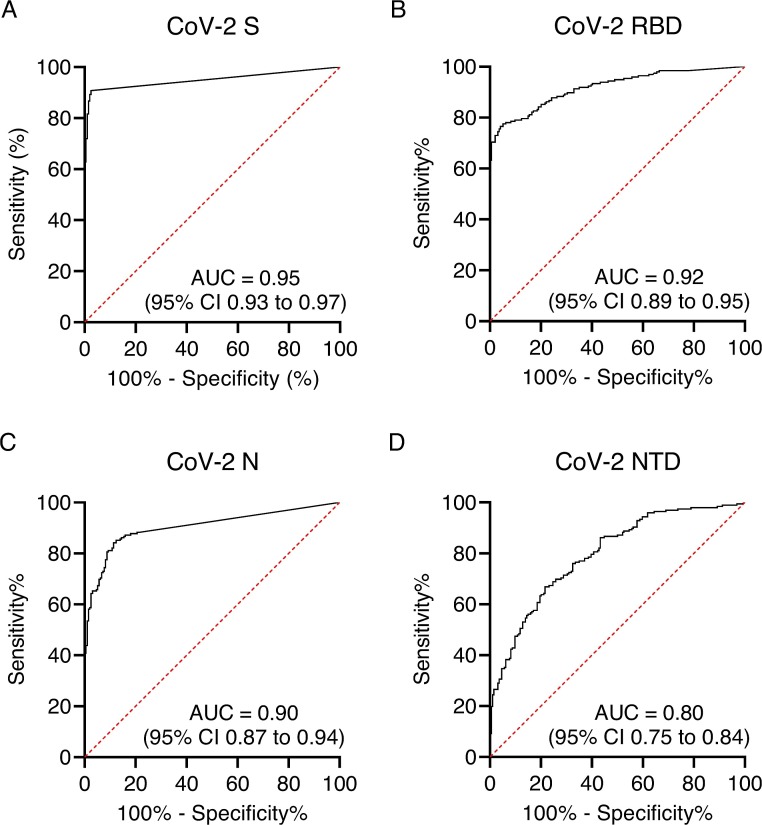
ROC curves were plotted to visualise the trade-off between sensitivity and specificity for each antigen (Fig. 2 A-D). The high area under the curve (AUC) values for S (0.95 %; 95 %CI 0.93 to 0.97), RBD (0.92 %, 0.89−0.95) and N (0.90 %, 0.87−0.94) indicates the high accuracy of these tests. Table 1 shows the Positive/negative cut-off values calculated from the ROC using LR > 10. NTD data was less consistent than the other SARS-CoV-2 antigens and demonstrated lower sensitivity and specificity so this antigen was not evaluated further.
Fig. 2.
Receiver Operating Characteristic (ROC) curves for each SARS-CoV-2 antigen.
Sensitivity and specificity were calculated using each value in the data table as a cut-off value (n = 390). Graphs show the sensitivity vs 100 %-specificity of SARS-CoV-2 antigen (a) spike (S), (b) receptor binding domain (RBD), (c) nucleocapsid (N) and (d) N terminal domain (NTD). The area under curve (AUC) and 95 % CI is also shown for each antigen.
The specificity for S, RBD and N assays are shown in Table 2 . Assay sensitivity was initially calculated on the entire COVID-19 cohort; S antigen had the highest AUC and was the most sensitive and specific at 90.8 % and 97.4 % respectively.
Table 2.
Assay specificity calculated for each SARS-CoV-2 antigen from the control cohort.
| Antigen | n | Positive | Negative | Specificity (95 % CI) (%) |
|---|---|---|---|---|
| CoV-2 S | 194 | 5 | 189 | 97.4 % (94.1 to 98.9) |
| CoV-2 RBD | 194 | 15 | 179 | 92.3% (87.6 to 95.3) |
| CoV-2 N | 194 | 14 | 180 | 92.8 % (88.2 to 95.7) |
3.5. Evaluation of sensitivity according to time since onset of symptoms
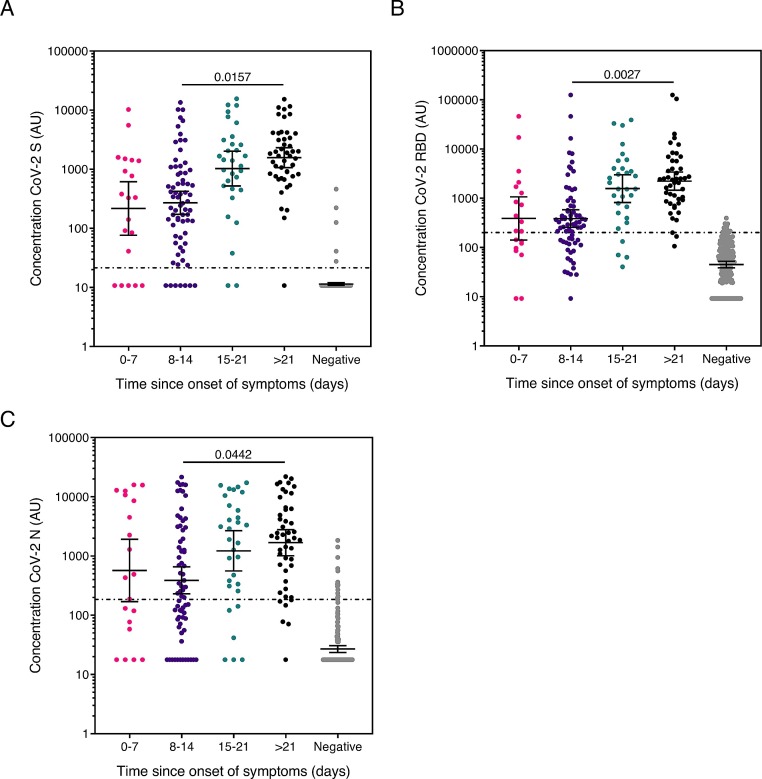
Fig. 3 shows the anti-S, RBD and N IgG concentration split into time since onset of symptom intervals of 0−7 days, 8–14 days, 15–21 days and over 21 days. For all three antigens, the median antibody concentration increased significantly with time since symptom onset (SARS-CoV-2 S, Spearman correlation (r) = 0.453; SARS-CoV-2 RBD, ; SARS-CoV-2 N, r = 0.392, all p=<0.0001 (Supplementary Fig. 2A-C)) and at all time points were higher than controls (p=<0.0001) (Fig. 3A-C).
Fig. 3.
Anti-SARS-CoV-2 IgG concentration according to time since onset of symptoms.
Graphs show the concentration of SARS-CoV-2 antibody against (a) spike (S), (b) receptor binding domain (RBD) and (c) nucleocapsid (N) in arbitrary units (AU) (based on the calibrated internal standard serum) of the COVID-19 cohort split in to intervals of 0−7 days, 8–14 days, 15–21 days and over 21 (>21) days since symptom onset (to sample collection). Error bars show geometric mean with 95 % CI, line shows positive/negative discrimination cut-off, *p < 0.05, ** p < 0.01 determined by Dunn’s multiple comparisons test. Comparisons across interval groups had p < 0.0001 by one-way ANOVA Kruskal-Wallis test. The assay sensitivity at each time point is shown in Table 3.
Sensitivity and specificity was calculated for groups 0−7d, >7d, >14d and >21d since the onset of symptom The S antigen was the most sensitive of the three, with a sensitivity of 96.2 % and 97.9 % >14 days and >21 days respectively (Table 3 ).
Table 3.
Assay sensitivity by time since onset of symptoms for each SARS-CoV-2 antigen calculated using the COVID-19 cohort with verified time between onset of symptoms and blood sampling. Time was divided into 0-7 days, over 7 days, over 14 days and over 21 days since the onset of symptoms.
| Antigen | Group | n | Positive | Negative | Sensitivity (95 % CI) (%) | |
|---|---|---|---|---|---|---|
| CoV-2 S | Total | 196 | 178 | 18 | 90.8 % (86.0–94.1) | |
| Time since onset of symptoms | 0−7 days | 20 | 15 | 5 | 75.0% (53.1–88.8) | |
| Over 7 days | 148 | 138 | 10 | 93.2% (88.0–96.3) | ||
| Over 14 days | 78 | 75 | 3 | 96.2 % (89.3–99.0) | ||
| Over 21 days | 47 | 46 | 1 | 97.9 % (88.8–99.9) | ||
| CoV-2 RBD | Total | 196 | 153 | 43 | 78.1% (71.8–83.3) | |
| Time since onset of symptoms | 0−7 days | 20 | 12 | 8 | 60.0% (38.7–78.1) | |
| Over 7 days | 148 | 119 | 29 | 80.4% (73.3–86.0) | ||
| Over 14 days | 78 | 71 | 7 | 91.0% (82.6–95.6) | ||
| Over 21 days | 47 | 44 | 3 | 93.6% (82.8–97.8) | ||
| CoV-2 N | Total | 196 | 143 | 53 | 73.0% (66.3–78.7) | |
| Time since onset of symptoms | 0−7 days | 20 | 12 | 8 | 60.0% (38.7–78.1) | |
| Over 7 days | 148 | 106 | 42 | 71.6% (63.9–78.3) | ||
| Over 14 days | 78 | 66 | 12 | 84.6% (75.0–91.0) | ||
| Over 21 days | 47 | 41 | 6 | 87.2% (74.8–94.0) | ||
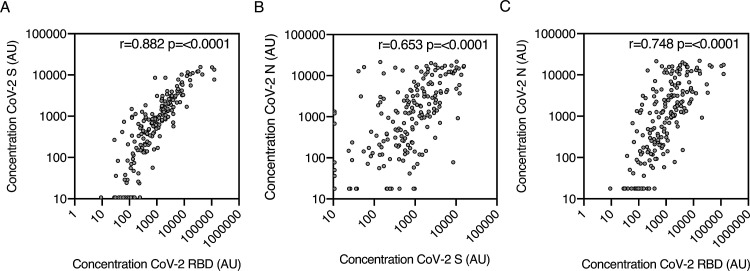
3.6. Antibody concentration relationship between antigens
The concentration of anti-S, RBD and N all correlated significantly with each other (p < 0.0001; Fig. 4 A-C), the strongest association was between S and RBD (r = 0.882) (Fig. 4A). Our two-class latent class model built using binary S, RBD and N antigen results predicted known status with 81.1 % (95 %CI 74.8–86.2) sensitivity and 99.0 % (95 %CI 95.9–99.8) specificity. It therefore had lower sensitivity and no meaningful improvement in specificity, compared to using the concentration of S antibody alone, with the 21.54 AU cut-off.
Fig. 4.
IgG concentration relationship between antigens.
Correlation between anti-SARS-CoV-2 antibody concentration of all COVID-19 group samples (n = 196) (a) S vs RBD, (b) S vs N and (c) N vs RBD. r and p value were determined by Spearman correlation. p values of <0.05 were considered as significant.
3.7. Pseudo-neutralisation
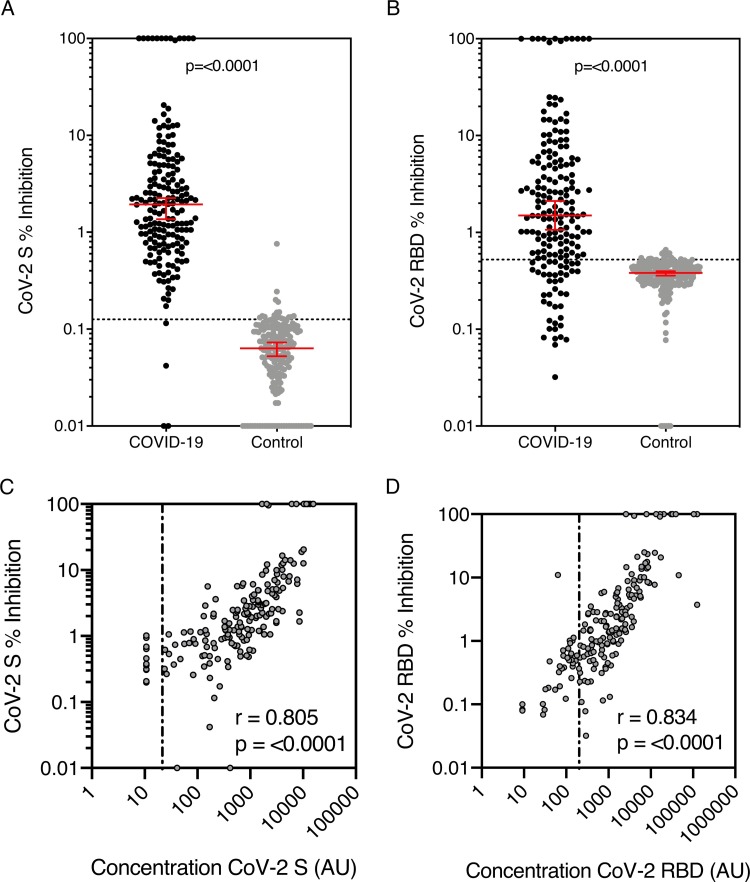
183 COVID-19 cohort samples with sufficient volume and 194 control group samples were evaluated in the pseudo-neutralisation assay. The percentage inhibition of ACE-2 receptor binding to the S and RBD antigens for the COVID-19 cohort was significantly higher than the controls (S, median 1.94 % (95 %CI 1.36–2.25) vs 0.063 % (95 %CI 0.053−0.073), p=<0.0001; RBD, 1.50 % (95 %CI 1.064–2.11) vs 0.38 % (95 %CI 0.36−0.39); p=<0.0001) (Fig. 5 A-B) and correlated with IgG concentration for both S and RBD antigens (Spearman correlation (r) = 0.805 and r = 0.834 respectively, p=<0.0001) (Fig. 5C-D).
Fig. 5.
Percentage inhibition by anti-SARS-CoV-2 S and RBD antibody measured by MSD pseudo-neutralisation assay.
Inhibition of ACE-2 binding by SARS-CoV-2 antibody against (a) spike (S) and (b) receptor binding domain (RBD) was measured using the MSD coronavirus pseudo-neutralisation assay. 183 COVID-19 cohort samples and 194 control samples were analysed. Graphs show median and 95 % CI with a line showing neutralisation assay positive/negative discrimination cut-off determined by ROC. The correlation between antibody concentration and percentage inhibition of (c) S and (d) RBD antigens in all positive group samples was assessed and r and p was determined by Spearman correlation, line shows binding assay positive/negative discrimination cut-off.
Cut-offs (LR > 10) were 0.162 % for S and 0.524 % for RBD (shown by the dotted line on Fig. 5A-B). Sensitivity and specificity for S were 97.8 % and 97.9 % respectively but lower for RBD (77.2 % and 92.8 % respectively). In the COVID-19 cohort there were some IgG positive sera that did not demonstrate neutralisation (below cut-off, n = 4 for S and 36 for RBD). These sera were predominantly those taken soon after the onset of symptoms; 22 between 0−7 days, 9 over 14 days and 5 over 21 days.
4. Discussion
Accurate tests of SARS-CoV-2 antibodies are critical for reliably evaluating exposure to the virus causing COVID-19. Despite a large number of assays rapidly becoming available, many have not undergone rigorous evaluation. In this study we describe a novel assay that can measure antibody to several SARS-CoV-2 antigens simultaneously as well as evaluating the functional capacity of anti-Spike antibodies.
The assay we used is based on existing technology developed by Meso Scale Discovery. We decided to evaluate IgG only as the kinetics of IgM responses appear to mimic those of IgG and thus add little value [4].
Unlike the majority of studies published to date, we were able to utilise a panel of COVID-19 convalescent plasma recently distributed by WHO to calibrate an internal standard made from pooled convalescent serum. This allowed us to express titres in arbitrary units that can then be compared to other assays that report values calibrated against the WHO panel. The assays performed reliably and consistently over the period of study and passed all the performance criteria expected for a solid-phase based assay.
Using a carefully defined cohort of known SARS-CoV-2 exposed individuals and relevant controls we were able to show the sensitivity and specificity of the assay for the four antigens of interest. Comparing the performance of S and RBD assays in a recently published systematic review and metanalysis of the diagnostic accuracy of serological tests for COVID-19 [10] the S assay we evaluated had superior sensitivity to all of the assays included in the review while RBD performance was superior to most. The reason for this could be related to the technical aspects of the assay itself including the integrity of the antigen used and the sensitivity of the detection platform but also the use of a well-defined cohort of individuals with known exposure to SARS-CoV-2. Only the N terminal domain of the spike protein did not perform well in this assay with poor sensitivity due to the overlap in antibody titres between the COVID-19 cohort and controls.
The assay format permitted the measurement of antibody against spike protein derived from SARS-1, MERS and two seasonal coronaviruses, but the results of antibody binding to these antigens could not be assessed in the same way as for the SARS-CoV-2 antigens due to the absence of defined negative and positive serum sets.
An advantage of this assay is its ability to measure antibody induced inhibition of ACE-2 receptor-spike interaction thought to be the major mechanism by which SARS viruses, including SARS-CoV-2 attach to host cell surfaces [11,12]. In the COVID-19 cohort, there was a good correlation between anti-S and anti-RBD IgG and function although a few sera bound antigen but did not neutralize. These were dominated by sera taken soon after infection and as recently described, could be non-neutralising and targeting epitopes outside the RBD [13]. Few of the control cohort sera had any pseudo-neutralisation activity despite pre-existing IgG to seasonal Coronavirus spike proteins suggesting season Coronavirus exposure is unlikely to modify interaction with SARS-CoV-2. Other cross reactive immunological mechanisms (eg T cells) cannot be ruled out and may explain the varied clinical response following exposure to SARS-CoV-2 [14]. This pseudo-neutralisation assay has been shown to correlate well with neutralisation assays using live SARS-CoV-2 (MSD, personal communication).
In summary, the MSD multiplexed coronavirus panel assay evaluated in this study is highly reproducible, specific and sensitive for the detection of anti-SARS-CoV-2 antibody over 14 days since the onset of COVID-19 symptoms. The assay can be adapted to measure antibody function which corelated well with spike protein antibody concentration.
Funding: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declarations of Competing Interest
None
Acknowledgements
The study team would like to thank Meso Scale Discovery for the donation of the plates and reagents that allowed us to complete the work, the COSTARS study team at Great Ormond Street Children’s Hospital, staff in the Great Ormond Street Children’s Hospital Clinical Immunology Laboratory for additional support and the NIHR UCL Great Ormond Street Biomedical Research Centre for underpinning infrastructure support that facilities translation research at GOSH.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104572.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.World Health Organisation . 2020. Coronavrius Disease 2019 (COVID-19) Situation Report - 51. [Google Scholar]
- 2.Salazar E., Kuchipudi S.V., Christensen P.A., Eagar T.N., Yi X., Zhao P., Jin Z., Long S.W., Olsen R.J., Chen J., Castillo B., Leveque C., Towers D.M., Lavinder J., Gollihar J.D., Cardona J., Ippolito G.C., Nissly R.H., Bird I.M., Greenawalt D., Rossi R.M., Gontu A., Srinivasan S., Poojary I.B., Cattadori I.M., Hudson P.J., Joselyn N., Prugar L., Huie K., Herbert A., Bernard D.W., Dye J., Kapur V., Musser J.M. Relationship between anti-spike protein antibody titers and SARS-CoV-2 in vitro virus neutralization in convalescent plasma. bioRxivorg. 2020 doi: 10.1101/2020.06.08.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome Coronavirus-2. Arch. Med. Res. 2020 doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C., Cai J.P., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C., Poon R.W., Luo C.T., Cheng V.C., Chan J.M.F., Hung I.F., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng M.P., Yansouni C.P., Basta N.E., Desjardins M., Kanjilal S., Paquette K., Caya C., Semret M., Quach C., Libman M., Mazzola L., Sacks J.A., Dittrich S., Papenburg J. Serodiagnostics for severe acute respiratory syndrome-related Coronavirus-2: a narrative review. Ann. Intern. Med. 2020 doi: 10.7326/M20-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollan M., Perez-Gomez B., Pastor-Barriuso R., Oteo J., Hernan M.A., Perez-Olmeda M., Sanmartin J.L., Fernandez-Garcia A., Cruz I., Fernandez de Larrea N., Molina M., Rodriguez-Cabrera F., Martin M., Merino-Amador P., Leon Paniagua J., Munoz-Montalvo J.F., Blanco F., Yotti R., Group E-CS Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020 doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrnes J.R., Zhou X.X., Lui I., Elledge S.K., Glasgow J.E., Lim S.A., Loudermilk R., Chiu C.Y., Wilson M.R., Leung K.K., Wells J.A. A SARS-CoV-2 serological assay to determine the presence of blocking antibodies that compete for human ACE2 binding. medRxiv. 2020 doi: 10.1101/2020.05.27.20114652. [DOI] [Google Scholar]
- 8.Kruttgen A., Cornelissen C.G., Dreher M., Hornef M., Imohl M., Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deeks J.J., Altman D.G. Diagnostic tests 4: likelihood ratios. BMJ. 2004;329:168–169. doi: 10.1136/bmj.329.7458.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A., Menzies D., Benedetti A., Ahmad Khan F. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn J.H., Li W., Choe H., Farzan M. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cell. Mol. Life Sci. 2004;61:2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.045. 894-904 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.H., Feng J., Nelson R.E., Singh S., Cohen K.W., McElrath M.J., Englund J.A., Chu H.Y., Pancera M., McGuire A.T., Stamatatos L. Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual. bioRxivorg. 2020 doi: 10.1101/2020.05.12.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020 doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.