Abstract
Coronavirus Disease 2019 (COVID-19), caused by SARS-CoV-2 (Severe Acute Respiratory Syndrome - Coronavirus-2) of the family Coronaviridae, appeared in China in December 2019. This disease was declared as posing Public Health International Emergency by World Health Organization on January 30, 2020, attained the status of a very high-risk category on February 29, and now having a pandemic status (March 11). COVID-19 has presently spread to more than 215 countries/territories while killing nearly 0.75 million humans out of cumulative confirmed infected asymptomatic or symptomatic cases accounting to almost 20.5 million as of August 12, 2020, within a short period of just a few months. Researchers worldwide are pacing with high efforts to counter the spread of this virus and to design effective vaccines and therapeutics/drugs. Few of the studies have shown the potential of the animal-human interface and zoonotic links in the origin of SARS-CoV-2. Exploring the possible zoonosis and revealing the factors responsible for its initial transmission from animals to humans will pave ways to design and implement effective preventive and control strategies to counter the COVID-19. The present review presents a comprehensive overview of COVID-19 and SARS-CoV-2, with emphasis on the role of animals and their jumping the cross-species barriers, experiences learned from SARS- and MERS-CoVs, zoonotic links, and spillover events, transmission to humans and rapid spread, and highlights the new advances in diagnosis, vaccine and therapies, preventive and control measures, one health concept along with recent research developments to counter this pandemic disease.
Keywords: COVID-19, SARS-Cov-2, Bat coronavirus, Zoonosis, Spillover, Expanding host range, Diagnosis, Prevention, Control, One health
1. Introduction
In the 21st century, we have faced a few deadly disease outbreaks caused by pathogenic viruses such as Bird flu caused by Avian influenza virus H5N1, Swine flu caused by reassorted influenza virus H1N1 pandemic 2009 (H1N1pdm2009), Severe Acute Respiratory Syndrome (SARS) caused by SARS-CoV (coronavirus), the Middle East respiratory syndrome (MERS) caused by MERS-CoV [[1], [2], [3]], Ebola [4], Zika [5,6], Nipah virus infections, and the most recent threat [7], Coronavirus Disease 2019 (COVID-19) that has been posed by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) of the family Coronaviridae, genus Betacoronavirus [[8], [9], [10], [11], [12], [13], [14]]. The SARS-CoV-2 virus emerged from the city of Wuhan, Hubei province, China, during December 2019, was declared as Public Health International Emergency by the World Health Organization (WHO) on January 31, 2020. Consequently, it was categorized in a high-risk category on February 29, 2020, and gained the pandemic status on March 11, 2020. The disease emerged from Wuhan, China, as its epicentre, which moved later to Italy, then the USA, and Brazil. Subsequently, within a short time interval of six months, it has affected nearly 215 countries/territories and claimed near to 0.75 million human deaths out of cumulative confirmed infected asymptomatic or symptomatic cases accounting to almost 20.5 million. SARS-CoV-2 has very adversely affected the USA, Brazil, India, Russia, South Africa, Peru, Mexico, Chile, Spain, the United Kingdom (UK), Iran, Pakistan, Saudi Arabia, Italy and other countries. The disease incidences are lower in children than adults but exhibit all symptoms of a disease like adults [15]. The lessons learned from earlier threats of SARS, MERS and the present COVID-19 pandemic situations warrants designing and implementing some modified plans and strategies to combat emerging and zoonotic pathogens that could pose pandemic threats/risks while taking away many human lives [11,[16], [17], [18], [19], [20], [21], [22]].
Researchers and health agencies across the world are putting high efforts to contain/restrain the spread of this deadly disease. They are pacing to develop potential vaccines and therapeutics/drugs [23,24]. Evidence from the initial outbreak indicates earlier cases had links to Huanan Wholesale Seafood Market in China [25] and further isolation of SARS-CoV-2 from different samples of the area (people, animals, birds, discharges, soil, structures) suggests the involvement of intermediate hosts [26]. Recently, a literature of review has pointed out the possible potential role of the animal-human interface, zoonotic links and spillover events towards the origin of SARS-CoV-2/COVID-19 [11,20,[27], [28], [29], [30], [31]].
In the past couple of decade's animal origin viral diseases, especially bats-linked, have increased many folds in humans with noted cross-species transmissions. Although many of the illnesses are linked with bats still information on their ecological behaviour, molecular aspects are limited, which could lead to more viral outbreaks shortly [32]. The ongoing COVID-19 pandemic has emphasized the importance of understanding the evolution of natural hosts in response to viral pathogens. In a recent study on ACE2 receptors, the gene was found under intense selection pressure in bats and positive selection in other selected mammalian hosts [33]. The SARS-CoV-2 is also thought to have originated from bats, just like SARS-CoV and MERS-CoV. Civets and dromedary camels are considered as the intermediate host of SARS- and MERS-CoV, respectively, from where they were transmitted to humans [34]. The understanding of genomic signatures of SARS-CoV-2 with other CoVs is must for strategic planning through identifying natural or intermediate hosts. Using genomic and protein data in a Natural Vector method (alignment-free approach), phylogenetic analysis revealed the possible transmission path originates from bats to pangolins to humans [35]. However, the likely source of virus origin and the intermediate host of SARS-CoV-2 are yet to be identified. Initially, when the novel virus emerged in China, a hypothesis was put forward, claiming the recent recombination event as the cause of the SARS-CoV-2 emergence.
Nevertheless, the phylogenetic and recombination analysis performed within the subgenus of Sarbecovirus demonstrated that the novel virus shows discordant clustering with Bat-SARS-like coronavirus (RaTG13) sequences thus rejecting the possibility of a recent recombination event [36]. Previously, it was found that the continuous passaging of MERS-CoV in non-susceptible cells that express viral receptors led to the accumulation of mutations in the spike protein gene. This paid attention to the potential of coronaviruses like MERS-CoV to undergo mutations that enhance viral entry into novel animal species, thus resulting in cross-species transmission [37]. The COVID-19 outbreak is still associated with several unanswered questions like the possibility of shedding of the virus before the onset of clinical signs, whether the transmission is limited to only through respiratory droplets, the possibility of an intermediate host that is responsible for zoonotic spillover, and the possible transmission characteristics [20,38]. Hitherto studies report that the spillover risk remains high from zoonotic viruses and on the same lines a study from North America proposed a hypothesized conceptual model demonstrating SARS-CoV-2 spillover from humans to naive wildlife host species through the gastrointestinal route where stool from COVID-19 infected patient contaminates water bodies and reaches to wildlife hosts [39].
Besides, the pandemic imposed a massive blow on the Chinese economy, which is not going to heal soon [40]. Instead of the current situation, Singapore's Prime Minister Lee Hsien Loong rightly said that the virus might have started in China. However, it does not respect nationality or race. It does not check your passport before it goes into your body, and anybody can be infected. Hence, all suspected people need to be tested and quarantined [41].
Further research exploring the SARS-CoV-2 associated zoonosis and mechanisms accounting for its initial transmission from animals to humans, will lead to sort out the spread of this virus as well as design and develop appropriate prevention and control strategies to counter COVID-19. The present comprehensive manuscript presents an overview on COVID-19, an emerging SARS-CoV-2 infectious disease while focusing mainly on the events and circumstantial evidences with regards to this virus jumping the species barriers, sharing a few lessons learned from SARS- and MERS-CoVs, zoonotic spillover events (zoonosis), acquiring transmission ability to infect humans, and adopting appropriate preventive and control measures [42]. It also highlights the recent advances in SARS-CoV-2 diagnosis (see Supplement Material S1), vaccine, drugs and therapies (see Supplement Material S2), which could aid to counter and restrain this emerging virus at the face of pandemic situations, as well as prevention and control (see Supplement Material S3).
2. The virus (SARS-CoV-2)
SARS-CoV-2 is an enveloped virus measuring approximately 50–200 nm in diameter with a single strand positive¬sense RNA genome ranging from 26 to 32 kilobases in length [43,44]. It has club¬shaped glycoprotein spikes in the envelope, giving it a crown¬like or coronal appearance [25]. The genome SARS-CoV-2 is comprised of 5′ untranslated region (5′ UTR) that includes 5′ leader sequence, open reading frame (ORF) 1a/b (replicase genes), spike (S) protein, envelop (E) protein, membrane/matrix (M) protein, and accessory proteins (orf 3,6,7a, 7b, 8 and 9b), nucleoprotein (N), and 3′ untranslated region (3′ UTR) in their sequence [45]. It has a 50% genetic identity to MERS-CoV and 80% to SARS-CoV [43,46]. The receptor-binding domain (RBD) of virus Spikes helps in binding to cellular receptor angiotensin-converting enzyme 2 (ACE-2) [47,48]. ORF and RBD of SARS-CoV-2 may have a role in elucidating cellular interactions and cross-species transmission mechanisms [49]. Receptor binding motifs (RBM) have a role in interaction with human receptors, human to human transmission, and cross-species transmission as Gln493 provides favourable interaction, and Asn501 shows compatibility with human ACE-2 [47]. Besides, SARS-CoV-2 has superior transmission competence in comparison to the SARS-CoV, leading to a continuously increasing number of confirmed cases [50]. SARS-CoV-2 has the potential to survive in the environment for several days [51]. Though believed to be sensitive to environmental factors and alcohol-based sanitizers, bleach, and chloroform, the SARS-CoV-2 can survive in wet surroundings for days and in closed air conditions up to 12 h [26,52]. Survival of SARS-CoV-2 varies with the nature of the surface (glass, fabric, metal, plastic, or paper), environment, and virus load. It can survive on surfaces for hours to several days. It can survive in aerosols for up to 3 h and on plastic for up to 72 h [26,53].
3. The disease (COVID-19)
The initial clinical picture of the COVID-19 was pneumonia of unknown origin as the first clinical cases were presented with signs of pneumonia [38]. Later it was diagnosed as SARS-CoV-2 infection that was associated with severe pneumonia. Hence, initially named as novel coronavirus pneumonia (NCP) [54]. As the outbreak proceeded, a series of cases were produced, developing a wide range of clinical signs with few remaining asymptomatic being in the early incubation stage of the disease. Thus COVID-19 is characterized by three major patterns of the clinical course of infection, including mild illness producing upper respiratory signs, non-life-threatening pneumonia, and severe pneumonia with Acute Respiratory Distress Syndrome (ARDS) [55,56]. Initially, mild signs appear for 7–8 days, followed by rapid deterioration and ARDS. It can be mild to moderate in 80% of affected cases, including pneumonia and non-pneumonia cases.
In comparison, 13.8% are severe cases, including dyspnea, respiratory distress, hemoptysis, gastrointestinal infection, liver, central nervous system, and lung damage cases [57,58]. Critical cases account for 6.1% and include respiratory failure, septic shock, and multiple organ failure/dysfunction cases. Few cases remain asymptomatic and include cases that can become any of the above during infection [26]. Thus, the symptoms can be non-specific and can range from no symptoms (asymptomatic) to severe pneumonia [26]. In this context, a study concluded that the COVID-19 is probably overestimated, as around 2.6 million people succumb to respiratory diseases every year in comparison to approximately 400,000 deaths due to the SARS-CoV-2 infection [59].
The typical clinical signs of COVID-19 are fever, chills, cough, fatigue, and chest distress [60,61]. Fever and cough are considered as the most common symptoms in COVID-19 patients [62], followed by headache, dyspnea, sore throat, hemoptysis, myalgia, diarrhoea, nausea, and vomiting are also observed [61,62]. Some patients have shown rhinorrhea, chest pain [25], nasal congestion [26], anorexia, pharyngalgia, and abdominal pain [63]. Furthermore, neurological symptoms like anosmia and ageusia are also reported as significant clinical symptoms of COVID-19 [64]. The characteristic of COVID-19 is attacking the lower respiratory tract and producing signs of upper respiratory distress, including rhinorrhea, sneezing, and sore throat [49]. The clinical presentation of individuals infected with SARS-CoV-2 revealed upper respiratory tract infection, viremia, viral shedding from the nasopharynx, and stool along with the development of nausea, vomiting or diarrhoea after antiviral treatment [65]. On diagnostic imaging using computed tomography (CT scan) and radiography (X-ray) bilateral pneumonia, ground-glass opacity, multiple mottling, pneumothorax, infiltration, consolidation or bronchoinflation sign has been noted in many cases of COVID-19 [25,49,66]. Previously, SARS-CoV was found to infect the brainstem heavily [67]. Even though fever is considered as the most common symptom associated with COVID-19 infection, a large proportion of the patients do not express fever during the initial hospital admission [68].
The different transmission routes of SARS-CoV-2 infections have not yet been entirely ascertained, and are still under investigation. Both direct and indirect pathways of transmission are being explored [69]. Similar to SARS and MERS, SARS-CoV-2 is predominantly spread via the respiratory route [70]. Person to person transmission is the main reason for community and global spread. The initial estimated reproduction number (Ro-value) of COVID-19 was assessed to be from 0.8 to 2.4 in December 2019, which later has been increased to a mean value of 2.6 (range 2.1–5.1) [71]. Human-to-human transmission is by face-to-face contact with a sneeze or cough, or from contact with secretions of infected people [56,70].
Nevertheless, the infectivity of other secretions and excretions are not fully understood and may require further study [45]. Aerosol and plastic surfaces can sustain virus for hours to days [53]. Travelling of infected people is considered as the main reason for the global spread of COVID-19 [20,56]. Although the asymptomatic and mild cases are the major hurdle in the evaluation of the real number of infected people, the genuine data on travellers returning from affected countries or areas may prove crucial in estimating the disease incidence [72]. The possible occurrence of super-spreading events is very high at large gatherings, and suspension of gathering during a pandemic may prove crucial in reducing the overall transmission [73].
Close contact with any person within 6 feet of the COVID-19 patient or anyone having direct contact with secretions of COVID-19 patients [74] may set up the infection. Unlike SARS-CoV, most transmissions in COVID-19 are during the prodromal period when the infected individuals produce large quantities of virus in the upper respiratory tract, move/travel, and usually work thus spreading virus before illness develops [56]. The rapid spread of COVID-19 among the susceptible population can be due to the wide variation in illness degrees that results in a missed diagnosis. Heavy viral load in asymptomatic cases and nosocomial transmission is spreading COVID-19 unknowingly [75]. Recently, high viral load was detected in the sputum of convalescent patients, pointing out the possibility of prolonged shedding of SARS-CoV-2 even after recovery. This finding, along with the fact that asymptomatic persons can also act as the potential source of infection, may warrant a reassessment in the transmission dynamics of the COVID-19 outbreak [76]. The presence of viral nucleic acids in faeces is an important finding, thereby increasing the possibility of faecal-oral transmission. However, symptoms may or may not be manifested [77]. This was found to be the unique feature of COVID-19 and was lacking in the previous SARS and MERS outbreaks. In a study conducted by the Chinese CDC, it was found that the majority of patients (80.9%) infected with COVID-19 infection were either asymptomatic or had mild pneumonia [56,66]. Furthermore, all forms of sexual contacts have been reported to pose a significantly high risk of disease transmission through respiratory aerosols and fomites. However, to date, no evidence of sexual transmission is available, and further investigation is required in this direction [78].
Disease severity is higher in older individuals, especially males with immunocompromised conditions and comorbidities like diabetes, asthma, or cardiovascular diseases [26,66]. These are considered to be vulnerable to the SARS-CoV-2 infection. Predisposition increases under risk environments where transmission of the virus from affected persons or contaminated fomites to unaffected ones becomes feasible. It was earlier noted that CoVs are not common to affect immunocompromised patients like other some viral infections (Influenza, Rhinovirus, Adenoviruses, to name a few). The current pandemic has shown SARS-CoV-2 to affect more lethally than young patients, mainly destroying the lung tissues [79]. Till now, evidence regarding the higher susceptibility of pregnant women in comparison to non-pregnant women lacks in COVID-19. Also, there is no evidence of vertical transmission (mother to fetus/baby transmission) of COVID-19 infection [80]. A case study reporting the birth of a healthy infant by a SARS-CoV-2 infected woman suggests that mother-to-child transmission is unlikely in the case of COVID-19. The study also pointed out that on the delivery day, all the samples tested negative except for sputum, which proved positive [81]. However, as per one most recent report, neonates have been found positive for SARS-CoV-2, indicating the possibility of vertical transmission from infected mothers to their progeny, thus rendering newborns into a high-risk group owing to their immature immune system [82].
Individuals harbouring SARS-CoV-2 may remain asymptomatic for the incubation period [83]. Different from SARS-CoV and MERS-CoV infection, the median incubation period of COVID-19 was found to be four days [68]. The median period from the development of signs to death was 14 days [84]. The case fatality rate (CFR) of COVID-19 was found to be lower than MERS and SARS [62]. However, current disease dynamics with the involvement of many more countries or areas may change the future mortality rate. The recent analysis suggests that the total fatality rate of COVID-19 is calculated at 3.46% [75]. However, Italy experienced the worst CFR of more than 9% with older people and males suffering from multiple comorbidities as primary victims [85].
SARS-CoV-2 has shown characteristics of efficient replication in the upper respiratory tract, causing the less abrupt onset of clinical signs just like the common cold and unlike SARS-CoV [70]. It can also replicate in the lower respiratory tract as has been noted in cases without pneumonia but having lesions in the lungs on radiological examination [70]. The pathogenesis mechanisms of COVID-19 are yet to be fully elucidated. However, both cellular and humoral immune responses against SARS-CoV-2 or its antigenic structures like spike protein (S) are believed to be of importance [86,87] with disturbed levels of inflammatory mediators playing a mediating role [66]. Following receptor binding with angiotensin-converting enzyme 2 (ACE2) through receptor binding motif (RBM) of the receptor-binding domain (RBD) of S1 subunit of the SARS-CoV-2 spike glycoprotein (S), virus gains entry in host cells [47,88,89]. S2 subunit helps in the fusion of viral and hosts cell membranes [47,90]. SARS-CoV-2 produces cytopathic effects in respiratory and gastrointestinal surface epithelial cells [91]. These include multinucleated syncytial cells, abnormally enlarged pulmonary cells, infiltration with mononuclear cells, lymphocytes infiltration in pulmonary organs, fibrinous exudation, and hyaline deposition [66]. Cytokine storm is believed to be involved in this inflammatory pathophysiology of the COVID-19 patients producing lung lesions and systemic symptoms [66]. Elevated levels of TNF-α, IL1B, IFNγ, IP10, GCSF, MIP1A, and MCP1, may have stimulated T-helper-1 (Th1) cells leading to this inflammatory cascade [66]. However, levels of anti-inflammatory mediators (IL4, IL10) were also increased, indicating T-helper-2 (Th2) stimulation, which suppresses inflammation, unlike what happens in SARS [66]. A study documented that the nucleic acid of SARS-CoV-2 detected in the faecal samples was as accurate as of that of pharyngeal samples obtained from infected patients.
Moreover, the patients tested positive for SARS-CoV-2 in stool showed no gastrointestinal symptoms and had no relation to the severity of lung infections [92]. One of the significant clinical signs of COVID-19 patients during the initial presentations was gastrointestinal symptoms. Hence, the involvement of GIT in pathogenesis needs to be explored. The significant laboratory findings include lymphopenia, increased values of erythrocyte sedimentation rate, C-reactive protein, lactate dehydrogenase, and decreased oxygenation index [61]. An increase in proinflammatory cytokine and a decrease in anti-inflammatory cytokines have also been noted [66]. Viral isolation has been achieved from bronchoalveolar lavage of affected persons; however, in the case of pregnant women, serum, faeces, urine, breast milk, umbilical cord blood, placenta, and amniotic fluid were found to be negative for SARS-CoV-2 [81]. At the same time, the sputum was tested positive [82]. The presence of abnormal coagulation parameters in patients with severe novel coronavirus pneumonia was associated with poor prognosis. The non-survivor patients had higher levels of D-dimer, and fibrin degradation product (FDP) along with longer activated partial thromboplastin time and prothrombin time compared to survivors at the time of admission [93].
Though clinical manifestations, pathological changes, and diagnostic laboratory findings can unravel the disease nature helping in devising therapeutic modalities, however, for epidemiological aspects and future prevention and control, simultaneous tracing of the origin and explaining the spillover events can prove beneficial.
4. Virus jumping the species barrier, zoonotic spillover, transmission to humans
4.1. Virus jumping the species barrier
The SARS-CoV-2 has first been reported from the pneumonia patients of the Wuhan city in Hubei province of China. These patients were involved in trading at a wet animal market in the Huanan area. It is believed that SARS-CoV-2 is introduced from the animal kingdom to human populations during November or December 2019, as revealed from the phylogeny of the genomic sequences from the initially reported cases [94]. The spillover of SARS-CoV-2 from animals to humans took place at the beginning of December 2019 [56], and the clinical cases appeared around ending December [46,95]. Genetic analysis showed that this novel virus is closely related to bat CoVs and is similar but distinct from the SARS virus [43]. Several evidences based on genome sequences, the homology of the ACE2 receptor, and the presence of single intact ORF on gene 8 indicate bats as a natural reservoir of these viruses. However, an unknown animal is yet to be unravelled as an intermediate host [43,46,47,49,56]. Initial investigations on animal source origin of SARS-CoV-2 have inconclusively revealed snakes [27], pangolins, and turtles [96]. The rapid spread of COVID-19 followed the initial animal to human spillover through human-to-human transmission. Genetic epidemiology had revealed that the spread from the beginning of December when the first cases were retrospectively traced in Wuhan was mainly by a human-to-human transmission and not due to continued spillover [56]. These species cross jumping, spillover, and rapid transmission events are linked to viral characteristics, host diversity, and environmental feasibility.
Coronaviruses being RNA viruses have high mutation rates that, besides creating new strains, enable them to adapt to a wide range of hosts. Hence, based on genome sequences, all known human CoVs have emerged from animal sources [97]. This seventh member of the human CoV has also been isolated initially from the pneumonia patients who were having direct or indirect links to the Huanan seafood market in Wuhan China, wherein other animals were also being sold [98]. These include a 49-year-old lady retailer in this wet animal market, a 61-year-old frequent visitor to this market, and a 32-year-old man [74,98]. Further, isolation of the SARS-CoV-2 from the environmental samples around this market, including people, animals, soil, discharges, or structures, strengthens the claims of involvement of hosts either as a reservoir or intermediate [26,52,99].
Recently, a Pomeranian dog as a probable intermediate host was identified; however, such reports are yet to be validated, and research is underway to explore the emergence of this infectious disease at the animal-human interface [29,99]. Multiple substitutions were observed in ACE2 receptors of a dog [96]. In this context, the Pomeranian dog of the infected owner found positive for COVID-19 suggest the permissiveness of the species for SARS-CoV-2 as a result of species jumping [99,100]. Among the fifteen dogs tested from different households with confirmed human COVID-19 cases in Hong Kong SAR, two dogs were found to be infected with SARS-CoV-2. The diagnosis was made using quantitative RT–PCR, serology, and viral genome sequencing. Virus isolation was also done from the samples obtained from one dog. The genetic sequences of viruses obtained from the two dogs were identical to the ones that were detected from their human cases indicating human-to-dog transmission [101].
Moreover, a study reported that the SARS-CoV-2 might infect the cats and further transmitted by the infected cat to other cats [102]. One cat was tested positive for SARS-CoV-2 in France that showed mild respiratory and digestive signs. The cat was tested positive by RT-qPCR on the rectal swab, and serological analysis identified the presence of antibodies against SARS-CoV-2. Genome analysis further confirmed that the SARS-CoV-2 isolated from the cat belongs to the phylogenetic clade A2a seen in French human indicating human-to-animal transmission [103]. This is not the first time that a domestic cat has been found susceptible to zoonotic coronavirus. During the 2003 SARS-CoV outbreak, domestic cats were tested positive for SARS-CoV that were living near SARS infected humans [21,104]. Even though experimental evidence indicates the possibility of SARS-CoV-2 transmission from infected to a susceptible cat close, SARS-CoV-2 transmission between cats or cat-to-humans are not reported under natural conditions [21].
Furthermore, along with dogs and cats, the zoo animals like tigers and lions were also reported to get the SARS-CoV-2 infection and exhibit clinical signs such as vomiting, diarrhoea, dry cough, breathing difficulty and wheezing [105,106]. Spillover of SARS-CoV-2 was also reported in mink farms of Netherlands, further increasing the concern of transmission to humans. Outbreaks of SARS-CoV-2 were reported in two mink farms holding 12,000 and 7500 animals. The virus is suspected to be introduced by a farmworker having COVID-19 [107,108].
Host-pathogen interactions and pathogenesis determine the severity and expression of disease [30,[109], [110], [111]]. Adaptation over time reduces the severity of infection as happened with HCoVs; however, the emergence of novel viruses or strains due to genetic alterations or recombinations can enhance hardness producing novel diseases like COVID-19 [111,112]. Evolutionarily, the balance of viral-human interaction and immune response against virus enables adaptation, thereby persistence in a host without severe or symptomatic disease when the aggravated pathogenesis results in mortality. Hence, loss of sustainable hosts and transmission to novel hosts becomes inevitable for future sustainability [111,112]. A pathogen cannot kill all its hosts, and for future sustainability, it adapts to some suitable host or spills over to a new host.
4.2. Zoonotic spillover
SARS-CoV-2 has been implicated to be originated from animals, and associated with animal linkages, spillover events, cross-species barrier jumping and zoonosis [9,20,27,[29], [30], [31],113]. Since the beginning of 2002 till the end of 2019, three coronaviruses viz. SARS-CoV, MERS-CoV, and SARS-CoV-2 have caused havoc in the human population globally and will continue to do so. Earlier identified betacoronaviruses (SARS-CoV and MERS-CoV) were reported in Guangdong province of China in November 2002 and Saudi Arabia in 2012, respectively [114]. SARS-CoV-2 is the third zoonotic betacoronaviruses recognized in this century.
However, the CFR of the SARS-CoV-2 is lower to date when compared with SARS and MERS. It should not be overlooked as many asymptomatic cases may remain undiagnosed due to the unavailability of diagnostic kits in China. With nearly 0.62 million deaths till the preparation of the manuscript, SARS-CoV-2 is proven to be deadliest as far as the number of deaths is concerned in comparison with SARS-CoV and MERS-CoV with 774 and 858 associated deaths, respectively [115,116]. Earlier, COVID-19 was linked with the exposure to the Huanan seafood market. However, individuals with no history of exposure above were also diagnosed with the illness, further supporting the human to human spread through droplets produced by cough and sneeze [66]. The spread of COVID-19 that occurred with a high pace and lack of transparency in reporting the disease by the Chinese Health Ministry and failure in the timely implementation of preventive measures has been considered as the primary contributor as stated earlier in SARS [19,117]. Both SARS-CoV and SARS-CoV-2 showed prominent similarities in their pathogenesis and epidemics. In both cases, bats were considered as the natural host, and the cold temperature and low humidity in cold, dry winter provided conducive environmental conditions that promoted the survival of the virus in the environment [62]. Further, Moriyama et al. [118] assessed the significance of the environmental factor on host immune system targeting innate and adaptive both responses in the respiratory tract.
Zoonotic spillover is the transmission of pathogens to humans from vertebrate animals [119]. At present, these spillovers are of significant concern as in the past, many spillovers in the form of Nipah, Hendra, Ebola, SARS, MERS, and ongoing COVID-19 involving many animal species like pigs, horses, monkeys, camels, civets, among others, were documented. Bovine CoVs have been reported to infect children and thus possess zoonotic potential [9,13,120,121]. Spillover is governed by the interaction of viral-specific proteins like S protein and host ACE2 receptor [30,31,111,112]. These S proteins have RBD in CoVs, which contain receptor binding motifs (RBM) that help in specific binding to host ACE2 receptors [47,122]. Mutations in amino acid sequences of RBDs results in a change in specificity of a receptor, interaction and binding, hence alteration in transmissibility, pathogenicity and cross-species jumping with a predisposition to novel and more severe diseases [110,123]. In the case of SARS-CoV-2, RBD of S protein has 10–15 times affinity ACE2R [88,110]. It has furin recognition sequence “RRAR” at the S1–S2 cleaving site that represents a functional site for the cellular serine protease TMPRSS2 thus increasing the efficiency of transmission and contagiousness [88,90,110]. In addition to enhanced binding affinity, electrostatic complementarity and hydrophobic interactions are critical to enhancing receptor binding and escaping antibody recognition by the RBD of SARS-CoV-2, thereby further increasing transmission capability and contagiousness [110].
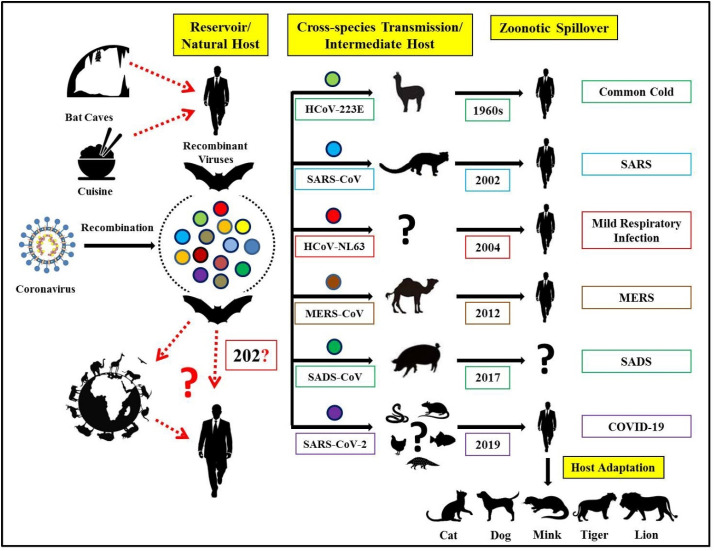
A detailed investigation regarding the emergence of new coronavirus, host range, and transmissibility is crucial to understand such pandemics shortly. The literature revealed that before the appearance of SARS-CoV and MERS-CoV, human coronavirus (HCoV) strains like HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1 were the CoVs strains producing mild infections in humans. However, their natural ancestral hosts were of animal origin, like bats for HCoV-NL63, and HCoV-229E and rodents were natural hosts for HCoV-OC43 and HKU1. These four HCoVs were initially of low pathogenicity. To enhance the pathogenicity, they used intermediate hosts such as cattle for HCoV-OC43 (natural host was rodent), and alpacas for HCoV-229E (bats were natural host) and this way acquired the ability to infect human beings with serious health hazards [124].
Added to the involvement of bats and pangolins, the recent reports revealing SARS-CoV-2 infection in cats, dogs, tigers, lions and minks have raised concerns over this virus affecting multiple animal species, and also points out towards the incidences of reverse zoonosis [31,102,106,108,125,126]. The ferrets, cats, and primates are suggested to be good candidates for susceptibility to SARS-CoV-2 [123,127]. COVID-19 research and surveillance in companion and pet animals, livestock animals, zoo animal species, wildlife animal species as well as their handlers, veterinarians, and owners need to be enhanced during the pandemic, which would help to follow better integrated one health strategies [128] and appropriate preventive and mitigation to counter SARS-CoV-2 effectively [29,126,[129], [130], [131], [132], [133]]. Significance of COVID-19 monitoring and implementation of suitable public health measures among workers involved in meat and poultry processing facilities/industries has been emphasized, which would protect them as well as aid in preserving the critical meat and poultry production infrastructure and the meat products [134].
4.3. Transmission to humans
The involvement of intermediate hosts in maintaining and transmitting the virus to susceptible host predisposes humans to novel CoVs leading to the emergence of new diseases in humans. The currently ongoing SARS-CoV-2/COVID-19 pandemic has put on hold the entire world [111,135]. The CoVs have frequently been associated with animal and human diseases and have a zoonotic interface [97,111]. Usually, one or more types of animal hosts are involved in the transmission cycle of CoVs to humans [30,97]. That can be natural host, reservoir host, intermediate host or definitive host [31]. Bats have been the natural hosts for human CoVs of Alphacoronavirus (HCoV-NL63, HCoV-229E) and Betacoronavirus (SARS-CoV, MERS-CoV, SARS-CoV-2) genera whereas for Betacoronavirus members HCoV-OC43 and HCoV-HKU1, rodents are the natural hosts. Genome sequence analysis has revealed bats as a natural host for SARS-CoV-2 [30,97]. In natural or reservoir hosts, CoVs adapts well, however, being unstable RNA viruses, they keep multiplying continuously without producing disease thereby enabling persistence or survivability and accumulation of mutations over the time resulting in the emergence of newer and novel strains of viruses [97,111,136]. These unique strains or viruses occasionally spill over to other species including animals or humans, adapting to their body systems and hence broaden the biological host range for evolutionary sustainability; however, results in epidemiological widening of disease sphere as well [25,31]. This transmission and adaptation scenario initiates a host-pathogen response resulting in the novel usually severe diseases that can at times be fatal in initial stages or over extended periods until virus pathogen adapts to host or the host develops sufficient immune defence [137,138].
It has been reported that almost all HCoVs have originated from animals like bats (SARS-CoV, MERS-CoV, HCoV-NL63, and HCoV-229E) and rodents (HCoV-OC43 and HKU1) [139,140]. Additionally, CoVs have been reported to infect several species of domestic and wild animals either clinically or subclinically [27,28,30,141]. Cattle, horses, camels, swine, dogs, cats, birds, rabbits, rodents, ferrets, mink, bats, snakes, frogs, marmots, hedgehogs, Malayan pangolin along with other wild animals may serve as a reservoir host of coronavirus [9,27,44,124,[142], [143], [144], [145], [146], [147]]. In the context of SARS-CoV-2, snakes, pangolins and bats have been suspected as intermediate hosts since the first cases of COVID-19 had links to Huanan Sea Food Market where different animals, birds, and wild animals were being sold along with seafood items [27,28,43,109,147,148]. Coronaviruses have been reported to cause salivary, enteric and respiratory infections in laboratory animals (mice, rat, guinea pig, and rabbit) and urinary tract infection, respiratory illness and reproductive disorder in poultry [9,142]. In bovine, canine, feline and swine CoVs infections have resulted in diarrhoea, enteritis, respiratory illness, gastro-intestinal affections and nervous symptoms [120,121,[149], [150], [151]]. Coronavirus, namely- SW1, has been reported in captive beluga whale using a panviral microarray method [152]. Among all the assumptions on animal hosts as the intermediate host, genomic and evolutionary information from pangolins reveals the highest closeness to the SARS-CoV-2 than any other host CoVs isolates [153]. The spike protein, the main target of many studies searching for a cure of COVID-19, has been found highly similar to SARS-CoV-2 and, thus, could serve as a surrogate system for further evaluations [153].
Bats are the natural reservoir host of many CoVs. As reported earlier, 7 out of 11 alphacoronaviruses and 4 out of 9 betacoronaviruses as per the International Committee on Taxonomy of Viruses (ICTV) classification were solely originated from bats [97,154]. According to the literature, bats have been regarded as a potential wildlife reservoir whereas civets and dromedary camels as intermediate hosts of SARS-CoV and MERS-CoV, respectively [155,156]. The bat coronavirus, BatCoV RaTG13, has shown higher relatedness to SARS-CoV-2 at the whole genome level and spike gene in particular [153]. Coexistence and frequent recombination between highly diversified and prevalent bat SARS-related coronaviruses (SARSr-CoV) and coronaviruses may suggest the probable emergence of novel viruses shortly [157,158]. Benvenuto et al. [159] analyzed the whole genome sequences of different CoVs using Fast Unconstrained Bayesian AppRoximation (FUBAR) to understand the evolutionary and molecular epidemiology of SARS-CoV-2. The authors concluded that SARS-CoV-2 clustered with sequences of bat SARS-like CoVs with a few mutations in nucleocapsid and spike glycoprotein, suggesting its probable transmission from the bats [159]. Bats, especially horseshoe bats (Rhinolophus spp.), are considered to be the known reservoirs of SARS-related CoVs. Since the bat origin, CoVs have always caused outbreaks in humans, studying the diversity and distribution of coronavirus populations in the bats will help to mitigate future outbreaks in humans and animals [160].
Interestingly, bats play a crucial role in all the spillovers mentioned above, indicating their importance in the emergence of new viruses. The reason behind the emergence and broad host range of CoVs in the past and present might be due to unstable RNA-dependent RNA polymerase (RdRp), lack of proof-reading ability, high frequency of mutations in the receptor-binding domain of spike gene and genetic recombination [140,161,162]. Bat CoVs have high diversity and great potential of spillover in different animal species, as reported earlier in civet cat and dromedary camel, leading to well-known pandemics SARS and MERS, respectively along with the recent spillover in pigs resulted in swine acute diarrhoea syndrome (SADS). However, spillover resulted in the emergence of SADS-CoV, which showed a 95% genomic identity with bat coronavirus, which led to severe mortality with 24,693 deaths in neonatal piglets [160]. Fortunately, it did not excel in the form of the third pandemic, and no human cases were reported till date. The spillover responsible for ongoing COVID-19 is still under investigation and a matter of great concern for the researchers all around the globe.
Based on Resampling Similarity Codon Usage (RSCU), snakes (Bungarus multicinctus and Naja atra) were suggested as wildlife reservoirs of SARS-CoV-2 and reported to be associated with the cross-species transmission [27] and later it was disapproved by other researchers [163,164]. Unfortunately, to date, the intermediate host of the SARS-CoV-2 is abstruse what results in its escalation in the human population around the globe. In this context, analyzing the interaction between the Asn501 site in RBD of spike glycoprotein of SARS-CoV-2 and the residue at 41 sites of ACE2 receptor of different hosts (pangolins, turtle, mouse, dog, cat, hamster and bat) revealed that tyrosine has higher receptor binding affinity than histidine suggesting pangolins and turtle be closer than bats to humans and maybe the probable intermediate hosts of SARS-CoV-2 [96]. However, this hypothesis was also contradicted by Li et al. [28] based on an insertion of the unique peptide (PRRA) in the SARS-CoV-2 virus, which was lacking in CoVs from pangolins. Moreover, SARS-CoV-2 showed higher similarity to the BetaCoV/bat/Yunnan/RaTG13/2013 compared to the ones that were isolated from the pangolins, thereby denied the direct link of the virus from pangolins. However, further studies are required to confirm the role of pangolins in SARS-CoV-2 spread to humans.
The receptor-binding domain of the spike protein of SARS-CoV interacts with the host receptor ACE2 facilitating its potential of cross-species, as well as human-to-human transmission [47]. Similarly, the spike protein of SARS-CoV-2 was reported to recognize ACE2 receptors expressed in fish, amphibians, reptiles, birds, and mammals and has a more robust binding capacity (affinity) in comparison to SARS-CoV [89]. This suggests their involvement as probable natural and intermediate hosts [161], which may further help in the selection of animal models for epidemic investigation and preventing its spread [47]. Bat origin CoVs have been found to cross the species barrier that favoured their transmission via recombination/mutations in the RBD. The evidence of a virus outbreak that occurred in Chinese pig farms suggests its possible cross-species [160,165]. Also, murine cells were found permissive for SARS-CoV after substitution of His353 with Lys353 in the ACE2 receptor of a mouse, which suggests the role of residue changes in the cross-species and human-to-human transmission [166]. Mutation in residues at position 479 and 487 of receptor binding motif (RBM) of SARS-CoV was reported to play a role in civet-to-human and human-to-human transmission, respectively [167,168].
The CoVs are more prone to recombination and mutations leading to variable host range, and resemblance of receptors in various hosts results in cross-species jumping [31,112,169]. Genetic divergence due to these genetic alterations results in the evolution of newer viral strains having altered virulence, tissue tropism, and host range [170,171]. Moreover, the presence of threonine at position 487 was reported to enhance the binding affinity of RBM for the ACE2 receptor of civet and humans [172]. However, many SARS-related coronaviruses (SARSr-CoV) have been reported in bats and used ACE2 receptors for entry into a host cell, which showed its potential to infect humans directly without any intermediate host [173]. In addition to this, no direct transmission of SARSr-CoV is reported from bats to humans to date. However, seropositivity on a serological investigation of individuals without prior exposure to SARS-CoV residing near bat caves in China revealed likely infection of humans by bat SARSr-CoV and related viruses [174]. Besides, the interspecies transmission potential of SARSr-CoVs is due to the ORF8 gene [175].
As per reports, the SARS-CoV emerged via recombination of bat SARSr-CoVs, was transmitted to farmed civets along with other mammals, and these infected civets spread the virus to market civets. The virus was reported to undergo mutations in infected market civets before its spillover to humans. Similarly, the MERS-CoV circulated for 30 years in camels before the pandemic [97,176] supporting the hypothesis that after species jumping the exogenous viruses opted for adaptation to the environment and host before spillover to humans [177]. Moreover, the possible spillover of other circulating bat SARSr-CoVs to humans from mammalian hosts soon is highly anticipated.
The cross-species jumping and adaptation are determined by the presence of specific receptors on host tissues (like ACE 2 receptor for HCoV-NL63, SARS-CoV and SARS-CoV-2, dipeptidyl peptidase-4 for MERS-CoV, human aminopeptidase N for HCoV-229E, 9-O-acetylsialic acids for HCoV-OC43, HCoV-HKU1) which help in binding and entry of the virus into host cells [30,31]. These receptors are present in various body systems in animals and humans, including respiratory and gastrointestinal systems [30]. Reservoir host animals including bats and rodents possess these receptors which are similar to those present in camels, masked palm civets (Paguma larvata), or bovines, that act as an intermediate host for different CoVs [30,110,111]. Presence of some of these receptors in humans like ACE2 or DPP4 makes them vulnerable to CoV infection like SARS-CoV and MERS-CoV causing SARS and MERS infections, respectively [31,168]. The MERS-CoV spike was found to possess the capacity for adapting to species variation in the host receptor DPP4 [37]. The mechanism expressed by MERS-CoV in adapting to infect cells of new species might be present in the other coronaviruses. ACE2 has also been found as a binding receptor for SARS-CoV-2 [47]. The species-specific variations in the host receptors limit the interaction with CoV spike protein, and this is responsible for the development of the species barrier that prevents spillover infection. Snakes, civets, and pangolins are considered as the potential intermediate hosts of COVID-19. However, further confirmation is required by tracking the origin of the virus. This is critical for preventing additional exposure to this fatal virus [178].
The probability of the SARS-CoV-2 spread during incubation and convalescent period has been suggested [76]. As per reports, presence of CoVs has been observed in respiratory droplets, body fluids and inanimate objects with the ability to remain infectious for nine days on contaminated surfaces resulting in its risk of self-inoculation via mucous membranes of the eyes, mouth or nose [[179], [180], [181]]. Nosocomial, as well as human-to-human transmission, have been reported to occur via virus-laden aerosols, contaminated hands or surfaces, and close community contact with an infected person [66,182,183]. The ocular route has been reported in the human-to-human transmission of SARS-CoV-2, as observed in SARS-CoV, suggesting the involvement of different ways other than the respiratory tract [184,185]. Later on, the probability of the faecal-oral route for potential transmission of the virus was also suggested [186]. The metatranscriptome sequencing of SARS-CoV-2 in the bronchoalveolar lavage fluid (BALF) of infected individuals resulted in polymorphism in few intra-hosts variants, suggesting the in vivo evolution of the virus thereby affecting its virulence, transmissibility, and infectivity [187].
An overview of coronaviruses jumping the cross-species barriers, zoonotic CoVs transmitted from bats to animals before spillover to humans, and possible prospects for further transmission to mammalian hosts is depicted in Fig. 1 .
Fig. 1.
Cross-species transmission of known zoonotic coronaviruses from bats to animals before spillover to humans and probable prospects of further transmission to mammalian hosts.
5. One health approach
The first two decades (2000–2020) of 21st century have proven a nightmare for the countries around the globe considering the coronavirus zoonosis, including the ongoing crisis of COVID-19 which has involved entire fields of global [188,189]. The countries affected severely by previous CoVs were even not evolved entirely from the effects of SARS and MERS when the COVID-19 struck almost the entire world. Novel coronavirus SARS-CoV-2 has shaken all the sectors of the countries irrespective of being developed or underdeveloped including healthcare system, economics, trade, infrastructure, service and production sectors [190,191].
Being a zoonotic disease with still unknown intermediate host, undisclosed features of a novel viral pathogen, unclear modes of transmission and ecological aspects, less explored pathogenesis and substantial morbidity and considerable mortality, the safety of all is a matter of great concern, and thus the involvement of various authorities was sought since the inception of disease [26,31,192,193]. The first time the need for One Health concept has risen to a level that authorities in various countries implemented coordinated approaches between medical, veterinary, public health, wildlife, food safety, environmental departments and so on [[193], [194], [195]]. That involved acquiring suggestions, diagnosis and prevention and treatment measures and their implementation in collaboration. Non-medical staff in association with the medical staff was employed for initial screening, quarantine, contact tracing when the expertise of molecular biologists or technicians from various disciplines was used in the laboratory diagnosis. Medical staff provided the cure and management of the patients when the public health departments, including public health engineering, municipality, food and supplies ensured sanitation, hygiene, food supply and safety. Imposing of lockdown was provided by security personnel's and the transport department facilitated the movement of stranded people. Thus, this crises management strategy involved various agencies directly or indirectly. However, as the animal, human and environmental health is linked to one another, the prime and future efforts should primarily focus on all these aspects. In addition to regular hand hygiene, respiratory etiquette, social/physical distancing, use of personnel protective equipment (PPE) and food safety recommendations, One Health approach encompasses the role of veterinary, medical and environmental specialists for the prevention and control of current COVID-19 crises and investigating the animal origin of COVID-19, regulating and limiting the sale and farming of wildlife species for food and taking a One Health approach to food systems feeding the world for the prevention of future pandemics [193,196,197].
Considering the contagiousness of the virus, discouraging the working of affected individuals, public health hygiene strategies, and social distancing has been recommended as preventive measures [26,198]. Food hygiene and safety, as recommended by OIE [193] and USDA [199], should be followed. As the viral survivability has been demonstrated on various surfaces [53] hence disinfection by using recommended disinfectants is necessary [200]. Environmental hygiene and cleanliness are also essential [200]. Interaction with animals and improper utilization of animal products during an outbreak should be avoided [193]. Though the One Health involves mainly public health, animal health and environmental experts, however, for the successful management of current crises and future prevention and control requires the participation of all concerned sectors having a role in public health measures, identifying clinical cases, diagnosis, contact tracing, proper infection control in various settings, isolation, quarantine, cure and management, public awareness, facilitation of infrastructure and other facilities through local administrations [26,195]. As the human COVID-19 cases are on the rise due to efficient human-to-human transmission, there is a subsequent rise in the natural infections of COVID-19 among the companion and wild animal species owing to the spillover. This is mainly because of the specific biological and virological characteristics of coronaviruses that gives them the ability to easily cross-species barriers [130]. Even though animal-to-human transmission is not reported in COVID-19, ‘one health’ approach is necessary to control this pandemic virus.
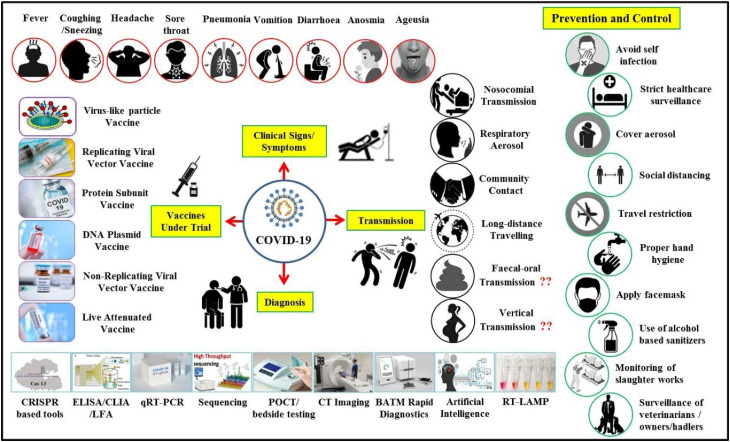
A schematic illustration of COVID-19 clinical signs, modes of transmission, important diagnostic methods, and advances in vaccine development along with salient prevention and control strategies are presented in Fig. 2 .
Fig. 2.
Schematic illustration of clinical signs, modes of transmission, advances in diagnosis, and vaccine designing along with salient prevention and control strategies to counter COVID-19.
6. Conclusions
With the rising number and worldwide spread of COVID-19, the need for global efforts rely heavily on the investigations carried out at infection sites to trace different aspects of this novel coronavirus outbreak. One of the critical facets and the earliest research must involve determining the root cause, origin, and source of this emerging infectious disease. Shreds of evidence have revealed various cross-species jumping or spillover from animals to humans of these zoonotic coronaviruses. Detailed serological investigation of all domestic and wild animals residing in the proximity to humans is of utmost necessity to know and prevent likely spillover of many other bat-related CoVs in the future. Rapid detection of spillovers above will only be possible by the implementation of an effective and robust surveillance system for circulating viruses with high zoonotic potential in animals. Besides, detection of a pathogen while crossing the species barrier to start circulation among humans and prevention of human-to-human transmission in early-stage may prove crucial in termination of a probable epidemic or pandemic. Application of ‘One Health’ concept involving medical, veterinary, wildlife, public health, and other related professionals may help in infection tracing, exploring risk factors and predisposition, minimizing risk to susceptible ones, and finally devising better prevention and control strategies.
In the initial stages of the COVID-19 outbreak, the steps taken for implementing stringent control and preventive measures have bought us some time. This time has to be efficiently utilized for developing SARS-CoV-specific therapeutic drugs and vaccines that can prevent the further spread of this fatal pathogen. For the time being early detection, isolation, and management of COVID-19 infected cases and for the immediate future awareness and implementation of the necessary steps of prevention and control to under risk and predisposed groups may help in long term prevention and control goals. However, it is clearly understood that relying exclusively on public health measures will not resolve the threat caused by COVID-19. Hence efforts to curb this emerging zoonosis at all levels need to be enforced under the One Health approach.
Author contributions
All the authors substantially contributed to the conception, design, analysis, and interpretation of data, checking and approving the final version of the manuscript, and agree to be accountable for its contents.
Funding
None.
Declaration of competing interest
None.
Acknowledgements
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tmaid.2020.101830.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Momattin H., Al-Ali A.Y., Al-Tawfiq J.A. A systematic review of therapeutic agents for the treatment of the Middle East respiratory syndrome coronavirus (MERS-CoV) Trav Med Infect Dis. 2019;30:9–18. doi: 10.1016/j.tmaid.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baharoon S., Memish Z.A. MERS-CoV as an emerging respiratory illness: a review of prevention methods. Trav Med Infect Dis. 2019:101520. doi: 10.1016/j.tmaid.2019.101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfaraj S.H., Al-Tawfiq J.A., Assiri A.Y., Alzahrani N.A., Alanazi A.A., Memish Z.A. Clinical predictors of mortality of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infection: a cohort study. Trav Med Infect Dis. 2019;29:48–50. doi: 10.1016/j.tmaid.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardona-Ospina J.A., Giselle-Badillo A., Calvache-Benavides C.E., Rodriguez-Morales A.J. Ebola virus disease: an emerging zoonosis with importance for travel medicine. Trav Med Infect Dis. 2014;12:682–683. doi: 10.1016/j.tmaid.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Nishiura H., Mizumoto K., Villamil-Gomez W.E., Rodriguez-Morales A.J. Preliminary estimation of the basic reproduction number of Zika virus infection during Colombia epidemic, 2015-2016. Trav Med Infect Dis. 2016;14:274–276. doi: 10.1016/j.tmaid.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Morales A.J. Zika and microcephaly in Latin America: an emerging threat for pregnant travelers? Trav Med Infect Dis. 2016;14:5–6. doi: 10.1016/j.tmaid.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Morales A.J., Ramirez-Vallejo E., Alvarado-Arnez L.E., Paniz-Mondolfi A., Zambrano L.I., Ko A.I. Fatal Zika virus disease in adults: a critical reappraisal of an under-recognized clinical entity. Int J Infect Dis. 2019;83:160–162. doi: 10.1016/j.ijid.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Dhama K., Karthik K., Khandia R., Chakraborty S., Munjal A., Latheef S.K., Kumar D., Ramakrishnan M.A., Malik Y.S., Singh R., Malik S.V.S., Singh R.K., Chaicumpa W. Advances in designing and developing vaccines, drugs, and therapies to counter Ebola virus. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.01803. 1803-03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhama K., Khan S., Tiwari R., Sircar S., Bhat S., Malik Y.S., Singh K.P., Chaicumpa W., Bonilla-Aldana D.K., Rodriguez-Morales A.J. Coronavirus disease 2019–COVID-19. Clin Microbiol Rev. 2020;33 doi: 10.1128/CMR.00028-20. e00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhama K., Verma A.K., Rajagunalan S., Deb R., Karthik K., Kapoor S., Mahima, Tiwari R., Panwar P.K., Chakraborty S. Swine flu is back again: a review. Pakistan J Biol Sci. 2012;15:1001–1009. doi: 10.3923/pjbs.2012.1001.1009. [DOI] [PubMed] [Google Scholar]
- 11.Malik Y.S., Sircar S., Bhat S., Sharun K., Dhama K., Dadar M., Tiwari R., Chaicumpa W. Emerging novel coronavirus (2019-nCoV)-current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q. 2020;40:68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munjal A., Khandia R., Dhama K., Sachan S., Karthik K., Tiwari R., Malik Y.S., Kumar D., Singh R.K., Iqbal H.M.N., Joshi S.K. Advances in developing therapies to combat Zika virus: current knowledge and future perspectives. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01469. 1469-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh R.K., Dhama K., Chakraborty S., Tiwari R., Natesan S., Khandia R., Munjal A., Vora K.S., Latheef S.K., Karthik K., Singh Malik Y., Singh R., Chaicumpa W., Mourya D.T. Nipah virus: epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies - a comprehensive review. Vet Q. 2019;39:26–55. doi: 10.1080/01652176.2019.1580827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein R.A. The 2019 coronavirus: learning curves, lessons, and the weakest link. Int J Clin Pract. 2020;74 doi: 10.1111/ijcp.13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranabothu S., Onteddu S., Nalleballe K., Dandu V., Veerapaneni K., Veerapandiyan A. Spectrum of COVID-19 in children. Acta Paediatr. 2020 doi: 10.1111/apa.15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azamfirei R. The 2019 novel coronavirus: a crown jewel of pandemics? Journal of critical care medicine. 2020;6:3–4. doi: 10.2478/jccm-2020-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen J., Kupferschmidt K. Strategies shift as coronavirus pandemic looms. Science. 2020;367:962. doi: 10.1126/science.367.6481.962. [DOI] [PubMed] [Google Scholar]
- 18.Khan S., Siddique R., Ali A., Xue M., Nabi G. Novel coronavirus, poor quarantine, and the risk of pandemic. J Hosp Infect. 2020;104:449–450. doi: 10.1016/j.jhin.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peeri N.C., Shrestha N., Rahman M.S., Zaki R., Tan Z., Bibi S., Baghbanzadeh M., Aghamohammadi N., Zhang W., Haque U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int J Epidemiol. 2020 doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez-Morales A.J., Bonilla-Aldana D.K., Balbin-Ramon G.J., Rabaan A.A., Sah R., Paniz-Mondolfi A., Pagliano P., Esposito S. History is repeating itself: probable zoonotic spillover as the cause of the 2019 novel Coronavirus Epidemic. Infez Med. 2020;28:3–5. [PubMed] [Google Scholar]
- 21.Rodriguez-Morales A.J., Dhama K., Sharun K., Tiwari R., Bonilla-Aldana D.K. Susceptibility of felids to coronaviruses. Vet Rec. 2020;186:e21. doi: 10.1136/vr.m1671. [DOI] [PubMed] [Google Scholar]
- 22.Watts C.H., Vallance P., Whitty C.J.M. Coronavirus: global solutions to prevent a pandemic. Nature. 2020;578:363. doi: 10.1038/d41586-020-00457-y. [DOI] [PubMed] [Google Scholar]
- 23.Dhama K., Sharun K., Tiwari R., Dadar M., Malik Y.S., Singh K.P., Chaicumpa W. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccines Immunother. 2020;16:1232–1238. doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik Y.S.K.N., Sircar S., Kaushik R., Bhat S., Dhama K., Gupta P., Goyal K., Singh M.P., Ghoshal U., El Zowalaty M.E., O R.V., Yatoo M.I., Tiwari R., Pathak M., Patel S.K., Sah R., Rodriguez-Morales A.J., Ganesh B., Kumar P., Singh R.K. Coronavirus disease pandemic (COVID-19): challenges and a global perspective. Pathogens. 2020;9 doi: 10.3390/pathogens9070519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia Ja, Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Organization Wh . 2020. Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19) pp. 1–40. [Google Scholar]
- 27.Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Zai J., Zhao Q., Nie Q., Li Y., Foley B.T., Chaillon A. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J Med Virol. 2020;92:602–611. doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murdoch D.R., French N.P. COVID-19: another infectious disease emerging at the animal-human interface. N Z Med J. 2020;133:12–15. [PubMed] [Google Scholar]
- 30.Salata C., Calistri A., Parolin C., Palù G. Coronaviruses: a paradigm of new emerging zoonotic diseases. Pathogens and Disease. 2020;77 doi: 10.1093/femspd/ftaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiwari R., Dhama K., Sharun K., Iqbal Yatoo M., Malik Y.S., Singh R., Michalak I., Sah R., Bonilla-Aldana D.K., Rodriguez-Morales A.J. COVID-19: animals, veterinary and zoonotic links. Vet Q. 2020;40:169–182. doi: 10.1080/01652176.2020.1766725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letko M., Seifert S.N., Olival K.J., Plowright R.K., Munster V.J. Bat-borne virus diversity, spillover and emergence. Nat Rev Microbiol. 2020;18:461–471. doi: 10.1038/s41579-020-0394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank H.K., Enard D., Boyd S.D. bioRxiv; 2020. Exceptional diversity and selection pressure on SARS-CoV and SARS-CoV-2 host receptor in bats compared to other mammals. 2020.04.20.051656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park M., Thwaites R.S., Openshaw P.J.M. COVID-19: lessons from SARS and MERS. Eur J Immunol. 2020;50:308–311. [Google Scholar]
- 35.Dong R., Pei S., Yin C., He R.L., Yau S.S.T. Analysis of the hosts and transmission paths of SARS-CoV-2 in the COVID-19 outbreak. Genes (Basel) 2020;11:637. doi: 10.3390/genes11060637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letko M., Miazgowicz K., McMinn R., Seifert S.N., Sola I., Enjuanes L., Carmody A., van Doremalen N., Munster V. Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 2018;24:1730–1737. doi: 10.1016/j.celrep.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu H., Stratton C.W., Tang Y.-W. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franklin A.B., Bevins S.N. Spillover of SARS-CoV-2 into novel wild hosts in North America: a conceptual model for perpetuation of the pathogen. Sci Total Environ. 2020;733 doi: 10.1016/j.scitotenv.2020.139358. 139358-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayittey F.K., Ayittey M.K., Chiwero N.B., Kamasah J.S., Dzuvor C. Economic impacts of Wuhan 2019-nCoV on China and the world. J Med Virol. 2020;92:473–475. doi: 10.1002/jmv.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren S.-Y., Gao R.-D., Chen Y.-L. Fear can be more harmful than the severe acute respiratory syndrome coronavirus 2 in controlling the corona virus disease 2019 epidemic. World J Clin Cases. 2020;8:652–657. doi: 10.12998/wjcc.v8.i4.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paniz-Mondolfi A.E., Sordillo E.M., Marquez-Colmenarez M.C., Delgado-Noguera L.A., Rodriguez-Morales A.J. The arrival of SARS-CoV-2 in Venezuela. Lancet. 2020;395:e85–e86. doi: 10.1016/S0140-6736(20)31053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Inf Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han Q., Lin Q., Jin S., You L. Coronavirus 2019-nCoV: a brief perspective from the front line. J Infect. 2020;80:373–377. doi: 10.1016/j.jinf.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren L.-L., Wang Y.-M., Wu Z.-Q., Xiang Z.-C., Guo L., Xu T., Jiang Y.-Z., Xiong Y., Li Y.-J., Li X.-W., Li H., Fan G.-H., Gu X.-Y., Xiao Y., Gao H., Xu J.-Y., Yang F., Wang X.-M., Wu C., Chen L., Liu Y.-W., Liu B., Yang J., Wang X.-R., Dong J., Li L., Huang C.-L., Zhao J.-P., Hu Y., Cheng Z.-S., Liu L.-L., Qian Z.-H., Qin C., Jin Q., Cao B., Wang J.-W. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl). 2020;133:1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94 doi: 10.1128/JVI.00127-20. e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu P., Hao X., Lau E.H.Y., Wong J.Y., Leung K.S.M., Wu J.T., Cowling B.J., Leung G.M. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Euro Surveill. 2020;25:2000044. doi: 10.2807/1560-7917.ES.2020.25.3.2000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. 102433-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian H.Y. [2019-nCoV: new challenges from coronavirus] Zhonghua yu fang yi xue za zhi [Chinese journal of preventive medicine] 2020;54:E001. doi: 10.3760/cma.j.issn.0253-9624.2020.0001. [DOI] [PubMed] [Google Scholar]
- 51.Control ECfDPa . 2020. Interim guidance for environmental cleaning in non-healthcare facilities exposed to SARS-CoV-2. Stockholm. [Google Scholar]
- 52.Prevention CfDCa . 2020. Coronavirus disease 2019 (COVID-19) [Google Scholar]
- 53.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y.-H., Dong J.-H., An W.-M., Lv X.-Y., Yin X.-P., Zhang J.-Z., Dong L., Ma X., Zhang H.-J., Gao B.-L. Clinical and computed tomographic imaging features of novel coronavirus pneumonia caused by SARS-CoV-2. J Infect. 2020;80:394–400. doi: 10.1016/j.jinf.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dhama K., Patel S.K., Pathak M., Yatoo M.I., Tiwari R., Malik Y.S., Singh R., Sah R., Rabaan A.A., Bonilla-Aldana D.K., Rodriguez-Morales A.J. An update on SARS-CoV-2/COVID-19 with particular reference to its clinical pathology, pathogenesis, immunopathology and mitigation strategies. Trav Med Infect Dis. 2020 doi: 10.1016/j.tmaid.2020.101755. 101755-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heymann D.L., Shindo N., Scientific WHO, Technical Advisory Group for Infectious H COVID-19: what is next for public health? Lancet. 2020;395:542–545. doi: 10.1016/S0140-6736(20)30374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Military Medical Research. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roussel Y., Giraud-Gatineau A., Jimeno M.-T., Rolain J.-M., Zandotti C., Colson P., Raoult D. SARS-CoV-2: fear versus data. Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105947. 105947-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bastola A., Sah R., Rodriguez-Morales A.J., Lal B.K., Jha R., Ojha H.C., Shrestha B., Chu D.K.W., Poon L.L.M., Costello A., Morita K., Pandey B.D. The first 2019 novel coronavirus case in Nepal. Lancet. 2020;20:279–280. doi: 10.1016/S1473-3099(20)30067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y.Y., Wang W.N., Lei Y., Zhang B., Yang J., Hu J.W., Ren Y.L., Lu Q.F. [Comparison of the clinical characteristics between RNA positive and negative patients clinically diagnosed with coronavirus disease 2019] Zhonghua jie he he hu xi za zhi = Zhonghua jiehe he huxi zazhi = Chinese journal of tuberculosis and respiratory diseases. 2020;43:427–430. doi: 10.3760/cma.j.cn112147-20200214-00095. [DOI] [PubMed] [Google Scholar]
- 62.Sun P., Qie S., Liu Z., Ren J., Jianing Xi J. Clinical characteristics of 50466 patients with 2019-nCoV infection. medRxiv. 2020 2020.02.18.20024539. [Google Scholar]
- 63.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in wuhan, China. J Am Med Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ollarves-Carrero M.F., Rodriguez-Morales A.G., Bonilla-Aldana D.K., Rodriguez-Morales A.J. Anosmia in a healthcare worker with COVID-19 in Madrid, Spain. Trav Med Infect Dis. 2020;35 doi: 10.1016/j.tmaid.2020.101666. 101666-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., Ng O.-T., Marimuthu K., Ang L.W., Mak T.M., Lau S.K., Anderson D.E., Chan K.S., Tan T.Y., Ng T.Y., Cui L., Said Z., Kurupatham L., Chen M.I.C., Chan M., Vasoo S., Wang L.-F., Tan B.H., Lin R.T.P., Lee V.J.M., Leo Y.-S., Lye D.C. For the Singapore novel coronavirus outbreak research T. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. J Am Med Assoc. 2020;323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y.-C., Bai W.-Z., Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol. 2020;92:552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guan W-j, Ni Z-y, Hu Y., Liang W-h, Ou C-q, He J-x, Liu L., Shan H., Lei C-l, Hui D.S.C., Du B., Li L-j, Zeng G., Yuen K.-Y., Chen R-c, Tang C-l, Wang T., Chen P-y, Xiang J., Li S-y, Wang J-l, Liang Z-j, Peng Y-x, Wei L., Liu Y., Hu Y-h, Peng P., Wang J-m, Liu J-y, Chen Z., Li G., Zheng Z-j, Qiu S-q, Luo J., Ye C-j, Zhu S-y, Zhong N-s. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cai J., Sun W., Huang J., Gamber M., Wu J., He G. Indirect virus transmission in cluster of COVID-19 cases, wenzhou, China, 2020. Emerg Infect Dis. 2020;26:1343–1345. doi: 10.3201/eid2606.200412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S., Tsoi H.-W., Lo S.K.-F., Chan K.-H., Poon V.K.-M., Chan W.-M., Ip J.D., Cai J.-P., Cheng V.C.-C., Chen H., Hui C.K.-M., Yuen K.-Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lai A., Bergna A., Acciarri C., Galli M., Zehender G. Early phylogenetic estimate of the effective reproduction number of SARS-CoV-2. J Med Virol. 2020;92:675–679. doi: 10.1002/jmv.25723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoehl S., Rabenau H., Berger A., Kortenbusch M., Cinatl J., Bojkova D., Behrens P., Böddinghaus B., Götsch U., Naujoks F., Neumann P., Schork J., Tiarks-Jungk P., Walczok A., Eickmann M., Vehreschild M.J.G.T., Kann G., Wolf T., Gottschalk R., Ciesek S. Evidence of SARS-CoV-2 infection in returning travelers from wuhan, China. N Engl J Med. 2020;382:1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Y., Eggo R.M., Kucharski A.J. Secondary attack rate and superspreading events for SARS-CoV-2. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30462-1. e47-e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yee J., Unger L., Zadravecz F., Cariello P., Seibert A., Johnson M.A., Fuller M.J. Novel coronavirus 2019 (COVID-19): emergence and implications for emergency care. JACEP. 2020;1:63–69. doi: 10.1002/emp2.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y., Wang Y., Chen Y., Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. 2020;92:568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao Q.Y., Chen Y.X., Fang J.Y. Novel coronavirus infection and gastrointestinal tract. Journal of Digestive Diseases. 2019;21:125–126. doi: 10.1111/1751-2980.12851. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turban J.L., Keuroghlian A.S., Mayer K.H. Sexual health in the SARS-CoV-2 era. Ann Intern Med. 2020:M20–M2004. doi: 10.7326/M20-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.D'Antiga L. Coronaviruses and immunosuppressed patients: the facts during the third epidemic. Liver Transplant. 2020;26:832–834. doi: 10.1002/lt.25756. [DOI] [PubMed] [Google Scholar]
- 80.Yang H., Wang C., Poon L.C. Novel coronavirus infection and pregnancy. Ultrasound Obstet Gynecol. 2020;55:435–437. doi: 10.1002/uog.22006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Y., Zhao R., Zheng S., Chen X., Wang J., Sheng X., Zhou J., Cai H., Fang Q., Yu F., Fan J., Xu K., Chen Y., Sheng J. Lack of vertical transmission of severe acute respiratory syndrome coronavirus 2, China. Emerg Infect Dis. 2020;26:1335–1336. doi: 10.3201/eid2606.200287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang L., Shi Y., Xiao T., Fu J., Feng X., Mu D., Feng Q., Hei M., Hu X., Li Z., Lu G., Tang Z., Wang Y., Wang C., Xia S., Xu J., Yang Y., Yang J., Zeng M., Zheng J., Zhou W., Zhou X., Zhou X., Du L., Lee S.K., Zhou W., Working Committee on P. Neonatal Management for the P, Control of the Novel Coronavirus I. Chinese expert consensus on the perinatal and neonatal management for the prevention and control of the 2019 novel coronavirus infection (First edition) Ann Transl Med. 2020;8 doi: 10.21037/atm.2020.02.20. 47-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lei J., Li J., Li X., Qi X. CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295 doi: 10.1148/radiol.2020200236. 18-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. J Am Med Assoc. 2020;323:1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- 86.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microb Infect. 2020;22:72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoffmann M., Kleine-Weber H., Krüger N., Müller M., Drosten C., Pöhlmann S. bioRxiv; 2020. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. 2020.01.31.929042. [Google Scholar]
- 89.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Habibzadeh P., Stoneman E.K. The novel coronavirus: a bird's eye view. Int J Occup Environ Med. 2020;11:65–71. doi: 10.15171/ijoem.2020.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J Med Virol. 2020;92:680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemostasis. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.GISAID . 2020. Genomic epidemiology of BetaCoV 2019-2020. [Google Scholar]
- 95.Du Toit A. Outbreak of a novel coronavirus. Nat Rev Microbiol. 2020;18 doi: 10.1038/s41579-020-0332-0. 123-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.OIE . 2020. Animal and environmental investigations to identify the zoonotic source of the COVID-19 Virus. [Google Scholar]
- 100.diseases PIsfi . DOG SUSPECTED; 2020. Coronavirus disease 2019 update (22): companion animal. REQUEST FOR INFORMATION. [Google Scholar]
- 101.Sit T.H.C., Brackman C.J., Ip S.M., Tam K.W.S., Law P.Y.T., To E.M.W., Yu V.Y.T., Sims L.D., Tsang D.N.C., Chu D.K.W., Perera R.A.P.M., Poon L.L.M., Peiris M. Infection of dogs with SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.S M. Coronavirus can infect cats — dogs, not so much. Nature. 2020 doi: 10.1038/d41586-020-00984-8. [DOI] [PubMed] [Google Scholar]
- 103.Sailleau C., Dumarest M., Vanhomwegen J., Delaplace M., Caro V., Kwasiborski A., Hourdel V., Chevaillier P., Barbarino A., Comtet L., Pourquier P., Klonjkowski B., Manuguerra J.-C., Zientara S., Le Poder S. First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transboundary and Emerging Diseases. 2020 doi: 10.1111/tbed.13659. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martina B.E.E., Haagmans B.L., Kuiken T., Fouchier R.A.M., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S.M., Lim W., Osterhaus A.D.M.E. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425 doi: 10.1038/425915a. 915-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.M C. The Brussels Times; 2020. Coronavirus: Belgian cat infected by owner. [Google Scholar]
- 106.WCSNewsroom . 2020. A tiger at bronx zoo tests positive for COVID-19; the tiger and the zoo's other cats are doing well at this time. [Google Scholar]
- 107.Enserink M. Coronavirus rips through Dutch mink farms, triggering culls. Science. 2020;368:1169. doi: 10.1126/science.368.6496.1169. [DOI] [PubMed] [Google Scholar]
- 108.Oreshkova N., Molenaar R.-J., Vreman S., Harders F., Munnink B.B.O., Hakze R., Gerhards N., Tolsma P., Bouwstra R., Sikkema R., Tacken M., de Rooij M.M.T., Weesendorp E., Engelsma M., Bruschke C., Smit L.A.M., Koopmans M., van der Poel W.H.M., Stegeman A. bioRxiv; Netherlands: 2020. SARS-CoV2 infection in farmed mink. April 2020. 2020.05.18.101493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang W., Tang J., Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J Med Virol. 2020;92:441–447. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y., Liu M., Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc Natl Acad Sci Unit States Am. 2020;117:13967. doi: 10.1073/pnas.2008209117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ye Z.-W., Yuan S., Yuen K.-S., Fung S.-Y., Chan C.-P., Jin D.-Y. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Menachery V.D., Graham R.L., Baric R.S. Jumping species—a mechanism for coronavirus persistence and survival. Current Opinion in Virology. 2017;23:1–7. doi: 10.1016/j.coviro.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Malik Y.S., Sircar S., Bhat S., Vinodhkumar O.R., Tiwari R., Sah R., Rabaan A., Rodriguez-Morales A., Dhama K. Emerging coronavirus disease (COVID-19), a pandemic public health emergency with animal linkages: current status update. Indian J Anim Sci. 2020;90 [Google Scholar]
- 114.Bonilla-Aldana D.K., Quintero-Rada K., Montoya-Posada J.P., Ramírez-Ocampo S., Paniz-Mondolfi A., Rabaan A.A., Sah R., Rodríguez-Morales A.J. SARS-CoV, MERS-CoV and now the 2019-novel CoV: have we investigated enough about coronaviruses? - a bibliometric analysis. Trav Med Infect Dis. 2020;33 doi: 10.1016/j.tmaid.2020.101566. 101566-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Organization Wh . 2020. Middle East respiratory syndrome coronavirus (MERS-CoV) [Google Scholar]
- 116.Wang L.F., Eaton B.T. Bats, civets and the emergence of SARS. Curr Top Microbiol Immunol. 2007;315:325–344. doi: 10.1007/978-3-540-70962-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smith R.D. Responding to global infectious disease outbreaks: lessons from SARS on the role of risk perception, communication and management. Soc Sci Med. 2006;63:3113–3123. doi: 10.1016/j.socscimed.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moriyama M., Hugentobler W.J., Iwasaki A. Seasonality of respiratory viral infections. Annual Review of Virology. 2020 doi: 10.1146/annurev-virology-012420-022445. [DOI] [PubMed] [Google Scholar]
- 119.Plowright R.K., Parrish C.R., McCallum H., Hudson P.J., Ko A.I., Graham A.L., Lloyd-Smith J.O. Pathways to zoonotic spillover. Nat Rev Microbiol. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Suzuki T., Otake Y., Uchimoto S., Hasebe A., Goto Y. Genomic characterization and phylogenetic classification of bovine coronaviruses through whole genome sequence analysis. Viruses. 2020;12:183. doi: 10.3390/v12020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang X.M., Herbst W., Kousoulas K.G., Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J Med Virol. 1994;44:152–161. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G., Shi Z.-L. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 2020.01.22.914952. [Google Scholar]
- 124.Lorusso A., Calistri P., Petrini A., Savini G., Decaro N. Novel coronavirus (SARS-CoV-2) epidemic: a veterinary perspective. Vet Ital. 2020;56:5–10. doi: 10.12834/VetIt.2173.11599.1. [DOI] [PubMed] [Google Scholar]
- 125.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020;368:1016. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.WJ D. Coronavirus SARS-CoV-2 (COVID-19) and companion animal pets. J Immuno Allerg. 2020;1:1–3. [Google Scholar]
- 127.Gollakner R., Capua I. Is COVID-19 the first pandemic that evolves into a panzootic? Vet Ital. 2020;56:7–8. doi: 10.12834/VetIt.2246.12523.1. [DOI] [PubMed] [Google Scholar]
- 128.Bonilla-Aldana D.K., Holguin-Rivera Y., Perez-Vargas S., Trejos-Mendoza A.E., Balbin-Ramon G.J., Dhama K., Barato P., Lujan-Vega C., Rodriguez-Morales A.J. Importance of the one health approach to study the SARS-CoV-2 in Latin America. One Health. 2020;10:100147. doi: 10.1016/j.onehlt.2020.100147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bhatia R. Need for integrated surveillance at human-animal interface for rapid detection & response to emerging coronavirus infections using One Health approach. Indian J Med Res. 2020;151:132–135. doi: 10.4103/ijmr.IJMR_623_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Leroy E.M., Ar Gouilh M., Brugère-Picoux J. The risk of SARS-CoV-2 transmission to pets and other wild and domestic animals strongly mandates a one-health strategy to control the COVID-19 pandemic. One Health. 2020 doi: 10.1016/j.onehlt.2020.100133. 100133-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McNamara T., Richt J.A., Glickman L. A critical needs assessment for research in companion animals and livestock following the pandemic of COVID-19 in humans. Vector Borne Zoonotic Dis. 2020;20:393–405. doi: 10.1089/vbz.2020.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nations FaAOotU . 2020. Mitigating the impacts of COVID-19 on the livestock sector. Rome. [Google Scholar]
- 133.Nations FaAOotU . 2020. Guidelines to mitigate the impact of the COVID-19 pandemic on livestock production and animal health. Rome. [Google Scholar]
- 134.Dyal J.W., Grant M.P., Broadwater K., Bjork A., Waltenburg M.A., Gibbins J.D., Hale C., Silver M., Fischer M., Steinberg J., Basler C.A., Jacobs J.R., Kennedy E.D., Tomasi S., Trout D., Hornsby-Myers J., Oussayef N.L., Delaney L.J., Patel K., Shetty V., Kline K.E., Schroeder B., Herlihy R.K., House J., Jervis R., Clayton J.L., Ortbahn D., Austin C., Berl E., Moore Z., Buss B.F., Stover D., Westergaard R., Pray I., DeBolt M., Person A., Gabel J., Kittle T.S., Hendren P., Rhea C., Holsinger C., Dunn J., Turabelidze G., Ahmed F.S., deFijter S., Pedati C.S., Rattay K., Smith E.E., Luna-Pinto C., Cooley L.A., Saydah S., Preacely N.D., Maddox R.A., Lundeen E., Goodwin B., Karpathy S.E., Griffing S., Jenkins M.M., Lowry G., Schwarz R.D., Yoder J., Peacock G., Walke H.T., Rose D.A., Honein M.A. COVID-19 among workers in meat and poultry processing facilities - 19 states, april 2020. Morb Mortal Wkly Rep. 2020;69 doi: 10.15585/mmwr.mm6918e3. [DOI] [PubMed] [Google Scholar]
- 135.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int J Biol Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.FW . In: Medical microbiology. Galveston. S B., editor. 1996. Viral genetics. [Google Scholar]
- 137.Brook C.E., Boots M., Chandran K., Dobson A.P., Drosten C., Graham A.L., Grenfell B.T., Müller M.A., Ng M., Wang L.-F., van Leeuwen A. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. Elife. 2020;9 doi: 10.7554/eLife.48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen Z.S.R. 2020. COVID-19 and Emerging Viral Diseases: the journey from animals to humans. [Google Scholar]
- 139.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li X., Song Y., Wong G., Cui J. Bat origin of a new human coronavirus: there and back again. Sci China Life Sci. 2020;63:461–462. doi: 10.1007/s11427-020-1645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dhama K., Pawaiya R.V.S., Chakraborty S., Tiwari R., Saminathan M., Verma A.K. Coronavirus infection in equines: a review. Asian J Anim Vet Adv. 2014;9:164–176. [Google Scholar]
- 143.Dhama K., Singh S.D., Barathidasan R., Desingu P.A., Chakraborty S., Tiwari R., Kumar M.A. Emergence of Avian Infectious Bronchitis Virus and its variants need better diagnosis, prevention and control strategies: a global perspective. Pakistan J Biol Sci. 2014;17:751–767. doi: 10.3923/pjbs.2014.751.767. [DOI] [PubMed] [Google Scholar]
- 144.Gravinatti M.L., Barbosa C.M., Soares R.M., Gregori F. Synanthropic rodents as virus reservoirs and transmitters. Rev Soc Bras Med Trop. 2020;53 doi: 10.1590/0037-8682-0486-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Monchatre-Leroy E., Boué F., Boucher J.-M., Renault C., Moutou F., Ar Gouilh M., Umhang G. Identification of alpha and beta coronavirus in wildlife species in France: bats, rodents, rabbits, and hedgehogs. Viruses. 2017;9:364. doi: 10.3390/v9120364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.World Health O . World Health Organization; Geneva: 2003. Consensus document on the epidemiology of severe acute respiratory syndrome (SARS) [Google Scholar]
- 147.Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.-J., Li N., Guo Y., Li X., Shen X., Zhang Z., Shu F., Huang W., Li Y., Zhang Z., Chen R.-A., Wu Y.-J., Peng S.-M., Huang M., Xie W.-J., Cai Q.-H., Hou F.-H., Liu Y., Chen W., Xiao L., Shen Y. Isolation and characterization of 2019-nCoV-like coronavirus from malayan pangolins. bioRxiv. 2020 2020.02.17.951335. [Google Scholar]
- 148.Jalava K. First respiratory transmitted food borne outbreak? Int J Hyg Environ Health. 2020;226 doi: 10.1016/j.ijheh.2020.113490. 113490-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Erles K., Brownlie J. Canine respiratory coronavirus: an emerging pathogen in the canine infectious respiratory disease complex. Vet Clin Small Anim Pract. 2008;38:815–viii. doi: 10.1016/j.cvsm.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Licitra B.N., Duhamel G.E., Whittaker G.R. Canine enteric coronaviruses: emerging viral pathogens with distinct recombinant spike proteins. Viruses. 2014;6:3363–3376. doi: 10.3390/v6083363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tekes G., Thiel H.J. Feline coronaviruses: pathogenesis of feline infectious peritonitis. Adv Virus Res. 2016;96:193–218. doi: 10.1016/bs.aivir.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mihindukulasuriya K.A., Wu G., Leger J., St, Nordhausen R.W., Wang D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J Virol. 2008;82:5084. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30 doi: 10.1016/j.cub.2020.03.063. 1346-51.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M., Zheng B.-J., Chan K.-H., Yuen K.-Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Drosten C.K.P., Memish Z.A. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;371:1359–1360. doi: 10.1056/NEJMc1409847. [DOI] [PubMed] [Google Scholar]
- 156.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S.M., Poon L.L.M. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 157.Nagy P.D., Simon A.E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 158.Rowe C.L., Fleming J.O., Nathan M.J., Sgro J.Y., Palmenberg A.C., Baker S.C. Generation of coronavirus spike deletion variants by high-frequency recombination at regions of predicted RNA secondary structure. J Virol. 1997;71:6183–6190. doi: 10.1128/jvi.71.8.6183-6190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Benvenuto D., Giovanetti M., Ciccozzi A., Spoto S., Angeletti S., Ciccozzi M. The 2019-new coronavirus epidemic: evidence for virus evolution. J Med Virol. 2020;92:455–459. doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhou P., Fan H., Lan T., Yang X.-L., Shi W.-F., Zhang W., Zhu Y., Zhang Y.-W., Xie Q.-M., Mani S., Zheng X.-S., Li B., Li J.-M., Guo H., Pei G.-Q., An X.-P., Chen J.-W., Zhou L., Mai K.-J., Wu Z.-X., Li D., Anderson D.E., Zhang L.-B., Li S.-Y., Mi Z.-Q., He T.-T., Cong F., Guo P.-J., Huang R., Luo Y., Liu X.-L., Chen J., Huang Y., Sun Q., Zhang X.-L.-L., Wang Y.-Y., Xing S.-Z., Chen Y.-S., Sun Y., Li J., Daszak P., Wang L.-F., Shi Z.-L., Tong Y.-G., Ma J.-Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem Biophys Res Commun. 2020;525:135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Patel A., Jernigan D.B. Initial public health response and interim clinical guidance for the 2019 novel coronavirus outbreak - United States, december 31, 2019-february 4, 2020. Morbidity and mortality weekly report. 2020;69:140–146. doi: 10.15585/mmwr.mm6905e1. nCo VCDCRT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Boni M.F., Lemey P., Jiang X., Lam T.T.-Y., Perry B., Castoe T., Rambaut A., Robertson D.L. bioRxiv; 2020. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. 2020.03.30.015008. [DOI] [PubMed] [Google Scholar]
- 164.Callaway Ewen., Cyranoski D. China coronavirus: six questions scientists are asking. Nature. 2020 doi: 10.1038/d41586-020-00166-6. [DOI] [PubMed] [Google Scholar]
- 165.Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Wang M., Lam C.S.F., Xu H., Guo R., Chan K.-H., Zheng B.-J., Yuen K.-Y. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 167.Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J Virol. 2008;82:6984. doi: 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Song H.-D., Tu C.-C., Zhang G.-W., Wang S.-Y., Zheng K., Lei L.-C., Chen Q.-X., Gao Y.-W., Zhou H.-Q., Xiang H., Zheng H.-J., Chern S.-W.W., Cheng F., Pan C.-M., Xuan H., Chen S.-J., Luo H.-M., Zhou D.-H., Liu Y.-F., He J.-F., Qin P.-Z., Li L.-H., Ren Y.-Q., Liang W.-J., Yu Y.-D., Anderson L., Wang M., Xu R.-H., Wu X.-W., Zheng H.-Y., Chen J.-D., Liang G., Gao Y., Liao M., Fang L., Jiang L.-Y., Li H., Chen F., Di B., He L.-J., Lin J.-Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.-J., Spiga O., Guo Z.-M., Pan H.-Y., He W.-Z., Manuguerra J.-C., Fontanet A., Danchin A., Niccolai N., Li Y.-X., Wu C.-I., Zhao G.-P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2430. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shang W., Yang Y., Rao Y., Rao X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. Vaccines. 2020;5:18. doi: 10.1038/s41541-020-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Brownlie J., Sibley D. What can animal coronaviruses tell us about emerging human coronaviruses? Vet Rec. 2020;186:446. doi: 10.1136/vr.m1463. [DOI] [PubMed] [Google Scholar]
- 171.Li Y., Wang H., Tang X., Ma D., Du C., Wang Y., Pan H., Zou Q., Zheng J., Xu L., Farzan M., Zhong G. bioRxiv; 2020. Potential host range of multiple SARS-like coronaviruses and an improved ACE2-Fc variant that is potent against both SARS-CoV-2 and SARS-CoV-1. 2020.04.10.032342. [Google Scholar]
- 172.Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.-K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ge X.-Y., Li J.-L., Yang X.-L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.-J., Luo C.-M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.-Y., Wang L.-F., Daszak P., Shi Z.-L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang N., Li S.-Y., Yang X.-L., Huang H.-M., Zhang Y.-J., Guo H., Luo C.-M., Miller M., Zhu G., Chmura A.A., Hagan E., Zhou J.-H., Zhang Y.-Z., Wang L.-F., Daszak P., Shi Z.-L. Serological evidence of bat SARS-related coronavirus infection in humans, China. Virol Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fan Y., Zhao K., Shi Z.-L., Zhou P. Bat coronaviruses in China. Viruses. 2019;11:210. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Müller M.A., Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Bosch B.-J., Lattwein E., Hilali M., Musa B.E., Bornstein S., Drosten C. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983-1997. Emerg Infect Dis. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ellwanger J.H., Chies J.A.B. Zoonotic spillover and emerging viral diseases time to intensify zoonoses surveillance in Brazil. Braz J Infect Dis. 2018;22:76–78. doi: 10.1016/j.bjid.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Amodio E., Vitale F., Cimino L., Casuccio A., Tramuto F. Outbreak of novel coronavirus (SARS-Cov-2): first evidences from international scientific literature and pending questions. Healthcare. 2020;8:51. doi: 10.3390/healthcare8010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dowell S.F., Simmerman J.M., Erdman D.D., Wu J.-S.J., Chaovavanich A., Javadi M., Yang J.-Y., Anderson L.J., Tong S., Ho M.S. Severe acute respiratory syndrome coronavirus on hospital surfaces. Clin Infect Dis. 2004;39:652–657. doi: 10.1086/422652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Olofsson S., Kumlin U., Dimock K., Arnberg N. Avian influenza and sialic acid receptors: more than meets the eye? Lancet Infect Dis. 2005;5:184–188. doi: 10.1016/S1473-3099(05)01311-3. [DOI] [PubMed] [Google Scholar]
- 181.Otter J.A., Donskey C., Yezli S., Douthwaite S., Goldenberg S.D., Weber D.J. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: the possible role of dry surface contamination. J Hosp Infect. 2016;92:235–250. doi: 10.1016/j.jhin.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lee P.-I., Hsueh P.-R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J Microbiol Immunol Infect. 2020;53:365–367. doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Belser J.A., Rota P.A., Tumpey T.M. Ocular tropism of respiratory viruses. Microbiol Mol Biol Rev. 2013;77:144–156. doi: 10.1128/MMBR.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lu C.-W., Liu X.-F., Jia Z.-F. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30313-5. e39-e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., Zhou Z., Yang J., Zhong J., Yang D., Guo L., Zhang G., Li H., Xu Y., Chen M., Gao Z., Wang J., Ren L., Li M. Genomic diversity of SARS-CoV-2 in coronavirus disease 2019 patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa203. ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Anonymous Emerging zoonoses: a one health challenge. Clin Med. 2020;19 doi: 10.1016/j.eclinm.2020.100300. 100300-00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Charfi M.R. Covid-19: first severe pandemic of the 21st century. La Tunisie medicale. 2020;98:255–257. [PubMed] [Google Scholar]
- 190.Noronha G.S.Y., Bureau E.T. 2020. Coronavirus outbreak will set back India's growth recovery. [Google Scholar]
- 191.Qamar M.A. COVID-19: a look into the modern age pandemic. J Publ Health. 2020 doi: 10.1007/s10389-020-01294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ahmad T., Haroon Dhama K., Sharun K., Khan F.M., Ahmed I., Tiwari R., Musa T.H., Khan M., Bonilla-Aldana D.K., Rodriguez-Morales A J., Hui J. Biosafety and biosecurity approaches to restrain/contain and counter SARS-CoV-2/COVID-19 pandemic: a rapid-review. Turkish J Biol. 2020;44:132–145. doi: 10.3906/biy-2005-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.OIE . 2020. Questions and answers on the COVID-19. [Google Scholar]
- 194.Amuasi J.H., Walzer C., Heymann D., Carabin H., Huong L.T., Haines A., Winkler A.S. Calling for a COVID-19 one health research coalition. Lancet. 2020;395:1543–1544. doi: 10.1016/S0140-6736(20)31028-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.El Zowalaty M.E., Järhult J.D. From SARS to COVID-19: a previously unknown SARS- related coronavirus (SARS-CoV-2) of pandemic potential infecting humans – call for a One Health approach. One Health. 2020;9:100124. doi: 10.1016/j.onehlt.2020.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.OHC . 2020. COVID-19 and one health. [Google Scholar]
- 197.Rabinowitz PM GG . Seattle Times; 2020. 3 steps to help prevent another animal-to-human virus pandemic. [Google Scholar]
- 198.Prevention CfDCa . 2020. COVID-19 and animals. [Google Scholar]
- 199.USDA . 2020. USDA ensures food safety during COVID-19 outbreak. [Google Scholar]
- 200.EPA . 2020. List N: disinfectants for use against SARS-CoV-2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.