Abstract
Innate immunity driven by pattern recognition receptor (PRR) protects the host from invading pathogens. Aquatic animals like fish where the adaptive immunity is poorly developed majorly rely on their innate immunity modulated by PRRs like toll-like receptors (TLR) and NOD-like receptors (NLR). However, current development to improve the fish immunity via TLR/NLR signaling is affected by a poor understanding of its mechanistic and structural features. This review discusses the structure of fish TLRs/NLRs and its interaction with pathogen associated molecular patterns (PAMPs) and downstream signaling molecules. Over the past one decade, significant progress has been done in studying the structure of TLRs/NLRs in higher eukaryotes; however, structural studies on fish innate immune receptors are undermined. Several novel TLR genes are identified in fish that are absent in higher eukaryotes, but the function is still poorly understood. Unlike the fundamental progress achieved in developing antagonist/agonist to modulate human innate immunity, analogous studies in fish are nearly lacking due to structural inadequacy. This underlies the importance of exploring the structural and mechanistic details of fish TLRs/NLRs at an atomic and molecular level. This review outlined the mechanistic and structural basis of fish TLR and NLR activation.
Keywords: TLR, NLR, NOD, Innate immunity, LRR
1. Introduction
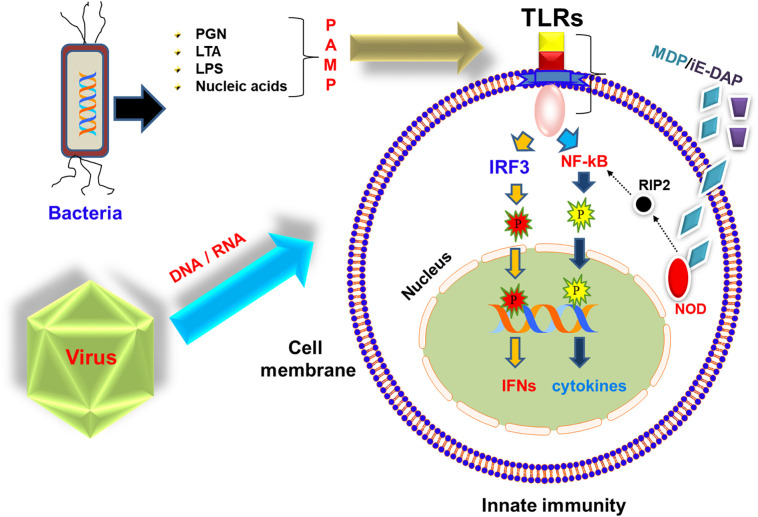
Innate immunity plays a very crucial role in protecting both lower and higher eukaryotes from endogenous and exogenous pathogenic invasion [[1], [2], [3], [4]]. While adaptive immunity is more pronounced in higher eukaryotes, in lower eukaryotes such as fish and amphibians, it is less pronounced. Thus, lower eukaryotes that are more exposed to microorganisms evolved and equipped with a better innate immunity, which play major role in protecting them against pathogenic infections [5,6]. Although, there exists a correlation between innate and adaptive immunity where the former guides the later [7], aquatic organisms majorly rely on the innate immunity during their developmental stage [8]. The innate immune system is the primary defense against infectious diseases with the contribution of various cell types including monocytes and macrophages, dendritic cells, neutrophils and natural killer cells [[9], [10], [11], [12]]. The mechanism of action of innate immunity involves a family of proteins characterized by a highly specialized structure often termed as pattern recognition receptors (PRRs) [13]. PRRs play an essential role in innate immunity by recognizing different conserved microbial motifs viz., carbohydrates (lipopolysaccharide, mannose, fructose, sucrose etc.) [14], nucleic acids (DNA/RNA) [15], peptides (flagellin) [16], peptidoglycans (PGN), lipoteichoic acids (LTA), lipopolysaccharides (LPS), muramyl dipeptide (MDP), γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP), N-formylmethionine, lipoproteins and glucans or endogenous substances collectively known as microbial/pathogen/danger-associated molecular patterns (MAMPs/PAMPs/DAMPs) [2,17]. PRRs are outspread in extracellular, membrane and cytoplasmic compartments and are classified according to their ligand specificity, function and cellular localization. Functionally, PRRs are divided into two categories: endocytic PRRs and signaling PRRs. Signaling PRRs include large families of membrane-bound toll-like receptors (TLRs) [[18], [19], [20]] and cytoplasmic NOD-like receptors (NLRs) [21,22]; and other receptors viz., retinoic acid-inducible gene I-like receptors (RLRs) [23,24], C-type lectin receptors (CLRs), AIM2-like receptor (ALRs) and OAS-like receptors (OLRs) [25,26]. PRRs following the detection of PAMPs trigger the signaling cascade and activate the innate immune response via stimulation of an array of downstream signaling molecules that include chemokines, cytokines, antimicrobial peptides and interferons (Fig. 1 ) [27,28].
Fig. 1.
A simplified schematic representation of the molecular pathway of fish innate immune TLR and NLR receptor. TLR senses PAMPs/MAMPs derived from bacteria (PGN, LTA, LPS etc.), virus (genetic material), fungi etc. and transduces the signal via its cytoplasmic domain to trigger downstream molecules following activation of cytokines and interferons (IFN). Intracellular PAMPs such as MDP and iE-DAP are recognized by NOD receptors following activation of effector molecules and conversion of pro-cytokines to active cytokines.
Aquatic animals especially fish; a chief resource for food and a source of micronutrients and essential acids has shown to be greatly affected by microbial diseases. Fish is one of the most commercially important agricultural product that directly influence the economy [29]. Thus, the protection of fish production by controlling the disease is a major area in aquaculture research. Fish that are exposed to bacteria, fungi and viruses in an aquatic medium largely protect themselves through their innate immunity. The innate immunity in fish is majorly contributed by the signaling PRRs i.e. TLRs and NLRs. TLRs were the first class of PRRs [30] that are extensively studied and characterized in fish and several other vertebrates [31]. Next to TLRs, NLRs are the second class of PRRs that are subjected to considerable investigation [32]. A key difference between TLRs and NLRs are the cellular localization and PAMPs selectivity. TLRs are a class of extracellular transmembrane PRRs, whereas NLRs belong to the class of intracellular cytoplasmic receptors [26]. In TLRs, the periplasmic extracellular domain (ECD) senses the PAMP following signal transduction to the cytoplasmic components via a single transmembrane (TM) domain (Fig. 1) [33]. Structural characterization of TLRs showed a conserved tripartite domain architecture consisting of ECD, a single-pass TM and a cytoplasmic Toll/IL-1 receptor (TIR) domain. The ECD of TLRs are composed of a tandem of evolutionary conserved leucine-rich repeats (LRRs) [34] that selectively recognizes PAMPs and varies in number from species-to-species. Like TLRs, NLRs also present a tripartite domain architecture characterized by a N-terminal effector domain, a central nucleotide binding domain (NBD) and a C-terminal LRR domain [35]. The function of the effector domain is similar to the cytoplasmic TIR domain of TLRs that interacts with downstream effector molecules following activation of signaling cascade involved in innate immune defense. The effector domain also classifies NLRs to subfamilies that include NLRA, NLRB, NLRC and NLRP [22,35]. The NBD domain (NACHT: NAIP, CIITA, HET-E and TP1) is responsible for ATPase activity and mediates oligomerization [36]. The LRR domain in NLR resemble to the topology of TLR-ECD domain and recognizes PAMPs, which crosses the plasma membrane or endogenous DAMPs (Fig. 1) [37]. Unlike TLRs, NLRs show a broad category of function that include signal transduction, autophagy, apoptosis regulation, inflammasome assembly and transcription activation. Readers interested to know more about the TLR and NLR domain architecture, signaling molecular pathway and function in human and other higher eukaryotes are referred to published review articles [21,33,[38], [39], [40], [41]]. This review specifically discusses the domain classification (ECD, TIR, CARD and NACHT), structure and function of fish TLR and NLR domains that include TLR2, TLR3, TLR22, NOD1 and NOD2 and downstream molecules like MyD88-TIR, TRIF and RIP2-CARD.
2. Domain architecture of fish TLRs and NLRs
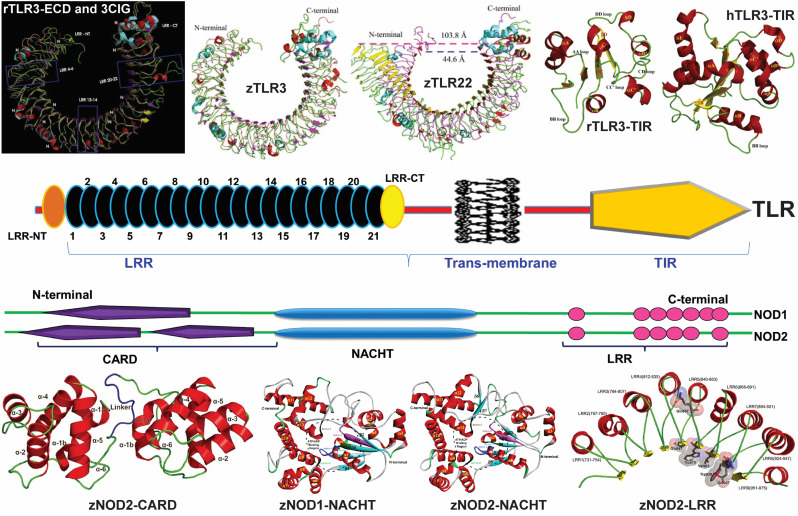
In fish several TLRs have been identified that share a close homology with other eukaryotes including humans (reviewed in [42,43]). Fish that are exposed to an array of microbes with rudimentary adaptive immunity possess more TLRs as compared to humans. While in mammals only 13 TLRs (10 TLRs in human:TLR1–TLR10; and 12 TLRs in mouse: TLR1–TLR9, TLR11–TLR13) [39] have been identified, in teleost, over 20 TLRs were discovered highlighting their importance in providing the first line of defense in fish [42]. Unlike higher vertebrates, in fish TLR6 and TLR10–13 are absent. But, in fish duplicate and multiple copies of TLRs (TLR3, TLR4, TLR5, TLR7, TLR8, TLR20 and TLR22) are identified that are shown to be involved in the fish development [43]. Fish TLRs are structurally homologous to human TLRs and share a sequence identity ~30–70% in the PAMP interacting LRR domains [[44], [45], [46]]. As shown in Fig. 2 , fish TLRs possess a signal peptide (<30 amino acids, aa) that gets cleaved from the mature protein. These mature TLRs composed of ~800–900 amino acids (TLR2, TLR3, TLR4, TLR5 and TLR22). The ECD consists of a variable number of LRR domains that share a substantial sequence identity with the homologous human or mouse TLRs. The number of LRR repeats varies across species and TLR class. In zebrafish TLR3, 24 LRR are identified, whereas Indian carp TLR3 (rohu) presents 27 LRR [44,45]. The amino acid composition of LRR motifs (xLxxLxLxxNxLxxLxxxxFxxLx; where L = leucine/isoleucine/valine/phenylamine; x = any amino acid residue; N = asparagine/threonine/serine/cysteine, and F = phenylalanine) varies across the TLRs making them selective to recognize a target PAMP [47]. Rohu TLR2 and zebrafish TLR22 are composed of 23 and 26 LRR domains, respectively, and each LRR has ~20 amino acids [44,46]. It should be noted that the position and number of LRR varies depending on the algorithm used for LRR motif prediction [[48], [49], [50], [51]]. In general, the secondary structure of LRR domains is evolutionarily highly conserved, but varies in primary structure (Fig. 2). The TM domain is a single-pass helix composed of ~20 amino acids that connects the cytoplasmic TIR domain with the ECD. The TIR domain is comparatively more conserved in fish and mammals than the LRR domain [52]. In general, like LRR motifs, the TIR domains are highly conserved in different species, sharing an average sequence identity >70% for the functional secondary structure units α/β that mediates a homotypic TIR–TIR interactions [46].
Fig. 2.
A schematic diagram showing the domain architectures of zebrafish TLRs and NLRs. Zebrafish TLR shares a tripartite domain architecture that includes an extracellular domain (ECD) composed of an array of leucine-rich-repeats (LRRs), a single-pass transmembrane (TM) domain and a cytosolic TIR domain. The right panel shows the presence of multiple sub-domains in zebrafish NOD1 and NOD2 (zNOD1 and zNOD2). Zebrafish NOD1 contain only one CARD domain at the N-terminus, whereas zNOD2 show two CARD domains (CARDa and CARDb). The central NACHT domain is characterized by five different functional motifs as indicated. The sequence conservations (* as a conserved residue) in zebrafish, human and mouse NACHT domains are accessed using multiple sequence alignment. The potential ATP binding sites in NACHT are highlighted and conserved in all three species. The C-terminal LRR domain in zebrafish NODs is shown in green and is small in number as compared to TLRs. The figures are reproduced from Ref. 44 and 68.
Unlike the TLR domain organization, the cytoplasmic NOD subgroup in fish NLRs share a structure that lacks signal peptide and TM domain. Nevertheless, both TLR and NOD receptors in fish have a common three-domain organization [53]. Importantly, the effector domain of fish NOD receptors consist of one or two caspase-activation and recruitment domains (CARD) that distinguish them from other NLR family proteins containing pyrin or baculovirus inhibitor of apoptosis protein repeat domain [22]. The five major NOD receptors NOD1–5 have been identified in different fish species that include zebrafish, Indian carps, catfish, Japanese flounder, Atlantic salmon etc. and are homologous to mammals [[54], [55], [56], [57], [58], [59], [60], [61], [62], [63]]. The NOD proteins in fish are comparatively larger than TLRs and composed of ~940 and ~980 amino acids (aa) in NOD1 and NOD2, respectively [54,58,60,[64], [65], [66]]. In zebrafish, the NOD1 and NOD2 proteins contain one (94 aa) and two CARD domains (96 and 90 aa) at the N-terminus that mediate homotypic CARD-CARD interactions with downstream signaling molecules (Fig. 2). The central domain in NOD proteins is rather complex and their functions are poorly understood. Sequence analysis of the central domain highlighted three conserved domains in fish that include NBD, two helical domains (HD) flanking a winged helix domain (WHD) [67,68]. These domains are connected through a variable length flexible linker allowing multiple functions such as dNTP binding, oligomerization and signal transduction. The NBD domain is divided into subdomains Walker A, Walker B and Sensor 1. The functional motif in Walker-A ‘G-D/E-A-G-S/V-G-K-S’ and Walker-B ‘L/F-T-F-D-G-L/F/Y-D-E’ subdomains that binds to ATP is highly conserved in fish, human and mouse NOD1/NOD2. Similarly, the functional motifs ‘G/S-L-C-G/H/S-I/L/V-P-L/V-F’ and ‘F/L/Y-E-F-F/L-H’ in HD and WHD subdomains, respectively, are conserved in NOD1 and NOD2 proteins highlighting a similar mode of function and dNTP binding. The LRR domain in fish NODs is rather smaller (~250 aa) as compared to TLRs (~700 aa). The difference in the length of LRR domain between TLRs and NLRs is due to their selective PAMP binding and homo- or hetero-oligomerization. Homo and heterodimers of TLR-ECD are known to mediate their function and dimerization is required to recognize long-sized PAMPs like RNA and DNA of viruses [[69], [70], [71], [72], [73]]. However, the comparatively short LRR domain having less number of LRR motifs present in fish NOD receptor is not involved in homo- or hetero-oligomerization.
3. PAMP/MAMP/DAMP specificity of fish TLRs and NLRs
Water is a great source of pathogenic microorganisms and provides a suitable environment for the growth of major aquatic microbes such as bacteria, fungi, virus, algae, and protozoa. While these microorganisms are a part of the food chain, several of them are known to cause severe diseases in fish. As an example, bacteria species such as Pseudomonas [74], Vibriosis [75], and Aeromonas [76]; viruses like viral hemorrhagic septicemia virus (VHSV), grass-carp reovirus (GCRV), betanodavirus are deadly and causes severe disease to both seawater and freshwater fish (reviewed in [77,78]). This not only directly affects the faster growing aquaculture industry, but also poses threat to human health due to a poor understanding of the evolution of novel aquatic viruses and their diversity. Such possible threats to human life can be realized from the recent COVID-19 pandemic that threatens millions of human life worldwide with an unknown origin.
In fish, TLRs are the most studied PRRs and their PAMP selectivity is comparatively best characterized. Common bacterial components recognized by fish TLR include PGN, LTA, LPS and flagellin (Fig. 1) [79]. TLR2 expression in fish has shown to be up-regulated in response to PGN and LTA [80]. TLR4 and TLR5 respectively have been identified to recognize LPS and flagellin in higher eukaryotes, but some fishes lacks TLR4 and do not recognize LPS and have multi-copies of TLR5 indicating their PAMP selectivity [[81], [82], [83]]. Viral and bacterial RNA is shown to be a molecular target for TLR3, TLR9, TLR13 and TLR22 in fish [31,[84], [85], [86], [87], [88]]. PAMPs for a major class of fish TLRs such as TLR1, TLR7, TLR8, TLR13-R20 and TLR23–27 remain unexplored. In addition to natural bacterial/viral components, synthetic ligands for fish TLRs have been tested that include poly(I:C), CpG DNA, CpG ODN and triacylated lipopeptide. Readers are referred to check previous review articles [42,43] to compare PAMP specificity between fish and mammal TLRs.
Like TLRs, fish NLRs recognize an array of ligands derived from microorganisms. NLRs trigger the activation of downstream molecules by sensing either PAMPs or DAMPs. Bacterial components such as LPS, PGN and its degraded products MDP and iE-DAP are shown to activate NOD1 and/or NOD2 expression (Fig. 1) [59,60,89]. Major bacterial/fungal components and synthetic ligands like LTA, poly(I:C), mannan, toxins, glucan or β-1,3-glucan tested to have no effect on NOD2 (zebrafish), but are potential ligands for other class of fish NLRs (for example poly(I:C) activate NLR-C3 and NOD1) [61,64]. Bacterial flagellin is also shown to activate NLR-C in common carp [90]. The effect of intracellular activators such as cholesterols, dNTPs, metabolites, and antimicrobial peptides, on NLRs are less explored in fish. Nevertheless, to date studies of the effect of other aquatic parasites such as protozoans, fungi, algae and their molecular components modulating TLR/NLR activation in fish remain mostly undiscovered. Furthermore, the reported stipulated number of ligands that are majorly identified in mammals and tested on fish limit our present understanding of the major classes of PAMPs that activate fish immunity via TLRs/NLRs recognition. To this end, lack of structural and mechanistic insights into the ligand recognition and specificity by fish TLRs/NLRs and innate immune activation further limits the development of novel synthetic molecules to boost fish innate immunity. While deriving such structural and mechanistic information is challenging, recently some studies shed light on the mechanism of PAMP interaction and downstream molecule interaction with TLR/NLR receptors using in silico structural biology approaches and are summarized in the following sections.
4. Comparative tertiary structure modeling of fish TLRs/NLRs
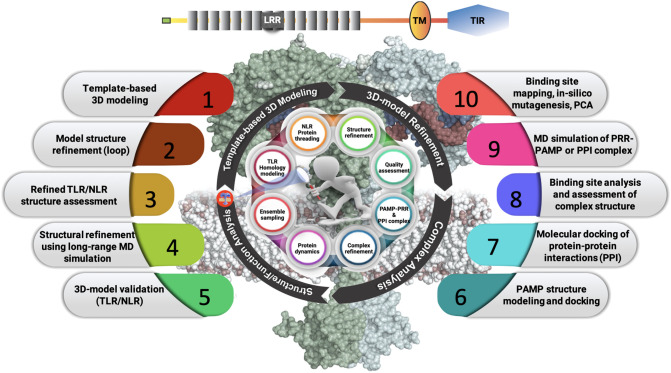
Establishment of a crosstalk between structure-to-function has been a bottleneck in the field of structural biology and drug-discovery research. Unfortunately, even after nearly a few decades since TLRs/NLRs are identified in zebrafish [91,92], experimental structure for these proteins is not available for most of the proteins except zebrafish TLR5 (ECD) [93] to understand their mechanism of action. The complex topology and molecular pathways are among the major roadblocks for the structural investigation. Nevertheless, the unavoidable limitations associated with experimental techniques add more challenges to solve these biological complex questions. For example, association of TLRs with membrane limits the application of X-ray crystallography technique to solve full-length TLR structure; the high-molecular size of TLRs/NLRs ~100 kDa (excluding homo-/hetero-oligomers) make it difficult to study using nuclear magnetic resonance (NMR). However, recent developments in cryo-EM that prefer large-molecular size protein and can be studied in a membrane environment look promising to study full-length TLR/NLR structure. In addition, advance membrane mimetic tools like nanodiscs and bicelles to study membrane proteins provide an promising alternative to study functional TLR domains by NMR [94,95]. To this end, computational techniques are an alternative and attractive approach that bridge the structure-to-function executing a coordinated sequence of structural bioinformatics tools. In this coordinated sequence, the first tool is building a tertiary model structure for fish TLRs/NLRs using a comparative/ab-initio molecular modeling approach as shown in Fig. 3 .
Fig. 3.
A schematic showing an in silico strategy for the structural and functional investigation of fish TLR and NLR proteins. Step (1–5) shows 3D structure modeling and refinement strategy for fish TLR/NLR proteins. Step (6–10) shows strategy to build TLR/NLR-PAMP or protein-protein complex following structural and functional analysis using long-range MD simulations.
In silico molecular modeling approach builds a 3D model structure with accuracy using the amino acid sequence of the target protein (query sequence) [96,97]. Using a homology based approach, the query protein sequence is scanned over experimentally solved structures (template) deposited in the public protein data bank (PDB) [98] database that presently contain >160,000 structures. Upon satisfying a threshold similarity/identity between the query sequence and template structure(s) using computational algorithms, 3D coordinates can be generated for the target protein using multiple steps as illustrated in Fig. 3 (reviewed in [97]). The quality of the initial 3D model structure mostly depends on the degree of sequence identity/similarity between the query and template protein. Comparative model building using templates available in the current PDB database shows a good reliability for fish TLRs and NLRs as summarized in Table 1 . Zebrafish as a model organism to study fish TLR/NLR is considered for searching for available templates. As listed in Table 1, most of the zebrafish TLRs show a sequence identity (≥30%) except for TLR18–20. It should be noted that the identity listed in Table 1 is obtained from BLASTp search against the current PDB database for the full-length zebrafish TLR/NLR proteins. Special attention for obtaining a maximum query coverage (minimum gap in the query-template alignment) and sequence identity need to be considered for the modeling of functional domains. TLR-ECD such as rohu TLR3-ECD, zebrafish TLR3-ECD and TLR22-ECD have shown a query-template sequence identity over ~25–30% which is considered to be reliable for homology modeling (Fig. 3) [97]. For instance, rohu TLR3-ECD shows an identity and similarity of 48 and 65%, respectively, with mouse TLR3-ECD; and rohu TLR3-TIR domain shares 33 and 56% sequence identity and similarity, respectively, with human TLR3-TIR [45]. Similarly, zebrafish TLR3 (identity/similarity: 48/64%)/TLR22-ECD (identity/similarity:28 and 43%) share a reasonable homology with human TLR3-ECD for comparative structure modeling [44]. Rohu TLR2-ECD (identity/similarity: 35/52%), TLR2-TIR (identity 71%) and common carp downstream molecule MyD88-TIR (identity 78%) also share a very good homology with mouse TLR and human MyD88 crystal structures (template) [46].
Table 1.
Current available templates in Protein Data Bank for zebrafish TLR and NLR model building.
| Zebrafish | Uniprot ID | Template (PDB ID) | Description | Organism | Identity (%) |
|---|---|---|---|---|---|
| TLR1 | B3DIW3 | 1FYV | TLR1 | Human | 57.52 |
| TLR2 | F1R1U3 | 5D3I | TLR 2 | Mouse | 34.03 |
| TLR3 | B8JIL3 | 3CIG | TLR3 | Mouse | 47.88 |
| TLR4aa | B3U3W0 | 3FXI | TLR4 | Human | 35.21 |
| TLR5bb | B3DIN1 | 3V44/3V47/5GY2 | TLR5b | zebrafish | n/a |
| TLR6c | Absent | n/a | n/a | n/a | n/a |
| TLR7 | F1QY64 | 5GMF | TLR7 | Rhesus macaque | 56.79 |
| TLR8aa | F1R2P4 | 3W3G | TLR8 | Human | 41.66 |
| TLR9 | F1QY61 | 3WPB | TLR9 | Horse | 35.52 |
| TLR10–13c | Absent | n/a | n/a | n/a | n/a |
| TLR18d | B3DKG5 | 3A79/3WPB | TLR6/TLR9 | Mouse/Horse | ~23 |
| TLR19d | A0A2R8Q6Z1 | 3J0A | TLR5 | Human | ~23 |
| TLR20d | F1QRG0 | 5ZSA | TLR7 | Rhesus macaque | ~23 |
| TLR21 | F1QMN8 | 4Z0C | TLR13 | Mouse | 30.52 |
| TLR22 | B3DJL6 | 3J0A | TLR5 | Human | 29.36 |
| NOD1 | X1WGQ4 | 5IRM/5IRL | NOD2 | Rabbit | 32.49 |
| NOD2aa | F8W3K2 | 5IRM/5IRL | NOD2 | Rabbit | 50.67 |
Represents TLR/NLR present in zebrafish with multiple copies (templates are shown for only one copy).
Crystal structure is available.
Represents a few TLRs absent in zebrafish, but are present in mouse/human.
TLRs with very low sequence identity (templates shown are based on the highest query coverage; whereas other TLR/NLR templates listed in the table are considered based on the maximum BLASTp score).
As compared to fish TLRs, NOD domains share a good sequence homology with the rabbit NOD2 crystal structure (Table 1) [99]. Previously, advanced techniques like ab-initio modeling [100] and protein threading [101] that employ a multi-template modeling approach was used to construct reliable 3D-model structures for rohu NOD1 and NOD2 LRR domains (Fig. 3) due to the unavailability of an experimental structure for NOD receptor [102,103]. However, at present the BLASTp search of rohu NOD1 LRR against PDB data bank resulted four hits that include the PDB IDs: 5IRM (34.66% identity), 5IRL (34.66% identity), 4R5D (29.63% identity) and 4R6G (28.64% identity). This suggests that, the better query-template identity between fish and rabbit NOD proteins [99] could provide an improved 3D model structure for rohu/zebrafish NOD proteins. Similarly, models for other NLR domains such as N-terminal CARD and C-terminal NACHT and effector molecules RIP2-CARD that have been successfully designed and tested could be further refined using rabbit NOD2 as a template [68,104]. The reliability of the previously reported model structures of fish TLR/NLR after successive and careful refinements [45,67,102,104] using several web-based validation programs are found good. These methods are also routinely used to assess the structural quality of experimentally solved structures. Interestingly, refined structures obtained using a comparative modeling approach for rohu and zebrafish TLRs/NLRs show a reliable validation score with a very minimum structural errors as shown in Fig. 4 . Such errors can be further refined using atomistic simulation techniques on a time-scale ranging from nanoseconds-microseconds as highlighted in Fig. 3.
Fig. 4.
3D model structures of fish TLR and NLR domains built using comparative modeling and protein threading approaches. The rohu and zebrafish TLR and NOD proteins are denoted as rTLR/zTLR and rNOD/zNOD. Superimposed structure of rohu TLR3-ECD and mouse TLR3-ECD (PDB ID: 3CIG) is shown on the top left. Zebrafish TLR3 structures before and after MD simulations are shown on the top center. The domain architecture of TLR and NOD proteins are shown in the center and the corresponding 3D model domain structures are shown on the top (TLR) and bottom (NOD). LRR domains are numbered and N- and C-terminal LRR domains are represented as LRR-NT and LTT-CT, respectively. The 3D model structures of the tripartite domain (CARD, NACHT and LRR) in zebrafish NOD1 and NOD2 are shown in the bottom. The linker connecting CARDa and CARDb (see Fig. 2) is colored in blue. NOD-LRR repeats shown on right bottom shows less number of LRR repeats when compared to TLR LRR motifs as shown on top left. The 3D model structures are reproduced from Ref. 44, 45, 67, and 68.
5. Structural refinement using molecular dynamics simulations
Structural assessment of modeled structure requires substantial refinement and spatial rearrangement of the initial template structure to improve accuracy [105]. Clashes between side-chain atoms, bonds, dihedral angles etc. can be refined using web-based programs and disordered loops can be refined using advanced modeling approaches (Fig. 3). However, optimization of domain structure and its spatial rearrangement and folding require dynamics over a threshold time-period under a physiological environment. For example, several proteins undergo conformational dynamics and structure rearrangement under a threshold temperature, salt, pH etc. Molecular dynamics (MD) simulations provide a platform to optimize the structural dynamics and folding under a physiological condition [106,107]. Classical MD simulations using all-atom and coarse-grained models have been successfully employed in studying biomolecule structure, dynamics, conformational change, ligand binding etc. [108] This makes it a very powerful tool and has also been applied in refining experimentally obtained protein, DNA and other biomolecule structures. As mentioned in Section 4, a poor homology between the query and template (low sequence identity) provides a poor quality model structure. In general, the target protein regions for which the template does not have a structure (low identity/similarity or due to low sequence coverage between query and template) are often built with structural errors that require substantial refinement. An example in fish TLRs are the loops connecting each LRR repeats or the linker (shown in blue, Fig. 4, bottom left) connecting the CARD domains in fish NOD2. It should be noted that the flexible loops in LRR are functionally very important and involved in the PAMP interaction. To this end, a homology model of fish TLR/NLR requires a threshold time-scale of all-atom MD simulation for structural optimization as presented in the schematic workflow (Fig. 3). In this review, the application of MD simulations are discussed in line with studying fish TLRs/NLRs, and readers who wish to know a broad application of MD simulation in studying biomolecules are referred to check previous review articles [[108], [109], [110]].
A major application of MD simulation in studying fish innate immune receptors is its use in tracking the interaction between PRRs and PAMPs in real-time under a physiological condition. In this approach, the target PRR molecule(s) and the PAMP(s) are initially placed at a certain distance from each other so that there exist no intermolecular interactions. Both molecules are equilibrated in an explicit solvent environment at physiological conditions and the intermolecular interactions are monitored over a time-scale ranging from nano- to micro-seconds. Such a technique is referred to as “blind docking” due to a lack of any prior knowledge of the PRR active site that binds to the PAMP. Such method is very useful for studying fish TLRs/NLRs that share <50% homology (identity) with human/mouse for which experimentally PAMP binding sites are resolved. The low sequence homology indicates differential binding pockets and PAMP specificity in fish that are very poorly understood. That said an assumption for a conserved PAMP binding site in both fish and human/mouse TLRs/NLRs need complex structure assessment (Fig. 3). To this end, MD simulations showed promising application in evaluating the fish PRR-PAMP complex stability (Fig. 3). In this method, a prior knowledge of PAMP (for example poly(I:C)) binding pocket in the targeted PRR (for example human TLR3) [111] is first retrieved. Molecular docking simulation is carried out allowing the poly(I:C) to find the optimal binding orientation around a predefined binding pocket in fish TLR3 referring to the prior knowledge obtained from human TLR3-poly(I:C) complex. The spatial arrangement of poly(I:C) in fish TLR3 yielding the lowest free energy is predicted to be the key binding site. As mentioned earlier, such exercises require further structural assessment and complex stability analysis in a physiological environment. All-atom MD simulations of the complex assist in structure refinement [112], validating the complex stability, monitoring change in conformation upon PAMP interaction and any possible dissociation/rearrangement of the PAMP around the protein active site (Fig. 3). Taken together MD simulations have proven to be very helpful in retrieving structural information in fish PRRs for which no experimental evidence is available. In the next section, a few structural studies on fish TLRs/NLRs using molecular docking and MD simulations are summarized.
6. Tertiary structure analysis of TLR/NLR domains
The validation and assessment of the quality of the modeled structures in fish are very important prior to their consideration for PAMP-PRR and/or protein-protein interaction analysis. Sahoo et al., for the first time built a 3D model structure for an Indian carp (Labeo rohita; rohu) TLR3 ECD and TIR domain and presented a comprehensive PAMP and protein-protein interaction analysis using model structure of zebrafish TRIF (TIR-domain-containing adapter-inducing interferon-β) [45]. The tertiary folding of rohu (rTLR3)-ECD model having a horseshoe shape structure show good resemblance with its template mouse TLR3-ECD crystal structure [69] with an average Cα root mean square deviation (RMSD) of ~0.5 Å (Fig. 4, top left). The reported rTLR3-ECD model shows very good validation scores in Ramachandran plot (99.5% residues in allowed regions), a high 3D profiling score (97.2% residues with good compatibility score), acceptable coarse packing quality and planarity [45]. Similarly, rTLR3-TIR model structure consisting of four β-sheets and α-helices align well with the human TLR1-TIR crystal structure [113] (Fig. 4, top right) yielding a Cα atom RMSD of ~0.86 Å. Validation reports of the rTLR3-TIR model structure satisfy the acceptable scores for structural analysis.
In another study, Sahoo et al., reported the modeled 3D structures for zebrafish TLR3 and TLR22 ECD domain (zTLR3 and zTLR22) that recognize a conserve PAMP (virus double-stranded RNA) [44]. Interestingly, they observed a significant difference between these two receptors post MD simulation. The zTLR3-ECD model structure closely resembles (horseshoe shape) with the rohu TLR3-ECD 3D model and human/mouse TLR3-ECD crystal structures. On the other hand, post MD simulation, zTLR22-ECD presents a substantial structural rearrangement measuring a backbone RMSD ~12 Å. The structural comparison provides several interesting findings. First, note that zTLR3-ECD and TLR22-ECD share a good sequence homology (25/42%; identity/similarity). Second, as TLR22 is absent in human/mouse, no specific homologues template structure (experimental structure of TLR22-ECD in any organism) is present. Therefore, Sahoo et al., generated 3D models of zTLR3-ECD and zTLR22-ECD using crystal structure of mouse/human TLR3-ECD as template(s) that initially show a less-flattened horseshoe shape structure (Fig. 4, top center). As discussed in the previous section, the powerful application of MD in optimizing the conformation and dynamics of modeled structure is demonstrated in this study using the model zTLR22-ECD structure. The initial compact zTLR22-ECD model undergo a significant conformational change (twist at both terminus; ~15° and 30° twisting at N- and C-terminus, respectively) during the 50 ns MD simulation presenting a flattened horseshoe structure measuring a distance of ~104 Å between N- and C-terminus as compared to ~45 Å before MD simulation (Fig. 4, top center) [44]. This conformational change and flattening of zTLR22-ECD is hypothesized to be crucial to recognize long-sized dsRNA molecules. Structural models of rohu TLR2-ECD, TLR2-TIR and common carp MyD88-TIR constructed using mouse/human TLR2-ECD and MyD88-TIR as templates have shown a very good secondary structure conservation between the target and template. The rohu TLR2-ECD also presents a horseshoe shape as observed for rohu TLR3-ECD composed of 23 LRR domains. Like mouse/human TLR2- or TLR3-ECD, fish TLR2/TLR3-ECD (rohu and zebrafish) LRR domain consist of an array of parallel β-strands facing the concave surface and are connected via loops. This arrangement from a stable hydrophobic core stabilized by hydrogen bonds (H-bond) between the neighboring β-strands and form a horseshoe shape that resembles TLR-ECD structure as observed in higher eukaryotes [33]. The outer or convex surface of the fish TLRs on the other hand is majorly compose of disordered loops with a few regions having short α-helices.
A key difference in fish TLR-ECD structure as compared to NOD-LRR is the secondary structure topology of convex surface. As illustrated in Fig. 4 (compare between zTLR3 and zNOD2-LRR), all fish TLR-ECD LRR domains are mostly unstructured in the convex surface, whereas structured (α-helix) in fish NOD-LRR (Fig. 4, bottom right). It should be noted that this structural difference between fish TLR- and NLR-LRR domain is not unique in fish, rather well conserved in higher eukaryotes including human [114]. The modeled structure of TLR27 consists of 19 LRR domains claimed to be exist only in three fishes by Wang et al. [115] The reported TLR27-ECD structure resembled the rohu TLR22-ECD horseshoe shape structure with the C-terminal region being slightly twisted yielding a flattened structure [115]. A heterodimeric model of TLR1-TLR2 complex is built using comparative modeling in common carp that shares a good fitting with the template human TLR1-TLR2 complex crystal structure [116]. Horseshoe structure is also observed for other fish TLRs that include Tibet fish TLR4 [117], zebrafish TLR5a and TLR5b [82] and miiuy croaker TLR28 [118]. The modeled zebrafish TLR5a-TLR5b heterodimer structure was reported to have a very similar structure when compared with the homodimer TLR5-flagellin complex structure resolved using X-ray crystallography [82,93].
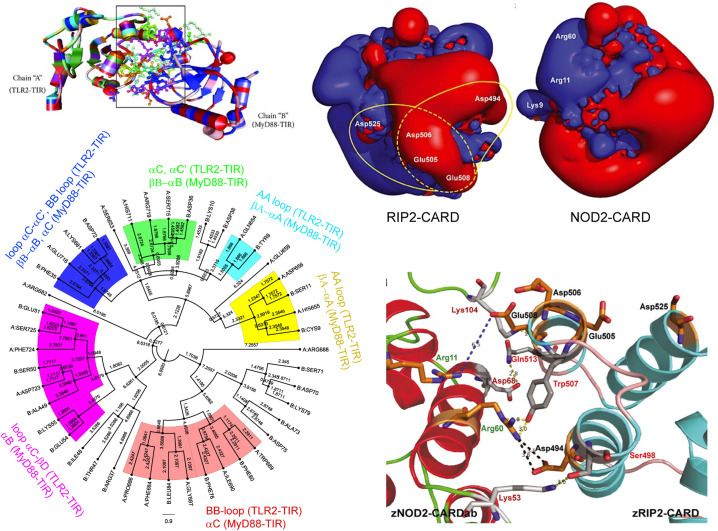
The structural investigation of fish NLR have been carried out for all individual domains i.e. CARD, NACHT and LRR in Indian carps or zebrafish [68,102,104]. Because of a low sequence homology, Maharana et al., employed a multi-template modeling approach to build zebrafish NOD1- and NOD2-CARD and RICK(RIP2)-CARD structure for protein-protein interaction study (Fig. 5 ) [104]. The initial model structures of zebrafish NOD1/2-CARD(a,b) and RIP2-CARD showed a good secondary structure conservation with each CARD composed of six α-helices (α1-α6) with 100% residues fall in the allowed regions of Ramachandran plot. Except α1 that shows a kink structure, but conserved in NOD and RIP2-CARD, all other five α-subdomains in the modeled structure show a regular shape. Importantly, electrostatic surface potential mapping highlighted a significant difference between zebrafish NOD-CARD and RIP2-CARD. The positive (basic) surface potential (blue) in NOD1/2-CARD is less as compared to RIP2-CARD or the acidic surface potential (red) is higher in NOD1/2-CARD as compared to RIP2 (Fig. 5) [104]. This observation underlie NOD-CARD require an exposed negative surface potential to bind RIP2-CARD in order to transduce the downstream signaling cascade.
Fig. 5.
Tertiary structure of zebrafish NOD-CARD domains modeled using template-based modeling approach as indicated. The CARD modeled structures shown on the top composed of six α-helices (α1–6) and charged residues are highlighted. Sequence alignment of zebrafish NOD1/2-CARD with human and mouse is shown in the center with conserved residues highlighted in different colors. Electrostatic surface potential of zebrafish NOD-CARD and downstream interacting RIP2 CARD domains are shown in the bottom at the indicated colors.
The figures are reproduced from Ref. 104 with permission from The Royal Society of Chemistry.
The simulated zebrafish NOD2-CARD structure rearranged the bi-lobed globular structure representing subdomains CARDa and CARDb connected via a linker showing in blue (Fig. 4, bottom left) [104]. The simulated model structures of zebrafish CARD domain show a high resemblance with the high-resolution NMR structure of NOD1 CARD [119]. Maharana et al., in another study predicted the model structures of the zebrafish NOD1 and NOD2 NACHT domain using the crystal structure of mouse NLRC4 (PDB ID: 4KXF) as a template to explore the NOD mediated signal transduction [68]. It should be noted that these models can be further refined using the rabbit NOD2 crystal structure (PDB ID: 5IRM/5IRL) that gives a better query-template sequence-structure alignment. The modeled zebrafish NOD1-NACHT and NOD2-NACHT as shown in Fig. 4 (bottom center) presented ~97% residues within the allowed regions of Ramachandran plot. Yet again, structural comparison and interpretation between NOD1- and NOD2-NACHT in zebrafish do not show a substantial difference in secondary structure (5 conserved parallel β-sheets and 13 α-helices) and folding as per the key functional subdomains NBD, HD1 and WHD are concerned. A notable difference is the presence of two additional β-sheets that are present in NOD2-NACHT (C-terminus), but absent in NOD1-NACHT. As mentioned earlier, the reported NOD-LRR domains are comparatively short and differ from TLR-LRR having helices on the convex surface. In fish the structure for the NOD-LRR domain is reported only for rohu and zebrafish NOD1 and NOD2 although NOD proteins are identified in several other fish species.
7. Structural interactions between PAMP and PRR
The binding interactions between PAMP and PRR has been comprehensively investigated for both TLRs and NLRs using experimental and computational biophysical techniques. However, in fish these structural information are very poorly explored. In the absence of experimental PRR structures in fish (except TLR5-ECD) [93], modeled TLR/NLR proteins have been used for structural analysis. As mentioned in Section 5, molecular docking simulations and blind-docking using MD simulations have been adopted to reveal PAMP interaction with fish TLR/NLRs. In this section, studies in fish describing the structural interaction between PAMP and TLR or NOD are discussed.
Interaction of a variety of natural and synthetic PAMP that includes poly(I:C), flagellin, iE-DAP, MDP, LPS, PGN, LTA and dsRNA with fish TLR/NOD receptors is reported. Molecular docking simulations using programs like AutoDock [120], FlexX [121], Glide [122], GOLD [123] and ArgusLab [124] is used to predict the binding sites for small-size PAMPs containing less number of atoms (poly(I:C), iE-DAP, MDP LPS, zymosan, PGN and LTA). For molecules with a large number of atoms (e.g. dsRNA and LPS), programs like HADDOCK [125], HexServer [126] and PatchDock [127] are used to build the PAMP-PRR complex. It is important to note that, the use of a combination of docking programs for a target molecule binding with the receptor could minimize the probability of non-specific interaction sites. For example, Sahoo et al., compared the binding energy and interaction sites for poly(I:C) using two different programs (AutoDock and ArgusLab) by varying the grid size (box surrounding specific interaction sites as reported previously for homologous proteins, and box embedding whole protein that allows the PAMP to identify the best binding pose). The results obtained in this study showed both the programs predict a conserved binding pocket for poly(I:C) in rohu and zebrafish TLR3 and TLR22 proteins [44].
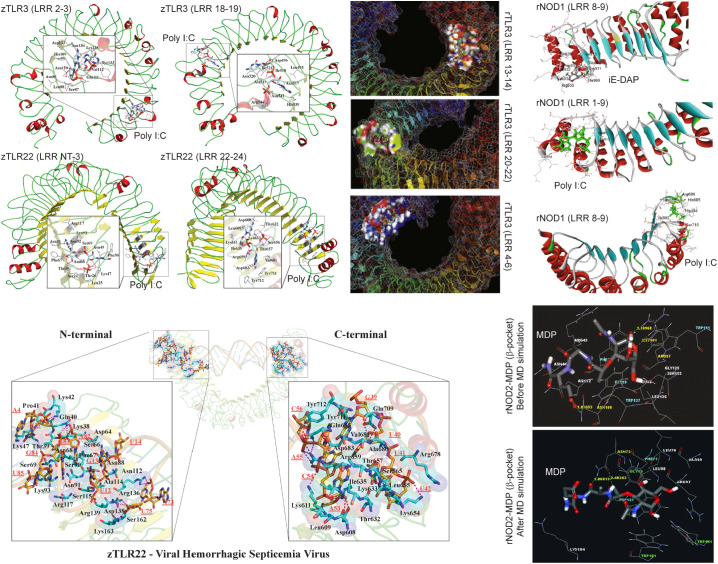
Two different binding sites are predicted for poly(I:C) in zebrafish TLR3 (LRR2–3 and LRR18–19) and TLR22 (LRRNT-3 and LRR22–24), and a comparatively high poly(I:C) binding affinity is shown for TLR22 (Fig. 6 , top left). An additional binding site is reported for poly(I:C) in rohu TLR3 (LRR4–6, LRR13–14 and LRR20–22) (Fig. 6, top center) [45]. Remarkably, this study further highlights the effect of mutation on the binding affinity of poly(I:C)with rohu TLR3. Selective mutations at LRR4–6 and LRR20–22 completely abolish poly(I:C) interactions, whereas LRR13–14 do not completely abolish poly(I:C) binding, but reduce the binding affinity [45]. Chakrapani et al., probed the binding interaction of poly(I:C) with a homology model structure of wild-type and mutant rohu TLR22 and observed complex instability in TLR22 mutant [128]. These findings highlights, docking simulation could be a powerful method to screen potential molecules for TLR activation. But, unfortunately, in fish no structure-based PAMP screening has been carried out yet. Voogdt et al., reported a heterodimeric complex structure for zebrafish TLR5 interacting with bacterial flagellin. They compared the structural differences in the TLR5-flagellin hetero-complex composed of two variable copies of TLR5 (TLR5a and TLR5b) discovered in zebrafish with the experimental homodimer TLR5 structure. This study identified that flagellin binding induces conformational change in the loop connecting LRR9–10, while the rest of the TLR5-ECD structure is highly similar to that of unbound flagellin structure [82]. Studies on rohu TLR2 interaction with different variants of PGN, LTA and zymosan also reported a conserved predicted binding site by AutoDock, FlexX and GOLD [46].
Fig. 6.
Structural interactions between different PAMPs and TLR and NOD receptors. Molecular docking simulations showing the binding sites of poly I:C in rohu and zebrafish TLR3-ECD and TLR22-ECD (referred to as rTLR and zTLR) predicted using AutoDock or GOLD programs. Molecular interaction of iE-DAP and poly I:C with rohu NOD1 (rNOD) predicted by molecular docking using the GOLD program. HADDOCK docking simulation illustrating the binding sites for the VHSV-dsRNA in the zebrafish TLR22-ECD. A comparative illustration of the MDP binding to rohu NOD2-LRR β-sheet pocket predicted from docking simulations (GOLD program) before and after MD simulations as indicated. The figures are reproduced from Ref. 44, 45, 102, and 103.
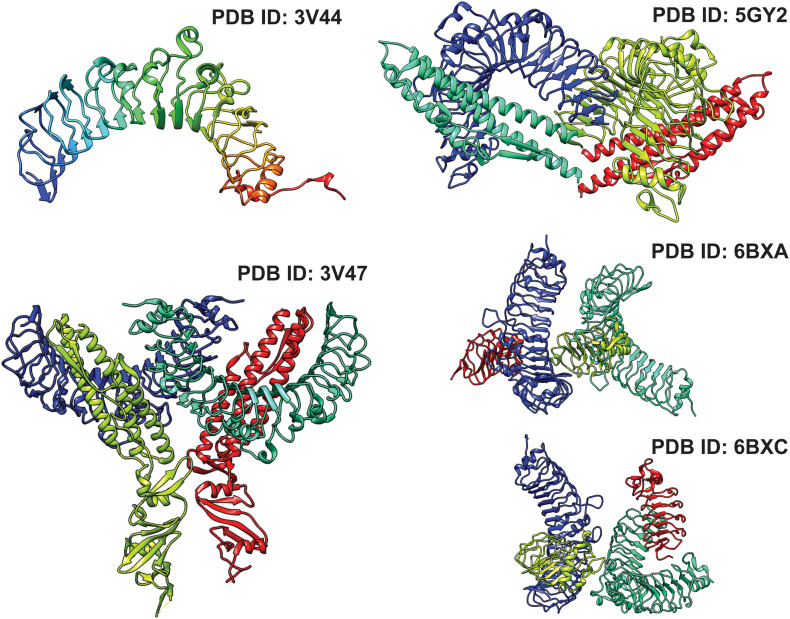
The interaction of bacterial flagellin with zebrafish TLR5-ECD and its complex has been solved using X-ray crystallography. The expression and purification of TLR has been a roadblock for structure determination. Yoon et al. successfully expressed and purified zebrafish TLR5 by truncating the C-terminal residues that generates N-terminal fragments having 6, 12 and 14 LRR motifs [93]. The apo structure of zebrafish orthologue TLR5 with 12 N-terminal LRR motifs (PDB ID: 3V44) solved with a resolution of 2.83 Å show a horseshoe shape structure (Fig. 7 ). For PAMP interaction analysis, three variable fragments (D1-D3) of Salmonella flagellin are generated that primarily form a 1:1 complex that further homodimerizes to generate a symmetric 2:2 complex with TLR5 N-terminal domain where the flagellin domain D1 is positioned on the lateral side of TLR5 ECD (Fig. 7). The high-resolution complex structure obtained at 2.47 Å reveals a unique binding activity of flagellin that mediates a tail-to-tail dimerization of TLR5 containing 14 LRR motifs (PDB ID: 3V47). The conserved domain D1, but not the hypervariable domain D2 in flagellin is identified to directly interact with zebrafish TLR5 (LRR-NT to LRR10). Interaction of Bacillus subtilis flagellin with zebrafish TLR5 is also studied using crystallography and a 2.1 Å complex structure (PDB ID:5GY2) is reported [129]. Unlike to Salmonella flagellin binding, the interaction of Bacillus subtilis flagellin composed of only two domains (D0 and D1) with zebrafish TLR5 (LRRNT-LRR14) show to form a 1:1 complex. Importantly, this study reported the binding of flagellin to TLR5 did not significantly alter the curvature of the LRR domain and its horseshoe shape (Fig. 7). Structural studies identify zebrafish TLR5 binding with the variable lymphocyte receptor (VLR) blocks flagellin interactions (Fig. 5, PDB IDs: 6BXA and 6BXC) [130]. VLRs are specialized adaptive immune receptor that binds to antigens via the TLR5 LRR domains. Two variable epitopes on TLR5 N-terminal ECD that are involved in flagellin binding are identified to interact with the VLR (Fig. 7).
Fig. 7.
Crystal structure of the N-terminal fragment of zebrafish TLR5 (ECD) complexed with bacterial flagellin (Salmonella enterica, 3V47), (Bacillus subtilis, 5GY2), and lamprey variable lymphocyte receptor 2 (VLR2, 6BXA) and VLR9 (6BXC).
The binding sites and affinity of poly(I:C), LPS and iE-DAP with rohu NOD1 receptor LRR domain was compared using four different docking programs (AutoDock, FlexX, GOLD and Glide) [102]. This study proposed two binding sites for poly(I:C) that are identified to be consistent in all the programs based on the binding score (Fig. 6, top right). An interesting observation from this study is the binding site predicted for poly(I:C) in rohu NOD1 differs from that of human NOD1 yielding a minimum docking score in all programs. On the other hand, a similar approach for iE-DAP interaction with rohu NOD1 using these programs predicted a binding site that is conserved in both human and rohu (Fig. 6, top right). The in silico structural findings could provide crucial mechanistic insights on the unique features of fish NLRs in recognizing PAMPs. In the absence of any prior knowledge to guide the docking simulations for novel PAMPs, ligand binding sites can be predicted using the model fish TLR/NLR structure. Following the predictions, grid boxes can be generated targeting the predicted pockets and binding affinity of the novel PAMP can be estimated. A summary of the interaction of fish TLR/NLR proteins with specific PAMPs and downstream signaling molecules are summarized in Table 2 .
Table 2.
A list of fish TLR/NLR structures and their interaction with PAMP or downstream molecules reported in silico or experimental methods.
| TLR/NLR | PAMP/Protein | Organism | Experimental method |
|---|---|---|---|
| TLR2 | PGN, LTA and Zymosan | Labeo rohita | Comparative modeling |
| TLR2 | MyD88-TIR | Labeo rohita | Comparative modeling |
| TLR3 | Poly(I:C) | Labeo rohita | Comparative modeling |
| TLR3 | dsRNA (AGCRV,VHSV and IHNV) | Labeo rohita | Comparative modeling |
| TLR3 | Poly(I:C) | Danio rerio | Comparative modeling |
| TLR3 | dsRNA (AGCRV,VHSV and IHNV) | Danio rerio | Comparative modeling |
| TLR3 | TRIF | Labeo rohita | Comparative modeling |
| TLR5 | Flagellin (Bacillus, Salmonella) | Danio rerio | X-ray crystallography |
| TLR5 | Variable lymphocyte receptor | Danio rerio | X-ray crystallography |
| TLR22 | Poly(I:C) | Danio rerio | Comparative modeling |
| TLR22 | dsRNA (AGCRV,VHSV and IHNV) | Danio rerio | Comparative modeling |
| TLR27 | n/a | Latimeria chalumnae | Protein threading |
| NOD1 | Poly(I:C), iE-DAP and LPS, ATP | Labeo rohita | Protein threading |
| NOD2 | MDP | Labeo rohita | Protein threading |
| NOD2 | MDP | Danio rerio | Protein threading |
| NOD1 | RIP2-CARD | Danio rerio | Protein threading |
| NOD2 | RIP2-CARD | Danio rerio | Protein threading |
Similarly, for large-size molecules (e.g. LPS), multiple docking servers could be helpful in revealing a consensus binding site as demonstrated for rohu NOD1-LPS complex using Hex and PatchDock server [102]. In addition to the docking simulation, as discussed in Section 5, it is crucial to decipher the stability of the complex in a physiological environment (solvent, pH, salt and temperature). Following docking simulation, a next recommended step is thus to evaluate the complex stability by MD simulations on a time-scale of nanoseconds to microseconds/milliseconds (Fig. 3). Often specific/non-specific binding of a targeted PAMP can be investigated looking at its atomistic interaction map before and after MD simulations. Maharana et al., compared the stability of the binding pocket for MDP in rohu NOD2 before (complex obtained from docking simulations) and after MD simulation (docked complex after undergoing multi-nanoseconds MD simulation) [103]. The comparative analysis showed a change in the binding site and number of H-bonds between MDP and rohu NOD2 before (Arg97 and Asn100 forms three H-bonds with MDP) and after MD simulations (Asn72 form two H-bonds with MDP), but do not alter the overall ligand binding pocket (Fig. 6, bottom right) [103]. In another approach, Maharana et al., probed the binding sites of ATP with zebrafish NOD1 or NOD2 NACHT domain using molecular docking and MD simulations. They compared the stability of ATP-NOD1/NOD2 complex in zebrafish obtained from AutoDock and a complex where ATP is manually placed ~3 Å from the active site following MD simulations [68].
Docking simulations integrated with all-atom MD simulations are proposed to be effective to probe interaction of virus dsRNA and TLRs. Unlike to small-ligand docking (blind docking), the dsRNA docking is driven by guiding the docking program with the interaction sites that are either derived from a homology analysis (human/mouse TLR3 crystal structure) [69,131], or obtained from poly(I:C) binding analysis which is a synthetic analog of dsRNA. HADDOCK docking program has shown to decipher the complex structure for TLR3/TLR22-ECD complex with dsRNA (Fig. 6, bottom left) derived from different viruses that include AGCRV, VHSV and Infectious Hematopoietic Necrosis Virus (IHNV). These large complex docked structures are simulated on a time-scale of nanoseconds to retrieve structural and mechanistic details at an atomic level. An interesting observation from MD simulation of dsRNA-TLR22 complex in zebrafish is an enhancement of the complex stability as compared to poly(I:C)-TLR22 complex [44]. The overall dynamics of the TLR22-ECD protein is shown to be greatly influenced by the dsRNA binding (Fig. 6, bottom left) which is investigated using an array of essential dynamics methods using the MD trajectory. The stability of the complex (both small-size and large-size PAMP) can be evaluated by computing the free binding energy between the receptor (TLR/NOD) and ligand (PAMP). Using this method, Sahoo et al. and Maharana et al., demonstrated the PAMP interaction specificity and stability in both rohu and zebrafish [44,67]. In this approach, from the MD trajectory of PAMP-PRR complex, a number of structures are retrieved at a specified time-interval. The driving force of PAMP interaction with fish TLR/NOD can be next computed using programs like MM/PBSA and MM/GBSA that provides a set of energetic parameters to better understand the molecular forces governing the PRR-PAMP interaction [132].
8. Probing downstream signaling via protein-protein interactions
The interaction between TLR/NOD receptors with downstream signaling molecules are important to understand the molecular process governing the innate immunity activation. Understanding this molecular process in real-time at an atomic-level is challenging due to the involvement of complex molecules that include PAMP, PRR, membrane and downstream effector molecules. However, this process can be dissected to reveal atomic information. To this end, unlike the PAMP-TLR/NOD interaction analysis, in fish the protein-protein interaction studies are miniscule. Moreover, to date no structural studies have been carried out for any fish TLRs in a membrane system.
The first reported study in fish reveals the protein-protein complex between rohu TLR3-TIR and zebrafish TRIF domain [45]. In the absence of any prior binding site information, the complex structure of TIR-TRIF is generated using the predicted protein-protein interactions sites by online web-servers (consPPISP [133] and InterProSurf [134]) [45]. Using a similar approach, rohu TLR2-TIR and its interaction with the downstream molecule myD88-TIR (common carp) domain is reported (Fig. 8 , top left) [46]. The TIR-TIR interface in rohu is predicted using web-programs like cons-PPISP, ProSurf and PatchDock, and a complex structural model of TIR-TIR is build using HADDOCK program. This study predicted key interface residues contributed by rohu TLR2-TIR and common carp MyD88-TIR domains that majorly include the loops and ∝-helices (Fig. 8, top left) [46]. The reported model structure of golden pompano MyD88 resembles common carp structure, however no protein-protein interactions has yet been reported [135]. Fish TLRs interaction with downstream protein molecules reported to date are listed in Table 2.
Fig. 8.
Structural insights into functional domain interactions between fish innate immune receptors and downstream signaling molecules. Simulated protein complex structure of rohu TLR2-TIR and common carp MyD88-TIR (left). The binding interface of the TIR-TIR complex is presented as a phylogeny where sub-domain interactions are highlighted in colors. The complex between zebrafish zNOD2-CARD and zRIP2-CARD domains is shown on the right. The surface electrostatic potential of both CARD domains are heighted in red and blue respectively representing a positive and negative surface potential. Atomistic interactions map for the NOD2-RIP2 protein complex show electrostatic interactions between positive and negative charge residues as indicated. The figures are reproduced from Ref. 46 and 67.
The interaction between zebrafish NOD2-CARD domain and CARD domain of the adaptor molecule RIP2 (serine–threonine kinase-2) is investigated using molecular dynamics simulation. The CARD-CARD interaction between zebrafish NOD1/NOD2 and RIP2 is suggested to be mediated by charge-charge interactions as both CARD domains share an opposite electrostatic surface potential (Fig. 5, Fig. 8) [67,104]. Maharana et al., probed the stability of NOD2-RIP2 CARD-CARD complex by generating two complexes by docking the positive surface of NOD2-CARD with two different negative surfaces (Complex-I: Asp494, Glu505, Glu508, and Asp506; Complex-II: Glu505, Glu508, Asp506, and Asp525) of RIP2-CARD [67]. The CARD-CARD protein complex is designed using only the CARDa domain of the zebrafish NOD2 (Fig. 2). From the MD simulation analysis, Complex-I is predicted to have a relatively more stable structure as compared to Complex-II and majorly stabilized by electrostatic interactions and salt-bridges. Together, the study proposed the acidic interface of zebrafish NOD1/NOD2-CARD binds to the basic interface of RIP2-CARD (Fig. 8). The study also anticipated a possible interaction of zebrafish CARDb with RIP-CARD, but are not yet investigated. Considering the importance of signal transduction mediated through CARD-CARD or TIR-TIR interactions, the reported in silico findings could provide insights into (i) specific CARD/TIR domain mutation and its association with fish disease; and (ii) strategies to modulate CARD/TIR domain interactions using small-molecule interventions.
9. Limitations of in silico structural analysis
In silico structural analysis of fish TLR/NLR proteins are often affected by a low sequence-structure homology with higher eukaryotes (Fig. 3), which eventually reflected on the comparative 3D model structure. As discussed in Section 4, model structure of fish TLR/NLR need structural re-modeling and refinement due to the periodic upgradation of the PDB databases. As an example, rohu/zebrafish NOD1/2 structures reported by Maharana et al., need further attention and re-modeling using available rabbit NOD2 crystal structure for future structural exercises. Structural errors in the initial model structure could affect the docking simulations and structural interpretations. Although long-range atomistic simulations show an alternative promising pathway to optimize the model structure (Fig. 3), more often an insufficient conformational sampling due to an expensive computational cost limits structural refinement. Modern computational resources that run in parallel for atomistic simulations have shown tremendous progress in past half-decades achieving multi-microseconds time-scale of all-atom MD simulations. However, to date the computation to probe fish TLRs/NLRs structure and function are carried out on a time-scale of multi-nanoseconds. In addition to that, the large-size structure of TLR/NLRs that require a MD simulation system comprising tens of thousands atoms is a major roadblock to achieve microseconds-millisecond MD simulations. An alternative to this is to build a coarse-grained model system that uses a united atom or sometimes referred to as extended atom to extend the time-scale of simulation, but with the cost of limiting the structural information. Moreover, importantly the structural interpretation of TLRs/NLRs are centered with protein-ligand interactions, and coarse-grained MD simulation of TLR/NLR-PAMP requires a proper parameterization of the ligand, which is often very challenging. Similarly, in the united atom model representation that usually excludes the explicit hydrogen atoms could affect the PRR-PAMP interactions. Nevertheless, complex systems like studying the protein-protein interactions and conformational dynamics of TLRs in a membrane bound and unbound state can be achieved on a time-scale of microseconds using the coarse-grained systems.
The inadequacy of experimental evidence for protein-protein complex for PRRs and its effector molecules often pose questions on the initial assessment of the complex structure. Prediction of protein-protein complexes with a native-like binding surface are challenging, and require experimental support and are still under development. Association of protein-protein complex with a non-native binding surface are found to be stable for hundreds of nanoseconds in MD simulation and required enhanced sampling techniques like “tempered binding” to obtain a native-like complex [136]. Thus, structural interpretation and stability analysis of the protein complex in fish TLRs/NLRs using MD simulation needs to be extended beyond the simplistic measurement of protein association and dissociation. In silico mutagenesis study using docking simulations and binding free energy computation provides important structural information in fish (rohu and zebrafish) [44,45]; however, requires experimental binding constants for structural validation. Lastly, structural interpretation in TLRs/NLRs are often carried out using a truncated functional sub-domain such as ECD, TIR, CARD and NACHT. The influence of connecting sub-domains or membrane (in case of TLRs) on the conformational dynamics (flexibility/rigidity) of the target domain of study are therefore not reflected in the structural analysis. This limits the overall in silico structural analysis both in apo and holo states. For example, the binding kinetics and interface between TIR-TIR in the absence of membrane and ECD can be influenced when studies are performed in a membrane interface using full-length TLR/NLR proteins.
10. Conclusion
A major progress in studying fish TLRs/NLRs has been witnessed over the past ten years. Many studies are centered on the identification and characterization of fish innate immune receptors. That said, very few studies to date have focused on exploring the structural and functional properties of fish innate immune receptors. On the other hand, in humans such studies have been carried out in depth with the development of several innate immune modulators that are under preclinical trials. Bearing in mind the rapid growth of world population, demand of fish in the food supply chain, and recent or past pandemic viral flus caused by viruses with animals as the reservoir hosts, it is crucial to explore fish innate immune receptors more in detail. Furthermore, our understanding of the fish TLR/NLR structure at an atomic level would enable in developing antagonist/agonist ligands to modulate fish immunity and disease, thus directly influencing the global economy. Fish, in particular zebrafish, is emerging as a valuable animal model for preclinical in vivo study and drug screening. This further highlights the importance of studying fish innate immune receptors to discover novel therapeutic strategies that can later be tested in higher organisms including humans.
Acknowledgements
The author thanks Dr. D.K. Tiwari, Ramalingaswami fellow at Goa University, India for his comments and suggestions in the preparation of the manuscript.
References
- 1.Akira S. Innate immunity and adjuvants. Philos. Trans. R. Soc. B Biol. Sci. 2011;366:2748–2755. doi: 10.1098/rstb.2011.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Beutler B. Innate immunity: an overview. Mol. Immunol. 2004;40:845–859. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Uthaisangsook S., Day N.K., Bahna S.L., Good R.A., Haraguchi S. Innate immunity and its role against infections. Ann. Allergy Asthma Immunol. 2002;88:253–265. doi: 10.1016/s1081-1206(10)62005-4. [DOI] [PubMed] [Google Scholar]
- 5.Buchmann K. Evolution of innate immunity: clues from invertebrates via fish to mammals. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinkevich B. Invertebrates versus vertebrates innate immunity: in the light of evolution. Scand. J. Immunol. 1999;50:456–460. doi: 10.1046/j.1365-3083.1999.00626.x. [DOI] [PubMed] [Google Scholar]
- 7.Iwasaki A., Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science (80-. ) 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uribe C., Folch H., Enriquez R., Moran G. Innate and adaptive immunity in teleost fish: a review. Vet. Med. (Praha) 2011;56:486–503. doi: 10.17221/3294-VETMED. [DOI] [Google Scholar]
- 9.Parihar A., Eubank T.D., Doseff A.I. Monocytes and macrophages regulate immunity through dynamic networks of survival and cell death. J. Innate Immun. 2010;2:204–215. doi: 10.1159/000296507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva M.T. When two is better than one: macrophages and neutrophils work in concert in innate immunity as complementary and cooperative partners of a myeloid phagocyte system. J. Leukoc. Biol. 2010;87:93–106. doi: 10.1189/jlb.0809549. [DOI] [PubMed] [Google Scholar]
- 11.Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 12.Costantini C., Cassatella M.A. The defensive alliance between neutrophils and NK cells as a novel arm of innate immunity. J. Leukoc. Biol. 2011;89:221–233. doi: 10.1189/jlb.0510250. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 14.Chow J.C., Young D.W., Golenbock D.T., Christ W.J., Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 15.Majer O., Liu B., Barton G.M. Nucleic acid-sensing TLRs: trafficking and regulation. Curr. Opin. Immunol. 2017;44:26–33. doi: 10.1016/j.coi.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tallant T., Deb A., Kar N., Lupica J., De Veer M.J., DiDonato J.A. Flagellin acting via TLR5 is the major activator of key signaling pathways leading to NF-κB and proinflammatory gene program activation in intestinal epithelial cells. BMC Microbiol. 2004;4 doi: 10.1186/1471-2180-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herwald H., Egesten A. On PAMPs and DAMPs. J. Innate Immun. 2016;8:427–428. doi: 10.1159/000448437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akira S., Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 19.Vijay K. Toll-like receptors in immunity and inflammatory diseases: past, present, and future. Int. Immunopharmacol. 2018;59:391–412. doi: 10.1016/j.intimp.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medzhitov R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y.K., Shin J.S., Nahm M.H. NOD-like receptors in infection, immunity, and diseases. Yonsei Med. J. 2016;57:5–14. doi: 10.3349/ymj.2016.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilmanski J.M., Petnicki-Ocwieja T., Kobayashi K.S. NLR proteins: integral members of innate immunity and mediators of inflammatory diseases. J. Leukoc. Biol. 2008;83:13–30. doi: 10.1189/jlb.0607402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onoguchi K., Yoneyama M., Fujita T. Retinoic acid-inducible gene-I-like receptors. J. Interf. Cytokine Res. 2011;31:27–31. doi: 10.1089/jir.2010.0057. [DOI] [PubMed] [Google Scholar]
- 24.Kato H., Takahasi K., Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhu J., Ghosh A., Sarkar S.N. OASL - a new player in controlling antiviral innate immunity. Curr. Opin. Virol. 2015;12:15–19. doi: 10.1016/j.coviro.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai T., Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thaiss C.A., Levy M., Itav S., Elinav E. Integration of innate immune signaling. Trends Immunol. 2016;37:84–101. doi: 10.1016/j.it.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 29.Peterman M.A., Posadas B.C. Direct economic impact of fish diseases on the East Mississippi catfish industry. N. Am. J. Aquac. 2019;81:222–229. doi: 10.1002/naaq.10090. [DOI] [Google Scholar]
- 30.Medzhitov R., Preston-Hurlburt P., Janeway C.A. A human homologue of the Drosophila toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 31.Nie L., Cai S.-Y., Shao J.-Z., Chen J. Toll-like receptors, associated biological roles, and signaling networks in non-mammals. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fritz J.H., Ferrero R.L., Philpott D.J., Girardin S.E. Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 33.Botos I., Segal D.M., Davies D.R. The structural biology of Toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell J.K., Mullen G.E.D., Leifer C.A., Mazzoni A., Davies D.R., Segal D.M. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–533. doi: 10.1016/S1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 35.Ting J.P.Y., Lovering R.C., Alnemri E.S., Bertin J., Boss J.M., Davis B.K., Flavell R.A., Girardin S.E., Godzik A., Harton J.A., Hoffman H.M., Hugot J.P., Inohara N., MacKenzie A., Maltais L.J., Nunez G., Ogura Y., Otten L.A., Philpott D., Reed J.C., Reith W., Schreiber S., Steimle V., Ward P.A. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koonin E.V., Aravind L. The NACHT family - a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem. Sci. 2000;25:223–224. doi: 10.1016/S0968-0004(00)01577-2. [DOI] [PubMed] [Google Scholar]
- 37.Proell M., Riedl S.J., Fritz J.H., Rojas A.M., Schwarzenbacher R. The nod-like receptor (NLR) family: a tale of similarities and differences. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meunier E., Broz P. Evolutionary convergence and divergence in NLR function and structure. Trends Immunol. 2017;38:744–757. doi: 10.1016/j.it.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 39.Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Zayat S.R., Sibaii H., Mannaa F.A. Toll-like receptors activation, signaling, and targeting: an overview. Bull. Natl. Res. Cent. 2019;43 doi: 10.1186/s42269-019-0227-2. [DOI] [Google Scholar]
- 41.Saxena M., Yeretssian G. NOD-like receptors: master regulators of inflammation and cancer. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J., Kong X., Zhou C., Li L., Nie G., Li X. Toll-like receptor recognition of bacteria in fish: ligand specificity and signal pathways. Fish Shellfish Immunol. 2014;41:380–388. doi: 10.1016/j.fsi.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 43.Palti Y. Toll-like receptors in bony fish: from genomics to function. Dev. Comp. Immunol. 2011;35:1263–1272. doi: 10.1016/j.dci.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Sahoo B.R., Dikhit M.R., Bhoi G.K., Maharana J., Lenka S.K., Dubey P.K., Tiwari D.K. Understanding the distinguishable structural and functional features in zebrafish TLR3 and TLR22, and their binding modes with fish dsRNA viruses: an exploratory structural model analysis. Amino Acids. 2015;47:381–400. doi: 10.1007/s00726-014-1872-2. [DOI] [PubMed] [Google Scholar]
- 45.Sahoo B.R., Basu M., Swain B., Maharana J., Dikhit M.R., Jayasankar P., Samanta M. Structural insights of rohu TLR3, its binding site analysis with fish reovirus dsRNA, poly I:C and zebrafish TRIF. Int. J. Biol. Macromol. 2012;51:531–543. doi: 10.1016/j.ijbiomac.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Sahoo B.R., Basu M., Swain B., Dikhit M.R., Jayasankar P., Samanta M. Elucidation of novel structural scaffold in rohu TLR2 and its binding site analysis with peptidoglycan, lipoteichoic acid and zymosan ligands, and downstream MyD88 adaptor protein. Biomed. Res. Int. 2013;2013 doi: 10.1155/2013/185282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng A.C.Y., Eisenberg J.M., Heath R.J.W., Huett A., Robinson C.M., Nau G.J., Xavier R.J. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4631–4638. doi: 10.1073/pnas.1000093107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bej A., Sahoo B.R., Swain B., Basu M., Jayasankar P., Samanta M. LRRsearch: an asynchronous server-based application for the prediction of leucine-rich repeat motifs and an integrative database of NOD-like receptors. Comput. Biol. Med. 2014;53:164–170. doi: 10.1016/j.compbiomed.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 49.Offord V., Coffey T.J., Werling D. LRRfinder: a web application for the identification of leucine-rich repeats and an integrative Toll-like receptor database. Dev. Comp. Immunol. 2010;34:1035–1041. doi: 10.1016/j.dci.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Matsushima N., Miyashita H., Mikami T., Yamada K. A new method for the identification of leucine-rich repeats by incorporating protein secondary structure prediction. Bioinforma. Genome Bioinforma. Comput. Biol. 2012:61–88. [Google Scholar]
- 51.Martin E.C., Sukarta O.C.A., Spiridon L., Grigore L.G., Constantinescu V., Tacutu R., Goverse A., Petrescu A.J. Lrrpredictor—a new LRR motif detection method for irregular motifs of plant NLR proteins using an ensemble of classifiers. Genes (Basel) 2020;11 doi: 10.3390/genes11030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mikami T., Miyashita H., Takatsuka S., Kuroki Y., Matsushima N. Molecular evolution of vertebrate Toll-like receptors: evolutionary rate difference between their leucine-rich repeats and their TIR domains. Gene. 2012;503:235–243. doi: 10.1016/j.gene.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 53.Benko S., Philpott D.J., Girardin S.E. The microbial and danger signals that activate nod-like receptors. Cytokine. 2008;43:368–373. doi: 10.1016/j.cyto.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 54.Swain B., Basu M., Samanta M. Molecular cloning and characterization of nucleotide binding and oligomerization domain-1 (NOD1) receptor in the Indian major carp, rohu (Labeo rohita), and analysis of its inductive expression and down-stream signalling molecules following ligands exposu. Fish Shellfish Immunol. 2012;32:899–908. doi: 10.1016/j.fsi.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 55.ying Gao F., cai Pang J., xin Lu M., le Yang X., ping Zhu H., li Ke X., gang Liu Z., meng Cao J., Wang M. Molecular characterization, expression and functional analysis of NOD1, NOD2 and NLRC3 in Nile tilapia (Oreochromis niloticus) Fish Shellfish Immunol. 2018;73:207–219. doi: 10.1016/j.fsi.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 56.Li J., Gao Y., Xu T. Comparative genomic and evolution of vertebrate NOD1 and NOD2 genes and their immune response in miiuy croaker. Fish Shellfish Immunol. 2015;46:387–397. doi: 10.1016/j.fsi.2015.06.026. [DOI] [PubMed] [Google Scholar]
- 57.Zhang L., Gao Z., Yu L., Zhang B., Wang J., Zhou J. Nucleotide-binding and oligomerization domain (NOD)-like receptors in teleost fish: current knowledge and future perspectives. J. Fish Dis. 2018;41:1317–1330. doi: 10.1111/jfd.12841. [DOI] [PubMed] [Google Scholar]
- 58.Bin Park S., ichi Hikima J., Suzuki Y., Ohtani M., Nho S.W., Cha I.S., Bin Jang H., Kondo H., Hirono I., Aoki T., Jung T.S. Molecular cloning and functional analysis of nucleotide-binding oligomerization domain 1 (NOD1) in olive flounder, Paralichthys olivaceus. Dev. Comp. Immunol. 2012;36:680–687. doi: 10.1016/j.dci.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 59.Bi D., Wang Y., Gao Y., Li X., Chu Q., Cui J., Xu T. Recognition of lipopolysaccharide and activation of NF-κB by cytosolic sensor NOD1 in teleost fish. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swain B., Basu M., Samanta M. NOD1 and NOD2 receptors in mrigal (Cirrhinus mrigala): inductive expression and downstream signalling in ligand stimulation and bacterial infections. J. Biosci. 2013;38:533–548. doi: 10.1007/s12038-013-9330-y. [DOI] [PubMed] [Google Scholar]
- 61.Zou P.F., Chang M.X., Li Y., Xue N.N., Li J.H., Chen S.N., Nie P. NOD2 in zebrafish functions in antibacterial and also antiviral responses via NF-κB, and also MDA5, RIG-I and MAVS. Fish Shellfish Immunol. 2016;55:173–185. doi: 10.1016/j.fsi.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 62.Chang M., Wang T., Nie P., Zou J., Secombes C.J. Cloning of two rainbow trout nucleotide-binding oligomerization domain containing 2 (NOD2) splice variants and functional characterization of the NOD2 effector domains. Fish Shellfish Immunol. 2011;30:118–127. doi: 10.1016/j.fsi.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 63.Xie J., Hodgkinson J.W., Katzenback B.A., Kovacevic N., Belosevic M. Characterization of three nod-like receptors and their role in antimicrobial responses of goldfish (Carassius auratus L.) macrophages to Aeromonas salmonicida and Mycobacterium marinum. Dev. Comp. Immunol. 2013;39:180–187. doi: 10.1016/j.dci.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 64.Paria A., Deepika A., Sreedharan K., Makesh M., Chaudhari A., Purushothaman C.S., Thirunavukkarasu A.R., Rajendran K.V. Identification of nod like receptor C3 (NLRC3) in Asian seabass, Lates calcarifer: characterisation, ontogeny and expression analysis after experimental infection and ligand stimulation. Fish Shellfish Immunol. 2016;55:602–612. doi: 10.1016/j.fsi.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 65.Gu T., Lu L., Wang J., Tian L., Wei W., Wu X., Xu Q., Chen G. The NOD1 and NOD2 in mandarinfish (Siniperca chuatsi): molecular characterization, tissue distribution, and expression analysis. BMC Genet. 2018;19 doi: 10.1186/s12863-018-0667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paria A., Makesh M., Chaudhari A., Purushothaman C.S., Rajendran K.V. Nucleotide-binding oligomerization domain-containing protein 1 (NOD1) in Asian seabass, Lates calcarifer: cloning, ontogeny and expression analysis following bacterial infection or ligand stimulation. Fish Shellfish Immunol. 2018;79:153–162. doi: 10.1016/j.fsi.2018.04.061. [DOI] [PubMed] [Google Scholar]
- 67.Maharana J., Patra M.C., De B.C., Sahoo B.R., Behera B.K., De S., Pradhan S.K. Structural insights into the MDP binding and CARD-CARD interaction in zebrafish (Danio rerio) NOD2: a molecular dynamics approach. J. Mol. Recognit. 2014;27:260–275. doi: 10.1002/jmr.2357. [DOI] [PubMed] [Google Scholar]
- 68.Maharana J., Sahoo B.R., Bej A., Jena I., Parida A., Sahoo J.R., Dehury B., Patra M.C., Martha S.R., Balabantray S., Pradhan S.K., Behera B.K. Structural models of zebrafish (Danio rerio) NOD1 and NOD2 NACHT domains suggest differential ATP binding orientations: insights from computational modeling, docking and molecular dynamics simulations. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu L., Botos I., Wang Y., Leonard J.N., Shiloach J., Segal D.M., Davies D.R. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science (80-. ) 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Botos I., Liu L., Wang Y., Segal D.M., Davies D.R. The Toll-like receptor 3:dsRNA signaling complex. Biochim. Biophys. Acta - Gene Regul. Mech. 2009;1789:667–674. doi: 10.1016/j.bbagrm.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee H.K., Dunzendorfer S., Tobias P.S. Cytoplasmic domain-mediated Dimerizations of Toll-like receptor 4 observed by β-lactamase enzyme fragment complementation. J. Biol. Chem. 2004;279:10564–10574. doi: 10.1074/jbc.M311564200. [DOI] [PubMed] [Google Scholar]
- 72.Ozinsky A., Underhill D.M., Fontenot J.D., Hajjar A.M., Smith K.D., Wilson C.B., Schroeder L., Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hajjar A.M., O’Mahony D.S., Ozinsky A., Underhill D.M., Aderem A., Klebanoff S.J., Wilson C.B. Cutting edge: functional interactions between Toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble Modulin. J. Immunol. 2001;166:15–19. doi: 10.4049/jimmunol.166.1.15. [DOI] [PubMed] [Google Scholar]
- 74.López J.R., Lorenzo L., Marcelino-Pozuelo C., Marin-Arjona M.C., Navas J.I. Pseudomonas baetica: pathogenicity for marine fish and development of protocols for rapid diagnosis. FEMS Microbiol. Lett. 2017;364 doi: 10.1093/femsle/fnw286. [DOI] [PubMed] [Google Scholar]
- 75.Ina-Salwany M.Y., Al-saari N., Mohamad A., Mursidi F.A., Mohd-Aris A., Amal M.N.A., Kasai H., Mino S., Sawabe T., Zamri-Saad M. Vibriosis in fish: a review on disease development and prevention. J. Aquat. Anim. Health. 2019;31:3–22. doi: 10.1002/aah.10045. [DOI] [PubMed] [Google Scholar]
- 76.Sugita H., Tanaka K., Yoshinami M., Deguchi Y. Distribution of Aeromonas species in the intestinal tracts of river fish. Appl. Environ. Microbiol. 1995;61:4128–4130. doi: 10.1128/aem.61.11.4128-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crane M., Hyatt A. Viruses of fish: an overview of significant pathogens. Viruses. 2011;3:2025–2046. doi: 10.3390/v3112025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kibenge F.S. Emerging viruses in aquaculture. Curr. Opin. Virol. 2019;34:97–103. doi: 10.1016/j.coviro.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 79.Ribeiro C.M.S., Hermsen T., Taverne-Thiele A.J., Savelkoul H.F.J., Wiegertjes G.F. Evolution of recognition of ligands from gram-positive bacteria: similarities and differences in the TLR2-mediated response between mammalian vertebrates and teleost fish. J. Immunol. 2010;184:2355–2368. doi: 10.4049/jimmunol.0900990. [DOI] [PubMed] [Google Scholar]
- 80.Samanta M., Swain B., Basu M., Panda P., Mohapatra G.B., Sahoo B.R., Maiti N.K. Molecular characterization of toll-like receptor 2 (TLR2), analysis of its inductive expression and associated down-stream signaling molecules following ligands exposure and bacterial infection in the Indian major carp, rohu (Labeo rohita) Fish Shellfish Immunol. 2012;32:411–425. doi: 10.1016/j.fsi.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 81.Sepulcre M.P., Alcaraz-Pérez F., López-Muñoz A., Roca F.J., Meseguer J., Cayuela M.L., Mulero V. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-κB activation. J. Immunol. 2009;182:1836–1845. doi: 10.4049/jimmunol.0801755. [DOI] [PubMed] [Google Scholar]
- 82.Voogdt C.G.P., Wagenaar J.A., van Putten J.P.M. Duplicated TLR5 of zebrafish functions as a heterodimeric receptor. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E3221–E3229. doi: 10.1073/pnas.1719245115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hwang S.D., Asahi T., Kondo H., Hirono I., Aoki T. Molecular cloning and expression study on Toll-like receptor 5 paralogs in Japanese flounder, Paralichthys olivaceus. Fish Shellfish Immunol. 2010;29:630–638. doi: 10.1016/j.fsi.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 84.Kongchum P., Hallerman E.M., Hulata G., David L., Palti Y. Molecular cloning, characterization and expression analysis of TLR9, MyD88 and TRAF6 genes in common carp (Cyprinus carpio) Fish Shellfish Immunol. 2011;30:361–371. doi: 10.1016/j.fsi.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 85.Liang Y., Ding X., Yu X., Wang Y., Zhou Y., He J., Shi Y., Zhang Y., Lin H., Lu D. Identification and functional characterization of Toll-like receptor 13 from orange-spotted grouper (Epinephelus coioides) Fish Shellfish Immunol. 2018;74:309–317. doi: 10.1016/j.fsi.2017.12.054. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y., Bi X., Chu Q., Xu T. Discovery of toll-like receptor 13 exists in the teleost fish: Miiuy croaker (Perciformes, Sciaenidae) Dev. Comp. Immunol. 2016;61:25–33. doi: 10.1016/j.dci.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 87.Samanta M., Swain B., Basu M., Mahapatra G., Sahoo B.R., Paichha M., Lenka S.S., Jayasankar P. Toll-like receptor 22 in Labeo rohita: molecular cloning, characterization, 3D modeling, and expression analysis following ligands stimulation and bacterial infection. Appl. Biochem. Biotechnol. 2014;174:309–327. doi: 10.1007/s12010-014-1058-0. [DOI] [PubMed] [Google Scholar]
- 88.Takano T., Kondo H., Hirono I., Endo M., Saito-Taki T., Aoki T. Molecular cloning and characterization of Toll-like receptor 9 in Japanese flounder, Paralichthys olivaceus. Mol. Immunol. 2007;44:1845–1853. doi: 10.1016/j.molimm.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 89.Bi D., Gao Y., Chu Q., Cui J., Xu T. NOD1 is the innate immune receptor for iE-DAP and can activate NF-κB pathway in teleost fish. Dev. Comp. Immunol. 2017;76:238–246. doi: 10.1016/j.dci.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 90.Li T., Shan S., Wang L., Yang G., Zhu J. Identification of a fish-specific NOD-like receptor subfamily C (NLRC) gene from common carp (Cyprinus carpio L.): characterization, ontogeny and expression analysis in response to immune stimulation. Fish Shellfish Immunol. 2018;82:371–377. doi: 10.1016/j.fsi.2018.08.045. [DOI] [PubMed] [Google Scholar]
- 91.Jault C., Pichon L., Chluba J. Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Mol. Immunol. 2004;40:759–771. doi: 10.1016/j.molimm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 92.Stein C., Caccamo M., Laird G., Leptin M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-11-r251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Il Yoon S., Kurnasov O., Natarajan V., Hong M., Gudkov A.V., Osterman A.L., Wilson I.A. Structural basis of TLR5-flagellin recognition and signaling. Science (80-. ) 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Civjan N.R., Bayburt T.H., Schuler M.A., Sligar S.G. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques. 2003;35:556–563. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- 95.Diller A., Loudet C., Aussenac F., Raffard G., Fournier S., Laguerre M., Grélard A., Opella S.J., Marassi F.M., Dufourc E.J. Bicelles: a natural “molecular goniometer” for structural, dynamical and topological studies of molecules in membranes. Biochimie. 2009;91:744–751. doi: 10.1016/j.biochi.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Werner T., Morris M.B., Dastmalchi S., Church W.B. Structural modelling and dynamics of proteins for insights into drug interactions. Adv. Drug Deliv. Rev. 2012;64:323–343. doi: 10.1016/j.addr.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 97.Fiser A. Template-based protein structure modeling. Methods Mol. Biol. 2010;673:73–94. doi: 10.1007/978-1-60761-842-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berman H., Henrick K., Nakamura H., Markley J.L. The worldwide protein data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maekawa S., Ohto U., Shibata T., Miyake K., Shimizu T. Crystal structure of NOD2 and its implications in human disease. Nat. Commun. 2016;7 doi: 10.1038/ncomms11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu D., Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins Struct. Funct. Bioinforma. 2012;80:1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lemer C.M.-., Rooman M.J., Wodak S.J. Protein structure prediction by threading methods: evaluation of current techniques. Proteins Struct. Funct. Bioinforma. 1995;23:337–355. doi: 10.1002/prot.340230308. [DOI] [PubMed] [Google Scholar]
- 102.Sahoo B.R., Swain B., Dikhit M.R., Basu M., Bej A., Jayasankar P., Samanta M. Activation of nucleotide-binding oligomerization domain 1 (NOD1) receptor signaling in labeo rohita by ie-dap and identification of ligand-binding key motifs in nod1 by molecular modeling and docking. Appl. Biochem. Biotechnol. 2013;170:1282–1309. doi: 10.1007/s12010-013-0263-6. [DOI] [PubMed] [Google Scholar]
- 103.Maharana J., Swain B., Sahoo B.R., Dikhit M.R., Basu M., Mahapatra A.S., Jayasankar P., Samanta M. Identification of MDP (muramyl dipeptide)-binding key domains in NOD2 (nucleotide-binding and oligomerization domain-2) receptor of Labeo rohita. Fish Physiol. Biochem. 2013;39:1007–1023. doi: 10.1007/s10695-012-9758-2. [DOI] [PubMed] [Google Scholar]
- 104.Maharana J., Dehury B., Sahoo J.R., Jena I., Bej A., Panda D., Sahoo B.R., Patra M.C., Pradhan S.K. Structural and functional insights into CARDs of zebrafish (Danio rerio) NOD1 and NOD2, and their interaction with adaptor protein RIP2. Mol. BioSyst. 2015;11:2324–2336. doi: 10.1039/c5mb00212e. [DOI] [PubMed] [Google Scholar]
- 105.Heo L., Feig M. Experimental accuracy in protein structure refinement via molecular dynamics simulations. Proc. Natl. Acad. Sci. U. S. A. 2018;115:13276–13281. doi: 10.1073/pnas.1811364115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adiyaman R., McGuffin L.J. Methods for the refinement of protein structure 3D models. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20092301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raval A., Piana S., Eastwood M.P., Dror R.O., Shaw D.E. Refinement of protein structure homology models via long, all-atom molecular dynamics simulations. Proteins Struct. Funct. Bioinforma. 2012;80:2071–2079. doi: 10.1002/prot.24098. [DOI] [PubMed] [Google Scholar]
- 108.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 109.Dror R.O., Dirks R.M., Grossman J.P., Xu H., Shaw D.E. Biomolecular simulation: a computational microscope for molecular biology. Annu. Rev. Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 110.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pirher N., Ivičak K., Pohar J., Benčina M., Jerala R. A second binding site for double-stranded RNA in TLR3 and consequences for interferon activation. Nat. Struct. Mol. Biol. 2008;15:761–763. doi: 10.1038/nsmb.1453. [DOI] [PubMed] [Google Scholar]
- 112.Guterres H., Lee H.S., Im W. Ligand-binding-site structure refinement using molecular dynamics with restraints derived from predicted binding site templates. J. Chem. Theory Comput. 2019;15:6524–6535. doi: 10.1021/acs.jctc.9b00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu Y., Tao X., Shen B., Horng T., Medzhitov R., Manley J.L., Tong L. Structural basis for signal transduction by the toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 114.Matsushima N., Miyashita H., Enkhbayar P., Kretsinger R.H. Comparative geometrical analysis of leucine-rich repeat structures in the NOD-like and Toll-like receptors in vertebrate innate immunity. Biomolecules. 2015;5:1955–1978. doi: 10.3390/biom5031955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang J., Zhang Z., Liu J., Li F., Chang F., Fu H., Zhao J., Yin D. Structural characterization and evolutionary analysis of fish-specific TLR27. Fish Shellfish Immunol. 2015;45:940–945. doi: 10.1016/j.fsi.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 116.Fink I.R., Pietretti D., Voogdt C.G.P., Westphal A.H., Savelkoul H.F.J., Forlenza M., Wiegertjes G.F. Molecular and functional characterization of Toll-like receptor (Tlr)1 and Tlr2 in common carp (Cyprinus carpio) Fish Shellfish Immunol. 2016;56:70–83. doi: 10.1016/j.fsi.2016.06.049. [DOI] [PubMed] [Google Scholar]
- 117.Tong C., Lin Y., Zhang C., Shi J., Qi H., Zhao K. Transcriptome-wide identification, molecular evolution and expression analysis of Toll-like receptor family in a Tibet fish, Gymnocypris przewalskii. Fish Shellfish Immunol. 2015;46:334–345. doi: 10.1016/j.fsi.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 118.Wang Y., Li J., Han J., Shu C., Xu T. Identification and characteristic analysis of TLR28: a novel member of the TLR1 family in teleost. Dev. Comp. Immunol. 2016;62:102–107. doi: 10.1016/j.dci.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 119.Manon F., Favier A., Núñez G., Simorre J.P., Cusack S. Solution structure of NOD1 CARD and mutational analysis of its interaction with the CARD of downstream kinase RICK. J. Mol. Biol. 2007;365:160–174. doi: 10.1016/j.jmb.2006.09.067. [DOI] [PubMed] [Google Scholar]
- 120.Goodsell D.S., Morris G.M., Olson A.J. Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit. 1996;9:1–5. doi: 10.1002/(SICI)1099-1352(199601)9:1<1::AID-JMR241>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 121.Rarey M., Kramer B., Lengauer T., Klebe G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 122.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 123.Verdonk M.L., Cole J.C., Hartshorn M.J., Murray C.W., Taylor R.D. Improved protein-ligand docking using GOLD. Proteins Struct. Funct. Genet. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 124.Thompson M.A. ACS Meet. 2004. Molecular docking using ArgusLab, an efficient shape-based search algorithm and the AScore scoring function; p. 172. [Google Scholar]
- 125.Dominguez C., Boelens R., Bonvin A.M.J.J. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 126.Macindoe G., Mavridis L., Venkatraman V., Devignes M.D., Ritchie D.W. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chakrapani V., Rasal K.D., Kumar S., Mohapatra S.D., Sundaray J.K., Jayasankar P., Barman H.K. In silico analysis of nsSNPs of carp TLR22 gene affecting its binding ability with poly I:C. Interdiscip. Sci. Comput. Life Sci. 2018;10:641–652. doi: 10.1007/s12539-017-0247-2. [DOI] [PubMed] [Google Scholar]
- 129.Song W.S., Jeon Y.J., Namgung B., Hong M., Il Yoon S. A conserved TLR5 binding and activation hot spot on flagellin. Sci. Rep. 2017 doi: 10.1038/srep40878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gunn R.J., Herrin B.R., Acharya S., Cooper M.D., Wilson I.A. VLR recognition of TLR5 expands the molecular characterization of protein antigen binding by non-Ig-based antibodies. J. Mol. Biol. 2018 doi: 10.1016/j.jmb.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bell J.K., Askins J., Hall P.R., Davies D.R., Segal D.M. The dsRNA binding site of human Toll-like receptor 3. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8792–8797. doi: 10.1073/pnas.0603245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discovery. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen H., Zhou H.X. Prediction of interface residues in protein-protein complexes by a consensus neural network method: test against NMR data. Proteins Struct. Funct. Genet. 2005;61:21–35. doi: 10.1002/prot.20514. [DOI] [PubMed] [Google Scholar]
- 134.Negi S.S., Schein C.H., Oezguen N., Power T.D., Braun W. InterProSurf: a web server for predicting interacting sites on protein surfaces. Bioinformatics. 2007;23:3397–3399. doi: 10.1093/bioinformatics/btm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Qi Z., Sun B., Zhang Q., Meng F., Xu Q., Wei Y., Gao Q. Molecular cloning, structural modeling, and expression analysis of MyD88 and IRAK4 of golden pompano (Trachinotus ovatus) Dev. Comp. Immunol. 2017;74:19–24. doi: 10.1016/j.dci.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 136.Pan A.C., Jacobson D., Yatsenko K., Sritharan D., Weinreich T.M., Shaw D.E. Atomic-level characterization of protein–protein association. Proc. Natl. Acad. Sci. U. S. A. 2019;116:4244–4249. doi: 10.1073/pnas.1815431116. [DOI] [PMC free article] [PubMed] [Google Scholar]