Abstract
Purpose
To evaluate the role of ensemble learning techniques with deep learning in classifying diabetic retinopathy (DR) in optical coherence tomography angiography (OCTA) images and their corresponding co-registered structural images.
Methods
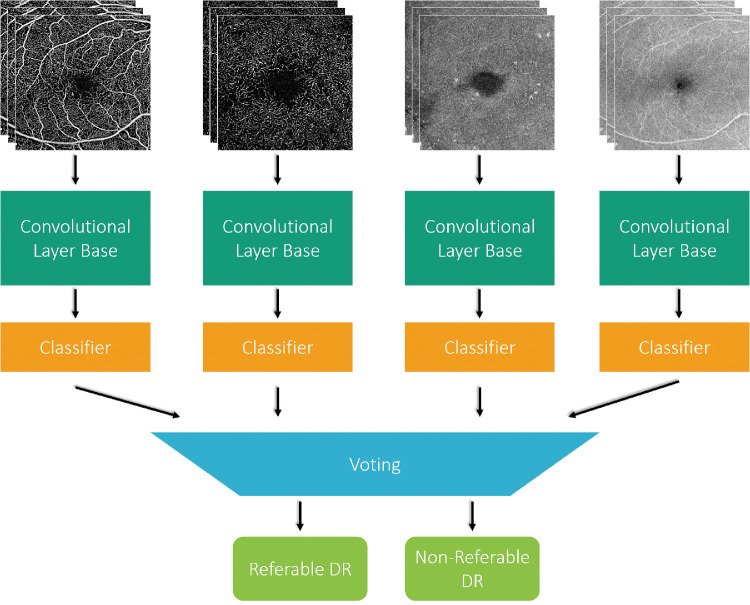
A total of 463 volumes from 380 eyes were acquired using the 3 × 3-mm OCTA protocol on the Zeiss Plex Elite system. Enface images of the superficial and deep capillary plexus were exported from both the optical coherence tomography and OCTA data. Component neural networks were constructed using single data-types and fine-tuned using VGG19, ResNet50, and DenseNet architectures pretrained on ImageNet weights. These networks were then ensembled using majority soft voting and stacking techniques. Results were compared with a classifier using manually engineered features. Class activation maps (CAMs) were created using the original CAM algorithm and Grad-CAM.
Results
The networks trained with the VGG19 architecture outperformed the networks trained on deeper architectures. Ensemble networks constructed using the four fine-tuned VGG19 architectures achieved accuracies of 0.92 and 0.90 for the majority soft voting and stacking methods respectively. Both ensemble methods outperformed the highest single data-type network and the network trained on hand-crafted features. Grad-CAM was shown to more accurately highlight areas of disease.
Conclusions
Ensemble learning increases the predictive accuracy of CNNs for classifying referable DR on OCTA datasets.
Translational Relevance
Because the diagnostic accuracy of OCTA images is shown to be greater than the manually extracted features currently used in the literature, the proposed methods may be beneficial toward developing clinically valuable solutions for DR diagnoses.
Keywords: machine learning, diabetic retinopathy, deep learning, optical coherence tomography, optical coherence tomography angiography
Introduction
Diabetic retinopathy (DR) is a leading cause of blindness in the working age population1 and of an estimated 285 million people worldwide with diabetes mellitus, more than one-third have signs of DR.2 Because patients with DR may be asymptomatic, even in late stages of the disease, it is recommended that any patient diagnosed with diabetes be screened regularly for signs of DR to palliate visual loss.3 Retinal microvasculopathy, such as microaneurysms and capillary occlusion and nonperfusion, is generally observed first4,5 followed by secondary inner retinal degeneration.6,7 Optical coherence tomography angiography (OCTA) is an emerging technology that is able to provide both vascular information for detecting signs of microvasculopathy and structural information through its inherently co-registered optical coherence tomography (OCT) volumes to detect neurodegeneration. The majority of publications investigating the diagnostic capability of OCTA as it relates to DR focus on manually created parameters based on a priori knowledge of the disease pathophysiology. Such morphometric and functional parameters can be quite useful in classifying diseased and nondiseased retinas and techniques using manually engineered features are considered traditional machine learning. However, in recent years a trend in deep learning has been to identify where potential information from the images that may be discarded (or not readily observed by human perception) otherwise can be detected and used for classification with convolutional neural networks (CNNs).
CNNs learn through stochastic optimization, hence they are inherently limited in performance due to the high variance in predictions that results from sensitivity to small fluctuations in the training set leading to overfitting.8 As such, large datasets are generally desired. Although large diabetic retinopathy databases (DRIVE,9 STARE,10 etc.) are publically available, they are comprised of fundus photographs, an imaging modality that does not have the ability of OCT/OCTA images to provide depth resolved images of the various retinal layers. An alternative approach to reducing the variance is to combine multiple, diverse, and accurate models to achieve greater predictive accuracy.11 This is termed ensembling, and in general, a neural network ensemble is constructed in two steps: training a number of component neural networks and then combining the component predictions.12 For training component neural networks, the most prevalent ensemble approaches are Bootstrap aggregating (bagging) and Boosting, which are algorithms that determine the training sets of component networks. Bagging13 is a method based on bootstrap sampling14 (sampling with replacement) that generates a number of training sets from an original training set and trains a component neural network on each sampled dataset. Boosting15–17 generates a series of component neural networks whose training sets are determined by the performance of previous ones. Incorrect predictions are more heavily emphasized in the training of later networks. The networks are then combined typically by majority voting, which can be used for segmentation networks18 as well as classification networks. Another method of combining multiple networks is stacking, whereby the networks are combined by a meta-classifier. This meta-classifier is typically a fully connected neural network and allows for more complex, nonlinear combinations of the network features.
In health care applications, identifying the underlying features through which the algorithm classifies disease, in addition to the quantitative algorithmic performance, is important to promote physician acceptance.19 As such, methods to visualize the areas of images most responsible for the CNNs classification are gaining popularity. Class activation maps (CAMs)20 are a common method where a heat map is generated by projecting the class specific weights of the output classification layer back to the feature maps of the last convolutional layer, thereby highlighting important regions for predicting a particular class. This method has been used in ophthalmic application previously to confirm CNN decision was based off the anterior chamber angle in categorizing angle closure,21 areas of OCT B-scans associated with various diagnoses22,23 and areas of segmentation error,24 and area of OCT enface images associated with the diagnosis of glaucoma.25 There exists several variants of this method that build off of the original CAM paper,20 including: Grad-Cam,26 Guided Grad-Cam,26 Guided Grad-Cam++,27 and GAIN.28
In this paper, we use ensemble learning techniques together with CNNs to classify referable DR, using OCT and OCTA images. The results of the deep learning algorithms will be compared to manually extracted features. Additionally, we will show how CAMs can be used to aid in the interpretation of the CNN classification.
Methods
Patients
This study adhered to the tenets of the Declaration of Helsinki and was approved by the Research Ethics Boards of the University of British Columbia and Simon Fraser University. Patients with diabetes mellitus type 1 or 2, and any diabetic retinopathy severity level, as well as controls were included in the study. Patients were excluded if they had substantial media opacity that would preclude successful imaging, active inflammation, structural damage to the center of the macula, substantial nondiabetic intraocular pathologies, or any intraocular surgery with the exception of cataract surgery. A total of 380 eyes were examined from 242 subjects. A total of 224 eyes were classified as nonreferable DR, and the other 156 were classified as having referable DR by a trained ophthalmologist. Referable DR was classified as having more than mild nonproliferative DR or any stage DR with diabetic macular edema. The mean age of patients with referable and nonreferable DR was 59.3 ± 11.7 years and 58.8 ± 17.4 years, respectively.
Optical Coherence Tomography Data
Patients were imaged using the Zeiss PlexEite 9000 (Zeiss Meditec Inc, Germany) with an A-scan rate of 100 Khz. OCTA was computed using the OCT microangiography complex algorithm. Data were acquired as a 3 × 3-mm volume centered on the foveal avascular zone (FAZ). A sampling rate of 300 × 300 was used that corresponds to a distance of 10 microns between scanning locations. Each B-scan location was scanned a total of four times. The A scan depth is reported as 3 mm in tissue with an optical axial resolution of 6.3 µm and a transverse resolution of 20 µm. Scans were only included in the study if the system specified signal strength was 7 or higher. Figure 1 shows representative OCT and OCTA enface images for a severe NPDR subject.
Figure 1.

Comparison of clinical features seen on both OCT and OCTA enface images of a proliferative DR patient. Dilated capillaries/microaneurysms (blue circles) are clearly visible in the superficial and deep capillary plexus of both OCT and OCTA images. Areas of capillary dropout (outlined in yellow) are more clearly seen in OCTA images, though the deep structural OCT image also shows areas of lower intensities in the larger area of nonperfusion.
Manual Feature Extraction
En face OCTA images were extracted from the superficial and deep vascular complexes and projection artifacts were removed using the in-built system software before being exported. Images were then segmented using a separate vessel segmentation DNN (Deep Neural Network) (Lo J, Heisler M, Vanzan V, et al., submitted, 2019) to calculate the handcrafted features. Methods for feature extraction have been previously reported,29–31 but are explained here in brief for completeness. Seven FAZ morphometric parameters were calculated from the vessel segmentation network results: area, perimeter, acircularity index, maximum and minimum diameter, axis ratio, and eccentricity. The FAZ was found as the largest connected nonvessel area. The centroid for this area was then used to determine the perimeter, and maximum and minimum diameter. Acircularity index was defined as the ratio of the perimeter of the FAZ to the perimeter of a circle with equal area. Axis ratio was the ratio of the maximum FAZ diameter to the minimum FAZ diameter, and eccentricity was calculated as the eccentricity of the ellipse made by the minimum and maximum diameters.
Five vascular parameters were also extracted from the superficial vessel segmentation network results: whole image density, inner density, central density, skeleton density, and fractal dimension. Before quantification, the vessel segmentation network result was binarized using a threshold of 0.5. Whole image density was then calculated as the proportion of measured area occupied by pixels that were classified by the algorithm as a vessel. Central density was calculated as the density within the center 1-mm circle, and inner density as the density in the ring between 1 and 3 mm from the center.
Diagnostic Network Architectures
Three different CNN architectures were used in this paper: VGG19, ResNet50, and DenseNet. Each network was loaded with the pretrained weights on the ImageNet dataset and truncated at the deepest convolutional layer. A global average pooling layer was then appended followed by a dense layer with two outputs.
For inputs, each base was trained with four different single data-type en face images extracted from the OCTA and OCT volumes. From the OCTA, both enface superficial and deep plexus images were extracted. Similarly, from the structural OCT volume, both enface superficial and deep plexus images were extracted.
The various networks were then combined to be of the configurations in Figures 2 and 3 to evaluate voting and stacking. For voting as in Figure. 2, we implemented a majority soft voting scheme by averaging out the probabilities calculated by individual networks. Although voting is the most common aggregation method in classification tasks, it only considers linear relationships among classifiers. Stacking as in Figure. 3, is another ensemble technique where the meta-classifier is able to learn complex associations. As ensemble networks perform best when the networks are diverse and accurate, the best performing trained network for each input type was chosen for this architecture. The input to the meta-classifier was the concatenation of last convolutional layer of each chosen component network. This was followed by a global average pooling layer, a dense layer with 1024 nodes and relu activation, a dense layer with 512 nodes and relu activation, and a final dense layer with 2 nodes for classification. The weights of all the trained convolutional bases were frozen while the meta-classifier was trained.
Figure 2.
Example of the majority voting ensemble method for combining classification results from multiple component networks. The component networks were previously trained on superficial and deep plexus enface images of OCT and OCTA volumes separately.
Figure 3.
Example of the Stacking Ensemble Method for combining classification results from multiple component networks. The component networks were previously trained on superficial and deep plexus enface images of OCT and OCTA volumes separately and the weights were frozen while the meta-classifier was trained.
To compare the diagnostic capability of a feature agnostic CNN to the manually extracted features, the 12 manual features were also fed into a classifier. The classifier comprised a multilayer perceptron with 2 hidden layers of 12 and 6 nodes, respectively, and 1 binary output for referable DR or nonreferable DR. A threshold of 50% was applied to the output probabilities to determine the classification.
Experimental Settings
The CNN-based detection method was implemented in Keras using the Tensorflow backend and Python 3.5.4. We ran the algorithm on a desktop PC with an i7-6700K CPU at 4.0 GHz, 16 GB of RAM, and a GeForce GTX 1060 GPU. Five-fold cross-validation was performed on each configuration, where the data were split 60% for training, 20% for validation, and 20% for test. Care was taken to ensure eyes from the same subject were only included in one of either the training, validation or testing datasets. Initially, all weights in the convolutional layer base were frozen and just the two new layers comprising the classifier were trained. This was done for 10 epochs, with a learning rate of 0.00001, batch size of 8, and 2 callback functions set to only save the best network and to stop training if the validation loss had not improved after 5 epochs. Then, all weights were unfrozen and the network was retrained for 20 epochs with the same callbacks and learning rate. As suggested in the literature,22 training from scratch on OCT images may be preferable as many of the low-level filters in networks pretrained on natural images are tuned to colors and OCT images are monochromatic. However, retraining the entire network preinitialized on ImageNet provided us with better performance than training from uninitialized weights, likely due to our significantly smaller dataset. Data augmentation techniques were also used with random rotations ([−5°, +5°]), zoom (≤ 20%), height and width shift (≤ 10%), and both horizontal and vertical flipping set. In response to the unbalanced classes used for training, class weights were also assigned to the loss function to mitigate any undue bias toward the class with more training data.
Model Visualization
In this paper, we will compare two class activation maps: the original class activation map20 and a variant termed Grad-Cam.26 Class activation maps were used to help visualize the areas of the image which were most helpful in determining the classification. The original CAM method did this by modifying the network architecture to add a global average pooling layer, followed by a dense layer to the convolutional network base. Then, then CAM was calculated as a weighted sum of the feature maps per class. Grad-CAM performs a similar function by using the gradients of any class, flowing into the final convolutional layer to produce the localization map. For our purpose, we have chosen to only propagate positive gradients for positive activations.32
Results
Manual Features
The mean values of the manually extracted features are shown in Table 1. Fifteen volumes were removed from the manual feature analysis because of poor segmentation resulting in inaccurate parameterization. Two-tailed t-tests indicate that all the manually extracted features in the dataset are statistically different between means, except the axis ratio. As such, all features except axis ratio were included in the manual feature classifier.
Table 1.
Mean Values of Manually Extracted Parameters
| Nonreferable DR (± SD) | Referable DR (± SD) | P Value | |
|---|---|---|---|
| FAZ parameters | |||
| Area (mm2) | 1.340 (0.825) | 1.915 (1.204) | < 0.01 |
| Perimeter (mm) | 7.591 (5.070) | 10.199 (5.741) | < 0.01 |
| Acircularity index | 1.835 (0.661) | 2.072 (0.665) | < 0.01 |
| Minimum diameter (mm) | 0.997 (0.242) | 1.088 (0.293) | < 0.01 |
| Maximum diameter (mm) | 1.545 (0.341) | 1.867 (0.534) | < 0.01 |
| Axis ratio | 1.654 (0.822) | 1.762 (0.429) | 0.11 |
| Eccentricity | 0.562 (0.152) | 0.625 (0.147) | < 0.01 |
| Vascular parameters | |||
| Vessel density | 0.449 (0.041) | 0.384 (0.044) | < 0.01 |
| Inner density | 0.463 (0.042) | 0.395 (0.046) | < 0.01 |
| Central density | 0.261 (0.062) | 0.207 (0.065) | < 0.01 |
| Skeleton density | 0.062 (0.007) | 0.051 (0.007) | < 0.01 |
| Fractal dimension | 1.883 (0.014) | 1.861 (0.017) | < 0.01 |
Diagnostic Network Results
Table 2 shows the accuracy of the single input networks. Interestingly, the VGG19 networks achieve the best accuracy for all four inputs. Additionally, the superficial structural images achieve the worst performance when compared with the other input image types. Conversely, the deep structural images achieve the highest accuracy out of all the single networks when using the VGG19 architecture.
Table 2.
Accuracy of Single Input Networks
| Deep | Superficial | Deep | Superficial | |
|---|---|---|---|---|
| Structural (± SD) | Structural (± SD) | Vascular (± SD) | Vascular (± SD) | |
| VGG19 | 87.45 (2.98) | 77.57 (2.57) | 85.56 (2.33) | 85.76 (2.86) |
| ResNet50 | 77.76 (5.72) | 67.81 (6.46) | 79.25 (3.76) | 76.92 (5.18) |
| DenseNet | 71.70 (1.83) | 64.51 (4.35) | 76.07 (5.54) | 81.70 (5.68) |
Table 3 reports the accuracy, sensitivity, and specificity of the ensembled networks as well as the network classifier the manual features. Both ensemble methods achieved higher accuracy than the manual feature classifier.
Table 3.
Comparison of Ensembled Networks to Manual Feature Classifier
| Majority Voting | Stacking | Manual Features | |
|---|---|---|---|
| Accuracy | 92.00 (1.92) | 89.86 (2.55) | 83.10 (4.89) |
| Sensitivity | 90.41 (6.23) | 87.38 (5.85) | 69.26 (9.02) |
| Specificity | 93.33 (5.18) | 92.09 (5.16) | 78.42 (6.32) |
Table 4 reports the accuracy, sensitivity, and specificity of a three-channel input ensemble network and three-channel input VGG19 network. As the superficial structural images showed the lowest diagnostic accuracy in the single networks (Table 2), and we used networks including pretrained ImageNet weights which require a three-channel input, the networks in these results did not include the superficial structural images. Majority voting outperformed the stacking method, and was chosen for comparison to a standard single VGG19 network.
Table 4.
Comparison of 3 Channel Input Networks
| Majority Voting | VGG-19 | |
|---|---|---|
| Accuracy | 90.71 (1.65) | 87.70 (3.41) |
| Sensitivity | 93.32 (5.34) | 94.20 (3.13) |
| Specificity | 87.74 (4.88) | 80.53 (6.60) |
Model Visualization
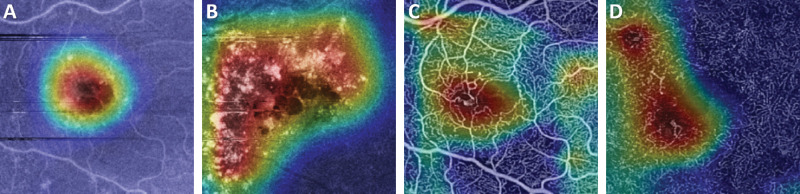
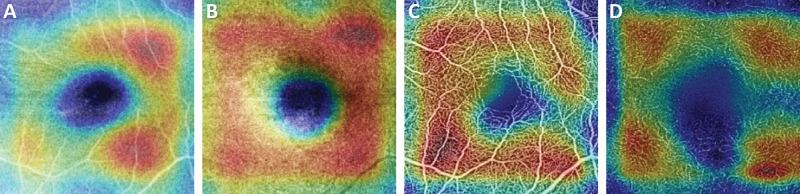
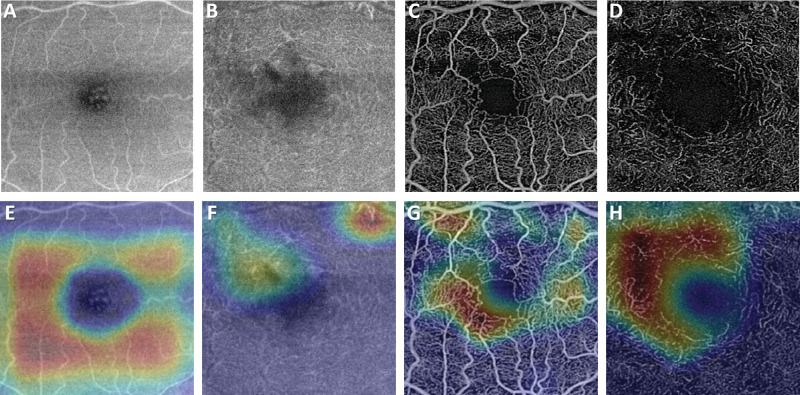
A comparison of representative CAM and Grad-CAM visualizations are shown in Figure 4 for the case of a subject with DR. As shown, the Grad-CAM image is better able to focus on features associated with the diseased regions. The vessel thickening above the FAZ, and region of capillary dropout to the left of the FAZ are shown to be more predictive of disease than the relatively normal looking vasculature more peripherally. An additional diabetic patient (severe DR) is shown in Figure 5 along with the Grad-Cam images for the superficial and deep plexus of both structural OCT and OCTA images. These images highlight that for each input image, the networks are searching for distinctly different features for classification. In the structural images (Figure 5), the most attention is given to regions of fluid and the hard exudates surrounding that region. For the OCTA images, the region of greatest influence is centered on the larger microaneurysms in the images. Conversely, for the control images shown in Figure 6, the whole parafoveal zone is shown to be influential.
Figure 4.
(A) A deep plexus enface image of a DR subject and the corresponding heat maps using the (B) original CAM method and (C) Grad-CAM. As shown by the smaller, more focal regions of warmer colors, the Grad-CAM image is able to localize on areas of disease better than the original CAM method. The vessel thickening above the FAZ, and region of capillary dropout to the left of the FAZ are shown to be more predictive of disease than the relatively normal looking vasculature more peripherally.
Figure 5.
Grad-CAMs for (A) the superficial structural image, (B) deep structural image, (C) superficial angiography image, (D) and deep vasculature image of a severe DR patient. Hard exudates and regions of fluid are highlighted in the structural images. Microaneurysms and regions of capillary dropout are highlighted in the vascular images.
Figure 6.
Grad-CAMs for (A) the superficial structural image, (B) deep structural image, (C) superficial angiography image, and (D) deep vasculature image of a control patient. Regions of higher uniform intensity in the structural images and regions of normal vasculature tend to have a greater effect on the classification.
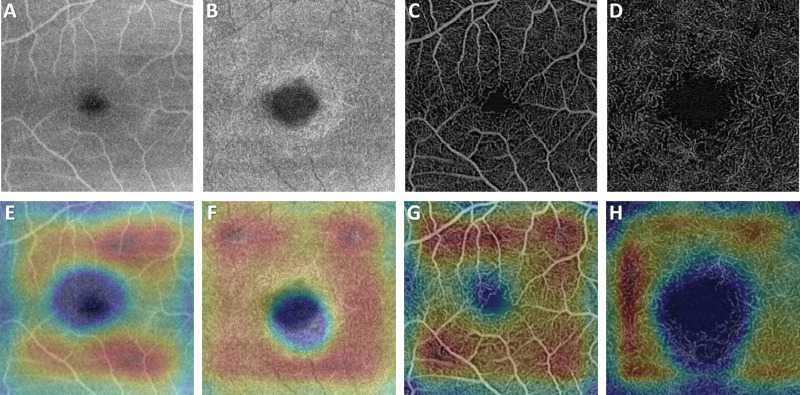
In cases of misclassification, the results from Grad-CAM resembled the pattern more typical of the incorrect classified category. For a referable NPDR patient, as in Figure 7, the Grad-CAMs show the characteristic pattern for nonreferable DR, having a brighter area in the parafoveal zone with the FAZ being less influential. For a case of mild DR incorrectly classified as referable DR, more focal regions of potential vessel dropout on the temporal side of the fovea are shown to be of high importance and contributing to the misclassification (Figs. 8F–8H).
Figure 7.
Representative Grad-CAM results for a NPDR patient that was misclassified as nonreferable. The (A) superficial structural image, (B) deep structural image, (C) superficial angiography image and (D) deep vasculature image of the referable NPDR patient who was misclassified as nonreferable DR and (E-H) the corresponding Grad-CAMs. The typical nonreferable DR pattern of a brighter parafoveal region is observed in all Grad-CAMs.
Figure 8.
Representative Grad-CAM results for a mild DR patient that was misclassified as referable DR. The (A) superficial structural image, (B) deep structural image, (C) superficial angiography image, and (D) deep vasculature image of the nonreferable mild DR patient who was misclassified as referable DR and (E-H) the corresponding Grad-CAMs. (F-H) Focal regions of potential vessel dropout on the temporal side of the fovea are shown to be of high importance in the images which were misclassified as referable DR, whereas the superficial structural image shows the characteristic parafoveal pattern because it was the only image correctly classified.
Discussion
As retinal imaging systems continue to improve, so does our ability to see hallmark features of DR. Optical coherence tomography angiography allows clinicians to view depth-resolved sections of the retina for both structural clues, as well as vascular. As a result, OCTA may enable accurate detection of DR if the right features are used for classification. In this paper, we compare the classification accuracy of features automatically learned from single plexus enface images, combinations of these learned features, and hand-crafted features. Insight to the features learned by the CNN are highlighted through CAM heat maps.
It is shown in Tables 2 and 3 that a combination of diverse component networks provide a higher accuracy than single component networks alone. Both majority voting and stacking techniques yield higher accuracies. Although meta-learners, like the one in the stacking technique, typically outperform simpler algorithms such as majority voting, the relationship between our four inputs was likely more straightforward than typical problems requiring meta-learners. Different configurations for the meta-learner were investigated, but none was able to beat the simple majority voting approach for this dataset. For the component networks, which were trained on enface images from a single plexus, the shallower network of VGG19 performed better on this task than the deeper state-of-the-art networks. While there is some precedent for this,33 it is expected that the ResNet or DenseNet architectures would perform better with more data. Although pretrained ResNet18 ImageNet weights are not currently available for use with Keras, this architecture may perform similarly or better than the VGG19 architecture. The superficial structural images had the worst performance, which may be due to their relatively homogenous appearance, even in diseased states. This lack of texture can make it difficult for CNNs to learn features. Additionally, the deep structural images appear to achieve accuracies on par or better than the vascular images, suggesting that OCT structural information should also be taken into account while analyzing OCTA volumes.
There exists strong evidence in the literature that hand-crafted features extracted from OCT and OCTA images are able to differentiate between grades of DR.34–36 One study37 that looked at combining hand-crafted features from both the superficial and deep plexus resulted in an overall accuracy, sensitivity, and specificity of 94.3, 97.9, and 87.0, when classifying between controls and mild DR patients. This paper extracts parameters for the vessel density, blood vessel caliber, and width of the FAZ and used an SVM for the classifier. Another recent paper38 uses both deep learning and manually extracted features to detect DR in OCT. For their network, they combined both handcrafted features and learned features to differentiate between grades 0 and 1 DR with an accuracy, sensitivity and specificity of 0.92, 0.90, and 0.95, respectively. It is important to note that with manually extracted features there is the ability for errors to propagate where errors which arise during the segmentation and parameterization phase to affect the classification. Future work could use an ensemble of both hand-crafted features and learned features to enhance performance.
To compare whether ensemble techniques achieved better performance than a standard CNN, the three grayscale images with the highest diagnostic accuracy in Table 2 were chosen to create an RGB image as input for a VGG-19 network. When compared with an ensemble network with the same inputs, the ensemble network achieved a higher accuracy and specificity as shown in Table 4. Additionally, although the superficial structural image had a significantly worse diagnostic accuracy on its own, the fact that the four-channel input ensemble network outperformed the three-channel input shows that it still holds important information for the classification of DR and should be considered in future works.
The CAMs, and Grad-CAM in particular, showed good localization of the biomarkers associated with DR, thereby increasing the interpretability of the CNN results. In correctly classified referable DR images, areas of hard exudates, DME, microaneurysms, and capillary dropout show higher activation. The interpretability, and subsequent physician trust in the networks, could also be increased by utilizing Grad-CAM in the choice of layers in which to ensemble as done in a recent paper by Liu et al.39 Grad-CAMs were created for each feature layer in networks trained to detect pseudo-progression of glioblastoma multiform, and a team of three specialists chose the most discriminating layer from each network with which to ensemble. This technique could be used in future work to increase both algorithm performance and physician uptake of the innovation.
Although this study demonstrates the ability of CNNs to classify DR in OCTA with high accuracy, there are notable limitations. First, the use of ensemble learning methods greatly increases the computational cost as it requires the training of multiple networks. This increases both training time as well as the data size of the final model, though this could be partially alleviated through training the component networks in parallel. Another limitation includes the restricted dataset size; however, this limitation was mitigated through the use of fine-tuning, data augmentation, and class-weighting of the loss function. The authors note that improved performance could be achieved through a larger dataset. Furthermore, the dataset only included images with a signal strength of 7 or above, which is sometimes infeasible in patients with pathology. The dataset also consists of images from only one machine, thereby potentially limiting the network's performance on other OCTA machines. Future work could endeavor to further stratify the DR classification.
Acknowledgments
Disclosure: M. Heisler, None; S. Karst, None; J. Lo, None; Z. Mammo, None; T. Yu, None; S. Warner, None; D. Maberley, None; M.F. Beg, None; E.V. Navajas, None; M.V. Sarunic, None
References
- 1. Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010; 376(9735): 124–136. doi: 10.1016/S0140-6736(09)62124-3. [DOI] [PubMed] [Google Scholar]
- 2. Yau JWY, Rogers SL, Kawasaki R, et al.. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care. 2012; 35(3): 556–564. doi: 10.2337/dc11-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antonetti DA, Klein R, Gardner TW. Diabetic retinopathy. N Engl J Med. 2012; 366: 1227–1239. doi: 10.1056/NEJMra1005073. [DOI] [PubMed] [Google Scholar]
- 4. Friedenwald J, Day R. The vascular lesions of diabetic retinopathy. Bull Johns Hopkins Hosp. 1950; 86(4): 253–254. [PubMed] [Google Scholar]
- 5. Friedenwald JS. Diabetic retinopathy. Am J Ophthalmol. 1950; 33(8): 1187–1199. doi: 10.1016/0002-9394(50)90988-3. [DOI] [PubMed] [Google Scholar]
- 6. Cogan DG, Toussaint D, Kuwabara T. Retinal vascular patterns. IV. Diabetic retinopathy. Arch Ophthalmol (Chicago, Ill 1960). 1961; 66: 366–378. doi: 10.1001/archopht.1961.00960010368014. [DOI] [PubMed] [Google Scholar]
- 7. Demirkaya N, van Dijk HW, van Schuppen SM, et al.. Effect of age on individual retinal layer thickness in normal eyes as measured with spectral-domain optical coherence tomography. Invest Ophthalmol Vis Sci. 2013; 54(7): 4934–4940. doi: 10.1167/iovs.13-11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rajaraman S, Jaeger S, Antani SK. Performance evaluation of deep neural ensembles toward malaria parasite detection in thin-blood smear images. PeerJ. 2019. doi: 10.7717/peerj.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Staal JJ, Abramoff MD, Niemeijer M, Viergever MA, van Ginneken B. Ridge based vessel segmentation in color images of the retina. IEEE Trans Med Imaging. 2004; 23(4): 501–509. [DOI] [PubMed] [Google Scholar]
- 10. Hoover A. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans Med Imaging. 2000; 19(3): 203–210. doi: 10.1109/42.845178. [DOI] [PubMed] [Google Scholar]
- 11. Hansen LK, Salamon P. Neural network ensembles. IEEE Trans Pattern Anal Mach Intell. 1990; 12(10): 993–1001. doi: 10.1109/34.58871. [DOI] [Google Scholar]
- 12. Zhou Z-H, Wu J, Tang W. Ensembling neural networks: many could be better than all. Artif Intell. 2002; 137(1): 239–263. doi: 10.1016/S0004-3702(02)00190-X. [DOI] [Google Scholar]
- 13. Breiman L. Bagging predictors. Mach Learn. 1996; 24: 123–140. [Google Scholar]
- 14. Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Monographs on Statistics and Applied Probability, No. 57. Chapman and Hall, London; 1993. [Google Scholar]
- 15. Schapire RE. The Strength of Weak Learnability. Mach Learn. 1990; 5: 197–227. doi: 10.1023/A:1022648800760. [DOI] [Google Scholar]
- 16. Freund Y, Schapire RE. A decision-theoretic generalization of on-line learning and an application to boosting.. Journal of Computer and System Sciences In: Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); 1995; 55(1): 119–139, 10.1006/jcss.1997.1504.. [DOI] [Google Scholar]
- 17. Freund Y. Boosting a weak learning algorithm by majority. Inf Comput. 1995; 121(2): 256–285. doi: 10.1006/inco.1995.1136. [DOI] [Google Scholar]
- 18. Ji Z, Chen Q, Niu S, Leng T, Rubin DL. Beyond retinal layers: a deep voting model for automated geographic atrophy segmentation in SD-OCT images. Transl Vis Sci Technol. 2018; 7(1): 1. doi: 10.1167/tvst.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ting DSW, Pasquale LR, Peng L, et al.. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019103: 167–175. doi: 10.1136/bjophthalmol-2018-313173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning Deep Features for Discriminative Localization. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition.; 2016. doi: 10.1109/CVPR.2016.319. [DOI]
- 21. Shi G, Jiang Z, Deng G, et al.. Automatic classification of anterior chamber angle using ultrasound biomicroscopy and deep learning. Transl Vis Sci Technol. 2019; 8(4): 25. doi: 10.1167/tvst.8.4.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mehta P, Lee A, Lee C, Balazinska M, Rokem A. Multilabel multiclass classification of OCT images augmented with age, gender and visual acuity data. bioRxiv. 2018. doi: 10.1101/316349. [DOI] [Google Scholar]
- 23. Lu W, Tong Y, Yu Y, Xing Y, Chen C, Shen Y. Deep learning-based automated classification of multi-categorical abnormalities from optical coherence tomography images. Transl Vis Sci Technol. 2018; 7(6): 41. doi: 10.1167/tvst.7.6.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jammal AA, Thompson AC, Ogata NG, et al.. Detecting retinal nerve fibre layer segmentation errors on spectral domain-optical coherence tomography with a deep learning algorithm. Sci Rep. 2019; 9(1): 9836. doi: 10.1038/s41598-019-46294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maetschke S, Antony B, Ishikawa H, Wollstein G, Schuman J, Garnavi R. A feature agnostic approach for glaucoma detection in OCT volumes. PLoS One. 2019; 14(7: e0219126. doi: 10.1371/journal.pone.0219126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision.; 2017. doi: 10.1109/ICCV.2017.74. [DOI]
- 27. Chattopadhay A, Sarkar A, Howlader P, Balasubramanian VN. Grad-CAM++: Generalized gradient-based visual explanations for deep convolutional networks. In: Proceedings - 2018 IEEE Winter Conference on Applications of Computer Vision, WACV 2018.; 2018. doi: 10.1109/WACV.2018.00097. [DOI]
- 28. Li K, Wu Z, Peng KC, Ernst J, Fu Y. Tell me where to look: guided attention inference network. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition.; 2018. doi: 10.1109/CVPR.2018.00960. [DOI]
- 29. Heisler M, Chan F, Mammo Z, et al.. Deep learning vessel segmentation and quantification of the foveal avascular zone using commercial and prototype OCT-A platforms. 2019.
- 30. Nesper PL, Roberts PK, Onishi AC, et al.. Quantifying microvascular abnormalities with increasing severity of diabetic retinopathy using optical coherence tomography angiography. Invest Ophthalmol Vis Sci. 2017; 58(6): BIO307–BIO315. doi: 10.1167/iovs.17-21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Carlo TE, Chin AT, Bonini Filho MA, et al.. Detection of microvascular changes in eyes of patients with diabetes but not clinical diabetic retinopathy using optical coherence tomography angiography. Retina. 2015; 35(11): 2364–2370. doi: 10.1097/IAE.0000000000000882. [DOI] [PubMed] [Google Scholar]
- 32. Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M. CoRR. 2014; abs/1412.6806. [Google Scholar]
- 33. Rajaraman S, Candemir S, Xue Z, et al.. A novel stacked generalization of models for improved TB detection in chest radiographs. Conf Proc . Annu Int Conf IEEE Eng Med Biol Soc IEEE Eng Med Biol Soc Annu Conf. 2018; 2018: 718–721. doi: 10.1109/EMBC.2018.8512337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhanushali D, Anegondi N, Gadde SGK, et al.. Linking retinal microvasculature features with severity of diabetic retinopathy using optical coherence tomography angiography. Invest Ophthalmol Vis Sci. 2016; 57(9): OCT519–OCT525. doi: 10.1167/iovs.15-18901. [DOI] [PubMed] [Google Scholar]
- 35. Johannesen SK, Viken JN, Vergmann AS, Grauslund J. Optical coherence tomography angiography and microvascular changes in diabetic retinopathy: a systematic review. Acta Ophthalmol. 2019; 97(1): 7–14. doi: 10.1111/aos.13859. [DOI] [PubMed] [Google Scholar]
- 36. Lu Y, Simonett JM, Wang J, et al.. Evaluation of automatically quantified foveal avascular zone metrics for diagnosis of diabetic retinopathy using optical coherence tomography angiography. Investig Ophthalmol Vis Sci. 2018; 59(6): 2212–2221. doi: 10.1167/iovs.17-23498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sandhu HS, Eladawi N, Elmogy M, et al.. Automated diabetic retinopathy detection using optical coherence tomography angiography: a pilot study. Br J Ophthalmol. 2018; 102(11): 1564–1569. doi: 10.1136/bjophthalmol-2017-311489. [DOI] [PubMed] [Google Scholar]
- 38. Li X, Shen L, Shen M, Tan F, Qiu CS. Deep learning based early stage diabetic retinopathy detection using optical coherence tomography. Neurocomputing. August 2019; 369: 134–144. doi: 10.1016/J.NEUCOM.2019.08.079. [DOI] [Google Scholar]
- 39. Liu X, Chan MD, Zhou X, Qian X. Transparency guided ensemble convolutional neural networks for stratification of pseudoprogression and true progression of glioblastoma multiform. 2019;arXiv 1902.09921.