Abstract
Purpose
To compare performance of independently developed deep learning algorithms for detecting glaucoma from fundus photographs and to evaluate strategies for incorporating new data into models.
Methods
Two fundus photograph datasets from the Diagnostic Innovations in Glaucoma Study/African Descent and Glaucoma Evaluation Study and Matsue Red Cross Hospital were used to independently develop deep learning algorithms for detection of glaucoma at the University of California, San Diego, and the University of Tokyo. We compared three versions of the University of California, San Diego, and University of Tokyo models: original (no retraining), sequential (retraining only on new data), and combined (training on combined data). Independent datasets were used to test the algorithms.
Results
The original University of California, San Diego and University of Tokyo models performed similarly (area under the receiver operating characteristic curve = 0.96 and 0.97, respectively) for detection of glaucoma in the Matsue Red Cross Hospital dataset, but not the Diagnostic Innovations in Glaucoma Study/African Descent and Glaucoma Evaluation Study data (0.79 and 0.92; P < .001), respectively. Model performance was higher when classifying moderate-to-severe compared with mild disease (area under the receiver operating characteristic curve = 0.98 and 0.91; P < .001), respectively. Models trained with the combined strategy generally had better performance across all datasets than the original strategy.
Conclusions
Deep learning glaucoma detection can achieve high accuracy across diverse datasets with appropriate training strategies. Because model performance was influenced by the severity of disease, labeling, training strategies, and population characteristics, reporting accuracy stratified by relevant covariates is important for cross study comparisons.
Translational Relevance
High sensitivity and specificity of deep learning algorithms for moderate-to-severe glaucoma across diverse populations suggest a role for artificial intelligence in the detection of glaucoma in primary care.
Keywords: glaucoma, artificial intelligence, optic disc, machine learning, imaging
Introduction
Early diagnosis of glaucoma is essential to halting irreversible vision loss and preventing blindness. An assessment of two-dimensional fundus photographs centered on the optic disc historically has been one of the most frequently used basic ophthalmologic tools for the detection of glaucomatous optic neuropathy. However, the reproducibility of detecting glaucomatous optic neuropathy from fundus photographs by clinicians can be limited.1,2 In 2013, the US Preventative Services Task Force reported that population-based screening for glaucoma cannot be recommended based on several issues, including a lack of a consistent definition of glaucoma and insufficient sensitivity and specificity for detecting glaucoma using fundus photographs.3 The use of automated evaluation of fundus photographs may be the key to addressing these issues and developing practical glaucoma detection programs. To address this issue, various groups around the world, using different photographic techniques and deep learning strategies,4 have proposed methods for automated evaluation of fundus photographs to detect glaucoma.5–13 We recently have shown that these approaches can be effective despite differences in fundus camera resolution capability or sensor type.5 Most important, deep learning-based methods have been shown to achieve high accuracy on unseen datasets, thanks to their ability to use complex visual features in fundus photographs for the assessment.5–13
Currently, however, little is known about how generalizable these deep learning algorithms for glaucoma detection are to new and independent patient populations and how well different models perform on the same datasets. Often, models are developed and trained on data collected from a specific geographic region or from a homogeneous population and their performance on different populations (e.g., patients from racial groups that are not represented in the training data) is not well-characterized.14 This is important for detecting glaucoma from information available in fundus photographs because it has been widely reported that there are considerable differences in the shape and appearance of optic discs across different races and ethnicities. For instance, differences in optic disc size, shape, and cup-to-disc ratio have been reported across races in healthy,15–19 glaucomatous,20,21 and ocular hypertensive individuals.22 Differences in retinal appearance based on pigment in the retinal pigment epithelium also exist across populations and the effect of these morphologic differences on the accuracy of automated glaucoma detection are an important consideration, especially when deploying these tools throughout the world. For example, if a model is trained using primarily data from patients of European descent collected in the United States, is that model effective at classifying data from patients of Japanese or African descent collected elsewhere in the world? What clinical and demographic characteristics influence model performance? An objective of this report is to exchange test datasets between two institutions, the University of California, San Diego (UCSD), and the University of Tokyo (UTokyo), that have developed deep learning models using datasets from their own countries in order to directly address this question.
One advantage of deep learning over many traditional machine learning methods is the ability to incorporate new data to improve already existing models. A deep learning model trained for one task is used as a starting point and weights are updated by additional training for a different task.23,24 Transfer learning approaches like this can help to address issues related to limited training data, can decrease training time, and can improve performance. Indeed, our research teams from UCSD and UTokyo recently reported the benefits of transfer learning using Imagenet in glaucoma detection from fundus photographs as well as diagnosing early stage glaucoma from optical coherence tomography (OCT) images.5,6,25,26 In both cases, diagnostic performance of the deep learning model was significantly improved by pretraining using transfer learning.5,6 Updating the trained model with new images for the same specialized task can also improve performance. This goal can be accomplished by adding new similar images to the existing model with or without freezing weights of specific layers of the original model. One can also retrain the model from scratch by creating a new training set by combining the original and new data. A quantitative comparison of different strategies for incorporating new data in deep learning models for application to independent patient populations has not been explored previously in the diagnosis of glaucoma using deep learning analysis of fundus photographs. Another objective of this report is to quantitatively compare strategies for incorporating new data into deep learning models using datasets from the United States and Japan.
Thus, the current study had three primary goals with respect to detecting glaucoma in fundus photographs using deep learning: (1) to compare the performance of different deep learning algorithms developed on independent datasets, (2) to characterize model performance stratified by relevant demographic and clinical covariates collected from diverse patient populations, and (3) to evaluate strategies for incorporating new data obtained from independent populations into existing models.
Methods
The fundus photographs used for training and initial evaluation of the deep learning models were taken from two independent datasets collected in the United States and Japan: The Diagnostic Innovations in Glaucoma Study/African Descent and Glaucoma Evaluation Study (DIGS/ADAGES) dataset and the Matsue Red Cross Hospital (MRCH) dataset. In addition to fundus photographs, several demographic and clinical variables including race, axial length (AL), and/or spherical equivalent (SE), and standard automated perimetry visual field (VF) testing results also were collected for each participant. Several additional, independent testing datasets were used to evaluate the models to help estimate their generalizability. All study and data collection protocols adhered to the Declaration of Helsinki. Table 1 summarizes the datasets used in the analysis.
Table 1.
Description of the Datasets Used in the Deep Learning Models
| DIGS/ADAGES | MRCH | Iinan | Hiroshima | ACRIMA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Normal | Glaucoma | Normal | Glaucoma | Normal | Glaucoma | Normal | Glaucoma | Normal | Glaucoma | |
| Training dataa | 1381/2601/8706 | 678/1293/5357 | 1768 | 1364 | — | — | — | — | — | — |
| Testing dataa | 180/319/483 | 90/150/276 | 49 | 61 | 110 | 95 | 75 | 91 | 309 | 396 |
| High myopia (%) | 0 | 2.9 | 27.5 | 35.6 | 0.9 | 3.2 | 0 | 0 | — | — |
| VF MD (dB)(95% CI) | −0.80(−1.02 to −0.58) | −3.32(−3.96 to −2.69) | — | −10.47(−12.60 to −8.34) | — | −4.31(−5.42 to −3.19) | −1.28(−2.04 to −0.53) | −13.54(−15.38 to −11.71) | — | — |
| Age (years)(95% CI) | 54.1(52.7 to 55.5) | 63.1(61.7 to 64.4) | 55.2(50.1 to 60.4) | 65.6(63.1 to 68.1) | 76.4(74.8 to 78.1) | 78.7(77.1 to 80.4) | 51.7(46.2 to 57.1) | 66.3(63.6 to 69.0) | — | — |
| Sex (% female) | 61.9 | 60.1 | 56.9 | 59.3 | 56.4 | 68.4 | 50.70 | 45.10 | — | — |
| Race (%) | ||||||||||
| African descent | 45.5 | 31.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Japanese descent | 1.2 | 5.4 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 |
| European descent | 51.6 | 62.7 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 |
| Other/unreported | 1.7 | 0.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
—, a measurement was unavailable for a given dataset.
The UCSD training and testing data are presented as the number of patients/eyes/scans.
The DIGS/ADAGES Dataset6
All participants were recruited as part of the UCSD-based DIGS or the multicenter ADAGES.27 DIGS (clinicaltrials.gov identifier: NCT00221897) and ADAGES (clinicaltrials.gov identifier: NCT00221923) are prospective studies designed to evaluate longitudinal changes in glaucoma. ADAGES participants were recruited as part of a multicenter collaboration that included the UCSD Hamilton Glaucoma Center (San Diego, CA), Columbia University Medical Center Edward S. Harkness Eye Institute (New York, NY), and the University of Alabama at Birmingham Department of Ophthalmology (Birmingham, AL). Informed consent was obtained from all participants. At study entry, participants had open angles on examination and a best-corrected visual acuity of 20/40 or better, at least one good quality stereophotograph, and at least two reliable VF tests with less than 33% fixation losses and false negatives and less than 15% false positives with no evidence of test artifacts. Participants with high myopia, defined as a SE of −6 diopters or lower, were excluded. Recruitment and data collection protocols were approved by the institutional review boards at each institution and adhered to the Health Insurance Portability and Accountability Act.
The DIGS/ADAGES study population and protocol has been described previously.27 The fundus photographs used in this analysis were captured on film between 2000 and 2011 as simultaneous stereoscopic photographs using the Nidek Stereo Camera Model 3-DX (Nidek Inc., Palo Alto, CA). Photographs were digitized and stored as high resolution (∼2200 × ∼1500) TIFF images. The entire dataset consisted of 7411 stereo pairs split into 14,822 individual images collected from 2920 normal and 1443 glaucoma eyes of 1561 normal and 768 patients with glaucoma. Multiple photographs per eye acquired at different study visits were included if available. The methods for photograph grading have been described previously.27,28 In brief, stereoscopic images were classified as glaucoma or normal by two independent graders, with all disagreements resolved by consensus (discussion among the graders to come to an agreed upon decision) or adjudication by a third senior grader if no consensus after discussion by graders (majority rule). All graders were trained and certified for photograph grading according to standard protocols of UCSD Optic Disc Reading Center. Senior graders had at least 2 years grading experience. Graders consisted of faculty in the UCSD Department of Ophthalmology and ophthalmologists who were completing glaucoma fellowships at UCSD. Photographs were excluded from the analysis only when graders determined that the photograph quality was insufficient to make a classification. No good quality photograph was excluded owing to an inability to classify the photograph. Neither VF testing or OCT imaging were used to diagnose glaucoma for this dataset. This dataset was randomly split by patient into training, validation, and testing subsets using an 85–5–10 percentage split.
The VF mean deviation (MD) was based on Humphrey Field Analyzer 24-2 SITA standard VF testing. AL was measured using the IOLMaster (Carl Zeiss Meditec, Dublin, CA).27
The MRCH Dataset
Data were collected at the MRCH and the study protocol was approved by the Research Ethics Committee of the MRCH and the Faculty of Medicine at UTokyo.5,11 The requirement of informed consent was waived and the study protocol was posted in the clinic to inform patients about the research in accordance with the regulations of the Japanese Guidelines for Epidemiologic Study issued by the Japanese Government.
The MRCH protocol has been previously described.5,11 The fundus images were digitally captured using the Nonmyd WX fundus camera (Kowa Company Ltd., Aichi, Japan). Fundus images had a field of view of 45 degrees and were stored as 2144 × 1424 pixel JPEG images. The training dataset consisted of 3132 images collected from 1768 normal and 1364 glaucoma eyes of 1768 normal and 1364 patients with glaucoma. Training images were classified as glaucoma based on review by a single glaucoma specialist based on the recommendations of the Japan Glaucoma Society Guidelines for Glaucoma.29 This training dataset was randomly split into training and validation subsets using an 95–5 percentage split. The testing dataset was collected from a nonoverlapping cohort and consisted of 110 images collected from 49 normal and 61 glaucoma eyes of 49 normal and 61 patients with glaucoma. For the testing cohort, glaucoma classification was based on expert review of both fundus and OCT images (RS-3000, Nidek, Gamagori, Japan). A glaucomatous classification required visible glaucomatous features in the fundus and OCT imaging, whereas a normal classification required the absence of glaucomatous features or signs of other retinal pathologies. More specifically, as detailed in our previous reports,5,11 the labeling of glaucoma was performed according to the recommendations of the Japan Glaucoma Society Guidelines for Glaucoma29; signs of glaucomatous changes were judged comprehensively, such as focal rim notching or generalized rim thinning, large cup-to-disc ratio with cup excavation with or without a laminar dot sign, retinal nerve fiber layer defects with edges at the optic nerve head margin, and disc edge hemorrhages. Other optic nerve head pathologies, such as optic nerve/optic nerve head hypoplasia and optic nerve pit, and other retinal pathologies such as retinal detachment, age-related macular degeneration, myopic macular degeneration, macular hole, diabetic retinopathy, and arterial and venous obstruction were carefully excluded, but mild epiretinal membrane (without any apparent retinal traction) and mild drusen (without any apparent degeneration) were not excluded. Fundus photographs free of signs of glaucoma and other optic nerve head/retinal pathologies were assigned to the normative dataset. Review was performed independently by three glaucoma specialists (M.T., H.M., and R.A.). and required unanimous agreement. Photographs were excluded if the diagnoses of the three examiners did not agree or if any grader classified the photograph as ungradable.
The SE was measured using RC-5000 refract-keratometer (Tomey GmbH, Nuremberg, Germany). VF testing was performed using the 30-2 test pattern of the Humphrey Field Analyzer (Carl Zeiss Meditec) and standard quality control protocols.
External Testing Datasets
The deep learning models were evaluated using an additional three independent, external datasets. The first external dataset (Iinan Dataset) was collected from patients visiting the glaucoma clinic at Iinan Hospital (Iinan Town, Japan) and included 215 images collected from 110 normal and 95 glaucoma eyes of 110 normal and 95 patients with glaucoma. The second external dataset (Hiroshima Dataset) was collected by the Department of Ophthalmology, Hiroshima University (Hiroshima, Japan) and included 171 images collected from 78 normal and 93 glaucoma eyes of 78 normal and 95 patients with glaucoma. In both of these datasets, fundus imaging, circumpapillary OCT imaging, refractive error, and 30-2 VF testing were collected. Like the MRCH dataset, classification was based on review of fundus and OCT images (RS-3000, Nidek, Gamagori, Japan) by three expert ophthalmologists (M.T., H.M., and R.A.) and required unanimous agreement with photos excluded in the case of disagreement or classified as ungradable.11 The glaucoma and normal groups were defined in the same manner with the MRCH dataset.
The third external dataset was the publicly available ACRIMA dataset.7 This dataset was collected as part of an initiative by the government of Spain and consisted of 705 images collected from 309 normal and 396 glaucoma eyes of 309 normal and 396 patients with glaucoma. Classification was based on review by a single experienced glaucoma expert (personal communication with Dr Diaz-Pinto, Nov 23, 2019). Classification was based solely on fundus photo review and no other clinical information was considered. Images were excluded if they did not provide a clear view of the optic nerve head region.
Deep Learning Strategies
Investigators at the UTokyo and UCSD independently developed deep learning systems to detect glaucoma from fundus images of the optic nerve head and have been described previously.6,11 In developing these models, much of the deep learning model development strategies were similar (both used ResNet30 architectures and horizontal flipping to augment data), but some differences did exist (layer depth and training hyperparameters).24 However, there were small differences in implementation, such as the preprocessing of the photographs (how photographs were cropped, and data augmentation strategies used). These variations in the implementation of the deep learning methods resulted in relatively small differences in results.
In addition to evaluating these original UTokyo and UCSD models, we explored how the incorporation of additional training datasets affects model performance and generalizability. Specifically, we defined two additional strategies: sequential and combined training. In the sequential approach, each model was first trained on an initial training dataset and then further optimized on the other dataset by updating the weights of the deep model layers. That is, the sequential UTokyo model was first trained on the MRCH dataset and weights were then updated by additional training on the DIGS/ADAGES dataset (and vice versa for the sequential UCSD model) without freezing weights of any layer. In the combined approach, all training data (DIGS/ADAGES + MRCH) were pooled together and a single round of training on this combined training dataset was performed without using initializing weights from either dataset. Table 2 summarizes these models and training strategies. Applying these strategies to both the UTokyo and UCSD models resulted in six combinations (an original, sequential, and combined for each) that were evaluated using the available testing datasets. The combined models developed at UCSD and UTokyo use the same combined dataset, but development of the deep learning models was done independently using the deep learning algorithms and strategies developed at each institution (Table 2), resulting in two different combined models.
Table 2.
Description of the Deep Learning Models and Training Approaches at UCSD and UTokyo
| Parameter | UCSD Deep Learning Model | UTokyo Deep Learning Models |
|---|---|---|
| Network architecture | ResNet5030 | ResNet3430 |
| Weight initialization | Pretraining on ImageNet24 | Pretraining on ImageNet24 |
| Data augmentation | Translation, horizontal flipping | Translation, scaling, rotation, and horizontal flipping |
| Training datasets | ||
| Original | DIGS/ADAGES, (ImageNet pretraining) | MRCH, (ImageNet pretraining) |
| Sequential | DIGS/ADAGES, weights updated using MRCH, (ImageNet pretraining) | MRCH, weights updated using DIGS/ADAGES, (ImageNet pretraining) |
| Combined | DIGS/ADAGES & MRCH (ImageNet pretraining) | MRCH & DIGS/ADAGES (ImageNet pretraining) |
Model Evaluation
Models were first evaluated using the independent testing subsets of the DIGS/ADAGES and MRCH datasets. To characterize the effects of demographic and clinical covariates on model predictions, performance stratified by race, myopia status, and disease severity was computed. For this analysis, performance by race was computed for three groups based on participant self-reported race: African descent, Japanese descent, and European descent. Participants without a self-reported race were excluded from this analysis. Myopia status was based on AL and SE. Where AL was available, eyes with an AL of 26 mm or greater were classified as high myopes. If AL was not available, eyes with an SE of less than −6.0 diopters were classified as high myopes. If neither was available, the eye was excluded from the analysis of high myopes. Disease severity was characterized based on VF MD. Glaucoma eyes with an MD of greater than −6.0 dB were classified as mild glaucoma and glaucoma eyes with an MD of −6.0 dB of less were classified as moderate-to-severe glaucoma.
Statistical Analysis
Model evaluation was performed using sensitivity, specificity, and area under the receiver operating characteristic curve (AUC). For all analyses, the AUC with 95% confidence intervals (CIs) and sensitivity at fixed levels of specificity (80%, 85%, 90%, and 95%) was computed. DeLong's test was used to assess the statistical significance of differences in AUC values. Because the DIGS/ADAGES testing set contained multiple images of the same eye, a clustered bootstrap approach was adopted to compute AUCs, bias-corrected confidence intervals, and conduct hypothesis tests.31 An additional evaluation also was performed on the independent, external datasets to estimate the ability of the models to generalize to other study populations.
Results
The demographic and clinical characteristics of the study populations and datasets are presented in Table 1. The DIGS/ADAGES dataset consisted of patients who were of European and African descent, whereas the MRCH, Iinan, and Hiroshima datasets included photographs from Japanese individuals exclusively. The ACRIMA dataset consisted of individuals from Spain. The healthy participants of both the DIGS/ADAGES and MRCH populations were generally younger than the patients with glaucoma by approximately 10 years. DIGS/ADAGES patients with glaucoma had less severe VF damage than the MRCH dataset (mean VF MD of −4.1 dB and −10.5 dB, respectively). The myopia patients were from the MRCH dataset almost exclusively because DIGS/ADAGES excluded high myopia from its study population.
The AUC and sensitivities at fixed specificities (80%, 85%, 90%, and 95%) of all models on the DIGS/ADAGES and MRCH testing data are summarized in Tables 3 and 4. Using the DIGS/ADAGES testing data, the UCSD original model and combined models performed significantly better (P = .002 and P = .014, respectively) than the sequential model (AUCs 0.92, 95% CI 0.89–0.94; AUC 0.90, 95% CI 0.87–0.93; and AUC 0.83, 95% CI 0.79–0.87, respectively). The UTokyo sequential, and combined models that included DIGS/ADAGES data in the training set performed significantly better (P = .002 and P < .001, respectively) on the DIGS/ADAGES dataset than the original model which was based exclusively on Japanese data AUCs of 0.88 (95% CI 0.84–0.92), 0.90 (95% CI 0.87–0.93), and 0.79 (95% CI 0.74, 0.83), respectively. On the MRCH testing data, the original, sequential, and combined UCSD and UTokyo models achieved similar diagnostic accuracy with AUCs ranging between 0.94 and 0.96 for the UCSD models and between 0.95 and 0.97 for the UTokyo models.
Table 3.
Performance of UCSD and UTokyo Models Using the Original, Sequential, and Combined Strategies on the DIGS/ADAGES Testing Dataset
| Sensitivity @ | |||||
|---|---|---|---|---|---|
| Model | AUC (95% CI) | 80% Specificity | 85% Specificity | 90% Specificity | 95% Specificity |
| UCSD | |||||
| Original | 0.92 (0.89–0.94) | 0.87 | 0.83 | 0.76 | 0.49 |
| Sequential | 0.83 (0.78–0.87) | 0.74 | 0.67 | 0.58 | 0.32 |
| Combined | 0.90 (0.87–0.93) | 0.86 | 0.81 | 0.71 | 0.53 |
| UTokyo | |||||
| Original | 0.79 (0.74–0.83) | 0.61 | 0.54 | 0.49 | 0.42 |
| Sequential | 0.88 (0.84–0.92) | 0.82 | 0.77 | 0.72 | 0.55 |
| Combined | 0.90 (0.87–0.93) | 0.85 | 0.82 | 0.72 | 0.57 |
Table 4.
Performance of UCSD and UTokyo Models Using the Original, Sequential, and Combined Strategies on the MRCH Testing Dataset
| Sensitivity @ | |||||
|---|---|---|---|---|---|
| Model | AUC (95% CI) | 80% Specificity | 85% Specificity | 90% Specificity | 95% Specificity |
| UCSD | |||||
| Original | 0.96 (0.94–0.99) | 0.95 | 0.95 | 0.92 | 0.85 |
| Sequential | 0.94 (0.91–0.99) | 0.92 | 0.90 | 0.88 | 0.86 |
| Combined | 0.94 (0.92–0.99) | 0.92 | 0.92 | 0.86 | 0.81 |
| UTokyo | |||||
| Original | 0.97 (0.93–1.00) | 0.95 | 0.95 | 0.93 | 0.88 |
| Sequential | 0.96 (0.93–1.00) | 0.95 | 0.90 | 0.90 | 0.90 |
| Combined | 0.95 (0.91–0.99) | 0.88 | 0.86 | 0.86 | 0.86 |
At a fixed specificity of 90% and 95% on the MRCH testing dataset, the best UCSD models achieved sensitivity of 92% and 86%, respectively, and the best UTokyo models achieved sensitivity of 93% and 90%, respectively. At a fixed specificity of 90% and 95% on the DIGS/ADAGES testing dataset, the best UCSD models achieved a sensitivity of 76% and 53%, respectively, whereas the best UTokyo models achieved a sensitivity of 72% and 57%, respectively.
The results of model performance by race are presented in Table 5. The eyes of individuals of Japanese descent had significantly worse glaucomatous VF damage than the eyes of individuals of African and European descent (mean VF MD of −8.73 dB, −3.93 dB, and −3.06 dB, respectively). The diagnostic accuracy of the UCSD and UTokyo models tended to perform better in the individuals of Japanese and African descent than of European descent, although these differences did not reach statistical significance.
Table 5.
Performance of the UCSD and UTokyo Models Stratified by Race on the Combined DIGS/ADAGES and MRCH Testing Datasets
| AUC (95% CI) | |||
|---|---|---|---|
| Model | African Descent (n = 306) | Japanese Descent (n = 131) | European Descent (n = 422) |
| Mean glaucoma VF MD (dB) | −3.93 (−5.03 to −2.82) | −8.73 (−10.59 to −6.86) | −3.06 (−3.86 to −2.23) |
| UCSD | |||
| Original | 0.95 (0.91 to 0.98) | 0.94 (0.88 to 0.97) | 0.90 (0.86 to 0.93) |
| Sequential | 0.89 (0.83 to 0.94) | 0.92 (0.86 to 0.96) | 0.81 (0.74 to 0.86) |
| Combined | 0.93 (0.87 to 0.96) | 0.93 (0.88 to 0.97) | 0.88 (0.84 to 0.92) |
| UTokyo | |||
| Original | 0.87 (0.80 to 0.93) | 0.92 (0.86 to 0.97) | 0.74 (0.67 to 0.80) |
| Sequential | 0.94 (0.90 to 0.97) | 0.91 (0.81 to 0.97) | 0.86 (0.80 to 0.91) |
| Combined | 0.95 (0.90 to 0.97) | 0.90 (0.82 to 0.96) | 0.87 (0.82 to 0.92) |
The model performance of eyes with and without high myopia is provided in Table 6. Eyes with high myopia had significantly worse glaucomatous VF damage than eyes without high myopia (mean VF MD of −10.48 dB and −4.18 dB, respectively). In general, both the UCSD models and UTokyo models had high diagnostic accuracy for detecting glaucoma in the eyes with high myopia (the AUC ranged between 0.94 and 0.98) and lower diagnostic accuracy in eyes without high myopia (the AUC ranged between 0.81 and 0.92), likely owing to the more severe glaucoma in the eyes with high myopia.
Table 6.
Performance of the UCSD and UTokyo Models by High Myopia Status on the Combined DIGS/ADAGES and MRCH Testing Datasets
| AUC (95% CI) | ||
|---|---|---|
| Model | High Myopia (n = 43) | Not High Myopia (n = 761) |
| Mean Glaucoma VF MD (dB) | −10.48 (−13.56, −7.41) | −4.18 (−4.92, −3.44) |
| UCSD | ||
| Original | 0.95 (0.82 to 1.00) | 0.92 (0.89 to 0.95) |
| Sequential | 0.97 (0.89 to 1.00) | 0.84 (0.80 to 0.88) |
| Combined | 0.98 (0.90 to 1.00) | 0.90 (0.87 to 0.93) |
| UTokyo | ||
| Original | 0.97 (0.88 to 1.00) | 0.81 (0.76 to 0.86) |
| Sequential | 0.94 (0.83 to 1.00) | 0.90 (0.87 to 0.93) |
| Combined | 0.97 (0.88 to 1.00) | 0.91 (0.88 to 0.94) |
As expected, both the UCSD and UTokyo model performance was higher in eyes with moderate-to-severe disease compared with eyes with mild disease (Table 7). Specifically, in the mild glaucoma eyes, the diagnostic accuracy of the UCSD model (AUCs between 0.89 and 0.91) was similar to the UTokyo models (AUCs between 0.78 and 0.89) and lower than the UCSD and Tokyo models for detecting moderate to severe disease (AUCs between 0.94 and 0.98).
Table 7.
Performance of the UCSD and UTokyo Models by Glaucoma Severity on the Combined DIGS/ADAGES and MRCH Testing Data Sets.
| AUC (95% CI) | |||
|---|---|---|---|
| Model | Any Glaucoma (n = 318) | Mild (n = 231) | Moderate-to-Severe (n = 87) |
| Mean Glaucoma VF MD (dB) | −4.76 (−5.48 to −4.04) | −1.34 (−1.57 to −1.10) | −13.86 (−15.09 to −12.63) |
| UCSD | |||
| Original | 0.92 (0.90 to 0.94) | 0.91 (0.88 to 0.93) | 0.98 (0.96 to 0.99) |
| Sequential | 0.85 (0.81 to 0.88) | 0.82 (0.77 to 0.86) | 0.95 (0.93 to 0.97) |
| Combined | 0.91 (0.88 to 0.93) | 0.89 (0.85 to 0.92) | 0.98 (0.96 to 0.99) |
| UTokyo | |||
| Original | 0.82 (0.77 to 0.86) | 0.78 (0.73 to 0.83) | 0.94 (0.90 to 0.98) |
| Sequential | 0.90 (0.86 to 0.93) | 0.88 (0.84 to 0.92) | 0.97 (0.94 to 0.99) |
| Combined | 0.91 (0.88 to 0.93) | 0.89 (0.86 to 0.92) | 0.98 (0.94 to 0.99) |
The best UCSD and UTokyo models also performed similarly on the external datasets (Table 8) with AUCs of 0.94 and 0.97, respectively, on the Iinan dataset, AUCs of 0.99 on the Hiroshima dataset, and AUCs of 0.84 and 0.86, respectively, on the ACRIMA dataset.
Table 8.
The Performance of UCSD and UTokyo Models on External Testing Datasets
| AUC (95% CI) | |||
|---|---|---|---|
| Model | Iinan (n = 205) | Hiroshima (n = 186) | ACRIMA (n = 705) |
| Mean glaucoma VF MD (dB) | −4.31 (−5.42 to −3.19) | −13.54 (−15.38 to −11.71) | — |
| UCSD | |||
| Original | 0.94 (0.91 to 0.97) | 0.96 (0.93 to 0.99) | 0.84 (0.81 to 0.87) |
| Sequential | 0.91 (0.87 to 0.95) | 0.99 (0.98 to 1.00) | 0.75 (0.72 to 0.79) |
| Combined | 0.91 (0.87 to 0.95) | 0.97 (0.95 to 0.99) | 0.80 (0.80 to 0.84) |
| UTokyo | |||
| Original | 0.95 (0.92 to 0.97) | 0.99 (0.99 to 1.0) | 0.82 (0.79 to 0.85) |
| Sequential | 0.97 (0.94 to 0.99) | 0.99 (0.99 to 0.99) | 0.86 (0.83 to 0.89) |
| Combined | 0.90 (0.86 to 0.94) | 0.95 (0.95 to 0.98) | 0.85 (0.82 to 0.88) |
The Figure shows a heat map comparing the glaucoma probability predictions of the UCSD and UTokyo combined models. To characterize cases of model disagreement, we identified images with good and poor levels of agreement between glaucoma probabilities for the two combined models. The combined UCSD and UTokyo models agree on the vast majority of cases (see the high density in the lower left and upper right corners of Fig. A). Example photographs where the models agree on correct predictions of glaucoma and normal images (Figs. B and C), as well as examples where the models disagree (Figs. D and E) are provided. In these cases of agreement, the images display typical signs of glaucomatous (Fig. B) or normal (Fig. C) optic discs. In these cases of disagreement, the discs have a less clear diagnosis (Fig. D) or have image quality issues (Fig. E). To help understand how these models make their decisions, we used class activation maps to identify the most informative regions of the fundus photos.32
Figure.
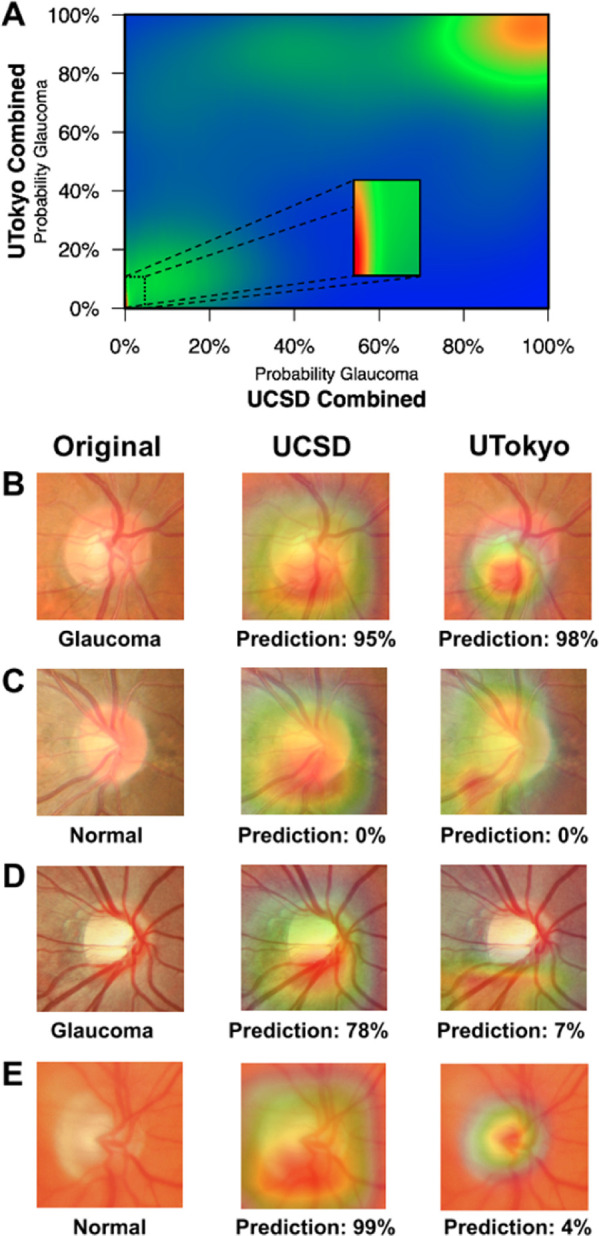
Case examples illustrating good and poor levels of agreement between the UCSD and UTokyo combined deep learning models. (A) A heat map showing the density of predictions from the combined UCSD and UTokyo models. Examples of both models agreeing on a correct classification of a glaucoma (B) and normal (C) images are provided along with the predicted probabilities of glaucoma from the combined models. Similar examples are shown for cases when the models disagreed about a glaucoma (D) and normal (E) image. In (B–E), the original fundus image (left) is shown along with a class activation map identifying informative regions used by the UCSD combined (middle) and UTokyo combined (right) models.
Discussion
This report compared several deep learning model training strategies and quantified the impact of demographic and clinical covariates of the study populations on model performance for detection of glaucoma from optic disc photographs. To our knowledge, this study is the first to directly compare the performance of deep learning algorithms developed by two independent investigators to detect glaucoma in very different glaucoma populations. Overall, the deep learning models performed very well with a best performance of 0.92 (95% CI 0.90–0.94) in detecting any glaucoma, 0.91 (95% CI 0.88–0.93) in detecting mild glaucoma, and 0.98 (95% CI 0.96–0.99) in detecting moderate-to-severe glaucoma in the primary datasets (DIGS/ADAGES and MRCH) that consisted of a large, diverse cohort of patients of African, Japanese, and European descent collected in the United States and Japan. Moreover, the UCSD and UTokyo models for detecting glaucoma in the MRCH dataset had a high sensitivity of at least 90% at a fixed specificity of 90%. This high diagnostic accuracy was similar to that of the first deep learning based algorithm approved by the US Food and Drug Administration for the detection of referral diabetic retinopathy, which is based on testing in 10 primary care sites, had a sensitivity of 87.2% and specificity of 90.8%.33 This finding suggests that the UCSD and UTokyo deep learning algorithms for the detection of moderate-to-severe glaucoma may be ready for testing in primary care settings. Because patients with moderate to severe disease are at a high risk of visual impairment and blindness owing to glaucoma, and up to 50% of glaucoma goes undetected in the population, detection of this stage of disease is a public health priority.34 Placement of fundus cameras with automated glaucoma detection in primary care settings and/or in underserved areas can help to reach many individuals who do not receive regular eye examinations.
In the current study, we first evaluated the performance of existing deep learning models on independent datasets that included patient populations that were substantially different from the training data. We then used two additional strategies, sequential and combined for integrating new training data into model development. Because model performance was overall quite high in the mostly moderate-to-severe glaucoma MRCH dataset, it is more informative to compare the performance of the original, sequential, and combined modeling strategies on the DIGS/ADAGES testing dataset, which included mostly eyes with mild glaucoma. It was not surprising that adding Japanese training examples to the UCSD model did not improve its ability to detect glaucoma in the DIGS/ADAGES dataset, because these examples were not relevant to the task of detecting glaucoma in eyes of African and European descent. In contrast, adding African and European eyes to the training of the Japanese model significantly increased its ability to detect glaucoma in the DIGS/ADAGES testing dataset from the original model (AUC 0.79, 95% CI 0.74–0.83) compared with the combined model (AUC 0.90, 95% CI 0.87–0.93; P < .001) and the sequential model (AUC 0.88, 95% CI 0.84–0.92, P = .002).
A strength of this study is that we computed model performance stratified by disease severity, myopia status, and race using the combined DIGS/ADAGES and MRCH testing data. With respect to disease severity (Table 7), all models performed better at detecting moderate-to-severe glaucoma compared with detecting mild or any glaucoma. The current results also suggest that the diagnostic accuracy of the trained deep learning model may be poorer when there are differences in the disease severity between the training and testing datasets. For example, the original UCSD model did significantly better than the original UTokyo model in detecting mild disease, likely because the MRCH training dataset had fewer examples of mild glaucoma, likely a result of the MRCH grading protocol that excluded cases of disagreement. Similarly, the MRCH glaucoma eyes had significantly more severe disease (as measured by VF MD) than the DIGS/ADAGES glaucoma eyes. In fact, it is likely that disease severity in the MRCH data accounts, at least in part, for the strong performance of all models in this dataset. This finding may also explain why the addition of the MRCH data to the DIGS/ADAGES dataset did not improve the performance of the UCSD model; the UCSD model had a sufficient number of moderate-to-severe cases in its training set to correctly identify the Japanese eyes, which consisted of mostly moderate-to-severe glaucoma in the MRCH dataset. Moreover, the disease severity could also help to explain the high performance of both the UCSD and UTokyo models in eyes with high myopia, even though there were very few high myopes in the DIGS/ADAGES training dataset (Table 6). A similar problem exists in determining the impact of race on model performance. All eyes of Japanese descent came from the MRCH data and again had relatively severe disease, whereas the eyes of African descent and European descent came from the DIGS/ADAGES data and had relatively mild disease. The result is that the models tended to perform well on the eyes of Japanese descent, even when they were not represented in the corresponding training set (e.g., original UCSD). It is unclear why all the models performed better on the eyes of African descent than the eyes of European descent. These subsets both came from the DIGS/ADAGES data and had similar disease severity (VF MD −3.93 dB vs. −3.06 dB, Table 5).
Numerous studies have evaluated deep learning methods for detection of glaucoma in a variety of populations. The diagnostic accuracy reported is generally very good, but varies considerably with AUCs ranging from 0.87 to 0.99.5–13 Most studies train and test on the same source population, which could generally lead to better performance than if the algorithm was tested in an independent population. To address this issue, an increasing number of studies include an independent testing data from other populations and geographic regions.7,8,14 Many studies do not, however, report disease severity or other important clinical (e.g., myopia) and demographic variables (e.g., race).7,8,13,14 Because these variables can substantially impact diagnostic accuracy, it is extremely important to report model performance as in stratified analysis by these covariates. For example, in a stud by Liu et al,14 the deep learning model performed much better in the Chinese datasets (the AUC ranged from 0.964 to 0.997) than the HGC DIGS/ADAGES (0.923) or public website (0.823) images. It is unclear why diagnostic accuracy varies across these study populations, but reporting accuracy by disease severity, myopia status, race, or other relevant covariates would make it easier to compare models across different studies.33,35 Additionally, care should be taken when comparing AUC values between models and datasets owing to differing healthy/glaucoma observation ratios (class imbalance).36 To this end, we have provided sensitivities at fixed specificities (Tables 3 and 4) as an additional performance metric and race-stratified sensitivities/specificities for all datasets (Supplementary Table S1).
With respect to the external testing datasets, performance was best across all models on the Hiroshima dataset (AUCs of 0.95–0.99), followed by the Iinan dataset (AUCs of 0.90–0.97), and was worst on the ACRIMA dataset (AUCs of 0.75–0.86). The relatively good performance on the Hiroshima dataset is likely due primarily to the very severe glaucoma included in the dataset (mean VF MD of −13.54 dB), which makes the detection task relatively easy. The models had the most trouble with the ACRIMA dataset, which differed not only in population (it was collected from a Spanish cohort), but also in glaucoma definition. For the MRCH datasets, glaucoma labeling was performed by three expert graders assessing fundus photographs as well as additional clinical information (e.g., VF and OCT imaging). When these experts disagreed on the labeling, that eye was excluded. This process would lead to these datasets being depleted of difficult cases, resulting in better model performance on the MRCH dataset, regardless of deep learning strategy. In the ACRIMA (and DIGS/ADAGES) datasets, no similar exclusion based on disagreement was performed, leading to datasets with more difficult cases and generally poorer model performance. Our highest AUC on ACRIMA (0.86) was, however, higher than previously reported results for this dataset (0.77).7 With respect to the original, sequential, and combined training strategies, there was no clear best performing strategy across these external datasets. Moreover, the original UCSD and UTokyo models performed similarly on each of these three external datasets.
For automated assessment of fundus photographs using deep learning algorithms to be accepted and used in clinical settings, it is important to open the “black box” and provide some insight into the inner workings of the model and to evaluate the possible reasons for algorithm failure. In the current study, we generated a heat map to compare the predictions of the UCSD and UTokyo combined models and qualitatively evaluated several example fundus photographs. Although the overall diagnostic accuracy of the UCSD and UTokyo algorithms was similar, there were cases in which they disagreed. This finding suggests that, although the algorithms perform similarly, the algorithms may fail on different eyes. We manually reviewed several cases and presented selected, illustrative cases in the Figures B–E. The Figure B provides an example of agreement on a glaucomatous image demonstrating inferior–temporal neuroretinal rim thinning. Similarly, the models strongly agreed on the example case provided in the Figure C, which was a typical normal optic disc. It is interesting to note that even when the UCSD and UTokyo models agreed on the correct predictions (Figs. B and C) and gave special attention to the inferior neuroretinal rim region, the size and location of the informative regions differed. In the one case of disagreement (Fig. D), the optic disc may have been challenging because the participant was a high myope (SE = −6.75 diopters) with relatively mild disease (MD = −4.25 dB). The informative regions for the models were similar (inferior neuroretinal rim region), and it is unclear why the UCSD model correctly classified this eye and the UTokyo model did not. In the final case (Fig. E), although reviewers accepted this image for grading, there may have been image quality issues that likely led to an incorrect prediction by the UCSD model, suggesting that the quality of fundus photographs should be considered when applying these models. The UTokyo model may have correctly predicted this case because it was highly focused on the optic disc, whereas the UCSD model was not. These cases illustrate the need for more research into understanding how the models use the images in their decision-making processes.
One of the limitations of the current study is the discrepancy in glaucoma definitions and labelling used in the different datasets, in particular between DIGS/ADAGES, MCRH/Iinan/Hiroshima, and ACRIMA datasets. For example, in the Japanese datasets, photographs were excluded if there was not complete agreement among the three graders. This factor could have the effect of excluding more difficult and/or early cases from the dataset, resulting in an easier task for the deep learning models. In addition, OCT was used to confirm the diagnosis in the Japanese dataset. Alternatively, the differences in severity of disease, definitions, and labelling from the published studies included in the current analysis can be considered a study strength. Specifically, it allows quantitative estimates of how diagnostic performance can vary depending on the reference standard or labelling strategy used. Another limitation is that this study only considered the impact of a small number of covariates on model performance (disease severity, myopia status, and race). Many additional demographic and clinical variables could impact model performance and additional work is needed to quantify their impact. Finally, there are numerous considerations that can affect the choice of a particular deep learning strategy for a prediction task. The choice of network architecture, hyperparameters, transfer learning versus training from scratch (and more) can affect not only the final accuracy, but also the number of learned parameters, training time and/or data needed, and the types of errors made by the model.37 In our previous work, we performed more extensive analyses to optimize our models with respect to these concerns.5,6,11 This study focused on diagnostic accuracy and how it changed across datasets and populations, because this is directly relevant to translating deep learning results into improvements in clinical care.
In conclusion, the current results suggest that the detection of glaucoma by trained deep learning models can achieve high accuracy across diverse populations, and provides quantitative comparisons of how model performance can vary across datasets consisting of glaucoma of different disease severity and ethnicity. Moreover, the results also show that consideration must be given to the selection of training data, labeling, severity of disease, and training strategies. Finally, the high sensitivity and specificity of these models for detection of moderate-to-severe glaucoma is similar to that of systems approved by the US Food and Drug Administration for the detection of referable diabetic retinopathy,12,33 which suggests a role for artificial intelligence in the detection of glaucoma in primary care.
Supplementary Material
Acknowledgments
Supported by NEI Grants EY11008, P30 EY022589, EY026590, EY022039, EY021818, EY023704, EY029058, T32 EY026590, R21 EY027945, and an unrestricted grant from Research to Prevent Blindness (New York, NY).
Disclosure: M. Christopher, None; K. Nakahara, None; C. Bowd, None; J.A. Proudfoot, None; A. Belghith, None; M.H. Goldbaum, None; J. Rezapour, German Research Foundation (DFG, research fellowship grant RE 4155/1-1) (F), German Ophthalmological Society (DOG) (F); R.N. Weinreb, Aerie Pharmaceuticals (C), Allergan (C), Eyenovia (C), Implantdata (C), Unity (C), Heidelberg Engineering (F), Carl Zeiss Meditec (F), Centervue (F), Bausch & Lomb (F), Genentech (F), Konan Medical (F), National Eye Institute (F), Optos (F), Optovue Research to Prevent Blindness (F); M.A. Fazio, National Eye Institute (F), EyeSight Foundation of Alabama (F), Research to Prevent Blindness (F), Heidelberg Engineering (F); C.A. Girkin, National Eye Institute (F), EyeSight Foundation of Alabama (F), Research to Prevent Blindness (F), Heidelberg Engineering (F); J.M. Liebmann, Aerie Pharmaceuticals, Alcon, Allergan, Bausch & Lomb, Carl Zeiss Meditec, Eyenovia, Galimedix, Heidelberg Engineering, Zeiss Meditec, National Eye Institute, Novartis, Research to Prevent Blindness, Inc.; G. De Moraes, Novartis (C), Galimedix (C), Belite (C), Reichert (C), Carl Zeiss Meditec (C), Heidelberg Engineering (F), Topcon (F); H. Murata, The Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant number 25861618) (F); K. Tokumo, None; N. Shibata, None; Y. Fujino, The Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant number 20768254) (F); M. Matsuura, The Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant number 00768351) (F); Y. Kiuchi, None; M. Tanito, None; R. Asaoka, The Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant number 18KK0253, 19H01114 and 17K11418) (F), Daiichi Sankyo Foundation of Life Science, Suzuken Memorial Foundation, The Translational Research program, Japan Agency for Medical Research and Development (AMED, Strategic Promotion for Practical Application of Innovative Medical Technology [TR-SPRINT]); L.M. Zangwill, Carl Zeiss Meditec (F), Heidelberg Engineering (F), National Eye Institute (F), BrightFocus Foundation (F), Optovue (F), Topcon Medical System Inc. (F)
References
- 1. Constantinou M, Ferraro JG, Lamoureux EL, Taylor HR. Assessment of optic disc cupping with digital fundus photographs. Am J Ophthalmol. 2005; 140: 529–531. [DOI] [PubMed] [Google Scholar]
- 2. Tielsch JM, Katz J, Quigley HA, Miller NR, Sommer A. Intraobserver and interobserver agreement in measurement of optic disc characteristics. Ophthalmology. 1988; 95: 350–356. [DOI] [PubMed] [Google Scholar]
- 3. Moyer VA, US Preventive Services Task Force. Screening for glaucoma: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2013; 159: 484–489. [DOI] [PubMed] [Google Scholar]
- 4. Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. 2006; 18: 1527–1554. [DOI] [PubMed] [Google Scholar]
- 5. Asaoka R, Tanito M, Shibata N, et al.. Validation of a deep learning model to screen for glaucoma using images from different fundus cameras and data augmentation. Ophthalmol Glaucoma. 2019; 2: 224–231. [DOI] [PubMed] [Google Scholar]
- 6. Christopher M, Belghith A, Bowd C, et al.. Performance of deep learning architectures and transfer learning for detecting glaucomatous optic neuropathy in fundus photographs. Sci Rep. 2018; 8: 16685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diaz-Pinto A, Morales S, Naranjo V, Kohler T, Mossi JM, Navea A. CNNs for automatic glaucoma assessment using fundus images: an extensive validation. Biomed Eng Online. 2019; 18: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomez-Valverde JJ, Anton A, Fatti G, et al.. Automatic glaucoma classification using color fundus images based on convolutional neural networks and transfer learning. Biomed Opt Express. 2019; 10: 892–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Z, He Y, Keel S, Meng W, Chang RT, He M. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology. 2018; 125: 1199–1206. [DOI] [PubMed] [Google Scholar]
- 10. Liu S, Graham SL, Schulz A, et al.. A deep learning-based algorithm identifies glaucomatous discs using monoscopic fundus photographs. Ophthalmol Glaucoma. 2018; 1: 15–22. [DOI] [PubMed] [Google Scholar]
- 11. Shibata N, Tanito M, Mitsuhashi K, et al.. Development of a deep residual learning algorithm to screen for glaucoma from fundus photography. Sci Rep. 2018; 8: 14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ting DSW, Cheung CY, Lim G, et al.. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017; 318: 2211–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Son J, Shin JY, Kim HD, Jung KH, Park KH, Park SJ. Development and validation of deep learning models for screening multiple abnormal findings in retinal fundus images. Ophthalmology. 2020; 127: 85–94. [DOI] [PubMed] [Google Scholar]
- 14. Liu H, Li L, Wormstone IM, et al.. Development and validation of a deep learning system to detect glaucomatous optic neuropathy using fundus photographs. JAMA Ophthalmol. 2019; 137: 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beck RW, Messner DK, Musch DC, Martonyi CL, Lichter PR. Is there a racial difference in physiologic cup size? Ophthalmology. 1985; 92: 873–876. [DOI] [PubMed] [Google Scholar]
- 16. Caprioli J, Miller JM.. Optic disc rim area is related to disc size in normal subjects. Arch Ophthalmol. 1987; 105: 1683–1685. [DOI] [PubMed] [Google Scholar]
- 17. Chi T, Ritch R, Stickler D, Pitman B, Tsai C, Hsieh FY. Racial differences in optic nerve head parameters. Arch Ophthalmol. 1989; 107: 836–839. [DOI] [PubMed] [Google Scholar]
- 18. Lee RY, Kao AA, Kasuga T, et al.. Ethnic variation in optic disc size by fundus photography. Curr Eye Res. 2013; 38: 1142–1147. [DOI] [PubMed] [Google Scholar]
- 19. Varma R, Tielsch JM, Quigley HA, et al.. Race-, age-, gender-, and refractive error-related differences in the normal optic disc. Arch Ophthalmol. 1994; 112: 1068–1076. [DOI] [PubMed] [Google Scholar]
- 20. Martin MJ, Sommer A, Gold EB, Diamond EL. Race and primary open-angle glaucoma. Am J Ophthalmol. 1985; 99: 383–387. [DOI] [PubMed] [Google Scholar]
- 21. Seider MI, Lee RY, Wang D, Pekmezci M, Porco TC, Lin SC. Optic disk size variability between African, Asian, white, Hispanic, and Filipino Americans using Heidelberg retinal tomography. J Glaucoma. 2009; 18: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zangwill LM, Weinreb RN, Berry CC, et al.. Racial differences in optic disc topography: baseline results from the confocal scanning laser ophthalmoscopy ancillary study to the ocular hypertension treatment study. Arch Ophthalmol. 2004; 122: 22–28. [DOI] [PubMed] [Google Scholar]
- 23. Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? Adv Neural Inf Process Syst. 2014; 27: 3320–3328. [Google Scholar]
- 24. Russakovsky O, Deng J, Su H, et al.. ImageNet large scale visual recognition challenge. Int J Comput Vision. 2015; 115: 211–252. [Google Scholar]
- 25. Asaoka R, Murata H, Hirasawa K, et al.. Using deep learning and transfer learning to accurately diagnose early-onset glaucoma from macular optical coherence tomography images. Am J Ophthalmol. 2019; 198: 136–145. [DOI] [PubMed] [Google Scholar]
- 26. Christopher M, Bowd C, Belghith A, et al.. Deep learning approaches predict glaucomatous visual field damage from OCT optic nerve head En face images and retinal nerve fiber layer thickness maps. Ophthalmology. 2019; 127: 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sample PA, Girkin CA, Zangwill LM, et al.. The African Descent and Glaucoma Evaluation Study (ADAGES): design and baseline data. Arch Ophthalmol. 2009; 127: 1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Medeiros FA, Zangwill LM, Bowd C, Vasile C, Sample PA, Weinreb RN. Agreement between stereophotographic and confocal scanning laser ophthalmoscopy measurements of cup/disc ratio: effect on a predictive model for glaucoma development. J Glaucoma. 2007; 16: 209–214. [DOI] [PubMed] [Google Scholar]
- 29. Japan Glaucoma Society. Available at: www.ryokunaisho.jp/english/guidelines.html. Accessed April 14, 2019.
- 30. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. The IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016: 770–778.
- 31. Ren SQ, Lai H, Tong WJ, Aminzadeh M, Hou XZ, Lai SH. Nonparametric bootstrapping for hierarchical data. J Appl Stat. 2010; 37: 1487–1498. [Google Scholar]
- 32. Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. 2016 IEEE Conference on Computer Vision and Pattern Recognition (Cvpr). 2016;2921–2929.
- 33. Ting DSW, Carin L, Abramoff MD. Observations and lessons learned from the artificial intelligence studies for diabetic retinopathy screening. JAMA Ophthalmol. 2019 Jun 13 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 34. Leske MC, Connell AM, Schachat AP, Hyman L. The Barbados Eye Study. Prevalence of open angle glaucoma. Arch Ophthalmol. 1994; 112: 821–829. [DOI] [PubMed] [Google Scholar]
- 35. Ting DSW, Lee AY, Wong TY. An ophthalmologist's guide to deciphering studies in artificial intelligence. Ophthalmology. 2019; 126: 1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berrar D, Flach P.. Caveats and pitfalls of ROC analysis in clinical microarray research (and how to avoid them). Brief Bioinform. 2012; 13: 83–97. [DOI] [PubMed] [Google Scholar]
- 37. Raghu M, Chiyuan Z, Kleinberg J, Bengio S. Transfusion: understanding transfer learning for medical imaging. 33rd Conference on Neural Information Processing System (NeurIPS 2019). Vancouver, Canada; 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


