Highlights
-
•
Four-way pooling correctly identified SARS-CoV-2 in 94 % of positive samples (n = 30/32) tested.
-
•
Low viral loads, corresponding with late CTs, may be missed by the pooling process.
-
•
1:4 pooling is associated with expected 2 CT loss in analytical sensitivity.
-
•
All individually negative samples (n = 128) were also negative by 4-way pooling.
Keywords: SARS-CoV-2, RT-PCR, COVID-19, Pooling, Diagnostics, Disease surveillance
Abstract
Background
SARS-CoV-2 testing demand has outpaced its supply. Pooling samples for lower risk populations has the potential to accommodate increased demand for SARS-CoV-2 molecular testing.
Objective
To evaluate the sensitivity, specificity, and reproducibility of 4-way pooling of SARS-CoV-2 specimens for high-throughput RT-PCR.
Study design
Individual samples were pooled 1:4 through automated liquid handling, extracted, and assayed by our emergency use authorized CDC-based RT-PCR laboratory developed test. Positive samples were serially diluted and theoretical and empirical PCR cycle thresholds were evaluated. Thirty-two distinct positive samples were pooled into negative specimens and individual CTs were compared to pooled CTs. Low positive samples were repeated for reproducibility and 32 four-way pools of negative specimens were assayed to determine specificity.
Results
Four-way pooling was associated with a loss of sensitivity of 1.7 and 2.0 CTs for our N1 and N2 targets, respectively. Pooling correctly identified SARS-CoV-2 in 94 % (n = 30/32) of samples tested. The two low positive specimens (neat CT > 35) not detected by pooling were individually repeated and detected 75 % (n=6/8) and 37.5 % (n = 3/8) of the time, respectively. All specimens individually determined negative were also negative by pooling.
Conclusion
We report that 1:4 pooling of samples is specific and associated with an expected 2 CT loss in analytical sensitivity. Instead of running each sample individually, pooling of four samples will allow for a greater throughput and conserve scarce reagents.
1. Introduction
Widespread COVID-19 infections have placed extraordinary demand on molecular diagnostics. COVID-19 cases continue to rise and laboratory capacities to detect SARS-CoV-2 RNA are becoming increasingly strained, causing delays in testing turnaround times [[1], [2], [3]]. To accommodate demand for increased testing volumes, on July 18th, 2020, the FDA issued its first Emergency Use Authorization (EUA) for sample pooling in diagnostic testing [4].
Early testing during the COVID-19 pandemic focused primarily on symptomatic individuals, but as we expand the populations tested to asymptomatic patients, overall positivity rate declines and pooling methods become increasingly favorable [5]. Sample pooling provides improved benefits as SARS-CoV-2 incidence rates decline; higher incidence rates (i.e. > 10 %) provide little advantage, but pooling with incidence rates <5% can substantially increase testing capacity [6,7].
Modeling suggests that for asymptomatic or mild cases based on overall lower SARS-CoV-2 incidence, scaled pooling of up to 30 samples can provide substantial benefit in accommodating increased testing demands for low-risk populations [[8], [9], [10], [11], [12]]. Theoretical calculations of pooling are useful; however, here we describe the practical utility of 4-way pooling of our EUA laboratory developed test (LDT) and evaluate its clinical application.
2. Methods
We programmed a HAMILTON Microlab STARlet Automated Liquid Handler (Atlantic Lab Equipmant, Beverly, MA) to perform 4-way pooling on our CDC-based Washington state EUA SARS-CoV-2 RT-PCR assay targeting N1 and N2 as previously described [13,14]. Fifty μL of each specimen was pipetted into a 96-well deep well plate yielding 200 μL of total viral transport media (VTM) per well. The MagNA Pure 96 platform (Roche, Basel, Switzerland) was utilized for total nucleic acid extraction, eluting into 50 μL elution buffer.
Initial water and VTM templates were used to confirm pipetting accuracy with 384 samples into a 96-well deep well plate. Artificial sample IDs were attributed to each respective pool and the eluted volume was manually confirmed for accuracy by pipette. Prior to pooling, neat samples were assayed by LDT and stored at 4 °C for <24 h. HeLa cells were included as a negative extraction control and water as a negative PCR template on every run.
We created a web application for converting Hamilton output files into plate maps that can be imported into the Applied Biosystems 7500 software. It concatenates the container IDs that were mixed in each well into sample names for the 7500 application so that they can be tracked. The web application also provides a form where the experiment output from the Applied Biosystems 7500 software is uploaded and edited to produce extracts for other downstream systems and processes.
These extracts include a. pdf print-out highlighting positive and inconclusive samples that need to be individually tested along with their rack location, a. json file with raw CT values that is loaded into our data warehouse, and a. csv file with the negative samples that can be imported into our Sunquest laboratory information system.
3. Results
To evaluate lower positivity levels of pooled samples, we first ran an initial 10-fold dilution series on a positive nasopharyngeal swab with an initial mean cycle threshold CT of 31.4 corresponding to 2,500–5,000 copies/mL by our LDT. The sample was serially diluted with phosphate-buffered saline 1:10, then 1:100, and then 1:1,000, corresponding to 1:40, 1:400, and 1:4,000 respective dilutions for pooling.
Next, we verified that low positive samples with CTs ≥33 – indicative of a low viral load – were not missed by pooling of samples (Table 1 ). Our data indicate that even though individual samples are diluted by the process of pooling four separate specimens together, the effect is not substantial enough to push borderline positives beyond the limit of detection.
Table 1.
CT comparison of low positives by pooling.
| Pool | Original N1 CT | Pooled N1 CT | Original N2 CT | Pooled N2 CT |
|---|---|---|---|---|
| 1 | 33.6 | 35.0 | 37.5 | 36.2 |
| 2 | 34.9 | 36.8 | 37.5 | 37.7 |
| 3 | 36.0 | 36.8 | 36.6 | 38.0 |
| 4 | 33.2 | 34.6 | 37.3 | 36.6 |
Abbreviations: CT, cycle threshold, NDET, not detected, N1 and N2 are SARS-CoV-2 nucleocapsid gene targets for PCR. Each pool contains 4 specimens: 1 unique positive sample pooled into 3 distinct negative samples. Low positive samples are defined as having a CT>33.0.
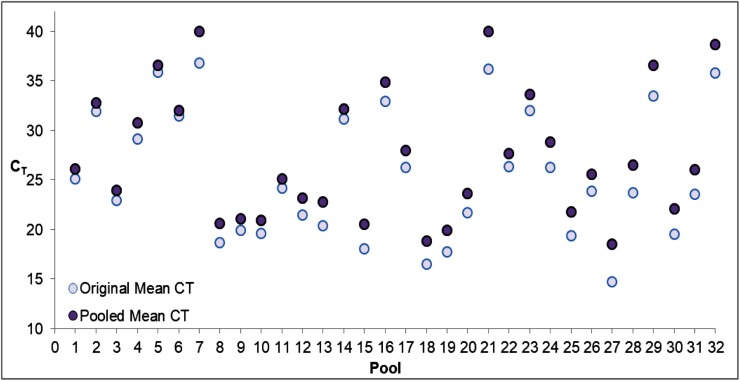
We expanded this experiment to include 32 additional SARS-CoV-2 positive specimens by the CDC-based Washington state EUA assay pooled into negative samples. Our results confirm the empirical CTs of pooled samples are not dissimilar from the theoretically calculated values (Table 2 ). As expected, diluted samples with theoretical CTs beyond our limit of detection were not detected (NDET) by our LDT [13]. The average delay of N1 and N2 target CTs associated with 1:4 pooling was 1.7 (95 % CI:1.12−2.32) and 2.0 (95 % CI: 1.29–2.66), respectively. Thirty out of 32 (94 %) positive samples pooled into negative specimens were detected by pooling (Fig. 1 , Supplementary Table 1). The only two missed samples by pooling had CTs ≥35, corresponding to an absolute quantification of approximately 250–500 genomic copies/mL based on our quantitative RT-PCR, with RNA standard values quantified by droplet digital RT-PCR [14]. Two low positives (CT >33.5) were inconclusive by pooling (one target positive), and according to our EUA protocol would be considered positive, with individual specimens repeated. These inconclusive results were confirmed as low positives when repeated from neat sample.
Table 2.
Positive sample serial dilution.
| Sample Dilution | Expected N1 CT | Actual N1 CT | Expected N2 CT | Actual N2 CT |
|---|---|---|---|---|
| Neat | 30.8 | 31.9 | 30.3 | 30.9 |
| 1:10 (1:40) | 35.4 | 35.4 | 34.9 | 35.3 |
| 1:100 (1:400) | 38.7 | NDET | 38.2 | NDET |
| 1:1,000 (1:4,000) | 42.0 | NDET | 41.5 | NDET |
Abbreviations: CT, cycle threshold, NDET, not detected, N1 and N2 are SARS-CoV-2 nucleocapsid gene targets for PCR. Parentheticals denote respective pooling dilutions.
Fig. 1.
CTs of 32 distinct positives pooled 1:4 into negative samples.
Abbreviations: CT, cycle threshold.
For the two samples that were not detected by pooling, CT values of 40 were artificially designated for pool 7 and pool 21. CT values are averaged from both N1 and N2 targets for the original neat samples and the pooled samples.
For reproducibility, the negative and inconclusive pools were repeated 8 additional times, and 3 additional times on neat positive specimens (Supplementary Table 2). Our data demonstrate we would have detected the first specimen (mean CT = 36.5) in 6 out of 8 pools (75 %). The second specimen (mean CT = 37.3) was detected in 3 out of 8 pools (37.5 %). To confirm specificity, we combined 128 unique patient specimens determined negative by the Washington state EUA assay into 32 pools. All 32 four-way pools were negative, demonstrating 100 % specificity.
4. Discussion
Here, we show the potential for four-way pooling to responsibly increase testing throughput with an expected 2 CT loss in sensitivity. We acknowledge that pooling multiple specimens together results in a mild - but expected - drop in PCR cycle thresholds, similar to other research [15]. According to Yelin et al. (2020), as pool size increases, each respective sample and potential SARS-CoV-2 RNA are diluted, corresponding with an observed linear CT increase of 1.24 for every twofold dilution [16]. Most theoretical calculations estimate pools between 4–5 samples optimize assay benefits by limiting the false negative rate and maintaining efficiency [[17], [18], [19]].
Our data indicate that the practical utility of pooling at this scale has applications for systematic community surveillance, testing of low-risk populations, and in resource-limited settings. The utilization of high-throughput and broadly available lab instruments such as Roche’s MP96 extraction platform and HAMILTON liquid handler allow concrete scalability of this EUA-authorized four-way pooling assay.
Borderline patients with low viral loads may be missed by the pooling process [20]. Notably however, a low positive PCR test result may correspond with the detection of SARS-CoV-2 RNA, but not necessarily infectious virus [21,22]. Specimens with viral loads of CT > 35 have been shown to not be routinely culturable in vitro [23].
Here, four-way pooling correctly identified SARS-CoV-2 in 94 % of samples, only missing low viral load specimens with CT >35. We report that 1:4 pooling of samples is associated with an acceptable 2 CT loss in analytical sensitivity. Pooling samples for SARS-CoV-2 molecular detection can be performed efficiently without sacrificing substantial accuracy or specificity. Optimized for lower positivity ratios (i.e. < 8%) [20,24,25], four-way pooling has considerable potential to accommodate increased demand for diagnostic testing of low-risk populations.
Funding
This work was supported by the Department of Laboratory Medicine and Pathology at the University of Washington Medical Center.
Declaration of Competing Interest
The authors declare no conflict of interest.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104570.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Meyer Robinson A.C.M. The Atlantic; 2020. A Dire Warning From COVID-19 Test Providers.https://www.theatlantic.com/science/archive/2020/06/us-coronavirus-testing-could-fail-again/613675/ [Google Scholar]
- 2.T.A.J.-C. J. Scott Trubey, Georgia sees testing system strained with rise in COVID-19 cases, Ajc. (n.d.). https://www.ajc.com/news/state--regional/georgia-sees-testing-system-strained-with-rise-covid-cases/pyb5miiLT74o8FUP9BsCAK/.
- 3.Quest Diagnostics Media Statement about COVID-19 Testing, Quest Diagnostics Newsroom. (n.d.). https://newsroom.questdiagnostics.com/COVIDTestingUpdates.
- 4.O. of the Commissioner . FDA; 2020. Coronavirus (COVID-19) Update: FDA Issues First Emergency Authorization for Sample Pooling in Diagnostic Testing.https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-first-emergency-authorization-sample-pooling-diagnostic [Google Scholar]
- 5.Hogan C.A., Sahoo M.K., Pinsky B.A. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA. 2020;323:1967. doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdalhamid B., Bilder C.R., McCutchen E.L., Hinrichs S.H., Koepsell S.A., Iwen P.C. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am. J. Clin. Pathol. 2020;153:715–718. doi: 10.1093/ajcp/aqaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherif A., Grobe N., Wang X., Kotanko P. Simulation of pool testing to identify patients with coronavirus disease 2019 under conditions of limited test availability. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres I., Albert E., Navarro D. Pooling of nasopharyngeal swab specimens for SARS‐CoV‐2 detection by RT‐PCR. J. Med. Virol. 2020 doi: 10.1002/jmv.25971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinnott-Armstrong N., Klein D., Hickey B. Evaluation of group testing for SARS-CoV-2 RNA. Infect. Diseases (except HIV/AIDS) 2020 doi: 10.1101/2020.03.27.20043968. [DOI] [Google Scholar]
- 10.Lohse S., Pfuhl T., Berkó-Göttel B., Rissland J., Geißler T., Gärtner B., Becker S.L., Schneitler S., Smola S. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30362-5. S1473309920303625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabrera J.J., Rey S., Pérez S., Martínez-Lamas L., Cores-Calvo O., Torres J., Porteiro J., García-Comesaña J., Regueiro B. 2020. Pooling for SARS-CoV-2 Control in Care Institutions; p. 15. (n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallapaty S. The mathematical strategy that could transform coronavirus testing. Nature. 2020 doi: 10.1038/d41586-020-02053-6. d41586-020-02053–6. [DOI] [PubMed] [Google Scholar]
- 13.Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer/probe sets and one assay kit. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00557-20. JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perchetti G.A., Nalla A.K., Huang M.-L., Zhu H., Wei Y., Stensland L., Loprieno M.A., Jerome K.R., Greninger A.L. Validation of SARS-CoV-2 detection across multiple specimen types. J. Clin. Virol. 2020 doi: 10.1016/j.jcv.2020.104438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shental N., Levy S., Skorniakov S., Wuvshet V., Shemer-Avni Y., Porgador A., Hertz T. Efficient high throughput SARS-CoV-2 testing to detect asymptomatic carriers. Infect. Diseases (except HIV/AIDS) 2020 doi: 10.1101/2020.04.14.20064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yelin I., Aharony N., Shaer Tamar E., Argoetti A., Messer E., Berenbaum D., Shafran E., Kuzli A., Gandali N., Shkedi O., Hashimshony T., Mandel-Gutfreund Y., Halberthal M., Geffen Y., Szwarcwort-Cohen M., Kishony R. Evaluation of COVID-19 RT-qPCR test in multi-sample pools. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanel R., Thurner S. 2020. Boosting Test-Efficiency by Pooled Testing Strategies for SARS-CoV-2, ArXiv:2003.09944 [q-Bio, Stat]http://arxiv.org/abs/2003.09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma R., Goyal S., Bist P. Optimal sample pooling: an efficient tool against SARS-CoV-2. Health Policy. 2020 doi: 10.1101/2020.07.03.20145953. [DOI] [Google Scholar]
- 19.Mulu A., Alemayehu D.H., Alemu F., Tefera D.A., Wolde S., Aseffa G., Seyoum T., Habtamu M., Abdissa A., Bayih A.G., Beyene G.T. Evaluation of sample pooling for screening of SARS-CoV-2. Genetic Genomic Med. 2020 doi: 10.1101/2020.06.10.20123398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Ami R., Klochendler A., Seidel M., Sido T., Gurel-Gurevich O., Yassour M., Meshorer E., Benedek G., Fogel I., Oiknine-Djian E., Gertler A., Rotstein Z., Lavi B., Dor Y., Wolf D.G., Salton M., Drier Y., Klochendler A., Eden A., Klar A., Geldman A., Arbel A., Peretz A., Shalom B., Ochana B.L., Avrahami-Tzfati D., Neiman D., Steinberg D., Ben Zvi D., shpigel Etai, Atlan G., Klein H., Chekroun H., Shani H., Hazan I., Ansari I., Magenheim I., Moss J., Magenheim J., Peretz L., Feigin L., Saraby M., Sherman M., Bentata M., Avital M., Kott M., Peyser M., Weitz M., Shacham M., Grunewald M., Sasson N., Wallis N., Azazmeh N., Tzarum N., Fridlich O., Sher R., Condiotti R., Refaeli R., Ben Ami R., Gallili R.Z., Helman R., Ofek S., Tzaban S., Piyanzin S., Anzi S., Dagan S., Lilenthal S., Sido T., Licht T., Friehmann T., Kaufman Y., Pery A., Saada A., Dekel A., Yeffet A., Shaag A., Gayego A.M., Shay E., Arbib E., Onallah H., Ben-Meir K., Levinzon L., Cohen-Daniel L., Natan L., Hamdan M., Rivkin M., Shwieki M., Vorontsov O., Barsuk R., Abramovitch R., Gutorov R., Sirhan S., Abdeen S., Yachnin Y., Daitch Y. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.06.009. S1198743X20303499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 22.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323:2249. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 23.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., Gautret P., Raoult D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutesa L., Ndishimye P., Butera Y., Souopgui J., Uwineza A., Rutayisire R., Musoni E., Rujeni N., Nyatanyi T., Ntagwabira E., Semakula M., Musanabaganwa C., Nyamwasa D., Ndashimye M., Ujeneza E., Mwikarago I.E., Muvunyi C.M., Mazarati J.B., Nsanzimana S., Turok N., Ndifon W. 2020. A Strategy for Finding People Infected with SARS-CoV-2: Optimizing Pooled Testing at Low Prevalence, ArXiv:2004.14934 [q-Bio]http://arxiv.org/abs/2004.14934 [Google Scholar]
- 25.Eliaz Y., Danovich M., Gasic G.P. Poolkeh finds the optimal pooling strategy for a population-wide COVID-19 testing (Israel, UK, and US as test cases) Public Global Health. 2020 doi: 10.1101/2020.04.25.20079343. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.