Abstract
Schizophrenia occurs in about one in four individuals with 22q11.2 deletion syndrome (22q11.2DS). The aim of this International Brain and Behavior 22q11.2DS Consortium (IBBC) study was to identify genetic factors that contribute to schizophrenia, in addition to the ~20-fold increased risk conveyed by the 22q11.2 deletion. Using whole-genome sequencing data from 519 unrelated individuals with 22q11.2DS, we conducted genome-wide comparisons of common and rare variants between those with schizophrenia and those with no psychotic disorder at age ≥25 years. Available microarray data enabled direct comparison of polygenic risk for schizophrenia between 22q11.2DS and independent population samples with no 22q11.2 deletion, with and without schizophrenia (total n=35,182). Polygenic risk for schizophrenia within 22q11.2DS was significantly greater for those with schizophrenia (padj=6.73×10−6). Novel reciprocal case-control comparisons between the 22q11.2DS and population-based cohorts showed that polygenic risk score was significantly greater in individuals with psychotic illness, regardless of the presence of the 22q11.2 deletion. Within the 22q11.2DS cohort, results of gene-set analyses showed some support for rare variants affecting synaptic genes. No common or rare variants within the 22q11.2 deletion region were significantly associated with schizophrenia. These findings suggest that in addition to conferring a greatly increased risk to schizophrenia, the risk is higher when the 22q11.2 deletion and common polygenic risk factors that contribute to schizophrenia in the general population are both present.
INTRODUCTION
There is accumulated evidence that the recurrent 22q11.2 deletion associated with 22q11.2 deletion syndrome (22q11.2DS) represents a clinically relevant cause of schizophrenia, and is one of the strongest known risk factors for schizophrenia (1). Schizophrenia occurs in about one in four individuals with 22q11.2DS, representing a more than 20-fold increase over the general population risk of 1% (1, 2). Identifying additional genetic modifiers of expressing schizophrenia in addition to the deletion is of scientific and clinical interest (1, 3, 4).
Discovering such additional genetic factors is a primary goal of the International 22q11.2DS Brain and Behavior Consortium (IBBC) (3). We recently reported evidence from genome-wide microarray data that other rare copy number variations (CNVs) outside of the 22q11.2 deletion region increase risk to schizophrenia in 22q11.2DS (5). A key question is whether other genetic contributions to schizophrenia, known to exert risk in the general population, are modifying risk of schizophrenia within the 22q11.2DS subgroup.
Schizophrenia is polygenic, with risk variants spanning the full spectrum of population frequencies. Common alleles identified from genome-wide association studies (GWAS) each contribute only a small effect to disease risk, but their aggregate effect can confer a far greater risk, quantifiable as a polygenic risk score (PRS). Analyzing by aggregating genes can also be a means to observe relevant associations (6). Studies of rare protein-coding variants implicate functional gene-sets relevant to neuronal functions and synaptic components including the activity-regulated cytoskeleton-associated protein (ARC) complex, amongst others (7–12).
The current study used whole genome sequencing and microarray data from the IBBC (3) to investigate the contribution of common (single nucleotide polymorphisms, SNPs) and rare genetic variants (comprising mostly single nucleotide variants (SNVs) in protein-coding regions of genes), i.e., sequence variant classes with prior evidence from idiopathic schizophrenia (8, 9, 13). To improve power, we examined collective signals imparted by the schizophrenia PRS for SNPs (6, 13), and functional gene-sets (10) for rare variants. We examined these variant effects both within the 22q11.2 deletion region and genome-wide, comparing individuals with schizophrenia and related major psychotic disorders to those with no psychotic disorder and aged ≥25 years, therefore largely past the age at greatest risk for onset of psychotic illness.
METHODS
Sample collection and diagnostic classification
The IBBC ascertainment and assessment methods have been previously detailed (3, 5). Briefly, individuals with a presumed 22q11.2 deletion, who were recruited from 22 international sites in studies approved by local institutional research ethics boards, and for whom existing DNA samples were available, were ascertained for the study. The focus of this study was on unrelated individuals with sufficient data to discern psychotic illness status, who were molecularly confirmed to have a typical hemizygous deletion at 22q11.2, i.e., that spanned the LCR22A-LCR22B region, most commonly extending from LCR22A-LCR22D (1, 3). As before, individuals were eligible for inclusion in this study if they met DSM diagnostic criteria for a major psychotic disorder (mostly schizophrenia) at any age, or had no history of any psychotic illness when assessed at age ≥25 years (5). Subjects with affective psychosis were excluded. A case consensus procedure assured stringent diagnostic categorization (3). Additional details are provided in the Supplement.
Whole-genome sequencing (WGS) and processing of DNA samples
Raw Illumina WGS data were mapped with PEMapper (14) to genome build GRCh38/hg38 with a median sequencing depth of 39X. The 22q11.2 deletion region was called based on sequencing depth, and variants were called independently with PECaller (14) for the genome-wide diploid and 22q11.2 haploid regions of the genomes. Routine QC metrics were performed (Table S1) and resulted in a multi-ethnic sample of 519 subjects (259 schizophrenia, 260 no psychotic illness). Using principal component analysis anchored with HapMap reference samples to infer ancestry, we further defined a European subsample of 435 subjects (214 schizophrenia, 221 no psychotic illness) based on subjects whose principal components were within 1 standard deviation of the average PC value for HapMap3 CEU subjects. Additional details on principal components and related QC are provided in the Supplement. These samples were then used for subsequent variant annotation and analyses.
Additional samples and array-based genotyping for polygenic risk assessment
An important aspect of our study was to assess the contribution of polygenic risk to schizophrenia in 22q11.2DS given previous results for schizophrenia in the general population (13).
To achieve this, we used an idiopathic schizophrenia case-control study (CLOZUK/WTCCC) (15) where samples were independent of those from the schizophrenia case-control training dataset used for PRS construction (6). To allow comparisons between the PRS derived from this dataset and our 22q11.2DS cohort, we used available Affymetrix Genome-Wide Human SNP 6.0 array genotype data (5, 16). Analyses were restricted to samples of European ancestry and autosomal SNPs. Identity by descent analysis identified three subjects from the 22q11.2DS cohort who were also present in the CLOZUK cohort. After excluding these individuals there were 432 (212 with schizophrenia; 220 non-psychotic controls) individuals from the 22q11.2DS cohort included for the primary PRS and rare variant analyses.
After alignment (HRCv1.1) (17) and phasing with Eagle2 (18), genotype imputation of the 22q11.2DS cohort was performed using Minimac3 (19). SNPs with common variants (MAF >10%), low missingness (<1%), HWE mid p >10−5 and high imputation quality (INFO >0.9) were retained for PRS analysis. PRS results obtained from imputed SNP data were compared to those obtained from WGS data using a sub-sample of 365 individuals (171 with schizophrenia; 194 non-psychotic) that had passed QC for both methods. Identity by descent analysis ensured correspondence between samples. Only overlapping SNPs that had passed QC in both datasets were kept for this analysis.
To establish a homogenous dataset that would enable a joint analysis of the array-based data from 22q11.2DS and CLOZUK/WTCCC, we initially conducted a QC step that used SNPs that were shared between both datasets (see Supplement for additional details). Following principal component analysis, samples >6 standard deviations from the mean of first 20 principal components were excluded to ensure homogeneity of the merged 22q11.2DS and CLOZUK/WTCCC datasets of European ancestry. This QC resulted in a dataset of 322 individuals with 22q11.2DS (147 with schizophrenia; 175 non-psychotic controls), 10,791 schizophrenia CLOZUK cases and 24,391 WTCCC controls for joint PRS analyses.
Statistical analyses
Single-variant association analysis of common variants
We performed association analyses of WGS data using PLINK 1.9. Variants were defined as common if sample MAF was either ≥1% outside the 22q11.2 deletion, or ≥2% within the 22q11.2 deletion (given the inherent reduction in the number of haploid variants observed). For analysis within the 22q11.2 deletion region, we only considered hemizygous SNPs, ie SNPs located in the deleted region. We therefore performed two sets of analyses: one where all individuals were included and where we considered SNPs located in the region spanning LCR22A-LCR22B, as this region is deleted in all study participants; and one where only individuals with the larger LCR22A-LCR22D deletion were included, using SNPs spanning that entire interval. We used logistic regression to regress disease status on a variant (additive coding), adjusting for sex and top principal components of ancestry. For analyses of the entire sample (n=519: 259 schizophrenia, 260 no psychotic illness), we adjusted for top 5 principal components derived from this sample plus HapMap reference samples (Fig. SF1). For analyses of the European subset (n=435: 214 schizophrenia, 221 no psychotic illness), we adjusted for the top 3 principal components derived from this subset. Given the possibility of unreliable asymptotic results for variants with sample MAF of 1–5%, we further studied these variants using Fisher’s Exact Test.
Polygenic risk score (PRS) analyses
We used PRSice software (PRSice-2) (20) to construct PRS in the European subset. We compared WGS and array data to assess robustness of results. Two different datasets were used as the discovery (training) sample. For the 22q11.2DS-only analyses (n=432) that used WGS data, we used summary results from the second Psychiatric Genomics Consortium mega-analysis of schizophrenia (PGC2 SCZ) (13). For the analysis of 22q11.2DS (n=322) and CLOZUK/WTCCC cohorts (n=10,791/24,391) that used array data, we used a version of the PGC2 SCZ data that excluded the CLOZUK/WTCCC samples (6). Only autosomal SNPs with high imputation quality (INFO > 0.9) were kept in these discovery datasets. In all target datasets, we used only variants with MAF >10% and excluded variants from the X chromosome, the 22q11.2 deletion region, and the Major Histocompatibility Complex (due to complex linkage disequilibrium patterns in the region) to calculate PRS. We also excluded ambiguous SNPs (A/T and C/G) that posed challenges in matching between discovery and validation sets. Specific coordinates of excluded regions are described in the Supplement.
Using PRSice, we clumped SNPs from the discovery datasets (R2 >0.1, 250 kb window), and then selected clumped SNPs at 13 P-value thresholds (ranging from <10−6 to 1). For each P-value threshold, PRSice constructed a PRS for each individual from the target datasets as an average of the per-allele score for each eligible SNP. The per-allele score for a SNP was calculated as half the product of the number of reference alleles (0, 1 or 2) and the estimated log odds ratio for the SNP in the discovery dataset.
For the 22q11.2DS-only analysis using WGS data, we used logistic regression to test for association between disease status and standardized PRS adjusting for the same covariates (sex, top 3 principal components for ancestry) as for the analyses of individual common variants above. For the 22q11.2DS-CLOZUK/WTCCC joint analysis, we adjusted for sex and the top 3 principal components from the array data. For the PRS comparison between WGS and array data the same number of covariates were used (top 3 principal components for ancestry and sex) but using the principal components from each respective dataset. For all models, we used PRSice-2 to calculate both odds ratios per standard deviation of PRS as well as Nagelkerke’s pseudo-R2 values to estimate the proportion of variance in schizophrenia explained by the PRS. We adjusted for testing of multiple correlated PRSs under different p-value thresholds using the P_ACT method (21).
Rare variant analyses
Rare variant analyses were conducted on the 432 subjects in the European subset used for PRS analysis. We defined rare variants as those with gnomAD global frequency ≤1% and 22q11.2DS sample frequency ≤1% for all variants, but analyzed those from the 22q11.2 deletion region separately (Table S2). We also performed analyses using 5 different sets of filtered variants, based on: 1) any variant with Combined Annotation Dependent Depletion, CADD,(17) >30, 2) any variant with CADD >15, 3) any missense variant, 4) any missense variant with CADD >15, and 5) any predicted loss-of-function (LOF) mutation (defined as stop-gains, frameshift, core splice site alterations, stop-loss, or start-loss). For each filtered set, we first performed single-variant analysis using Fisher’s Exact Test within PLINK 1.9. We then used the R package SKAT (22) to perform burden and variance-component (i.e., SKAT) tests within genes. For each filtered variant set, we considered only those genes with ≥5 rare variants observed in the 22q11.2DS European sample. We performed both unweighted and weighted analyses in a gene with the latter upweighting effects of extremely rare variants using a beta (0.5,0.5) distribution. For all analyses, we adjusted for sex, top 3 principal components of ancestry, total number of genome-wide variants, and PRS based on WGS data.
In addition to gene-based analyses, we performed weighted and unweighted gene-set burden analyses with 30 functional pathways (Table S3) previously used to study CNV enrichment in schizophrenia, involving genes relevant to neuronal functions and synaptic components, or genes with homologues in mouse genes grouped by organ system (10). These analyses used the same filtering scheme described above and adjusted for multiple correlated tests using P_ACT (21). As a complementary analysis, we constructed logistic-regression models that, for a given pathway, assumed distinct effects for burden of LOF mutations and burden of missense variants passing or not passing CADD >15. We then used a likelihood-ratio statistic to test the null hypothesis that coefficients for all burden variables were equal to 0. These logistic-regression models used the same rare-variant weighting schemes and covariate adjustment as above. To adjust for multiple testing, we estimated the false discovery rate (FDR) based on outcome-label permutations balanced on covariates using the R package BiasedUrn (23). Finally, we also performed genome-wide and deletion region-wide burden tests of global rare variation (i.e., regardless of gene function) between cases and controls using the same filtering schemes, variant-weighting schemes, and covariate adjustments as above.
RESULTS
22q11.2DS sample
The main sample comprised 435 unrelated individuals of European ancestry with a 22q11.2 deletion: 214 with schizophrenia (mean age at onset 22.1 (SD 8.3) years; mean age at assessment 30.9 (SD 12.8) years) and 221 with no lifetime diagnosis of psychotic disorder and age ≥25 years (mean age at assessment 36.4 (SD 10.0) years) (5). Sample characteristics are representative of those for all ethnicities (Table S4). Results for 22q11.2 deletion sizes showed the expected ~90% of subjects with the typical 2.5 Mb LCR22A–LCR22D 22q11.2 deletion (Fig. 1) (1, 5) and no significant difference in distribution between the two phenotypic subgroups (Χ2=0.0024, df=1, p=0.96).
Figure 1: IBBC 22q11.2 DS cohort overview and schematic of data generation and analyses performed.
The full IBBC cohort and European subset are illustrated as yellow circles, broken down by numbers with or without schizophrenia, as well as sex and 22q11.2 deletion extent for individuals of European Ancestry. Whole-genome sequencing methods and data are represented by blue boxes and microarray data represented by green boxes (lighter for IBBC 22q11.2DS data, darker for schizophrenia case-control study data (15)). i to vi represent the comparisons presented in Fig. 3 for novel reciprocal case-control comparisons of schizophrenia PRS between the 22q11.2DS and population-based cohorts.
Genome-wide schizophrenia polygenic risk score (PRS)
Using the PRS to analyze common variants collectively in 22q11.2DS showed that the PRS was significantly higher in those with schizophrenia than in those with no psychotic illness; significance was greatest using a p-value threshold (pT) of 0.05 to select individual SNPs from the training set (13) (Nagelkerke’s pseudo-R2 0.077, padjusted=6.73 × 10−6). The risk of schizophrenia in individuals with 22q11.2DS increased by 1.77-fold (95% CI 1.41–2.22) for every incremental standard deviation in PRS (Fig. 2a and 2b; Table S5 shows results for other pT values). Using pT=0.05, individuals within the highest quartile of PRS had a 3.42-fold (95% CI: 1.88–6.22) increased risk of schizophrenia compared to those in the lowest quartile (Fig. SF2).
Figure 2. Polygenic risk score analyses of schizophrenia in 22q11.2DS.
Analysis of schizophrenia PRS in 432 individuals of European ancestry with 22q11.2DS, with (n=212) or without (n=220) schizophrenia. Variants from the 22q11.2 deletion, MHC regions and X chromosome were excluded and results adjusted for sex, top 3 ancestry PCs. P-values were adjusted using P-ACT for the 13 p-value thresholds (pT) used for PRS construction (13). Details on the number of SNPs used (maximum 68,966 SNPs for polygenic risk scoring at pT=1, minimum 202 SNPs at pT=1×10−6) are shown in Table S9. Figure 2a. The proportion of variance in schizophrenia explained by PRS (Nagelkerke’s R2) across different pT thresholds. At pT=0.05, Nagelkerke’s pseudo-R2= 0.077 (adjusted p=6.73×10−6). Figure 2b. Odds ratio and 95% confidence interval bars per one standard deviation increase in PRS across different pT thresholds. At pT=0.05 a one standard deviation increase in PRS corresponded to a 1.77-fold higher odds for schizophrenia in 22q11.2DS.
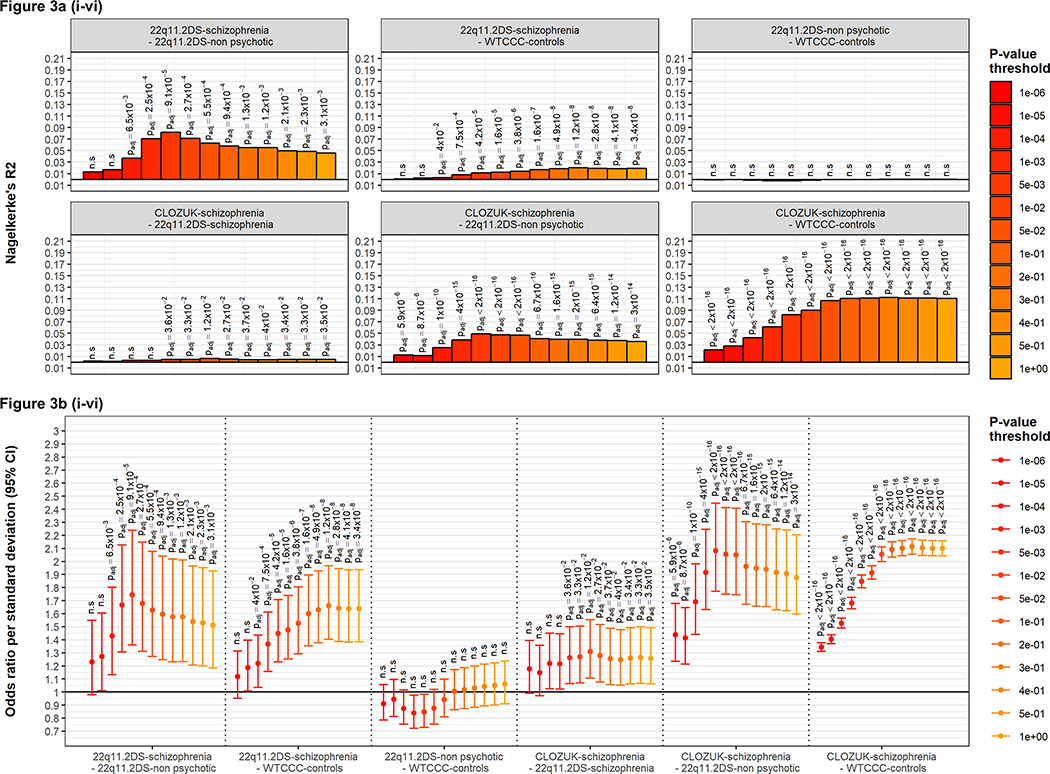
Using a subset of samples for which imputed data were available from microarray-based genotypes allowed us to directly compare schizophrenia PRS between 22q11.2DS (n=322; 147 with schizophrenia, 175 without psychotic illness) and an idiopathic schizophrenia case-control cohort with no 22q11.2 deletion (10,791 schizophrenia CLOZUK cases, 24,391 WTCCC controls) (6). We first ensured the concordance of genotypes obtained by array and WGS (mean [median] variant concordance of 99.4% [99.6%]) and observed similar PRS distributions across methods (Fig. SF3); see Supplementary results for more details. Using this smaller sample and imputed data we again found significant differences in PRS for the 22q11.2DS schizophrenia group compared to those with no psychotic illness (pT=0.05: R2=0.063, padjusted=5.5×10−4, OR=1.63, 95% CI:1.27–2.08; Fig. 3a(i) and 3b(i)), confirming that the PRS results were comparable to those using WGS data. We also confirmed the significant difference in PRS between the CLOZUK-schizophrenia and the WTCCC-control samples, which was comparable to the results obtained when using larger case-control datasets (6, 13): pT=0.05: R2=0.11, padjusted <2×10−16, OR=2.05, 95% CI: 2.00–2.11; Fig. 3a(vi) and 3b(vi).
Figure 3. Polygenic risk score analyses in 22q11.2DS individuals with or without schizophrenia, and a case-control cohort without 22q11.2DS.
PRS analyses were performed for 322 individuals with 22q11.2DS (147 with schizophrenia, 175 without) and 35,182 individuals with no 22q11.2 deletion (10,791 schizophrenia CLOZUK cases, 24,391 WTCCC controls) (6); all subjects were of European ancestry.
Six comparisons of schizophrenia PRS were considered (Figure 1) between: (i) individuals with 22q11.2DS, with and without schizophrenia (ii) 22q11.2DS-schizophrenia and WTCCC controls (iii) 22q11.2DS-non-psychotic and WTCCC controls (iv) the CLOZUK schizophrenia cohort and 22q11.2DS-schizophrenia, (v) the CLOZUK schizophrenia cohort and 22q11.2DS-non-psychotic, and (vi) the CLOZUK schizophrenia cohort and WTCCC controls.
Figure 3a shows the proportion of variance explained by PRS (Nagelkerke’s R2) for each of the six comparisons (clockwise: i to vi). Figure 3b shows the odds ratio and 95% confidence interval bars per PRS standard deviation constructed for these six PRS analyses (left to right: i to vi). Taken as a whole, these results demonstrate that the PRS in the CLOZUK-schizophrenia is significantly greater than that seen in 22q11.2DS individuals with schizophrenia, which itself is significantly greater than that seen in the WTCCC-controls and the 22q11.2DS non-psychotic groups.
We then proceeded to examine novel reciprocal case-control comparisons between the 22q11.2DS and population-based cohorts (Fig. 1). Individuals with 22q11.2DS-schizophrenia had a significantly higher PRS than WTCCC-controls (pT=0.05: R2=0.014, padjusted 3.8×10−6, OR=1.53, 95% CI: 1.29–1.81; Fig. 3a(ii) and 3b(ii)). Similarly, the CLOZUK-schizophrenia group had a significantly higher PRS than the 22q11.2DS-non-psychotic group (pT=0.05: R2=0.047, padjusted <2×10−16, OR=2.05, 95% CI: 1.74–2.41; Fig. 3a(v) and 3b(v)). These results indicate that PRS was significantly greater in individuals with psychotic illness, regardless of the presence of the 22q11.2 deletion.
Within diagnostic group analyses revealed that the PRS observed in schizophrenia cases from the CLOZUK cohort was significantly greater that that seen in those with schizophrenia and 22q11.2DS (pT=0.05: R2=0.0065, padjusted 0.01, OR=1.31, 95% CI:1.10–1.55; Fig. 3a(iv) and 3b(iv)). The PRS results for the non-psychotic 22q11.2DS group were not significantly different than those for WTCCC controls (Fig. 3a(iii) and 3b(iii)); however, the 22q11.2DS sample size was underpowered for this comparison given the low effect size (Table S6).
We assessed the consistency of these results with a liability threshold model that assumed additive effects of the 22q11.2 deletion and PRS on risk for schizophrenia; details of our methodology and results are provided in the Supplemental methods and Table S7. The model that best fit the observed data (where the sum of the absolute difference in observed and expected PRS was the lowest (0.005) across all 4 cohorts) estimated that the PRS contributes to 5.2% (range 4.9% to 5.5%) of the variance of liability for schizophrenia. This best-fit model also provided an estimate of the shift in liability attributable to the 22q11.2 deletion (1.0, range 0.2–1.9) from population-based expectations of schizophrenia prevalence (2.33, based on top 1% CI of the normal distribution). In comparison, an increase in the PRS by one standard deviation corresponds to a shift in liability of 0.23. Working back from the estimated shift in liability attributable to the 22q11.2 deletion (i.e., 2.33–1.00=1.33) provides a predicted prevalence of schizophrenia in 22q11.2DS of 9.2% (range 1.7% to 33.7%) (Table S7). Using this best-fit liability threshold model to calculate the sample sizes required to provide 80% power to detect a nominally significant difference in PRS at pT=0.05, indicated that apart from the comparison of PRS between 22q11.2DS individuals without psychosis and WTCCC controls, all other primary analyses of polygenic risk were sufficiently powered (Table S6).
Common variants - intact 22q11.2 allele and genome-wide association (GWAS)
Given previous evidence for a genetic modifier locus in studies of cardiac phenotypes in 22q11.2DS (24), we considered the possibility that individual genetic risk factors could confer an elevated effect size in association analyses of 22q11.2DS that compared those with and without schizophrenia. We examined this possibility by conducting association analyses of individual SNPs, restricted to the 22q11.2 deletion region (3,252 SNPs for full dataset, 3,236 SNPs for European ancestry) and genome-wide (4,313,064 SNPs outside this deletion region). None of these analyses identified genome-wide significant associations with schizophrenia in either the European cohort (n=435) or total cohort (n=519) (Fig. SF4 to SF9, Tables S8 to S11). Genome-wide, the top SNP was rs12824373 (p=6.91×10−7), located in an intron of the CRADD gene. This SNP was not significantly associated with schizophrenia in the PGC GWAS (13). Also, no SNP previously reported to reach genome-wide-significant levels of association with schizophrenia (13) yielded suggestive evidence of association in this GWAS of 22q11.2DS (minimum unadjusted p-value=7.4×10−3; Table S12). Results were also non-significant when restricting to 724,010 SNPs in 2925 LOF-intolerant genes (6) (minimum p=4.88×10−5).
Rare variants - individual, gene-based and gene-set analyses
Similar to common variant results, there was no significant association of schizophrenia with any individual rare variant, nor for individual gene-based rare variant analyses, within or outside the 22q11.2 deletion region (Fig. SF10, Table S13–15). There were few LOF variants within the 22q11.2 deletion region (i.e., in effect, knock-out mutations) (Fig. 4).
Figure 4. Representation of 46 genes spanning the 22q11.2 LCR22A-LCR22D deletion region annotated for contextual information and results from the current study:
Representation of 46 genes spanning the 22q11.2 low copy repeat (LCR) LCR22A-LCR22D deletion region annotated for contextual information and, on grey background, results from the current study. The sections numbered 1–11 indicate the following:
1) Gene expression in brain, ranging from none to low (white) to low-medium (yellow) to medium-high (orange) to very high (red) according to BrainSpan (BrainSpan: Atlas of the Developing Human Brain [[http://developinghumanbrain.org]]http://developinghumanbrain.org]) and as previously processed by (10);
2 & 3) Exome Aggregation Consortium (ExAC) (48) calculated probability (ranging from 0 to 1.00, shown as a spectrum from white to deep violet) that the gene is intolerant to (2) two LOF variants (recessive; pRec), or (3) a single LOF variant (haploinsufficient; pLI);
4 & 5) Neuro-phenotype data availability, (4) from mouse in the form of mouse homologues from MGI (49) as a union of two MPO-based gene-sets [MP:0005386 behavior/neurological phenotype, MP:0003631 nervous system phenotype] as used in (10), and (5) for human neurologic disease genes from the Clinical Genomic Database (CGD) (CGD: Clinical Genomic Database. [https://research.nhgri.nih.gov/CGD/]);
6) Clusters of small rare deletions in the general population per the Database of Genomic Variants (DGV) (50), indicating presumed tolerance to hemizygosity, where an aqua circle/oval represents a single deletion cluster of ≤1% frequency;
7) Rare LOF variants found in this 22q11.2DS cohort, thus representing a presumed null mutation, each variant observed in a single individual either within the schizophrenia (magenta) or non-psychotic (green) subgroups; number indicates the number of individuals (1, 2, or 3) in the schizophrenia or non-psychotic group with a LOF variant identified;
8) Genes in the 22q11.2 deletion region that show nominal significance for rare variants (in yellow, p-values ranged between 7.92E-03 and 4.75E-02 based on SKAT or Burden test) or for common variants (in red, p-values ranged between 3.63E-03 and 4.23E-02 based on SKAT test) in gene-based association tests for schizophrenia in this study using data for the subset of European ancestry. All results for the SKAT gene-based test for common variants can be found in Table S19. Note that all genes with nominal significance for rare variants are indicated in yellow, but that only CDC45, PI4KA, CLTCL1 and TRMT2A comply with the n≥5 rare variants criterion (see Table S15 for exact number of rare variants).
9) Schematic of gene positions in the 22q11.2 region.
10) Schematic of the positions, relative to genes, of the main 22q11.2 deletion region LCR22s A, B, C, and D.
11) Approximate genomic extents of the two most common 22q11.2 deletion sizes.
NA = Data not available.
Analyzing genome-wide rare variants collectively in functional gene-sets (Table S3), comparable to methods used previously (5, 10), but with the addition of accounting for polygenic risk in 22q11.2DS, showed nominally significant results (Table 1, Table S16). The top four results (unadjusted p values 5.9×10−3 – 5.5×10−4) implicated the Kirov_ARC gene-set (11, 25). Though none remained significant after adjustment for multiple testing, one result was borderline significant (p=0.053) after correcting for 300 tests (Table 1). Overall genome-wide rare-variant burden (i.e., regardless of gene function) showed no significant relationship to schizophrenia (unadjusted p values: genome-wide 0.35–0.79, 22q11.2 deletion region 0.02–0.79; Tables S17–18). Similarly, there was no significant enrichment of heterozygous truncating variants (p=0.76) in a gene-set comprising 2,925 genes considered intolerant to LOF variation (gnomAD 2.1 observed/expected LOF, upper 95% CI <0.35).
Table 1.
Gene-set burden association results for rare variants in Kirov ARC pathway and schizophrenia in 432 European subjects with 22q11.2DS
| Burden (SKAT) | ||||
| Variant group and weight | P-value | Adjusted P-value (FWER)* | ||
| Missense – any | 0.00055 | 0.053 | ||
| Missense – any, weighted for rarity | 0.0028 | 0.22 | ||
| Any with CADD > 15 | 0.0045 | 0.31 | ||
| Missense with CADD > 15 | 0.0059 | 0.37 | ||
| Burden (Logistic Regression)*** | ||||
| Variant group | Coefficient point estimate | Coefficient Ward P-value | Global P-value | Adjusted P-value (permutation-FDR)** |
| LOF Indel | NA | NA | 0.0012 | 0.25 |
| LOF SNV | 0.17 | 0.12 | ||
| Missense with CADD > 15 | −0.30 | 0.0065 | ||
| Missense other | −0.22 | 0.034 | ||
Adjusted p-value based on multiple test correction using P_ACT for the 300 tests conducted (30 gene-sets (10) x 5 variant filters x 2 weighting schemes)
Adjusted p-value based on multiple test correction using permutation-based FDR for 1000 iterations of 360 tests (30 gene-sets (10) x 2 variant sets x 3 frequency weighting schemes x 2 PRS and no PRS)
Burden result based on a logistic regression model combines effects from different types of rare variants (LOF Indel, LOF SNV, Missense with CADD > 15 and Missense other) with PRS as one of the covariates
Kirov_ARC = 28 genes in an activity-regulated cytoskeleton-associated protein (ARC) complex curated by George Kirov in 2012 (11).
Using a complementary multivariate method to assess the functional genome-wide rare variant burden supported the Kirov_ARC gene-set (11) as the most promising (permutation-based FDR ~25%) of the 30 gene-sets tested, but revealed possible heterogeneity of findings (Fig. SF11). For rare missense variants in this gene-set, the coefficients were negative, indicating the variant burden is enriched in the non-psychotic 22q11.2DS group. (Wald p-value, coefficient: 2.6×10−3, −0.35 for missense CADD >15 variants; 0.04, −0.22 for other missense variants). In contrast, the contribution of LOF SNVs was positive (Wald p-value 0.21, coefficient 0.16), though non-significant, suggesting that for the Kirov_ARC gene-set, the LOF rare variant burden may be somewhat enriched in those with schizophrenia. The main gene driving the putatively protective rare missense burden signal for the Kirov_ARC gene-set was DLGAP2 (gene-level nominal p-value 0.01, CADD >15 missense variants).
DISCUSSION
The results of this study provide new data relevant to our understanding of the genetic architecture underlying the exceptionally increased risk for developing schizophrenia and related psychotic illnesses imparted by the 22q11.2 deletion (1). We demonstrate for the first time that individuals with 22q11.2DS who develop schizophrenia have a significantly greater schizophrenia polygenic risk score than those with no psychotic illness, suggesting that both the 22q11.2 deletion and the aggregate risk conferred by common genetic risk factors outside the deleted region contribute to schizophrenia in 22q11.2DS. There was no evidence for contributions from common or rare variants at the individual variant or gene-based level. There was however some suggestive evidence that rare coding variants in genes involved in ARC synaptic gene complexes, previously reported to be involved in large population-based schizophrenia samples (8–12), may contribute to modifying risk of schizophrenia in 22q11.2DS.
Contribution of genome-wide polygenic risk
The PRS results showing that collective common genetic risk factors independent of the 22q11.2 deletion have a substantive role in increasing risk of schizophrenia in 22q11.2DS are consistent with research indicating the importance of polygenic risk in developing schizophrenia in large general population studies (6, 13). They are also consistent with findings for individuals with other CNVs that increase risk for neuropsychiatric and neurodevelopmental disorders (26–28). They differ somewhat from those of a study examining CNVs ascertained as part of the PGC that reported small PRS effects in 56 individuals with schizophrenia and a 22q11.2 deletion, in contrast to greater PRS effects for CNVs with low risk for schizophrenia that were also present in control samples (26). Availability of non-psychotic control groups and the sample sizes available could have contributed to the strong effect of schizophrenia PRS observed in our study.
The ability to directly compare PRS between individuals with 22q11.2DS and cohorts of schizophrenia and controls with no 22q11.2 deletion allowed us to determine that the contribution of polygenic risk to schizophrenia was slightly but significantly lower in those with, than in those without a 22q11.2 deletion (6). This suggests that when our results are considered according to the liability threshold model, the presence of the 22q11.2 deletion may lower to some extent the degree of background polygenic risk involved in developing schizophrenia.
Broad confidence intervals precluded our ability to determine additive or other relationships between the PRS and 22q11.2 deletion. Nonetheless, assuming an additive liability model, the proportion of variance liability for schizophrenia in 22q11.2DS attributable to the PRS was estimated to be 5.2% (range 4.9%−5.5%), which is in line with the estimates of 7% and 5.7% reported previously for idiopathic schizophrenia (6, 13). Based on this model there were wide ranges for the estimated shift in liability attributed to the 22q11.2 deletion (from the expected 1% population prevalence of schizophrenia) (Table S7) and estimated schizophrenia prevalence in 22q11.2DS (1.7% to 33.7%). The latter values would therefore embrace the low estimates reported in other studies using statistical models (29, 30) and the actual observed prevalence of psychotic illness in 22q11.2DS of >20% (1, 31, 32).
Contribution of genome-wide rare variant functional gene-sets
Complementary to the polygenic risk results, and in innovative analyses that accounted for PRS effects, the results also indicated a possible role for rare variants in the gene-set representing the ARC postsynaptic protein complex (8–12, 25). Consistent with our results in 22q11.2DS, effect sizes are expected to be modest for individual rare SNVs and related gene-set results in schizophrenia (25). It is therefore encouraging that our results for 22q11.2DS provided suggestive evidence in line with multiple studies that have reported rare variants, both SNVs and CNVs, affecting coding sequence of genes from the ARC and closely related synaptic gene-sets to be significantly associated with idiopathic schizophrenia (8–12, 29). Higher powered studies will be needed to evaluate the precise genomic architecture, including direction of effect patterns for rare variants (11, 25, 33, 34).
Role of the 22q11.2 deletion and possible genetic mechanisms
We have proposed that the presence of the 22q11.2 deletion may lower an individual’s tolerance for the expression of multiple biological mechanisms and/or pathways involved in developing schizophrenia (3, 5), and anticipate that this is likely to be related to the collective effect of lowered multi-gene dosage at the 22q11.2 locus. Importantly, we did not find evidence for risk attributable to the 22q11.2 deletion to be mediated through common or rare variants of genes located on the intact 22q11.2 allele, suggesting that it is unlikely that such variants have a large effect on schizophrenia risk. This included the few LOF (in effect null) variants that we identified (Fig. 4) and are now the focus of an in-depth reverse phenotyping effort. These negative results appear consistent with a recent study of idiopathic schizophrenia reporting the absence of detectable individual gene effects for 22q11.2 deletion genes, in contrast to genes in other CNV regions (35).
Establishing the exact biological mechanisms through which the 22q11.2 deletion confers increased risk to schizophrenia is beyond the scope of this study. However, given the profile of the genes in this region, including those encoding transcription factors, mitochondrial proteins, and others, and the fact that most are expressed in brain (Fig. 4), the 22q11.2 deletion region may be particularly prone to general neurodevelopmental perturbation, and impaired regulatory mechanisms. For example, haploinsufficiency of DGCR8 affects miRNA buffering genome-wide, thus could reveal or enhance effects of common and rare variants elsewhere in the genome (36–41). Many of the genes in the ARC gene-set, for example, are predicted to be affected by DGCR8 haploinsufficiency (36, 41).
Large GWAS of schizophrenia indicate that the multiple SNPs involved at each significant locus are mostly intergenic or intronic, tag protein-coding and miRNA genes, and increase or decrease risk in about equal proportions, each with a very small effect size (6, 13). Thus the common variant signal measured by polygenic risk may be consistent with a role for non-coding and/or regulatory mechanisms in schizophrenia, including in 22q11.2DS (6, 13, 37, 41, 42). The precise complement of common and rare variants leading to schizophrenia may be personal to the individual but nonetheless observable in aggregate across individuals (43, 44). Further investigations designed to determine the exact nature of the primary 22q11.2 deletion-related biological insult therefore promises to improve our understanding of the mechanisms underlying schizophrenia in both 22q11.2DS and the general population.
Advantages and limitations
We had available the largest sample of 22q11.2DS subjects with and without a psychotic disorder ever brought together to study the impact of common and rare SNVs to schizophrenia risk. The signal imparted by additional polygenic common variant risk, similar to previous studies using idiopathic schizophrenia case-control samples several orders of magnitude larger (6, 13), produced clearly significant results. The results support our design, using the non-psychotic 22q11.2DS subgroup as the main comparison for 22q11.2DS cases with schizophrenia subgroup within 22q11.2DS. A power analysis indicated that our primary analyses of polygenic risk using the 22q11.2DS schizophrenia group were adequately powered for comparisons with 22q11.2DS without psychotic illness and with the large population-based case-control samples. Also, to our knowledge, this was the first study of schizophrenia genetics to account for polygenic risk while analyzing rare variant burden.
Despite these advantages, power was limited by the available cohort size (Fig. SF12) and would be affected by the possibility that some 22q11.2DS subjects in the non-psychotic group may go on to develop schizophrenia. It is probable that some of the genome-wide polygenic risk associated with schizophrenia may also be involved in other neurodevelopmental disorders (45). This may make the current results all the more remarkable, given the presence of one or more neurodevelopmental phenotypes in most individuals with 22q11.2DS (46).
With respect to rare variants, the current study focused primarily on the signal derived from the exome where findings are more likely to be biologically interpretable and potentially clinically relevant than that from non-coding variants (5, 33). Our conservative design, e.g., for each filtered variant set, considering only those genes with 5 or more rare variants observed in the 22q11.2DS sample of European ancestry, and using 30 gene-sets thus correcting for 300 tests, may have affected power. Also, as in other sequencing studies (25), certain variant types, e.g., indels, were imperfectly captured (14). Recruiting larger samples of individuals with schizophrenia and older adults with no psychotic illness from the 22q11.2DS population could provide the means to further delineate the strength of the relationship between the factors identified, including rare variants in the small ARC gene-set, and the risk for schizophrenia. This would also be essential to enable investigation of other mechanisms with lower effect sizes. In future studies, larger samples of 22q11.2DS may also assist in determining the role of X chromosome variants in the context of a 22q11.2 deletion, and provide sufficient data to assess those of non-European ancestry (5).
Although we were unable to use these WGS data to simultaneously assess CNVs or other structural variants (14), we have previously demonstrated in a smaller but overlapping sample that additional rare CNVs overlapping genes outside of the 22q11.2 deletion region contribute to schizophrenia risk in 22q11.2DS (5). Also, the IBBC proband-only 22q11.2DS sample provides no capability of examining inheritance, parent-of-origin, or segregation data for the 22q11.2 deletion or any other variants, or the family history of schizophrenia, all of which are factors of interest in disease expression (1, 3, 5). In smaller samples, the parental origin and inheritance status of the 22q11.2 deletion have not appeared to play a role in schizophrenia, however (47), and one can expect that >90% of the 22q11.2 deletions would arise as de novo mutations (1).
CONCLUSIONS
Our results indicate that, despite its high impact, the risk conferred by the 22q11.2 deletion is even greater when present with the genome-wide risk factors that also have a key role in schizophrenia risk in the general population (6, 13, 25). Given the magnitude of the risk of the 22q11.2 deletion relative to common genetic risk factors, including polygenic risk, understanding the biological consequences of this deletion will be an important next step in appreciating the underlying pathology of schizophrenia. The overall results further illustrate the value of using 22q11.2DS as a model for delineating the genetic architecture of schizophrenia. The direction of effect, and the type and precise position of the additional variants will be important factors to consider. Lessons learned from 22q11.2DS may be applicable to other recurrent CNVs that contribute to risk for schizophrenia in the general population (10). Future analyses of raw whole-genome sequencing data will allow for more detailed assessment of variants and pathways implicated, and the overall genetic architecture and proposed mechanisms of schizophrenia, incorporating those related to structural variants (5). These observations have important implications for research, including functional genomic studies using induced pluripotent stem cells from individuals with 22q11.2DS to model schizophrenia, and potentially for clinical genetic practice. Given the magnitude of the effect size of the 22q11.2 deletion, prospective studies using algorithms generated from such sequence data together with phenotypic data relevant to schizophrenia risk could lead to findings of clinical relevance to the individual (3, 48).
Supplementary Material
Acknowledgements
The IBBC is supported by the National Institute of Mental Health grants U01MH101719, U01MH0101720, U01MH0101723, U01MH101722, and U01MH101724. Other grant support includes: Brain and Behavior Foundation Young Researcher grant (formerly NARSAD, grant n. 21278) to Dr. Armando, Fondecyt-Chile Grant 1171014 to Drs. Fritsch, Ornstein, and Repetto, grant FWO G.0E11.17N to Dr. Vermeesch, MRC Centre grant MR/L010305/1, National Institute of Mental Health grant U01MH101724, and Welsh Government funded Research Centre 514032 to Dr. Owen, NARSAD (Brain and Behavior Research Foundation) to Dr. Ousley, National Institute of Mental Health grant MH064824 to Drs. Antshel, Fremont, Kates, and Simon, NIH grant R01GM117946 to Drs. Duncan and Voss, NIH grant U54NS091859 to Drs. Chopra, Epstein, Johnston, and Warren, NIH grants R01MH100917 and R01GM117946 to Drs. Cutler, Epstein, Kotlar, Mulle, and Zwick, NIMH K01 MH112774 to Dr. Jalbrzikowski, NIMH R01 MH085953 and NIH U54 EB020403 to Drs. Bearden and Kushan, NIMH U01MH101724 to Dr. van den Bree, P01HD070454 to Drs. Crowley, Diacou, Emanuel, Guo, Jin, Lilley, McDonald-McGinn, McGinn, E.M. Moss, Morrow, Salmons, Sharkus, Tran, Unolt, Zackai, and Zhao, P50MH09689 and R01-MH-1072351 to Dr. R.E. Gur, P50MH096891 to Drs. Calkins and R.C. Gur, National Institute of Mental Health R01MH085953 to Drs. C. Murphy and D. Murphy, R01-MH-107235 to Dr. R.C. Gur, Swiss National Science Foundation (Grant number 324730_144260) and National Center of Competence in Research (NCCR) “Synapsy-The Synaptic Bases of Mental Diseases” (Grant number 51NF40-185897) to Dr. Eliez, T32 MH019112 to Dr. Tang, the Binational Science Foundation, grant 2017370 to Dr. Gothelf, U01 MH087626 to Drs. Calkins, Gallagher, R.C. Gur, and R.E. Gur, U54HD090260 to Drs. Diacou, Guo, Morrow, and Zhao, UO1-MH191719 and R01 MH087636-01A1 to Drs. Crowley, Emanuel, Jin, Lilley, McDonald-McGinn, McGinn, E.M. Moss, Salmons, Sharkus, Tran, Unolt, and Zackai, Van de Werf fund for cardiovascular research to Dr. Breckpot, IWT 131625 to Dr. Demaerel, and Wellcome Trust grant 102428/Z/13/Z to Drs. Monfeuga and Williams.
The authors thank all individuals with 22q11.2 deletion syndrome and their families for their generous contributions to this and related research studies, and are grateful to support charities including Max Appeal, The 22Crew and Unique. This work included participants of the NCMH study, a collaboration between Cardiff, Swansea and Bangor Universities. We thank the following members of the Division of Psychological Medicine and Clinical Neurosciences at the Cardiff School of Medicine for technical assistance: S. Chawner, A. Cunningham, C. Eaton, A. Evans, S. Morrison, H. Moulding, P. Birch, M. Tong, S. Bowen, L. Benger, M. Thomas, L. Reed, C.T. Carmichael, L. Bates, B. Lugonja, J. Morgan, N. Vinh, A. Evans, L. Hopkins, L. Tram, S. Jacques, S. Lewis.
Footnotes
Conflict of Interest: Dr. Merico is a full-time employee and is entitled to a stock option of Deep Genomics Inc., a start-up developing oligonucleotide therapeutics for rare genetic disorders. Dr. Tassone received funding from Zynerba and Asuragen Inc. for her work on Fragile X syndrome. All other authors report no financial relationships with commercial interests.
REFERENCES
- 1.McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JA, et al. 22q11.2 deletion syndrome. Nat Rev Dis Primers. 2015;1:15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fung WL, Butcher NJ, Costain G, Andrade DM, Boot E, Chow EW, et al. Practical guidelines for managing adults with 22q11.2 deletion syndrome. Genet Med. 2015;17(8):599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gur RE, Bassett AS, McDonald-McGinn DM, Bearden CE, Chow E, Emanuel BS, et al. A neurogenetic model for the study of schizophrenia spectrum disorders: the International 22q11.2 Deletion Syndrome Brain Behavior Consortium. Mol Psychiatry. 2017;22(12):1664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Insel TR. Rethinking schizophrenia. Nature. 2010;468(7321):187–93. [DOI] [PubMed] [Google Scholar]
- 5.Bassett AS, Lowther C, Merico D, Costain G, Chow EWC, van Amelsvoort T, et al. Rare Genome-Wide Copy Number Variation and Expression of Schizophrenia in 22q11.2 Deletion Syndrome. Am J Psychiatry. 2017;174(11):1054–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardinas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh T, Walters JTR, Johnstone M, Curtis D, Suvisaari J, Torniainen M, et al. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet. 2017;49(8):1167–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landen M, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19(11):1433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49(1):27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17(2):142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston HR, Chopra P, Wingo TS, Patel V, International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome, Epstein MP, et al. PEMapper and PECaller provide a simplified approach to whole-genome sequencing. Proc Natl Acad Sci U S A. 2017;114(10):E1923–E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees E, Kendall K, Pardinas AF, Legge SE, Pocklington A, Escott-Price V, et al. Analysis of Intellectual Disability Copy Number Variants for Association With Schizophrenia. JAMA Psychiatry. 2016;73(9):963–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo T, Diacou A, Nomaru H, McDonald-McGinn DM, Hestand M, Demaerel W, et al. Deletion size analysis of 1680 22q11.2DS subjects identifies a new recombination hotspot on chromosome 22q11.2. Hum Mol Genet. 2018;27(7):1150–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh PR, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane HK, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48(11):1443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31(9):1466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conneely KN, Boehnke M. So many correlated tests, so little time! Rapid adjustment of P values for multiple correlated tests. Am J Hum Genet. 2007;81(6):1158–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Epstein MP, Duncan R, Jiang Y, Conneely KN, Allen AS, Satten GA. A permutation procedure to correct for confounders in case-control studies, including tests of rare variation. Am J Hum Genet. 2012;91(2):215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mlynarski EE, Sheridan MB, Xie M, Guo T, Racedo SE, McDonald-McGinn DM, et al. Copy-Number Variation of the Glucose Transporter Gene SLC2A3 and Congenital Heart Defects in the 22q11.2 Deletion Syndrome. Am J Hum Genet. 2015;96(5):753–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rees E, Carrera N, Morgan J, Hambridge K, Escott-Price V, Pocklington AJ, et al. Targeted Sequencing of 10,198 Samples Confirms Abnormalities in Neuronal Activity and Implicates Voltage-Gated Sodium Channels in Schizophrenia Pathogenesis. Biol Psychiatry. 2019;85(7):554–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergen SE, Ploner A, Howrigan D, CNV Analysis Group and the Schizophrenia Working Group of the Psychiatric Genomics Consortium, O’Donovan, Smoller JW, et al. Joint Contributions of Rare Copy Number Variants and Common SNPs to Risk for Schizophrenia. Am J Psychiatry. 2019;176(1):29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tansey KE, Rees E, Linden DE, Ripke S, Chambert KD, Moran JL, et al. Common alleles contribute to schizophrenia in CNV carriers. Mol Psychiatry. 2016;21(8):1085–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niemi MEK, Martin HC, Rice DL, Gallone G, Gordon S, Kelemen M, et al. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature. 2018;562(7726):268–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirov G, Rees E, Walters JT, Escott-Price V, Georgieva L, Richards AL, et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry. 2014;75(5):378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoeffding LK, Trabjerg BB, Olsen L, Mazin W, Sparso T, Vangkilde A, et al. Risk of Psychiatric Disorders Among Individuals With the 22q11.2 Deletion or Duplication: A Danish Nationwide, Register-Based Study. JAMA Psychiatry. 2017;74(3):282–90. [DOI] [PubMed] [Google Scholar]
- 31.Fung SJ, Sivagnanasundaram S, Weickert CS. Lack of change in markers of presynaptic terminal abundance alongside subtle reductions in markers of presynaptic terminal plasticity in prefrontal cortex of schizophrenia patients. Biol Psychiatry. 2011;69(1):71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider M, Debbane M, Bassett AS, Chow EW, Fung WL, van den Bree M, et al. Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry. 2014;171(6):627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Multiple Sclerosis Genetics Consortium. Low-Frequency and Rare-Coding Variation Contributes to Multiple Sclerosis Risk. Cell. 2018;175(6):1679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganna A, Satterstrom FK, Zekavat SM, Das I, Kurki MI, Churchhouse C, et al. Quantifying the Impact of Rare and Ultra-rare Coding Variation across the Phenotypic Spectrum. Am J Hum Genet. 2018;102(6):1204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coe BP, Stessman HAF, Sulovari A, Geisheker MR, Bakken TE, Lake AM, et al. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet. 2019;51(1):106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40(6):751–60. [DOI] [PubMed] [Google Scholar]
- 37.Forstner AJ, Degenhardt F, Schratt G, Nothen MM. MicroRNAs as the cause of schizophrenia in 22q11.2 deletion carriers, and possible implications for idiopathic disease: a mini-review. Front Mol Neurosci. 2013;6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schofield CM, Hsu R, Barker AJ, Gertz CC, Blelloch R, Ullian EM. Monoallelic deletion of the microRNA biogenesis gene Dgcr8 produces deficits in the development of excitatory synaptic transmission in the prefrontal cortex. Neural Dev. 2011;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brzustowicz LM, Bassett AS. miRNA-mediated risk for schizophrenia in 22q11.2 deletion syndrome. Front Genet. 2012;3:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merico D, Costain G, Butcher NJ, Warnica W, Ogura L, Alfred SE, et al. MicroRNA Dysregulation, Gene Networks, and Risk for Schizophrenia in 22q11.2 Deletion Syndrome. Front Neurol. 2014;5:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merico D, Zarrei M, Costain G, Ogura L, Alipanahi B, Gazzellone MJ, et al. Whole-Genome Sequencing Suggests Schizophrenia Risk Mechanisms in Humans with 22q11.2 Deletion Syndrome. G3 (Bethesda). 2015;5(11):2453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warnica W, Merico D, Costain G, Alfred SE, Wei J, Marshall CR, et al. Copy number variable microRNAs in schizophrenia and their neurodevelopmental gene targets. Biol Psychiatry. 2015;77(2):158–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costain G, McDonald-McGinn DM, Bassett AS. Prenatal genetic testing with chromosomal microarray analysis identifies major risk variants for schizophrenia and other later-onset disorders. Am J Psychiatry. 2013;170(12):1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrison PJ. Recent genetic findings in schizophrenia and their therapeutic relevance. J Psychopharmacol. 2015;29(2):85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smeland OB, Bahrami S, Frei O, Shadrin A, O’Connell K, Savage J, et al. Genome-wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swillen A, Moss E, Duijff S. Neurodevelopmental outcome in 22q11.2 deletion syndrome and management. Am J Med Genet A. 2018;176(10):2160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bassett AS, Marshall CR, Lionel AC, Chow EW, Scherer SW. Copy number variations and risk for schizophrenia in 22q11.2 deletion syndrome. Hum Mol Genet. 2008;17(24):4045–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van L, Boot E, Bassett AS. Update on the 22q11.2 deletion syndrome and its relevance to schizophrenia. Curr Opin Psychiatry. 2017;30(3):191–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.