Abstract
Background and Aim:
Rabbit hemorrhagic disease (RHD) is an economically important disorder of rabbits, where infection results in severe losses to the meat and fur industries. Our goal was to characterize the RHD virus (RHDV) strains currently circulating in different regions of Egypt.
Materials and Methods:
Fifty rabbits suspected of harboring RHDV from 15 Egyptian governorates were evaluated. Diseased rabbits were identified by clinical signs and postmortem lesions. RHDV was confirmed through hemagglutination assay (HA) and polymerase chain reaction (PCR). Partial sequencing of the VP60 gene was performed for genotyping.
Results:
From 50 rabbits, we identified 16 cases of RHDV (32%) by HA and PCR, including seven males and nine females. We identified two distinct genotypes through sequencing of an amplified fragment of the virus VP60 gene. One group is composed of those circulating primarily in upper Egypt, which is closely related to the classical G3-G5 virus strains, and the second group, circulating predominantly in lower Egypt, was more closely related to the RHDV2 variant. The overall nucleotide sequence identity ranged from 78.4% to 100%, and identity with the vaccine strains ranged from 78.8% to 91.1%.
Conclusion:
Our results constitute important documentation of RHDV strains currently circulating in Egypt. The findings suggest that there may be a limit to the effectiveness of currently applied vaccine strains as this formulation may not cover all circulating strains. A wider investigation that includes both domestic and wild rabbits will be needed to identify appropriate control measures for this disease.
Keywords: Egypt, native rabbits, prevalence, rabbit hemorrhagic disease virus, VP60
Introduction
The first case of rabbit hemorrhagic disease virus (RHDV) in Egypt was recorded in Sharkia Province in 1991, and this disease has since spread to other Egyptian governorates [1]. Despite active vaccination programs, RHDV remains a threat to farms that produce rabbit meat for popular consumption [2] because of high disease-associated morbidity and mortality.
Rabbit hemorrhagic disease (RHD) is a rapidly fatal viral disease of adult rabbits. The first recorded clinical cases of RHD were in China [3]. The disease spread rapidly through Asia and Europe and became endemic within just few years [4-6]. The etiological agent, RHDV, is a member of the Calicivirus family. Typically, RHDV kills more than 90% of susceptible adult animals within 2-3 days of infection [6]. In 2010, a new strain, RHDV2, was identified in association with infection of rabbits of different ages and populations in France [7]. RHDV2 spread rapidly throughout Europe [8,9] and was detected in Australia in 2015 [10] and in Canada in 2016 [11]. RHDV2 is now considered endemic in Europe [12]. The disease associated with RHDV2 includes symptoms that are similar to those of classic RHDV, although mortality rates are comparatively low [13]. Transmission of RHDV can be through oral, nasal, and conjunctival routes, and transmission through a vector-like insect has been reported [14]. Transmission of RHDV may also occur through direct contact with an infected animal as infected rabbits may shed the virus in their excretions [15]. The pathogenesis of RHD includes development of petechial hemorrhages in multiple systemic organs as a result of virus-induced hyper-coagulopathy, and the most severe form of these lesions appear in the liver, trachea, and lungs [16]. RHDV also promotes fatal hepatitis, most notably in adult rabbits [7]. RHDV2 can result in fatal infections of rabbits at different ages, including those as young as 11 days old [17]. This new lagovirus has also been detected in wild hares [18,19]. The combination of classical and variant viruses by definition has resulted in increased diversity in this virus species [20,21]. The genomic structures of RHDV and RHDV2 are similarly arranged, and both include two open reading frames (ORFs). ORF1 encodes the nonstructural proteins, including the RNA-dependent RNA polymerase and the major capsid protein (VP60). The second ORF encodes a minor structural protein known as vp10 [22,23]. Many polymerase chain reaction (PCR)-based methods are available for the identification of viruses, including rabbit viruses, and these methods are substantially more rapid than traditional virus isolation as they offer the possibility of high throughput with improved sensitivity and specificity. PCR also facilitates virus gene sequencing so that all vaccine and wild-type virus strains can be fully typed and differentiated. PCR can also be used for the detection of pathogens that are difficult to be isolated using traditional methods and those that are present in low titer in test samples [24]. The virus VP60 gene has been selected as a target [25] for reverse-transcription PCR (RT-PCR) assay which is currently used as a general screening tool for rabbit and hare caliciviruses. The VP60 gene was also used for the development of a universal RT-PCR method for detecting lagoviruses that uses primers that span a highly conserved region [26].
The aim of the current study was to identify circulating RHDV strains found in distinct populations of rabbits in Egypt. Our goal is to use molecular analyses to determine the relationship of any new isolates to the strains currently used for the critical vaccination strategies.
Materials and Methods
Ethical approval
This study does not contain any experimental studies with animals. Diseased rabbits were euthanized before sampling in accordance with the regulations of the General Organization for Veterinary Services and Animal Health Research Institute.
Study period and location
The study was performed during 2018-2019 along 15 representative Governorates.
Sample collection
Lung and liver samples were collected from 10 rabbits per flock after intravenous administration of 3-5 mg/kg xylazine and 35-40 mg/kg ketamine to facilitate euthanasia. All tissue samples were collected from all freshly killed rabbits. The surveyed populations were of various ages and sexes. All samples were collected aseptically, placed in sterile bags, and transported to the lab on dry ice for further laboratory evaluation. The samples were collected from rabbits in 15 different Egyptian governorates, as listed in Table-1.
Table-1.
The sources of examined samples for RHDV.
| No. | Egyptian governorates | Total no of rabbits in flocks | No of examined suspected flock | Source of samples | |||
|---|---|---|---|---|---|---|---|
| Male | Vaccination status | Female | Vaccination status | ||||
| 1. | Cairo | 100 | 4 | 1 | unvacc | 3 | unvacc |
| 2. | Giza | 209 | 6 | 1 | unvacc | 5 | unvacc |
| 3. | Fayom | 74 | 4 | 1 | unvacc | 3 | unvacc |
| 4. | Kaliobia | 36 | 2 | 1 | unvacc | 1 | unvacc |
| 5. | Behira | 205 | 3 | 0 | 0 | 3 | 2 vacc |
| 6. | Sharkia | 385 | 5 | 2 | unvacc | 3 | 3 vacc |
| 7. | Gharbia | 290 | 7 | 2 | unvacc | 5 | 1 vacc |
| 8. | Menofia | 23 | 2 | 1 | unvacc | 1 | unvacc |
| 9. | khafr el Sheikh | 60 | 2 | 1 | unvacc | 1 | unvacc |
| 10. | Alex | 40 | 1 | 0 | 0 | 1 | unvacc |
| 11. | Dakhlia | 107 | 4 | 0 | 0 | 4 | unvvacc |
| 12. | Menia | 96 | 5 | 3 | unvacc | 2 | unvacc |
| 13. | Sohag | 60 | 1 | 0 | 0 | 1 | unvacc |
| 14. | Assuit | 71 | 3 | 1 | unvacc | 2 | unvacc |
| 15. | Qina | 20 | 1 | 0 | 0 | 1 | unvacc |
| Total | 1776 | 50 | 14 | - | 36 | 6 vacc | |
RHDV=Rabbit hemorrhagic disease virus
Hemagglutination assay (HA)
An HA was performed according to previously published methods [27]. Briefly, a fragment of liver tissue was mechanically homogenized in 10-20% (w/v) saline solution (pH 7.2), followed by clarification of the supernatant by centrifugation at 5000 g for 10 min. Type O human red blood cells (RBCs; VACSERA, Cairo, Egypt) were washed in phosphate-buffered saline (PBS; pH 6.5) and then subjected to centrifugation at 500 g for 10 min. The sedimented erythrocytes were re-suspended at 0.75% concentration in PBS (pH 7.2). HA was performed with serial two-fold dilutions of clarified liver homogenate in 50 ml PBS, pH 7.2, and positive and negative controls were also included. Fifty microliters of 0.75% washed type O human RBCs were added to each well, followed by incubation at 4°C for 1 h. The HA titer was determined according to the reciprocal of the highest dilution capable of generating hemagglutination of the RBCs.
RNA extraction
RNA extraction from the clarified tissue homogenates was performed using the QIAamp viral RNA Mini kit (Qiagen, Gmbh, Germany) according to the manufacturer’s instructions. Briefly, 140 µl of the supernatant was incubated with 560 µl of AVL lysis buffer and 5.6 µl of carrier RNA at room temperature for 10 min. After incubation, 560 µl of 100% ethanol was added. The sample was then washed and centrifuged 2 times. RNA was eluted with 60 µl of elution buffer.
PCR amplification
Oligonucleotide primers (Metabion, Germany) that were designed [28] to amplify 538 bp of the VP60 gene (P33: CCACCACCAACACTTCAGGT and P34: CAGGTTGAACACGAGTGTGC) were used in a 25 µl reaction containing 12.5 µl of Quantitect probe RT-PCR buffer (Qiagen, Gmbh, Germany), 1 µl of each of the primers at a concentration of 20 pmol, 0.25 µl of reverse transcriptase, 7.25 µl of water, and 3 µl of RNA template. The reaction was performed in a Biometra thermal cycler. Reverse transcription was carried out at 50°C for 30 min, followed by a primary denaturation step at 95°C for 15 min, 35 cycles of 94°C for 30 s, 56°C for 40 s, and 72°C for 45 s. A final extension step was performed at 72°C for 10 min.
Analysis of the PCR products
Fifteen microliters of the amplified VP60 PCR products were evaluated by gel electrophoresis using ultrapure 1.5% agarose (Invitrogen, Thermo Fisher Scientific, Germany) in 1×Tris-borate-EDTA (TBE) buffer at room temperature. Gelpilot 100 bp DNA ladder (Qiagen, Gmbh, Germany) was used to determine fragment sizes. PCR-amplified bands were detected by imaging using a gel documentation system (Alpha Innotech, Biometra). Data were analyzed using Automatic Image Capture Software (Protein Simple, formerly Cell Biosciences, San Jose, CA, USA).
Gene sequencing
Genetic and phylogenetic analyses
PCR products were purified using a QIAquick PCR Product extraction kit (Qiagen, Gmbh, Germany). Sequence reactions were performed using a Bigdye Terminator V3.1 cycle sequencing kit (Perkin-Elmer), and purification was performed using Centri-Sep spin columns (Thermo Fisher, Germany). VP60 sequences were obtained using a 3130 genetic analyzer (Applied Bio-systems, Life technologies, Thermo Fisher, Germany). Basic Local Alignment Search Tool (BLAST®) [29] alignment was performed to establish sequence similarities to the sequences deposited in the GenBank database. The MegAlign module of Lasergene DNA-Star version 12.1 was used to determine phylogenetic distances among the analyzed strains [30], and MEGA6 was used to create a phylogenetic tree using maximum composite likelihood with 1000 bootstrap replications, neighbor-joining, and maximum parsimony [31].
Results
In this study, the diagnosis of suspected RHDV cases was determined based on disease-associated clinical signs, postmortem lesions, HA activity, conventional PCR, and sequencing of the RHDV VP60 gene. A total of 50 rabbit herds were initially identified as suspected RHDV, and this suspicion was based on records of sudden death in rabbit flocks associated with neurologic and respiratory signs together with liver necrosis and generalized petechial hemorrhages in multiple tissues as revealed by postmortem examination. These animals surveyed from 15 different governorates in Egypt. Only six flocks of female rabbits had been vaccinated against RHDV, and all 16 rabbits ultimately diagnosed for RHDV were from flocks that were unvaccinated (Table-1, Supplementary Tables-S1 and S2 and Figure-1).
Supplementary Table-1.
Collection data for positive samples.
| Sample | Age | Governorate | Sample | Collection date | Sex | Quantitative plate HA result | HA titer | Vaccination status | No. of rabbits |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 weeks | Kaliobia | Lung and Liver | March 2018 | Male | + | 212 | Unvacc | 20 |
| 2 | 12 weeks | Faium | Liver | July 2019 | Female | + | 210 | Unvacc | 12 |
| 3 | 8 weeks | Menia | Lung and Liver | July 2019 | Male | + | 214 | Unvacc | 30 |
| 4 | 6 weeks | Sharkia | Liver | June 2018 | Male | + | 211 | Unvacc | 35 |
| 5 | 4 weeks | Sharkia | Liver | June 2018 | Male | + | 213 | Unvacc | 50 |
| 6 | 7 weeks | Menia | Liver | July 2019 | Male | + | 212 | Unvacc | 18 |
| 7 | 6 weeks | Assiut | Lung and Liver | July 2019 | Female | + | 216 | Unvacc | 20 |
| 8 | 2 weeks | Menofia | Lung and Liver | July 2019 | Male | + | 212 | Unvacc | 5 |
| 9 | 8 weeks | Gharbia | Liver | February 2019 | Female | + | 29 | Unvacc | 10 |
| 10 | 10 weeks | Qena | Lung and Liver | April 2019 | Female | + | 214 | Unvacc | 20 |
| 11 | 18 days | Dakahlia | Lung and Liver | April 2019 | Female | + | 211 | Unvacc | 30 |
| 12 | 9 weeks | Assiut | Lung and Liver | January 2019 | Male | + | 211 | Unvacc | 40 |
| 13 | 10 weeks | Sohag | Liver | August 2018 | Female | + | 213 | Unvacc | 60 |
| 14 | 12 weeks | Behira | Liver | February 2019 | Female | + | 214 | Unvacc | 25 |
| 15 | 6 weeks | Faium | Liver | December 2018 | Female | + | 210 | Unvacc | 20 |
| 16 | 8 weeks | Menia | Liver | August 2018 | Female | + | 212 | Unvacc | 9 |
HA=Hemagglutination assay
Supplementary Table-2.
Collection data for negative samples.
| Sample | Age | Governorate | Sample | Collection date | Sex | Vaccination status | No. of rabbits |
|---|---|---|---|---|---|---|---|
| 1 | 4 weeks | Cairo | Lung and Liver | May 2019 | Male | Unvacc | 50 |
| 2 | 10 weeks | Cairo | Lung and Liver | May 2019 | Female | Unvacc | 30 |
| 3 | 3 weeks | Cairo | Lung and Liver | May 2019 | Female | Unvacc | 10 |
| 4 | 1 month | Cairo | Lung and Liver | May 2019 | Female | Unvacc | 10 |
| 5 | 3 weeks | Giza | Lung and Liver | December 2018 | male | Unvacc | 15 |
| 6 | 19 days | Giza | Lung and Liver | November 2018 | Female | Giza-2006 | 100 |
| 7 | 2 months | Assiut | Liver | July 2019 | Female | Unvacc | 11 |
| 8 | 1 month | Behira | Lung and Liver | February 2019 | Female | Giza-2006 | 120 |
| 9 | 3 months | Kaliobia | Lung and Liver | March 2018 | Female | Unvacc | 16 |
| 10 | 2 months | Menia | Lung and Liver | August 2018 | Female | Unvacc | 27 |
| 11 | 1 month | Menia | Lung and Liver | August 2018 | Male | Unvacc | 12 |
| 12 | 7 weeks | Gharbia | Liver | February 2019 | Male | Unvacc | 40 |
| 13 | 3 months | Gharbia | Liver | February 2019 | Female | Giza-2006 | 150 |
| 14 | 1 month | Giza | Lung and Liver | May 2019 | Female | Unvacc | 40 |
| 15 | 5 weeks | Giza | Lung and Liver | May 2019 | Female | Unvacc | 30 |
| 16 | 2 weeks | Giza | Lung and Liver | May 2019 | Female | Unvacc | 9 |
| 17 | 3 months | Giza | Lung and Liver | May 2019 | Female | Unvacc | 15 |
| 18 | 2 months | Kafrelsheikh | Lung and Liver | January 2019 | male | Unvacc | 20 |
| 19 | 5 weeks | Behira | Lung and Liver | April 2019 | Female | Giza-2006 | 60 |
| 20 | 6 weeks | Kafrelsheikh | Lung and Liver | March 2019 | Female | Unvacc | 50 |
| 21 | 9 weeks | Alex | Lung and Liver | June 2019 | Female | Unvacc | 40 |
| 22 | 2 weeks | Menofia | Lung and Liver | February 2019 | Female | Unvacc | 18 |
| 23 | 17 days | Gharbia | Liver | January 2019 | Female | Unvacc | 22 |
| 24 | 3 months | Gharbia | Liver | January 2019 | male | Unvacc | 31 |
| 25 | 1 month | Gharbia | Liver | January 2019 | Female | Unvacc | 20 |
| 26 | 6 weeks | Gharbia | Liver | January 2019 | Female | Unvacc | 7 |
| 27 | 7 weeks | Dakahlia | Lung and Liver | October 2018 | Female | Unvacc | 27 |
| 28 | 2 months | Dakahlia | Lung and Liver | October 2018 | Female | Unvacc | 30 |
| 29 | 1 month | Dakahlia | Lung and Liver | October 2018 | Female | Unvacc | 20 |
| 30 | 5 weeks | Sharkia | Lung and Liver | June 2018 | Female | Giza-2006 | 90 |
| 31 | 3 months | Sharkia | Lung and Liver | June 2018 | Female | Giza-2006 | 70 |
| 32 | 20 days | Sharkia | Lung and Liver | June 2018 | Female | Giza-2006 | 200 |
| 33 | 9 weeks | Faium | Lung and Liver | December 2018 | Female | Unvacc | 18 |
| 34 | 5 weeks | Faium | Lung and Liver | December 2018 | Male | Unvacc | 24 |
Figure-1.
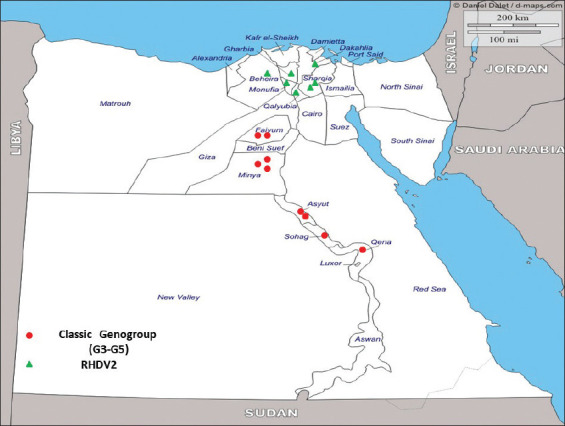
Egyptian epidemiological map for rabbit hemorrhagic disease virus (RHDV) according to the geographical distribution during (2018-2019). The classical form of RHDV virus appeared as red dots, while RHDV2 appeared green triangle and mainly distributed in the lower Egypt [Source: https://www.d-maps.com].
As shown in Figure-1, the flocks that were RHDV-positive were from the upper Egyptian governorates, and those diagnosed with RHDV2 were from the lower Egyptian provinces.
VP60 PCR
A specific 538 bp band encoding a fragment of the virus VP60 gene was amplified by PCR in 16 of the 50 rabbit herds tested (32%). Fifty percent of the males tested were found to be RHDV-positive, in contrast to only 25% of females tested (Table-2).
Table-2.
Incidence of RHDV in both examined sexes.
| Sum of no. of cases | Cases | |||
|---|---|---|---|---|
| Negative | Positive | Total | Positive RHDV percentage | |
| Female | 27 | 9 | 36 | 25 |
| Male | 7 | 7 | 14 | 50 |
| Grand total | 34 | 16 | 50 | 32 |
RHDV=Rabbit hemorrhagic disease virus
HA activity
All of the 16 samples from RHDV-PCR-positive rabbits also tested positive for HA activity. HA titers varied from 29 to 216.
DNA sequencing
DNA sequencing of the VP60 amplicons revealed a range of 78.4-100% nucleotide sequence identity. Sequence analysis indicated that two distinct RHDV types were circulating in different districts in Egypt. A sequence from one set of amplicons was closely related to that of the classical G3-G5 strains, and the other set of amplicons was more closely related to RHDV2 (Figure-2). The new sequences were assigned GenBank accession numbers MN295014-MN295029. In 2018, both RHDV genotypes (RHDV group G3-G5 and RHDV2) were detected equally. In 2019, RHDV2 was identified more frequently, as shown in Table-3. The overall nucleotide sequence identity with the Egyptian RHDV-vaccine strains (GenBank accessions numbers KX133721.1 and JQ995154.1) ranged from 78.8% to 91.1%.
Figure-2.
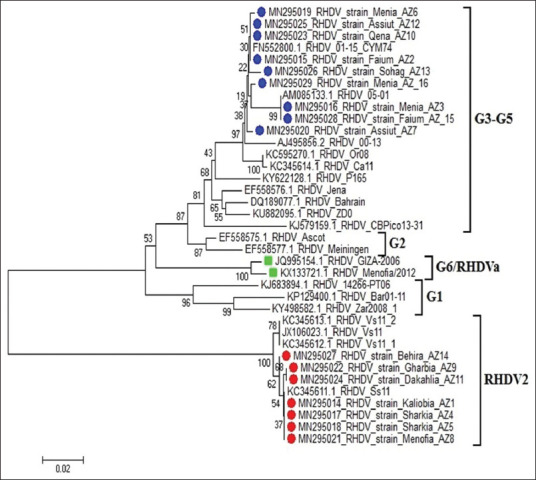
The phylogenetic tree of sequenced strains (AZ1 and AZ16) and other randomly selected strains form GenBank (MEGA 6 – neighbor-joining and maximum parsimony tool) [31].
Table-3.
A collective data for the positive cases with reference to the year of detection, governorates and strain type.
| Flock no | Age | Age group | Governorate | Sex | Collection date | Genbank acc. no. and code | Genogroup | Quantitative plate HA result | HA titer |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 weeks | <2 months | Kaliobia | Male | March 2018 | MN295014 AZ1 | RHDV2 | + | 212 |
| 2 | 12 weeks | 2 months and more | Faium | Female | July 2019 | MN295015 AZ2 | Classic RHDV Genogroup5 | + | 210 |
| 3 | 8 weeks | 2 months and more | Menia | Male | July 2019 | MN295016 AZ3 | Classic RHDV Genogroup5 | + | 214 |
| 4 | 6 weeks | <2 months | Sharkia | Male | June 2018 | MN295017 AZ4 | RHDV2 | + | 211 |
| 5 | 4 weeks | <2 months | Sharkia | Male | June 2018 | MN295018 AZ5 | RHDV2 | + | 213 |
| 6 | 7 weeks | <2 months | Menia | Male | July 2019 | MN295019 AZ6 | Classic RHDV Genogroup5 | + | 212 |
| 7 | 6 weeks | <2 months | Assiut | Female | July 2019 | MN295020 AZ7 | Classic RHDV Genogroup5 | + | 216 |
| 8 | 2 weeks | <2 months | Menofia | Male | July 2019 | MN295021 AZ8 | RHDV2 | + | 212 |
| 9 | 8 weeks | 2 months and more | Gharbia | Female | February 2019 | MN295022 AZ9 | RHDV2 | + | 29 |
| 10 | 10 weeks | 2 months and more | Qena | Female | April 2019 | MN295023 AZ10 | Classic RHDV Genogroup5 | + | 214 |
| 11 | 18 days | <2 months | Dakahlia | Female | April 2019 | MN295024 AZ11 | RHDV2 | + | 211 |
| 12 | 9 weeks | 2 months and more | Assiut | Male | January 2019 | MN295025 AZ12 | Classic RHDV Genogroup5 | + | 211 |
| 13 | 10 weeks | 2 months and more | Sohag | Female | August 2018 | MN295026 AZ13 | Classic RHDV Genogroup5 | + | 213 |
| 14 | 12 weeks | 2 months and more | Behira | Female | February 2019 | MN295027 AZ14 | RHDV2 | + | 214 |
| 15 | 6 weeks | <2 months | Faium | Female | December 2018 | MN295028 AZ15 | Classic RHDV Genogroup5 | + | 210 |
| 16 | 8 weeks | 2 months and more | Menia | Female | August 2018 | MN295029 AZ16 | Classic RHDV Genogroup5 | + | 212 |
RHDV=Rabbit hemorrhagic disease virus, HA=Hemagglutination assay, PCR=Polymerase chain reaction
Most cases of RHDV were detected during the summer months. In 2018, infections were detected at the highest frequency (50%) during the month of August. By contrast, in 2019, 83% of RHDV infections were detected in July.
The phylogenetic tree is notable for clear clustering of the two distinct virus genotypes, and these groups can be distinguished from the two aforementioned vaccine strains, as shown in Figure-2.
Epidemiological findings
The sequence data revealed that there are presently two genotypes of RHDV circulating in distinct regions and governorates within Egypt. As shown in Figure-1, the geographical distribution of the circulating strains of the RHDV virus showed that genotypes associated with the classical RHDV strain (G3-G5) were concentrated mainly in upper Egypt, and those associated with RHDV2 circulated in lower Egypt.
The RHDV-positive samples were divided into two groups according to the age of the rabbit host (<2 months vs. ≥2 months of age). As shown in Table-4, classical RHDV genotype samples represented 75% of the older group, while RHDV2 samples represented 62.5% of the younger group.
Table-4.
The difference between age groups in relation to the detected RHDV strains.
| Age groups | Classic RHDV Genogroup (5) | RHDV2 (variant) | Total positive | % of classical | % of variant RHDV-2 |
|---|---|---|---|---|---|
| 2 months and more (8 weeks or more) | 6 | 2 | 8 | 6/8=75 | 2/8=25 |
| <2 months (<8 weeks) | 3 | 5 | 8 | 3/8=37.5 | 5/8=62.5 |
| Total | 9 | 7 | 16 | 9/16=56.3 | 7/16=43.7 |
RHDV=Rabbit hemorrhagic disease virus, HA=Hemagglutination assay, PCR=Polymerase chain reaction
Discussion
According to an 2017 report from the Food and Agriculture Organization of the United Nations, Egypt is one of the five major producers of rabbit meat, where Italy was the fifth largest producer of rabbit meat in the world, following China, the Democratic People’s Republic of Korea, Spain, and Egypt [32]. In the early 1990s, RHDV was classified as a member of the family Caliciviridae [33,34], genus Lagovirus [6]. The virus is typically highly contagious among rabbits of different species and ages [35,36], and infection with the virus can result in sudden death that is typically associated with liver necrosis and acute hepatitis [37,38] with generalized congestion and hemorrhage [39,40]. In this study, we identified 16 cases that were positive for RHDV using PCR, and these 16 cases were hemagglutinin-positive, with titers ranging from 29 to 216. These results are consistent with those reported previously [13] that indicated that RHDV2 efficiently agglutinates human type O RBCs and confirmed the use of HA as a routine diagnostic tool for the detection of RHDV2 in infected samples. Conventional RT-PCR was used to detect RNA of RHDV from liver and lung samples, and 16 out of 50 cases (32%) of suspected RHDV were revealed as positive by this method. The sampled cases were from 15 Egyptian governorates and were collected in 2018-2019. The classical RHDV strain was identified primarily in localities within upper Egypt, and RHDV2 was detected primarily within the governorates of the lower Egypt. Overall, the detection of the circulating strains of both RHDV and RHDV-2 is consistent with findings reported previously [12] regarding RHDV2 as the dominant circulating strain in both wild and domestic rabbits. Interestingly, the disease associated with the RHDV2 variant manifests in a similar fashion to that caused by classical RHDV, although the mortality rates may be more variable and tend to be lower [13].
An initial analysis of genetic diversity among these strains was initiated by comparing partial sequences from a few RDHVs isolated from European countries [41,42]. Our results are consistent with those reported in a previous study [43], in which rabbits <28 days old were found to be susceptible to RHDV disease. In this study, the samples were collected from rabbits at different ages and during different seasons. The group that tested positive was divided into those <2 months of age and those ≥2 months of age, as shown in Table-3. Classical RHDV was detected in three out of eight cases of the younger age group, while five cases tested positive for RHDV2. However, in the older age group, six of the eight rabbits (75%) tested positive for classical RHDV. An earlier publication, [44] discussed the fact that many clinical cases have been recorded from different governorates within Egypt, with clinical signs consistent with acute RHD, and these results suggested that the two strains might have comparatively equal virulence during natural RHDV outbreaks. Regarding the seasonality of this disease, most of the cases in our study were detected during the summer season, most notably in the month of July (Figure-3). By contrast, other reports [45] noted that RHDV was identified in rabbits during the spring of 1991 in Sharkia Province associated with 90% losses. RHDV was also reported during the winter and spring of 1993 in upper Egypt (Minya, Assiut, and Sohag Provinces), with mortality rates from 26.7% to 100% among 14-16 week-old rabbits. Amplification, sequencing, and phylogenies of the VP60 gene from all incidents, in addition to the immunological typing of the virus material from three cases, confirmed the presence of RHDV2 [43].
Figure-3.
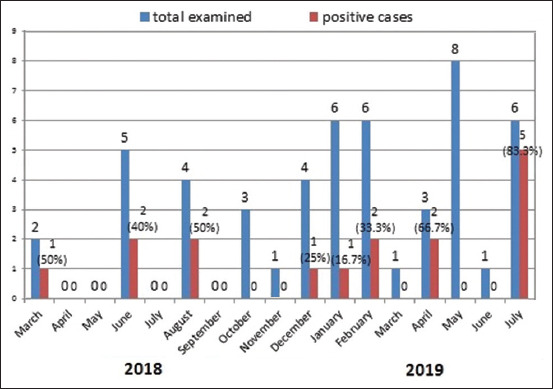
The distribution of positive cases in proportion to the total examined with relation to the month and year of detection (2018-2019) in Egypt.
In this study, we amplified a 538 bp fragment of the VP60 gene, and this has been reported to be highly conserved among RHDV variants. Our findings are consistent with those reported earlier [25] regarding the efficacy of VP60 as a target for PCR assays and as a general screening tool for rabbit and hare caliciviruses. The VP60 gene was used to develop universal Lagovirus PCR primers that span a highly conserved region within RHDV [26]. Results from another study [46] revealed that the viral VP60 gene was targeted to explore all rabbit lagoviruses circulating in Australia. Here, we used these primers exclusively for the primary screening of animals for suspected RHDV infections. Among our findings, the VP60 sequences from our study were most closely related to those from 2010 to 2012 RHDV2 isolates from France, Spain, and Portugal with 97%-98% nucleotide sequence identity [47]. Interestingly, RT-PCR primers targeting the VP60 gene sequence were also used for the detection of several distinct species of lagoviruses, including the European brown hare syndrome virus and rabbit calicivirus [26].
The study analyzed the phylogeny of 16 RHDV strains detected in 2018 and 2019 from 15 different provinces in Egypt, and amplification and sequencing of a fragment of the VP60 capsid gene from virus cDNA resulted in the identification of novel virus strains with nucleotide identities ranging from 78.4% to 100%. Among our main conclusions, we found that the two major RHDV strains, namely, RHDV and RHDV2, were circulating in different regions of Egypt. One of these strains was the classical G3-G5 RHDV, and the other strain was variant-type RHDV2 (GenBank accessions numbers MN295014-MN295029).
The rabbit industry is of importance in the production of both meat and fur, and as such, it needs to focus on ways to prevent viral, bacterial, and parasitic diseases [22,48,49]. Our results confirm infections with different strains of RHDV that is circulating concurrently within Egypt, and these viruses infect both male and female rabbits within various age groups. Our results also suggest that the current vaccine strain may not be sufficient in protecting against all strains currently in circulation. As such, research into future vaccination strategies is stressed here.
In the current study, RT-PCR amplification of a fragment of the VP60 gene confirmed the presence of RHDV RNA in liver and lung samples of 16 cases out of 50 total suspected cases (32%), suggesting a high incidence of this disease. These findings are similar to those reported in earlier Egyptian studies [50] that focused on the examination and detection of the RHDV VP60 gene from 20 liver homogenates from infected rabbits during an earlier virus outbreak in Egypt (2015 to 2016). In this earlier study, four isolates were among the classical RHDV strains, and only one isolate was identified as a variant. The cases were identified in various Egyptian governorates in 2018-2019. Our findings revealed a high distribution and significant circulation of both the classical and variant virus strains within Egyptian rabbit populations.
DNA sequence analysis resulted in the classification into two genotypes, specifically classic RHDV and variant RHDV2. To the best of our knowledge, these variant RHDV2 strains have not been detected previously in Egypt. As such, this presents a new challenge and threat to the rabbit industry in Egypt. The classical strain was circulating primarily in localities within upper Egypt. However, the new European RHDV2 variant was identified within the governorates of lower Egypt. This result is somewhat to be expected as the lower Egyptian governorates are much closer to some of the European countries such as France, where the variant strain was first recorded. Some authors claim that RHDV2 has become the dominant strain circulating in Europe, and it may have already replaced older RHDV strains and accounts for most of the reported cases in wild and domestic rabbits [12]. The disease associated with RHDV2 variant includes similar manifestations as those seen in response to an infection with one of the classical strains, although mortality rates are variable [13].
The initial characterization of RHDV genetic diversity was initiated by sequencing and comparing partial VP60 sequences with related isolates from European countries. One research group [16] reported that the classic virus strains typically cause disease in rabbits older than 2 months of age. The results of our study revealed that both genotypes can infect rabbits of different ages. Another previous report [44] noted that clinical cases of acute RHDV from different Egyptian governorates demonstrated similar virulence to cases of previously reported natural RHDV outbreaks. Most of our cases were detected in the summer season, most notably in the month of July, as shown in Figure-3. An earlier report [45] mentioned that RHD had been reported during the spring of 1991 in Sharkia Province, with 90% losses, and also during the winter and spring of 1993 in upper Egypt (Minia and Sohag governorates), with mortality rates of 26.7%-100% in 14-16-old rabbits. Subsequent outbreaks have occurred in other governorates, including Kaliobia [51] and Assuit [52].
Conclusion
In this study, we reported the detection of two distinct genotypes of RHDV among rabbits of different ages and both genders. We are quite concerned to confirm the emergence and near-dominance of the RHDV2 variant in rabbit populations throughout Egypt. These findings underscore the urgent need to launch a new vaccine that is capable of protection against both circulating genotypes.
Authors’ Contributions
AME designed this study and applied the molecular analysis. AGS performed molecular biology tests and HA test. Both authors collected samples, drafted, revised the manuscript, analyzed the data, and approved the final manuscript. Both authors read and approved the final manuscript.
Acknowledgments
The authors thank the rabbit farm owners’ for their collaboration during sampling. This work was funded by the Reference Laboratory for Veterinary Quality Control on Poultry Production, Animal Health Research Institute, Egypt. Great thanks for Dr. Neveen R. Bakry for her support in the epidemiological data analysis
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published map and institutional affiliation.
References
- 1.Ghanem I.A, Ismail A.N. Occurrence of rabbit hemorrhagic disease in Sharkiaa province. Zag. Vet. Med. J. 1992;20(4):491–502. [Google Scholar]
- 2.Alboghdady M.A, Alashry M.K. The demand for meat in Egypt:An almost ideal estimation. Afr. J. Agric. Resour. Econ. 2010;4(1):70–81. [Google Scholar]
- 3.XU W.Y. Viral hemorrhagic disease of rabbits in the people's republic of China:Epidemiology and virus characterization. Rev. Sci. Tech. 1991;10(2):393–408. [PubMed] [Google Scholar]
- 4.Le Gall-Recule G, Zwingelstein F, Laurent S, de Boisseson C, Portejoie Y, Rasschaert D. Phylogenetic analysis of rabbit haemorrhagic disease virus in France between 1993 and 2000, and the characterisation of RHDV antigenic variants. Arch. Virol. 2003;148(1):65–81. doi: 10.1007/s00705-002-0908-1. [DOI] [PubMed] [Google Scholar]
- 5.Alda F, Gaitero T, Suarez M, Merchan T, Rocha G, Doadrio I. Evolutionary history and molecular epidemiology of rabbit haemorrhagic disease virus in the Iberian Peninsula and Western Europe. BMC Evol. Biol. 2010;10(1):347. doi: 10.1186/1471-2148-10-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrantes J, van der Loo W, Le Pendu J, Esteves P.J. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV):A review. Vet. Res. 2012;43(1):12. doi: 10.1186/1297-9716-43-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Gall-Recule G, Zwilgenstaein F, Boucher S, Le Normand B, Plassiart G, Portejoie Y, Decors A, Bertagnoli S, Guerin J.L, Marchandeau S. Detection of a new variant of haemorrhagic disease virus in France. Vet. Rec. 2011;168(5):137–138. doi: 10.1136/vr.d697. [DOI] [PubMed] [Google Scholar]
- 8.Almeida T, Lopes A.M, Magalhaes M.J, Neves F, Pinheiro A, Goncalves D, Leitao M, Esteves P.J, Abrantes J. Tracking the evolution of the G1/RHDVb recombinant strains introduced from the Iberian Peninsula to the Azores Islands, Portugal. Infect. Genet. Evol. 2015;34(2015):307–313. doi: 10.1016/j.meegid.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Duarte M, Carvalho C, Bernardo S, Barros S.V, Benevides S, Flor L, Monteiro M, Marques I, Henriques M, Barros S.C, Fagulha T, Ramos F, Luis T, Fevereiro M. Rabbit haemorrhagic disease virus 2 (RHDV2) outbreak in Azores:Disclosure of common genetic markers and phylogenetic segregation within the European strains. Infect. Genet. Evol. 2015;35(2015):163–171. doi: 10.1016/j.meegid.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Hall R.N, Mahar J.E, Haboury S, Stevens V, Holmes E.C, Strive T. Emerging rabbit hemorrhagic disease virus 2 (RHDVb), Australia. Emerg. Infect. Dis. 2015;21(12):2276–2278. doi: 10.3201/eid2112.151210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Organization of Animal Health. Rabbit Haemorrhagic Disease, Canada-immediate Notification Report. Paris, France: World Organization of Animal Health; 2016. Available from: http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_referMapFullEventReport&reportid20799?. Retrieved on 03-06-2020. [Google Scholar]
- 12.Mahar J.E, Hall R.N, Peacock D, Kovaliski J, Piper M, Mourant R, Huang N.N, Campbell S, Gu X.N, Read A, Urakova N, Cox T, Holmes E.C, Strive T. Rabbit hemorrhagic disease virus 2 (RHDV2;GI.2) is replacing endemic strains of RHDV in the Australian landscape within 18 months of its arrival. J. Virol. 2018;92(2):e01374–17. doi: 10.1128/JVI.01374-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Gall-Recule G, Lavazza A, Marchandeau S, Bertagnoli S, Zwilgenstaein F, Cavadini P, Martinelli N, Lombardi G, Guerin J.L, Lemaitre E, Décors A, Boucher S, Le Normand B, Capucci L. Emergence of a new lago virus related to rabbit haemorrhagic disease virus. Vet. Res. 2013;44(1):81. doi: 10.1186/1297-9716-44-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asgari S, Hardy J.R, Sinclair R.G, Cooke B.D. Field evidence for mechanical transmission of rabbit haemorrhagic disease virus (RHDV) by flies (Diptera Calliphoridae) among wild rabbits in Australia. Virus Res. 1998;54(2):123–132. doi: 10.1016/s0168-1702(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 15.Ohlinger V.F, Haas B, Thiel H.J. Rabbit haemorrhagc disease (RHD):Characterization of the causative calicivirus. Vet. Res. 1993;24(2):103–116. [PubMed] [Google Scholar]
- 16.OIE. Terrestrial manual. In:Rabbit Haemorrhagic Disease. 2010 Ch. 2.6.2. OIE Paris. France. [Google Scholar]
- 17.Dalton K.P, Nicieza I, Balseiro A, Muguerza M.A, Rosell J.M, Casais R, Alvarez A.L, Parra F. Variant rabbit hemorrhagic disease virus in young rabbits, Spain. Emerg. Infect. Dis. 2012;18(12):2009–2012. doi: 10.3201/eid1812.120341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velarde R, Cavadini P, Neimanis A, Cabezon O, Chiari M, Gaffuri A, Lavin S, Grilli G, Widen D.G, Lavazza A, Capucci L. Spillover events of infection of brown hares (Lepus europaeus) with rabbit haemorrhagic disease Type 2 virus (RHDV2) caused sporadic cases of an European brown hare syndrome-like disease in Italy and Spain. Transbound. Emerg. Dis. 2016;64(6):1750–1761. doi: 10.1111/tbed.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall R.N, Peacock D.E, Kovaliski J, Mahar J.E, Mourant R, Piper M, Strive T. Detection of RHDV2 in European brown hares (Lepus europaeus) in Australia. Vet. Rec. 2017;180(5):121. doi: 10.1136/vr.104034. [DOI] [PubMed] [Google Scholar]
- 20.Lopes A.M, Blanco-Aguiar J, Martin-Alonso A, Leitao M, Foronda P, Mendes M, Goncalves D, Abrantes J, Esteves P.J. Full genome sequences are key to disclose RHDV2 emergence in the Macaronesian Islands. Virus Genes. 2018;54(1):1–4. doi: 10.1007/s11262-017-1523-2. [DOI] [PubMed] [Google Scholar]
- 21.Silvério D, Lopes A.M, Melo-Ferreira J, Magalhães M.J, Monterroso P, Serronha A, Maio E, Alves P.C, Esteves P.J, Abrantes J. Insights into the evolution of the new variant rabbit haemorrhagic disease virus (GI.2) and the identification of novel recombinant strains. Transbound. Emerg. Dis. 2018;65(4):983–992. doi: 10.1111/tbed.12830. [DOI] [PubMed] [Google Scholar]
- 22.Dalton K.P, Nicieza I, de Llano D, Gullón J, Inza M, Petralanda M, Arroita Z, Parra F. Vaccine breaks:Outbreaks of myxomatosis on Spanish commercial rabbit farms. Vet. Microbiol. 2015;178(3-4):208–216. doi: 10.1016/j.vetmic.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Meyers G, Wirblich C, Thiel H.J, Thumfart J.O. Rabbit hemorrhagic disease virus:Genome organization and polyprotein processing of a calicivirus studied after transient expression of cDNA constructs. Virology. 2000;276(2):349–363. doi: 10.1006/viro.2000.0545. [DOI] [PubMed] [Google Scholar]
- 24.Kwit E, Rzeżutka A. Molecular methods in detection and epidemiologic studies of rabbit and hare Viruses:A review. J. Vet. Diagn. Invest. 2019;31(4):497–508. doi: 10.1177/1040638719852374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Gall-Reculé G, Lemaitre E, Bertagnoli S, Hubert C, Top S, Decors A, Marchandeau S, Guitton J.S. Large-scale lago virus disease outbreaks in European brown hares (Lepus europaeus) in France caused by RHDV2 strains spatially shared with rabbits (Oryctolagus cuniculus) Vet. Res. 2017;48(1):70. doi: 10.1186/s13567-017-0473-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strive T, Wrighta J.D, Robinson A.J. Identification and partial characterization of a new lago virus in Australian wild rabbits. Virology. 2009;384(1):97–105. doi: 10.1016/j.virol.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Chasey D, Lucas M.H, Westcott D.G, Sharp G, Kitching A, Hughes S.K. Development of diagnostic approach to the identification of rabbit haemorrhagic disease. Vet. Rec. 1995;137(7):158–160. doi: 10.1136/vr.137.7.158. [DOI] [PubMed] [Google Scholar]
- 28.Fahmy H.A, Arafa A, Mahmoud A.H. Molecular diagnosis of rabbit hemorrhagic disease virus (RHDV) Egypt. J. Comp. Path. Clin. Path. 2010;23(1):85–101. [Google Scholar]
- 29.Altschul S.F, Gish W, Miller W, Myers E.W, Lipmanl D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J.D, Higgins D.G, Gibson T.J. CLUSTAL W:Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6:Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Food and Agriculture Organization of the United Nations. FAO Database. 2019. Available from: http://www.faostat.fao.org. Retrieved on 05-03-2019.
- 33.Meyers G, Wirblich C, Thiel H.J. Rabbit hemorrhagic disease virus-molecular cloning and nucleotide sequencing of a calicivirus genome. Virology. 1991;184(2):664–676. doi: 10.1016/0042-6822(91)90436-f. [DOI] [PubMed] [Google Scholar]
- 34.Moussa A, Chasey D, Lavazza A, Capucci L, Smid B, Meyers G, Rossi C, Thiel H.J, Vlasak R, Ronsholt L, Nowotny N, McCullough K, Gavier-Widenm D. Haemorrhagic disease of lagomorphs:Evidence for a calicivirus. Vet. Microbiol. 1992;33(1-4):375–381. doi: 10.1016/0378-1135(92)90065-2. [DOI] [PubMed] [Google Scholar]
- 35.Miao O, Qi R, Veldkamp L, Ijzer J, Kik M.L, Zhu J.M, Tang A, Dong D, Shi Y, Van Oers M.M, Liu G, Pijlman G.P. Immunogenicity in rabbits of virus-like particles from a contemporary rabbit haemorrhagic disease virus Type 2 (GI.2/RHDV2/b) isolated in the Netherlands. Viruses. 2019;11(6):553. doi: 10.3390/v11060553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urakova N, Hall R, Strive T, Frese M. Restricted host specificity of rabbit hemorrhagic disease virus supported by challenge experiments in immune-compromised mice (Mus musculus) J. Wildl. Dis. 2019;55(1):218–222. doi: 10.7589/2018-03-067. [DOI] [PubMed] [Google Scholar]
- 37.Alonso C, Oviedo J.M, Martin-Alonso J.M, Diaz E, Boga J.A, Parra F. Programmed cell death in the pathogenesis of rabbit hemorrhagic disease. Arch. Virol. 1998;143(2):321–332. doi: 10.1007/s007050050289. [DOI] [PubMed] [Google Scholar]
- 38.Park J.H, Lee Y.S, Itakura C. Pathogenesis of acute necrotic hepatitis in rabbit hemorrhagic disease. Lab. Anim. Sci. 1995;45(4):445–449. [PubMed] [Google Scholar]
- 39.Ueda K, Park J.H, Ochiai K, Itakura C. Disseminated intravascular coagulation (DIC) in rabbit haemorrhagic disease. Jpn. J. Vet. Res. 1992;40(4):133–141. [PubMed] [Google Scholar]
- 40.Marques R.M, Costa E.S.A, Aguas A.P, Teixeira L, Ferreira P.G. Early acute depletion of lymphocytes in calicivirus-infected adult rabbits. Vet. Res. Commun. 2010;34(8):659–668. doi: 10.1007/s11259-010-9437-7. [DOI] [PubMed] [Google Scholar]
- 41.Milton I.D, Vlasak R, Nowotny N, Rodak L, Carter M.J. Genomic 3'terminal sequence comparison of three isolates of rabbit haemorrhagic disease virus. FEMS Microbiol. Lett. 1992;72(1):37–42. doi: 10.1016/0378-1097(92)90486-8. [DOI] [PubMed] [Google Scholar]
- 42.Parra F, Boga J.A, Marin M.S, Casais R. The amino terminal sequence of VP60 from rabbit hemorrhagic disease virus supports its putative subgenomic origin. Virus Res. 1993;27(3):219–228. doi: 10.1016/0168-1702(93)90034-k. [DOI] [PubMed] [Google Scholar]
- 43.Neimanis A.S, Ahola H, Zohari S, Pettersson U.K, Bröjer C, Capucci L, Gavier-Widén D. Arrival of rabbit haemorrhagic disease virus 2 to Northern Europe:Emergence and outbreaks in wild and domestic rabbits (Oryctolagus cuniculus) in Sweden. Transbound. Emerg. Dis. 2018;65(1):213–220. doi: 10.1111/tbed.12650. [DOI] [PubMed] [Google Scholar]
- 44.Awad N.F, Kotb G.K. Genetic characterization of rabbit hemorrhagic disease virus from naturally infected rabbits in Sharkia Governorate. Egypt. J. Virol. Sci. 2018;3(1):10–19. [Google Scholar]
- 45.El-Zanaty K. Some investigation on rabbit viral haemorrhagic disease in Upper Egypt. Assiut. Vet. Med. J. 1994;30(60):293–305. [Google Scholar]
- 46.Hall R.N, Mahar J.E, Read A.J, Mourant R, Piper M, Huang N, Strive T. A strain-specific multiplex RT-PCR for Australian rabbit haemorrhagic disease viruses uncovers a new recombinant virus variant in rabbits and hares. Transbound. Emerg. Dis. 2018;65(2):e444–e456. doi: 10.1111/tbed.12779. [DOI] [PubMed] [Google Scholar]
- 47.Nowotny N, Bascunana C.R, Ballagi-Pordany A, Gavier-Widen D, Uhlen M, Belak S. Phylogenetic analysis of rabbit haemorrhagic disease and European brown hare syndrome viruses by comparison of sequences from the capsid protein gene. Arch. Virol. 1997;142(4):657–673. doi: 10.1007/s007050050109. [DOI] [PubMed] [Google Scholar]
- 48.Carvalho C.L, Silva S, Gouveia P, Costa M, Duarte E.L, Henriques A.M, Barros S.S, Luís T, Ramos F, Fagulha T, Fevereiro M, Duarte M.D. Emergence of rabbit haemorrhagic disease virus 2 in the archipelago of Madeira, Portugal (2016-2017) Virus Genes. 2017;53(6):922–926. doi: 10.1007/s11262-017-1483-6. [DOI] [PubMed] [Google Scholar]
- 49.Isomursu M, Neimanis A, Karkamo V, Nylund M, Holopainen R, Nokireki T, Gadd T. An outbreak of rabbit hemorrhagic disease in Finland. J. Wildl Dis. 2018;54(4):838–842. doi: 10.7589/2017-11-286. [DOI] [PubMed] [Google Scholar]
- 50.Magouzi A.F, Elsayed E.A, Metwally A.Y. Detection and characterization of rabbit hemorrhagic disease virus strains circulating in Egypt. Bulg. J Vet Med. 2017;22(4):409–418. [Google Scholar]
- 51.Sharawi S.S.A. Studies on the Virus Causing Haemorrhagic Septicemia in Rabbits Master Thesis. Zagazig, Egypt: Zagazig University; 1992. [Google Scholar]
- 52.Salem B, El Ballal S.S. The occurrence of rabbit viral haemorrhagic disease (RVHD) in Egypt. Assiut. Vet. Med. J. 1992;27(53):295–304. [Google Scholar]


