Abstract
Background
Non‐small cell lung cancer (NSCLC) is one of the leading causes of cancer‐related death globally. This study aimed to disclose the role of circular RNA circ_0072088 in NSCLC and illustrate its potential mechanism.
Methods
Quantitative real‐time polymerase chain reaction (qRT‐PCR) was applied to detect the expression of circ_0072088, zinc finger RNA binding protein (ZFR), microRNA‐377‐5p (miR‐377‐5p) and NOVA alternative splicing regulator 2 (NOVA2). The viability, colony formation, cell cycle, migration and invasion of NSCLC cells were measured by cell counting kit‐8 (CCK8) assay, colony formation assay, flow cytometry, wound healing assay and transwell invasion assay. Western blot assay was employed to examine the protein levels of proliferating cell nuclear antigen (PCNA), Cyclin D1, matrix metallopeptidase 2 (MMP2), MMP9 and NOVA2. The downstream targets of circ_0072088 and miR‐377‐5p were searched through using circular RNA Interactome and TargetScan databases, and the interaction between miR‐377‐5p and circ_0072088 or NOVA2 was confirmed by dual‐luciferase reporter assay and RNA immunoprecipitation (RIP) assay. in vivo tumor growth assay was used to evaluate the functions of circ_0072088 in the progression of NSCLC in vivo.
Results
GSE101586 dataset and the analysis of tissue specimens showed that circ_0072088 was aberrantly upregulated in tumor tissues of lung cancer and NSCLC. Circ_0072088 interference caused marked suppression on the proliferation and motility of NSCLC cells. Circ_0072088 could negatively regulate miR‐377‐5p through direct combination. Circ_0072088 contributed to the progression of NSCLC through sponging miR‐377‐5p. MiR‐377‐5p could directly interact with NOVA2, and the overexpression of NOVA2 overturned miR‐377‐5p‐mediated influence on NSCLC cells. Circ_0072088 facilitated the progression of NSCLC in vivo.
Conclusions
Circ_0072088 facilitated the proliferation and metastasis of NSCLC cells through upregulating NOVA2 via functioning as a competitive endogenous RNA (ceRNA) for miR‐377‐5p.
Keywords: Circ_0072088, miR‐377‐5p, NOVA2, NSCLC, proliferation
Introduction
The conventional therapeutic methods for non‐small cell lung cancer (NSCLC) patients are surgery and chemotherapy. However, due to the chemoresistance and metastasis, the prognosis of NSCLC patients is still dismal. 1 , 2 Finding effective biomarkers and uncovering the molecular mechanism behind the progression of NSCLC are crucial for improving the prognosis of NSCLC patients.
Non‐coding RNAs (ncRNAs) are unable to code proteins, and they are involved in many cellular processes, including proliferation, motility and apoptosis. 3 , 4 Circular RNAs (circRNAs) are ncRNAs that feature a closely circular structure. 5 Many circRNAs have been found to be novel markers of multiple cancers, including NSCLC. 6 , 7 , 8 Zhang et al. claimed that circ_0072088 (circ_103809/circZFR) facilitated the development of NSCLC through miR‐101‐3p/CUL4B axis. 9 Nevertheless, the working mechanism of circ_0072088 in NSCLC remains to be illustrated.
MicroRNAs (miRNAs) are small ncRNAs 18–24 nucleotides in length. 10 CircRNAs have been found to function as miRNAs sponges, a mechanism referred to as competing endogenous RNA (ceRNA). 11 Accumulating articles have alluded to the crucial roles of miRNA in cancers. For example, Wang et al. found that miR‐384 impeded the development of papillary thyroid cancer via PRKACB. 12 Wu et al. proved that miR‐9 accelerated the proliferation of bladder cancer cells through activating Notch signaling. 13 Wu et al. claimed that miR‐377‐5p restrained the progression of lung cancer through AKT1 pathway. 14 We focused on the expression pattern and the signal network of miR‐377‐5p in NSCLC.
NOVA alternative splicing regulator 2 (NOVA2) is a member of the NOVA family, which plays an important role in the survival and development of motor neurons. 15 , 16 NOVA1 has been previously reported to be involved in the tumorigenesis of multiple cancers, including lung cancer. 17 , 18 , 19 However, the role of NOVA2 in cancers is hardly known. Xiao et al. claimed that miR‐7‐5p inhibited the malignant behaviors of NSCLC cells via NOVA2. 20 Herein, NOVA2 was found to be a novel target of miR‐377‐3p, and the working mechanism of NOVA2 in NSCLC was further explored.
In this study, circ_0072088 was found to be dysregulated in NSCLC. The functions of circ_0072088 on the proliferation and motility of NSCLC cells are disclosed, and the downstream signal axis was explored.
Methods
Patients and cell lines
A total of 45 pairs of NSCLC tumor specimens and adjacent normal specimens were collected from NSCLC patients who were diagnosed at the Affiliated Hospital of Shandong University of Traditional Chinese Medicine. Written informed consent was provided by all participants. The present study was approved by the Ethics Committee of the Affiliated Hospital of Shandong University of Traditional Chinese Medicine. Normal human lung epithelial cell line BEAS‐2B and NSCLC cell lines (NCI‐H1299 and A‐549) were purchased from BeNa Culture Collection (Beijing, China). All cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Biowest, Loire Valley, France), 10% penicillin (100 U/mL) and 10% streptomycin (100 μg/mL) at 37°C with 5% CO2.
Quantitative real‐time polymerase chain reaction (qRT‐PCR)
After extracting RNA with Trizol reagent (Life Technologies, Carlsbad, CA, USA), Transcriptor First Strand cDNA Synthesis Kit (Takara, Dalian, China) and All‐in‐One miRNA First stand cDNA Synthesis Kit (GeneCopoeia, Rockville, MD, USA) were used to synthesize complementary DNA (cDNA) from messenger RNA (mRNA) and miRNA. The specific primers used in this study were shown as below: circ_0072088 (Forward, 5′‐ATGGTCTGCAGTCCTGTGTG‐3′; Reverse, 5′‐TGGTGGCATGTTTTGTCATT‐3′), zinc finger RNA binding protein (ZFR; Forward, 5′‐TCACACAGTTACTGCTGCCT‐3′; Reverse, 5′‐GGGGGTGGTGGTGGTGGTGC‐3′), miR‐377‐5p (Forward, 5′‐AUCACACAAAGGCAACUUUUGU‐3′; Reverse, 5′‐ACAAAAGTTGCCTTTGTGTGTU‐3′), NOVA2 (Forward, 5′‐GGGTTCCCATAGACCTGGAC‐3′; Reverse, 5′‐CGCTCAGTAGTACCTGGGTAA‐3′), U6 (a housekeeping gene for miRNA; Forward, 5′‐CTCGCTTCGGCAGCACA‐3′; Reverse, 5′‐AACGCTTCACGAATTTGCGT‐3′), glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH; a housekeeping gene for mRNA; Forward, 5′‐TGTTCGTCATGGGTGTGAAC‐3′; Reverse, 5′‐ATGGCATGGACTGTGGTCAT‐3′). The quantification was performed through using the 2−ΔΔCt method.
RNase R treatment
RNA samples were exposed to 3 U/mg RNase R (Epicenter Technologies, Madison, WI, USA) for 30 minutes. The abundance of circ_0072088 and its matching mRNA (ZFR) was detected using qRT‐PCR.
Cell transfection
Three small interfering RNAs targeting circ_0072088 (si‐hsa_circ_0072088#1, si‐hsa_circ_0072088#2 and si‐hsa_circ_0072088#3), siRNA negative control (si‐NC), hsa_circ_0072088 overexpression plasmid (hsa_circ_0072088), circRNA‐NC (circ‐NC), short hairpin RNA targeting hsa_circ_0072088 (sh‐hsa_circ_0072088), shRNA‐NC (sh‐NC), miRNA‐377‐5p mimics (miR‐377‐5p), miRNA‐NC (NC), miR‐377‐5p inhibitor (anti‐miR‐377‐5p), anti‐NC, NOVA2 overexpression plasmid (NOVA2) and empty vector (vector) were purchased from Genepharma (Shanghai, China). After being grown to 70%–80% confluence, transfection was carried out using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Cell counting kit‐8 (CCK8) assay
NSCLC cells were seeded into 96‐well plates and transfected with relevant plasmid or RNA. After transfection for indicated time point, 10 μL CCK‐8 (Sigma, St. Louis, MO, USA) was added into the wells of 96‐well plates for four hours. The optical density value (450 nm) in different groups was examined by a microplate reader (Nikon, Tokyo, Japan).
Colony formation assay
NSCLC cells were seeded into six‐well plates at the concentration of 150 cells per well. Two weeks later, the colonies were washed using phosphate‐buffered saline (PBS) and stained with hematoxylin. The colonies with less than 50 cells were excluded from this study.
Flow cytometry
NSCLC cells were collected and fixed using 70% cold ethanol solution at −20°C overnight. Then, 10 μM RNase was utilized to remove RNA. DNA was marked with 20 mg/mL propidine iodide (PI; Solarbio, Beijing, China) for 20 minutes at 37°C. The percentage of NSCLC cells at different phases was assessed using a flow cytometer (BD Biosciences, San Jose, CA, USA).
Wound healing assay
We then seeded 5 × 105 NSCLC cells into six‐well plates. When the confluence reached about 90%, a scratch was made using a 200 μL pipette tip. The detached NSCLC cells were removed using PBS. The scratch was photographed at 0 and 24 hours. Each experiment was repeated three times.
Transwell invasion assay
After transfection for 24 hours, NSCLC cells were suspended in 100 μL serum‐free medium and seeded into the upper chambers precoated with 50 μL Matrigel (Sigma), and 600 μL medium with 10% FBS was added to the lower chambers. After 24 hours incubation, the invasive NSCLC cells were fixed using 4% paraformaldehyde and stained using 0.05% gentian violet. Invasive cell number was obtained through counting invasive NSCLC cells in five random fields.
Western blot assay
After relevant treatment, NSCLC cell lysate was prepared. The cell lysate was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) gel and blotted to the polyvinylidene fluoride (PVDF) membrane. After blocking, the membrane was then incubated with antiproliferating cell nuclear antigen (anti‐PCNA; ab92552; Abcam, Cambridge, MA, USA), anti‐Cyclin D1 (ab16663; Abcam), anti‐matrix metallopeptidase 2 (MMP2; ab215986; Abcam), anti‐MMP9 (ab219372; Abcam), anti‐NOVA2 (ab51004; Abcam) or anti‐GAPDH (ab181602; Abcam) followed by incubation with the secondary antibody (ab205718; Abcam). The protein signal was detected using the enhanced chemiluminescent (ECL) system (Beyotime, Shanghai, China).
Dual‐luciferase reporter assay
Circ_0072088‐miRNAs interactions and miR‐377‐5p‐mRNAs interactions were explored by circular RNA Interactome and TargetScan websites, respectively. The binding sites with miR‐377‐5p in the sequence of circ_0072088 or NOVA2 were cloned into pGL3 luciferase reporter vector (Promega, Madison, WI, USA), termed as hsa_circ_0072088‐wt or NOVA2‐wt. Meanwhile, these sites were mutated and inserted to the same vector, termed as hsa_circ_0072088‐mut or NOVA2‐mut. NSCLC cells were cotransfected with the recombinant luciferase reporter vector and miR‐377‐5p or NC. The luciferase activity was measured using the dual‐luciferase reporter assay system (Promega).
RNA immunoprecipitation (RIP) assay
Magna RI RNA‐Binding Protein Immunoprecipitation kit (Millipore, Billerica, MA, USA) was applied in this study. Sepharose beads (Bio‐Rad, Hercules, CA, USA) were incubated with Argonaute‐2 antibody (Anti‐Ago2) or Immunoglobulin G antibody (Anti‐IgG). NSCLC cells were disrupted using RIP buffer (Millipore), and then incubated with precoated sepharose beads. The expression of relevant RNAs was measured by qRT‐PCR.
In vivo tumor growth assay
BALB/c mice (five weeks; Chinese Academy of Medical Sciences, Beijing, China) were used to perform the in vivo tumor growth assay to explore the influence of circ_0072088, miR‐377‐5p and NOVA2 on the growth of NSCLC tumors in vivo. Animal manipulates were approved by the Animal Research Committee of Affiliated Hospital of Shandong University of Traditional Chinese Medicine. A‐549 cells stably expressing sh‐NC or sh‐hsa_circ_0072088 were harvested and adjusted to the concentration of 2 × 106 cells/100 μL PBS. Then, 100 μL cell suspension was subcutaneously injected into the left flank of nude mice. The tumor volume was recorded every week using the formula of length × width2 × 0.5. The mice were sacrificed after five weeks by injection, and the tumors were dissected and weighed. The abundance of hsa_circ_0072088, miR‐377‐5p, PCNA and NOVA2 in tumor specimens was detected by qRT‐PCR or western blot assay.
Statistical analysis
Data in this study are displayed as mean ± standard deviation (SD). Student's t‐test was utilized to assess the differences between two groups, and the differences among three groups were assessed by one‐way analysis of variance (ANOVA) followed by Tukey's test. The linear correlation was evaluated using Spearman's correlation coefficient. A P‐value less than 0.05 was identified as statistically significant.
Results
Circ_0072088 identified as an NSCLC‐associated circRNA
The expression profile of circRNAs in lung cancer tissues (n = 5) and adjacent normal tissues (n = 5) was analyzed according to GEO database (GSE101586). The top 10 up‐ and downregulated circRNAs in lung cancer tissues compared with that in normal tissues are shown in Fig 1a. Hsa_circ_0072088 (hsa_circ_103809/circZFR) was selected for further study. In GSE101586 dataset, circ_0072088 was highly expressed in lung cancer tissues than that in normal tissues (Fig 1b), implying that circ_0072088 might serve as a pivotal regulator in NSCLC progression. To further verify the expression pattern of circ_0072088 in NSCLC, we detected the level of circ_0072088 in NSCLC tissues (n = 45) and matching nontumor tissues (n = 45). As shown in Fig 1c, there was a significant upregulation in the expression of circ_0072088 in NSCLC tissues in comparison with that in adjacent normal tissues. In addition, circ_0072088 was also found to be markedly upregulated in NSCLC cells compared with that in BEAS‐2B cells (Fig 1d). RNase R was used to confirm the circular structure of circ_0072088, and its matching linear mRNA (ZFR mRNA) served as a control. As shown in Fig 1e, circ_0072088 was resistant to RNase R, while the level of its matching linear mRNA was dramatically decreased in RNase R treatment group, suggested that circ_0072088 possessed the loop structure. These results suggested that circ_0072088 might participate in the progression of NSCLC.
Figure 1.

Circ_0072088 is identified as a NSCLC‐associated circRNA. (a) The differentially expressed circRNAs in lung cancer tissues and normal tissues, including 10 highly expressed and 10 low expressed circRNAs, are shown as a heat map. (b) The abundance of circ_0072088 in normal tissues and lung cancer tissues of GSE101586 dataset is shown. (c) qRT‐PCR was used to detect the expression of circ_0072088 in adjacent normal tissues (n = 45) and NSCLC tissues (n = 45). (d) The expression of circ_0072088 was measured in normal human lung epithelial cells BEAS‐2B and NSCLC cell lines (NCI‐H1299 and A‐549) by qRT‐PCR. (e) The levels of circ_0072088 and its matching linear mRNA were examined in NSCLC cells treated with RNase R by qRT‐PCR NCI‐H1299 ( ) RNase R−, and (
) RNase R−, and ( ) RNase R+; and A‐549 (
) RNase R+; and A‐549 ( ) RNase R−, and (
) RNase R−, and ( ) RNase R+. *P < 0.05.
) RNase R+. *P < 0.05.
Circ_0072088 functions as an oncogene in NSCLC
To illustrate the functions of circ_0072088 in NSCLC cells, three circ_0072088 specific siRNAs were used. As indicated in Fig 2a, the expression of circ_0072088 was markedly reduced in NSCLC cells with the transfection of circ_0072088 siRNAs, especially in si‐hsa_circ_0072088#1 group. CCK8 assay showed that the viability of NSCLC cells was significantly suppressed in si‐hsa_circ_0072088#1 group in comparison with that in si‐NC group (Fig 2b). The colony formation ability of NSCLC cells was also inhibited with the interference of circ_0072088 (Fig 2c). The percentage of NSCLC cells in G1 phase was increased in si‐hsa_circ_0072088#1 group, suggested that the cell cycle was arrested in G1/S transition in si‐hsa_circ_0072088#1 group (Fig 2d). The migration rate of NSCLC cells was markedly reduced with the silencing of hsa_circ_0072088 (Fig 2e). The results of the transwell invasion assay showed that the invasive cell number was dramatically downregulated with the transfection of si‐hsa_circ_0072088#1 (Fig 2f), demonstrated that circ_0072088 depletion inhibited the invasion of NSCLC cells. The proliferation‐related proteins (PCNA and Cyclin D1) and metastasis‐related proteins (MMP2 and MMP9) were both downregulated in si‐hsa_circ_0072088#1 group compared with that in si‐NC group (Fig 2g), suggesting the oncogenic role of circ_0072088 in the proliferation and metastasis of NSCLC cells. Taken together, hsa_circ_0072088 promoted the proliferation and motility of NSCLC cells.
Figure 2.

Circ_0072088 functions as an oncogene in NSCLC. (a) The abundance of circ_0072088 was detected in NCI‐H1299 and A‐549 cells transfected with si‐NC, si‐hsa‐circ_0072088#1, si‐hsa‐circ_0072088#2 or si‐hsa‐circ_0072088#3 by qRT‐PCR. (B‐G) NSCLC cells were transfected with si‐NC or si‐circ_0072088#1. (b) The viability of NSCLC cells was detected by CCK8 assay NCI‐H1299 ( ) si‐NC, and (
) si‐NC, and ( ) si‐hsa_circ_0072088#1; A‐549 (
) si‐hsa_circ_0072088#1; A‐549 ( ) si‐NC, and (
) si‐NC, and ( ) si‐hsa_circ_0072088#1. (c) The proliferation of NSCLC cells was examined by colony formation assay (
) si‐hsa_circ_0072088#1. (c) The proliferation of NSCLC cells was examined by colony formation assay ( ) si‐NC, and (
) si‐NC, and ( ) si‐hsa_circ_0072088#1. (d) The cycle of NSCLC cells was evaluated by flow cytometry (
) si‐hsa_circ_0072088#1. (d) The cycle of NSCLC cells was evaluated by flow cytometry ( ) S, (
) S, ( ) G, and (
) G, and ( ) G2. (e) The migratory rate of NSCLC cells was assessed by wound healing assay. (f) Transwell invasion assay was applied to detect the invasion of NSCLC cells (
) G2. (e) The migratory rate of NSCLC cells was assessed by wound healing assay. (f) Transwell invasion assay was applied to detect the invasion of NSCLC cells ( ) si‐NC, and (
) si‐NC, and ( ) si‐hsa_circ_0072088#1. (g) Western blot assay was employed to detect the protein expression of proliferation‐related proteins and metastasis‐related proteins in NSCLC cells NCI‐H1299 (
) si‐hsa_circ_0072088#1. (g) Western blot assay was employed to detect the protein expression of proliferation‐related proteins and metastasis‐related proteins in NSCLC cells NCI‐H1299 ( ) si‐NC, and (
) si‐NC, and ( ) si‐hsa_circ_0072088#1; A‐549 (
) si‐hsa_circ_0072088#1; A‐549 ( ) si‐NC, and (
) si‐NC, and ( ) si‐hsa_circ_0072088#1. *P < 0.05.
) si‐hsa_circ_0072088#1. *P < 0.05.
MiR‐377‐5p is a downstream target of circ_0072088 in NSCLC cells
The target relationship between circ_0072088 and miR‐377‐5p was predicted by circular RNA Interactome website, and the binding sites are shown in Fig 3a. The results of dual‐luciferase reporter assay suggested that the transfection of miR‐377‐5p markedly downregulated the luciferase activity in hsa_circ_0072088‐wt group, while the luciferase activity remained unaffected in hsa_circ_0072088‐mut group (Fig 3b). RIP assay showed that hsa_circ_0072088 and miR‐377‐5p were all pulled‐down when using Ago2 antibody compared with that in anti‐IgG group (Fig 3c). To address the regulatory relationship between hsa_circ_0072088 and miR‐377‐5p in NSCLC cells, we overexpressed and silenced hsa_circ_0072088 using hsa_circ_0072088 and si‐hsa_circ_0072088#1, respectively. As depicted in Fig 3d, the ectopic expression of hsa_circ_0072088 downregulated the expression of miR‐377‐5p, while the interference of circ_0072088 increased the level of miR‐377‐5p in NSCLC cells. The expression pattern of miR‐377‐5p was explored in 45 pairs of NSCLC tumor specimens and normal specimens. As indicated in Fig 3e, a marked downregulation of miR‐377‐5p was observed in NSCLC tumor tissues when compared with that in normal tissues. The level of miR‐377‐5p was negatively correlated with the expression of circ_0072088 in NSCLC tumor tissues (Fig 3f). As mentioned in Fig 3g, the level of miR‐377‐5p was also found to be downregulated in NSCLC cells than that in BEAS‐2B cells. These findings showed that miR‐377‐5p was a direct target of circ_0072088 in NSCLC cells.
Figure 3.

MiR‐377‐5p is a downstream target of circ_0072088 in NSCLC cells. (a) The target sequence between circ_0072088 and miR‐377‐5p was predicted by circular RNA Interactome website. (b) Dual‐luciferase reporter assay was employed to confirm the target relationship between circ_0072088 and miR‐377‐5p in NCI‐H1299 and A‐549 cells NCI‐H1299 ( ) NC, and (
) NC, and ( ) miR‐377‐5p; A‐549 (
) miR‐377‐5p; A‐549 ( ) NC, and (
) NC, and ( ) miR‐377‐5p. (c) RIP assay was performed to verify the combination between circ_0072088 and miR‐377‐5p in these two NSCLC cell lines NCI‐H1299 (
) miR‐377‐5p. (c) RIP assay was performed to verify the combination between circ_0072088 and miR‐377‐5p in these two NSCLC cell lines NCI‐H1299 ( ) Anti‐IgG, and (
) Anti‐IgG, and ( ) Anti‐Ago2; A‐549 (
) Anti‐Ago2; A‐549 ( ) Anti‐IgG, and (
) Anti‐IgG, and ( ) Anti‐Ago2. (d) The regulatory relationship between circ_0072088 and miR‐377‐5p was explored in NSCLC cells transfected with circ‐NC, hsa_circ_0072088, si‐NC or si_hsa_circ_0072088#1 by qRT‐PCR. (e) The enrichment of miR‐377‐5p was detected in 45 pairs of normal tissues and NSCLC tumor tissues by qRT‐PCR. (f) The correlation between the levels of circ_0072088 and miR‐377‐5p in NSCLC tissues (n = 45) was evaluated by Spearman's correlation coefficient. (g) qRT‐PCR was employed to examine the enrichment of miR‐377‐5p in BEAS‐2B and NSCLC cells. *P < 0.05.
) Anti‐Ago2. (d) The regulatory relationship between circ_0072088 and miR‐377‐5p was explored in NSCLC cells transfected with circ‐NC, hsa_circ_0072088, si‐NC or si_hsa_circ_0072088#1 by qRT‐PCR. (e) The enrichment of miR‐377‐5p was detected in 45 pairs of normal tissues and NSCLC tumor tissues by qRT‐PCR. (f) The correlation between the levels of circ_0072088 and miR‐377‐5p in NSCLC tissues (n = 45) was evaluated by Spearman's correlation coefficient. (g) qRT‐PCR was employed to examine the enrichment of miR‐377‐5p in BEAS‐2B and NSCLC cells. *P < 0.05.
Circ_0072088 acts as an oncogenic molecule through sponging miR‐377‐5p in NSCLC cells
Si‐hsa_circ_0072088#1 and anti‐miR‐377‐5p were cotransfected into NSCLC cells to explore whether circ_0072088 functioned through sponging miR‐377‐5p. The transfection of anti‐miR‐377‐5p counteracted the promoting effect of si‐hsa_circ_0072088#1 on the level of miR‐377‐5p in NSCLC cells (Fig 4a). As shown in Fig 4b,c, si‐hsa_circ_0072088#1‐mediated inhibitory effect on the proliferation of NSCLC cells was overturned by the addition of anti‐miR‐377‐5p. The results of flow cytometry showed that the inhibitory impact caused by the interference of circ_0072088 on the cell cycle of NSCLC cells was counteracted by the transfection of anti‐miR‐377‐5p (Fig 4d). To address the influence of circ_0072088 and miR‐377‐5p on the metastasis of NSCLC cells, wound healing assay and transwell invasion assay were carried out. As mentioned in Fig 4e,f, the migration and invasion abilities of NSCLC cells were recovered in si‐hsa_circ_0072088#1 and the anti‐miR‐377‐5p cotransfected group. The influence of miR‐377‐5p and circ_0072088 on the proliferation and metastasis of NSCLC cells was also confirmed by western blot assay. The expression of PCNA, Cyclin D1, MMP2 and MMP9 was downregulated in NSCLC cells with the silencing of circ_0072088, and the addition of anti‐miR‐377‐5p recovered the levels of these proteins (Fig 4g,h). Collectively, circ_0072088 was involved in the tumorigenesis of NSCLC through acting as a sponge of miR‐377‐5p.
Figure 4.

Circ_0072088 acts as an oncogenic molecule through sponging miR‐377‐5p in NSCLC cells. (a–h) NSCLC cells were transfected with si‐NC, si‐hsa_circ_0072088#1, si‐hsa_circ_0072088#1 + anti‐NC or si‐hsa_circ_0072088#1 + anti‐miR‐377‐5p. (a) The level of miR‐377‐5p was examined in NSCLC cells by qRT‐PCR ( ) si‐NC, (
) si‐NC, ( ) si‐hsa_circ_0072088#1, (
) si‐hsa_circ_0072088#1, ( ) si‐hsa_circ_0072088#1+anti‐NC, and (
) si‐hsa_circ_0072088#1+anti‐NC, and ( ) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p. (b) CCK8 assay was employed to detect the proliferation of NSCLC cells NCI‐H1299 (
) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p. (b) CCK8 assay was employed to detect the proliferation of NSCLC cells NCI‐H1299 ( ) si‐NC, (
) si‐NC, ( ) si‐hsa_circ_0072088#1, (
) si‐hsa_circ_0072088#1, ( ) si‐hsa_circ_0072088#1+anti‐NC, and (
) si‐hsa_circ_0072088#1+anti‐NC, and ( ) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p; A‐549 (
) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p; A‐549 ( ) si‐NC, (
) si‐NC, ( ) si‐hsa_circ_0072088#1, (
) si‐hsa_circ_0072088#1, ( ) si‐hsa_circ_0072088#1+anti‐NC, and (
) si‐hsa_circ_0072088#1+anti‐NC, and ( ) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p. (c) The colony formation ability of NSCLC cells was assessed by colony formation assay (
) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p. (c) The colony formation ability of NSCLC cells was assessed by colony formation assay ( ) si‐NC, (
) si‐NC, ( ) si‐hsa_circ_0072088#1, (
) si‐hsa_circ_0072088#1, ( ) si‐hsa_circ_0072088#1+anti‐NC, and (
) si‐hsa_circ_0072088#1+anti‐NC, and ( ) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p. (d) Flow cytometry was used to measure the cell cycle of NSCLC cells (
) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p. (d) Flow cytometry was used to measure the cell cycle of NSCLC cells ( ) G1, (
) G1, ( ) S, and (
) S, and ( ) G2. (e and f) The migration and invasion of NSCLC cells were assessed by wound healing assay and transwell invasion assay (e) (
) G2. (e and f) The migration and invasion of NSCLC cells were assessed by wound healing assay and transwell invasion assay (e) ( ) si‐NC, (
) si‐NC, ( ) si‐hsa_circ_0072088#1, (
) si‐hsa_circ_0072088#1, ( ) si‐hsa_circ_0072088#1+anti‐NC, and (
) si‐hsa_circ_0072088#1+anti‐NC, and ( ) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p; (f) (
) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p; (f) ( ) si‐NC, (
) si‐NC, ( ) si‐hsa_circ_0072088#1, (
) si‐hsa_circ_0072088#1, ( ) si‐hsa_circ_0072088#1+anti‐NC, and (
) si‐hsa_circ_0072088#1+anti‐NC, and ( ) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p. (g and h) Western blot assay was employed to measure the expression of PCNA, Cyclin D1, MMP2 and MMP9 in NSCLC cells NCI‐H1299 (
) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p. (g and h) Western blot assay was employed to measure the expression of PCNA, Cyclin D1, MMP2 and MMP9 in NSCLC cells NCI‐H1299 ( ) si‐NC, (
) si‐NC, ( ) si‐hsa_circ_0072088#1, (
) si‐hsa_circ_0072088#1, ( ) si‐hsa_circ_0072088#1+anti‐NC, and (
) si‐hsa_circ_0072088#1+anti‐NC, and ( ) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p; A‐549 (
) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p; A‐549 ( ) si‐NC, (
) si‐NC, ( ) si‐hsa_circ_0072088#1, (
) si‐hsa_circ_0072088#1, ( ) si‐hsa_circ_0072088#1+anti‐NC, and (
) si‐hsa_circ_0072088#1+anti‐NC, and ( ) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p. *P < 0.05.
) si‐hsa_circ_0072088#1+anti‐miR‐377‐5p. *P < 0.05.
NOVA2 could bind to miR‐377‐5p in NSCLC cells
To uncover the working mechanism of circ_0072088/miR‐377‐5p axis in NSCLC, the other crucial genes were explored. According to TargetScan database, NOVA2 was a possible target of miR‐377‐5p (Fig 5a). Luciferase activity was dramatically reduced in miR‐377‐5p and NOVA2‐wt group, and the transfection of miR‐377‐5p had no influence on the luciferase activity of NOVA2‐mut group (Fig 5b), suggesting the direct interaction between miR‐377‐5p and NOVA2. As indicated in Fig 5c, miR‐377‐5p and NOVA2 were enriched in Anti‐Ago2 group other than Anti‐IgG group. MiR‐377‐5p and circ_0072088 were cotransfected into NSCLC cells to illustrate the regulatory relationship among circ_0072088, miR‐377‐5p and NOVA2 in NSCLC cells. MiR‐377‐5p overexpression downregulated the level of NOVA2, and the addition of circ_0072088 recovered the expression of NOVA2 in NSCLC cells (Fig 5d). There was a significant increase in the mRNA and protein expression of NOVA2 in NSCLC tissues in comparison with that in adjacent normal tissues (Fig 5e,f). Meanwhile, the protein expression of NOVA2 was also found to be increased in NSCLC cells than that in BEAS‐2B cells (Fig 5g). As mentioned in Fig 5h,i, the level of NOVA2 was positively correlated with the expression of circ_0072088, while there was an inverse correlation between the abundance of miR‐377‐5p and NOVA2. These results revealed that NOVA2 was a target of miR‐377‐5p in NSCLC cells.
Figure 5.
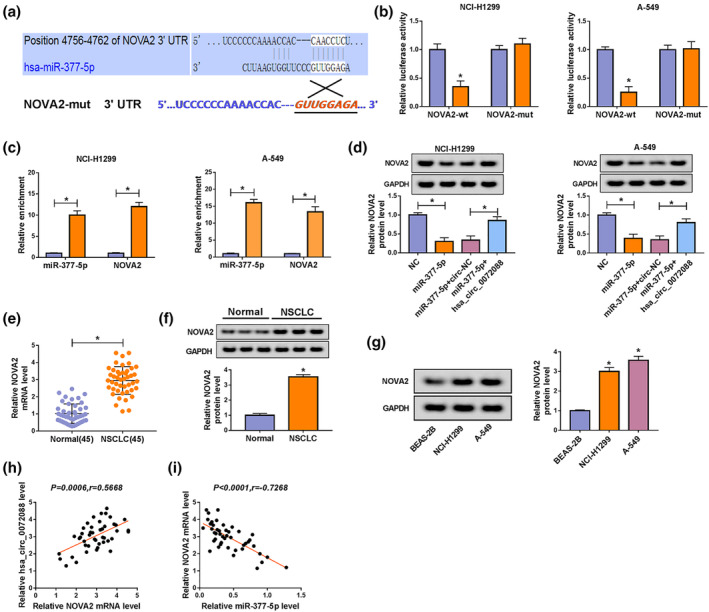
NOVA2 could bind to miR‐377‐5p in NSCLC cells. (a) TargetScan website was used to search the targets of miR‐377‐5p. (b and c) Dual‐luciferase reporter assay and RIP assay were employed to confirm the combination between miR‐377‐5p and NOVA2 (b) NCI‐H1299 ( ) NC, and (
) NC, and ( ) miR‐377‐5p; A‐549 (
) miR‐377‐5p; A‐549 ( ) NC, and (
) NC, and ( ) miR‐377‐5p (c) NCI‐H1299 (
) miR‐377‐5p (c) NCI‐H1299 ( ) Anti‐IgG, and (
) Anti‐IgG, and ( ) Anti‐Ago2; A‐549 (
) Anti‐Ago2; A‐549 ( ) Anti‐IgG, and (
) Anti‐IgG, and ( ) Anti‐Ago2. (d) Western blot assay was carried out to detect the expression of NOVA2 in NSCLC cells transfected with NC, miR‐377‐5p, miR‐377‐5p + circ‐NC or miR‐377‐5p + hsa_circ_0072088. (e and f) qRT‐PCR and Western blot assay were used to measure the mRNA and protein expression of NOVA2 in NSCLC tumor specimens and corresponding normal specimens. (g) The protein expression of NOVA2 was examined in NSCLC cells and BEAS‐2B cells by western blot assay. (h and i) The linear relationship between the expression of NOVA2 and circ_0072088 or miR‐377‐5p was evaluated using Spearman's correlation coefficient. *P < 0.05.
) Anti‐Ago2. (d) Western blot assay was carried out to detect the expression of NOVA2 in NSCLC cells transfected with NC, miR‐377‐5p, miR‐377‐5p + circ‐NC or miR‐377‐5p + hsa_circ_0072088. (e and f) qRT‐PCR and Western blot assay were used to measure the mRNA and protein expression of NOVA2 in NSCLC tumor specimens and corresponding normal specimens. (g) The protein expression of NOVA2 was examined in NSCLC cells and BEAS‐2B cells by western blot assay. (h and i) The linear relationship between the expression of NOVA2 and circ_0072088 or miR‐377‐5p was evaluated using Spearman's correlation coefficient. *P < 0.05.
NOVA2 overexpression overturns miR‐377‐5p‐mediated inhibition on the malignant potential of NSCLC cells
MiR‐377‐5p‐mediated downregulation of NOVA2 was overturned by the introduction of NOVA2 ectopic expression plasmid (Fig 6a). MiR‐377‐5p accumulation caused significant inhibition on the proliferation and colony formation of NSCLC cells, and the inhibitory influence was counteracted by the addition of NOVA2 (Fig 6b,c). Cell cycle was suppressed by the overexpression of miR‐377‐5p, and it was recovered in miR‐377‐5p and NOVA2 cotransfection group (Fig 6d). The introduction of NOVA2 also reversed the suppressive effects of miR‐377‐5p transfection on the migration and invasion of NSCLC cells (Fig 6e,f). The results of western blot assay revealed that miR‐377‐5p transfection downregulated the expression of PCNA, Cyclin D1, MMP2 and MMP9, while the addition of NOVA2 regained the expression of these proteins (Fig 6g,h). Taken together, miR‐377‐5p exerted an antitumor role through inhibiting NOVA2 in NSCLC.
Figure 6.

NOVA2 overexpression overturns miR‐377‐5p‐mediated inhibition on the malignant potential of NSCLC cells. (a–h) NCI‐H1299 and A‐549 cells were transfected with NC, miR‐377‐5p, miR‐377‐5p + vector or miR‐377‐5p + NOVA2. (a) The abundance of NOVA2 protein in NSCLC cells was detected by western blot assay. (b) The proliferation capacity of NSCLC cells was assessed by CCK8 assay NCI‐H1299 ( ) NC, (
) NC, ( ) miR‐377‐5p, (
) miR‐377‐5p, ( ) miR‐377‐5p+vector, and (
) miR‐377‐5p+vector, and ( ) miR‐377‐5p+NOVA2; A‐549 (
) miR‐377‐5p+NOVA2; A‐549 ( ) NC, (
) NC, ( ) miR‐377‐5p, (
) miR‐377‐5p, ( ) miR‐377‐5p+vector, and (
) miR‐377‐5p+vector, and ( ) miR‐377‐5p+NOVA2. (c) The proliferation of NSCLC cells was evaluated by colony formation assay (
) miR‐377‐5p+NOVA2. (c) The proliferation of NSCLC cells was evaluated by colony formation assay ( ) NC, (
) NC, ( ) miR‐377‐5p, (
) miR‐377‐5p, ( ) miR‐377‐5p+vector, and (
) miR‐377‐5p+vector, and ( ) miR‐377‐5p+NOVA2. (d) The cell cycle of NSCLC cells was analyzed by flow cytometry (
) miR‐377‐5p+NOVA2. (d) The cell cycle of NSCLC cells was analyzed by flow cytometry ( ) G1, (
) G1, ( ) S, and (
) S, and ( ) G2. (e and f) The metastasis of NSCLC cells was evaluated by wound healing assay and transwell invasion assay (e) (
) G2. (e and f) The metastasis of NSCLC cells was evaluated by wound healing assay and transwell invasion assay (e) ( ) NC, (
) NC, ( ) miR‐377‐5p, (
) miR‐377‐5p, ( ) miR‐377‐5p+vector, and (
) miR‐377‐5p+vector, and ( ) miR‐377‐5p+NOVA2; (f) (
) miR‐377‐5p+NOVA2; (f) ( ) NC, (
) NC, ( ) miR‐377‐5p, (
) miR‐377‐5p, ( ) miR‐377‐5p+vector, and (
) miR‐377‐5p+vector, and ( ) miR‐377‐5p+NOVA2. (g and h) The proliferation and motility of NSCLC cells were measured by western blot assay (g) NCI‐H1299 (
) miR‐377‐5p+NOVA2. (g and h) The proliferation and motility of NSCLC cells were measured by western blot assay (g) NCI‐H1299 ( ) NC, (
) NC, ( ) miR‐377‐5p, (
) miR‐377‐5p, ( ) miR‐377‐5p+vector, and (
) miR‐377‐5p+vector, and ( ) miR‐377‐5p+NOVA2; (h) A‐549 (
) miR‐377‐5p+NOVA2; (h) A‐549 ( ) NC, (
) NC, ( ) miR‐377‐5p, (
) miR‐377‐5p, ( ) miR‐377‐5p+vector, and (
) miR‐377‐5p+vector, and ( ) miR‐377‐5p+NOVA2. *P < 0.05.
) miR‐377‐5p+NOVA2. *P < 0.05.
Circ_0072088 accelerates the tumor growth of NSCLC in vivo
To illustrate the role of circ_0072088 in vivo, A‐549 cell lines that stably transfected with sh‐NC or sh‐hsa_circ_0072088 were built. Nude mice were randomly divided into two groups that injected with A‐549 cells stably expressing sh‐NC or sh‐hsa_circ_0072088, respectively. As mentioned in Fig 7a, mice that in sh‐hsa_circ_0072088 group possessed smaller tumors compared with that in sh‐NC group. The xenograft tumors were dissected from mice after five‐week injection, and the tumors were weighed. As indicated in Fig 7b, a marked reduction in tumor weight was found in sh‐hsa_circ_0072088 group than that in sh‐NC group. qRT‐PCR and Western blot assay were performed to detect the expression of crucial molecules. As mentioned in Fig 7c,d, the expression of circ_0072088, PCNA and NOVA2 was reduced, and the level of miR‐377‐5p was markedly upregulated. The expression of circ_0072088 was positively related to the protein abundance of PCNA and NOVA2, while there was a negative association between the expression of circ_0072088 and miR‐377‐5p. These findings demonstrated that circ_0072088 promoted the tumor growth of NSCLC in vivo.
Figure 7.

Circ_0072088 accelerates the tumor growth of NSCLC in vivo. (a) The tumor volume in nude mice injected with A‐549 cells stably expressing sh‐NC or sh‐hsa_circ_0072088 was measured every week ( ) sh‐NC, and (
) sh‐NC, and ( ) sh‐hsa_circ_0072088. (b) The tumor weight was detected after injection for five weeks. (c and d) The expression of hsa_circ_0072088, miR‐377‐5p, PCNA protein and NOVA2 protein in tumor tissues was examined by qRT‐PCR and western blot assay (c) (
) sh‐hsa_circ_0072088. (b) The tumor weight was detected after injection for five weeks. (c and d) The expression of hsa_circ_0072088, miR‐377‐5p, PCNA protein and NOVA2 protein in tumor tissues was examined by qRT‐PCR and western blot assay (c) ( ) sh‐NC, and (
) sh‐NC, and ( ) sh‐hsa_circ_0072088; (d) (
) sh‐hsa_circ_0072088; (d) ( ) sh‐NC, and (
) sh‐NC, and ( ) sh‐hsa_circ_0072088. *P < 0.05.
) sh‐hsa_circ_0072088. *P < 0.05.
Discussion
CircRNAs have been shown to play important roles in human cancers and diseases. 21 , 22 , 23 Circ‐ITCH has been reported to suppress the progression of cervical cancer through miR‐93‐5p/FOXK2 axis. 24 Circ‐MAPK4 impeded the apoptosis of glioma cells via miR‐125a‐3p signaling. 25 She et al. claimed that circ_0062389 accelerated the development of NSCLC cells through miR‐103a‐3p/CCNE1 axis. 26 The abundance of circ_0072088 was found to be aberrantly increased in NSCLC tissues and cells in this study. Circ_0072088 acted as an oncogenic gene in a variety of cancers, such as bladder cancer, renal carcinoma, NSCLC and hepatocellular carcinoma. 9 , 27 , 28 , 29 For instance, Wang et al. proved that circ_0072088 depletion restrained the proliferation and motility of renal carcinoma cells via miR‐206. 28 Zhang et al. claimed that circ_0072088 promoted the development of NSCLC through enhancing the level of CUL4B via sequestering miR‐101‐3p. 9 In the present study, loss‐of‐function experiments were conducted to evaluate the biological role of circ_0072088 in NSCLC cells. The results revealed that circ_0072088 silencing inhibited the proliferation and metastasis of NSCLC cells, suggesting the protumor role of circ_0072088 in NSCLC cells.
In addition to miR‐101‐3p, 9 miR‐377‐5p has been confirmed as a target of circ_0072088 in NSCLC cells. MiR‐377‐5p could suppress the growth, invasion and cell cycle of lung cancer cells through regulating AKT1 signaling. 14 However, the function and mechanism of miR‐377‐5p in NSCLC still remain to be revealed. MiR‐377‐5p was negatively regulated by circ_0072088 in NSCLC cells. In contrary to circ_0072088, miR‐377‐5p was markedly downregulated in NSCLC tissues and cells compared with normal tissues and BEAS‐2B cells. CeRNA mechanism is an important manner by which circRNAs function. 11 For example, circ_0063809‐mediated paclitaxel resistance of ovarian cancer cells through upregulating FOXR2 via sequestering miR‐1252. 30 Zhou et al. reported that circ_101064 contributed to the proliferation, migration and invasion of glioma cells through releasing PIWIL1 from miR‐154‐5p. 31 We speculated whether circ_0072088 functioned through acting as miR‐377‐5p sponge. Rescue experiments revealed that circ_0072088 exerted its oncogenic role in NSCLC via sponging miR‐377‐5p.
MiRNAs can regulate cellular functions through negatively regulating the expression of target genes to modulate the progression of multiple cancers. For example, Li et al. claimed that miR‐608 accelerated the apoptosis of pancreatic ductal adenocarcinoma cells through downregulating BRD4. 32 Xiang et al. reported that miR‐23a‐3p facilitated the cell cycle of hepatocellular carcinoma cells through promoting G1/S transition via negatively modulating protocadherin 17. 33 MiR‐876‐5p impeded the progression of colorectal cancer through downregulating RASAL2 and inhibiting YAP signaling. 34 NOVA2 was a candidate target of miR‐377‐5p. And the results of dual‐luciferase reporter assay and RIP assay verified that NOVA2 was a direct target of miR‐377‐5p. NOVA2 has been reported to be a protumor protein in NSCLC, and miR‐7‐5p inhibited the metastasis of NSCLC cells through down‐regulating NOVA2. 20 NOLVA2 was modulated by circ_0072088/miR‐377‐5p axis in this study. Meanwhile, NOVA2 accumulation recovered the malignant behaviors of NSCLC cells inhibited by the transfection of miR‐377‐5p mimics. These findings suggested that circ_0072088 promoted the proliferation and motility of NSCLC cells through elevating the level of NOVA2 via sponging miR‐377‐5p in vitro.
Animal experiments showed that circ_0072088 promoted the tumor growth of NSCLC in vivo. The expression of miR‐377‐5p and NOVA2 revealed the negative or positive association with circ_0072088 level, respectively. Nevertheless, more experiments are needed to illustrate if miR‐377‐5p/NOVA2 axis is essential for the function of circ_0072088 in vivo.
Collectively, circ_0072088 has been confirmed as an oncogene in NSCLC. Circ_0072088 accelerated the proliferation, migration and invasion of NSCLC cells through miR‐377‐5p/NOVA2 axis. Circ_0072088/miR‐377‐5p/NOVA2 axis might be an underlying target for NSCLC therapy.
Disclosure
The authors declare that they have no financial conflicts of interest.
Acknowledgment
None.
Contributor Information
Zhaofeng Tan, Email: ilznmmp@163.com.
Lei Xia, Email: jkoiau@163.com.
References
- 1. Reck M. Pembrolizumab as first‐line therapy for metastatic non‐small‐cell lung cancer. Immunotherapy 2018; 10: 93–105. [DOI] [PubMed] [Google Scholar]
- 2. Nagano T, Tachihara M, Nishimura Y. Mechanism of resistance to epidermal growth factor receptor‐tyrosine kinase inhibitors and a potential treatment strategy. Cell 2018; 7(11): 212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beermann J, Piccoli MT, Viereck J, Thum T. Non‐coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev 2016; 96: 1297–325. [DOI] [PubMed] [Google Scholar]
- 4. de Almeida RA, Fraczek MG, Parker S, Delneri D, O'Keefe RT. Non‐coding RNAs and disease: The classical ncRNAs make a comeback. Biochem Soc Trans 2016; 44: 1073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Curry‐Hyde A, Ueberham U, Arendt T et al Neural circular transcriptomes across mammalian species. Genomics 2020; 112(2): 1162–1166. [DOI] [PubMed] [Google Scholar]
- 6. Song Z, Zhang Q, Zhu J, Yin G, Lin L, Liang C. Identification of urinary hsa_circ _0137439 as potential biomarker and tumor regulator of bladder cancer. Neoplasma 2020; 67: 137–46. [DOI] [PubMed] [Google Scholar]
- 7. Pan B, Qin J, Liu X et al Identification of serum Exosomal hsa‐circ‐0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet 2019; 10: 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cui J, Li W, Liu G et al A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non‐small cell lung cancer cell proliferation and migration by regulating miR‐520f. Artif Cells Nanomed Biotechnol 2019; 47: 810–21. [DOI] [PubMed] [Google Scholar]
- 9. Zhang H, Wang X, Hu B, Zhang F, Wei H, Li L. Circular RNA ZFR accelerates non‐small cell lung cancer progression by acting as a miR‐101‐3p sponge to enhance CUL4B expression. Artif Cells Nanomed Biotechnol 2019; 47: 3410–6. [DOI] [PubMed] [Google Scholar]
- 10. Wu CL, Ho JY, Chou SC, Yu DS. MiR‐429 reverses epithelial‐mesenchymal transition by restoring E‐cadherin expression in bladder cancer. Oncotarget 2016; 7: 26593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhong Y, Du Y, Yang X et al Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018; 17: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Wang B, Zhou H et al MicroRNA‐384 inhibits the progression of papillary thyroid cancer by targeting PRKACB. BioMed Res Int 2020; 2020: 4983420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu W, Jing Y, Xu Q et al MiR‐9 promotes proliferation and inhibits apoptosis of bladder cancer cells via notch signaling pathway. Panminerva Med 2020. 10.23736/S0031-0808.19.03818-7. [DOI] [PubMed] [Google Scholar]
- 14. Wu H, Liu HY, Liu WJ et al miR‐377‐5p inhibits lung cancer cell proliferation, invasion, and cell cycle progression by targeting AKT1 signaling. J Cell Biochem 2018. 10.1002/jcb.28091. [DOI] [PubMed] [Google Scholar]
- 15. Ule J, Stefani G, Mele A et al An RNA map predicting Nova‐dependent splicing regulation. Nature 2006; 444: 580–6. [DOI] [PubMed] [Google Scholar]
- 16. Yano M, Hayakawa‐Yano Y, Mele A, Darnell RB. Nova2 regulates neuronal migration through an RNA switch in disabled‐1 signaling. Neuron 2010; 66: 848–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen B, Zhang Y, Yu S et al MicroRNA‐339, an epigenetic modulating target is involved in human gastric carcinogenesis through targeting NOVA1. FEBS Lett 2015; 589: 3205–11. [DOI] [PubMed] [Google Scholar]
- 18. Zhi F, Wang Q, Deng D et al MiR‐181b‐5p downregulates NOVA1 to suppress proliferation, migration and invasion and promote apoptosis in astrocytoma. PLOS One 2014; 9: e109124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu X, Zheng H. NOVA1 acts as an oncogene in melanoma via regulating FOXO3a expression. J Cell Mol Med 2018; 22: 2622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiao H. MiR‐7‐5p suppresses tumor metastasis of non‐small cell lung cancer by targeting NOVA2. Cell Mol Biol Lett 2019; 24: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu J, Li D, Luo H, Zhu X. Circular RNAs: The star molecules in cancer. Mol Aspects Med 2019; 70: 141–52. [DOI] [PubMed] [Google Scholar]
- 22. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: A novel type of biomarker for cancer. Breast Cancer 2018; 25: 1–7. [DOI] [PubMed] [Google Scholar]
- 23. Gao JL, Chen G, He HQ, Wang J. CircRNA as a new field in human disease research. Zhongguo Zhong Yao Za Zhi 2018; 43: 457–62. (In Chinese.). [DOI] [PubMed] [Google Scholar]
- 24. Li J, Guo R, Liu Q, Sun J, Wang H. Circular RNA Circ‐ITCH inhibits the malignant behaviors of cervical cancer by microRNA‐93‐5p/FOXK2 Axis. Reprod Sci 2020; 27: 860–8. [DOI] [PubMed] [Google Scholar]
- 25. He J, Huang Z, He M et al Circular RNA MAPK4 (circ‐MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR‐125a‐3p in gliomas. Mol Cancer 2020; 19: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. She Y, Han Y, Zhou G et al hsa_circ_0062389 promotes the progression of non‐small cell lung cancer by sponging miR‐103a‐3p to mediate CCNE1 expression. Cancer Genet 2019; 241: 12–9. [DOI] [PubMed] [Google Scholar]
- 27. Zhang WY, Liu QH, Wang TJ, Zhao J, Cheng XH, Wang JS. CircZFR serves as a prognostic marker to promote bladder cancer progression by regulating miR‐377/ZEB2 signaling. Biosci Rep 2019; 39(12): BSR20192779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang M, Gao Y, Liu J. Silencing circZFR inhibits the proliferation, migration and invasion of human renal carcinoma cells by regulating miR‐206. Onco Targets Ther 2019; 12: 7537–50. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR‐3619‐5p/CTNNB1 axis and activating Wnt/beta‐catenin pathway. Arch Biochem Biophys 2019; 661: 196–202. [DOI] [PubMed] [Google Scholar]
- 30. Zhang S, Cheng J, Quan C et al circCELSR1 (hsa_circ_0063809) contributes to paclitaxel resistance of ovarian cancer cells by regulating FOXR2 expression via miR‐1252. Mol Ther Nucleic Acids 2019; 19: 718–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou H, Zhang Y, Lai Y et al Circ_101064 regulates the proliferation, invasion and migration of glioma cells through miR‐154‐5p/ PIWIL1 axis. Biochem Biophys Res Commun 2020; 523(3): 608–614. [DOI] [PubMed] [Google Scholar]
- 32. Li M, Li T, Ma W, Wang X, Zhao G. MicroRNA‐608 promotes apoptosis via BRD4 downregulation in pancreatic ductal adenocarcinoma. Oncol Lett 2020; 19: 1418–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xiang Y, Yang Y, Lin C et al MiR‐23a‐3p promoted G1/S cell cycle transition by targeting protocadherin17 in hepatocellular carcinoma. J Physiol Biochem 2020; 76(1): 123–134. [DOI] [PubMed] [Google Scholar]
- 34. Ren L, Zhang Z, Feng Y, Luo M, Hao Z. MicroRNA‐876‐5p represses the cell proliferation and invasion of colorectal cancer through suppressing YAP signaling via targeting RASAL2. Clin Exp Pharmacol Physiol 2020; 47: 867–76. [DOI] [PubMed] [Google Scholar]


