Abstract
Long noncoding RNAs (lncRNAs) are >200‐bp molecules that do not generally code for proteins. Human lncRNAs have well‐characterized roles in gene expression regulation, particularly with regard to protein‐coding genes, and their dysregulation has been linked to disease. Here, we set out to investigate changes in the expression of lncRNAs related to apoptosis and autophagy in the peripheral blood mononuclear cells (PBMCs) of rheumatoid arthritis (RA). In addition, we aimed to correlate lncRNA expression profiles with clinical indexes and self‐perception of patients (SPP). To this end, we employed RNA sequencing of lncRNAs in PBMCs from three patients with RA and three healthy controls. We used bioinformatics to screen several dysregulated lncRNAs related to apoptosis and autophagy. To validate key lncRNA candidates, we performed quantitative reverse transcriptase–PCR on 20 patients with RA and 20 healthy controls. We found the expression of seven lncRNAs (MAPKAPK5‐AS1, ENST00000619282, C5orf17, LINC01189, LINC01006, DSCR9 and MIR22HG) was significantly altered in PBMCs of patients with RA. Receiver operating characteristic curve analysis suggested that MIR22HG [area under the curve (AUC) = 0.846, P = 0.000], DSCR9 (AUC = 0.783, P = 0.005), LINC01189 (AUC = 0.677, P = 0.034), MAPKAPK5‐AS1 (AUC = 0.644, P = 0.025) and ENST00000619282 (AUC = 0.636, P = 0.043) are potential biomarkers of RA. Spearman's correlation analysis revealed selected lncRNAs correlated with clinical indexes and SPP. Therefore, we highlight that some lncRNAs related to apoptosis and autophagy may serve as potential biomarkers for diagnosis and monitoring of RA progression, which also correlate with several clinical indexes and SPP.
Keywords: apoptosis, autophagy, lncRNAs, rheumatoid arthritis, RNA‐seq
We investigated changes in the expression of lncRNAs related to apoptosis and autophagy in the peripheral blood mononuclear cells (PBMCs) of rheumatoid arthritis (RA). Expression of seven lncRNAs (MAPKAPK5‐AS1, ENST00000619282, C5orf17, LINC01189, LINC01006, DSCR9, and MIR22HG) was significantly altered in PBMCs of RA patients. Squares, triangles, and circles represent lncRNAs, miRNAs, and mRNAs, respectively. Red nodes: upregulation; blue nodes: downregulation.
Abbreviations
- AUC
area under the curve
- C
complement
- DEG
differentially expressed gene
- GO
Gene Ontology
- IgA
immunoglobulin A
- IgG
immunoglobulin G
- IgM
immunoglobulin M
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- lncRNA
long noncoding RNA
- MH
mental health
- PBMC
peripheral blood mononuclear cell
- PF
physical function
- RA
rheumatoid arthritis
- RE
role emotional
- RF
rheumatoid factor
- ROC
receiver operating characteristic
- RP
role physical
- qRT‐PCR
quantitative reverse transcriptase–PCR
- SAS
Anxiety Self‐Assessment Scale
- SD
standard deviation
- SDS
Depression Self‐Assessment Scale
- SF
social function
- SPP
self‐perception of patients
- VAS
Visual Analogue Scale
- VT
vitality
Rheumatoid arthritis (RA) is a chronic inflammatory autoimmune disorder that affects peripheral joints and causes injury to secondary organs, including interstitial lung tissue and cardiac tissue [1, 2]. Misdiagnosis of RA can cause joint deformity and loss of ability to work [3]. However, the mechanism underlying the pathogenesis of RA is yet to be elucidated [4, 5]. Many studies have reported that apoptosis and autophagy play an important role in the pathogenesis of RA [6]. Early diagnosis and proper treatment are critical for improving the quality of life of patients and their families [7]. Nevertheless, current diagnostic methods suggest that the early diagnosis and treatment of RA remain a challenge [8, 9]. Therefore, new biomarkers are needed for improving the treatment and prognosis of RA [10].
More than 80% of the human genome is transcribed into RNA transcripts with little or no protein‐coding ability [11]. However, recent studies have revealed that although long noncoding RNAs (lncRNAs) do not generally code for proteins [12], they play a role in regulating the expression of human protein‐coding genes [13, 14]. Numerous studies have revealed that lncRNAs participate in the initiation and development of multiple diseases, including various cancers [15, 16], gynecological diseases [17, 18] and various autoimmune disorders [19, 20]. They act as miRNA sponges by sequestering miRNAs through base pair complementarity, thus keeping them away from their target mRNAs [21]. A recent study based on microarray analysis reported that the lncRNA ENST00000456270 was highly expressed in the peripheral blood mononuclear cells (PBMCs) of patients with RA and may serve as a potential biomarker for the diagnosis of RA [22]. Although the study demonstrated that lncRNAs are important biological molecules for studying the molecular mechanisms underlying the pathogenesis of RA, different aspects, such as apoptosis, autophagy, immune inflammation and oxidative stress, must be explored to gain a comprehensive understanding of the pathogenesis mechanism.
The objective of this study was to investigate the role of lncRNAs associated with apoptosis and autophagy as serological biomarkers for RA. We believe that our study will contribute to the development of effective strategies for the early diagnosis, treatment and prevention of RA.
Materials and methods
Sample collection
In this study, 20 patients with RA who were diagnosed at the Department of Rheumatology and Immunology in the First Affiliated Hospital of Anhui University of Traditional Chinese Medicine from November 2019 to December 2019 were included in the RA group. In addition, 20 age‐ and sex‐matched healthy subjects who underwent routine physical examinations in the same hospital during the same period served as the healthy control group (Table 1). All patients with RA fulfilled the criteria proposed by the American College of Rheumatology [23]. Patients with severe liver or kidney dysfunction, patients receiving immunosuppressive agents and pregnant women were excluded from the study. The research protocol was in accordance with the tenets of the Declaration of Helsinki and was approved by the Ethics Committee of First Affiliated Hospital of Anhui University of Traditional Chinese Medicine, and all patients signed an informed consent form.
Table 1.
Characteristics and clinical features of RA patient samples used in RNA sequencing. F, female; M, male; NA, Not Applicable.
| Index | RA (n = 20) | Control (n = 20) | P |
|---|---|---|---|
| Sex (M/F) | 4/16 | 4/16 | 1.000 |
| Age (years) a | 55.4 ± 14.50 | 54.12 ± 13.29 | 0.872 |
| Silk time (years) | 10.02 ± 9.80 | NA | NA |
| Erythrocyte sedimentation rate (mm·h−1) | 46.65 ± 17.51 | NA | NA |
| High‐sensitivity C‐reactive protein (mg·L−1) | 21.38 ± 19.51 | NA | NA |
| RF (U·mL−1) | 132.77 ± 114.43 | NA | NA |
| Cyclic citrullinated peptide (U·mL−1) | 264.32 ± 265.03 | NA | NA |
| IgA (g·L−1) | 3.04 ± 0.52 | NA | NA |
| IgG (g·L−1) | 13.54 ± 3.34 | NA | NA |
| IgM (g·L−1) | 1.64 ± 0.72 | NA | NA |
| C3 (g·L−1) | 1.03 ± 0.15 | NA | NA |
| C4 (g·L−1) | 0.23 ± 0.10 | NA | NA |
| Disease Activity Score in 28 joints score | 6.46 ± 1.04 | NA | NA |
| VAS score | 6.89 ± 1.30 | NA | NA |
| SAS score | 57.86 ± 4.03 | NA | NA |
| SDS score | 56.80 ± 6.67 | NA | NA |
| Physical function score | 48.00 ± 13.17 | NA | NA |
| Role physical score | 15.00 ± 12.25 | NA | NA |
| Body pain score | 56.70 ± 13.21 | NA | NA |
| General health score | 45.25 ± 8.29 | NA | NA |
| Vitality score | 40.80 ± 7.16 | NA | NA |
| SF score | 51.88 ± 13.85 | NA | NA |
| RE score | 31.67 ± 24.67 | NA | NA |
| MH score | 48.60 ± 11.42 | NA | NA |
Mean ± SD. The significance difference between two groups was tested via analysis of Student's t‐test.
Biochemical measurements
The clinical indexes were sex, age, course of disease, erythrocyte sedimentation rate, high‐sensitivity C‐reactive protein, rheumatoid factor (RF), anti‐cyclic citrullinated peptide Ig, immunoglobulin A (IgA), immunoglobulin G (IgG), immunoglobulin M (IgM), complement (C) 3 (C3) and C4. Self‐perception of patients (SPP) was measured using the following tools: Disease Activity Score in 28 joints, Visual Analogue Scale (VAS), Anxiety Self‐Assessment Scale (SAS) and Depression Self‐Assessment Scale (SDS) scores. The 36‐Item Short Form Health Survey was composed of eight dimensions: physical function, role physical, body pain, general health, vitality, social function (SF), role emotional (RE) and mental health (MH). All of the participants enrolled were asked to fill in the 36‐Item Short Form Health Survey scale under the guidance of two clinical doctors.
RNA sequencing and bioinformatics analysis
Total RNA from each sample was used to construct cDNA libraries using the VAHTSTM Total RNA‐Seq (H/M/R) for high‐throughput sequencing. In brief, the RNA was reverse transcribed into first‐strand cDNA, followed by second‐strand cDNA synthesis, end repair, dA tailing and adaptor ligation. The digested products were purified using VAHTSTM DNA Clean Beads, and PCR amplified and sequenced using Illumina HiSeq 2500 (San Diego, CA, USA). The tagged cDNA libraries were paired‐end sequenced using an Illumina HiSeq 2500 with 51 plus 7 cycles by Dianxi Biotechnology, Shanghai, China. To fully understand the biological functions of lncRNAs related to immunity and inflammation, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. Coexpression network analysis was performed using Comparative Co‐Expression Network Construction and Visualization Tool (CoExpNetViz, Chicago, IL, USA).
PBMC preparation and total RNA extraction
PBMCs were isolated by Ficoll density centrifugation of 5 mL of blood layered on 6 mL of 1.077 g·mL−1 Ficoll‐Paque PLUS (GE Healthcare, Uppsala, Sweden). Cell concentration was adjusted between 5 × 106 and 7 × 106 cells·mL−1. Total RNA was extracted from PBMCs using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The quantity of RNA was measured using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, NC, USA).
Quantitative reverse transcriptase–PCR analysis
cDNA was synthesized using the RT reagent kit with gDNA Eraser (TaKaRa, Shiga, Japan). The primers used in quantitative reverse transcriptase–PCR (qRT‐PCR) are shown in Table 2. β‐Actin expression was used as an internal control. Relative expression values were calculated for each of the three experiments as 2−ΔΔCT.
Table 2.
Specific lncRNA primers used for qRT‐PCR analysis. F, forward; R, reverse.
| Gene name | Sequence (5′–3′) | bp |
|---|---|---|
| GAPDH | F: GGAGCGAGATCCCTCCAAAAT | 500 |
| R: GGCTGTTGTCATACTTCTCATGG | ||
| MAPKAPK5‐AS1 | F: GCGGAAAGTGACCAAGAG | 2390 |
| R: CTTCTCCAGAGCCTGGTCAC | ||
| ENST00000619282 | F: CCTGGTGGGAGAGAATTGAA | 651 |
| R: ATGAGAGCCAAGCAAGAGGA | ||
| C5orf17 | F: CCACACCGACACCTATACCC | 2003 |
| R: TCGACTCTCCACTGTGATGC | ||
| LINC01189 | F: GTCTGCCCAGCTACTCCAAG | 1193 |
| R: CTCCTACCGCTCCTGTTGAG | ||
| LINC01006 | F: GTGTGTCAGGCATTGTACCG | 3350 |
| R: GCCCTGTTTCCAAAAGATCA | ||
| DSCR9 | F: ATTCCCTCCCCTATCACCAG | 1681 |
| R: CCTAGCATACGCTGGAGGAC | ||
| MIR22HG | F: TGGAGGAGGGGGTTAGAGTT | 2659 |
| R: GGGGATCACATACCACCTTG |
Statistical analysis
All data are presented as the means ± standard deviation (SD). Comparison between two groups was analyzed by the Student's t‐test or nonparametric Mann–Whitney–Wilcoxon test. The difference among more than two groups was analyzed by the one‐way ANOVA, followed by Tukey's post hoc test. A P value <0.05 was considered statistically significant. All statistical analyses were performed using the graphpad prism 8.0 (GraphPad Software Inc., San Diego, CA, USA) and spss 19.0 software (SPSS, Chicago, IL, USA).
Results
Screening of abnormal expression of lncRNAs in PBMCs from patients with RA and healthy controls
We performed high‐throughput sequencing of lncRNAs in PBMCs from three patients with RA (KB12, KB13 and KB14) and three healthy controls (YZ22, YZ31 and YZ47). A total of 341 lncRNAs were detected. The volcano plots in Fig. 1A show the variations in the expression of lncRNAs related to apoptosis and autophagy between patients with RA and healthy controls. Hierarchical clustering further revealed differential expression of lncRNAs related to apoptosis and autophagy between patients with RA and healthy controls (Fig. 1B)
Fig. 1.
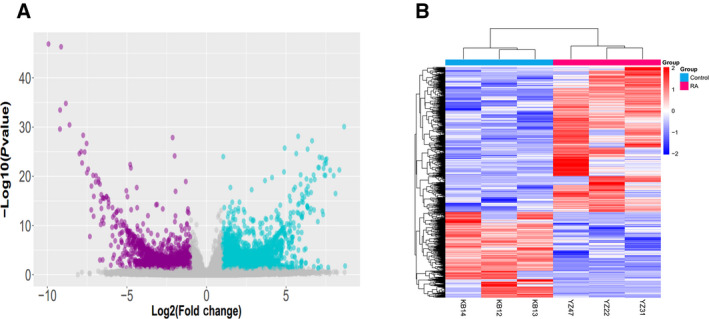
Expression profiling of lncRNAs. (A) Differentially expressed lncRNAs between patients with RA and healthy controls are displayed by volcano plots. Differentially expressed lncRNAs between two groups were compared by Student's t‐test. (B) Each column represents PBMCs from a patient with RA or a healthy control. The red lines represent up‐regulated lncRNAs.
The correlation matrix revealed very poor correlation between the samples (Fig. 2).
Fig. 2.
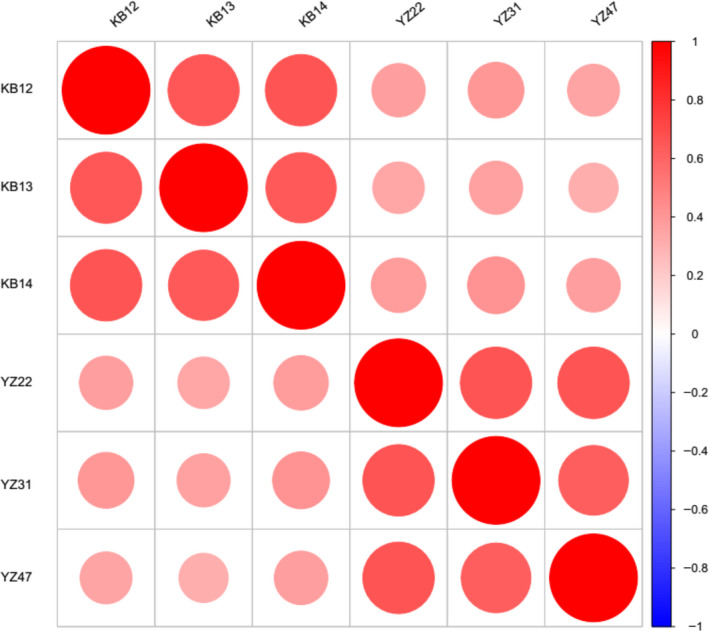
Correlation matrix showing Pearson's coefficients between the samples. Red represents positive correlation, and blue represents negative correlation; the larger the circle, the stronger the correlation. YZ (21, 22, 47) represents three healthy control samples, and KB (13, 12, 14) represents three RA samples.
The top two up‐regulated and five down‐regulated lncRNAs are presented in Table 3.
Table 3.
Basic characteristics of the seven differentially expressed lncRNAs.
| lncRNAs | Gene ID | P | Fold change | Regulation | Gene symbol |
|---|---|---|---|---|---|
| MAPKAPK5‐AS1 | 51275 | 0.022 | −2.50 | Down‐regulation | TMEM116 |
| ENST00000619282 | ENST00000619282.1 | 0.000 | 1.80 | Up‐regulation | P2RX7 |
| C5orf17 | 439936 | 0.028 | −2.01 | Down‐regulation | NBPF14 |
| LINC01189 | 643648 | 0.044 | −2.16 | Down‐regulation | ACSL1 |
| LINC01006 | 100506380 | 0.030 | −2.48 | Down‐regulation | RNF32 |
| DSCR9 | 257203 | 0.039 | −1.94 | Down‐regulation | TTC3 |
| MIR22HG | 84981 | 0.031 | 1.66 | Up‐regulation | SMYD4 |
Bioinformatics analysis of lncRNAs
We performed GO enrichment analysis for the differentially expressed genes (DEGs), with particular attention to GO biological processes and molecular function. According to GO analysis, the DEGs were mainly involved in the regulation of autophagy and apoptosis (Fig. 3)
Fig. 3.
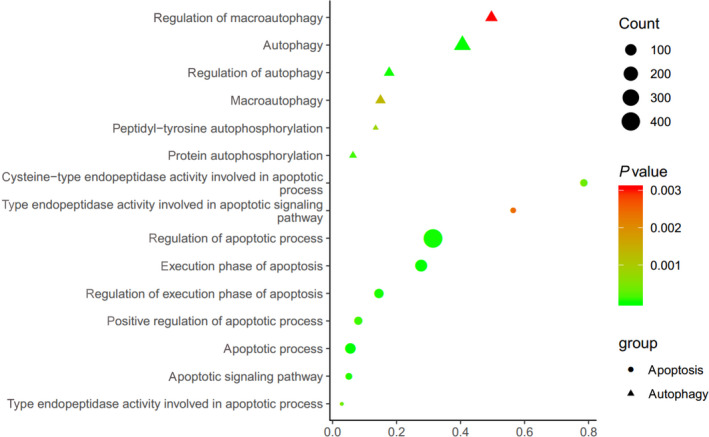
GO analysis. Ranking of top 15 biological processes based on GO analysis of differentially expressed lncRNAs. The bar plot shows the top enrichment score of the significant pathway. The intensity of the color indicates the level of difference; darker shades represent greater difference. The circle represents the relationship; larger circles represent a stronger relationship between the gene and the pathway.
Moreover, KEGG pathway analysis showed that these lncRNAs were mainly involved in the nuclear factor‐κB, phosphatidylinositol 3‐kinase (PI3K)‐Akt and Adenosine 5'‐monophosphate (AMP)‐activated protein kinase signaling pathways (Fig. 4A,B)
Fig. 4.
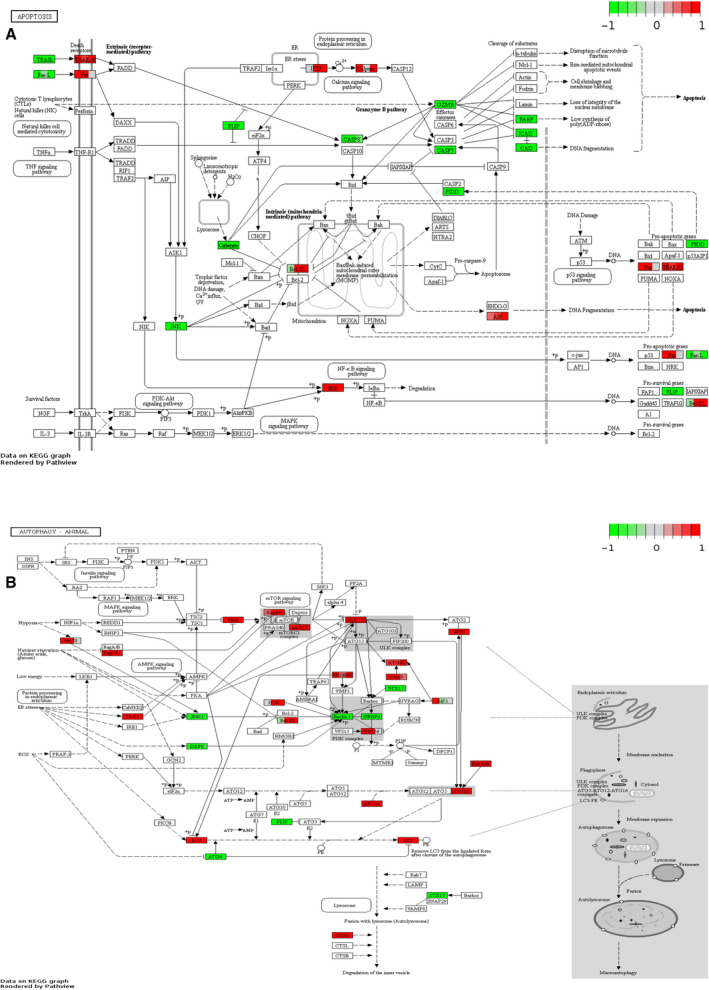
Expression of DEGs involved in the apoptosis and autophagy signaling pathways. (A) Expression of DEGs involved in the apoptosis signaling pathway. (B) Expression of DEGs involved in the autophagy signaling pathway.
To identify the possible modulating mechanisms of the lncRNAs, lncRNA–mRNA and miRNA–mRNA interaction networks were established according to lncRNA sequences and miRNA target genes (Fig. 5A,B), and a lncRNA–miRNA–mRNA co‐expression network was constructed (Fig. 6).
Fig. 5.
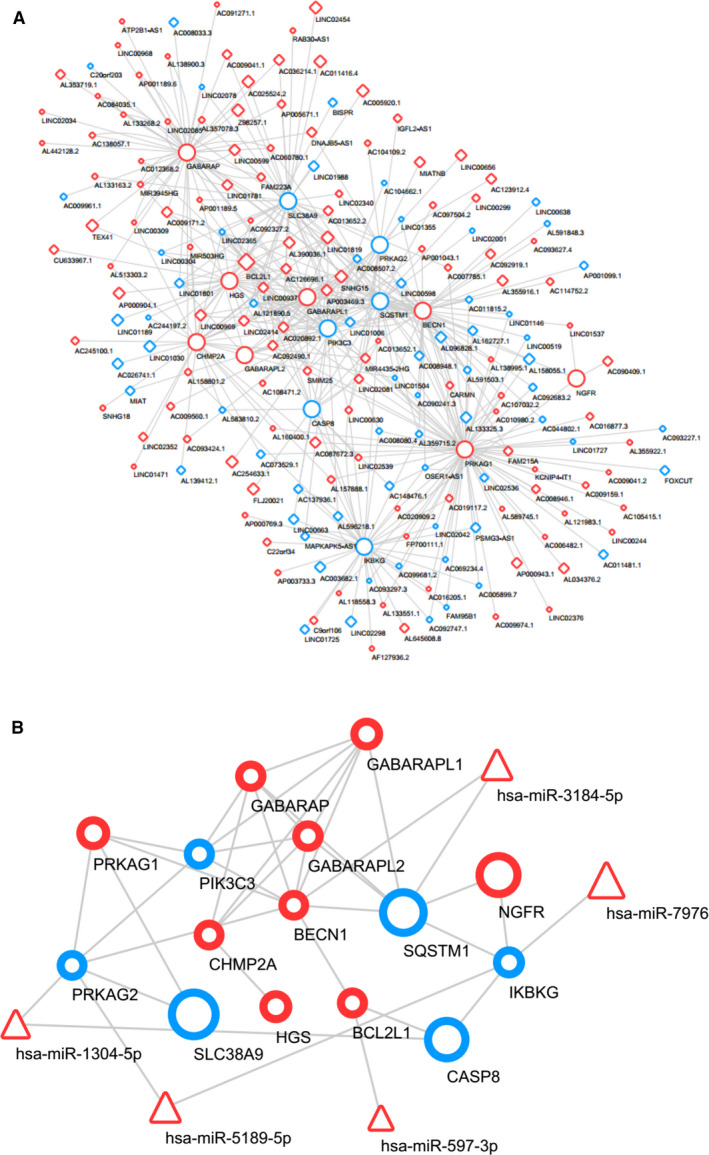
lncRNA–mRNA (A) and miRNA–mRNA (B) interaction networks established based on lncRNA sequences and miRNA target genes. Diamond shapes, triangles and circles represent lncRNAs, miRNAs and mRNAs, respectively. Red represents up‐regulation, and blue represents down‐regulation. Node size represents the degree.
Fig. 6.
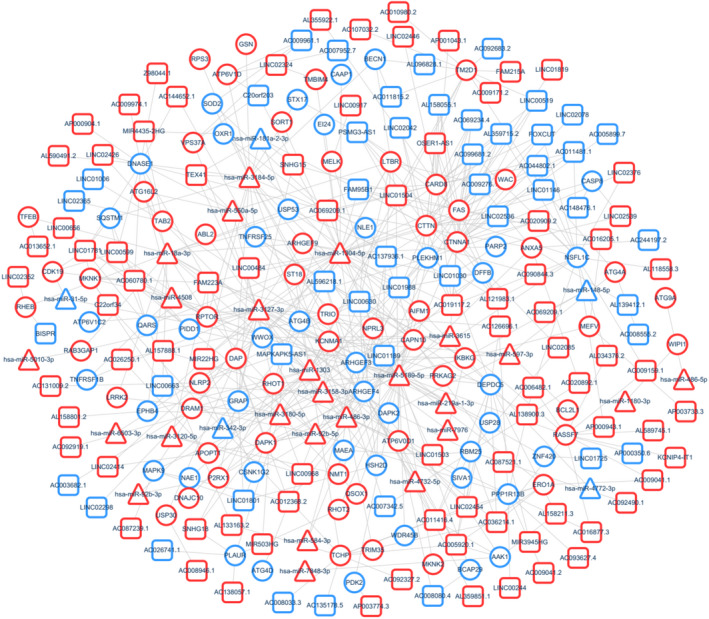
lncRNA–miRNA–mRNA coexpression network. Squares, triangles and circles represent lncRNAs, miRNAs and mRNAs, respectively. Red represents up‐regulation, and blue represents down‐regulation. Node size represents the degree.
qRT‐PCR validation of differentially expressed lncRNAs
To validate our RNA sequencing data, we selected two up‐regulated lncRNAs (ENST00000619282 and MIR22HG) and five down‐regulated lncRNAs (MAPKAPK5‐AS1, C5orf17, LINC01189, LINC01006 and DSCR9) related to apoptosis and autophagy. qRT‐PCR analyses were performed to validate candidate gene expression using an independent set of samples from 20 patients with RA and 20 healthy controls. qRT‐PCR showed that the average expression levels of ENST00000619282, LINC01006 and MIR22HG were significantly higher (Fig. 7A–C) and that of MAPKAPK5‐AS1, LINC01189 and DSCR9 were lower (Fig. 7D–F) in the PBMCs of patients with RA as compared with that in the PBMCs of healthy controls. There were no significant differences in the expression levels of C5orf17 between the RA group and the healthy control group.
Fig. 7.
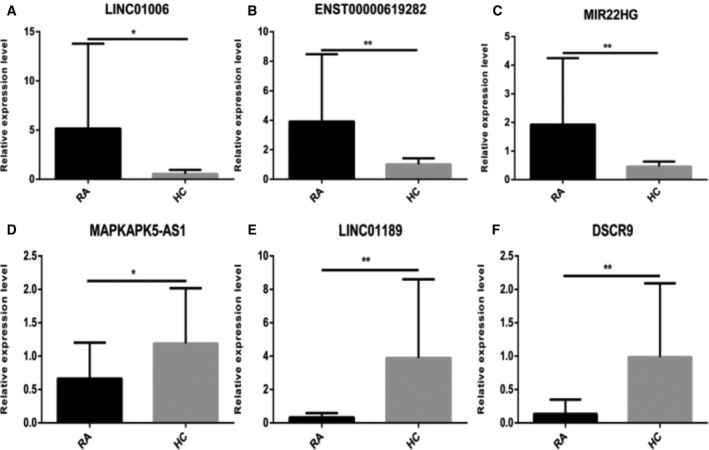
Relative expression levels of lncRNAs. (A–C) The relative expression of ENST00000619282, LINC01006 and MIR22HG was up‐regulated. (D–F) The relative expression of MAPKAPK5‐AS1, LINC01189 and DSCR9 was down‐regulated. All of the results were presented as the mean ± SD based on three replicates involving three samples. The significance difference between two groups was tested via analysis of Student's t‐test. *P < 0.05, **P < 0.01, ***P < 0.001.
Receiver operating characteristic curve analysis of confirmed PMBC lncRNAs of patients with RA
Receiver operating characteristic (ROC) curve analysis was performed to evaluate the biological functions and diagnostic value of the four lncRNAs. ROC curves of confirmed lncRNAs showed that the levels of MIR22HG [area under the curve (AUC) = 0.846, P = 0.000], DSCR9 (AUC = 0.783, P = 0.005), LINC01189 (AUC = 0.677, P = 0.034), MAPKAPK5‐AS1 (AUC = 0.644, P = 0.025) and ENST00000619282 (AUC = 0.636, P = 0.043), as shown in Fig. 8A–E.
Fig. 8.
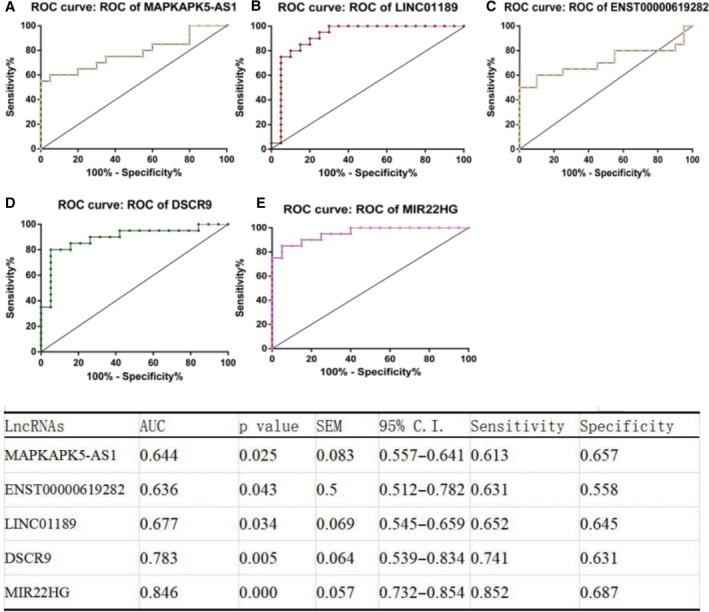
ROC curve analysis of lncRNAs: (A) MIR22HG, (B) DSCR9, (C) LINC01189, (D) MAPKAPK5‐AS1 and (E) ENST00000619282.
Spearman's correlation test of clinical indexes, SPP and confirmed lncRNAs in PBMCs of patients with RA
To determine whether the differentially expressed lncRNAs related to inflammation and immunity in the PBMCs of patients with RA were relevant biomarkers for the severity of RA, we performed the Spearman's correlation test to assess the correlation of MIR22HG, DSCR9, LINC01189, MAPKAPK5‐AS1 and ENST00000619282 with clinical indexes and SPP. Results of the correlation analysis revealed a positive correlation between MAPKAPK5‐AS1 and IgM (Fig. 9A). LINC01189 positively correlated with C3, SDS and MH (Fig. 9B–D), and negatively correlated with RF, IgA, IgG and SF (Fig. 9E–H). ENST00000619282 positively correlated with RF and SF (Fig. 9I,J), and negatively correlated with SAS (Fig. 9K). DSCR9 positively correlated with RF, RE and MH (Fig. 9L–N). MIR22HG positively correlated with the course of disease and IgG (Fig. 9O,P).
Fig. 9.
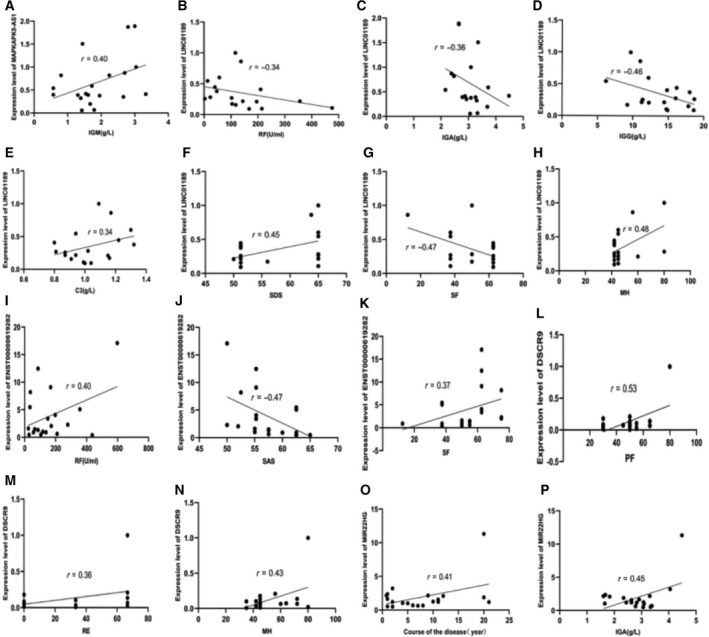
Correlation of lncRNAs with clinical indices and SPP. (A) Correlation between MAPKAPK5‐AS1 and IgM. (B–H) Correlation between LINC01189 and C3, SDS, MH, RF, IgA, IgG and SF. (I–K) Correlation between ENST00000619282 and RF, SF and SAS. (L, M, N) Correlation between DSCR9 and RF, RE and MH. (O, P) Correlation between MIR22HG and the course of disease, IgG.
Discussion
lncRNAs are important competing endogenous RNAs [24]. They are a class of noncoding RNAs that regulate diverse physiological processes by controlling gene expression [25, 26]. Intensive studies have been carried out to elucidate the relationship between lncRNAs and diseases, especially cancer [27]. Researchers have discovered many lncRNAs that are considered as potential diagnostic biomarkers of cancers. For example, the lncRNA LINC00173 enhances triple‐negative breast cancer progression by suppressing miR‐490‐3p expression [28]. The lncRNA NORAD regulates lung cancer cell proliferation, apoptosis, migration and invasion via the miR‐30a‐5p/ADAM19 axis [9]. So far, two case reports have shown that the expression of lncRNAs is significantly dysregulated in the PBMCs of patients with RA as compared with that in healthy control PBMCs, which suggests that lncRNAs have the potential to serve as biomarkers of RA [22, 29]. Therefore, changes in the expression of lncRNAs may reflect various underlying diseases.
In this study, we revealed that lncRNAs related to apoptosis and autophagy were differentially expressed in the PBMCs of patients with RA as compared with healthy controls. A total of 341 differentially expressed lncRNAs were detected, of which 231 were up‐regulated and 110 were down‐regulated. Several lncRNAs related to apoptosis and autophagy were aberrantly expressed in patients with RA, suggesting that these lncRNAs may play an important role in the pathogenesis of RA. According to GO analysis, these genes were mainly involved in the regulation of autophagy and apoptosis. Moreover, KEGG pathway analysis showed that these lncRNAs were mainly involved in the nuclear factor‐κB, PI3K‐Akt and Adenosine 5'‐monophosphate (AMP)‐activated protein kinase signaling pathways.
RA is an inflammatory autoimmune disease, in which the immune system of the body mistakenly attacks the joints and other organs [30, 31]. RA affects the flexibility or extensibility of the joints, as well as organ function. The function of lncRNAs in RA provides new insights into the understanding of RA [32]. However, little is known about the diagnostic value of lncRNAs in RA [33]. lncRNAs remove the inhibitory effect of miRNAs on their target mRNAs, indirectly up‐regulating these target mRNAs [34].
In this study, we found that the average expression levels of ENST00000619282, LINC01006 and MIR22HG in the PBMCs of patients with RA were significantly higher than that in the PBMCs of healthy controls (Fig. 7A–C), whereas the expression of MAPKAPK5‐AS1, LINC01189 and DSCR9 was significantly lower in patients with RA as compared with healthy controls. Among these lncRNAs, MIR22HG, DSCR9, LINC01189, MAPKAPK5‐AS1 and ENST00000619282 showed higher ROC AUC and may serve as potential diagnostic biomarkers of RA. In addition, the Spearman's correlation test showed that MAPKAPK5‐AS1 positively correlated with IgM; LINC01189 positively correlated with C3, SDS and MH, and negatively correlated with RF, IgA, IgG and SF; ENST00000619282 positively correlated with RF and SF, and negatively correlated with SAS; DSCR9 positively correlated with RF, RE and MH; and MIR22HG positively correlated with the course of disease and IgG. These results suggest that these lncRNAs are of great importance for practical applications.
However, there are certain limitations to the current study that must be addressed in future research. For example, no significant differences in the expression levels of C5orf17 were observed between the RA and healthy control groups, and C5orf17 had no correlation with clinical indexes and SPP. This discrepancy probably arose because of the relatively small sample size. In addition, the individual differences between patients with RA, including socioeconomic status, disease severity and disease duration, could probably affect the accuracy of the results. Understanding the exact mechanism of lncRNAs in apoptosis and autophagy will be the goal of our next study.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
J. Wen, JL, HJ, LW, Yue Sun and LX contributed to the study design. J Wen analyzed the data, wrote the first draft and revised the manuscript. Yanqui Sun, YZ and PZ contributed to specimen and data collection. XD, XW and J. Wang supervised the project and contributed to manuscript revision. All authors reviewed and accepted the content of the final manuscript.
Acknowledgements
This work was supported by grants from the Ministry of Science and Technology, National Key Research and Development Program, Chinese Medicine Modernization Research Key Project (2018YFC1705204); National Nature Fund Program (81973655); National Key Research and Development Program, Foreign Science and Technology Cooperation Project of Anhui (201904b11020011); Anhui Provincial Quality Engineering Teaching and Research Project (2018jyxm1068); Anhui Famous Traditional Chinese Medicine Liu Jian Studio Construction Project (Traditional Chinese Medicine Development Secret [2018] No. 11); National Key Innovative Talents Training Project of Traditional Chinese Medicine [National Education Letter of Traditional Chinese Medicine (2019) No. 128]; Key Research and Development Plan Project of Anhui Province (201904a07020004); Anhui Provincial Laboratory of Applied Basis and Development of Internal Medicine and Modern Traditional Chinese Medicine (2016080503B041); and 12th batch of ‘115’ Innovation Team of Anhui Province [Anhui Talent Office (2019) No. 1].
Data accessibility
The data used to support the findings of this study are available from the corresponding author upon request.
References
- 1. Coutant F and Miossec P (2020) Evolving concepts of the pathogenesis of rheumatoid arthritis with focus on the early and late stages. Curr Opin Rheumatol 32, 57–63. [DOI] [PubMed] [Google Scholar]
- 2. Farquhar H, Vassallo R, Edwards AL and Matteson EL (2019) Pulmonary complications of rheumatoid arthritis. Semin Respir Crit Care Med 40, 194–207. [DOI] [PubMed] [Google Scholar]
- 3. Goekoop‐Ruiterman YP, de Vries‐Bouwstra JK, Allaart CF, van Zeben D, Kerstens PJ, Hazes JM, Zwinderman AH, Ronday HK, Han KH, Westedt ML et al (2008) Clinical and radiographic outcomes of four different treatment strategies in patients with early rheumatoid arthritis (the BeSt study): a randomized, controlled trial. Arthritis Rheum 58, 194–207. [DOI] [PubMed] [Google Scholar]
- 4. Croia C, Bursi R, Sutera D, Petrelli F, Alunno A and Puxeddu I (2019) One year in review 2019: pathogenesis of rheumatoid arthritis. Clin Exp Rheumatol 37, 347–367. [PubMed] [Google Scholar]
- 5. Calabresi E, Petrelli F, Bonifacio AF, Puxeddu I and Alunno A (2018) One year in review 2018: pathogenesis of rheumatoid arthritis. Clin Exp Rheumatol 36, 175–184. [PubMed] [Google Scholar]
- 6. Vomero M, Manganelli V, Barbati C, Colasanti T, Capozzi A, Finucci A, Spinelli FR, Ceccarelli F, Perricone C, Truglia S et al (2019) Reduction of autophagy and increase in apoptosis correlates with a favorable clinical outcome in patients with rheumatoid arthritis treated with anti‐TNF drugs. Arthritis Res Ther 21, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Madav Y, Barve K and Prabhakar B (2020) Current trends in theranostics for rheumatoid arthritis. Eur J Pharm Sci 145, 105240. [DOI] [PubMed] [Google Scholar]
- 8. Goekoop‐Ruiterman YP and Huizinga TW (2010) Rheumatoid arthritis: can we achieve true drug‐free remission in patients with RA? Nat Rev Rheumatol 6, 68–70. [DOI] [PubMed] [Google Scholar]
- 9. Li J, Xu X, Wei C, Liu L and Wang T (2020) Long noncoding RNA NORAD regulates lung cancer cell proliferation, apoptosis, migration, and invasion by the miR‐30a‐5p/ADAM19 axis. Int J Clin Exp Pathol 13, 1–13. [PMC free article] [PubMed] [Google Scholar]
- 10. Karami J, Aslani S, Tahmasebi MN, Mousavi MJ, Sharafat Vaziri A, Jamshidi A, Farhadi E and Mahmoudi M (2020) Epigenetics in rheumatoid arthritis; fibroblast‐like synoviocytes as an emerging paradigm in the pathogenesis of the disease. Immunol Cell Biol 98, 171–186. [DOI] [PubMed] [Google Scholar]
- 11. Cech TR and Steitz JA (2014) The noncoding RNA revolution‐trashing old rules to forge new ones. Cell 157, 77–94. [DOI] [PubMed] [Google Scholar]
- 12. McDonel P and Guttman M (2019) Approaches for understanding the mechanisms of long noncoding RNA regulation of gene expression. Cold Spring Harb Perspect Biol 11, a032151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uthaya Kumar DB and Williams A (2020) Long non‐coding RNAs in immune regulation and their potential as therapeutic targets. Int Immunopharmacol 81, 106279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kazimierczyk M, Kasprowicz MK, Kasprzyk ME and Wrzesinski J (2020) Human long noncoding RNA interactome: detection, characterization and function. Int J Mol Sci 21, 1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ren Z, Liu X, Si Y and Yang D (2020) Long non‐coding RNA DDX11‐AS1 facilitates gastric cancer progression by regulating miR‐873‐5p/SPC18 axis. Artif Cells Nanomed Biotechnol 48, 572–583. [DOI] [PubMed] [Google Scholar]
- 16. Jiang W, Kai J, Li D, Wei Z, Wang Y and Wang W (2020) lncRNA HOXB‐AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K‐AKT pathway. J Cell Physiol 10 10.1002/jcp.29618 [DOI] [PubMed] [Google Scholar]
- 17. Liu W, Zhuang R, Feng S, Bai X, Jia Z, Kapora E and Tan W (2020) Long non‐coding RNA ASB16‐AS1 enhances cell proliferation, migration and invasion via functioning as a ceRNA through miR‐1305/Wnt/β‐catenin axis in cervical cancer. Biomed Pharmacother 125, 109965. [DOI] [PubMed] [Google Scholar]
- 18. Xue F, Xu YH, Shen CC, Qin ZL and Zhou HB (2020) Non‐coding RNA LOXL1‐AS1 exhibits oncogenic activity in ovarian cancer via regulation of miR‐18b‐5p/VMA21 axis. Biomed Pharmacother 125, 109568. [DOI] [PubMed] [Google Scholar]
- 19. Wang JB, Li J, Zhang TP, Lv TT, Li LJ, Wu J, Leng RX, Fan YG, Pan HF and Ye DQ (2019) Expression of several long noncoding RNAs in peripheral blood mononuclear cells of patients with systemic lupus erythematosus. Adv Med Sci 64, 430–436. [DOI] [PubMed] [Google Scholar]
- 20. Wang JB, Li J, Zhang TP, Lv TT, Li LJ, Wu J, Leng RX, Fan YG, Pan HF and Ye DQ (2019) Long non‐coding RNAs modulate Sjögren's syndrome associated gene expression and are involved in the pathogenesis of the disease. J Clin Med 8, 2569–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang Y (2018) The novel regulatory role of lncRNA‐miRNA‐mRNA axis in cardiovascular diseases. J Cell Mol Med 22, 5768–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yuan M, Wang S, Yu L, Qu B, Xu L, Liu L, Sun H, Li C, Shi Y and Liu H (2017) Long noncoding RNA profiling revealed differentially expressed lncRNAs associated with disease activity in PBMCs from patients with rheumatoid arthritis. PLoS One 12, e0186795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, Birnbaum NS, Burmester GR, Bykerk VP, Cohen MD et al (2010) 2010 Rheumatoid arthritis classification criteria: an american college of rheumatology/european league against rheumatism collaborative initiative. Arthritis Rheum 62, 2569–2581. [DOI] [PubMed] [Google Scholar]
- 24. Jiang H, Ma R, Zou S, Wang Y, Li Z and Li W (2017) Reconstruction and analysis of the lncRNA‐miRNA‐mRNA network based on competitive endogenous RNA reveal functional lncRNAs in rheumatoid arthritis. Mol Biosyst 13, 1182–1192. [DOI] [PubMed] [Google Scholar]
- 25. Ji Z, Song R, Regev A and Struhl K (2015) Many lncRNAs, 5'UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4, e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ponting CP, Oliver PL and Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136, 629–641. [DOI] [PubMed] [Google Scholar]
- 27. Luo L, Wang M, Li X, Tian J, Zhang K, Tan S and Luo C (2020) Long non‐coding RNA LOC285194 in cancer. Clin Chim Acta 502, 1–8. [DOI] [PubMed] [Google Scholar]
- 28. Fan H, Yuan J, Li X, Ma Y, Wang X, Xu B and Li X (2020) LncRNA LINC00173 enhances triple‐negative breast cancer progression by suppressing miR‐490‐3p expression. Biomed Pharmacother 125, 109987. [DOI] [PubMed] [Google Scholar]
- 29. Song J, Kim D, Han J, Kim Y, Lee M and Jin EJ (2015) PBMC and exosome‐derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med 15, 121–126. [DOI] [PubMed] [Google Scholar]
- 30. Metsios GS, Moe RH, van der Esch M, van Zanten JJCSV, Fenton SAM, Koutedakis Y, Vitalis P, Kennedy N, Brodin N, Bostrom C et al (2020) The effects of exercise on cardiovascular disease risk factors and cardiovascular physiology in rheumatoid arthritis. Rheumatol Int 40, 347–357. [DOI] [PubMed] [Google Scholar]
- 31. Sidiropoulos PI, Goulielmos G, Voloudakis GK, Petraki E and Boumpas DT (2008) Inflammasomes and rheumatic diseases: evolving concepts. Ann Rheum Dis 67, 1382–1389. [DOI] [PubMed] [Google Scholar]
- 32. Mousavi MJ, Jamshidi A, Chopra A, Aslani S, Akhlaghi M and Mahmoudi M (2018) Implications of the noncoding RNAs in rheumatoid arthritis pathogenesis. J Cell Physiol 234, 335–347. [DOI] [PubMed] [Google Scholar]
- 33. Deviatkin AA, Vakulenko YA, Akhmadishina LV, Tarasov VV, Beloukhova MI, Zamyatnin AA and Lukashev AN (2020) Emerging concepts and challenges in rheumatoid arthritis gene therapy. Biomedicines 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heward JA and Lindsay MA (2014) Long non‐coding RNAs in the regulation of the immune response. Trends Immunol 35, 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


