Abstract
CD226, a member of the immunoglobulin superfamily, is a functional protein initially expressed on natural killer and T cells. In recent years, the function of CD226 has been increasingly realized and researched. Accumulating evidence shows that CD226 is closely related to the occurrence of autoimmune diseases, infectious diseases, and tumors. Because of the CD226’s increasing importance, the author herein discusses the structure, mechanism of action, and role of CD226 in various pathophysiological environments, allowing for further understanding of the function of CD226 and providing the basis for further research in related diseases.
Keywords: CD226, adhesion factor, immune, autoimmune disease, tumor
The Structure of CD226
Discovery and Naming
CD226, namely T lineage specific activation antigen 1 (TLisA1), platelet and T cell antigen 1 (PTA-1), or DNAM-1, is a member of the immunoglobulin superfamily. The molecular membrane contains two V-like domains of immunoglobulin (Shibuya et al., 1996; Sherrington et al., 1997). Burns et al. first found that CD226 was expressed on the surface of T cells in 1985, documenting that it was related to the activation of CTL, so it was named TLisA1 (Burns et al., 1986). Later studies revealed that CD226 was also expressed in platelets and involved in platelet activation aggregation, so it was called PTA1 (Shibuya et al., 1996). In 1997, Burns et al. confirmed that TLisA1 and DNAM-1 are actually the same molecule (Sherrington et al., 1997). In 2000, at the Seventh International Conference of human leukocyte differentiation antigen collaboration group, the molecule was officially named CD226 (Mason et al., 2001).
Gene and Structure
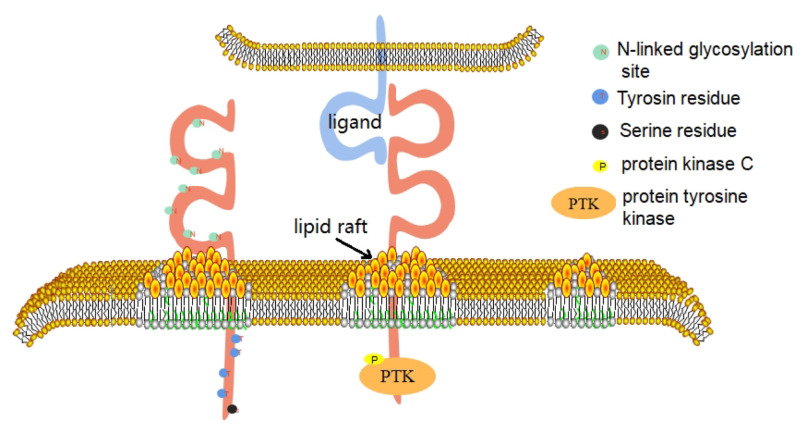
CD226 is a conserved sequence in human and mouse genes, which are located in the 18q22.3 and 18E4 bands of the chromosome, respectively (Jinlong et al., 2002). In 2002, the CD226 gene of mice was successfully identified, which has a total length of 2,487 BP. It contains an open reading frame of 1002 BP and encodes a leading sequence of 18 amino acids and a mature CD226 protein of 315 amino acids (Xinhai et al., 2002). In 2006, the promoter sequence of human CD226 gene was identified (Jian et al., 2006). The gene has at least two promoters, which are located at −810 to −287 bp and +33 to +213 bp, that have distinct tissue-specific roles and are physically separated by a negative regulatory element (Jian et al., 2006). Human CD226 gene contains 7 exons and 6 introns, of which exon 7 encodes 41 amino acids in the cytoplasmic region (Jinlong et al., 2002). Bioinformatics analysis shows that, within this CD226 gene region, there are putative binding sites for transcription factors AP-1, Sp1, PEA3, and Ets-1 (Jian et al., 2006). The full length of human CD226 cDNA is 2,664 BP, which contains an open reading frame and encodes a leading sequence of 18 amino acids and a mature CD226 protein of 318 amino acids. The extracellular domain contains 230 amino acids, including 2 immunoglobulin V-like domains, and 8 N-linked glycosylation sites and it is thus extremely susceptible to degradation of its glycan residues; deglycosylation yields a 35 kDa protein (Shibuya et al., 1996). The transmembrane domain contains 28 amino acids. The intracellular domain contains 60 amino acids. The intracellular domain contains 4 tyrosine residues and 1 serine residue, which can collect signal proteins after phosphorylation. This reaction is based on the interaction between CD226 and its ligand (Jun et al., 2012). The first domain outside the envelope of CD226 molecule is its structural basis for recognition of ligands, adhesion, immune synapse formation, and cytotoxic effect (Shengke et al., 2014; Figure 1).
FIGURE 1.
CD226 consists of three domains. The extracellular domain includes 2 immunoglobulin V-like domains and 8 N-linked glycoprotein sites. The first domain outside the envelope of CD226 molecule is its structural basis for recognition of ligands, adhesion, immune synapse formation and cytotoxic effect. The intracellular domain contains 4 tyrosine residues and 1 serine residue. When CD226 binds to its ligand, the CD226 molecule directionally moves to lipid rafts on the cell membrane and recruits intracellular signaling molecules such as PTK and PKC, which phosphorylate on the four residues and activate the cell.
There are three kinds of single nucleotide mutations in the exon of CD226 gene, including CD226 rs763361, rs34794968, and rs727088 (Bossini-Castillo et al., 2012). These three single nucleotide polymorphisms of CD226 gene have been confirmed to be related to the susceptibility of a variety of autoimmune diseases (Bossini-Castillo et al., 2012). CD226 rs763361/gly307ser non-synonymous mutations are associated with susceptibility to autoimmune diseases such as type 1 diabetes (T1D), rheumatoid arthritis (RA), multiple sclerosis (MS), autoimmune thyroid disease (AITD), and systemic sclerosis (SSc; Todd et al., 2007; Qiu et al., 2008; Smyth et al., 2008; Dieudé et al., 2011). Because the non-synonymous mutation encoded by this allele can encode the cytoplasmic tail (exon 7) of CD226 protein, this mutation may affect the function of T cells or other cells (Todd et al., 2007). Indeed, T cell and other cell dysfunction is closely associated with the onset and development of autoimmune diseases (Li et al., 2013; Maogen et al., 2014; Yaoyao et al., 2018). Other hypotheses suggest that this mutation may disrupt the binding sites of enhancers and/or silencers, thereby changing RNA splicing (Todd et al., 2007). In addition, the replacement of rs763361 with serine may also interfere with the phosphorylation of 322 tyrosine and 329 serine sites of CD226, as well as post-translational modifications in downstream signaling pathways (Todd et al., 2007; Boquan and Zhuwei, 2010). CD226 rs727088, a single nucleotide polymorphism, may influenc the expression of CD226 at the transcription level, which has been found to be highly correlated with tumor susceptibility (Löfgren et al., 2010). The single nucleotide mutation of CD226 rs34794968 alone does not affect the occurrence and development of the disease, but it can play a synergistic role with the above two mutations (Bossini-Castillo et al., 2012).
Expression and Distribution
The expression patterns of CD226 are varied (Ralston et al., 2004). In the peripheral blood, CD226 is expressed on T cells, NK cells, NK T cells, B cells, monocyte/macrophage, dendritic cells (DC), megakaryocyte/platelet lineage, and hematopoietic precursor cells (Burns et al., 1986; Scott et al., 1989; Kojima et al., 2003; Shibuya et al., 2003; Reymond et al., 2004; Dongchu et al., 2005). Endothelial cells also display low quantities of this protein in resting conditions, yet expression is significantly enhanced upon their stimulation (Lihua et al., 2003). CD226 is also expressed on mature mast cell and on bone marrow-derived CD34 + progenitor cells, but not on progenitors of the erythrocytic lineage (Dongchu et al., 2005; Bachelet et al., 2006). It was demonstrated that CD226 + NK cells play an important role in the recognition of several types of human tumors, such as myeloma, melanoma, and ovarian carcinoma, and Recent studies have shown that CD226 may be one of the markers of mature NK cells (Martinet et al., 2015). We previously reported that CD226 + NK cells are elevated in lupus and predominately infiltrated the lupus kidney. In addition, these activated NK cells mediated tissue injury by producing cytotoxic granules, eventually contributing to lupus nephritis (Huang et al., 2011).
Ligands of CD226
In 2003, Bottino et al. confirmed that the ligand of human CD226 was CD155 (nacl-5, PVR) and CD112 (nicotine-2) which have a similar molecular weight, 70 kD, and 65/60 KD. They belong to the members of the nicotine-like protein family and the nicotine protein family (Bottino et al., 2003; Wang et al., 2009). In 2005, Tahara-Hanaoka et al. (2005) demonstrated that the ligands of mouse CD226 molecules are also CD155 and CD112. CD155 and CD112 molecules are widely expressed in a variety of tissue cells, such as nerve cells, endothelial cells, epithelial cells, antigen-presenting cells, fibroblasts, pathogen infected cells, and a variety of tumor cells (Bottino et al., 2003; Tahara-Hanaoka et al., 2004; Dardalhon et al., 2005; Bryceson et al., 2006; Pende et al., 2006; Gilfillan et al., 2008; Kraus et al., 2016). The solid cancers, as well as hematological malignancies, have high levels of CD155 and CD112 making them good targets for CTL attack via CD226-specific binding (Castriconi et al., 2004; Pende et al., 2005; Moretta et al., 2006; Carlsten et al., 2007; El-Sherbiny et al., 2007). They are also expressed in immune cells such as monocytes, DCs, and activated T cells (Pende et al., 2006), and also affect some physiological processes.
The Function of CD226
CD226 Participates in the Function of CTL Cells and NK Cells
In human body, CD226 is highly expressed on the surface of NK cells and CD8 + T cells (Shibuya et al., 1996). In mice, 40–50% of NK cells and all CTL cells constitutively express CD226 (Dardalhon et al., 2005).
CD226, as an adhesion molecule, promotes the migration, activation, proliferation, differentiation, and function of CD8 + T cells. CTL cells need to migrate to the inflammatory site or tumor microenvironment through adhesion molecules to establish close contact (Bryceson et al., 2006). In the secondary lymphoid organs, adhesion molecules mediate the interaction between CTL cells and antigen-presenting cells, and finally activate, proliferate and differentiate them. The interaction of CD226/CD155 is very important for the proliferation of CD8 + T cells and the immune response of antigen-specific CD8 + T cells (Gilfillan et al., 2008). The co-stimulatory signaling pathway of CD8 + T cells mediated by CD226 was interrupted in the process of chronic HIV-1 infection. Therefore, the expression of CD226 on the surface of CD8 + T cells is down regulated, which impairs the effect of CTL (Cella et al., 2010).
More and more evidences show that CD226 is involved in the biological function of NK cells (Enqvist et al., 2015; Martinet and Smyth, 2015; Martinet et al., 2015). The combination of CD226 with CD155 and/or CD112, in cooperation with NKp30, can induce NK cells to dissolve immature DC and promote the proliferation of mature DC (Balsamo et al., 2009). DC maturation promotes immune response by enhancing adaptive immune system (Zheng et al., 2006; Ramalingam et al., 2012; Peng et al., 2020). The interaction between CD226 and its ligands is involved in the cross-linking of NK and T cells. In graft versus host disease (GVHD; Lozano et al., 2013) and other autoimmune diseases (Ardolino et al., 2011), NK cells can recognize and kill antigen stimulated T cells that have been activated/proliferated and can also promote the differentiation of helper T cells.
CD226, together with CD96, TIGIT, and CRTAM, is also involved in the regulation of NK cell function (Ralston et al., 2004; Chan et al., 2014; Nabekura et al., 2014; Shu-Bin et al., 2020). After LPS stimulation, the proportion of IFN-γ + NK cells in CD226 deficient mice was significantly lower than that in wild-type mice (Chan et al., 2014). CD96 is believed to inhibit the secretion of IFN-γ by NK cells. Therefore, CD96 and CD226 molecules reverse the secretion of IFN-γ in NK cells (Chan et al., 2014). TIGIT, CD96, and CRTAM are able to recognize the nectin and Necl family molecules and regulate the function of NK cells, which further complicates the research of CD226 in the biological function of NK cells (Chan et al., 2014). In recent years, it has been reported that the synergistic effect of CD226 and these three molecules balances the activation of NK cells in vivo (Martinet and Smyth, 2015).
CD226 Participates in the Function of CD4 + T Cells
In 2005, Dardalhon et al. (2005) stated that CD226 was a specific surface marker of Th1 cells in mice. In 2006, Shibuya et al. revealed that the freshly isolated CD4 + T cells in the mice also expressed low level CD226 molecules, that initial T cells expressed CD226 molecules, and that the most polarized Th1 and Th2 cells also expressed CD226 molecules (Shibuya et al., 2006). In 2012, Lozano et al. demonstrated that TIGIT can inhibit T cell functions by competing with CD226 (Lozano et al., 2012). In 2015, Fuhrman et al. found that CD226 was expressed in memory CD4 + T cells (Fuhrman et al., 2015). These abnormal cells and relative pro-inflammatory cytokines contribute to many immunologic diseases (Yang and Song Guo, 2016; Sujuan et al., 2018, 2019).
In 2013, Sutavani et al. (2013) proved that the surface marker of human type 1 regulatory T cells (Tr1) is CD4 + CD49b + LAG-3 + CD226+. Tr1 is a kind of chronic activated CD4 + T cells in the presence of IL-10 (Weishan et al., 2017; Fang et al., 2018), which has the functions of low proliferation, high secretion of IL-10, low expression of IL-2, and IL-4 (Gagliani et al., 2013). CD226 is highly expressed on the surface of human type I regulatory T cells and participates in the killing of myeloid antigen presenting cells (Magnani et al., 2011). Tr1 has the function of down regulating immune response and maintaining peripheral immune tolerance. It has the application prospect in the treatment of autoimmune diseases, tumors, and the rejection of allogeneic tissue and organ transplantation.
Recently, it has been found that CD226 and TIGIT molecules expressed on the surface of human CD4 + Foxp3 + Treg cells are related to their stability and inhibition. Similar to Tr1 cells, CD4 + Foxp3 + Treg are crucial for maintaining homeostasis and preventing autoimmune issues (Ya et al., 2014; Anping et al., 2016). The purity and inhibitory function of Treg subsets of CD226 + TIGIT- will be weakened after amplification, and with the increase of IL-10 and effector cytokines, it is suggested that the expression of CD226 affects the function of Treg (Fuhrman et al., 2015). Foxp3, Helios were highly expressed in CD226-TIGIT + Treg cells and with Treg-specific demethylated region. In vitro inhibition experiments show that TIGIT expression in Treg cells is related to its strong inhibitory activity. On the other hand, the expression level of CD226 in activated Treg cells is up-regulated. Therefore, in the process of looking for blocking CD226 to weaken the activity of traditional effector T cells, attention should be focused on appropriate doses to avoid the simultaneous reduction of Treg function (Fuhrman et al., 2015). However, in patients with RA, we recently demonstrated that while CD226 and TIGIT both showed elevated expression levels in CD4 + Foxp3 + cells, they were not associated with disease activity of RA patients (Mengru et al., 2019). Thus, CD226 doesn’t seem to be an ideal marker for human Treg cells.
CD226 Is Involved in the Function of Other Cells
CD226 is also expressed in platelets. The cross-linking of CD226 and mAb can induce platelet activation, allowing thrombin to induce CD226 tyrosine phosphorylation and mediate platelet adhesion (Scott et al., 1989). Subsequently, it is documented that CD226 mediates the adhesion of platelets and megakaryocytes to vascular endothelial cells (Kojima et al., 2003).
Human mast cells and eosinophils can simultaneously express CD226 and CD112. These cells play an important role in promoting allergic diseases (Wenru et al., 2012, 2014, 2015). In late and chronic stages of anaphylactic inflammation, mast cells and eosinophils with tissue infiltration form a regulatory unit. In the presence of eosinophils, mast cell FCεRI mediated degranulation is enhanced. This effect is partly caused by the interaction of CD226/CD112 triggering Fyn, LAT, and phospholipase Cγ2 signaling pathways involved in the above progress (Bachelet et al., 2006) (Figure 2).
FIGURE 2.
As an adhesion molecule, Binding of CD226 and CD155 can mediate the interaction between CTL cells and professional antigen presenting cells, ultimately enabling them to activate, proliferate and differentiate. CD226 also can induces platelet activation and mediates platelet adhesion by cross-linking with its mAb. The killing of tumor cells by NK cells mainly depends on the binding of CD226 to CD112 and CD155 expressed on tumor cells. In the presence of eosinophils, mast cell FCεRI mediated degranulation is enhanced. This effect is partly caused by the interaction of CD226/CD112.
CD226 and Clinical Diseases
CD226 and Autoimmune Diseases
Previous studies indicated that the expression of CD226 is negatively correlated with the inhibitory function of Foxp3 + Tregs. TIGIT, a co-inhibitory molecule on T cells, exerts immunosuppressive effects by competing with CD226 for the same CD155 ligand (Lozano et al., 2012). TIGIT and CD226 costimulatory axis plays an important role in the immunoregulatory function of Foxp3 + Tregs and is related to several autoimmune diseases. Several lines of evidence support that the suppressive capacity of CD226 + Tregs is inhibited (Ning et al., 2019), and that TIGIT + Tregs are highly suppressive and more so (Joller et al., 2014). Experimental autoimmune encephalomyelitis (EAE) contributes to a breakdown of self-tolerance and leads to Th17 cells infiltrating the central nervous system to mediate inflammation and neuronal injury (Rostami and Ciric, 2013). Zhang et al. reported that EAE susceptibility in mice treated with anti-CD226 pAb was markedly decreased via balancing the Th17/Treg ratio (Rong et al., 2016), and the enhanced suppressive capacity of Tregs during EAE is related to the absence of CD226 and the increased expression levels of TIGIT (Ning et al., 2019). Ulcerative colitis (UC) is a chronic inflammatory immune-related disease. Long et al. found that lack of CD226 expression on Foxp3 + Tregs play a positive role in the recovery of clinical remission from active stage in UC patients and TIGIT expression on CD226-Foxp3 + Tregs have a potential positive effect on the suppression of CD226-Foxp3 + Tregs (Yan et al., 2020).
In recent years, with the development of sequencing technology, the relationship between gene and disease is becoming more clear. The study of CD226 gene polymorphism and autoimmune disease susceptibility has entered a new stage.
CD226 rs763361 is 307 glycine replaced by serine. This mutation may be related to the development of a series of immune related diseases, such as SLE, SSc, T1D, and RA (Du et al., 2011; Avouac et al., 2013; Mattana et al., 2014; Elhai et al., 2015). SSc is a chronic autoimmune disease that affects connective tissue, characterized by dermal fibrosis and skin thickening. In the CD226-/- mice model, the characteristics of fibrosis were weakened when compared to the wild-type mice. It is possible that the expression of CD226 promotes the development of SSc (Avouac et al., 2013). In the in vitro model of RA, CD226, and CD226Ls were expressed in NK cells and fibroblast like synovitis (FLS) of RA patients, respectively, suggesting that RA-FLS cells can be recognized and killed by NK cells (Nielsen et al., 2014). Many studies have found that autoimmune diseases are related to the dysfunction of NK cells. It is speculated that the 307 glycine serine mutation of CD226 may cause the dysfunction of NK cells (Fogel et al., 2013). The activation of T cells requires the guanine nucleotide exchange factor VAV1 (Katzav et al., 1989). CD226 engagement triggered VAV1 activation through tyrosine phosphorylation and synergized with signaling through T cell receptor (TCR) to positively regulate cytokine production by CD4 + T cells. Moreover, co-engagement of the TCR, and CD226 rs763361 that is associated with autoimmunity further enhanced VAV1 activation and IL-17 production (Gaud et al., 2018). All of the above results suggest the feasibility of targeting CD226 in the treatment of autoimmune diseases.
CD226 and Tumors
CD226 is expressed on the surface of NK cells and CD8 + T cells. As an important NK cell activating receptor, CD226 is widely involved in various immune responses. Studies have shown that CD226 plays an important role in the killing of tumor cells by NK cells (Verhoeven et al., 2008; Lakshmikanth et al., 2009). Formation of stable conjugates with tumor cells is essential for NK cells to exert tumor killing effects, and CD226 enables prolonged stable interaction between NK and tumor cells (Kim et al., 2017). CD155 and CD112 are two important ligands of CD226. CD112 is down regulated in tumor tissue. It can combine with CD226 on the surface of NK cells, thereby activating NK cells to kill tumor cells (Xiaojun et al., 2014). Similarly, the decrease of CD155 in liver cancer tissue can reduce the cytotoxic effect mediated by NK cells (Jiuyu et al., 2014). An experiment in vivo has demonstrated that CD226 mediates phosphorylation of FOXO1 and activates NK cells through interaction with CD155-expressing tumor cells (Xiangnan et al., 2018). Tumor immunotherapy offers promising outcomes in patients with tumor. Concepción et al. discovered the importance of the CD226/immunoglobulin-like receptor ratio of NK cells induced by licensing interactions as critical determinants for solid cancer immune surveillance, and provides predictive biomarkers for patient survival that may also improve the selection of donors for NK cells immunotherapy (Guillamón et al., 2018). It is generally believed that Tregs in tumors impede T cell responses to tumors (Naturally, 2005). Among the current tumor immunotherapy regimens targeting Tregs, the biggest problem is the lack of highly selective drugs for Tregs. This is mainly due to the fact that highly specific markers have not yet been found in Tregs. Julien et al. indicated that a high TIGIT/CD226 ratio in Tregs regulates their suppressive function and stability in melanoma, and they suggested novel immunotherapies to activate CD226 in Tregs together with TIGIT blockade to counteract Treg suppression in cancer patients (Fourcade et al., 2018). In recent years, some researches began to pay attention to the expression of CD226 in tumor cells. These studies found that expression of CD226 on hepatoma cells was down-regulated and that expression was related to the survival rate and survival time of patients (Baoxing et al., 2017). In addition, CD226 gene polymorphism was found to be directly related to tumor risk (Shaoqing et al., 2013). All of the above results suggest CD226 may have anti-tumor effects. Thus, the role and mechanism of CD226 in various cancers warrants an in-depth study in the future.
CD226 and Viral Infections
In most virus infected diseases, NK cells can eliminate virus-infected and transformed cells (Cooper et al., 2001; Smyth et al., 2005). They recognize target cells via a germ-line–encoded repertoire of activating and inhibitory receptors (Bryceson et al., 2006). CD226 is an important activating receptors. NK cells require CD226 for recognition of HCV-infected hepatoma cells (Stegmann et al., 2012) and HCMV-infected myeloid DCs (Magri et al., 2011). Human immunodeficiency virus (HIV) infection causes dysfunction of the innate and adaptive immune systems, and disturbs NK cells and CD8 + T cells function and the surface expression levels of some receptors (Watzl et al., 2014; Zhuo et al., 2017). Yin et al. (2018) demonstrates that TIGIT expression is specifically elevated on CD226 + NK cells in HIV-infected individuals, and high levels of TIGIT can inhibit IFN-γ production by NK cells, while blockade of TIGIT can restore their function. Similar results were found on CD8 + T cells in HIV-infected individuals. HIV-specific CD8 + T cells were almost exclusively TIGIT+, and HIV-specific TIGIThi cells were negatively correlated with polyfunctionality and displayed a diminished expression of CD226 (Tauriainen et al., 2017). All these highlight the important role of TIGIT/CD226 axis in viral infections and suggest a potential new avenue for the development of therapeutic strategies toward a functional cure.
Conclusion
Since its discovery, CD226 has been demonstrated to be widely expressed on various immune cells and plays an important functional role in the immune system. Many aspects of the pathophysiological state of CD226 have not yet been clarified; however, current research on the function and mechanisms of this molecule is allowing for increased understanding of the molecule’s clinical relevance. Indeed, CD226, as a costimulatory factor, plays an important role in the development of various diseases. Thus, manipulation of CD226 expression and function may be a feasible therapeutic strategy for many immune related diseases and tumors.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
ZH and GQ designed the study and wrote the review. SZ conceived and designed the review. SZ and JM edited and revised review. All authors discussed and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported in part by The National Natural Science Foundation of China (Grant Nos. 81460411 and 81660450), Guangxi Natural Science Foundation Project (Nos: 2017AD23009, 2017JJD10037, and 2015jjAA40450), and Key Lab Development Project from Guangxi (all to GQ).
References
- Anping X., Ya L., Weiqian C., Julie W., Youqiu X., Feng H., et al. (2016). TGF-β induced regulatory T cells directly suppress B cell responses through a noncytotoxic mechanism. J. Immunol. 196 3631–3641. 10.4049/jimmunol.1501740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardolino M., Zingoni A., Cerboni C., Cecere F., Soriani A., Iannitto M. L., et al. (2011). DNAM-1 ligand expression on Ag-stimulated T lymphocytes is mediated by ROS-dependent activation of DNA-damage response: relevance for NK-T cell interaction. Blood 117 4778–4786. 10.1182/blood-2010-08-300954 [DOI] [PubMed] [Google Scholar]
- Avouac J., Elhai M., Tomcik M., Ruiz B., Friese M., Piedavent M., et al. (2013). Critical role of the adhesion receptor DNAX accessory molecule-1 (DNAM-1) in the development of inflammation-driven dermal fibrosis in a mouse model of systemic sclerosis. Ann. Rheum. Dis. 72 1089–1098. 10.1136/annrheumdis-2012-201759 [DOI] [PubMed] [Google Scholar]
- Bachelet I., Munitz A., Mankutad D., Levi-Schaffer F. (2006). Mast cell costimulation by CD226/CD112 (DNAM-1/Nectin-2): a novel interface in the allergic process. J. Biol. Chem. 281 27190–27196. 10.1074/jbc.M602359200 [DOI] [PubMed] [Google Scholar]
- Balsamo M., Zambello R., Teramo A., Pedrazzi M., Sparatore B., Scordamaglia F., et al. (2009). Analysis of NK cell/DC interaction in NK-type lymphoproliferative disease of granular lymphocytes (LDGL): role of DNAM-1 and NKp30. Exp. Hematol. 37 1167–1175. 10.1016/j.exphem.2009.06.010 [DOI] [PubMed] [Google Scholar]
- Baoxing J., Ludong T., Zhe J., Yan J., Yu F., Yahui L. (2017). MiR-892a promotes hepatocellular carcinoma cells proliferation and invasion through targeting CD226. J. Cell Biochem. 118 1489–1496. 10.1002/jcb.25808 [DOI] [PubMed] [Google Scholar]
- Boquan J., Zhuwei X. (2010). A novel interface consisting of homologous immunoglobulin superfamily members with multiple functions. Cell. Mol. Immunol. 7 11–19. 10.1038/cmi.2009.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossini-Castillo L., Simeon C. P., Beretta L., Broen J. C., Vonk M. C., Espinosa G., et al. (2012). A multicenter study confirms CD226 gene association with systemic sclerosis-related pulmonary fibrosis. Arthrit. Res. Ther. 12:R85. 10.1186/ar3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottino C., Castriconi R., Pende D., Rivera P., Nanni M., Carnemolla B., et al. (2003). Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J. Exp. Med. 198 557–567. 10.1084/jem.20030788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryceson Y. T., March M. E., Ljunggren H.-G., Long E. O. (2006). Activation, coactivation, and costimulation of resting human natural killer cells. Immunol. Rev. 214 73–91. 10.1111/j.1600-065X.2006.00457.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns G. F., Triglia T., Werkmeister J. A., Begley C. G., Boyd A. W. (1986). TLiSA1, a human T lineage-specific activation antigen involved in the differentiation of cytotoxic T lymphocytes and anomalous killer cells from their precursors. J. Exp. Med. 161 1063–1078. 10.1084/jem.161.5.1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsten M., Björkström N. K., Norell H., Bryceson Y. (2007). DNAX Accessory molecule-1 mediated recognition of freshly isolated ovarian carcinoma by resting natural killer cells. Cancer Res. 67 1317–1325. 10.1158/0008-5472.CAN-06-2264 [DOI] [PubMed] [Google Scholar]
- Castriconi R., Dondero A., Corrias M. V., Lanino E., Pende D., Moretta L., et al. (2004). Natural killer cell-mediated killing of freshly isolated neuroblastoma cells: critical role of DNAX accessory molecule-1-poliovirus receptor interaction. Cancer Res. 64 9180–9184. 10.1158/0008-5472.CAN-04-2682 [DOI] [PubMed] [Google Scholar]
- Cella M., Presti R., Vermi W., Lavender K., Turnbull E., Ochsenbauer-Jambor C., et al. (2010). Loss of DNAM-1 contributes to CD8+ T-cell exhaustion in chronic HIV-1 infection. Eur. J. Immunol. 40 949–954. 10.1002/eji.200940234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. J., Martinet L., Gilfillan S., Souza-Fonseca-Guimaraes F., Chow M. T., Town L., et al. (2014). The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 15 431–438. 10.1038/ni.2850 [DOI] [PubMed] [Google Scholar]
- Cooper M. A., Fehniger T. A., Caligiuri M. A. (2001). The biology of human natural killer-cell subsets. Trends Immunol. 22 633–640. 10.1016/s1471-4906(01)02060-9 [DOI] [PubMed] [Google Scholar]
- Dardalhon V., Schubart A. S., Reddy J., Meyers J. H., Monney L., Sabatos C. A., et al. (2005). CD226 is specifically expressed on the surface of Th1 cells and regulates their expansion and effector functions. J. Immunol. 175 1558–1565. 10.4049/jimmunol.175.3.1558 [DOI] [PubMed] [Google Scholar]
- Dieudé P., Guedg M. E., Wipff J., Revillod L., Riemekasten G., Matucci-Cerinic M., et al. (2011). Association of the CD226 Ser(307) variant with systemic sclerosis: evidence of a contribution of costimulation pathways in systemic sclerosis pathogenesis. Arthrit. Rheum. 63 1097–1105. 10.1002/art.30204 [DOI] [PubMed] [Google Scholar]
- Dongchu M., Yinghui S., Di L., Haoyang W., Bing D., Xinghai Z., et al. (2005). CD226 is expressed on the megakaryocytic lineage from hematopoietic stem cells/progenitor cells and involved in its polyploidization. Eur. J. Haematol. 74 228–240. 10.1111/j.1600-0609.2004.00345.x [DOI] [PubMed] [Google Scholar]
- Du Y., Tian L., Shen L. X., Wang F., Yu L. K., Song Y., et al. (2011). Association of the CD226 single nucleotide polymorphism with systemic lupus erythematosus in the Chinese Han population. Tissue Antig. 77 65–67. 10.1111/j.1399-0039.2010.01568.x [DOI] [PubMed] [Google Scholar]
- Elhai M., Chiocchia G., Marchiol C., Lager F., Renault G., Colonna M., et al. (2015). Targeting CD226/DNAX accessory molecule-1 (DNAM-1) in collagen-induced arthritis mouse models. J. Inflamm. 12:9. 10.1186/s12950-015-0056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sherbiny Y. M., Meade J. L., Holmes T. D., McGonagle D., Mackie S. L., Morgan A. W., et al. (2007). The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Res. 67 8444–8449. 10.1158/0008-5472.CAN-06-4230 [DOI] [PubMed] [Google Scholar]
- Enqvist M., Ask E. H., Forslund E., Carlsten M., Abrahamsen G., Béziat V., et al. (2015). Coordinated expression of DNAM-1 and LFA-1 in educated NK cells. J. Immunol. 194 4518–4527. 10.4049/jimmunol.1401972 [DOI] [PubMed] [Google Scholar]
- Fang L., Xiao Xue F., Shang Ling Z., Hong Y. H., Ying D. C., Yun F. P., et al. (2018). Sonic hedgehog signaling pathway mediates proliferation and migration of fibroblast-like synoviocytes in rheumatoid arthritis via MAPK/ERK signaling pathway. Front. Immunol. 9:2847. 10.3389/fimmu.2018.02847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel L. A., Yokoyama W. M., French A. R. (2013). Natural killer cells in human autoimmune disorders. Arthrit. Res. Ther. 15:216. 10.1186/ar4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourcade J., Zhaojun S., Chauvin J.-M., Ka M., Davar D., Pagliano O., et al. (2018). Cd226 opposes tigit to disrupt tregs in melanoma. JCI Insight. 3:e121157. 10.1172/jci.insight.121157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman C. A., Yeh W.-I., Seay H. R., Lakshmi P. S., Chopra G., Zhang L., et al. (2015). Divergent phenotypes of human regulatory T cells expressing the receptors TIGIT and CD226. J. Immunol. 195 145–155. 10.4049/jimmunol.1402381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliani N., Magnani C. F., Huber S., Gianolini M. E., Pala M., Licona-Limon P., et al. (2013). Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat. Med. 19 739–746. 10.1038/nm.3179 [DOI] [PubMed] [Google Scholar]
- Gaud G., Roncagalli R., Chaoui K., Bernard I., Familiades J., Colacios C., et al. (2018). The costimulatory molecule CD226 signals through VAV1 to amplify TCR signals and promote IL-17 production by CD4+T cells. Sci. Signal. 11:eaar3083. 10.1126/scisignal.aar3083 [DOI] [PubMed] [Google Scholar]
- Gilfillan S., Chan C. J., Cella M., Haynes N. M., Rapaport A. S., Boles K. S., et al. (2008). DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J. Exp. Med. 205 2965–2973. 10.1084/jem.20081752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillamón F. C., Martínez-sánchez V. M., MRowiec G. L. (2018). NK cell education in tumor immune surveillance: DNAM-1/KIR receptor ratios as predictive biomarkers for solid tumor outcome. Cancer Immunol. Res. 6 1537–1547. 10.1158/2326-6066 [DOI] [PubMed] [Google Scholar]
- Huang Z., Fu B., Zheng S. G., Li X., Sun R., Tian Z., et al. (2011). Involvement of CD226+ NK cells in immunopathogenesis of systemic lupus erythematosus. J. Immunol. 186 3421–3431. 10.4049/jimmunol.1000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian J. L., Zhu C. S., Xu Z. W., Ouyang W. M., Ma D. C., Zhang Y., et al. (2006). Identification and characterization of the CD226 gene promoter. J. Biol. Chem. 281 28731–28736. 10.1074/jbc.M601786200 [DOI] [PubMed] [Google Scholar]
- Jinlong K., Yong Z., Demin L., Weiming O., Boquan J. (2002). Analysis of promoter sequence and SNP site of human CD226 gene. Chin. J. Cell. Mol. Immunol. 18 203–207. 10.3321/j.issn:1007-8738.2002.03.002 30704229 [DOI] [Google Scholar]
- Jiuyu G., Liang F., Rongrong L., Ying W., Jinliang X., Yibing C., et al. (2014). UPR decreases CD226 ligand CD155 expression and sensitivity to NK cell-mediated cytotoxicity in hepatoma cells. Eur. J. Immunol. 44 3758–3767. 10.1002/eji.201444574 [DOI] [PubMed] [Google Scholar]
- Joller N., Lozano E., Burkett P. R., Patel B., Xiao S., Zhu C., et al. (2014). Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity 40 569–581. 10.1016/j.immuni.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun L., Xiaomin Q., Zhujun C., Xiang X., Feng G., Shujun Z., et al. (2012). Crystal structure of cell adhesion molecule nectin-2/CD112 and its binding to immune receptor DNAM-1/CD226. J. Immunol. 188 5511–5520. 10.4049/jimmunol.1200324 [DOI] [PubMed] [Google Scholar]
- Katzav S., Martin-Zanca D., Barbacid M. (1989). Vav, a novel human oncogene derived from a locus ubiquitously expressed in hematopoietic cells. EMBO J. 8 2283–2290. 10.1002/j.1460-2075.1989.tb08354.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Shin B. R., Lee H. K., Lee J. H., Kim K. H., Choi J. E., et al. (2017). Cd226-/- natural killer cells fail to establish stable contacts with cancer cells and show impaired control of tumor metastasis in vivo. Oncoimmunology 6:e1338994. 10.1080/2162402x.2017.1338994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H., Kanada H., Shimizu S., Kasama E., Shibuya K., Nakauchi H., et al. (2003). CD226 mediates platelet and megakaryocytic cell adhesion to vascular endothelial cells. J. Biol. Chem. 278 36748–36753. 10.1074/jbc.M300702200 [DOI] [PubMed] [Google Scholar]
- Kraus A. K., Chen J., Edenhofer I., Ravens I., Gaspert A., Cippà P. E., et al. (2016). The role of T cell costimulation via DNAM-1 in kidney transplantation. PLoS One 11:e0147951. 10.1371/journal.pone.0147951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmikanth T., Burke S., Ali T. H., Kimpfler S., Ursini F., Ruggeri L., et al. (2009). NCRs and DNAM-1 mediate NK Cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J. Clin. Invest. 119 1251–1263. 10.1172/JCI36022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Wang J. C., Liang T. H., Zhu M. H., Wang J. Y., Fu X. L., et al. (2013). Pathologic finding of increased expression of interleukin-17 in the synovial tissue of rheumatoid arthritis patients. Int. J. Clin. Exp. Pathol. 6 1375–1379. 10.1159/000354821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lihua C., Xin X., Xinhai Z., Wei J., Jinlong J., Chaojun S., et al. (2003). The expression, regulation and adhesion function of a novel CD molecule, CD226, on human endothelial cells. Life Sci. 73 2373–2382. 10.1016/s0024-3205(03)00606-4 [DOI] [PubMed] [Google Scholar]
- Löfgren S. E., Delgado-Vega A. M., Gallant C. J., Sánchez E., Frostegård J., Truedsson L., et al. (2010). A 3’-untranslated region variant is associated with impaired expression of CD226 in T and natural killer T cells and is associated with susceptibility to systemic lupus erythematosus. Arthrit. Rheum. 62 3404–3414. 10.1002/art.27677 [DOI] [PubMed] [Google Scholar]
- Lozano E., Dominguez-Villar M., Kuchroo V., Hafler D. A. (2012). The TIGIT/CD226 axis regulates human t cell function. J. Immunol. 188 3869–3875. 10.4049/jimmunol.1103627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano E., Joller N., Cao Y., Kuchroo V. K., Hafler D. A. (2013). The CD226/CD155 interaction regulates the proinflammatory (Th1/Th17)/anti-inflammatory (Th2) balance in humans. J. Immunol. 191 3673–3680. 10.4049/jimmunol.1300945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani C. F., Alberigo G., Bacchetta R., Serafini G., Andreani M., Roncarolo M. G., et al. (2011). Killing of myeloid APCs via HLA class I, CD2 and CD226 defines a novel mechanism of suppression by human Tr1 cells. Eur. J. Immunol. 41 1652–1662. 10.1002/eji.201041120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri G., Muntasell A., Romo N., Sáez-Borderías A., Pende D., Geraghty D. E., et al. (2011). NKp46 and DNAM-1 NK-cell receptors drive the response to human cytomegalovirus-infected myeloid dendritic cells overcoming viral immune evasion strategies. Blood 117 846–856. 10.1182/blood-2010-08-301374 [DOI] [PubMed] [Google Scholar]
- Maogen C., Xiaohong L., Ya L., Qiang L., Yiling D., Zhongmin L., et al. (2014). The function of BAFF on T helper cells in autoimmunity. Cytokine Growth Fact. Rev. 25 301–305. 10.1016/j.cytogfr.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet L., Andrade L. F. D., Guillerey C., Lee J. S., Liu J., Souza-Fonseca-Guimaraes F., et al. (2015). DNAM-1 expression marks an alternative program of NK cell maturation. Cell Rep. 11 85–97. 10.1016/j.celrep.2015.03.006 [DOI] [PubMed] [Google Scholar]
- Martinet L., Smyth M. J. (2015). Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol. 15 243–254. 10.1038/nri3799 [DOI] [PubMed] [Google Scholar]
- Mason D., André P., Bensussan A., Buckley C., Civin C., Clark E., et al. (2001). CD Antigens 2001. J. Leukoc. Biol. 211 685–690. 10.1006/cimm.2001.1831 [DOI] [PubMed] [Google Scholar]
- Mattana T. C. C., Santos A. S., Fukui R. T., Mainardi-Novo D. T. O., Costa V. S., Santos R. F., et al. (2014). CD226 rs763361 is associated with the susceptibility to type 1 diabetes and greater frequency of GAD65 autoantibody in a Brazilian cohort. Med. Inflamm. 2014:694948. 10.1155/2014/694948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengru Y., Yan L., Biyao M., Youqiu X., Congxiu Y., Yutong J., et al. (2019). Helios but not CD226, TIGIT and Foxp3 is a potential marker for CD4+ Treg cells in patients with rheumatoid arthritis. Cell. Physiol. Biochem. 52 1178–1192. 10.33594/000000080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta L., Bottino C., Pende D., Castriconi R., Mingari M. C., Moretta A. (2006). Surface NK receptors and their ligands on tumor cells. Semin. Immunol. 18:151. 10.1016/j.smim.2006.03.002 [DOI] [PubMed] [Google Scholar]
- Nabekura T., Kanaya M., Shibuya A., Fu G., Gascoigne N. R. J., Lanier L. L. (2014). Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity 40 225–234. 10.1016/j.immuni.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naturally S. S. (2005). arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat. Immunol. 6 345–352. 10.1038/ni1178 [DOI] [PubMed] [Google Scholar]
- Nielsen N., Pascal V., Fasth A. E. R., Sundström Y., Galsgaard E. D., Ahern D., et al. (2014). Balance between activating NKG2D, DNAM-1, NKp44 and NKp46 and inhibitory CD94/NKG2A receptors determine natural killer degranulation towards rheumatoid arthritis synovial fibroblasts. Immunology 142 581–593. 10.1111/imm.12271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning W., Shuang L., Jingyi J., Liang F., Qianli M., Xian W., et al. (2019). CD226 attenuates Treg suppressive capacity via CTLA-4 and TIGIT during EAE. Immunol. Res. 67 486–496. 10.1007/s12026-019-09112-9 [DOI] [PubMed] [Google Scholar]
- Pende D., Bottino C., Castriconi R., Cantoni C., Marcenaro S., Rivera P., et al. (2005). PVR (CD155) and Nectin-2 (CD112) as ligands of the human DNAM-1 (CD226) activating receptor: involvement in tumor cell lysis. Mol. Immunol. 42 463–469. 10.1016/j.molimm.2004.07.028 [DOI] [PubMed] [Google Scholar]
- Pende D., Castriconi R., Romagnani P., Spaggiari G. M., Marcenaro S., Dondero A., et al. (2006). Expression of the DNAM-1 ligands, Nectin-2 (CD112) and poliovirus receptor (CD155), on dendritic cells: relevance for natural killer-dendritic cell interaction. Blood 107 2030–2036. 10.1182/blood-2005-07-2696 [DOI] [PubMed] [Google Scholar]
- Peng Y.-Q., Qin Z.-L., Fang S.-B., Xu Z.-B., Zhang H.-Y., Chen D., et al. (2020). Effects of myeloid and plasmacytoid dendritic cells on ILC2s in patients with allergic rhinitis. J. Allergy Clin. Immunol. 145 855–867. 10.1016/j.jaci.2019.11.029 [DOI] [PubMed] [Google Scholar]
- Qiu Z., Zhang K., Qiu X., Zhou M., Li W. (2008). CD226 Gly307Ser association with multiple autoimmune diseases: a meta-analysi. Hum. Immunol. 74 249–255. 10.1016/j.humimm.2012.10.009 [DOI] [PubMed] [Google Scholar]
- Ralston K. J., Hird S. L., Xinhai Z., Scott J. L., Boquan J., Thorne R. F., et al. (2004). The LFA-1-associated molecule PTA-1 (CD226) on T cells forms a dynamic molecular complex with protein 4.1G and human discs large. J. Biol. Chem. 279 33816–33828. 10.1074/jbc.M401040200 [DOI] [PubMed] [Google Scholar]
- Ramalingam R., Larmonier C. B., Thurston R. D., Midura-Kiela M. T., Zheng S. G., Ghishan F. K., et al. (2012). Dendritic cell-specific disruption of TGF-β receptor II leads to altered regulatory T cell phenotype and spontaneous multiorgan autoimmunity. J. Immunol. 189 3878–3893. 10.4049/jimmunol.1201029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond N., Anne-Marie I., Devilard E., Fabre S., Chabannon C., Xerri L., et al. (2004). DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J. Exp. Med. 199 1331–1341. 10.1084/jem.20032206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Z., Hanyu Z., Yun Z., Kun C., Chunmei Z., Chaojun S., et al. (2016). CD226 ligation protects against EAE by promoting IL-10 expression via regulation of CD4+ T cell differentiation. Oncotarget 7 19251–19264. 10.18632/oncotarget.7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostami A., Ciric B. (2013). Role of Th17 cells in the pathogenesis of CNS inflammatory demyelination. J. Neurol. Sci. 333 76–87. 10.1016/j.jns.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. L., Dunn S. M., Jin B., Hillam A. J., Walton S., Berndt M. C., et al. (1989). Characterization of a novel membrane glycoprotein involved in platelet activation. J. Biol. Chem. 264 13475–13482. [PubMed] [Google Scholar]
- Shaoqing S., Bin Z., Kui Z., Lin Z. (2013). Association between two genetic variants of CD226 gene and cervical squamous cell carcinoma: a case-control study. Gene 519 159–163. 10.1016/j.gene.2012.11.039 [DOI] [PubMed] [Google Scholar]
- Shengke H., Kuikui G., Xiaodong Z., Haiming W., Rui S., Zhigang T. (2014). CD226 protein is involved in immune synapse formation and triggers natural killer (NK) cell activation via its first extracellular domain. J. Biol. Chem. 289 6969–6977. 10.1074/jbc.M113.498253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrington P. D., Scott J. L., Jin B., Simmons D., Dorahy D. J., Lloyd J., et al. (1997). TLiSA1 (PTA1) activation antigen implicated in T Cell differentiation and platelet activation is a member of the immunoglobulin superfamily exhibiting distinctive regulation of expression. J. Biol. Chem. 272 21735–21744. 10.1074/jbc.272.35.21735 [DOI] [PubMed] [Google Scholar]
- Shibuya A., Campbell D., Hannum C., Yssel H., Franz-Bacon K., McClanahan T., et al. (1996). DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 4 573–581. 10.1016/S1074-7613(00)70060-4 [DOI] [PubMed] [Google Scholar]
- Shibuya K., Lanier L. L., Phillips J. H., Ochs H. D. (1999). Physical and functional association of LFA-1 with DNAM-1 adhesion molecule. Immunity 11 615–623. 10.1016/S1074-7613(00)80136-3 [DOI] [PubMed] [Google Scholar]
- Shibuya K., Shibata K., Tahara-Hanaoka S., Shibuya A. (2006). Comment on “CD226 is specifically expressed on the surface of Th1 cells and regulates their expansion and effector functions”. J. Immunol. 176:3885. [DOI] [PubMed] [Google Scholar]
- Shibuya K., Shirakawa J., Kameyama T., Shin-Ichiro H., Tahara-Hanaoka S., Miyamoto A., et al. (2003). CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J. Exp. Med. 198 1829–1839. 10.1084/jem.20030958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu-Bin F., Hong-Yu Z., Cong W., Bi-Xin H., Xiao-Qing L., Xiang-Ci M., et al. (2020). Small extracellular vesicles derived from human mesenchymal stromal cells prevent group 2 innate lymphoid cell-dominant allergic airway inflammation through delivery of miR-146a-5p. J. Extracell. Ves. 9:1723260. 10.1080/20013078.2020.1723260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. J., Plagnol V., Walker N. M., Cooper J. D., Downes K., Yang J. H. M., et al. (2008). Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 359 2767–2777. 10.1056/NEJMoa0807917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth J., Cretney E., Kelly J., Westwood J., Street S., Yagita H., et al. (2005). Activation of NK cell cytotoxicity. Mol. Immunol. 42 501–510. 10.1016/j.molimm.2004.07.034 [DOI] [PubMed] [Google Scholar]
- Stegmann K. A., Björkström N. K., Ciesek S., Lunemann S., Jaroszewicz J., Wiegand J., et al. (2012). Interferon α–stimulated natural killer cells from patients with acute hepatitis C virus (HCV) infection recognize HCV-infected and uninfected hepatoma cells via DNAX accessory molecule-1. J. Infect. Dis. 205 1351–1362. 10.1093/infdis/jis210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujuan Y., Chichu X., Ye C., Julie W., Xiaoqing C., Zhengqi L., et al. (2019). Differential roles of TNFα-TNFR1 and TNFα-TNFR2 in the differentiation and function of CD4 + Foxp3 + induced treg cells in vitro and in vivo periphery in autoimmune Diseases. Cell Death Dis. 10:27. 10.1038/s41419-018-1266-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujuan Y., Julie W., David D. B., Song Guo Z. (2018). Role of TNF-TNF receptor 2 signal in regulatory T cells and its therapeutic implications. Front. Immunol. 9:784. 10.3389/fimmu.2018.00784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutavani R. V., Bradley R. G., Ramage J. M., Jackson A. M., Durrant L. G., Spendlove I. (2013). CD55 costimulation induces differentiation of a discrete T regulatory type 1 cell population with a stable phenotype. J. Immunol. 191 5895–5903. 10.4049/jimmunol.1301458 [DOI] [PubMed] [Google Scholar]
- Tahara-Hanaoka S., Miyamoto A., Hara A. (2005). Identification and characterization of murine DNAM-1 (CD226) and its poliovirus receptor family ligands. Biochem. Biophys. Res. Commun. 329 996–1000. 10.1016/j.bbrc.2005.02.067 [DOI] [PubMed] [Google Scholar]
- Tahara-Hanaoka S., Shibuya K., Onoda Y., Hua Z., Satoshi Y., Akitomo M., et al. (2004). Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int. Immunol. 16 533–538. 10.1093/intimm/dxh059 [DOI] [PubMed] [Google Scholar]
- Tauriainen J., Scharf L., Frederiksen J., Naji A., Ljunggren H.-G., Sönnerborg A., et al. (2017). Perturbed CD8+ T cell TIGIT/CD226/PVR axis despite early initiation of antiretroviral treatment in HIV infected individuals. Sci. Rep. 7:40354. 10.1038/srep40354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J. A., Walker N. M., Cooper J. D., Smyth D. J., Clayton D. G. (2007). Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39 857–864. 10.1038/ng2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven D. H. J., Hooge A. S., Mooiman E. C. K., Santos S. J., Dam M. M., Gelderblom H., et al. (2008). NK cells recognize and lyse Ewing sarcoma cells through NKG2D and DNAM-1 receptor dependent pathways. Mol. Immunol. 45 3917–3925. 10.1016/j.molimm.2008.06.016 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhang W., Wu D.-A., Chen C., Xu Q.-Z., Zhao B., et al. (2009). Molecular cloning, characterization and three-dimensional modeling of porcine nectin-2/CD112. Vet. Immunol. Immunopathol. 132 257–263. 10.1016/j.vetimm.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Watzl C., Urlaub D., Fasbender F., Claus M. (2014). Natural killer cell regulation-beyond the receptors. F1000Prime Rep. 6:87. 10.12703/P6-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weishan H., Solouki S., Koylass N., Song Guo Z. (2017). ITK signalling via the Ras/IRF4 pathway regulates the development and function of Tr1 cells. Nat. Commun. 8:15871. 10.1038/ncomms15871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenru S., Huimin F., Maogen C., Julie W., David B., Xiaoshun H., et al. (2012). Induced CD4+ forkhead box protein–positive T cells inhibit mast cell function and established contact hypersensitivity through TGF-β1. J. Allergy Clin. Immunol. 130 444–452. 10.1016/j.jaci.2012.05.011 [DOI] [PubMed] [Google Scholar]
- Wenru S., Qian W., Longhui H., Jingwen H., Xiaoqing C., Guihua C., et al. (2014). Doxycycline exerts multiple anti-allergy effects to attenuate murine allergic conjunctivitis and systemic anaphylaxis. Biochem. Pharmacol. 91 359–368. 10.1016/j.bcp.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Wenru S., Qian W., Longhui H., Jingwen H., Xiaoqing C., Guihua C., et al. (2015). Culture medium from TNF-α–stimulated mesenchymal stem cells attenuates allergic conjunctivitis through multiple antiallergic mechanisms. J. Allergy Clin. Immunol. 136 423–432. 10.1016/j.jaci.2014.12.1926 [DOI] [PubMed] [Google Scholar]
- Xiangnan D., Almeida P. D., Manieri N., Nagata D. T., Wu B. (2018). CD226 regulates natural killer cell antitumor responses via phosphorylation-mediated inactivation of transcription factor FOXO1. Proc. Natl. Acad. Sci.U.S.A. 115 E11731–E11740. 10.1073/pnas.1814052115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaojun H., Ping Q., Yibing C., Xingchun Z., Yousheng W., Fange L., et al. (2014). Low expression of CD112 is associated with poor overall survival in patients with hepatocellular carcinoma. Hum. Pathol. 45 1944–1950. 10.1016/j.humpath.2014.06.001 [DOI] [PubMed] [Google Scholar]
- Xinhai Z., Demin L., Weiming O. (2002). Gene cloning and heteromorphism of mouse CD226 (PTA1). Chin. J. Immunol. 6 371–375. [Google Scholar]
- Ya L., Qin L., Ling L., Maogen C., Zanxian X., Jilin M., et al. (2014). Phenotypic and functional characteristic of a newly identified CD8+Foxp3-CD103+ regulatory T cells. J. Mol. Cell Biol. 6 81–92. 10.1093/jmcb/mjt026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Chengbin W., Changsheng X., Xiaoxu L., Chunhong F., Xiaotao Z., et al. (2020). Recovery of CD226-TIGIT + FoxP3 + and CD226-TIGIT-FoxP3 + regulatory T cells contributes to clinical remission from active stage in ulcerative colitis patients. Immunol. Lett. 218 30–39. 10.1016/j.imlet.2019.12.007 [DOI] [PubMed] [Google Scholar]
- Yang L., Song Guo Z. (2016). Hall of fame among pro-inflammatory cytokines: interleukin-6 gene and its transcriptional regulation mechanisms. Front. Immunol. 7:604. 10.3389/fimmu.2016.00604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaoyao Z., Siqi X., Youjun X., Qian Q., Maohua S., Jingnan W., et al. (2018). Long noncoding RNA LERFS negatively regulates rheumatoid synovial aggression and proliferation. J. Clin. Invest. 128 4510–4524. 10.1172/JCI97965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Tingting L., Zhuo W., Meichen M., Jie L., Zining Z., et al. (2018). Expression of the inhibitory receptor TIGIT Is Up-regulated specifically on NK Cells with CD226 activating receptor from HIV-infected individuals. Front. Immunol. 9:2341. 10.3389/fimmu.2018.02341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S. G., Meng L., Wang J. H., Watanabe M., Barr M. L., Cramer D. V., et al. (2006). Transfer of regulatory T cells generated ex vivo modifies graft rejection through induction of tolerogenic CD4+CD25+ cells in the recipient. Int. Immunol. 18 279–289. 10.1093/intimm/dxh368 [DOI] [PubMed] [Google Scholar]
- Zhuo W., Tong W., Meichen M., Zining Z., Yajing F., Jing L., et al. (2017). Elevated interferon-gamma-induced protein 10 and its receptor CXCR3 impair NK cell function during HIV infection. J. Leukoc. Biol. 102 163–170. 10.1189/jlb.5A1016-444R [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.