Abstract
In plants, sugar transporters play an important role in the allocation of sugars from cells in source organs to cells in sink organs. Hence, an understanding of the molecular basis and regulation of assimilate partitioning by sugar transporters is essential. Leaves are the main source of photosynthetic products. In jujube (Ziziphus jujuba Mill.), the mechanisms regulating initial sugar unloading in leaves are still unclear. In this study, an expression profiling analysis showed that ZjSWEET2.2, encoding a sugar transporter in the SWEET family, is highly expressed in leaves. Over-expression of ZjSWEET2.2 increased carbon fixation in photosynthetic organs. Our analyses showed that ZjSWEET2.2 encodes a plasma membrane-localized sugar transporter protein. Its expression levels were found to be suppressed under drought stress and by high concentrations of exogenous sugars, but increased by low concentrations of exogenous sugars. Finally, DNA sequence analyses revealed several cis-elements related to sugar signaling in the promoter of ZjSWEET2.2. Together, these results suggest that ZjSWEET2.2 functions to mediate photosynthesis by exporting sugars from photosynthetic cells in the leaves, and its gene expression is regulated by sugar signals.
Keywords: Jujube (Ziziphus jujuba Mill.), SWEET, photosynthesis, sugar signal, sugar transporter
Introduction
In higher plants, photosynthates produced at the photosynthetic “source” (mainly mesophyll cells) are essential for growth and development, and are major nutritional components of fruits (Zhu et al., 2010). Photosynthate allocation occurs via transport from the source organ to the heterotrophic “sink” organs (fruits, roots, seeds) (Lalonde et al., 2004; Ayre, 2011). Adequate photosynthate production in the leaves can substantially increase the number of flowers and fruit weight. Improvements in allocation efficiency can increase the proportion of total biomass allocated to harvestable organs (Zhu et al., 2010; Paterson and Li, 2011). Sugar translocation depends on active transport by transmembrane proteins or passive transport via the plasmodesmata. In higher plants, sugar transporters play crucial roles in mediating carbohydrate fluxes (Patrick et al., 2001) and are linked with biomass gain. By now, sugar transporters found in plants mainly include three types: monosaccharide transporters (MST), sucrose transporters (SUT) and SWEET transporters (Sugars Will Eventually be Exported Transporters). Of these, MSTs and SUTs have typical structural characteristics of the major facilitator superfamily (MFS) with high hydrophobicity and generally include 12 transmembrane domains (Fang et al., 2020), which is the most important membrane transporter family. SUTs are crucial for the sucrose long-distance transportation in phloem to sink organs as sucrose is the important form of photosynthetic products (Chandran et al., 2003; Scofield et al., 2007). In the sink organs, some of the disaccharide can be degraded to monosaccharide and mediated by MSTs (Doidy et al., 2019), while in pea seeds, most carbohydrates transported into cotyledons cells in the form of sucrose by sucrose transporter PsSUT1 (Zhou et al., 2009). Besides, sucrose can be used as an important sugar signal to regulate plant growth and development (Osuna et al., 2007).
As a novel family of sugar transporters, the role of SWEET proteins was first identified in Arabidopsis thaliana (Chen et al., 2010). A protein structure prediction analysis indicated that SWEET transporter protein belonged to the MtN3 family and have seven transmembrane domains (TMSs) with two duplicated units of three TMSs and a linker element (the fourth TMS) (Chen et al., 2012; Xuan et al., 2013). SWEET proteins have sugar efflux and influx activity, and have diverse physiological functions (Jeena et al., 2019). Other studies sequenced and characterized SWEET family members in various plant species, such as tomato (Solanum lycopersicum), wheat, citrus, and pear (Pyrus bretschneideri) (Xu et al., 2013; Feng et al., 2015; Li et al., 2017; Gao et al., 2018). The results of those studies provided some details of the roles of SWEET transporters in regulating sugar transport and accumulation. SWEET transporters show low affinity for hexose and sucrose during sugar efflux or import. In Nicotiana, Arabidopsis, and brassicas, SWEET9 facilitates sucrose efflux for nectar secretion (Lin et al., 2014). SWEET15 mediates sugar export in the endosperm to promote embryo development (Wang et al., 2019b) or seed filling (Chen et al., 2015a). Other SWEET proteins function under different osmotic stress conditions or in pathogenic reactions (Chen et al., 2015b; Jeena et al., 2019). Some SWEET proteins are involved in the mobilization of carbohydrates in leaves (Chen et al., 2010), which may affect photosynthetic efficiency. In Arabidopsis, AtSWEET11 and 12 were shown to play crucial roles in sugar efflux from mesophyll cells to the apoplast (Chen et al., 2012); and in tomato, SlSWEET1a was found to play roles in glucose efflux from mature to young leaves (Ho et al., 2019). Nevertheless, the function of many SWEET sugar transporters, especially in perennial woody crops, remains unclear.
Jujube (Ziziphus jujuba Mill.), a member of the Rhamnaceae family, is an important dry fruit crop with a worldwide distribution (Qu and Wang, 1993). It is strongly resistant to drought and salinity stress. With a cultivation area of more than 2 million ha, jujube has become the primary source of income for 20 million farmers. Our previous studies identified that sugar transporters play significant roles in the accumulation of sugars in jujube fruit (Zhang et al., 2018). However, little is known about the mechanism of sugar efflux by sugar transporters in leaves. In this study, we identified ZjWEET2.2 as another member of the SWEET family in Z. jujuba, and conducted a preliminary evaluation of its functions. The gene encoding ZjWEET2.2 was found to be highly expressed in photosynthetic organs, and the protein was found to localize to the plasma membrane. Over-expression of this gene increased carbon fixation. Our results indicate that the expression of ZjSWEET2.2 and the activity of its encoded protein are mediated by sugar signals. The results of this study provide new insights into the function and regulation of SWEET sugar transporters.
Materials and Methods
Plant Materials
The Chinese dry jujube cultivar “No. 4 Jinsi,” and fresh jujube “Zaozhuangcuizao” and “Dongzao,” were grown at the Jujube Experimental Station, Shandong Institute of Pomology, Taian, China. The fruits of “No. 4 Jinsi” were collected at different developmental stages [young fruit at 10 days after anthesis (DAA), enlarged fruit at 40 DAA, white mature fruit at 80 DAA, half-red fruit at 100 DAA, and fully red fruit at 110 DAA]. Leaf, root, and phloem (scraped from the stem using a blade) samples were also collected.
Jujube young trees were grown in an incubator under a 16-h light/8-h dark photoperiod, and watered every five days to keep water content at more than 60% of maximum field capacity. For drought stress treatments, water was withheld for 2 weeks when the leaves showed wilting phenotype; while the control group was keep watering every five days. The leaves were harvested from water deficit-treated trees and well watered controls for analyses. In the exogenous sugar application experiment, the treatment groups were sprayed with 3% or 10% (w/v) glucose or sucrose, and the control group was sprayed with the same amount of distilled water. Leaves were collected at 6 h after treatment for analyses. For each treatment, three biological replicates were analyzed. All samples were directly frozen in liquid nitrogen and stored at −80°C until analysis.
Construction of Phylogenetic Tree
Jujube genes putatively encoding sugar transporter proteins were obtained from the Junzao genome (accession number: LPXJ00000000) (Huang et al., 2016). The BLASTP tool was used to retrieve loci encoding putative sugar transporters, using Arabidopsis sequences as queries with E-values ≤1e-5. To construct the phylogenetic tree, the sequences of transporters from jujube, Arabidopsis, and Solanum lycopersicum L. were aligned by ClustalW with MEGA5 (Tamura et al., 2011). The phylogenetic tree was constructed using the Neighbor-Joining method, and the phylogeny test was based on the bootstrap method with 1000 replications.
Vector Construction and Plant Transformation
The vector was constructed using the method described by Jiang et al. (2017b). The coding sequence of ZjSWEET2.2 was inserted downstream of the CaMV35S promoter in the plant expression vector pVBG23000-GFP, which includes the gene encoding green fluorescent protein (GFP). For the subcellular localization analysis, the positive vector CaMV35S-ZjSWEET2.2-GFP was transiently expressed in tobacco leaves after infiltration with Agrobacterium tumefaciens strain GV3101 containing the vector. The pRT101-AtPIP2A-red fluorescent protein (RFP) was used as a plasma membrane marker, and was co-transformed with CaMV35S-ZjSWEET2.2-GFP. The tobacco plants were incubated at 25°C for 48 to 72 h, and the fluorescence of GFP was observed under a high resolution laser confocal microscope (Zeiss, Jena, Germany).
For the transient overexpression analysis, jujube leaves were vacuum-infiltrated with Agrobacterium containing the vector along with 500 µl Tween 20. The CaMV35S-GFP construct was used as a control. The leaves were cultured on Murashige and Skoog (MS) medium for 48 to 72 h in an incubator under a 12-h dark/12-h light photoperiod. The presence and relative amount of the transgene in transgenic leaves were determined by qRT-PCR.
Analyses of mRNA Levels
Total RNA was extracted using a Plant RNA Extraction Kit (Foregene, Chengdu, China) following the manufacturer’s instructions. To analyze the transcript levels of ZjSWEET2.2 and genes related to carbon fixation [RBCS (encoding ribulose bisphosphate carboxylase/oxygenase), PGK (encoding phosphoglycerate kinase), RPI (encoding ribose-5-phosphate isomerase), and PRK (encoding phosphoribulokinase)], reverse-transcription quantitative real-time PCR (qRT-PCR) was performed using a SYBR Premix Ex Taq Kit (Vazyme, Nanjing, China) on a Bio-Rad IQ5 instrument (Bio-Rad, Hercules, CA, USA). The reaction volume was 20 µl. All qRT-PCRs were performed with three technical replicates and three biological replicates. The primers were designed from the coding sequences of jujube genes using Primer5 software and are listed in Supplemental Table 1 . Primer specificity was determined based on melting curve analyses. Expression data were analyzed using the 2-ΔΔCT method (Livak & Schmittgen, 2001). The selected reference gene was UBQ (Zhang et al., 2015).
Measurement of Sugar Contents
The contents of sucrose, glucose, and fructose were measured by high performance liquid chromatography (HPLC) according to Gao and Wang, (2013). All samples were extracted and analyzed in triplicate. For each sample, 1 g tissue was homogenized and measured. Three biological replicates were analyzed for each time point and variety.
Results
Sequence Analysis of ZjSWEET2.2
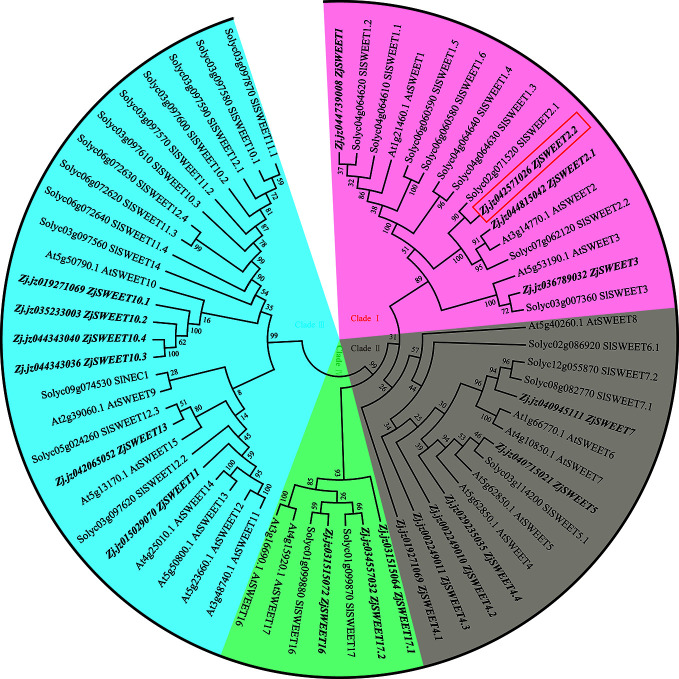
We identified 19 candidate ZjSWEET genes from the jujube genome. The phylogenetic tree was constructed using amino acid sequences of SWEET transporters from jujube, A. thaliana, and S. lycopersicum ( Supplemental Table 2 ). The SWEET sugar transporters from the three species could be classified into four clades ( Figure 1 ). Clade I includes the members of SWEET1, SWEET2, and SWEET3, subclades), clade II includes the members from SWEET4 to SWEET8, clade III includes the members from SWEET9 to SWEET 15, and clade IV includes SWEET16 and SWEET17. Consistent with the fact that a recent common whole genome duplication event has occurred in many species, there were duplicate copies of some SWEET genes in the genome. In our comparisons, ZjSWEET2.2 in clade I of the SWEET family showed the closest relationship with its paralog ZjSWEET2.1 and its ortholog AtSWEET2 from A. thaliana. Protein structure analyses of ZjSWEET2.2 revealed seven predicted TMDs in a protein comprising 266 amino acids ( Supplemental Figure 1 ).
Figure 1.
Maximum likelihood phylogeny of SWEET sugar transporters. Accession numbers for SWEET genes from Arabidopsis are those reported by Wei et al. (2014) and those for tomato genes are those reported by Feng et al. (2015).
Subcellular Localization of ZjSWEET2.2
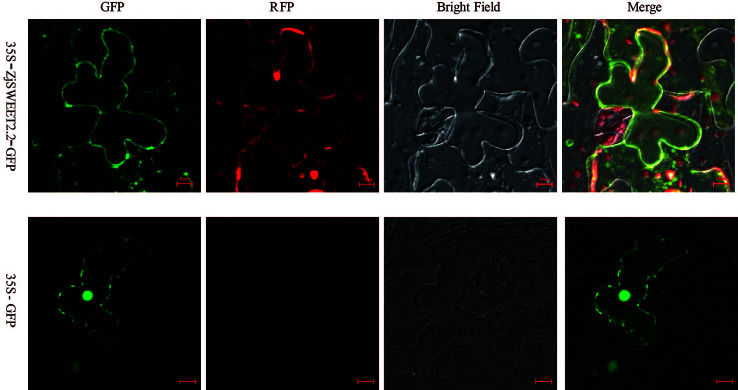
The coding sequence for ZjSWEET2.2 was fused to eGFP and transiently expressed in leaves of Nicotiana benthamiana to determine the subcellular localization of its encoded protein. The construct was co-expressed with the mRFP1-labeled plasma membrane marker. Confocal images showed that GFP fluorescence was localized to the plasma membrane, the cytoplasm and nucleus of the tobacco epidermal cells, and ZjSWEET2.2-GFP was co-located with the plasma membrane marker at the plasma membrane ( Figure 2 ).
Figure 2.
Subcellular localization of ZjSWEET2.2. 35S:GFP-ZjSWEET2.2 was transiently expressed in tobacco leaves and co-localized with transiently expressed plasma membrane marker RFP. Scale bars, 10 μm.
Tissue Expression Patterns of ZjSWEET2.2
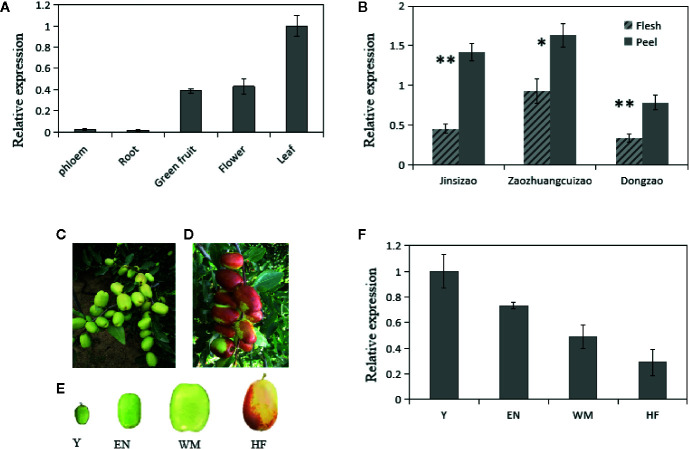
Our previous RNA sequencing results showed that ZjSWEET2.2 transcript levels were highest in leaves among various tested jujube tissues (spires, leaves, flowers, fruit, and stems) (Huang et al., 2016; PRJNA306374). To explore the putative functions of ZjSWEET2.2, we determined its transcript levels in different tissues ( Figure 3 ). The transcript levels of ZjSWEET2.2 were highest in the leaves followed by flowers and green fruit, while there were low transcript levels in the root and the phloem of branches ( Figure 3A ). We also determined its transcript levels in the flesh and green peel of three different cultivars ( Figure 3B ). In all cultivars, the ZjSWEET2.2 transcript levels were higher in the peel than in the flesh, consistent with the higher chlorophyll content in the peel than in the flesh. Furthermore, ZjSWEET2.2 transcript levels were analyzed in different development stages of fruit (peel) ( Figure 3F ). The transcript levels of ZjSWEET2.2 in fruit were high at the young stage and decreased as fruit ripened, alongside the degradation of chlorophyll.
Figure 3.
Transcript levels of ZjSWEET2.2 in different tissues of jujube and phenotypic characteristics of fruits at different development stages. (A) Transcript levels of ZjSWEET2.2 in phloem, root, flowers, leaves, and fruit. (B) Transcript levels of ZjSWEET2.2 in peel and flesh of three different jujube varieties. (C) White mature fruit and (D) half-red fruit on jujube tree. (E) Four different development stages of jujube fruits (Y, young fruit; EN, enlargement stage; WH, white mature stage; HF, half-red stage). (F) Changes in relative transcript levels of ZjSWEET2.2 during fruit development. Three biological replicates were analyzed. Error bars represent SE. Asterisks indicate significant difference as determined by Student’s t-test (**P < 0.01; *P < 0.05).
Overexpression of ZjSWEET2.2 in Transgenic Lines
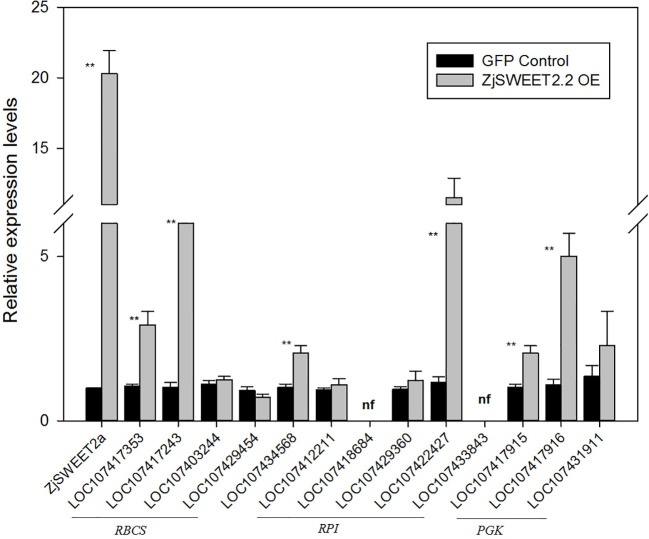
To explore the patterns of regulation of ZjSWEET2.2 in leaves, the CaMV 35S: ZjSWEET2a-GFP construct was transformed into jujube leaves for transient expression. We then determined the transcript levels of ZjSWEET2.2 and genes related to photosynthetic carbon assimilation by qRT-PCR. The transcript level of ZjSWEET2.2 in the ZjSWEET2.2-OE line was nearly 20 times higher than that in the GFP-expressing control. We determined the transcript levels of genes encoding RBCs, the key enzyme in carbon fixation in the leaf; RPIs, which function upstream of RBCs; and PGKs, which function downstream of this pathway ( Figure 4 ). Two genes in the RBCs family were significantly up-regulated in the ZjSWEET2.2-OE line compared with the control. Among the RPIs family, two members were expressed weakly and two showed no differences in their transcript levels between the ZjSWEET2.2-OE line and the control. However, the LOC107422427 transcript levels in leaves were higher in ZjSWEET2.2-OE than in the control. One PRK gene was up-regulated in the ZjSWEET2.2-OE line. In the PGKs gene family, PGK1 (LOC107431911) transcript levels were not affected in the ZjSWEET2.2-OE line, but the other two PGKs were up-regulated. Together, these results suggested that overexpression of ZjSWEET2.2 increased carbon fixation to some extent via increased export of assimilation products from photosynthetic tissues.
Figure 4.
Transcript levels of genes involved in carbon fixation in photosynthetic organ. RBCSs: ribulose bisphosphate carboxylases; PRKs, phosphoribulokinases, RPIs: ribose-5-phosphate isomerases, PGKs: phosphoglycerate kinases. Results are normalized against transcript level in control group, which was set to 1. Data are mean ± SD of three replicates. ** indicate significant difference (P < 0.01) as determined by Student's t-test.
Regulation of ZjSWEET2.2 by Sugars
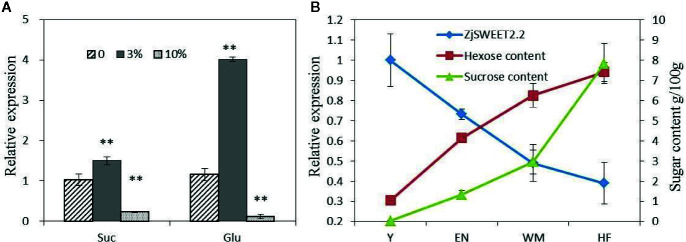
Exogenous sugars at two concentrations were sprayed onto the leaves of jujube plants, and then the transcript levels of ZjSWEET2.2 were monitored by qRT-PCR ( Figure 5A ). After treatment with 3% (w/v) exogenous glucose, the transcript level of ZjSWEET2.2 was significantly up-regulated by approximately three-fold, compared with that in the control. In the plants treated with 3% (w/v) exogenous sucrose, the transcript level was also increased, but only by 50% compared with that in the control. When the concentration of exogenous sugars was increased to 10% (w/v), the transcript level of ZjSWEET2.2 decreased to lower than that in the control, and this difference was more significant in the plants treated with glucose than in those treated with sucrose. These results showed that the expression of ZjSWEET2.2 was induced by low concentrations of sugars, but suppressed by high concentrations of sugars, and was more responsive to glucose than to sucrose.
Figure 5.
Transcript levels of ZjSWEET2.2 in leaves of plants treated with exogenous sugars (A), and correlation between transcript levels of ZjSWEET2.2 and monosaccharide sugar content (B). Three biological replicates were analyzed. Error bars represent SE. ** indicate significant difference (P < 0.01) as determined by Student’s t-test.
Next, we analyzed the correlation between the transcript levels of ZjSWEET2.2 and sugar contents in fruit during development. The contents of hexoses (fructose and glucose) increased steadily during fruit development, while sucrose began to accumulate rapidly from the white ripening stage (2.9 g/100 g fruit fresh weight), ultimately reaching 7.8 g/100 g ( Figure 5B ). A correlation analysis showed that the transcript level of ZjSWEET2.2 was significantly negatively correlated with hexose content (r = −0.985, P < 0.015).
To explore the regulation of ZjSWEET2.2 expression, the 2-kb promoter region upstream (5′) of the start codon of ZjSWEET2.2 was sequenced and analyzed ( Supplemental Figure 2 ). Tools at the PlantCare server were used to identify cis-elements in this sequence, and additional cis-elements that are known to be sugar regulation regions were also considered (Li et al., 2006; Baena-Gonzalez et al., 2007). In total, 12 cis-elements related to sugar repression and seven related to sugar induction were identified ( Table 1 ). Interestingly, most of these elements were located within the 1-kb region upstream of the start codon. The large number of sugar signaling-related cis-elements in close proximity to the start codon strongly suggested that sugar signals regulate the expression of ZjSWEET2.2.
Table 1.
Known cis-acting elements involved in sugar repression/induction in 2,000-bp fragment of ZjSWEET2.2 promoter.
| Cis-Elements | Sequence | Response | Copies | References |
|---|---|---|---|---|
| W-box | TTGACC | Sugar induce | 2 | Sakr et al., 2018; Chen et al., 2019 |
| W-box | TGACT | Sugar induce | 1 | Sakr et al., 2018 |
| SUCROSE BOX 3 | AAATCA.AA | Sugar induce | 4 | Hwang et al., 1998; Lu et al., 1998; |
| TATCCAOSAMY | TATCCA | Sugar suppress | 2 |
Lu et al., 2002; Sun et al., 2003
Baena-Gonzalez et al., 2007 |
| G box | CACGTG | Sugar suppress | 1 | Hwang et al., 1998 |
| CATCC | CATCC | Sugar suppress | 2 | Li et al., 2006 |
| I-BOX core | GATAA | Sugar suppress | 3 | Manzara et al., 1991 |
| AMYBOX1 | TAACAAA | Sugar suppress | 1 | Hwang et al., 1998 |
| EVENINGAT core | ATATCT | Sugar suppress | 2 | Li et al., 2006 |
| GATTA | GATTA | Sugar suppress | 1 | Li et al., 2006 |
Suppression of ZjSWEET2.2 Expression Under Drought Stress
We determined the transcript levels of ZjSWEET2.2 in leaves of jujube plants under drought stress. The results showed that the transcript level decreased to approximately one-quarter its pre-stress level under drought stress, suggesting that SWEET2 plays important roles in regulating cell osmotic potential by reversing sugar efflux. Consistent with this, the contents of fructose and glucose in leaves of jujube increased under drought stress ( Supplemental Figure 3 ).
Discussion
ZjSWEET2.2 Plays Roles in Mediating Sugar Export From Photosynthetic Organs
The SWEET sugar transporters from the three species could be classified into four clades, previously uncovered in Arabidopsis (Chen et al., 2010; Eom et al., 2015; Doidy et al., 2019). Our analyses showed that ZjSWEET2.2 is in the clade I SWEET gene family, and is phylogenetically closest to AtSWEET2. In Arabidopsis, AtSWEET2 encodes a glucose transporter (Xuan et al., 2013; Chen et al., 2015a). In this study, we found that ZjSWEET2.2 could be regulated by exogenous glucose, but its expression was relatively insensitive to exogenous sucrose. Therefore, we speculate that ZjSWEET2.2 also mainly exports glucose. We detected low transcript levels of ZjSWEET2.2 in sink organs of jujube, but high transcript levels in leaves, where photosynthates are produced and then continuously exported to sink organs. Although sucrose may account for the largest proportion of sugars exported from the leaf, it can also be hydrolyzed into glucose and fructose by cell wall invertases, thereby forming a sucrose gradient to facilitate outflow (Osorio et al., 2014). When the ZjSWEET2.2 protein was transiently expressed in tobacco leaves, it co-localized with a plasma membrane marker in the plasma membrane, suggesting that this is the primary site of apoplastic sugar transport. On the basis of our results, we suggest that the plasma membrane-localized sugar transporter ZjSWEET2.2 directly transports glucose and indirectly facilitates sucrose loading in the leaf. AtSWEET2 limits carbon sequestration in the roots in Arabidopsis, while LcSWEET2a in Litchi chinensis is involved in early seed development (Xie et al., 2019). In contrast, ZjSWEET2.2 is expressed abundantly in chlorenchyma cells. Besides, AtSWEET2 was characterized as a vacuolar transporter (Chen et al., 2015b), while the orthologous gene ZjSWEET2.2 is located at the plasma membrane. We speculate that the function of SWEET2 may differ among different species. Soluble sugars are known to be involved in the regulation of osmotic potential in cells of plants under salt and drought stress (Lemoine et al., 2013). In our study, the transcript level of ZjSWEET2.2 significantly decreased in jujube plants under drought stress. This led to decreased sugar export from cells, so that the content of hexose sugars increased. Together, our results suggest that ZjSWEET2.2 plays a key role in sugar export from source (photosynthetic) leaves.
Previous studies have shown that the over-accumulation of carbohydrates in leaves reduces the photosynthetic rate (Baker et al., 2016). In Arabidopsis, ATSWEET11 and 12 are located in the vascular tissues of leaves. Mutations of their encoding genes were shown to result in the accumulation of starch in leaves, which seriously reduced photosynthetic efficiency (Chen et al., 2012). Similar phenomena have been observed in Zea mays, in which photosynthesis was impaired in the genome-edited knock-out mutants of ZmSWEET13 paralogs (a, b, and c) (Bezrutczyk et al., 2018). In contrast, increasing sugar efflux can stimulate photosynthetic activity (Ainsworth and Bush, 2011). The maize protein CTS1, a homolog of ATSWEET1, increases photosynthesis by regulating the sugar content in subsidiary cells in maize (Wang et al., 2019a). In our study, overexpression of ZjSWEET2.2 resulted in increased transcript levels of genes related to carbon fixation and photosynthesis. We speculate that ZjSWEET2.2 indirectly improves photosynthesis by stimulating phloem loading and decreasing the carbohydrate levels in mesophyll cells.
Sugar Signals Mediate Expression of ZjSWEET2.2
Accumulation of free sugars can lead to down-regulation of photosynthesis through sugar signaling networks (Rolland et al., 2006). In this study, we found that the abundance of ZjSWEET2.2 transcripts was significantly negatively correlated with sugar accumulation in fruits, and was significantly reduced by exogenous sugars at a high concentration. These results suggested that high concentrations of soluble sugars may also down-regulate the expression of the sugar exporter ZjSWEET2.2. In contrast, a low concentration of exogenous glucose promoted ZjSWEET2.2 expression, indicating that it is induced by low-sugar signals. Both sugar-induction and sugar-repression cis-acting regions were identified in the promoter of ZjSWEET2.2, including three W boxes (TTGACC/T), which are indispensable for mediating sugar signaling (Sakr et al., 2018). The WRKY-type transcription factors specifically recognize W-Box cis-elements (Eulgem et al., 2000; Jiang et al., 2017a) and are known to be involved in sugar induction of gene expression (Chen et al., 2019). However, we detected more sugar-repression elements than sugar-induction elements in the promoter of ZjSWEET2.2. A previous study demonstrated that sugar regulatory motifs in the promoter of STP1 in Arabidopsis are involved in repression of gene expression by glucose (Cordoba et al., 2015). Six out of the seven different motifs identified in the promoter of STP1 were also detected in the promoter of ZjSWEET2.2. We detected two TATCCA elements in the 2-kb region of the ZjSWEET2.2 promoter. In rice, the TATCCAOSAMY motif is recognized by the MYB-type transcription factor OsMYBS2, which regulates expression of α-Amy3 in response to sugar signaling (Lu et al., 2002; Baena-Gonzalez et al., 2007). Consistent with this, we speculate that some transcription factors bind to the cis-acting regions in the ZjSWEET2.2 promoter to mediate its expression. Further research is needed to clarity the regulation mechanism of these putative orthologs.
Model of Regulation Mechanism of ZjSWEET2.2
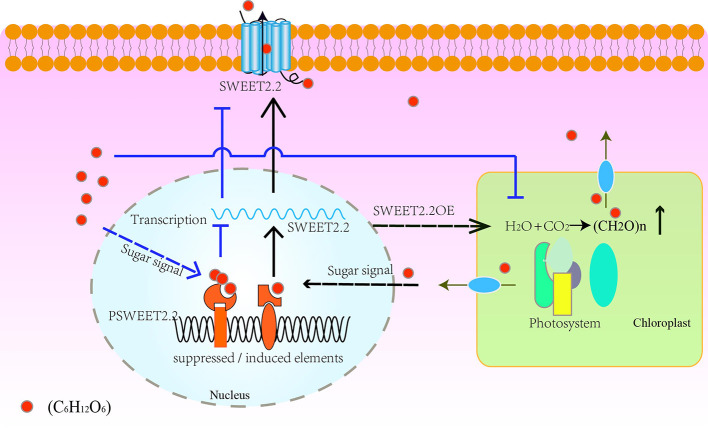
On the basis of the expression patterns of ZjSWEET2.2 in different tissues, we propose a hypothetical model for its regulation mechanism. As shown in Figure 6 , our results suggest that ZjSWEET2.2 plays a critical role in exporting sugars from leaves. Low sugar content may promote ZjSWEET2.2 expression. Over-expression of ZjSWEET2.2 increases carbon fixation into photosynthates by decreasing the carbohydrate content in mesophyll cells. When the sugar content increases in the cytosol, such as under drought stress or in ripening fruit, the expression of sugar-responsive transcription factors increases. These bind to sugar-suppressed elements in the promoter region of ZjSWEET2.2 to repress its expression. Besides, the over-accumulation of sugar reduces the photosynthetic rate (Baker et al., 2016). This model also explains the phenomenon of high expression levels of ZjSWEET2.2 in mature leaves, but decreasing levels in ripening fruit as sugars accumulate.
Figure 6.
Schematic model of regulation of ZjSWEET2.2 expression by sugar signals to mediate sugar transport. ZjSWEET2.2 exports photosynthates from leaves under normal conditions. Over-expression of ZjSWEET2.2 facilitates carbon fixation. Low sugar signals bind to inducing cis-elements in promoter of ZjSWEET2.2 to activate its transcription. High sugar signals decreases the ZjSWEET2.2 expression and reduces the photosynthetic rate.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Author Contributions
YG was responsible for the part of over-expression of SWEET2a and incubated jujube seedlings. MW measured the sugar content of jujube. CZ was accountable for design of the work, analysis, drafting the work.
Funding
This work was supported by the Chinese National Natural Science Foundation (Grant no. 31800573) and The National Key Research and Development Program of China (2018YFD1000607).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jennifer Smith, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01081/full#supplementary-material
Abbreviations
SWEET2a, sugars will eventually be exported transporter 2a; RBCS, ribulose bisphosphate carboxylase/oxygenase; PGK, phosphoglycerate kinase; RPI, ribose-5-phosphate isomerase; PRK, phosphoribulokinase.
References
- Ainsworth E. A., Bush D. R. (2011). Carbohydrate Export from the Leaf: A Highly Regulated Process and Target to Enhance Photosynthesis and Productivity. Plant Physiol. 155 (1), 64–69. 10.1104/pp.110.167684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayre B. G. (2011). Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 4, 377–394. 10.1093/mp/ssr014 [DOI] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J. M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. 10.1038/nature06069 [DOI] [PubMed] [Google Scholar]
- Baker R. F., Leach K. A., Boyer N. R., Swyers M. J., Benitez-Alfonso Y., Skopelitis T., et al. (2016). Sucrose transporter ZmSut1 expression and localization uncover new insights into sucrose phloem loading. Plant Physiol. 172, 1876–1898. 10.1104/pp.16.00884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezrutczyk M., Hartwig T., Horschman M., Char S. N., Yang J., Yang B., et al. (2018). Impaired phloem loading in zmsweet13a, b, c sucrose transporter triple knock-out mutants in Zea mays. New Phytol. 218, 594–603. 10.1111/nph.15021 [DOI] [PubMed] [Google Scholar]
- Chandran D., Reinders A., Ward J. M. (2003). Substrate specificity of the Arabidopsis thaliana sucrose transporter AtSUC2. J. Biol. Chem. 278 (45), 44320–44325. 10.1074/jbc.M308490200 [DOI] [PubMed] [Google Scholar]
- Chen L. Q., Hou B. H., Lalonde S., Takanaga H., Hartung M. L., Qu X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468 (7323), 527–532. 10.1038/nature09606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Q., Qu X. Q., Hou B. H., Sosso D., Osorio S., Fernie A. R., et al. (2012). Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 335 (6065), 207–211. 10.1126/science.1213351 [DOI] [PubMed] [Google Scholar]
- Chen L. Q., Lin I. W., Qu X. Q., Sosso D., McFarlane H. E., Londono A., et al. (2015. a). A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27 (3), 607–619. 10.1105/tpc.114.134585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Y., Huh J. H., Yu Y. C., Ho L. H., Chen L. Q., Tholl D., et al. (2015. b). The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 83 (6), 1046–1058. 10.1111/tpj.12948 [DOI] [PubMed] [Google Scholar]
- Chen Q., Xu X., Xu D., Zhang H., Zhang C., Li G. (2019). WRKY18 and WRKY53 Coordinate with HISTONE ACETYLTRANSFERASE1 to Regulate Rapid Responses to Sugar. Plant Physiol. 180 (4), 2212–2226. 10.1104/pp.19.00511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordoba E., Aceves-Zamudio D. L., Hernandez-Bernal A. F., Ramos-Vega M., Leon P. (2015). Sugar regulation of SUGAR TRANSPORTER PROTEIN 1 (STP1) expression in Arabidopsis thaliana. J. Exp. Bot. 66 (1), 147–159. 10.1093/jxb/eru394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doidy J., Vidal U., Lemoine R. (2019). Sugar transporters in Fabaceae, featuring SUT MST and SWEET families of the model plant Medicago truncatula and the agricultural crop Pisum sativum. PloS One 14 (9), e0223173. 10.1371/journal.pone.0223173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom J. S., Chen L. Q., Sosso D., Julius B. T., Lin I. W., Qu X. Q., et al. (2015). SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 25, 53–62. 10.1016/j.pbi.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 (5), 199–206. 10.1016/S1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Fang T., Peng Y., Rao Y., Li S. H., Zeng L. H. (2020). Genome-Wide Identification and Expression Analysis of Sugar Transporter (ST) Gene Family in Longan (Dimocarpus longan L.). Plants-Basel 9 (3), 342. 10.3390/Plants9030342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C. Y., Han J. X., Han X. X., Jiang J. (2015). Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 573 (2), 261–272. 10.1016/j.gene.2015.07.055 [DOI] [PubMed] [Google Scholar]
- Gao Q. H., Wang M. (2013). Response to Comment on Effect of drying of jujubes (Ziziphus jujuba Mill.) on the contents of sugars, organic acids, alpha-tocopherol, beta-carotene, and phenolic compounds. J. Agric. Food Chem. 61 (19), 4665. 10.1021/jf400098v [DOI] [PubMed] [Google Scholar]
- Gao Y., Wang Z. Y., Kumar V., Xu X. F., Yuan D. P., Zhu X. F., et al. (2018). Genome-wide identification of the SWEET gene family in wheat. Gene 642, 284–292. 10.1016/j.gene.2017.11.044 [DOI] [PubMed] [Google Scholar]
- Ho L. H., Klemens P. A. W., Neuhaus H. E., Ko H. Y., Hsieh S. Y., Guo W. J. (2019). SlSWEET1a is involved in glucose import to young leaves in tomato plants. J. Exp. Bot. 70 (12), 3241–3254. 10.1093/jxb/erz154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhang C. M., Zhao X., Fei Z. J., Wan K. K., Zhang Z., et al. (2016). The Jujube Genome Provides Insights into Genome Evolution and the Domestication of Sweetness/Acidity Taste in Fruit Trees. PloS Genet. 12 (12), ARTN e1006433. 10.1371/journal.pgen.1006433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y. S., Karrer E. E., Thomas B. R., Chen L., Rodriguez R. L. (1998). Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation. Plant Mol. Biol. 36, 331–341. 10.1023/A:1005956104636 [DOI] [PubMed] [Google Scholar]
- Jeena G. S., Kumar S., Shukla R. K. (2019). Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol. Biol. 100 (4-5), 351–365. 10.1007/s11103-019-00872-4 [DOI] [PubMed] [Google Scholar]
- Jiang J., Ma S., Ye N., Jiang M., Cao J., Zhang J. (2017. a). WRKY transcription factors in plant responses to stresses. J. Integ. Plant Biol. 59, 86–101. 10.1111/jipb.12513 [DOI] [PubMed] [Google Scholar]
- Jiang Y. H., Liu C. H., Yan D., Wen X. H., Liu Y. L., Wang H. J., et al. (2017. b). MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar ‘Granny Smith’. J. Exp. Bot. 68 (5), 1055–1069. 10.1093/jxb/erx029 [DOI] [PubMed] [Google Scholar]
- Lalonde S., Wipf D., Frommer W. B. (2004). Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu. Rev. Plant Biol. 55, 341–372. 10.1146/annurev.arplant.55.031903.141758 [DOI] [PubMed] [Google Scholar]
- Lemoine R., La Camera S., Atanassova R., Dédaldéchamp F., Allario T., Pourtau N., et al. (2013). Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 4, 272. 10.3389/fpls.2013.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lee K. K., Walsh S., Smith C., Hadingham S., Sorefan K., et al. (2006). Establishing glucose and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res. 16, 414–427. 10.1101/gr.4237406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. M., Qin M. F., Qiao X., Cheng Y. S., Li X. L., Zhang H. P., et al. (2017). A New Insight into the Evolution and Functional Divergence of SWEET Transporters in Chinese White Pear (Pyrus bretschneideri). Plant Cell Physiol. 58 (4), 839–850. 10.1093/pcp/pcx025 [DOI] [PubMed] [Google Scholar]
- Lin I. W., Sosso D., Chen L. Q., Gase K., Kim S. G., Kessler D., et al. (2014). Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 508 (7497), 546–549. 10.1038/nature13082 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25 (4), 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu C. A., Lim E. K., Yu S. M. (1998). Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 273, 10120–10131. 10.1074/jbc.273.17.10120 [DOI] [PubMed] [Google Scholar]
- Lu C. A., Ho T. H., Ho S. L., Yu S. M. (2002). Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alphaamylase gene expression. Plant Cell 14, 1963–1980. 10.1105/tpc.001735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzara T., Carrasco P., Gruissem W. (1991). Developmental and organspecific changes in promoter DNA-protein interactions in the tomato rbcS gene family. Plant Cell 3, 1305–1316. 10.1105/tpc.3.12.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S., Ruan Y. L., Fernie A. R. (2014). An update on source-to-sink carbon partitioning in tomato. Front. Plant Sci. 5, 516. 10.3389/fpls.2014.00516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna D., Usadel B., Morcuende R., Gibon Y., Blasing O. E., Hohne M., et al. (2007). Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 49 (3), 463–491. 10.1111/j.1365-313X.2006.02979.x [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Li Z.-K. (2011). Paleo-green revolution for rice. PANS 108, 10931–10932. 10.1073/pnas.1107959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick J. W., Zhang W., Tyerman S. D., Offler C. E., Walker N. A. (2001). Role of membrane transport in phloem translocation of assimilates and water. Funct. Plant Biol. 28, 697–709. 10.1071/PP01023 [DOI] [Google Scholar]
- Qu Z., Wang Y. (1993). Fruit tree records of China, Chinese jujube volume (Beijing: China Forestry Publ. House; ). [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. 10.1146/annurev.arplant.57.032905.105441 [DOI] [PubMed] [Google Scholar]
- Sakr S., Wang M., Dédaldéchamp F., Perez-Garcia M.-D., Ogé L., Hamama L., et al. (2018). The sugar-signaling hub: overview of regulators and interaction with the hormonal and metabolic network. Int. J. Mol. Sci. 19, 2367–2379, 2506. 10.1093/pcp/pcw157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield G. N., Hirose T., Aoki N., Furbank R. T. (2007). Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. J. Exp. Bot. 58 (12), 3155–3169. 10.1093/jxb/erm153 [DOI] [PubMed] [Google Scholar]
- Sun C., Palmqvist S., Olsson H., Boren M., Ahlandsberg S., Jansson C. (2003). A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15 (9), 2076–2092. 10.1105/tpc.014597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 (10), 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yan S., Xin H., Huang W., Zhang H., Teng S., et al. (2019. a). A Subsidiary Cell-localized Glucose Transporter Promotes Stomatal Conductance and Photosynthesis. Plant Cell 31, 1328–1343. 10.1105/tpc.18.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Yokosho K., Guo R., Whelan J., Ruan Y. L., Ma J. F., et al. (2019. b). The Soybean Sugar Transporter GmSWEET15 Mediates Sucrose Export from Endosperm to Early Embryo. Plant Physiol. 180 (4), 2133–2141. 10.1104/pp.19.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Liu F., Chen C., Ma F., Li M. (2014). The Malus domestica sugar transporter gene family: identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front. Plant Sci. 5, 569. 10.3389/fpls.2014.00569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H. H., Wang D., Qin Y. Q., Ma A. N., Fu J. X., Qin Y. H., et al. (2019). Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development. BMC Plant Biol. 19 (1), 499. 10.1186/s12870-019-2120-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Chen L. L., Ruan X., Chen D., Zhu A., Chen C., et al. (2013). The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 45 (1), 59–66. 10.1038/ng.2472 [DOI] [PubMed] [Google Scholar]
- Xuan Y. H., Hu Y. B., Chen L. Q., Sosso D., Ducat D. C., Hou B. H., et al. (2013). Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. U.S.A. 110 (39), E3685–E3694. 10.1073/pnas.1311244110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. M., Huang J., Li X. G. (2015). Identification of appropriate reference genes for RT-qPCR analysis in Ziziphus jujuba Mill. Sci. Hortic. 197, 166–169. 10.1016/j.scienta.2015.09.026 [DOI] [Google Scholar]
- Zhang C. M., Bian Y., Hou S. H., Li X. G. (2018). Sugar transport played a more important role than sugar biosynthesis in fruit sugar accumulation during Chinese jujube domestication. Planta 248 (5), 1187–1199. 10.1007/s00425-018-2971-1 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Chan K., Wang T. L., Hedley C. L., Offler C. E., Patrick J. W. (2009). Intracellular sucrose communicates metabolic demand to sucrose transporters in developing pea cotyledons. J. Exp. Bot. 60 (1), 71–85. 10.1093/jxb/ern254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. G., Long S. P., Ort D. R. (2010). Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61, 235–261. 10.1146/annurev-arplant-042809-112206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .