Abstract
A continuing challenge to crop production worldwide is the spectrum of diseases caused by geminiviruses, a large family of small circular single-stranded DNA viruses. These viruses are quite diverse, some containing mono- or bi-partite genomes, and infecting a multitude of monocot and dicot plants. There are currently many efforts directed at controlling these diseases. While some of the methods include controlling the insect vector using pesticides or genetic insect resistance (Rodríguez-López et al., 2011), this review will focus on the generation of plants that are resistant to geminiviruses themselves. Genetic resistance was traditionally found by surveying the wild relatives of modern crops for resistance loci; this method is still widely used and successful. However, the quick rate of virus evolution demands a rapid turnover of resistance genes. With better information about virus-host interactions, scientists are now able to target early stages of geminivirus infection in the host, preventing symptom development and viral DNA accumulation.
Keywords: geminivirus, resistance, genetic resistance, agriculture, genetic engineering
Introduction: Viral Proteins May Be Targets for Resistance
Geminiviruses are circular single-stranded DNA viruses that infect a wide range of plant species including many important crops. Damages attributed to geminiviruses include over $300 million in loss in the Indian bean industry (Patil et al., 2014), up to 100% loss of tomato crop in Italy and the Dominican Republic (Picó et al., 1996), and nearly $2 billion loss in African cassava production (Patil and Fauquet, 2009). The impact of geminiviruses is widespread and destructive. The family Geminiviridae has nine genera based on viral genome structure and insect vectors. In the case of begomoviruses, genomes can be mono- or bipartite, with each circular DNA (~2.5 Kb) packaged in a twinned icosahedral particle (Zerbini et al., 2017).
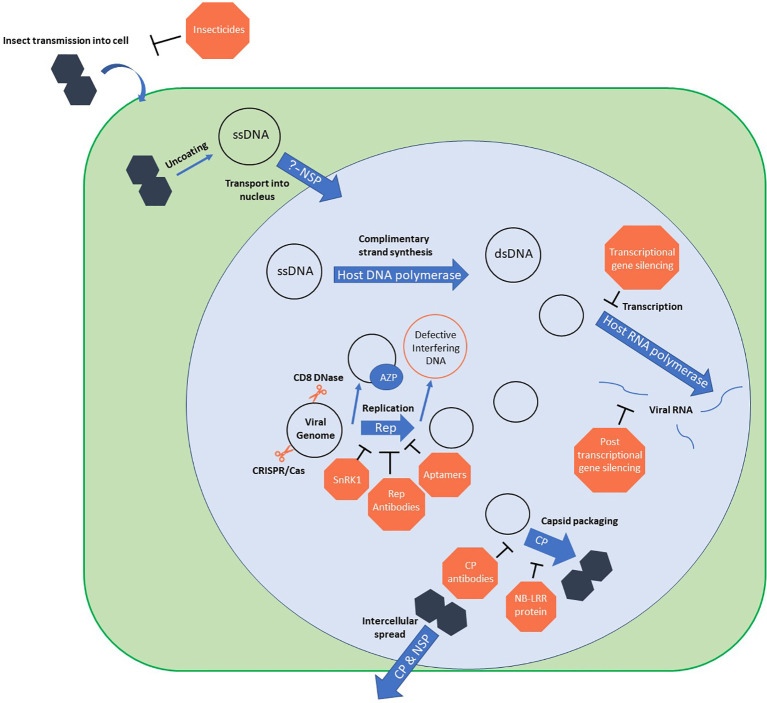
Geminivirus infection begins when an insect vector containing virions feeds on a host plant. The viral genome is deposited and unpackaged in the phloem cells. A complementary strand is synthesized, then the dsDNA is replicated and packaged into mini-chromosomes using host histones (reviewed in Jeske, 2009 and Hanley-Bowdoin et al., 2013). The viral genes are transcribed by host RNA Polymerase II. The viral genome is replicated by rolling-circle and recombination-dependent replication systems (Jeske et al., 2001). These processes require both host and viral proteins. See Figure 1 for an overview of the geminivirus life cycle.
Figure 1.
Geminivirus life cycle with points of resistance. Orange blocks show points where known resistance mechanisms interfere with the virus. ssDNA, single stranded DNA; dsDNA, double stranded DNA; NSP, nuclear shuttle protein; AZP, artificial zinc-finger protein; Rep, replication-associated protein; CP, coat protein.
The most critical geminiviral protein for virus replication is the replication initiator protein (Rep). It initiates replication by binding the origin sequence, nicking the DNA, and associating with host factors (Laufs et al., 1995; Kong et al., 2000; Arguello-Astorga et al., 2004; Desvoyes et al., 2006). The geminiviral transcriptional activator protein (TrAP) is involved in pathogenicity and suppresses both transcriptional and post transcriptional gene silencing (Sunter and Bisaro, 1992; Hong et al., 1996; Voinnet et al., 1999; Shivaprasad et al., 2005; Trinks et al., 2005; Wang et al., 2005; Chowda-Reddy et al., 2009; Castillo-González et al., 2015; Kumar et al., 2015). Furthermore, the replication enhancer protein (REn) contributes to increased viral replication and interacts with host factors and with Rep. Geminiviruses also encode proteins for packaging and movement. The coat protein (CP) comprises the viral capsid and is critical for vector specificity (Noris et al., 1998) and viral nuclear import (Liu et al., 1999) and is also required for symptom development. In bipartite geminiviruses, the nuclear shuttle protein (NSP) is responsible for nuclear import by binding ssDNA. It also aids in intercellular movement by interacting with the movement protein (MP) at the membrane. MP is responsible primarily for the intercellular spread of viral DNA (Noueiry et al., 1994) by increasing the size exclusion limit of plasmodesmata (Rojas et al., 2001). For excellent reviews in these subjects please see Hanley-Bowdoin et al., 2013; Czosnek et al., 2017; Rojas et al., 2018, and Martins et al., 2020.
Naturally Occurring Resistance
In the process of crop domestication, traits that are not directly beneficial are often bred out in favor of those increasing yield in particular conditions. When a new challenge arises, breeders search wild relatives for traits that increase survival. This is the case with geminiviruses: there are multiple geminivirus resistance genes from undomesticated relatives that are utilized in agriculture. Examples include many crops like beans (reviewed by Blair and Morales, 2008) or cotton (Zaidi et al., 2020). This minireview will cover resistance genes in tomato and cassava in detail. A selected list of resistance genes for these crops is presented in Table 1.
Table 1.
List of genes involved in geminivirus resistance and geminiviral suppressors of silencing.
| Gene | Protein | Plant | Virus | Reference |
|---|---|---|---|---|
| Ty-1, Ty-3 | RNA-dependent polymerase | S. lycopersicum | TYLCV | (Zamir et al., 1994; Verlaan et al., 2013; Butterbach et al., 2014) |
| Ty-2 | NB-LRR class protein | S. lycopersicum | TYLCV | (Yang et al., 2014; Yamaguchi et al., 2018) |
| Ty-4 | S. lycopersicum | TYLCV | (Ji et al., 2009) | |
| ty-5 | pelota | S. lycopersicum | TYLCV | (Hutton et al., 2012; Scott et al., 2015; Lapidot et al., 2015) |
| Ty-6 | S. lycopersicum | TYLCV | (Scott et al., 2015) | |
| CMD1, CMD3 | M. esculenta | ACMV | (Fregene et al., 2000; Akano et al., 2002; Okogbenin et al., 2012) | |
| CMD2 | M. esculenta | ACMV | (Akano et al., 2002; Kuria et al., 2017) | |
| CchGLP | MN-SOD | C. chinense | PHYVV, PepGMV | (León-Galván et al., 2011; Guevara-Olvera et al., 2012; Mejía-Teniente et al., 2015) |
| SlSnRK1 | SnRK1 | S. lycopersicum | TYLCV, CaLCuV | (Shen et al., 2011; Shen et al., 2014; Hulsmans et al., 2016) |
| Permease-1 like | S. habrochaites introgressed S. lycopersicum | TYLCV | (Eybishtz et al., 2009) | |
| LeHT1 | hexose transporter | S. lycopersicum | TYLCV | (Eybishtz et al., 2010; Sade et al., 2012) |
| SlVSRLip | lipocalin-like | S. lycopersicum | TYLCV | (Sade et al., 2012) |
| NIK | NSP-interacting kinase | A. thaliana, S. lycopersicum, G. max | begomoviruses | (Brustolini et al., 2015) |
| SGS3 | RNA binding protein in PTGS | N. benthamiana | TYLCV | (Li et al., 2017) |
| WRKY | Group III transcription factors | S. lycopersicum | TYLCV | (Huang et al., 2016) |
Solanum chilense is the most popular wild tomato relative for introgressing Tomato yellow leaf curl virus (TYLCV) resistance, as over 80% of its accessions are resistant (Yan et al., 2018). Many of the Ty family of resistance loci are from this plant. Ty-1 was introgressed into tomato from S. chilense and mapped to chromosome 6. It confers a tolerant symptomless response against TYLCV in homozygous plants (Zamir et al., 1994). Ty-1 is allelic with another resistance gene from S. chilense, Ty-3. These genes encode an RNA-dependent RNA polymerase (RDR) similar to RDRs 3, 4, and 5 in Arabidopsis thaliana, implying a role for RNA interference (Verlaan et al., 2013). A. thaliana itself may be a source for resistance as found by Reyes et al. (2017) in an accession showing immunity to Cabbage leaf curl virus (CbLCV) and Beet curly top virus (BCTV). Ty-1 plants have increased siRNA levels in comparison to non-resistant cultivars, corresponding with high levels of viral DNA methylation. This indicates transcriptional gene silencing (TGS) is likely involved in Ty-1/Ty-3 mediated resistance (Butterbach et al., 2014).
Ty-2 is a TYLCV resistance locus on tomato chromosome 11 introgressed from Solanum habrochaites (Yang et al., 2014). This locus hosts a gene known as TYNBS1 that encodes an Nucleotide Binding, Leucine Rich Repeat (NB-LRR) protein, known elsewhere to provide pathogen resistance (Yamaguchi et al., 2018). The recessive ty-5 locus from a hybrid tomato known as Tyking has been mapped to chromosome 4. It is closely tied to the Quantitative trait loci (QTL) marker SlNAC1 (Anbinder et al., 2009; Hutton et al., 2012). The gene responsible for ty-5 resistance is pelota, which encodes an mRNA surveillance factor homolog (Lapidot et al., 2015; Wang Y. et al., 2018). Ty-4 and Ty-6 are other resistances genes from S. chilense that are less understood. Ty-4 is a minor locus on tomato chromosome 3 (Ji et al., 2009). Ty-6 lies on chromosome 10, has incomplete dominance, and protects tomatoes against Tomato mottle virus (ToMoV) and TYLCV (Scott et al., 2015; Gill et al., 2019).
Cassava mosaic disease (CMD) is caused by multiple cassava mosaic geminiviruses, often in complexes. CMD has only three markers in cassava known to confer resistance. CMD2 is a single locus preferred by breeders due to its dominance (Akano et al., 2002). However, CMD2 is monogenic and thus at risk of viral evolution overcoming resistance. Monogenic resistance is not sufficient for long-term disease resistance, requiring constant innovation to keep ahead of viral evolution. The newest marker associated with CMD resistance is CMD3 (Okogbenin et al., 2012). It arose through crossbreeding of cultivars with the CMD2 locus and another recessive resistance locus, CMD1, and appears to provide the highest resistance level of the three (Kuria et al., 2017).
The CMD2-provided resistance is lost when the plants are regenerated through somatic embryogenesis (Beyene et al., 2016). The mechanism for this change is unknown. The other CMD resistance alleles (CMD1 and CMD3) are unaffected and still reliable for use. The loss of CMD2-based resistance could provide a method to predict if other traits will share the same phenomenon (Chauhan et al., 2018). There are several potential mechanisms for this effect including somaclonal variation, which is caused by epigenetic changes when the cell undergoes the disorganized regeneration phase in tissue culture (Bairu et al., 2011; Lee and Seo, 2018). However, virus evolution through pseudorecombination or recombination and not changes in the host can also be at play to break resistance in the field.
This ability to overcome genetic resistance is a very challenging aspect in geminivirus-plant interactions. One such example is the discovery of sequences enhancing geminivirus symptoms (SEGS) during CMD infection. These SEGS (encoded in the cassava genome) enabled the virus to cause symptoms in otherwise resistant cultivars, breaking the resistance (Ndunguru et al., 2016). Viruses themselves also have a high rate of mutation that can change the virus-host interactions to evade resistance. For TYLCV, the mean rate of genomic substitutions is estimated to be 2.88 x 10-4 nucleotide substitutions per site every year (Duffy and Holmes, 2008). This ability of geminiviruses to overcome genetic resistance has led to a resurgence of Cotton leaf curl disease (CLCuD) in south Asia after it had been nearly eradicated (Amrao et al., 2010).
Geminivirus-Plant Interactions and Understanding Host Resistance
Reverse genetics is a common method for identifying genes involved in viral infection. Eybishtz et al. used this method in 2009 to identify Permease I-like protein as a resistance factor against TYLCV. In knockout resistant plants, viral genomic titer increased and resulted in susceptibility (Eybishtz et al., 2009). Similarly, the hexose transporter LeHT1 was also identified as a resistance gene. In LeHT1-knockout plants, TYLCV accumulates and causes a necrotic response. This implies programmed cell death may be a factor in geminiviral response (Eybishtz et al., 2010). A downstream gene in the same LeHT1 pathway, a lipocalin-like gene, also results in a necrotic response to TYLCV when mutated from the resistant tomato (Sade et al., 2012).
A separate geminivirus resistance gene that also involves programmed cell death includes the germin-like CchGLP gene found in resistant peppers. This protein has manganese superoxide reductase activity which increases upon infection by Pepper golden mosaic virus or Pepper huasteco yellow vein virus in resistant plants only (León-Galván et al., 2011). Resistant peppers with knocked-out CchGLP develop symptoms, indicating this gene has an important role in defense (Mejía-Teniente et al., 2015). When transferred into susceptible tobacco plants, CchGLP conferred a mild symptom phenotype with increased reactive oxygen levels and expression of systemic acquired resistance-related genes (Guevara-Olvera et al., 2012). Reactive oxygen species promote programmed cell death, a general defense mechanism to prevent the spread of infectious entities such as bacteria or viruses (Lam, 2004). Tomato leaf curl New Delhi Virus (ToLCNDV) resistance protein SlRPT4, a 26S proteasome, is also shown to regulate programmed cell death and ROS production. This is in addition to its inhibitory binding of the viral genome (Sahu et al., 2016).
SnRK1 is a major regulator of energy and nutrients in the plant cell. It is also emerging with a role in plant response to biotic stress (Hulsmans et al., 2016). When challenged with geminivirus infection, A. thaliana SnRK1 has been shown to phosphorylate TrAP of CbLCV, delaying and attenuating symptoms (Shen et al., 2014). TrAP is an RNA silencing suppressor and acts as a transcription factor for two other geminiviral proteins (Fondong, 2013). In tomato, the analogue SlSnRK1 was shown to interact with the geminivirus satellite pathogenicity factor βC1. SlSnRK1 is shown to phosphorylate βC1, leading to symptom delay and viral DNA load reduction (Shen et al., 2011). Additionally, SnRK1 phosphorylates the Tomato golden mosaic virus Rep protein and interferes with its dsDNA binding function. This reduces viral DNA replication and symptoms (Shen et al., 2018). These features make SnRK1 an important factor in geminivirus defense. Other aspects of the host response like SUMOylation, senescence response and autophagy (reviewed by Kumar, 2019) may be used against the virus but viable resistance strategies have yet to be developed.
Another receptor kinase that is known to counteract geminivirus infection is the NSP-interacting kinase (NIK) family (Santos et al., 2010). A. thaliana NIKs are activated via oligomerization and autophosphorylation upon begomovirus infection (Brustolini et al., 2015). The downstream target of NIK is the ribosomal protein L10, which enters the nucleus and downregulates translation-related genes to slow down the infection (Santos et al., 2010; Brustolini et al., 2015). The begomovirus has evolved alongside NIK to suppress its antiviral activity by binding to the critical threonine 474, preventing the phosphorylation required for NIK activation (Santos et al., 2010). This same T474 when changed to phosphomimetic aspartate constitutively activates antiviral response genes, circumvents interference by NSP, and confers tolerance to begomovirus infection (Brustolini et al., 2015). Furthermore, a GTPase that interacts with NSP can also be conceived as a target to develop resistance against geminiviruses (Martins et al., 2020). These responses exemplify that understanding and changing genetic interactions can provide new solutions for disease management.
The status of the plant hormones in response to geminiviruses has not been exhaustively assessed. Hormone changes related to pathogen response fall within the salicylic acid (SA), jasmonic acid (JA), and ethylene pathways (ET), where SA pathway is upregulated during CbLCV infection in A. thaliana, JA is downregulated, and ET has both responses in a transcriptomic study (Ascencio-Ibáñez et al., 2008). Plants overexpressing SA (cpr1 mutants) displayed a substantial delay in symptom appearance upon infection, suggesting that plants with SA pathway always on may have partial protection against geminiviruses. On the other side, the CbLCV depletes the production of jasmonate. JA is known for deterring insects and reducing the transmissibility of geminiviruses (Escobar-Bravo et al., 2016) so it could be used to reduce infection and impact transmission of the virus (Sun et al., 2017). The issue is that SA and JA seemed to be antagonistic on the plants, which will make a plant producing both simultaneously at high levels not a viable alternative. Auxins, gibberellins, cytokinins, brassinosteroids, abscisic acid, and strigolactones are all involved in the plant responses. However, not enough information is available yet to derive a putative resistance path.
Engineered Resistance
Natural defense mechanisms are often utilized to engineer a plant with geminivirus resistance. This review will briefly cover selected examples of engineered geminivirus resistance that interfere with the viral replication cycle. Figure 1 puts these mechanisms in context with geminiviral life cycle. For a more detailed look at these approaches, see Loriato et al. (2020).
Interfering With Viral Proteins
Engineered resistance can act on proteins required for viral replication or on viral DNA itself. In an approach known as “immunomodulation”, transgenic plants express antibodies against viral proteins. Single-chain antibodies generated against the geminiviral coat protein were shown to bind and provide resistance in vivo (Zakri et al., 2012). Similarly, anti-Rep antibody expression can provide resistance, but the response varies between lines due to variable transgene expression (Safarnejad et al., 2009). DNase 3D8 is a recombinant antibody with single- and double-stranded non-specific DNase activity that has been tested against geminiviruses BCTV and Beet severe curly top virus. Though the expression levels had to be kept low to protect host nucleic acid, 3D8 expression prevented high levels of viral DNA accumulation (Lee et al., 2013).
Peptide aptamers are engineered peptide sequences to be expressed by the host to interfere with the activity of a protein of interest. These are significantly smaller than single-chain antibodies, but work similarly. They have been developed and tested to disrupt functions of geminivirus proteins, one of which is Rep from Tomato Golden Mosaic Virus. The aptamers bound Rep in vivo and lowered viral DNA production (Lopez-Ochoa et al., 2006). In transgenic tomato, peptide aptamers were also effective against TYLCV and ToMoV by reducing viral symptoms and viral load (Reyes et al., 2013). Another approach for replication interference is by competition, as has been shown with subgenomic DNA. However, this approach is highly virus-specific, making its practical use limited and inefficient (Stanley et al., 1990; Stenger, 1994).
Viral infection can also be impeded by sequestration of protein targets. Rep can be out-competed for origin binding by artificial zinc-finger proteins with specific DNA binding, and one has been shown with greater affinity for the TYLCV origin than Rep (Mori et al., 2013). A plant expressing it or a similar protein may inhibit geminivirus replication. Mendoza-Figueroa et al. found a globulin-derived peptide with high affinity for the TYLCV origin and that reduced viral load when applied to infected plants. This indicates that the peptide may not need to be expressed by the plant, but rather can be applied exogenously to interfere with Rep activity (Mendoza-Figueroa et al., 2018).
Viral Responses Against Gene Silencing Rendered by the Host
Gene silencing regulates host processes by eliminating mRNA before translation and is a defense against viral RNA. Dicer-like proteins cleave dsRNA sequences into short interfering RNA (siRNA) molecules 21–24 nt in length (Rey and Fondong, 2018). The siRNA is loaded into a complex with Argonaute proteins, which are then directed to complimentary DNA or RNA sequences. This results in several outcomes, including degradation or sequestration of existing RNAs (post-transcriptional gene silencing) or targeting DNA for methylation (transcriptional gene silencing) (Mathieu and Bender, 2004; Raja et al., 2010). RDRs spread silencing throughout the plant by multiplying the secondary siRNA signal (Rey and Fondong, 2018).
To counteract silencing, geminiviruses have evolved viral suppressors of RNA silencing which are often pathogenicity factors. For example, V2 of Tomato yellow leaf curl China virus interacts with and sequesters secondary siRNAs, hindering the spread of silencing. V2 of TYLCV suppresses gene silencing by interacting with host proteins suppressor of gene silencing 3 and histone deacetylase 6 (Fondong, 2013; Wieczorek and Obrępalska-Stęplowska, 2015; Li et al., 2017; Wang B. et al., 2018). Another example of RNA silencing suppression is TrAP of Mungbean yellow mosaic Indian virus. It slows siRNA production by blocking RDR6-mediated biogenesis of siRNA, binds to Argonaute 1 to prevent its action, and lowers global DNA methylation levels (Buchmann et al., 2009; Kumar et al., 2015). C4 protein of Cotton leaf curl Multan virus, Cassava mosaic viruses, Tomato leaf curl New Delhi virus, and BCTV is also known to suppress silencing (Vanitharani et al., 2004; Ismayil et al., 2018; Vinutha et al., 2018) and can be a symptom determinant in infection (Mills-Lujan and Deom, 2010).
Upon geminivirus challenge, an increase in siRNA is correlated with a decrease in symptoms (Chellappan et al., 2004). Resistance and recovery phenotypes are strongly associated with RNA silencing. The intergenic region of the BCTV is targeted by transcriptional gene silencing, evidenced by a greater proportion of siRNAs produced for this region along with a heavier methylation load in recovered plants (Yadav and Chattopadhyay, 2011; Coursey et al., 2018). There is a significant inverse relation between the viral DNA methylation and the disease progression, measured by symptom severity and viral titer (Rodríguez-Negrete et al., 2009; Yadav and Chattopadhyay, 2011).
Transgenic approaches have been developed in multiple systems to generate plants that produce antiviral siRNAs, artificial miRNAs, long non-coding RNAs, synthetic trans-acting small interfering RNAs, etc., with different levels of success (some reviewed in Kumar and Khan, 2019). This concept has been proven in transgenic beans against Bean golden mosaic virus (Aragão et al., 2013). In general, a viral gene is expressed in the host to initiate PTGS quicker and to a higher degree when challenged by the virus. The plants expressing the gene in hairpin form had the highest levels of resistance and siRNA, with some plants showing no symptoms at all (Leibman et al., 2015; Tomar et al., 2018). However, this is apparently dosage-dependent, as multiple studies have shown that resistance is inversely correlated with siRNA level and can be broken by high viral titer (Vanderschuren et al., 2009; Leibman et al., 2015). Furthermore, it has been described that the resistance is sequence dependent and only covers a single species of virus (Fuentes et al., 2016). This limits the use of the technology but does not preclude its application when the prevalent virus is a single species.
The recent advent of the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system has introduced new tools for generating resistance to geminiviruses. It has potential as a specific editing tool, and has shown to be effective against several different geminiviruses when expressed in plants (reviewed by Zaidi et al., 2016). In all cases, the presence of the Cas endonuclease and targeted short guide RNAs (sgRNAs) reduced viral titer and symptom development. Studies suggest that the level of Cas9 expression was a deciding factor in the level of symptom reduction (Baltes et al., 2015; Ji et al., 2015). Not only is this system effective in reducing geminivirus infection but confers multiple-virus resistance when the conserved nonanucleotide sequence was targeted. This was demonstrated in a dual infection with TYLCV and BCTV. (Ali et al., 2015).
CRISPR/Cas9 offers a great tool to integrate geminivirus resistance into susceptible plants, yet it is not perfect. Zhang et al. described in 2018 that their high-specificity gRNA was still showing off-target effects when expressed in A. thaliana. These were reduced by using a modified gRNA scaffold (expressing the gRNA adjacent to tRNA9met) and using the SpCas9 mutant (Zhang et al., 2018). Another unintended consequence of CRISPR-based viral resistance is the evolution of the virus to evade sgRNA. This adaptation renders the resistance useless (Mehta et al., 2019).
Conclusions
Although geminiviruses are very successful at infecting their hosts, plants also evolve to overcome or tolerate infection. Protective measures are introduced by humans as we seek to maintain our crop productivity. It is important to continue innovating to keep pace with the rapid viral evolution. Resistance based on DNA sequence is at a disadvantage as it relies on sequence to remain unchanged. These also tend to be very virus-specific, making it difficult to protect crops against mixed infections that are common. Antibody-based systems and peptide aptamers may hold better long-term resistance since they put more indirect evolutionary pressure on the geminivirus genome. Additionally, the peptide aptamers showed a broader base of effectiveness (Reyes et al., 2013), which may be effective at fighting mixed infections. Aptamers can be improved and mixed or re-designed using in silico approaches to increase its affinity. However, the long history of virus/host competitive evolution shows that protein-based resistance can still be overcome. To continue developing more resistant crops, we must gain a greater basic understanding of how these viruses infect the plants and how the plant responds to and harbors viral replication. An ideal resistance would prevent initial viral replication, eliminating the opportunity for the virus to evolve. Furthermore, a combinatorial or additive approach targeting both the virus and the vector may provide with a better opportunity to impair the infection and provide a longer lasting protection to crops.
Author Contributions
JA-I conceived and supervised the review topics. KB wrote the first draft. All authors contributed to the article and approved the submitted version.
Funding
This work was partially funded by the Bill and Melinda Gates Foundation (CPT005698) and by the T&E Biochemistry Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Drs. Niki Robertson, Wei Shen, and Maria Reyes for the critical review of the manuscript. We also apologize to any authors and work that was not cited due to restrictions in the size of the manuscript.
References
- Akano A. O., Dixon A. G. O., Mba C., Barrera E., Fregene M. (2002). Genetic Mapping of a Dominant Gene Conferring Resistance to Cassava Mosaic Disease. Theor. Appl. Genet. 105 (4), 521–525. 10.1007/s00122-002-0891-7 [DOI] [PubMed] [Google Scholar]
- Ali Z., Abulfaraj A., Idris A., Ali S., Tashkandi M., Mahfouz M. M. (2015). CRISPR/Cas9-Mediated Viral Interference in Plants. Genome Biol. 16, 238–248. 10.1186/s13059-015-0799-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrao L., Amin I., Shahid M.S., Briddon R. W., Mansoor S. (2010). Cotton Leaf Curl Disease in Resistant Cotton Is Associated with a Single Begomovirus That Lacks an Intact Transcriptional Activator Protein. Virus Res. 152 (1–2), 153–163. 10.1016/j.virusres.2010.06.019 [DOI] [PubMed] [Google Scholar]
- Anbinder I., Reuveni M., Azari R., Paran I., Nahon S., Shlomo H., et al. (2009). Molecular Dissection of Tomato Leaf Curl Virus Resistance in Tomato Line TY172 Derived from Solanum Peruvianum. Theor. Appl. Genet. 119, 519–530. 10.1007/s00122-009-1060-z [DOI] [PubMed] [Google Scholar]
- Aragão F. J. L., Nogueira E. O. P. L., Tinoco M. L. P., Faria J. C. (2013). Molecular Characterization of the First Commercial Transgenic Common Bean Immune to the Bean Golden Mosaic Virus. J. Biotechnol. 166 (1–2), 42–50. 10.1016/j.jbiotec.2013.04.009 [DOI] [PubMed] [Google Scholar]
- Arguello-Astorga G., Lopez-Ochoa L., Kong L.-J., Orozco B. M., Settlage S. B., Hanley-Bowdoin L. (2004). A Novel Motif in Geminivirus Replication Proteins Interacts with the Plant Retinoblastoma-Related Protein. J. Virol. 78 (9), 4817–4826. 10.1128/JVI.78.9.4817-4826.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio-Ibáñez J. T., Sozzani R., Lee T.-J., Chu T.-M., Wolfinger R. D., Cella R., et al. (2008). Global Analysis of Arabidopsis Gene Expression Uncovers a Complex Array of Changes Impacting Pathogen Response and Cell Cycle during Geminivirus Infection. Plant Physiol. 148 (1), 436–454. 10.1104/pp.108.121038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairu M. W., Aremu A. O., Staden J. V. (2011). Somaclonal Variation in Plants: Causes and Detection Methods. Plant Growth Regulation no 63, 147–173. 10.1007/s10725-010-9554-x [DOI] [Google Scholar]
- Baltes N. J., Hummel A. W., Konecna E., Cegan R., Bruns A. N., Bisaro D. M., et al. (2015). Conferring Resistance to Geminiviruses with the CRISPR–Cas Prokaryotic Immune System. Nat. Plants 1 (10), 15145. 10.1038/nplants.2015.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene G., Chauhan R. D., Wagaba H., Moll T., Alicai T., Miano D., et al. (2016). Loss of CMD2-Mediated Resistance to Cassava Mosaic Disease in Plants Regenerated through Somatic Embryogenesis. Mol. Plant Pathol. 17 (7), 1095–1110. 10.1111/mpp.12353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair M. W., Morales F. J. (2008). “Geminivirus Resistance Breeding in Common Bean,” in CAB Reviews: Perspectives in Agriculture, Veterinary Science, Nutrition and Natural Resources, (CABI Wallingford UK: ) vol. 3. 10.1079/PAVSNNR20083089 [DOI] [Google Scholar]
- Brustolini O. J. B., Machado J. P. B., Condori-Apfata J. A., Coco D., Deguchi M., Loriato V. A. P., et al. (2015). Sustained NIK-Mediated Antiviral Signalling Confers Broad-Spectrum Tolerance to Begomoviruses in Cultivated Plants. Plant Biotechnol. J. 13 (9), 1300–1311. 10.1111/pbi.12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmann R.C., Asad S., Wolf J. N., Mohannath G., Bisaro D. M. (2009). Geminivirus AL2 and L2 Proteins Suppress Transcriptional Gene Silencing and Cause Genome-Wide Reductions in Cytosine Methylation. J. Virol. 83 (10), 5005–5013. 10.1128/JVI.01771-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterbach P., Verlaan M. G., Dullemans A., Lohuis D., Visser R. G.F., Bai Y., et al. (2014). Tomato Yellow Leaf Curl Virus Resistance by Ty-1 Involves Increased Cytosine Methylation of Viral Genomes and Is Compromised by Cucumber Mosaic Virus Infection. Proc. Natl. Acad. Sci. United States America 111 (35), 12942–12947. 10.1073/pnas.1400894111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-González C., Liu X., Huang C., Zhao C., Ma Z., Hu T., et al. (2015). Geminivirus-Encoded TrAP Suppressor Inhibits the Histone Methyltransferase SUVH4/KYP to Counter Host Defense. ELife 4, e06671. 10.7554/eLife.06671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan R. D., Beyene G., Taylor N. J. (2018). Multiple Morphogenic Culture Systems Cause Loss of Resistance to Cassava Mosaic Disease. BMC Plant Biol. 18 (1), 132. 10.1186/s12870-018-1354-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P., Vanitharani R., Fauquet C. M. (2004). Short Interfering RNA Accumulation Correlates with Host Recovery in DNA Virus-Infected Hosts, and Gene Silencing Targets Specific Viral Sequences. J. Virol. 78 (14), 7465–7477. 10.1128/JVI.78.14.7465-7477.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowda-Reddy R. V., Dong W., Felton C., Ryman D., Ballard K., Fondong V. N. (2009). Characterization of the Cassava Geminivirus Transcription Activation Protein Putative Nuclear Localization Signal. Virus Res. 145 (2), 270–278. 10.1016/j.virusres.2009.07.022 [DOI] [PubMed] [Google Scholar]
- Coursey T., Regedanz E., Bisaro D. M. (2018). Arabidopsis RNA Polymerase V Mediates Enhanced Compaction and Silencing of Geminivirus and Transposon Chromatin during Host Recovery from Infection. J. Virol. 92 (7), e01320–e01317. 10.1128/JVI.01320-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czosnek H., Hariton-Shalev A., Sobol I., Gorovits R., Ghanim M. (2017). The Incredible Journey of Begomoviruses in Their Whitefly Vector. Viruses 9 (10), 273–291. 10.3390/v9100273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B., Ramirez-Parra E., Xie Qi, Chua N. H., Gutierrez C. (2006). Cell Type-Specific Role of the Retinoblastoma/E2F Pathway during Arabidopsis Leaf Development. Plant Physiol. 140 (1), 67–80. 10.1104/pp.105.071027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Holmes E. C. (2008). Phylogenetic Evidence for Rapid Rates of Molecular Evolution in the Single-Stranded DNA Begomovirus Tomato Yellow Leaf Curl Virus. J. Virol. 82 (2), 957–965. 10.1128/JVI.01929-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Bravo R., Alba J. M., Pons C., Granell A., Kant M. R., Moriones E., et al. (2016). A Jasmonate-Inducible Defense Trait Transferred from Wild into Cultivated Tomato Establishes Increased Whitefly Resistance and Reduced Viral Disease Incidence. Front. Plant Sci. 7:1732:1732. 10.3389/fpls.2016.01732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eybishtz A., Peretz Y., Sade D., Akad F., Czosnek H. (2009). Silencing of a Single Gene in Tomato Plants Resistant to Tomato Yellow Leaf Curl Virus Renders Them Susceptible to the Virus. Plant Mol. Biol. 71 (1–2), 157–171. 10.1007/s11103-009-9515-9 [DOI] [PubMed] [Google Scholar]
- Eybishtz A., Peretz Y., Sade D., Gorovits R., Czosnek H. (2010). Tomato Yellow Leaf Curl Virus Infection of a Resistant Tomato Line with a Silenced Sucrose Transporter Gene LeHT1 Results in Inhibition of Growth, Enhanced Virus Spread, and Necrosis. Planta 231 (3), 537–548. 10.1007/s00425-009-1072-6 [DOI] [PubMed] [Google Scholar]
- Fondong V. N. (2013). Geminivirus Protein Structure and Function. Mol. Plant Pathol. 14 (6), 635–649. 10.1111/mpp.12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregene M., Bernal A., Duque M., Dixon A., Tohme J. (2000). AFLP Analysis of African Cassava (Manihot Esculenta Crantz) Germplasm Resistant to the Cassava Mosaic Disease (CMD). Theor. Appl. Genet. 100, 678–685. 10.1007/s001220051339 [DOI] [Google Scholar]
- Fuentes A., Carlos N., Ruiz Y., Callard D., Anchez Y., Ia M., et al. (2016). Field Trial and Molecular Characterization of RNAi-Transgenic Tomato Plants That Exhibit Resistance to Tomato Yellow Leaf Curl Geminivirus. Mol. Plant-Microbe Interact. 29 (3), 197–209. 10.1094/MPMI-08-15-0181-R [DOI] [PubMed] [Google Scholar]
- Gill U., Scott J. W., Shekasteband R., Ogundiwin E., Schuit C., Francis D. M., et al. (2019). Ty-6, a Major Begomovirus Resistance Gene on Chromosome 10, Is Effective against Tomato Yellow Leaf Curl Virus and Tomato Mottle Virus. Theor. Appl. Genet. 132 (5), 1543–1554. 10.1007/s00122-019-03298-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Olvera L., Ruíz-Nito M. L., Rangel-Cano R. M., Torres-Pacheco I., Rivera-Bustamante R. F., Muñoz-Sánchez C. I., et al. (2012). Expression of a Germin-like Protein Gene (CchGLP) from a Geminivirus-Resistant Pepper (Capsicum Chinense Jacq.) Enhances Tolerance to Geminivirus Infection in Transgenic Tobacco. Physiol. Mol. Plant Pathol. 78, 45–50. 10.1016/J.PMPP.2012.01.005 [DOI] [Google Scholar]
- Hanley-Bowdoin L., Bejarano E. R., Robertson D., Mansoor S. (2013). Geminiviruses: Masters at Redirecting and Reprogramming Plant Processes. Nat. Rev. Microbiol. 11, 777–788. 10.1038/nrmicro3117 [DOI] [PubMed] [Google Scholar]
- Hong Y., Saunders K., Hartley M. R., Stanley J. (1996). Resistance to Geminivirus Infection by Virus-Induced Expression of Dianthin in Transgenic Plants. Virology 220, 119–127. 10.1006/viro.1996.0292 [DOI] [PubMed] [Google Scholar]
- Huang Y., Li M.-Y., Wu P., Xu Z.-S., Que F., Wang F., et al. (2016). Members of WRKY Group III Transcription Factors Are Important in TYLCV Defense Signaling Pathway in Tomato (Solanum Lycopersicum). BMC Genomics 17 (1), 788. 10.1186/s12864-016-3123-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmans S., Rodriguez M., Coninck B. De, Rolland F. (2016). The SnRK1 Energy Sensor in Plant Biotic Interactions. Trends Plant Sci. 21 (8), 648–661. 10.1016/J.TPLANTS.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Hutton S. F., Scott J. W., Schuster D. J. (2012). Recessive Resistance to Tomato Yellow Leaf Curl Virus from the Tomato Cultivar Tyking Is Located in the Same Region as Ty-5 on Chromosome 4. HortScience 47 (3), 324–327. 10.21273/HORTSCI.47.3.324 [DOI] [Google Scholar]
- Ismayil A., Haxim Y., Wang Y., Li H., Qian L., Han T., et al. (2018). Cotton Leaf Curl Multan Virus C4 Protein Suppresses Both Transcriptional and Post-Transcriptional Gene Silencing by Interacting with SAM Synthetase. PloS Pathog. 14 (8), e1007282. 10.1371/journal.ppat.1007282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske H., Lütgemeier M., Preiß W. (2001). DNA Forms Indicate Rolling Circle and Recombination-Dependent Replication of Abutilon Mosaic Virus. EMBO J. 20 (21), 6158–6167. 10.1093/emboj/20.21.6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske H. (2009). “Geminiviruses,” in TT Viruses. Current Topics in Microbiology and Immunology, vol. 331 . Eds. de Villiers E. M., Hausen H. (Berlin, Heidelberg: Springer; ), 185–226. 10.1007/978-3-540-70972-5_11 [DOI] [PubMed] [Google Scholar]
- Ji Y., Scott J. W., Schuster D. J., Maxwell D. P. (2009). Molecular Mapping of Ty-4, a New Tomato Yellow Leaf Curl Virus Resistance Locus on Chromosome 3 of Tomato. J. Am. Soc. Hortic. Sci. 134 (2), 281–288. 10.21273/JASHS.134.2.281 [DOI] [Google Scholar]
- Ji X., Zhang H., Zhang Yi, Wang Y., Gao C. (2015). Establishing a CRISPR–Cas-like Immune System Conferring DNA Virus Resistance in Plants. Nat. Plants 1 (10). 15144 10.1038/nplants.2015.144 [DOI] [PubMed] [Google Scholar]
- Kong L.-J., Orozco B. M., Roe J. L., Nagar S., Ou S., Feiler H. S., et al. (2000). A Geminivirus Replication Protein Interacts with the Retinoblastoma Protein through a Novel Domain to Determine Symptoms and Tissue Specificity of Infection in Plants. EMBO J. 19 (13), 3485–3495. 10.1093/emboj/19.13.3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Khan J. A. (2019). “Geminivirus Resistance Strategies,” in Geminiviruses. Ed. Kumar R. (Springer Nature Switzerland: Springer International Publishing; ), 197–218. 10.1007/978-3-030-18248-9_11 [DOI] [Google Scholar]
- Kumar V., Mishra S. K., Rahman J., Taneja J., Sundaresan G., Sanan Mishra N., et al. (2015). Mungbean Yellow Mosaic Indian Virus Encoded AC2 Protein Suppresses RNA Silencing by Inhibiting Arabidopsis RDR6 and AGO1 Activities. Virology 486, 158–172. 10.1016/J.VIROL.2015.08.015 [DOI] [PubMed] [Google Scholar]
- Kumar R.V. (2019). Plant Antiviral Immunity against Geminiviruses and Viral Counter-Defense for Survival. Front. Microbiol. 10 :1460:1460. 10.3389/fmicb.2019.01460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuria P., Ilyas M., Ateka E., Miano D., Onguso J., Carrington J. C., et al. (2017). Differential Response of Cassava Genotypes to Infection by Cassava Mosaic Geminiviruses. Virus Res. 227, 69–81. 10.1016/J.VIRUSRES.2016.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E. (2004). Controlled Cell Death, Plant Survival and Development. Nat. Rev. 5 (4), 305–315. 10.1038/nrm1358 [DOI] [PubMed] [Google Scholar]
- Lapidot M., Karniel U., Gelbart D., Fogel D., Evenor D., Kutsher Y., et al. (2015). A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota. PloS Genet. 11 (10), e1005538. 10.1371/journal.pgen.1005538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs J., Traut W., Heyraud F., Matzeit V., Rogers S. G., Schell J., et al. (1995). In Vitro Cleavage and Joining at the Viral Origin of Replication by the Replication Initiator Protein of Tomato Yellow Leaf Curl Virus. Proc. Natl. Acad. Sci. United States America 92, 3879–3883. 10.1073/pnas.92.9.3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Seo P. J. (2018). Dynamic Epigenetic Changes during Plant Regeneration. Trends Plant Sci. 23 (3), 235–247. 10.1016/j.tplants.2017.11.009 [DOI] [PubMed] [Google Scholar]
- Lee G., Shim H.-K., Kwon M.-H., Son S.-H., Kim K.-Y., Park E.-Y., et al. (2013). A Nucleic Acid Hydrolyzing Recombinant Antibody Confers Resistance to Curtovirus Infection in Tobacco. Plant Cell Tissue Organ Culture (PCTOC) 115 (2), 179–187. 10.1007/s11240-013-0357-4 [DOI] [Google Scholar]
- Leibman D., Prakash S., Wolf D., Zelcer A., Anfoka G., Haviv S., et al. (2015). Immunity to Tomato Yellow Leaf Curl Virus in Transgenic Tomato Is Associated with Accumulation of Transgene Small RNA. Arch. Virol. 160 (11), 2727–2739. 10.1007/s00705-015-2551-7 [DOI] [PubMed] [Google Scholar]
- León-Galván F., De A., Joaquín-Ramos J., Torres-Pacheco I., De La Rosa A. P.B., Guevara-Olvera L., et al. (2011). A Germin-Like Protein Gene (CchGLP) of Capsicum Chinense Jacq. Is Induced during Incompatible Interactions and Displays Mn-Superoxide Dismutase Activity. Int. J. Mol. Sci. 12, 7301–7313. 10.3390/ijms12117301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wang Y., Zhou X. (2017). SGS3 Cooperates with RDR6 in Triggering Geminivirus-Induced Gene Silencing and in Suppressing Geminivirus Infection in Nicotiana Benthamiana. Viruses 9 (9). 10.3390/v9090247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Boulton M., II, Thomas C. L., Prior D. A. M., Oparka K. J., Davies J. W. (1999). Maize Streak Virus Coat Protein Is Karyophyllic and Facilitates Nuclear Transport of Viral DNA. Mol. Plant-Microbe Interact. 12 (10), 894–900. 10.1094/MPMI.1999.12.10.894 [DOI] [PubMed] [Google Scholar]
- Lopez-Ochoa L., Ramirez-Prado J., Hanley-Bowdoin L. (2006). Peptide Aptamers That Bind to a Geminivirus Replication Protein Interfere with Viral Replication in Plant Cells. J. Virol. 80 (12), 5841–5853. 10.1128/JVI.02698-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loriato V. A. P., Martins L. G. C., Euclydes N. C., Reis P. A. B., Duarte C. E. M., Fontes E. P. B. (2020). Engineering Resistance against Geminiviruses: A Review of Suppressed Natural Defenses and the Use of RNAi and the CRISPR/Cas System. Plant Sci. 292:110410. 10.1016/j.plantsci.2020.110410 [DOI] [PubMed] [Google Scholar]
- Martins L. G. C., Raimundo G. A. S., Ribeiro N. G. A., Silva J. C. F., Euclydes N. C., Loriato V. A. P., et al. (2020). A Begomovirus Nuclear Shuttle Protein-Interacting Immune Hub: Hijacking Host Transport Activities and Suppressing Incompatible Functions. Front. Plant Sci. 11:398. 10.3389/fpls.2020.00398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O., Bender J. (2004). RNA-Directed DNA Methylation. J. Cell Sci. 117, 4881–4888. 10.1242/jcs.01479 [DOI] [PubMed] [Google Scholar]
- Mehta D., Stürchler A., Anjanappa R. B., Zaidi S. S.-e., Hirsch-Hoffmann M., Gruissem W., et al. (2019). Linking CRISPR-Cas9 Interference in Cassava to the Evolution of Editing-Resistant Geminiviruses. Genome Biol. 20 (1), 80. 10.1186/s13059-019-1678-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejía-Teniente L., Joaquin-Ramos A., Torres-Pacheco I., Rivera-Bustamante R., Guevara-Olvera L., Rico-García E., et al. (2015). Silencing of a Germin-Like Protein Gene (CchGLP) in Geminivirus-Resistant Pepper (Capsicum Chinense Jacq.) BG-3821 Increases Susceptibility to Single and Mixed Infections by Geminiviruses PHYVV and PepGMV. Viruses 7 (12), 6141–6151. 10.3390/v7122930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Figueroa J. S., Kvarnheden A., Méndez-Lozano J., Rodríguez-Negrete E.-A., Arreguín-Espinosa De Los Monteros R., Soriano-García M. (2018). A Peptide Derived from Enzymatic Digestion of Globulins from Amaranth Shows Strong Affinity Binding to the Replication Origin of Tomato Yellow Leaf Curl Virus Reducing Viral Replication in Nicotiana Benthamiana. Pesticide Biochem. Physiol. 145, 56–65. 10.1016/j.pestbp.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Mills-Lujan K., Deom C. M. (2010). Geminivirus C4 Protein Alters Arabidopsis Development. Protoplasma 239 (1–4), 95–110. 10.1007/s00709-009-0086-z [DOI] [PubMed] [Google Scholar]
- Mori T., Takenaka K., Domoto F., Aoyama Y., Sera T. (2013). Inhibition of Binding of Tomato Yellow Leaf Curl Virus Rep to Its Replication Origin by Artificial Zinc-Finger Protein. Mol. Biotechnol. 54 (2), 198–203. 10.1007/s12033-012-9552-5 [DOI] [PubMed] [Google Scholar]
- Ndunguru J., León L. D., Doyle C. D., Sseruwagi P., Plata G., Legg J. P., et al. (2016). Two Novel DNAs That Enhance Symptoms and Overcome CMD2 Resistance to Cassava Mosaic Disease. J. Virol. 90 (8), 4160–4173. 10.1128/JVI.02834-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noris E., Vaira A. M., Caciagli P., Masenga V., Gronenborn B., Accotto G. P. (1998). Amino Acids in the Capsid Protein of Tomato Yellow Leaf Curl Virus That Are Crucial for Systemic Infection, Particle Formation, and Insect Transmission. J. Virol. 72 (12), 10050–10057. 10.1128/JVI.72.12.10050-10057.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry A. O., Lucas W. J., Gilbertson R. L. (1994). Two Proteins of a Plant DNA Virus Coordinate Nuclear and Plasmodesmal Transport. Cell 76 (5), 925–932. 10.1016/0092-8674(94)90366-2 [DOI] [PubMed] [Google Scholar]
- Okogbenin E., Egesi C. N., Olasanmi B., Ogundapo O., Kahya S., Hurtado P., et al. (2012). Molecular Marker Analysis and Validation of Resistance to Cassava Mosaic Disease in Elite Cassava Genotypes in Nigeria. Crop Sci. 52 (6), 2576–2586. 10.2135/cropsci2011.11.0586 [DOI] [Google Scholar]
- Patil B. L., Fauquet C. M. (2009). Cassava Mosaic Geminiviruses: Actual Knowledge and Perspectives. Mol. Plant Pathol. 10 (5), 685–701. 10.1111/j.1364-3703.2009.00559.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil B. L., Kumar L., Hema M., Sreenivasulu P., Kumar P.L., Reddy D. V.R. (2014). Tropical Food Legumes: Virus Diseases of Economic Importance and Their Control. Adv. Virus Res. 90, 431–505. 10.1016/B978-0-12-801246-8.00009-3 [DOI] [PubMed] [Google Scholar]
- Picó B., Díez M. J., Nuez F. (1996). Viral Diseases Causing the Greatest Economic Losses to the Tomato Crop. II. The Tomato Yellow Leaf Curl Virus - A Review. Scientia Hortic. 67 (3–4), 151–196. 10.1016/S0304-4238(96)00945-4 [DOI] [Google Scholar]
- Raja P., Wolf J. N., Bisaro D. M. (2010). RNA Silencing Directed against Geminiviruses: Post-Transcriptional and Epigenetic Components. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 1799 (3–4), 337–351. 10.1016/J.BBAGRM.2010.01.004 [DOI] [PubMed] [Google Scholar]
- Rey M. E. C., Fondong V. N. (2018). “Mechanisms of Virus Resistance in Plants,” in Genes, Genetics and Transgenics for Virus Resistance in Plants. Ed. Patil B. L. (Norfolk, UK: Caister Academic Press; ), 1–23. 10.21775/9781910190814.01 [DOI] [Google Scholar]
- Reyes M., II, Nash T. E., Dallas M. M., Ascencio-Ibáñez J.T., Hanley-Bowdoin L. (2013). Peptide Aptamers That Bind to Geminivirus Replication Proteins Confer a Resistance Phenotype to Tomato Yellow Leaf Curl Virus and Tomato Mottle Virus Infection in Tomato. J. Virol. 87 (17), 9691–9706. 10.1128/JVI.01095-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes M., II, Flores-Vergara M. A., Guerra-Peraza O., Rajabu C., Desai J., Hiromoto-Ruiz Y. H., et al. (2017). A VIGS Screen Identifies Immunity in the Arabidopsis Pla-1 Accession to Viruses in Two Different Genera of the Geminiviridae. Plant J. 92 (5), 796–807. 10.1111/tpj.13716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-López M. J., Garzo E., Bonani J. P., Fereres A., Fernández-Muñoz R., Moriones E. (2011). Whitefly Resistance Traits Derived from the Wild Tomato Solanum Pimpinellifolium Affect the Preference and Feeding Behavior of Bemisia Tabaci and Reduce the Spread of Tomato Yellow Leaf Curl Virus. Virology 101 (10), 1191. 10.1094/PHYTO-01-11-0028 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Negrete E. A., Carrillo-Tripp J., Rivera-Bustamante R. F. (2009). RNA Silencing against Geminivirus: Complementary Action of Posttranscriptional Gene Silencing and Transcriptional Gene Silencing in Host Recovery. J. Virol. 83 (3), 1332–1340. 10.1128/JVI.01474-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M. R., Jiang H., Salati R., Xoconostle-Cá B., Sudarshana M. R., Lucas W. J., et al. (2001). Functional Analysis of Proteins Involved in Movement of the Monopartite Begomovirus, Tomato Yellow Leaf Curl Virus. Virology 291, 110–125. 10.1006/viro.2001.1194 [DOI] [PubMed] [Google Scholar]
- Rojas M. R., Macedo M. A., Maliano M. R., Soto-Aguilar M., Souza J. O., Rob W., et al. (2018). World Management of Geminiviruses. Annu. Rev. Phytopathol. 56 (1), 637–677. 10.1146/annurev-phyto-080615-100327 [DOI] [PubMed] [Google Scholar]
- Sade D., Eybishtz A., Gorovits R., Sobol I., Czosnek H. (2012). A Developmentally Regulated Lipocalin-like Gene Is Overexpressed in Tomato Yellow Leaf Curl Virus-Resistant Tomato Plants upon Virus Inoculation, and Its Silencing Abolishes Resistance. Plant Mol. Biol. 80 (3), 273–287. 10.1007/s11103-012-9946-6 [DOI] [PubMed] [Google Scholar]
- Safarnejad M. R., Fischer R., Commandeur U. (2009). Recombinant-Antibody-Mediated Resistance against Tomato Yellow Leaf Curl Virus in Nicotiana Benthamiana. Arch. Virol. 154 (3), 457–467. 10.1007/s00705-009-0330-z [DOI] [PubMed] [Google Scholar]
- Sahu P. P., Sharma N., Puranik S., Chakraborty S., Prasad M. (2016). Tomato 26S Proteasome Subunit RPT4a Regulates ToLCNDV Transcription and Activates Hypersensitive Response in Tomato. Sci. Rep. 6 (1), 1–12. 10.1038/srep27078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A. A., Lopes K. V. G., Apfata J. A. C., Fontes E. P. B. (2010). NSP-Interacting Kinase, NIK: A Transducer of Plant Defence Signalling. J. Exp. Bot. 61 (14), 3839–3845. 10.1093/jxb/erq219 [DOI] [PubMed] [Google Scholar]
- Scott J. W., Hutton S. F., Freeman J. H. (2015). Fla. 8638B and Fla. 8624 Tomato Breeding Lines with Begomovirus Resistance Genes Ty-5 Plus Ty-6 and Ty-6, Respectively. HortScience 50 (9), 1405–1407. 10.21273/HORTSCI.50.9.1405 [DOI] [Google Scholar]
- Shen Q., Liu Z., Song F., Xie Qi, Hanley-Bowdoin L., Zhou X. (2011). Tomato SlSnRK1 Protein Interacts with and Phosphorylates βC1, a Pathogenesis Protein Encoded by a Geminivirus β-Satellite. Plant Physiol. 157 (3), 1394–1406. 10.1104/pp.111.184648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Dallas M. B., Goshe M. B., Hanley-Bowdoin L. (2014). SnRK1 Phosphorylation of AL2 Delays Cabbage Leaf Curl Virus Infection in Arabidopsis. J. Virol. 88 (18), 10598–10612. 10.1128/JVI.00761-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Bobay B. G., Greeley L. A., Reyes M., II, Rajabu C. A., Blackburn R.K., et al. (2018). Sucrose Nonfermenting 1-Related Protein Kinase 1 Phosphorylates a Geminivirus Rep Protein to Impair Viral Replication and Infection. Plant Physiol. 178 (1), 372–389. 10.1104/pp.18.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P. V., Akbergenov R., Trinks D., Rajeswaran R., Veluthambi K., Hohn T., et al. (2005). Promoters, Transcripts, and Regulatory Proteins of Mungbean Yellow Mosaic Geminivirus. J. Virol. 79 (13), 8149–8163. 10.1128/jvi.79.13.8149-8163.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley J., Frischmuth T., Ellwood S. (1990). Defective Viral DNA Ameliorates Symptoms of Geminivirus Infection in Transgenic Plants. Proc. Natl. Acad. Sci. United States America 87 (16), 6291–6295. 10.1073/PNAS.87.16.6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger D. C. (1994). Strain-Specific Mobilization and AmpLification of a Transgenic Defective-Interfering DNA of the Geminivirus Beet Curly Top Virus. Virology 203 (2), 397–402. 10.1006/VIRO.1994.1501 [DOI] [PubMed] [Google Scholar]
- Sun H., Chen L., Li J., Hu M., Ullah A., He X., et al. (2017). The JASMONATE ZIM-Domain Gene Family Mediates JA Signaling and Stress Response in Cotton. Plant Cell Physiol. 58 (12), 2139–2154. 10.1093/pcp/pcx148 [DOI] [PubMed] [Google Scholar]
- Sunter G., Bisaro D. M. (1992). Transactivation of Geminivirus AR1 and BRI Gene Expression by the Viral AL2 Gene Product Occurs at the Leve1 of Transcription. Plant Cell 4, 1321–1331. 10.1105/tpc.4.10.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar G., Chakrabarti S. K., Sharma N. N., Jeevalatha A., Sundaresha S., Vyas K., et al. (2018). RNAi-Based Transgene Conferred Extreme Resistance to the Geminivirus Causing Apical Leaf Curl Disease in Potato. Plant Biotechnol. Rep. 12 (3), 195–205. 10.1007/s11816-018-0485-8 [DOI] [Google Scholar]
- Trinks D., Rajeswaran R., Shivaprasad P. V., Akbergenov R., Oakeley E. J., Veluthambi K., et al. (2005). Suppression of RNA Silencing by a Geminivirus Nuclear Protein, AC2, Correlates with Transactivation of Host Genes. J. Virol. 79 (4), 2517–2527. 10.1128/jvi.79.4.2517-2527.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderschuren H., Alder A., Zhang P., Gruissem W. (2009). Dose-Dependent RNAi-Mediated Geminivirus Resistance in the Tropical Root Crop Cassava. Plant Mol. Biol. 70 (3), 265–272. 10.1007/s11103-009-9472-3 [DOI] [PubMed] [Google Scholar]
- Vanitharani R., Chellappan P., Pita J. S., Fauquet C. M. (2004). Differential Roles of AC2 and AC4 of Cassava Geminiviruses in Mediating Synergism and Suppression of Posttranscriptional Gene Silencing Downloaded From. J. Virol. 78 (17), 9487–9498. 10.1128/JVI.78.17.9487-9498.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan M. G., Hutton S. F., Ibrahem R. M., Kormelink R., Visser R. G.F., Scott J. W., et al. (2013). The Tomato Yellow Leaf Curl Virus Resistance Genes Ty-1 and Ty-3 Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases. PloS Genet. 9 (3), e1003399. 10.1371/journal.pgen.1003399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinutha T., Kumar G., Garg V., Canto T., Palukaitis P., Ramesh S. V., et al. (2018). Tomato Geminivirus Encoded RNAi Suppressor Protein, AC4 Interacts with Host AGO4 and Precludes Viral DNA Methylation. Gene 678, 184–195. 10.1016/j.gene.2018.08.009 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Pinto Y. M., Baulcombe D. C. (1999). Suppression of Gene Silencing: A General Strategy Used by Diverse DNA and RNA Viruses of Plants. Proc. Natl. Acad. Sci. United States America 96 (24), 14147–14152. 10.1073/PNAS.96.24.14147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Buckley K. J., Yang X., Buchmann R.C., Bisaro D. M. (2005). Adenosine Kinase Inhibition and Suppression of RNA Silencing by Geminivirus AL2 and L2 Proteins. J. Virol. 79 (12), 7410–7418. 10.1128/JVI.79.12.7410-7418.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Yang X., Wang Y., Xie Y., Zhou X. (2018). Tomato Yellow Leaf Curl Virus V2 Interacts with Host Histone Deacetylase 6 To Suppress Methylation-Mediated Transcriptional Gene Silencing in Plants. J. Virol. 92 (18), e00036–e00018. 10.1128/jvi.00036-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jiang J., Zhao L., Zhou R., Yu W., Zhao T. (2018). Application of Whole Genome Resequencing in Mapping of a Tomato Yellow Leaf Curl Virus Resistance Gene. Sci. Rep. 8 (9592), JVI.00036–18. 10.1038/s41598-018-27925-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek P., Obrępalska-Stęplowska A. (2015). Suppress to Survive—Implication of Plant Viruses in PTGS. Plant Mol. Biol. Rep. 33 (3), 335–346. 10.1007/s11105-014-0755-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R. K., Chattopadhyay D. (2011). Enhanced Viral Intergenic Region–Specific Short Interfering RNA Accumulation and DNA Methylation Correlates with Resistance Against a Geminivirus. Mol. Plant-Microbe Interact. 24 (10), 1189–1197. 10.1094/MPMI-03-11-0075 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H., Ohnishi J., Saito A., Ohyama A., Nunome T., Miyatake K., et al. (2018). An NB-LRR Gene, TYNBS1, Is Responsible for Resistance Mediated by the Ty-2 Begomovirus Resistance Locus of Tomato. Theor. Appl. Genet. 131 (6), 1345–1362. 10.1007/s00122-018-3082-x [DOI] [PubMed] [Google Scholar]
- Yan Z., Pérez-de-Castro A., Díez M. J., Hutton S. F., Visser R. G. F., Wolters A. M. A., et al. (2018). Resistance to Tomato Yellow Leaf Curl Virus in Tomato Germplasm. Front. Plant Sci. 9 (1198), 9–1198. 10.3389/fpls.2018.01198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Caro M., Hutton S. F., Scott J. W., Guo Y., Wang X., et al. (2014). Fine Mapping of the Tomato Yellow Leaf Curl Virus Resistance Gene Ty-2 on Chromosome 11 of Tomato. Mol. Breed. 34 (2), 749–760. 10.1007/s11032-014-0072-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S. S.-e., Mansoor S., Ali Z., Tashkandi M., Mahfouz M. M. (2016). Engineering Plants for Geminivirus Resistance with CRISPR/Cas9 System. Trends Plant Sci. 21 (4), 279–281. 10.1016/J.TPLANTS.2016.01.023 [DOI] [PubMed] [Google Scholar]
- Zaidi S. S. e., Naqvi R. Z., Asif M., Strickler S., Shakir S., Shafiq M., et al. (2020). Molecular Insight into Cotton Leaf Curl Geminivirus Disease Resistance in Cultivated Cotton (Gossypium Hirsutum). Plant Biotechnol. J. 18 (3), 691–706. 10.1111/pbi.13236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakri A. M., Ziegler A., Commandeur U., Fischer R., Torrance L. (2012). In Vivo Expression and Binding Activity of ScFv-RWAV, Which Recognizes the Coat Protein of Tomato Leaf Curl New Delhi Virus (Family Geminiviridae). Arch. Virol. 157 (7), 1291–1299. 10.1007/s00705-012-1310-2 [DOI] [PubMed] [Google Scholar]
- Zamir D., Ekstein-Michelson I., Zakay Y., Navot N., Zeidan M., Sarfatti M., et al. (1994). Mapping and Introgression of a Tomato Yellow Leaf Curl Virus Tolerance Gene, TY-1. Theor. Appl. Genet. 88 (2), 141–146. 10.1007/BF00225889 [DOI] [PubMed] [Google Scholar]
- Zerbini F.M., Briddon R. W., Idris A., Martin D. P., Moriones E., Navas-Castillo J., et al. (2017). ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 98, 131–133. 10.1099/jgv.0.000738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Xing H.-L., Wang Z.-P., Zhang H.-Y., Yang F., Wang X.-C., et al. (2018). Potential High-Frequency off-Target Mutagenesis Induced by CRISPR/Cas9 in Arabidopsis and Its Prevention. Plant Mol. Biol. 96 (4–5), 445–456. 10.1007/s11103-018-0709-x [DOI] [PMC free article] [PubMed] [Google Scholar]