Abstract
Background
Streptococcus sanguinis is Gram-positive bacteria that contribute to caries. Many antibacterial agents are resistant against bacteria so that the discovery of new antibacterial agents is a crucial issue. Mechanism of antibacterial agents by disrupting cell wall bacteria is a promising target to be developed. One of the enzymes contributing to the cell wall is MurA enzyme. MurA is an enzyme catalyzing the first step of peptidoglycan biosynthesis in the cell wall formation. Inhibiting MurA is an effective and efficient way to kill the bacteria. Source of bioactive compounds including the antibacterial agent can be found in natural product such as herbal plant. Piper betle L. was reported to contain active antibacterial compounds. However, there is no more information on the antibacterial activity and molecular mechanism of P. betle’s compound against S. sanguinis.
Purpose
The study aims to identify antibacterial constituents of P. betle L. and evaluate their activities through two different methods including in vitro and in silico analysis.
Materials and Methods
The antibacterial agent was purified by bioactivity-guided isolation with combination chromatography methods and the chemical structure was determined by spectroscopic methods. The in vitro antibacterial activity was evaluated by disc diffusion and dilution methods while the in silico study of a compound binds on the MurA was determined using PyRx program.
Results
The antibacterial compound identified as allylpyrocatechol showed inhibitory activity against S. sanguinis with an inhibition zone of 11.85 mm at 1%, together with MIC and MBC values of 39.1 and 78.1 μg/mL, respectively. Prediction for molecular inhibition mechanism of allylpyrocatechols against the MurA presented two allylpyrocatechol derivatives showing binding activity of −5.4, stronger than fosfomycin as a reference with the binding activity of −4.6.
Conclusion
Two allylpyrocatechol derivatives were predicted to have a good potency as a novel natural antibacterial agent against S. sanguinis through blocking MurA activity that causes disruption of bacterial cell wall.
Keywords: Piper betle L., allylpyrocatechol, Streptococcus sanguinis, MurA enzyme
Introduction
The low awareness of oral health can cause interference with the human health body.1 More than 750 bacteria are living in the oral environment and uncontrolled growth of bacteria to form the plaque, then it causes some oral diseases like caries, gingivitis, and periodontitis.2 One of the original bacteria in the oral cavity of humans is S. sanguinis those important as key pioneering bacteria to form biofilms on the tooth surface called dental plaque.3–5 The bacteria will be receptor-anchoring salivary glycoprotein, which used to inhibit bacterial growth. The colonies of S. sanguinis can form a new environment to be localization other microorganisms, resulting in pathogenesis.6,7
Currently, some antibiotics used as treatment stages to inhibit the bacteria, but it does not work completely, especially for pathogenic bacteria.8 Some antibiotics make an abnormal environment that causes stress conditions to bacteria, then induce cell death.9 The stressful condition influences evolution with genetic modification, known as resistance to antibiotics.10
There has been a rising interest in the alternative for mechanical plaque removal with antibacterial agents, it can inhibit bacterial growth without genetic modification and become an effective and efficient treatment. One of the options is the disruption of the bacteria cell wall by inhibiting peptidoglycan biosynthesis. Many enzymes contributing to peptidoglycan biosynthesis include MurA (UDP-N-acetylglucosamine enolpyruvyl transferase). This enzyme catalyzes transfer enolpyruvyl to UDP-N-acetylglucosamine.11,12 By blocking the action of MurA enzyme, bacteria cell wall will disrupt, and finally, bacteria will die automatically. This makes the MurA enzyme a promising target for antimicrobial agent.13,14
Nowadays, the research for drug discovery programs by many scientists used natural product sources to search lead compounds of new antibacterial agents.15,16 Some antibacterial compounds against S. sanguinis have been found and reported from natural products, as kaurane diterpene from A. foliacea,17 eugenol from E. caryophyllata,18 berberine from B. vulgaris,19 sabinene from C. japonica,20 and β-caryophyllene from R. officinalis.21 Another edible medicinal plant that potency has antibacterial properties is a Plant from Piper genus which is Piper betle L.22 The ethyl acetate extract of this plant showed the highest antibacterial activity against Streptococcus pyogenes, Streptococcus aureus, Porphyromonas vulgaris, and Escherichia coli, respectively.23 Other paper reported antibacterial compounds were isolated from P. betle L. identified as phenolic compounds derivatives of eugenol, hydroxyl chavicol, and chavibetol showed active against S. marcescens, K. pneumonia, E. faecalis and B. subtilis, respectively,24 while the active antibacterial constituents of P. betle L. against S. sanguinis have no reported.
As well as discovering new lead active compounds is an important point, the structure–activity relationship (SAR) study is another critical aspect to determine the drugs candidates are selective, specific, and effective to a receptor of target diseases. To determine and evaluates the prospective molecular mechanism aspects of new active compounds can be conducted with in silico analysis by predicting ligand conformation and the site binding targets.25 This step will complement the in vitro and in vivo assay data, thus speeding up the process of drug discovery.26
In this study, the research is focused on determining active constituents of P. betle L. as lead antibacterial compounds, and then it will be evaluated for their activities as an antibacterial against pathogenic oral bacteria S. sanguinis together with a prediction of the mechanism of action of antibacterial compounds as an inhibitor of MurA enzyme will be predicted by in silico studies.
Materials and Methods
Materials
The leaf of P. betle L. was cultivated in March 2019, from a local farmer in Bandung, West Java – Indonesia. The specimen was identified and deposit at the Laboratory of Taxonomy, Department of Biology, Universitas Padjadjaran, Indonesia. The chemicals for extraction and purification were used distilled organic solvent of methanol, n-hexane, ethyl acetate, acetone, ethanol, and distilled water, while the chemicals for spectroscopic analyses were used pro-analyzed (p.a) grades. The column chromatography separation was carried out on Silica G 60 (0.063–0.200 mm and 0.200–0.500 mm) and ODS RP-18 (0.040–0.063 mm), and on Silica G 60 F254 and ODS RP-18 F254S for TLC together with 10% H2SO4 (v/v) in ethanol for chemical identification analysis.
The bacteria Streptococcus sanguinis ATCC 10566 used for antibacterial test on Muller Hinton broth and Muller Hinton agar as a medium, chlorhexidine as a positive control, and anaerobic jar for antibacterial assay (purchased from Merck Co. Ltd. and Sigma Aldrich).
The material for in silico, the 3D structure of the MurA used in this study was obtained from the Protein Data Bank (PDB) ID: 1UAE.27 The receptor used is a protein (enzyme) UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) from UniProt knowledgebase (http://www.uniprot.org/): UniProtKB – P0A749. Structure 3D MurA obtained using the RSCB program (https://www.rcsb.org) with 1UAE format PDB. The tested ligand is Allylpyrocatechol 1. (3-prop-2-enylbenzene- 1,2-diol) and its stereoisomer of Allylpyrocatechol 2 (4-prop-2-enylbenzene-1,2-diol). Both compounds were retrieved from PubChem with ID compounds that are ID 70775 (1) and ID 292101 (2). Ligand as positive control is fosfomycin (ID 446987). All this data was taken from the PubChem compound database (https://www.ncbi.nlm.nih.gov/pccompound).
Instruments
The structure of active compounds was determined by spectroscopic methods of ultra-violet (UV) by 8452A Diode Array, infra-red (IR) with FTIR Shimadzu 8400, NMR (1H-NMR, 13C-NMR, DEPT 135°, HMQC, 1H–1H COSY, HMBC) with JEOL type ECA 500 MHz, and mass spectrometry (MS) with Water Acquit UPLC type triquadrupole. The TLC plates visualized by UV detector lamps with wavelengths of λmax at 254 and 365 nm. Antibacterial activity assay used microplate 96 well, micropipettes, microtubes, incubators, paper disks, and Biochrom microplate readers.
Isolation Compound from Extract of P. betle L
The freshly P. betle L. leaf (0.58 kg) was extracted with methanol and then was subsequently partitioned between n-hexane-water and water-ethyl acetate, to get extracts of methanol (32.82 g), n-hexane (14.39 g), and ethyl acetate (1.08 g) and water (10.03 g), respectively. All extract adjusted in a series concentration for antibacterial activity assay and the most active fraction was further separated.
The active ethyl acetate extract (0.62 g) was gradually chromatographed on Silica G 60 (0.063–0.200 mm) eluted with n-hexane, ethyl acetate, and methanol with gradient 5% (v/v) to yield 21 fractions. All fractions were tested by antibacterial assay to determine which fraction the most active is. The most active fraction is fraction 1 (0.2353 g) so it was further purified subsequently by chromatographed on ODS RP-18 eluted with H2O-MeOH with a gradient of 5% (v/v) and on Silica G 60 eluted n-hexane-EtOAc of gradient 1% (v/v) to yielded compound 1 (18 mg).
Structure Determination of Active Compound 1
The structure of active compounds was determined by comprehensive analysis data of spectroscopic methods including ultra-violet (UV) spectrum, infra-red (IR) spectrum, 1D and 2D-NMR spectra (1H-NMR, 13C-NMR, DEPT 135°, HMQC, 1H-1H COSY, HMBC) and mass spectrometry (MS) spectrum. The Original spectra of UV, FTIR, NMR and MS can be seen in the Supplement material section.
Evaluation for the Antibacterial Activity from the Extract and Active Compound of P. betle L. leaf
Antibacterial effects of betle (P. betle L.) extracts evaluated against Streptococcus sanguinis ATCC 10566 and were observed using Kirby–Bauer disk diffusion. The assay procedure of the sensitivity or resistance of S. sanguinis to compounds test according to CLSI protocols (CLSI, 2012).28 The extracts of P. betle were adjusted in series concentrations of 2%, 4%, 6%, 8%, and 10% while compound 1 was diluted in various concentrations of 1–5% for antibacterial evaluations against S. sanguinis. Methanol and water were used as negative control while chlorhexidine 2% as a positive control. On the other hand, bacteria were prepared by growing 1 ose of bacteria in 5 mL of broth media. This solution was incubated for 24 h at 37°C. After incubation, the optical density of the solution was measured using a Microplate reader at 620 nm. This solution was diluted by broth media until making 0.5 Mc Farland of bacteria solution. This culture (100 μL) was swabbed onto the agar media. Then, paper discs (6 mm) were impregnated with 20 μL of each sample and then placed on the surface of the agar contained bacteria. Furthermore, the samples were incubated for 48 h at 37°C. These tests were performed in duplicate. Inhibitions zone value (in mm) was measured after the incubation.
The MIC and MBC evaluation of Allylpyrocatechol 1 against S. sanguinis ATCC 10566 were determined by the micro-dilution method in 96-well microplate.29 The bacterial cells were pre-cultured in Muller Hinton broth at 37°C under aerobic conditions and incubated in the presence of compounds with the concentrations obtained by serial two-fold dilution at 37°C without shaking in the same broth for 24 h on microplate wells. The optical density of the solution in microplate was measured using a microplate at 620 nm. The MICs were estimated as the lowest concentrations where the bacterial cells were observed by OD value. Then, each solution in the well was spread on the surface of the agar and incubated for 24 h at 37°C. The minimum concentration of sample that is no bacteria growth under colony counter is determined as MBC value.
In silico Characterization of the Allylpyrocatechol Compounds
Characteristics of allylpirocatechol were confirmed using an online program. The chemical structure of allylpyrocatechol was drawn using PubChem (https://pubchem.ncbi.nlm.nih.gov/compound/292101) to obtain Canonical SMILES (C = CCC1 = C (C (= CC = C1) O) O). Canonical SMILES is used to convert chemical structures into 3D using the OPEN BABEL 2.4.2 program in PDB format, and the Structure 3D of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) was obtained using the RSCB program (https://www.rcsb.org/structure/1UAE) PDB format.
Virtual screening and docking of ligand-protein used Autodock Vina in the open-source PyRx 0.8 software. Allylpyrocatechols 1 and 2 as a ligand were used to bind MurA as a protein target, ligand-free for blind docking. The selected conformation was the conformation with the lowest bond energy that has a bond energy score of less than 1.0Å in the mean square root deviation (RMSD). The results of docking are visualized using PYMOL and then analyzed by the online program proteins plus https://proteins.plus/. PYMOL program shows the docking position and ligand-residue interactions in the form of 3D molecules. The ligation position of each MurA-Allylpyrocatechol complex was compared with the 3D structure of MurA-Fosfomycin. This step was used to determine the similarity of the allylpirocatechols 1 and 2 ligation position with fosfomycin.
Results
Isolation of Compound from the Extract of P. betle L. leaf
The form of an Isolated compound is a yellow oil that can be diluted in methanol. This pure compound that was isolated from P. betle has Rf value 0.45 in Thin-Layer Chromatography (TLC) on the ODS silica with methanol-water 3:7, v/v. Meanwhile, it has an Rf value of 0.4 in the 2D-TLC with a combination solvent n-hexane-ethyl acetate and n-hexane-acetone 4:1, v/v.
Structural Characterization of Compound
Spectral data of compound 1 are UV: 203.5 and 283.0 nm. IR: 3348, 1281, 3078, 1638, and 2925 cm−1. MS (negative ion mode): (m/z): 149.37. 1H-NMR (CD3OD): δH 3.26 (2H, d, 7.0 Hz, H-1), 6.79 (1H, d, 8.0 Hz, H-2), 5.07 (2H, ddd, 1.5; 8.0; 16.5 Hz, H-3), 6.71 (1H, d, 2.5 Hz, H-4), 6.62 (1H, dd, 2.0; 8.0 Hz, H-5), 5.89 and 5.94 (1H, pd, 7.0; 8.0; 16.5 Hz, H-7). 13C-NMR (CD3OD): δC 39.6 (C-1), 115.6 (C-2), 115.7 (C-3), 115.9 (C-4), 121.2 (C-5), 133.4 (C-6), 137.7 (C-7), 141.7 (C-8), 143.5 (C-9).
Compound 1 was a yellow oil and the molecular weight (m/z) of 1 was 149.37 (C9H10O2) based on the [M+H]− peak in mass spectrometry ions. The IR spectra indicated the presence of hydroxyl and carbonyl group at absorptions of 3348 (O-H), 1281 (C-O), 3078 (C-H sp2), 1638 (C = C), and 2925 cm−1 (C-H sp3), respectively.24 Further supporting data from the UV-Vis spectrum showed absorption peaks at 203.5 nm correspond to the E band with an auxochrome substituent of a heteroatom and at 283.0 nm those suggested for an aromatic benzoic bond, respectively.30
The 13C-NMR spectrum of 1 revealed nine carbon signals including for one methylene carbon at δC 39.6 ppm, two alkene carbons at δC 137.7 and 115.7 ppm, four aromatic carbons at δC 115.6, 115.9, 121.2, 133.4; ppm together with two oxygenated quaternary carbons at 141.8 and 143.5 ppm, respectively.
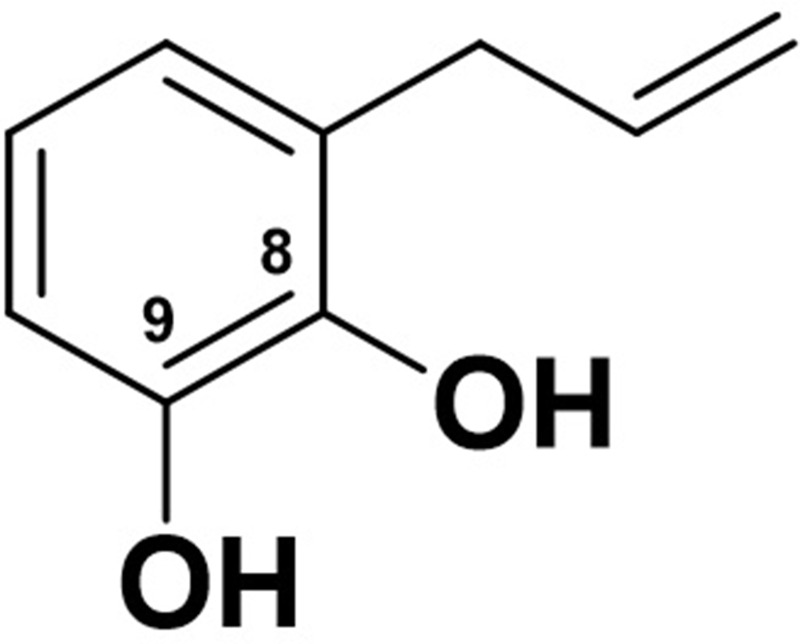
The 1H-NMR spectrum showed three proton signals for an aromatic skeleton at δH 6.71 (1H, d, J = 2.0 Hz); 6.62 (1H, dd, J = 2.0; 8.0 Hz); and 6.79 ppm (1H, d, J = 8.0 Hz) which indicated the meta and ortho positions. The other proton signals at δH 5.07 ppm (1H, dd, J = 1.5; 8.0 Hz dan 1H, dd, J = 1.5; 16.5 Hz) were considered as geminal proton,24 while the double bond group was identified by proton signals at δH 5.89 and 5.94 ppm (1H, ddd, J = 7.0; 8.0; 16.5 Hz), respectively. From the J values of 8.0 and 16.5 Hz, the configuration cis and trans of the geminal protons to double bond were identified. Further analysis of HMBC analysis showed a correlated signal of H-1 to C2, C3, C5, C6, and C7 formed of the alleged group. Other correlated signals showed connectivity of H-2 to C5, C6 and C9, H-3 to C1 and C7, H-4 to C1, C5 and C8, H5 to C1, C4, and C8, and H-7 to C1 and C6, respectively. According to the spectroscopic data, compound 1 suggested as a phenolic derivative attached to two hydroxyl groups at C9 and C10, respectively. Based on the analysis of spectral data and compare data with published papers, the compound was identified as 1,2-dihydroxylallylbenzene (Allylpyrocatechol), as shown in Figure 1.31,32
Figure 1.
The structure of compound 1.
Determination of the Antibacterial Activity of Extract and Compound
The data of antibacterial extracts are reported in Table 1 presented that ethyl acetate and n-hexane extracts active inhibit the growth of S. sanguinis at 4% and 10% with inhibitions zone values of 22.45 and 21.75 mm, respectively, those similar to inhibition zones of 22.06 by 2% Chlorhexidine as a positive control. Sub-fraction of ethyl acetate extract (F1-F21) were tested antibacterial assay. The three most active fractions are F1-F3 with inhibition zone values of 14.11, 9.97, and 7.18 mm, respectively. Meanwhile, chlorhexidine as positive control showed inhibition zone values of 17.80 mm.
Table 1.
The Inhibition Zones of P. betle L. Extracts Against S. sanguinis
| Samples | Inhibition Zone (mm) | ||||
|---|---|---|---|---|---|
| 2% | 4% | 6% | 8% | 10% | |
| Methanol | 9.6 | 12.05 | 15.4 | 16.75 | 19.05 |
| n-Hexane | 12.5 | 14.8 | 16.3 | 20.25 | 21.75 |
| Ethyl acetate | 14.8 | 22.45 | 23.15 | 23.45 | 27.9 |
| Water-methanol | 0 | 0 | 0 | 0 | 0 |
| Methanol (-) | 0 | 0 | 0 | 0 | 0 |
| Water (-) | 0 | 0 | 0 | 0 | 0 |
| Chlorhexidine (+) | 22.06 | - | - | - | - |
To evaluate the antibacterial activity of compound 1, the inhibition zone activity was measured by Kirby–Bauer. As shown in Table 2, compound 1 inhibits bacterial growth at 4% those equal with inhibition zones of chlorhexidine at 2% as a positive control. According to references of inhibition zones values, compound 1 at 1–5% with inhibitions zones of 11.85–255 classified as a strong antibacterial agent (i.z ≥6 mm: strong, 3–6 mm: moderate, ≤3 mm: weak).33
Table 2.
The Inhibition Zones of Allylpyrocatechol 1 Against S. sanguinis
| Allylpyrocatechol 1 | Inhibition Zone (mm) |
|---|---|
| 1% | 11.85 |
| 2% | 16.60 |
| 3% | 20.60 |
| 4% | 22.90 |
| 5% | 25.15 |
| Methanol (-) | 0 |
| Chlorhexidine (+) | 22.8 |
Further antibacterial activity evaluation, the minimum inhibition concentration (MIC) and minimum bactericidal concentration (MBC) were determined. The active compound 1 showed MIC and MBC values of 39.1 and 78.1 μg/mL, respectively. Based on the data in published papers, the MIC and MBC of 1 were categorized as good antibacterial activity.34 On the other hand, fosfomycin as a control showed the MIC value 62.5 μg/mL and does not have MBC value.35 It means that fosfomycin just inhibits the bacteria growth but not kill the bacteria. Since compound 2 was found in a published paper, the MIC and MBC values were not reported, the structure will be used as a molecule model as isomer compound of 1 for in silico study. The assay data as shown in Table 3, suggested that the antibacterial activity of compound 1 is better than fosfomycin.
Table 3.
Data of MIC and MBC of Allylpyrocatechols 1 and 2 Against S. sanguinis
| Compound | Concentration (μg/mL) | |
|---|---|---|
| MIC | MBC | |
| Allylpyrocatechols 1 | 39.1 | 78.2 |
| Allylpyrocatechols 2 | ND | ND |
| Chlorhexidine | 3.12 | 6.25 |
| Fosfomycin | 62.5 | None |
Antibacterial Activity Prediction of Allylpyrocatechols Through Molecular Interaction with UDP-N-Acetylglucosamine Enolpyruvyl Transferase (MurA)
Based analysis of in vitro antibacterial activity of allylpyrocatechol 1, the molecular mechanism prediction of allylpyrocatechols 1 and 2 as an inhibitor to inhibit MurA enzyme was evaluated by in silico study. The analysis of the result as shown in Table 4 presented the molecular docking compounds 1 and 2 with MurA indicated the strength of the interaction between each compound and MurA that able to be seen from the binding energy.
Table 4.
Prediction of Antibacterial Activity of MurA-Allylpyrocatechols 1 and 2
| Ligand | Binding Affinity |
|---|---|
| Allylpyrocatechols 1 | −5.4 |
| Allylpyrocatechols 2 | −5.4 |
| Fosfomycin | −4.6 |
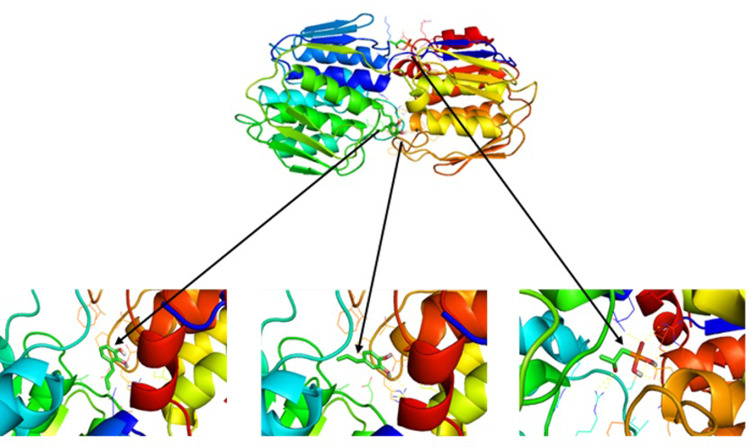
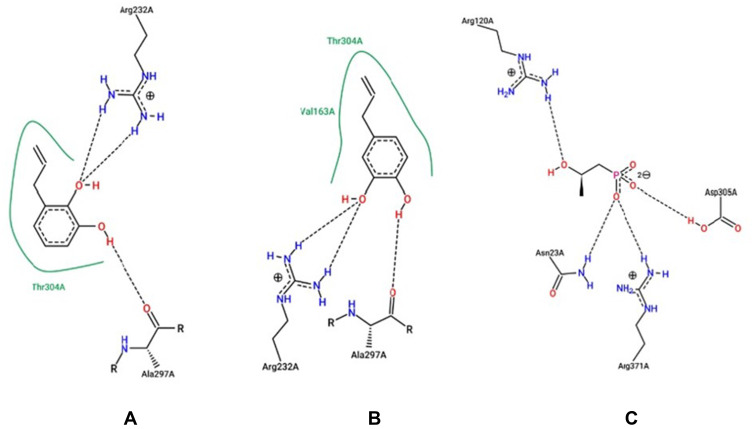
As shown in Figure 2, the bond position of each complex is different. The Allylpyrocatechols 1 and 2 located in a similar cavity; meanwhile, fosfomycin occupies a different position. Residues that interact with the ligands as in Table 5. Compounds 1 and 2 bind the same amino acid residues that are Arg232A dan Ala297A. Although they ligate the same residues, the structural conformation of them is different. On the other hand, fosfomycin binds different and more residues. Beside notice what residues bound to MurA, interaction type that happens in the complex can be known through online program protein plus (Figure 3).
Figure 2.
Ligation position of allylpyrocatechol on UDP-N-acetylglucosamine enolpyruvyl transferase (MurA).
Table 5.
Hydrogen Bond in MurA-Allylpyrocatechols 1 and 2
| Ligand | Residues Binding at Ligand-Protein Complex |
|---|---|
| Allylpyrocatechols 1 | Arg232, Ala997 |
| Allylpyrocatechols 2 | Arg232, Ala297 |
| Fosfomycin | Arg120A, Asn23A, Arg371A, Asp305A |
Figure 3.
Interaction ligand-MurA with the ligand compound 1 (A) 2 (B) and fosfomycin (C).
Discussion
Currently, discovery new prospective antibacterial agent those have selective, specific, effective, and appropriate to treat bacterial target is an interesting research focus to resolve some oral diseases caused by pathogenic bacteria. Bioactivity-guided purification of lead compounds as antibacterial constituents was subjected to some natural medicinal plants. Based on the ethnobotany and ethnopharmacology data, the edible herbal plant Sirih (Piper betle L.) was selected as sources to isolated new antibacterial agents against pathogenic oral bacteria.36 In the previous report, the extracts and active compounds of this plant were active as antibacterial,37 anti-fungal,38,39 anti-inflammation,40 anti-proliferative,41 anti-cancer,42 and anti-oxidant,43–45 respectively.
Separation and purification chemical constituents of P. betle L. extract guided by bioactivity assay against S. sanguinis ATCC 10566 lead to isolating antibacterial compound, and the structure was identified as allylpyrocatechol (1,2-dihydroxylallylbenzene). The structure is shown in Figure 1. The published paper showed that the extract of P. betle L. was active against S. mutans, while the active constituent as an antibacterial agent against S. sanguinis is not reported, yet.37
Allylpyrocatechol is an important natural small molecule because it used as a precursor compound to make other bioactive chemicals by chemicals synthesis reactions. Then, the isolation, characterization, and identification procedure of Allylpyrocatechol also was reported at some published papers.46
Based on the antibacterial assay, Allylpyrocatechol (1) is weaker than chlorhexidine but it is stronger than fosfomycin. It can be suggested that Allylpyrocatechol has potential as a new candidate antibacterial agent for S. sanguinis. Some natural antibacterial against S. sanguinis were reported at some paper,34,47 but the antibacterial activity of allylpyrocatechol against S. sanguinis is for the first time in this report. This study adds to the list of bacteria that can be inhibited by allylpyrocatechol which in previous studies mentioned that allylpyrocatechol is active against Staphylococcus aureus and Candida albicans.38,48
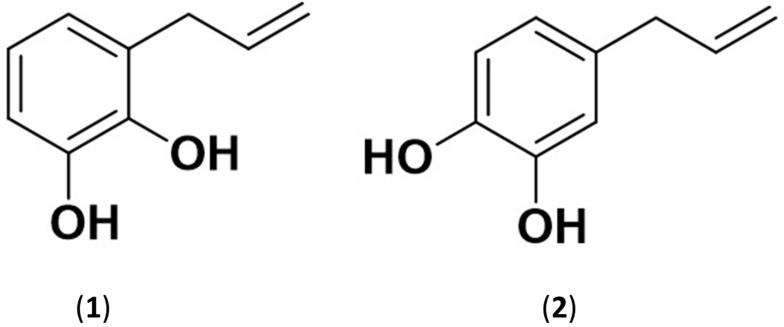
In this study, an in silico test of compound 1 and its isomer, compound 2 (Figure 4) is carried out to determine the stereoselective and regioselective factors of compounds 1 and 2 of the same target receptor. The parameters measured are binding affinity and the residues binding the MurA. The bond energy or binding affinity (ΔG) is the free energy of a bond. The value of ΔG is getting smaller or more negative indicates the stability and strength of the best bond and the bond formed is getting stronger.49 Free energy of compounds 1, 2, and fosfomycin is −5.4, −5.4, and −4.6 Kcal/mol, respectively. Fosfomycin which has been known as an inhibitor of MurA enzyme has the weakest of the free energy of the other ligands. It showed that the tested ligands have better stability when interacting with the MurA so that the P. betle L. compound has good potential as a MurA inhibitor.
Figure 4.
The structure of Allylpyrocatechol 1 and 2.
The protein residue of Asn23A, Arg120A, Asp305A, and Arg371A was ligating to fosfomycin. Those residues are part of residues that bond on the uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) that has been known as a substrate of MurA. In the complex of UDP-GlcNAc-MurA, Asn23A, and Asp305A residues are bound to the glucosamine group while the Arg120A residue is bound to a phosphate group.27 Therefore, it can be stated that compounds 1 and 2 are competitive with each other but two of them does not competitive with fosfomycin and UDP-GlcNA.50
Compounds 1 and 2 showed a similar hydrophilic interaction (interaction on Arg232A and Ala297A residues). Nevertheless, compound 2 has two hydrophobic interactions (Thr304A and Val163A residues) while compound 1 only has one hydrophobic interaction with Thr304A residue. Meanwhile, fosfomycin just has hydrophilic interaction (Asn23A, Arg120A, Asp305A, and Arg371A). By no hydrophilic interaction in the complex of MurA-fosfomycin, it deduces that the reason why the free energy of fosfomycin is the weakest. According to the result of in silico studies, compounds 1 and 2 have similar characteristics so it can be concluded that the stereoselective and regioselective factors do not significantly affect the antibacterial properties, especially in inhibition of the MurA enzyme.
Data of free energy through in silico are in line with antibacterial activity provided in vitro assay which binding energy and MIC value of compounds 1 and 2 are lower than fosfomycin. It deduces that allylpyrocatechol able to be alternative potential inhibitor MurA substituted fosfomycin.
Conclusion
This study gives more information about the phenolic compound list from P. betle leaf. Based on the characterization data, the isolated compound that is 1,2-dihydroxylallylbenzene (Allylpyrocatechol). This compound is classified as a moderate active antibacterial agent against S. sanguinis. This result is in line with the in silico evaluation. Docking simulations show that Allylpyrocatechols 1 (4-prop-2-enylbenzene-1,2-diol) has better stability than fosfomycin when interacting with the MurA. It deduces Allylpyrocatechols 1 potencies as a non-competitive inhibitor of MurA. This research suggests that P. betle L. can be a source of the antibacterial agents. Even though further research including in vitro, in vivo and clinical studies, are needed, this research provides basic information in drug discovery, especially the screening of antibacterial compounds from P. betle.
Acknowledgments
The authors are grateful to the Ministry of Research, Technology, and Higher Education of the Republic of Indonesia for Grant of Penelitian Magister 2020. This research was supported by Academic Leadership Grant 2020 from Universitas Padjadjaran, Sumedang Indonesia.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Avila M, Ojcius DM, Yilmaz Ö. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28(8):405–411. doi: 10.1089/dna.2009.0874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoo S, Murata RM, Duarte S. Antimicrobial traits of tea- and cranberry-derived polyphenols against Streptococcus mutans. Caries Res. 2011;45:327–335. doi: 10.1159/000329181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Truper GH, De’clari L. Taxonomic note: necessary correction of specific epithets formed as substantives (Nouns) “in Apposition”. Int J Syst Bacteriol. 1997;47:908–909. [Google Scholar]
- 4.Xu P, Ge X, Chen L, et al. Genome-wide essential gene identification in Streptococcus sanguinis. Sci Rep. 2011;1(125):1–9. doi: 10.1038/srep00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu B, Macleod LC, Kitten T, et al. Streptococcus sanguinis biofilm formation & interaction with oral pathogens. Future Microbiol. 2018;13(8):915–932. doi: 10.2217/fmb-2018-0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martini AM, Moricz BS, Ripperger AK, et al. Association of novel Streptococcus sanguinis virulence factors with pathogenesis in an native valve infective endocarditis model. Front Microbiol. 2020;11(10):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu B, Song L, Kong X, et al. A novel regulator modulates glucan production, cell aggregation and biofilm formation in Streptococcus sanguinis SK36. Front Microbiol. 2018;9(1154):1–14. doi: 10.3389/fmicb.2018.01154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossman TH. Tetracycline antibiotics and resistance. Cold Spring Harb Perspect Med. 2016;6(4):1–24. doi: 10.1101/cshperspect.a025387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cegelski L, Marshall G, Eldridge RG, Hultgren JS. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krašovec R, Jerman I. Bacterial multicellularity as a possible source of antibiotic resistance. Med Hypotheses. 2003;60(4):484–488. doi: 10.1016/S0306-9877(02)00394-8 [DOI] [PubMed] [Google Scholar]
- 11.Bugg THD, Walsh CT. lntracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat Prod Rep. 1992;9(3):199–215. doi: 10.1039/np9920900199 [DOI] [PubMed] [Google Scholar]
- 12.Epand RM, Walker C, Epand RF, et al. Molecular mechanism of membrane targeting antibiotics. Biochim Biophys Acta. 2016;1858(5):980–987. doi: 10.1016/j.bbamem.2015.10.018 [DOI] [PubMed] [Google Scholar]
- 13.Deepak SM, Patil PP, Aher SJ, et al. Mur-A: a critical target behind new antibacterial drug discovery. Indo Am J Pharm Res. 2014;4(1):220–225. [Google Scholar]
- 14.Dev A, Adil MT, Kumar P. In silico based approach to identify Mura as a potential drug target for leprosy. Acta Sci Microbiol. 2020;3(3):1–7. [Google Scholar]
- 15.Salehi B, Sharopov F, Martorell M, et al. Phytochemicals in Helicobacter pylori infections: what are we doing now? Int J Mol Sci. 2018;19(8):2361–2394. doi: 10.3390/ijms19082361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salehi B, Sener B, Kilic M, et al. Dioscorea plants: a genus rich in vital nutra-pharmaceuticals-A review. Iran J Pharm Res. 2019;18:68–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambrosio SR, Furtado NAJC, Oliveira DCR, et al. Antimicrobial activity of kaurane diterpenes against oral pathogens. Verlag der Zeitschrift für Naturforsch. 2008;63c:326–330. doi: 10.1515/znc-2008-5-603 [DOI] [PubMed] [Google Scholar]
- 18.Moon SE, Kim HY, Cha JD. Synergistic effect between clove oil and its major compounds and antibiotics against oral bacteria. Arch Oral Biol. 2011;56(9):907–916. doi: 10.1016/j.archoralbio.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 19.Freires IA, Denny C, Benso B, et al. Antibacterial activity of essential oils and their isolated constituents against cariogenic bacteria: a systematic review. Molecules. 2015;20(4):7329–7358. doi: 10.3390/molecules20047329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cha JD, Jeong MR, Jeong SI, et al. Chemical composition and antimicrobial activity of the essential oil of Cryptomeria japonica. Phytother Res. 2007;21(3):295–299. doi: 10.1002/ptr.1864 [DOI] [PubMed] [Google Scholar]
- 21.Bernardes WA, Lucarini R, Tozatti MG, et al. Antibacterial activity of the essential oil from Rosmarinus officinalis and its major components against oral pathogens. Zeitschrift Naturforshung. 2010;65c:588–593. [DOI] [PubMed] [Google Scholar]
- 22.Salehi B, Zakaria ZA, Gyawali R, et al. Piper species: a comprehensive review on their phytochemistry, biological activities and applications. Molecules. 2019;24(7):1364–1480. doi: 10.3390/molecules24071364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty D, Shah B. Antimicrobial, anti-oxidative, and anti-Hemolytic activity of Piper betle leaf extracts. Int J Pharm Pharm. 2011;3(3):192–199. [Google Scholar]
- 24.Patel N, Mohan JSS. Isolation and characterization of potential bioactive compounds from Piper betle varieties Banarasi and Bengali leaf extract. Int J Herb Med. 2017;5(5):182–191. [Google Scholar]
- 25.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3(11):935–949. doi: 10.1038/nrd1549 [DOI] [PubMed] [Google Scholar]
- 26.Srinivasan P, Arumugam DS, Manikandan R, Arulvasu C. Molecular docking studies of 1,2 disubstituted idopyranose from Vitex negundo with anti-diabetic activity of type 2 diabetes. Int J Pharma Bio Sci. 2011;2:68–83. [Google Scholar]
- 27.Skarzynski T, Mistry A, Wonacott A, et al. Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Structure. 1996;4(12):1465–1474. doi: 10.1016/S0969-2126(96)00153-0 [DOI] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute (CLSI - formerly NCCLS). Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard. 11th ed. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- 29.Clinical and Laboratory Standards Institute document M7-A8. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard. 9th ed. Wayne, PA, USA: Clinical and laboratory standards institute; 2012. [Google Scholar]
- 30.Blanco L, Garza M. Phenol degradation using glassy carbon electrodes modified with particles of Co-Mo alloy. Int J Electrochem Sci. 2013;8:5698–5709. [Google Scholar]
- 31.Abdullah NF, Anifah W, Hussain RM. Optimized method for purification of allylpyrocatechol from Piper Betle L. ethanolic extract using HPLC and H1-NMR. J Pharm Sci & Res. 2015;7(6):292–301. [Google Scholar]
- 32.Panda S, Sikdar M, Biswas S, et al. Allylpyrocatechol, isolated from betle leaf Ameliorates thyrotoxicosis in rats by altering thyroid peroxidase and thyrotropin receptors. Sci Rep. 2019;1:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan X, Chen F, Wu T, et al. The acid, bile tolerance, and antimicrobial property of Lactobacillus acidophilus NIT. Food Control. 2009;20(6):598–602. doi: 10.1016/j.foodcont.2008.08.019 [DOI] [Google Scholar]
- 34.Kuete V. Potential of Cameroonian plants and derived products against microbial infections: a review. Planta Med. 2010;76(14):1479–1491. doi: 10.1055/s-0030-1250027 [DOI] [PubMed] [Google Scholar]
- 35.Kurnia D, Apriyanti E, Soraya C, Satari MH. Antibacterial flavonoids against oral bacteria of Enterococcus faecalis ATCC 29212 from Sarang Semut (Myrmecodia pendans) and its inhibitor activity against enzyme MurA. Curr Drug Discov Technol. 2019;16(3):290–296. doi: 10.2174/1570163815666180828113920 [DOI] [PubMed] [Google Scholar]
- 36.Nagori K, Singh M, Alexander A, et al. Piper betle L.: a review on its ethnobotany, phytochemistry, pharmacological profile and profiling by new hyphenated technique DART-MS (Direct Analysis in Real Time Mass Spectrometry). J Pharm Res. 2011;4:2991–2997. [Google Scholar]
- 37.Nalina T, Rahim ZHA. The crude aqueous extract of Piper betle L. and its antibacterial effect towards Streptococcus mutans. Am J Biochem Biotechnol. 2007;3(1):10–15. doi: 10.3844/ajbbsp.2007.10.15 [DOI] [Google Scholar]
- 38.Ali I, Khan FG, Suri KA, et al. In vitro antifungal activity of hydroxychavicol isolated from Piper betle L. Ann Clin Microbiol Antimicrob. 2010;9(7):1–9. doi: 10.1186/1476-0711-9-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan S, Singh S, Gaikwad S, et al. Optimization of process parameters for the synthesis of silver nanoparticles from Piper betle leaf aqueous extract, and evaluation of their anti-phytofungal activity. Environ Sci Pollut Res. 2019. doi: 10.1007/s11356-019-05239-2 [DOI] [PubMed] [Google Scholar]
- 40.Sarkar D, Saha P, Gamre S, et al. Anti-inflammatory effect of allylpyrocatechol in LPS-induced macrophages is mediated by suppression of iNOS and COX-2 via the NF-κB pathway. Int Immunopharmacol. 2008;8:1264–1271. doi: 10.1016/j.intimp.2008.05.003 [DOI] [PubMed] [Google Scholar]
- 41.Bauri AK, Brodie PJ, Kingston DGI. Anti-proliferative allylic phenols from the methanol extract of Piper betle. Am J Pharm Pharmacol. 2018;5(3):13–18. [Google Scholar]
- 42.Chakraborty JB, Mahato SK, Joshi K, et al. Hydroxychavicol, a Piper betle leaf component, induces apoptosis of CML cells through mitochondrial reactive oxygen species-dependent JNK and endothelial nitric oxide synthase activation and overrides imatinib resistance. Cancer Sci. 2012;103(1):88–99. doi: 10.1111/j.1349-7006.2011.02107.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishiwaki K, Ohigashi K, Deguchi T, et al. Structure-activity relationships and docking studies of hydroxychavicol and its analogs as xanthine oxidase inhibitors. Chem Pharm Bull. 2018;66:741–747. doi: 10.1248/cpb.c18-00197 [DOI] [PubMed] [Google Scholar]
- 44.Abrahim NN, Kanthimathi MS, Abdul-Aziz A. Piper betle shows antioxidant activities, inhibits MCF-7 cell proliferation, and increases activities of catalase and superoxide dismutase. BMC Complement Altern Med. 2012;12:220. doi: 10.1186/1472-6882-12-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar D, Kundu S, De S, et al. The antioxidant activity of allylpyrocatechol is mediated via decreased generation of free radicals along with escalation of antioxidant mechanisms. Phytother. Res. 2013;27:324–329. doi: 10.1002/ptr.4720 [DOI] [PubMed] [Google Scholar]
- 46.Abdullah NF, Hussain RM. Isolation of allylpyrocatechol from Piper betle L. leaves by using high-performance liquid chromatography. J Liq Chromatogr Relat Technol. 2015;38(2):289–293. doi: 10.1080/10826076.2014.908782 [DOI] [Google Scholar]
- 47.Medina-Flores D, Ulloa-Urizar G, Camere-Colarossi R, et al. Antibacterial activity of Bixa orellana L. (achiote) against Streptococcus mutans and Streptococcus sanguinis. Asian Pac J Trop Biomed. 2016;6(5):400–403. doi: 10.1016/j.apjtb.2016.03.005 [DOI] [Google Scholar]
- 48.Abdullah NF, Hussain RM, Amom Z. Effect of APC on killing of Staphylococcus aureus by oxidative stress agents. Jurnal Teknologi. 2015;78(5):39–44. [Google Scholar]
- 49.Gupta A, Chaudhary N, Kakularam KR, et al. The augmenting effects of desolvation and conformational energy terms on the predictions of docking programs against mPGES-1. PLoS One. 2015;10(8):1–16. doi: 10.1371/journal.pone.0134472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Syahputra G, Ambarsari L, Sumaryada T. Simulasi docking kurkumin enol, bismetoksikurkumin dan analognya sebagai inhibitor enzim1,2-lipoksigenase. J Biofisika. 2014;10(1):55–67. [Google Scholar]