Summary
Bud tip progenitor cells give rise to all murine lung epithelial lineages (Rawlins et al., 2009; Yang et al., 2018), and have been described in the developing human lung (Danopoulos et al., 2018; Miller et al., 2018; Nikolić et al., 2017); however, the mechanisms controlling human bud tip differentiation into specific lineages are unclear. Here, we used homogeneous human bud tip organoid cultures and identified SMAD signaling as a key regulator of the bud tip-to-airway transition. SMAD-induction lead to differentiation of airway-like organoids possessing functional basal cells capable of clonal expansion and multilineage differentiation. To benchmark in vitro derived organoids, we developed a single-cell mRNA sequencing atlas of the human lung from 11.5–21 weeks gestation, which revealed high degrees of similarity between in vitro-derived and in vivo airway. Together, this work sheds light on human airway differentiation in vitro, and provides a single cell atlas of the developing human lung.
eTOC
Miller and Yu et al. show that transient SMAD activation followed by inhibition patterns human lung bud tip progenitor organoids to functional airway cell types, including basal cells. Comparison of organoids and in vivo developing human lung epithelium by scRNAseq demonstrates a high degree of transcriptomic similarity.
Introduction
Basal stem cells serve as epithelial stem cells in multiple organ systems, including lung, skin, esophagus, breast and prostate, where they contribute to organ homeostasis and repair after injury (Fletcher et al., 2011; Pardo-Saganta et al., 2015; Rock et al., 2009; Schnittke et al., 2015; Seery, 2002; Shackleton et al., 2006; Tumbar, 2004), and their misregulation has been implicated in disease (Van de Laar et al., 2014; Murata et al., 2007; Pardo-Saganta et al., 2015). In the lung, basal stem cells reside on the basolateral surface of the pseudostratified epithelium and are marked by the transcription factor TP63, which is essential for basal cell fate determination and function (Arason et al., 2014; Daniely et al., 2004). In mice, they are restricted to the trachea, whereas in humans, basal stem cells extend deep into the bronchial tree (Hong et al., 2004; Rock et al., 2009). Moreover, TP63+ basal-like cells have been identified in the distal airways of human patients with pulmonary fibrosis, suggesting these distal basal-like cells may contribute to repair and disease in humans (Murata et al., 2007). The critical role that basal cells play during lung homeostasis and repair has driven interest in in generating basal stem cells for regenerative medicine. However, the mechanisms governing airway and basal cell fate specification during development remain unclear, limiting our ability to efficiently generate basal stem cells in vitro from human pluripotent stem cells.
Lineage tracing experiments in mice have shown that a specialized population of lung epithelial progenitor cells, called bud tip progenitors, genetically marked using Sox9-creER, give rise to TP63+ cells (Yang et al., 2018). Similarly, human fetal bud tip progenitor cells give rise to putative TP63+ basal stem cells upon transplantation in vivo (Nikolić et al., 2017); however, the exploration of mechanisms regulating human airway and basal cell specification has been hindered by limited tissue availability.
Here, we interrogated the developmental mechanisms controlling the bud tip-to-airway and basal cell transition during human lung development. Using a homogeneous human bud tip organoid culture system, we performed a small-scale screen on developmentally relevant signaling pathways to determine the ability of different cell signaling modulators to induce airway or basal-like fates in vitro. We identified SMAD activation via TGFβ1 and BMP4 as potent inducers of TP63 expression in bud tip organoids. Induced organoids were subsequently expanded in airway expansion media similar to those previously described(Mou et al., 2016; Tadokoro et al., 2016). To determine how similar induced organoids were to the native human fetal lung airway epithelium, we generated single cell data for in vitro-derived airway/basal cell organoids and a single cell atlas of the developing human airway from 11.5–21 weeks gestation. Comparative analysis revealed that in vitro derived organoids recapitulate molecular features of their fetal counterparts. We further isolated in vitro-derived basal-like cells using fluorescent activated cell sorting (FACS) and showed that these cells are able to undergo expansion from a single cell, can self-renewal, and undergo multilineage differentiation in vitro. Collectively, this work provides a framework for deducing and validating key regulators of cell fate decisions and identified SMAD signaling as a critical regulator of newly born airway and basal cells in the lung.
Results
SMAD activation induces TP63 expression in bud tip progenitor organoids in vitro
Prior lineage tracing in mice suggests that TP63+ cells are specified very early from Sox9+ progenitor cells in mice (Yang et al., 2018). Interestingly, while TP63 is a marker of mature basal cells, lineage tracing in mice using the Tp63-creER has shown that Cre activation prior to E10.5 leads to labeling of the entire lung epithelium and labeling of TP63+ cells that are negative for the mature basal cell marker KRT5, suggesting that early TP63+/KRT5- cells prior to E10.5 are multipotent progenitors, not true basal cells (Yang et al., 2018). This data demonstrated that basal cells in the mouse tracheal epithelium are patterned during early development. As the lung develops and the airway extends, bud tip progenitors self-renew at the tips of branching buds, leaving cells behind that will contribute to the intrapulmonary airways (Figure 1A). However, humans and mice have differences in the distribution of basal cells throughout the bronchial tree, with the human bronchial tree containing basal cells in all but the smallest bronchioles. The extension of basal cells throughout the human intrapulmonary airway raises the possibility that bud tip progenitors differentiate into all airway lineages, including airway basal cells, and for a long window of time during human development.
Figure 1. SMAD activation induces TP63 expression in bud tip progenitor organoids.
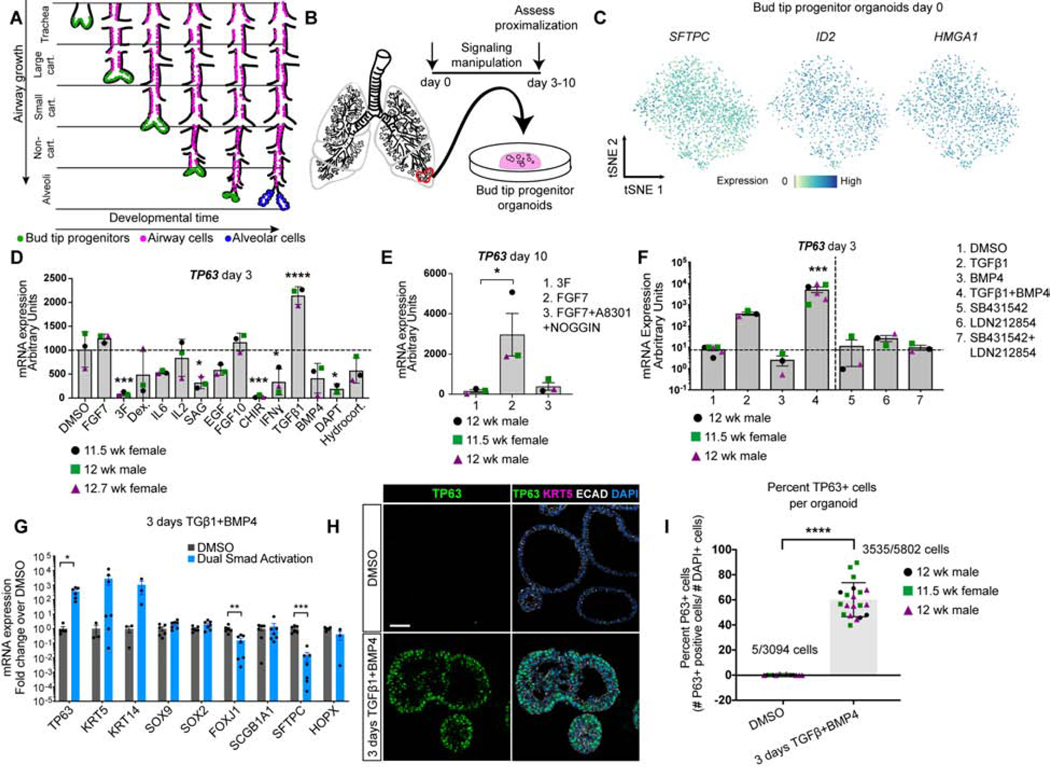
A) Schematic of lung epithelial development. As the airways extend, bud tip progenitors are maintained as progenitors in the tips of branching buds, and leave cells behind that give rise to the intrapulmonary airways. Late in development, remaining bud tip progenitors differentiate into alveolar cells. No bud tip progenitors are present in the adult lung. B) Schematic of creation of epithelium-only bud tip progenitor organoids from 12 week fetal lungs. C) Feature plots for bud tip progenitor marker genes SFTPC, ID2 and HMGA1 from scRNA-seq of day 0 bud tip progenitor organoids maintained for 3 weeks in culture. Additional feature plots from this same data set are shown in Figure 4A. D) mRNA expression by QRT-PCR of basal cell marker TP63 in bud tip progenitor organoids treated for 3 days with serum-free basal medium supplemented with DMSO (control) or with signaling factors known to be important for lung development and cellular differentiation. DMSO (1:1,000 dilution), FGF7 (10 ng/mL), ‘3F’ (FGF7 10ng/mL, CHIR99021 3μM, ATRA 50 nM), Dexamethasone (25 ng/mL), IL6 (10 ng/mL), IL2 (50 U/mL), Smoothened Agonist (SAG; 500 nM), EGF (100 ng/mL), FGF10 (500 ng/mL), CHIR99021 (2 μM), IFNγ (10 ng/mL), TGFβ1 (100 ng/mL), BMP4 (100 ng/mL), DAPT (10 μM), Hydrocortisone (100 ng/mL). Gene expression is reported as arbitrary units. Treatment with TGFβ1 led to a significant increase in the expression of TP63 (one-way Analysis of Variance (ANOVA) (alpha=0.05, p<0.0001, F=14.7. Dunnett’s test of multiple comparison’s compared the mean of each group to the mean of the DMSO control group.) Estimated p values are shown on the graph. Error bars are plotted to show mean +/− the standard error of the mean. N=3 independent biological specimens. Data is from a single experiment. E) Bud tip progenitor organoids were treated with FGF7 (10 ng/mL), a permissive environment for TP63 expression compared to maintenance in bud tip progenitor medium (‘3F’), or with FGF7 (10 ng/mL) plus factors to inhibit SMAD signaling (A8301 [1 μM] and NOGGIN [100 ng/mL]) and TP63 gene expression was evaluated by QRT-PCR after 10 days in culture. A one-way Analysis of Variance was used followed by Tukey’s multiple comparison test to compare the means of each group to the mean of every other group. Estimated p values are reported on the graph. Error bars are plotted to show mean +/− the standard error of the mean. N=3 independent biological specimens. Data is from a single experiment. F) Bud tip progenitor organoids were treated for 3 days with SMAD activation or inhibition conditions and expression of TP63 was evaluated by QRT-PCR for all treatment groups. All media contained 3F components (FGF7 10ng/mL, CHIR99021 3μM, ATRA 50 nM), with individual groups containing combinations of: DMSO (1:1000 dilution), TGFβ1 (100 ng/mL), BMP4 (100 ng/mL), SB431542 (10 μM), LDN212854 (200 nM). One-way ANOVA alpha=0.05, F=21.19, p<0.0001; Tukey’s multiple comparisons of the mean of each group versus the mean in all other groups, estimated p values are reported on the graph. 3 days TGFβ1 and BMP4 is referred to as ‘dual SMAD activation’, or ‘DSA’. Data is plotted as arbitrary units. Error bars are plotted to show mean +/− the standard error of the mean. N=3 independent biological specimens. Data is from a single experiment and is representative of n=3 experiments. G) QRT-PCR for markers of canonical differentiated lung epithelial cell types showing DMSO (gray bars) and DSA treated (blue bars) organoids after 3 total days of treatment. Data is plotted as fold change over DMSO controls. Two-sided Mann-Whitney Tests were performed to compare the mean of the DMSO group to the Dual Smad Activation group (alpha=0.05). Error bars represent the mean +/− the standard error of the mean. n=3 independent biological specimens, and data is from a single experiment and is representative of n=3 experiments. H) Protein staining of DMSO treated (control) fetal bud tip progenitor organoids (top row) and 3 days of DSA treatment (bottom row) for TP63+ protein (green), KRT5 (pink) and DAPI (blue). Scale bar represents 50 μm. I) Quantification of (g). Total number of TP63+ cells were counted for 3–9 individual organoids across 3 biological replicates. n=3 independent biological specimens. A two-sided Mann Whitney test (alpha=0.05) was used to compare the means of each sample. For all graphs, p values are reported as follows: * p<0.05; ** p<0.01, *** p<0.001, **** p<0.0001.
To test the ability of human bud tip progenitor cells to differentiate into different airway lineages, we leveraged human fetal bud tip progenitor organoid cultures (Miller et al., 2018). Isolation and expansion of bud tip progenitors in media possessing ‘3 Factors’ (‘3F media’), Fibroblast Growth Factor 7 (FGF7), the small molecule CHIR-99021, and all-trans Retinoic Acid (ATRA), sufficiced to maintain human fetal bud tip progenitors as a homogeneous population of organoids in vitro as reveled by single cell RNA sequencing (scRNA-seq) (Figure 1B). As is indicated by dimension reduction and t-distributed Stochastic Neighbor Embedding (tSNE) to visualize cell clustering, bud tip progenitor organoid cells exhibited low cellular heterogeneity and expressed human bud tip progenitor markers SFTPC, ID2 and HMGA1 (Figure 1C, additional feature plots in Figure 4a and S5A) (Miller et al., 2018; Nikolić et al., 2017).
Figure 4. In vitro-derived airway organoid cells recapitulate molecular features of native fetal airway cells.
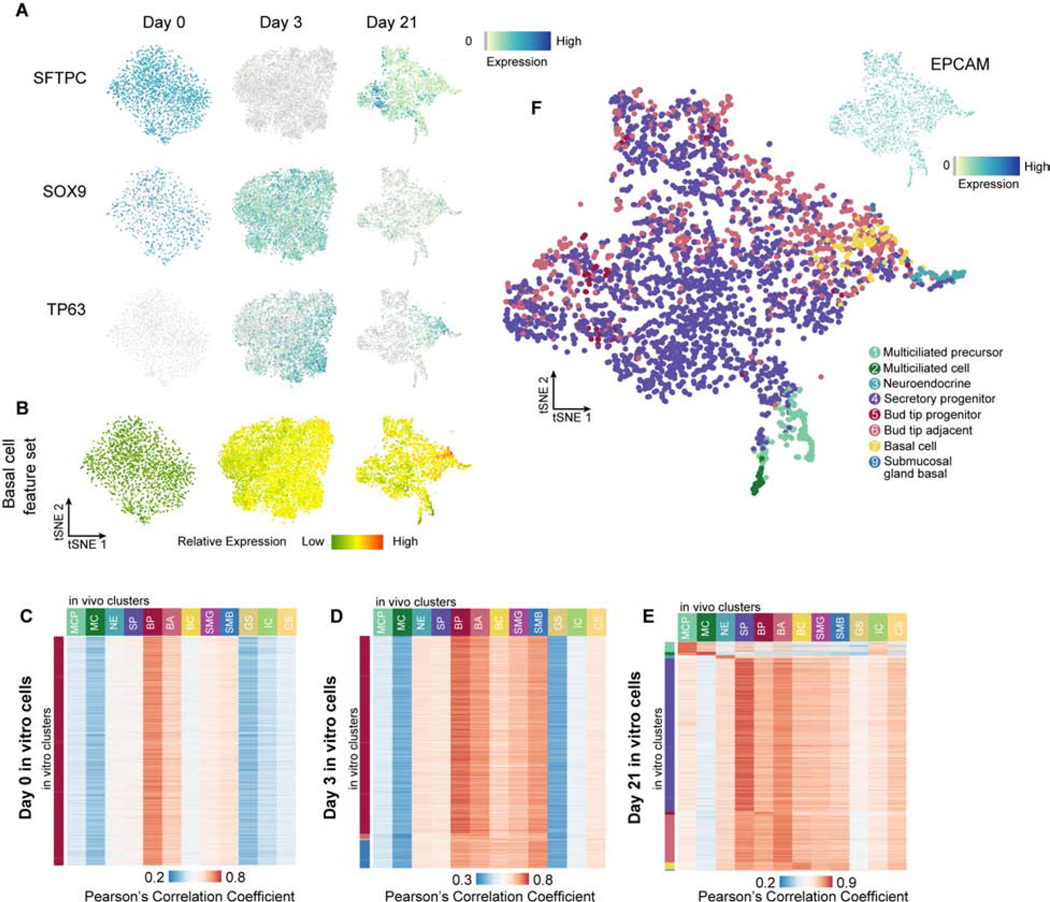
A) Feature plots showing expression of bud tip progenitor markers SFTPC and SOX9 and basal cell marker TP63 from scRNA-seq of in vitro cells at day 0 (bud tip progenitor maintenance), day 3 (after 3 days of DSA) and day 21 (3 days DSA, 18 days DSI). Additional feature plots of the Day 0 scRNA-seq data are shown in Figure 1C. B) tSNE projections from day 0, day 3 and day 21 in vitro derived cells showingthe sum of z-transformed expression levels of 331 fetal basal cell cluster markers, as identified from human fetal basal cells in Fig. 3, in day 0, day 3 and day 21 in vitro samples. C-E) Heatmap showing Pearson’s correlation coefficients (PCCs) of transcriptome between each of 2,000 randomly selected cell from each time point (day 0 [C], day 3 [D] and day 21 [E]) and the 12 fetal epithelial sub-clusters. PCCs were calculated using log-normalized expression levels of genes that are top 50 markers in any of the fetal epithelial sub-clusters and detected in in vitro cells. Colors represent range of PCCs for all cells in each time point. F) 21 day organoids were subjected to scRNA-seq and visualized by tSNE. Colors represent the fetal cell type identity as described in Fig 3 with the highest PCC to each individual 21 day cell. Top right corner: feature plot showing expression pattern of EPCAM across day 21 cells. C-F) MCP=multiciliated precursor, MC=multiciliated, NE=neuroendocrine, SP=secretory progenitor, BP=bud tip progenitor, BA=bud tip adjacent, BC=basal cell, SMG=submucosal gland, SMB=submucosal basal, GS=goblet-like secretory, IC=intermediate ciliated, CS=club-like secretory.
Next, we tested the ability of several signaling pathway activators and inhibitors known to be important for lung development to induce TP63 gene expression in bud tip progenitor organoids after 3 days (Figure 1D). Bud tip progenitor organoids grown in 3F media expressed low TP63. Many treatment groups showed modest increases in TP63 expression, comparable to the levels observed in DMSO controls, suggesting that expression in these groups may be due to stochastic differentiation occuring upon removal of 3F media. TGFβ1 treatment was the only condition that significantly induced TP63 mRNA expression above the DMSO control (Figure 1D). To test whether inhibition of SMAD signaling could block TP63 expression in a permissive environment, we treated bud tip progenitor organoids for 10 days with 3F media, with FGF7-only media, a group that showed modest TP63 expression following removal of CHIR99021 and ATRA from the media similar to DMSO controls at 3 days, or with FGF7 plus SMAD inhibitors A8301 and NOGGIN (Figure 1E). We included FGF7 in the control group to support organoid survival, which is poor in DMSO conditions. The FGF7-only group had significantly higher expression of TP63 relative to 3F media; however, the addition of SMAD inhibitors blocked TP63 induction, suggesting that activation of SMAD signaling is important for TP63 expression in this context.
The result that SMAD activation induced TP63 expression was interesting and unexpected, since others have shown SMAD inhibition is required to maintain mature adult basal stem cells in vitro (Mou et al., 2016; Tadokoro et al., 2016). As a follow-up experiment to the our screen, we tested the ability of multiple SMAD activators and inhibitors alone, or in combination, to regulate TP63 expression in bud tip progenitor organoids after 3 days of treatment (Figure 1F, Figure S1A–C). Consistent with our previous finding, TGFβ1 induced TP63 (Figure 1D). However, TGFβ1 plus BMP4 (herein referred to as ‘dual SMAD activation’; DSA) resulted in the most significant increase in TP63 expression by QRT-PCR (Figure 1F). In contrast, ‘dual SMAD inhibition’ (DSI) had no effect on TP63 when compared to controls (Figure 1F). 3 days of DSA also led to non-significant increases of KRT5 and KRT14 but showed no increase in markers for other lung epithelial cell types (Figure 1G). Protein staining revealed that 3,535 cells out of 5,802 DAPI+ cells counted (60.13% (+/−13.04%)) in DSA treated bud tip organoids expressed TP63 after 3 days, compared to 5 out of 3,094 DAPI+ cells (0.0016%) in controls (Figure 1H, I). Protein expression of the mature basal cell marker KRT5 was absent in TP63+ after 3 days of treatment (Figure 1H). DSA treatment led to more dense epithelial structures (Figure 1H; Figure S1C, D, F) with significantly reduced proliferation as measured by KI67 staining (Figure S1D, E) and increased apoptosis (S1F, G). DSA significantly increased TP63 expression both in the presence and absence of CHIR99021 (Figure S1H). Consistent with bud tip progenitor organoid experiments, treatment of explanted pieces of whole distal lung tissue (n=2, 10 and 11 weeks gestation) with DSA led to significant increases in TP63 protein and mRNA expression (Figure S1I–L), suggesting DSA is a potent inducer of TP63 in multiple contexts.
Nuclear SMAD localization in the developing human fetal airway and trachea
Given that SMAD activation induced TP63 expression in bud tip progenitor organoids, we sought to interrogate the status of active SMAD signaling in vivo by examining nuclear phospho-SMAD immunofluorescence and TP63 expression in 12 and 17 week fetal lungs. In 12 week fetal lungs, we found weak pSMAD staining in bud tip progenitors (Figure 2A, Figure S2A) whereas clear nuclear pSMAD staining was observed in TP63+ cells within the non-cartilaginous and small-cartilaginous airways (pSMAD2 - Figure 2A; pSMAD1,5,8 - Figure S2A; Isotype controls shown in Figure S2B). Interestingly, nuclear pSMAD2 and pSMAD1,5,8 staining was weak in tracheal TP63+ cells (Figure 2A; Figure S2A). TP63+ cells in 17 week fetal lungs lack strong nuclear pSMAD staining irrespective of anatomical location (Figure S2C, D). Consistent with previous studies in the adult trachea showing that nuclear pSMAD is low or non-detectable in mature basal cells (Hong et al., 2004; Mou et al., 2016; Tadokoro et al., 2016), pSMAD staining in the TP63+ cells of the fetal trachea is nearly undetectable, while luminal TP63- tracheal epithelial cells show strong pSMAD immunofluorescence (Figure 2A).
Figure 2. SMAD signaling in the native fetal lung and expansion of TP63+ organoids by SMAD inhibition.
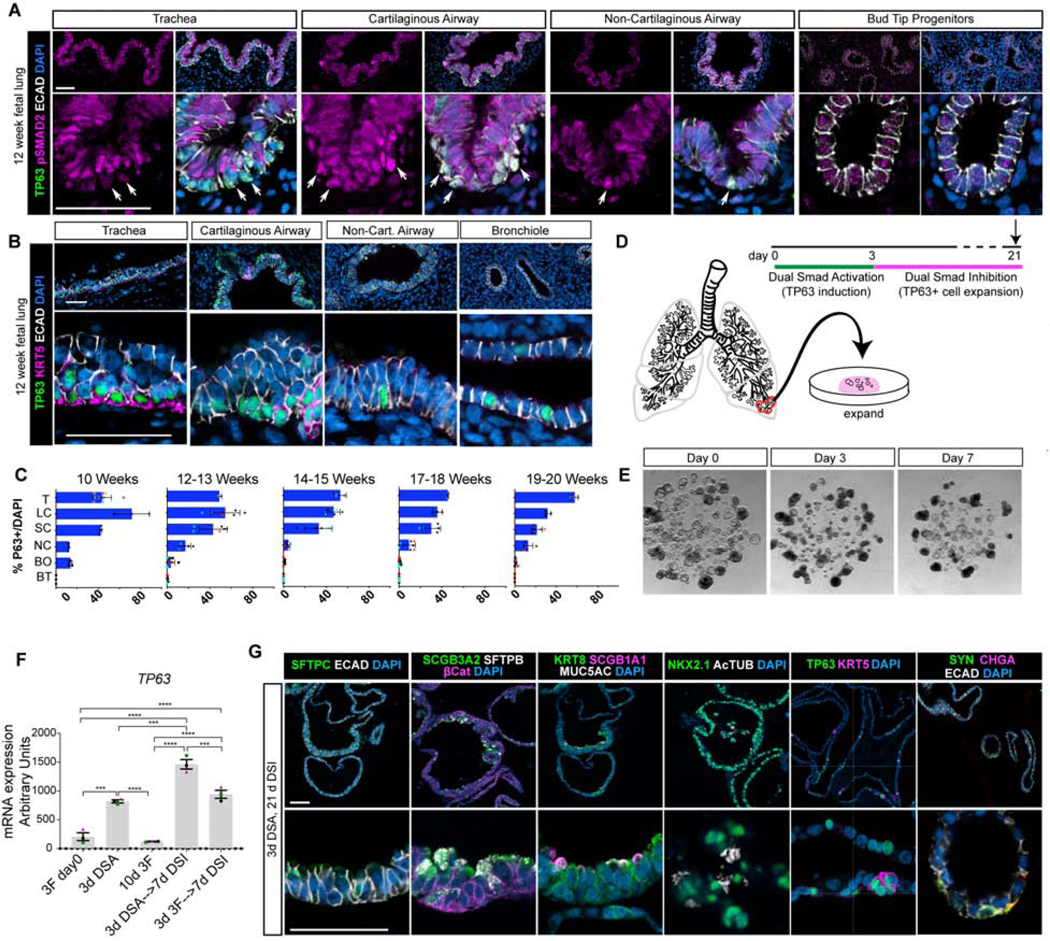
A) Protein staining for TP63 (green), phospho-SMAD2 (pSMAD2; pink), E-Cadherin (ECAD; white) and DAPI (blue) of 12 week fetal lungs along the proximal-distal axis. Representative images shown from n=3 independent specimens. Scale bars represent 50 μm. B) Protein staining of 12 week fetal lungs along the proximal-distal axis for TP63 (green) Keratin 5 (KRT5; pink), E-Cadherin (ECAD; white) and DAPI (blue). Representative images shown from n=3 independent specimens. Scale bars represent 50 μm. C) Quantification of TP63+ cells, defined as TP63+ cells within an airway region divided by 100 DAPI+ cells within that airway region, throughout the bronchial tree from 10–20 weeks gestation. 100 DAPI+ cells were counted for each replicate for each region. Biological replicates are plotted using cyan, red, and black dots. Trachea (T), Large Cartilaginous Airway (LC), Small Cartilaginous Airway (SC), Non-Cartilaginous Airway (NC), Bronchiole (BO), Bud Tip (BT). Data is plotted as mean +/− the SEM. D) Overview of experimental design. Bud tip progenitor organoids were treated with 3 days of DSA and then treated with Dual Smad Inhibition (DSI) to expand the organoids. E) Brightfield images of bud tip progenitor organoids at day 0, day 3 of DSA treatment, and after 3 days of DSA and 7 days of DSI (day 10). Scale bar represents 200 μm. F) QRT-PCR for TP63 in bud tip progenitor organoids cultured with 3F bud tip progenitor maintenance medium for 10 days compared to organoids either treated with 3 days of DSA followed by DSI or with 3F directly followed by DSI. Bud tip progenitor organoids were derived from 3 separate 12-week fetal specimens. Data is plotted as the mean +/− SEM. Statistically significant variation in means was calculated using a one-way ANOVA (alpha=0.05) followed by dunnett’s test of multiple comparisons of the mean of each group to the mean of every other group. Data is from a single experiment. Estimated p values are reported as follows: * p<0.05; ** p<0.01, *** p<0.001, **** p<0.0001. G) After 21 days in culture, organoids were interrogated for protein staining of the bud tip progenitor marker SFTPC (green), fetal secretory cells (SCGB3A2 [green], SFTPB [white]), club cells (SCGB1A1 [pink]), goblet cells (MUC5AC [white]), multiciliated cells (AcTUB [white]), basal-like cells (TP63 [green] and KRT5 [pink]), and neuroendocrine cells (CHGA [pink], Synaptophysin [green]). Scale bars represent 50 μm.
We next examined TP63+ cells throughout the airway at 12 and 17 weeks to determine whether they co-expressed KRT5, which is associated with mature basal cells (Figure 2B). At 12 weeks gestation, TP63+ cells within the trachea and primary bronchi co-express KRT5. In the cartilaginous airways, the majority of TP63+ cells were KRT5-, however, TP63+/KRT5+ cells were visible in distinct clusters within the airways (Figure 2B). In the non-cartilaginous airways and the distal bronchioles, TP63+ cells were KRT5- (Figure 2B). Quantification of TP63+ cell distribution throughout the airway over developmental time between 10 and 20 weeks gestation showed a drop in TP63+ cells in the smaller airways over time, with little to no change in the number of tracheal TP63+ cells (Figure 2C). By 17 weeks gestation, all TP63+ cells within the trachea and upper bronchi co-expressed TP63 and KRT5 (Figure S2F). However, even by 19 weeks gestation, rare TP63+/KRT5- cells were present in the distal bronchioles (Figure S2E). Together, these data suggest that nuclear pSMAD+/TP63+ cells are not bona fide basal cells since they lack KRT5 expression; rather, mature human fetal basal cells co-express TP63 and KRT5, and lack nuclear pSMAD expression. We hypothesize that pSMAD+/TP63+/KRT5-negative cells may be immature airway progenitor cells, similar to those in the early mouse airway (Yang et al., 2018), and after 3 days of dual SMAD activation in vitro.
Expansion of SMAD-induced airway organoids
Given that DSA-induced organoids stop proliferating, and other work showing basal cells can be cultured in vitro in dual SMAD inhibition conditions (Mou et al., 2016; Murata et al., 2007; Tadokoro et al., 2016), we reasoned that the TP63+/KRT5- cells in day 3 organoids (Figure 1) may overcome proliferation defects and differentiate into mature airway cells if cultured in appropriate expansion media (Figure 2D). In a screen for organoid expansion, we found that supplementation with FGF10 and Y27632, a RHO kinase inhibitor, in addition to inhibitors of TGFβ and BMP (‘DSI expansion medium’: FGF10, A8301, NOGGIN, Y27632) allowed for the recovery of some DSA-induced organoids as evidenced by an increase in diameter (Figure S3C); however, many organoids did not survive DSA treatment (Figure S3D). Treatment of bud tip progenitor organoids with DSA followed by 7 days of DSI led to some TP63+ cells co-expressing KRT5 (Figure S3E), and increased levels of other canonical airway cell markers, such as FOXJ1 (multiciliated cells) and SCGB1A1 (secretory cells) as assessed by QRT-PCR (Figure S3F–G). Bud tip progenitor organoids treated with DSA followed by DSI revealed that pre-treatment with DSA significantly increased TP63 expression after 10 days (Figure 2F). Consistent with organoids grown from the adult human lung (Sachs et al., 2019), we found induced organoids grown in DSI expansion medium for 21 days demonstrated protein expression for markers of differentiated airway cells. Organoids showed expression of fetal airway secretory cells (SCGB3A2+/SFTPB+, see also Figure 3) multiciliated cells (FOXJ1+, ACTUB+), club cells (SCGB1A1+), goblet cells (MUC5AC+) and neuroendocrine cells (CHGA+, SYN+), in addition to TP63+ cells and a subset of TP63+/KRT5+ basal-like cells (Figure 2G). Protein staining in DSA-DSI treated organoids showed a significant reduction in pSMAD staining in treated organoids versus 3F controls, although this was expected since treated organoids were being expanded in SMAD inhibiting media. Despite SMAD inhibitory media, low levels of pSMAD2 were still detectable in TP63- cells on the luminal side of the treated organoids (Figure S2G), whereas pSMAD1,5,8 was undetectable in DSA-DSI treated organoids (Figure S2G). After several days in culture, many beating multiciliated cells were present within organoids and the luminal contents within organoids appeared to swirl with directionality, showing that multiciliated cells were functional and able to propel luminal contents (Video S1). These results suggest that DSA-induction followed by DSI-expansion stimulates bud tip organoids to differentiate into a proximal airway phenotype.
Figure 3. Defining cell signatures of human fetal lung epithelial cells.
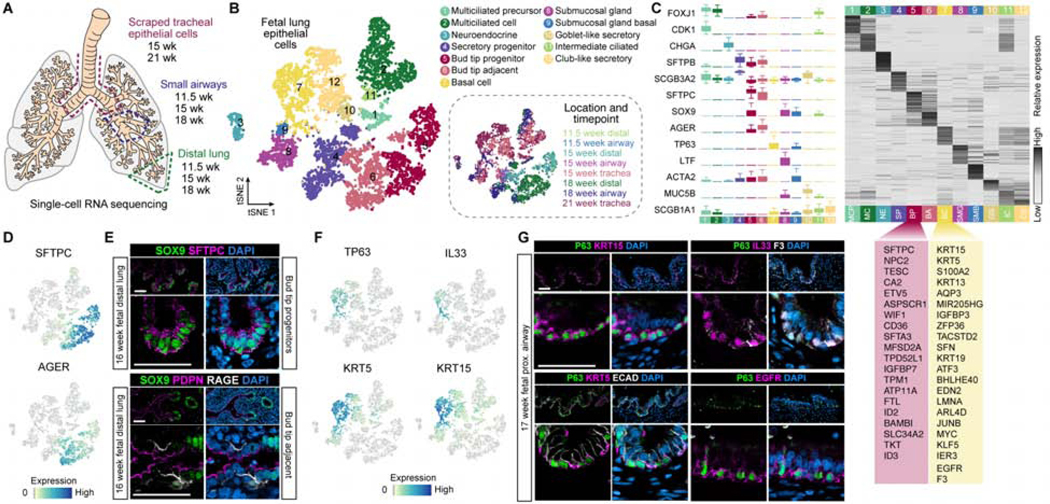
A) Schematic of the experimental setup and samples included in the analysis. B) A total of 8,443 EPCAM+ cells were computationally isolated and cellular transcriptome heterogeneity was visualized using a tSNE plot revealing 12 clusters of cells. C) Left panel: boxplots (interquartile range with minimum and maximum, outliers removed from plot) show expression level distributions of canonical cell type markers in each cluster. Right panel: heatmap shows z-transformed cluster average expression levels of top 50 cluster markers ranked by log-transformed fold change in expression levels in each cluster compared to other clusters in any of the clusters. Clusters were identified by expression of markers canonically associated with cell populations based on published literature, or given a new name (e.g. secretory progenitor, bud tip adjacent). A list of the upregulated genes in the bud tip progenitor cluster (BP) and the basal cell cluster (BC) with top fold change in expression levels relative to all other clusters is shown and provides a gene signature for these cell populations. Cell cluster names and colors correspond to those in Figure 1B. D) Feature plots of canonical bud tip progenitor markers SFTPC and AGER, a canonical marker of alveolar epithelial type 1 cells. E) Protein staining for SOX9 (green), SFTPC (pink), and DAPI (blue) in bud tips of a 16 week fetal lung specimen. Protein staining for SOX9 (green), PDPN (pink), RAGE (also known as AGER; white), and DAPI (blue) in cells adjacent to the bud tips in a 16 week fetal lung sample. Scale bars represent 50 μm. F) Feature plots showing expression of canonical basal cell markers TP63 and KRT5, as well as KRT15 and IL33, which are highly specific to cluster 7. G) Protein staining of tracheal TP63+ basal cells in 17 week fetal lungs for other basal cell markers identified in cluster 7 by scRNA-seq: KRT15 (pink), EGFR (pink), IL33 (pink), F3 (white) and PDPN (pink).
Single-cell transcriptomics defines bud tip progenitor and basal stem cell signatures in the developing human lung
To determine whether the cells generated in vitro were similar to cells in the human fetal lung, we performed scRNA-seq on dissociated whole distal lung, small airway, and scraped tracheal epithelial cells (Figure 3A, Figure S4A) from human fetal lung tissue ranging from 11.5 to 21 weeks gestation. We identified diverse immune, endothelial, mesenchymal, and epithelial populations (Figure S4B–C). We computationally extracted 8,443 epithelial cells based on clusters expressing canonical epithelial markers (e.g. EPCAM; Figure S4C–D; see Table S1 for numbers of cells per sample), then re-clustered these cells and visualized the heterogeneity using tSNE (Figure 3B). Based on this analysis, we identified 12 epithelial cell clusters, and cluster identities were assigned based on known markers, where possible (Figure 3C; Figure S4C, E; Table S1). We identified multiple known groups of lung epithelial cells including bud tip progenitors (BP - cluster 5), basal cells (BC - cluster 7) and differentiated cell types including multiciliated cells (MC - cluster 2), neuroendocrine cells (NE - cluster 3), and club-like and goblet-like secretory cells (CS, GS - clusters 10 and 12, respectively). We also identified two previously undescribed epithelial cell clusters, which we term ‘bud tip adjacent’ (BA - cluster 6) and ‘secretory progenitor cells’ (SP - cluster 4). Cluster 6 bud tip adjacent cells co-expressed a unique combination of genes including bud tip progenitor markers (low SFTPC, ID2) as well as canonical AECI markers such as HOPX, PDPN and AGER (Figure 3C–D). Protein staining of the human fetal lung at 16 weeks shows that these cells are physically located adjacent to the SOX9+ bud tips and are PDPN+/AGER+/SOX9- (Figure 3E; n=3 biological replicates). Cells in cluster 4 express high levels of SCGB3A2, SFTPB and CFTR, were molecularly distinct from other secretory cell populations; for example, they did not express the club cell marker SCGB1A1 (Figure 3C), and did not match any known cell type described in the adult human or murine lung.
We did not observe any clearly identifiable clusters for rare cell types such as tuft cells or ionocytes (Fig. S4F)(Montoro et al., 2018; Plasschaert et al., 2018). Based on the proportion of these cell types reported in the mouse airway (Montoro et al., 2018) and in human culture systems (Plasschaert et al., 2018), and based on the number of cells we analyzed here, we would expect to see these cells represented in the data if present in the human fetal lung. The absence of these cells in our dataset supports the idea that they are not yet present in the developing human lung. However unlikely, we are unable to rule out the possibility that tuft cells and ionocytes have a lower abundance in the fetal lung than in the adult mouse and human airway and these cells are therefore absent from our analysis due to number of cells sequenced.
We next identified unique gene signatures and validated protein markers for human fetal bud tip progenitors and basal stem cells. Expression of canonical markers of human bud tip progenitors, including SOX9, SFTPC, ETV5 and ID2 (Figure 3C–D; Table S1)(Miller et al., 2018; Nikolić et al., 2017; Rockich et al., 2013; Vaughan et al., 2015; Zuo et al., 2015), exhibited the strongest expression enrichment in cluster 5, indicating it is a bud tip progenitor cell cluster. Further, comparison of genes expressed in the bud tip progenitor cell cluster relative to expression in all other clusters allowed us to identify marker genes most highly enriched in bud tip progenitors, thereby defining a transcriptional signature of human fetal bud tip progenitors at 11.5–18 weeks gestation (Figure 3C, Figure S 4G, Table S1). Immunofluorescent staining in fetal lung tissue confirmed protein expression of several bud tip markers (Figure 3E; n=3 biological replicates).
To define a basal stem cell signature, we first identified basal cells as cells within cluster 7 based on expression of the canonical basal cell markers TP63 and KRT5 (Figure 3C, F). In addition, this cluster highly expressed a set of genes including KRT15, IL33, S100A2, F3, EGFR, and PDPN (Figure 3C, F; Table S1), and protein staining validated that KRT5, IL33, F3, EGFR, KRT15 and PDPN were expressed within TP63+ cells in the trachea at 17 weeks gestation (Figure 3G; Figure S4H; n=3 biological replicates). Of the basal cell enriched genes, EGFR and F3 were surface markers uniquely co-expressed in basal cells. We identified antibodies that allowed us to use fluorescence activated cell sorting (FACS) to purify human basal cells (Figure 3C, G), and validated that EPCAM+ sorting followed by EGFR+/F3+ sorting enriched TP63+ basal cells from primary fetal lung tissue (Figure S4H, I) compared to EPCAM+ sorted cells alone. Together, these data provide a reference atlas of epithelial cells with defined molecular signatures that are present in the human developing lung epithelium.
In vitro-derived airway cell transcriptional signatures correlate highly with native fetal airway epithelial cell signatures at single cell resolution
In order to understand the cell composition organoids and determine whether they matched in vivo cell types from the native fetal lung, we analyzed the transcriptomes of 2,106 bud tip organoid cells prior to differentiation (‘day 0’), 9,400 cells immediately after 3 days of DSA treatment (‘day 3’) and 3,755 cells from DSA-induced organoids treated for 3 days followed by 18 days of DSI expansion medium (‘day 21’ organoids) using scRNA-seq. tSNE dimension reduction of the data from each time point separately allowed visualization of cell heterogeneity and the expression patterns of cell markers for bud tip progenitors (SFTPC and SOX9) and Basal Cells (TP63, KRT5, KRT15), and other cell types (Figure 4A, Figure S5A). Day 0 cells have only one homogeneous cluster (Figure 1C, Figure 4A). SFTPC is initially expressed at day 0, is greatly reduced by day 3, but increases again in specific cells by day 21 (Figure 4A). Clustering of Day 3 and Day 21 cells identified 7 and 11 clusters, respectively (Figure S5B–D, Table S1). TP63 is absent from day 0 cells, but is detected in 26% of cells by day 3, and is detected in a more restricted subset cells (13%) by day 21 (Figure 4A). For a general characterization of the acquisition of basal cell transcriptome features throughout treatment, we summarized the expression of 331 in vivo fetal basal cell marker genes as the ‘basal cell signature’, and examined the average scaled expression of this gene set in in vitro-derived cells. Cells of day 3 and day 21 induced organoids upregulated basal cell marker genes (Figure 4B). Meanwhile, compared to day 3, day 21 organoids show even further upregulation of the basal cell signature in specific cells (Figure 4B).
To evaluate the transcriptomic similarity between in vitro derived cells and in vivo fetal lung epithelial cells, we quantified the transcriptome similarity of each cell to the in vivo epithelial cell cluster gene expression signatures as identified in Figure 3B–C using Pearson’s correlation coefficients (Figure 4C). All cells of in vitro bud tip progenitor cells (day 0) displayed the highest degree of gene expression similarity with in vivo bud tip progenitor cells (Figure 4C). The majority of cells from organoids treated with 3 days of DSA (day 3) are most similar to bud tip progenitors, while some of them show marginally higher similarity to bud tip adjacent, sub mucosal basal cells, or basal cells (Figure 4E). Consistent with the protein staining demonstrating that SMAD-induced cells have protein staining reminiscent of cell types found in the airway, organoids at day 21 contain cells that have transcriptomes most highly correlated with multiciliated cells, neuroendocrine cells, SCGB3A2+/SFTPB+/CFTR+ secretory cells, bud tip adjacent cells, and basal cells (Figure 4E, F). At day 21 cells, we visualized cells using tSNE, where each cell is colored by the best-correlated in vivo cluster (Figure 4F). Expression of individual marker genes for each population show consistent patterns (Figure S5A). In vitro-derived TP63+ cells also express KRT15 and low levels of KRT5. Together, this analysis suggests that DSA-induction followed by DSI-expansion of bud tip progenitor organoids and generates airway cells recapitulating molecular signatures of the native developing human airway.
It is appreciated in the field that the transcriptional signature of an in vitro derived cell is unlikely to be absolutely equivalent to an in vivo cell type, given that the environment is dramatically different in the two contexts. To further interrogate differences between in vitro and in vivo cell types, we performed a head-to-head comparison of these cells to examine the overlap between top marker genes of each fetal epithelial cluster and those of each in the day 3 and day 21 in vitro clusters (Figure S5E–G, Table S1). Collectively, this analysis showed that the correlation between in vitro and in vivo clusters is strong; however, even though these cells share genetic and functional similarities, there are still major differences driven by the in vitro or in vivo context. We next sought to gain insight into possible signaling mechanisms underlying the effects of SMAD activation and inhibition on bud tip progenitors. To identify putative transcription factors and other regulatory elements involved, we analyzed day 3 (SMAD activation stage) and day 21 (SMAD inhibition stage; Figure S5G, Table S1) single cell data using pySCENIC (Aibar et al., 2017), a software that enables inference of transcription factors, gene regulatory networks (‘regulons’) and cell types from single-cell RNA-seq data. To identify regulons associated the TP63+ cell gene features, we selected those targets which were overrepresented in TP63+ cells (Figure S5G). In the TP63+ cell associated regulons, no SMAD family members were identified transcription factors, but some of the transcription factors are predicted to be bound by SMADs according to MSigDB(Liberzon et al., 2011; Subramanian et al., 2005), suggesting that SMAD factors may play a regulatory role upstream of transcription factors that directly regulate TP63 expression. We further found that TP63+ cell associated regulons indirectly regulated by SMAD family differed between day 3 and day 21 time points (Figure S5G).
In vitro-derived TP63+ cells can be isolated and exhibit properties of single cell-expansion, self-renewal and multilineage differentiation
We sought to determine whether in vitro-derived TP63+ cells exhibited functional hallmarks of basal stem cells, including expansion of isolated cells, self-renewal and the ability to undergo multilineage differentiation into airway cell types such as multiciliated cells and secretory cells. To do this, we isolated TP63+ cells with FACS using the basal cell enriched cell surface proteins EGFR and F3, which were co-expressed in induced organoids based on co-immunofluorescent staining with TP63+ (Figure 3C, Figure 5A–C, Figure S6A–D). Bud tip organoids were treated with DSA for 3 days and expanded for 18 days in DSI expansion medium (Figure 5A). Organoids were subsequently dissociated and FACS was used to isolate EGFR+/F3+ cells (Figure 5C, Figure S6A–C). The percentage of EGFR+/F3+ double positive cells isolated for each biological replicate varied from 5.84% to 25.9%, reflecting variability between independent biological replicates. FACS isolated cells from all 3 biological replicates were immediately affixed to glass slides via cytospin and were stained for TP63 protein expression. This analysis showed that 92.09 (+/− 1.66)% of EGFR+/F3+ cells co-expressed nuclear TP63, whereas 8.09% of EGFR-/F3- cells expressed nuclear TP63 (Figure 5D, E; n=3 biological replicates). We also observed many EGFR+/F3- and EGFR-/F3+ cells in the sample (Fig. 5C, Fig. S6A–D), most of which were not TP63+ (Fig. 5D–E). After sorting, a fraction of the EGFR+/F3+ cells were re-plated in Matrigel and grown in vitro in DSI expansion medium. Organoids derived from a single cell suspension of EGFR+/F3+ cells contain TP63+ basal-like, MUC5AC+ goblet-like, SCGB1A1+ club-like and AcTUB+/FOXJ1+ multiciliated cells as shown by protein staining (Figure 5F, G, Figure S6G). At least 5 organoids for each stain from each biological replicate were counted. These cell types were not present in untreated controls (Figure S6E). Interestingly, no neuroendocrine cells were detected in EGFR+/F3+ expanded airway organoids, although they were present in DSA-DSI treated organoids that were unsorted (Figure 5F). This lack of neuroendocrine cells could possibly indicate that the specification of neuroendocrine cells occurs very early in development, directly from the bud tip progenitor cells. Additionally, organoids derived from sorted cells exhibited functional multiciliated cells that beat, along with swirling luminal contents (Video S2).
Figure 5. Isolated in vitro-derived basal-like cells give rise to clonal airway organoids.
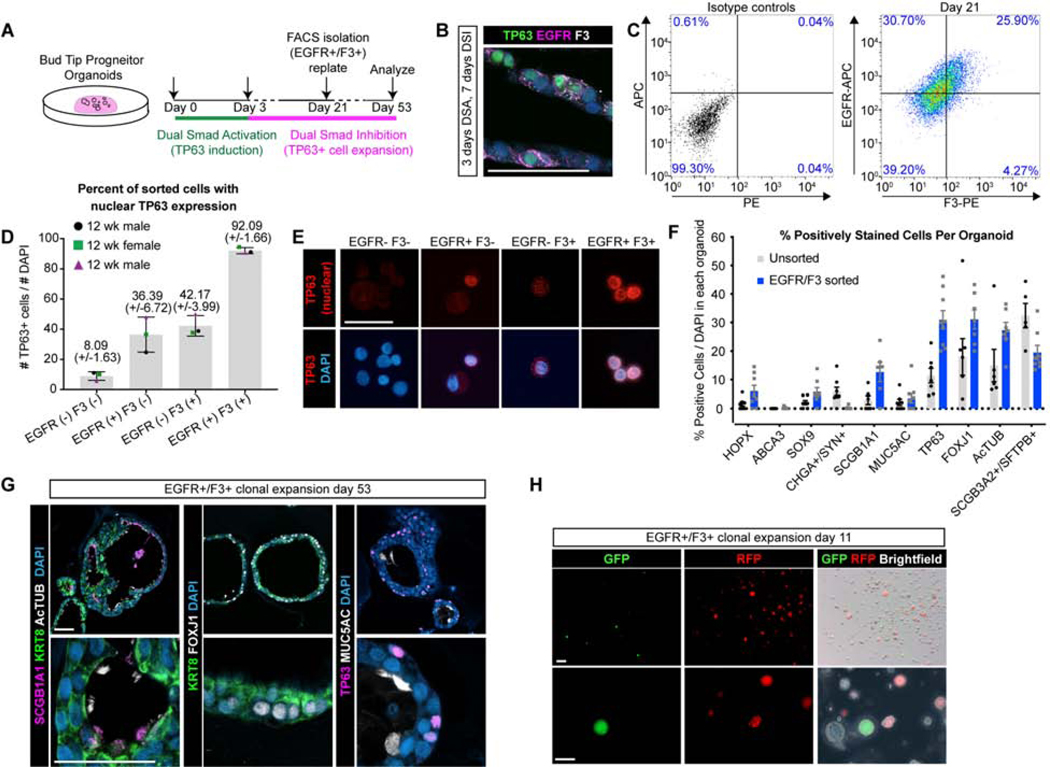
A) Overview of experimental design. B) Protein stain for TP63 (green), EGFR (pink), F3 (white) and DAPI (blue) in organoids treated for 3 days of DSA followed by 7 days of DSI. Scale bar represents 50 μm. C) EGFR/F3 and control FACS plots for 1 representative biological replicate (n=3 biological replicates). D-E) Protein staining for TP63 (red, nuclear) and quantification of TP63+ cells/total cells on sorted cells spun onto a glass slide using a Cytospin. Error bars represent the mean +/− the standard error of the mean. N=3 biological replicates, shown as 3 separate colors on the plot. F) Quantification of the percentage of cells in each organoid containing positive protein staining for canonical cell type markers in unsorted organoids versus organoids that were grown from isolated EGFR+/F3+ cells. N=3 biological replicates per group and n=3 technical replicates (individual organoids) per biological replicate. A total of 198 cells were counted for the EGFR-/F3- group, 110 cells counted for EGFR+/F3- group, 38 cells counted for the EGFR-/F3+ group, and 88 cells counted for the EGFR+/F3+ group. G) Protein staining of organoids derived from EGFR/F3 sorted cells for secretory marker SCGB1A1 (pink), multiciliated markers AcTUB+ and FOXJ1+ (white), epithelial marker KRT8 (green), basal cell marker TP63 (pink), goblet cell marker MUC5AC (white) and DAPI (blue). Data shown from a single biological replicate and is representative of N=3 biological replicates. Scale bars represents 50 μm. H)GFP (green), RFP (red) and brightfield images of whole organoids 11 days after re-plating and mixing EGFR/F3 sorted cells from GFP and RFP expressing groups. N=1 biological replicate with n=6 technical replicates (wells of mixed organoids). Data shown from a single experiment and is representative of n=3 experiments. Scale bar represents 200 μm (top row) and 100 μm (bottom row).
To test whether EGFR+/F3+ derived organoids cells exhibited clonal expansion, bud tip progenitor organoids were treated with 3 days of DSA followed by expansion with DSI. After 2 weeks of expansion in DSI, separate batches of organoids were infected with a lentivirus driving expression of GFP or with a lentivirus driving expression of mCherry, in order to track and visualize these cells over time. Organoids were allowed to expand for an additional 2 weeks in DSI, and EGFR+/F3+ cells were isolated using FACS (Figure S6I). Sorted cells from each condition (GFP-infected or mCherry-infected organoids), were plated alone or as a mixture to evaluate whether clonal populations formed. Of the resulting organoids, whole organoids are composed either entirely of GFP+ cells or RFP+ cell, or were negative for both fluorescent reporters, reflecting the fact that not all EGFR+/F3+ cells were labeled. No chimeric organoids, consisting of GFP/RFP, or of fluorescent and non-fluorescent cells, were observed. This suggested that individual organoids are derived from single cells, rather than from cell aggregation (Figure 5H). Together, this data demonstrates that basal cells from DSA induced organoids are functional.
Discussion
Here we show that human fetal bud tip progenitors are able to give rise to functional lung airway cells, including basal cells, and that SMAD signaling potently stimulates bud tip cells to differentiate into proximal airway epithelium.
Consistent with previous studies in mice, we found that homogeneous human bud tip progenitor organoids are able to give rise to airway cells (Rawlins et al., 2009). Cre-mediated lineage tracing in mice showed only early (<E9.5) Sox9+ epithelial progenitors give rise to tracheal epithelium and basal cells (Yang et al., 2018). While basal cells are restricted to the tracheal epithelium in mice, they extend far into the bronchial tree in humans, suggesting human bud tip progenitors give rise to intrapulmonary basal cells as the airway undergoes branching (Figure 1A). Our data from 12 week fetal lungs shows TP63+ cells are found along the length of the bronchiolar tree, and 12 week human bud tip progenitor cells grown in vitro retain the ability to give rise to multiple airway cells types, including basal cells. In addition, SMAD signaling robustly induced the expression of TP63 in bud tip progenitor organoids after 3 days, but did not initiate a full basal cell signature gene set at this time point. Instead, these cells behaved like airway progenitor cells, giving rise to organoids that contained multiple airway cell types, including functional multiciliated and secretory cells. Another possible explanation for this data is that dual SMAD activation provides a general signal to bud tip progenitors to initiate a proximal cell fate, with further lineage specification occurring later. This is supported by previous reports in the literature suggesting a role for both TGFβ1 and BMP4 in regulating proximal-distal specification of the mouse airway (Heine et al., 1990; Mahoney et al., 2014; Shu et al., 2005; Weaver et al., 1999).
In the developing mouse lung, multiciliated cells and secretory cells do not emerge until E14.5 or E15.5, respectively (Rawlins et al., 2007), suggesting that cells within the airway prior to differentiation are SOX2+ airway progenitors. Therefore, bud tip progenitors do not necessarily give rise directly to functionally mature airway cell types, rather, cells pass through intermediate progenitor states before becoming functionally mature. Our work suggests that intermediate progenitor cells may exist in the human fetal lung, but additional work is required to determine the lineage relationships. Our single cell transcriptomic analysis identified two cell populations that have not been reported previously. The first of these populations, ‘bud tip adjacent cells’, reside physically adjacent to the bud tip cells in vivo in 12–20 week fetal lungs, and these cells express canonical markers of alveolar cell types. The second of these populations shares a transcriptional profile with secretory cells, but also co-expresses a unique combination of genes/proteins that include high levels of SCGB3A2, SFTPB and CFTR. Interestingly, these cells are not likely ionocytes (Montoro et al., 2018; Plasschaert et al., 2018) as they lack FOXI1 expression, and they are transcriptomically distinct from club-like and goblet-like cell clusters. A population of SCGB3A2+/UPK3A+ secretory progenitors cells has been reported in the developing mouse lung (Guha et al., 2012), which were shown through lineage tracing to be precursors of club and multiciliated cells. The current study does not resolve the lineage relationships between SCGB3A2+/SFTPB+/CFTR+ and terminally differentiated cells; however, we also demonstrate that an analogous population of cells is present in DSA induced organoids.
Previous studies using murine genetics to study SMAD signaling in the mouse trachea/airway and studies using human tissue have shown that nuclear phosphorylated SMADs are absent from mature tracheal basal cells. Consistent with this, cultured basal cells require that SMAD signaling inhibition in order to maintain their proliferative and undifferentiated state in vitro (Mou et al., 2016; Tadokoro et al., 2016). The current work adds to this body of literature by describing a dynamic dual role for SMAD signaling in the airway. Consistent with reports in adult airway organoids (Sachs et al., 2019), but in contrast to previously published work in 2D culture showing that SMAD inhibition prevents differentiation of basal cells (Mou et al., 2016), we found SMAD inhibition in organoids allowed differentiation into airway-like cells, especially within cells lining the inner lumen of the organoids, and we observed some phospho-SMAD2 was maintained in the inner luminal cells, suggesting that the 3D environment may allow SMAD activation and differentiation within the organoids. Expansion and maturation of basal cells derived from bud tip organoids took place in an SMAD-inhibitory environment, which is consistent with previous studies (Mou et al., 2016; Tadokoro et al., 2016). It is noteworthy that we observed nuclear phosphorylated SMAD signaling in TP63+ cells in less mature cells lower in the human fetal airway, suggesting that SMAD activation may be involved in the generation of TP63+ cells in the intrapulmonary airways. However, it remains to be determined whether TP63+/KRT5-/pSMAD-high cells go on to give rise to bona fide basal cells in the adult airway, or whether they represent transient proximal airway progenitors.
Taken together, this work suggests SMAD activation induces TP63 expression and proximalizes undifferentiated bud tip progenitor organoids, which ultimately give rise to airway cell types, including basal cells. The resources provided here lay groundwork to further interrogate cellular relationships in the human fetal lung and to benchmark in vitro findings against the in vivo cell populations, and may inform methods to generate airway and basal cells from human pluripotent stem cells in vitro (Dye et al., 2015; Miller et al., 2018, 2019).
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Contact Jason R. Spence at spencejr@umich.edu for requests for materials.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Human Lung Tissue
Human tissue research was reviewed and approved by The University of Michigan Institutional Review Board (IRB). Human lung tissue was obtained from the University of Washington Laboratory of Developmental Biology. Tissue was shipped overnight in UW-Belzer’s solution on ice.
Bud Tip Progenitor Organoids
All experiments were carried out using tissue from 3 different biological specimens at approximately 12 weeks gestation. The peripheral portion of the lungs were enzymatically and mechanically disrupted to isolate bud tip epithelial cells, which were subsequently cultured in 3-dimensional Matrigel droplets with media conditions optimized to expand and maintain bud tip progenitor organoids, as previously described(Miller et al., 2018). Briefly, 1 cm2 segments of distal lung tissue were cut from the lung with a scalpel, tissue was dissociated using lung dispase (Corning) on ice for 30 minutes, followed by incubation in 100% Fetal Bovine Serum (ThermoFisher Scientific cat. no. 16000044; FBS) for 15 minutes. Tissue was then vigorously pipetted up and down with a p200 to separate the epithelial bud tips from the mesenchyme. Tissue was washed multiple times in sterile 1x PBS to remove mesenchymal cells and epithelium-enriched bud tips were plated in a Matrigel droplet. Buds tips could be frozen down immediately after isolation or at any time after culture in 10% DMSO, 10% FBS and 80% DMEM F12.
METHOD DETAILS
Paraffin processing, tissue preparation, protein staining and imaging
For all protein staining experiments, analysis was carried out on n=3 independent biological specimens, and representative images are shown in the figures. For protein analysis, tissue was immediately fixed in 4% Paraformaldehyde for 24 hours at 4°C. Tissue was washed in 3 washes of 1X PBS for a total of 2 hours, and then dehydrated by an alcohol series of each concentration diluted in 1x PBS, 30 minutes in each solution: 25% Methanol, 50% Methanol, 75% Methanol, 100% Methanol, 100% Ethanol, 70% Ethanol. Tissue was processed into paraffin blocks in an automated tissue processor (Leica ASP300) with 1 hour solution changes overnight. 7 μm-thick sections were cut from paraffin blocks and immunohistochemical staining was performed as previously described(Miller et al., 2018; Spence et al., 2009). A list of antibodies and concentrations can be found in Table S2. All images were taken on a NIKON A1 confocal and assembled using Photoshop Creative Suite 6. Imaging parameters were kept consistent for all images of the same experiment and any post-imaging manipulations were performed equally on all images from a single experiment.
In situ hybridization
For all in situ hybridization staining experiments, analysis was carried out on n=3 independent biological specimens, and representative images are shown in the figures. Human fetal lung tissue was fixed for 24 hours at room temperature in 10% Neutral Buffered Formalin (NBF), washed with DNAse/RNAse free water (Gibco) for 3 changes for a total of 2 hours. Tissue was dehydrated by an alcohol series diluted in DNAse/RNAse free sterile water for 30 minutes in each solution: 25% Methanol, 50% Methanol, 75% Methanol, 100% Methanol. Tissue was stored long-term in 100% Methanol at 4°C. Prior to paraffin embedding, tissue was equilibrated in 100% Ethanol, and then 70% Ethanol. Tissue was processed into paraffin blocks in an automated tissue processor (Leica ASP300) with 1 hour changes overnight. Paraffin blocks were sectioned to generate 7 μm-thick sections. All materials, including the microtome and blade, were sprayed with RNAse-away solution prior to use. Slides were sectioned freshly the night before the in situ hybridization procedure, baked for 1 hour in a 60°C dry oven, and stored overnight at room temperature in a slide box with a silicone desiccator packet, and with seams sealed using parafilm. The in situ hybridization protocol was performed according to the manufacturer’s instructions (ACDbio; RNAscope multiplex fluorescent manual protocol). The human TP63 probe was generated by ACDbio targeting 4309–1404 of TP63 (accession NM_001114982.1) and is commercially available (acdbio.com, catalog number 601891-C2). The human KRT5 probe was generated by ACDbio targeting 78–2053 of KRT5 (accessionNM_000424.3) and is commercially available (acdbio.com, catalog number 310241).
Infection of organoids with GFP-lentivirus or mCherry-lentivirus
Lentiviral particles were generated from a construct expressing GFP under the control of a PGK promoter with puromycin selection (Addgene plasmid #19070), or by a construct expressing mCherry under the control of a PGK promoter (Addgene plasmid #21217) by the University of Michigan Viral Vector Core. Under a dissecting microscope in a sterile hood, organoids were removed from Matrigel droplets by vigorous pipetting with a p200 pipette. Organoids were transferred to a 1.5 mL Eppendorf snap-cap tube with ~250 μL of regular culture medium. Cells were then passaged through a 27-gauge needle attached to a 1 mL syringe 2 times in order to shear the epithelial organoids into small fragments. Cells were spun down for 5 seconds at full speed using a mini centrifuge (similar to ThermoScientific mySPIN 6, cat. no. 75004061), and the remaining floating Matrigel and culture medium were removed from the tube using the needle and syringe under a dissecting microscope. 1 mL of regular culture medium was added to the Eppendorf tube containing the organoid cell fragments, and the cells were transferred to 1 well of a 12-well tissue culture plate. 10 μM of Y27630 was added to improve cell survival and 0.5 mL of high titer virus was added to the well with cell fragments. The plate was placed in a tissue culture incubator (37°C, 0.5% CO2) on a rocker for 6 hours. After 6 hours, the cells suspension was moved to a 15 mL conical tube, spun down at 300g for 5 minutes at 4°C, the supernatant was removed and treated with bleach solution, and the cells were washed and spun down (300g, 5 minutes, 4°C) 3X with DMEM. On the final wash, cells were resuspended in 100% Matrigel and plated as a 3-dimensional droplet, allowed to solidify, and then overlaid with bud tip progenitor maintenance medium.
Preparation of tissue for single cell RNA sequencing
Human Fetal Tissue
To dissociate human fetal tissue to single cells, tissue was first dissected into regions (trachea/bronchi, small airways, distal lung) using forceps and a scalpel in a petri dish filled with ice-cold 1X HBSS (with Mg2+, Ca2+). For harvesting cells from the trachea and bronchi, the airways were transferred to a fresh petri dish filled with ice-cold 1X HBSS and were opened longitudinally with spring-loaded microscissors. The epithelium was scraped with a scalpel and a p200 pipette tip, prewashed with 1% BSA in HBSS to reduce cells from sticking, was used to collect epithelial cells and place them in a 15 mL conical tube. For distal lung tissue, a section roughly 1cm2 was cut with a scalpel from the most distal lung regions and minced using a scalpel and forceps. This tissue was then transferred to a 15 mL conical tube.
Dissociation enzymes and reagents from the Neural Tissue Dissociation Kit (Miltenyi, cat. no. 130-092-628) were used, and all incubation steps were carried out in a refrigerated centrifuge pre-chilled to 10°C unless otherwise stated. All tubes and pipette tips used to handle cell suspensions were pre-washed with 1% BSA in HBSS to prevent adhesion of cells to the plastic. Tissue was treated for 15 minutes at 10°C with Mix 1 and then incubated for 10 minute increments at 10°C with Mix 2 interrupted by agitation by pipetting with a P200 pipette until fully dissociated. Cells were filtered through a 70 μm filter coated with 1% BSA in 1X HBSS, spun down at 500g for 5 minutes at 10°C and resuspended in 500μl 1X HBSS (with Mg2+, Ca2+). 1 mL Red Blood Cell Lysis buffer was then added to the tube and the cell mixture was placed on a rocker for 15 minutes in the cold room (4°C). Cells were spun down (500g for 5 minutes at 10°C), and washed twice by suspension in 2 mLs of HBSS + 1% BSA followed by centrifugation. Cells were then resuspended in 1% BSA in HBSS with 0.5 units/μL of RNAseI (ThermoFisher cat. no. AM2294) in order to reduce RNA present in the media/buffer. Cells were counted using a hemocytometer (ThermoFisher), then spun down and resuspended (if necessary) to reach a concentration of 700–1000 cells/μL and kept on ice. Single cell libraries were immediately prepared on the 10x Chromium at the University of Michigan Sequencing Core facility with a target of 10,000 cells. A full, detailed protocol of tissue dissociation for single cell RNA sequencing can be found at www.jasonspencelab.com/protocols.
Organoids
To dissociate organoids to single cell suspensions, organoids were removed from the Matrigel droplet by vigorous pipetting with a p200 in a small petri dish, then transferred to a 15 mL conical tube containing 8 mL of TrypLE express (ThermoFisher Scientific cat. no. 12605036) and placed in a tissue culture incubator at 37°C, 5% CO2 on a rocker for no longer than 30 minutes. Every 5 minutes, the tube was removed from the incubator and cells were agitated by pipetting the solution up and down with a P1000 pipette. Cells were confirmed to be at single cell suspension by microscope. After most cells were in a single cell suspension, cells were spun down at 300g for 5 minutes at 4°C, resuspended in 1% BSA in HBSS, filtered through a 70 μm filter (similar to Fisher Scientific cat. no. 087712) and spun down again at 300g for 5 minutes at 4°C. All tubes used to handle cell suspensions were pre-washed with 1% BSA in HBSS to prevent adhesion of cells to the plastic walls of tubes. Cells were then resuspended in 100 μL of 1% BSA in HBSS with 0.5 units/μL of RNAseI (ThermoFisher cat. no. AM2294), counted using a hemocytometer (ThermoFisher), then spun down and resuspended (if necessary) to reach a concentration of 700–1000 cells/μL and kept on ice. Single cell libraries were immediately prepared on the 10x Chromium at the University of Michigan Sequencing Core facility with a target of 10,000 cells.
RNA extraction and qRT-PCR analysis
At least 1 well, containing 20–50 organoids, for each biological replicate was collected and RNA was extracted for QRT-PCR analysis. More than 1 well of organoids was collected per biological replicate and served as a technical replicate when available. mRNA for QRT-PCR was isolated using the MagMAX-96 Total RNA Isolation Kit (Life Technologies). RNA quality and concentration was determined on a Nanodrop 2000 spectrophotometer (Thermo Scientific). 100 ng of RNA for each sample was used to generate a cDNA library using the VILO cDNA kit (Invitrogen). QRT-PCR was performed on a Step One Plus Real-Time PCR System (Life technologies) using SYBR Green Master Mix (Qiagen). Expression was calculated as a change relative to GAPDH expression using arbitrary units, calculated using the following equation: [2^(GAPDH Ct – Gene Ct)] × 10000. Some data were plotted as fold change of arbitrary expression value over a control. For this analysis, expression values for each gene for each sample, including controls, were divided by the average expression of that gene for the control group. Fold change was calculated as follows: [ExpressionGene / AverageExpressionControls]. A list of QRT-PCR primers used can be found in Table S3.
Fluorescence Activated Cell Sorting (FACS)
To dissociate organoids into single cell suspension, organoids were first removed from the Matrigel droplet by vigorous p200 pipetting in a small petri dish filled with basal medium. Whole organoids or epithelial fragments were then transferred to a 15 mL conical tube containing 8 mL of TrypLE Express (ThermoFisher Scientific cat. no. 12605036) and placed in a tissue culture incubator at 37°C, 5% CO2 on a rocker for no longer than 30 minutes. Every 5 minutes, the tube was removed from the incubator and cells were agitated by pipetting the solution up and down with a P1000 pipette. Cells were confirmed to be in single cell suspension by visualization under an inverted microscope. After cells were in single cell suspension, they were spun down at 300g for 5 minutes at 4°C, resuspended in 1% BSA in HBSS, filtered through a 70 μm filter to remove any cell clusters (similar to Fisher Scientific cat. no. 087712) and spun down again at 300g for 5 minutes at 4°C. All tubes used to handle cell suspensions were prewashed with 1% BSA in HBSS to prevent adhesion of cells to the plastic walls of tubes.
After the second wash, cells were resuspended in sorting buffer and evenly distributed in to several tubes. Cells were incubated with isotype antibody controls that were used to set FACS gates, or incubated with Anti-EGFR-APC, human (Milteny cat. no. 130-110-587, 1:50 dilution), Anti-EGFR-PE, human (Milteny cat. no. 130-110-528, 1:50 dilution), Anti CD142 (F3)-PE, human (Milteny cat. no. 130-098-743, 1:11 dilution), Anti CD142 (F3)-APC, human (Milteny cat. no. 130-115-685, 1:11 dilution), REA control IgG1-APC (Milteny 130-113-434) or mouse IgG1-PE (Milteny cat. no. 130-113-762) and sorted using a Sony Synergy SY3200 system. Data was analyzed using Winlist 8.0 and FlowJo version 10.5.3 for Mac. Cells were sorted into 1mL of 1%BSA in HBSS.
To dissociate primary lung tissue for FACS isolation of bud tip adjacent cells, 1cm2 regions of lung were mechanically removed from the lung and cut in to small pieces using a scalpel until nearly homogeneous. This tissue was then transferred to a 15mL conical tube containing 15 mL TrypLE Express, and the protocol continued as described above for organoid dissociation.
Cytospin analysis
20% of cells isolated from each group for FACS were used to evaluate the percentage of sorted cells expressing TP63. 200 μL of cell suspension (20% of 1 mL) was isolated in a separate 1.5 mL microcentrifuge tube and FBS was added to each aliquot to a final concentration of 5% vol/vol. Cells were placed in clean cytospin cones and spun at 600g for 5 minutes on a Shandon Scientific Cytospin on to charged glass slides. Slides were allowed to air dry for 5 minutes before being fixed in 100% ice cold Methanol for 10 minutes. Slides were air dried for another 10 minutes and were then washed with 2 changes of PBS for a total of 10 minutes on a rocker. The regular staining protocol for immunofluorescence was followed (blocking, primary antibody incubation overnight at 4°C, wash, secondary antibody incubation for 1 hour at room temperature, wash, coverslip, image).
Culture Media, Growth Factors and Small Molecules
All experiments utilized serum-free basal medium that has been previously described11,33. Briefly, serum-free basal medium consists of DMEM F12 (ThermoFisher Scientific cat. no. 21331020 or 21331-020) supplemented with 1X N2 supplement (ThermoFisher Scientific cat. no. 17502048), 1X B27 supplement (ThermoFisher Scientific cat. no. 17504044), 1X L Glutamine (200 mM), 1X Penicilin-Streptomycin (5000 U/mL, ThermoFisher Scientific cat. no. 15140122) and 0.05% Bovine Serum Albumin (Sigma-Aldrich cat. no. A9647). On the day of use, medium is supplemented with 0.4 μM Monothio-glycerol (Sigma-Aldrich, cat. no. M6145) and 50 μg/mL Ascorbic Acid (L-Ascorbic Acid, Sigma-Aldrich cat. no. A4544, CAS Number 50-81-7). To maintain bud tip progenitor organoids in a progenitor state, serum-free basal medium was further supplemented with FGF7 (10 ng/mL, Recombinant Human Fibroblast Growth Factor 7; R&D Systems cat. no. 251-KG/CF), CHIR99021 (3 μM, Stem Cell Technologies cat. no. 72054), and All Trans Retinoic Acid (ATRA; 50 nM, Stemgent cat. no. 04-0021, CAS Number 302-79-4). Basal cell expansion medium used the same basal medium, but was supplemented with FGF10, A8301, NOGGIN and Y27632. Growth factors and small molecules were used at the following concentrations: FGF10 (500 ng/mL, made in-house as previously described), A8301 (1 μM, Stem Cell Technologies cat. no. 72024), NOGGIN (100 ng/mL, R&D Systems, cat. no. 6057), Y27632 (APExBIO cat. no. A30008), LDN212854 (200 nM, R&D Systems cat. no. 6151/10), SB431542 (10 μM, Stemgent cat. no. 04–0010), TGFβ1(100 ng/mL, R&D systems cat. no. 240-B-002), BMP4 (100 ng/mL, R&D systems cat. no. 314-BP-050) Dexamethasone (25 ng/mL, Stem Cell Technologies cat. no. 72092), IL6 (10 ng/mL, R&D Systems, cat. no. 206-IL-010), IL2 (50 U/mL, R&D Systems, cat. no. 202-IL-010), Smoothened Agonist (SAG; 500 nM, R&D Systems, cat. no. 4366/1), EGF (100 ng/mL, R&D Systems cat. no. 236-EG-200), IFNγ (10 ng/mL, R&D Systems cat. no. CAA31639), DAPT (10 μM, R&D Systems cat. no. 2634/10), Hydrocortisone (100 ng/mL, Stem Cell Technologies cat. no. 74142).
Quantification and Statistical Analysis
All statistical analysis (quantification of immunofluorescent images and QRT-PCR data) was performed in GraphPad Prism 6 software. For quantification of protein stains in organoids, at least 3 independent organoids were counted (technical replicates) from n=3 separate biological specimens (biological replicates). All quantification of protein staining was done in a blinded fashion by an independent researcher. Statistical comparisons of data between two groups (e.g. control versus experimental condition) were made using unpaired two-sided Mann-Whitney rank-sum tests. A p-value of less than 0.05 was considered significant. For QRT-PCR analysis, n=3 biological replicates were used. For each biological replicate, 1–3 well of organoids containing 20–50 organoids per well (technical replicates) was collected for analysis. To determine significance differences across multiple groups, a one-way Analysis of Variance (ANOVA) was performed followed by Tukey’s multiple comparisons analysis comparing the mean of each group to the mean of every other group. A p-value of less than 0.05 was considered significant. On graphs, p-values for multiple comparisons after ANOVAs are reported as follows: * p<0.05; ** p<0.01, *** p<0.001, **** p<0.0001. For Figure 2C, 100 DAPI + cells were counted for each airway region for each sample, and results are plotted as a percentage of TP63+ (TP63+/100 DAPI+ cells).
Data and Code Availability
Data preprocessing and cluster identification
All single-cell RNA-sequencing was performed with an Illumina Hiseq 4000 by the University of Michigan DNA Sequencing core. Reads alignment to human reference genome and generation of gene expression matrix were done using 10x Genomics Cell Ranger v2.1.1–2.2.1 software with provided hg19 reference genome and transcriptome annotation. In each sample, to ensure high data quality for further analysis, cells with more than 20,000 or less than 1,500 genes or mitochondrial transcript fraction over 10% and genes expressed in less than 3 cells were excluded for further analysis. All fetal tissue samples were then combined. Gene expression levels were log-normalized by the total number of unique molecular identifier (UMI) per cell. Cell cycle phase was scored by expression levels of cell cycle related genes. Cellular variance of total number of UMI, mitochondrial transcript fraction and cell cycle phase was regressed out using linear regression. Non-cell cycle related genes showing highly variable expression levels (highly variable genes) were identified in each fetal tissue sample separately. For the combined fetal data, principle component analysis (PCA) was based on z-transformed expression levels of genes that were identified as highly variable genes in at least two samples. Graph-based clustering approach and tSNE dimension reduction were performed based on Euclidean distance between the top 20 PCs. Clusters were classified into epithelial, mesenchymal, endothelial, neuronal, immune cell lineages and red blood cell cluster based on expression patterns of canonical marker genes. To better resolve cellular heterogeneity of epithelial cells, cells with low expression levels of EPCAM in the epithelial lineage were further removed, sub-clustering on the remaining epithelial cells was performed. Identification of positive markers for each cluster was performed with Wilcoxon Rank Sum test. Hierarchical clustering of clusters based on average expression levels of cluster markers was performed. Combined with canonical cell type marker expression patterns, clusters with shared cell type identity were merged. Analyses mentioned in this section were mainly performed with Seurat(Butler et al., 2018).
Characterization of cellular transcriptome heterogeneity in in vitro samples
Each in vitro sample was analyzed separately. Data preprocessing of in vitro data followed the same procedure as fetal tissue data except that minimum number of detected genes in cells of day 0 is 1,000 and cellular variance of cell cycle phase was not regressed out. To quantify the transcriptome similarity between in vitro cells and fetal epithelial sub-clusters, Pearson’s correlation coefficients of log-normalized gene expression levels between in vitro cells and fetal epithelial cell sub-clusters were calculated using the genes for fetal tissue kNN network construction. Clustering and positive marker gene expression identification followed the same procedure as for fetal data. Gene ontology enrichment in cluster markers was performed using DAVID (v6.8)(Huang et al., 2009a, 2009b). Significant enrichment was defined as Benjamini-corrected P < 0.05.
Reconstruction of regulon network from in vitro scRNA-seq data
To reconstruct the regulons under SMAD activation and inhibition treatment, we ran pySCENIC(Aibar et al., 2017) on in vitro day 3 and day 21 scRNA-seq data separately. Required auxiliary datasets, i.e. ranking of human whole genome based on transcription factors (TFs) and TF enriched motif annotation, were downloaded from cisTarget databases. Regulons that contain targets being activated or repressed are both obtained. Two regulons were linked if targets of one regulon is the master TF of another regulon. Genes annotated to be bound by SMAD or SMAD3 or SMAD4 was downloaded from MSigDB(Liberzon et al., 2011; Subramanian et al., 2005).
Detailed methods, including code used to process raw data, can be found at https://github.com/qianhuiyu/miller_lung.
Accession numbers
Raw scRNA-seq data associated with this study will be deposited in the EMBL-EBI ArrayExpress database (Accession number: E-MTAB-8221).
Supplementary Material
Supplemental Table 1, related to Figures 1, 3 and 4
Number of cells in fetal and in vitro sc-RNAseq samples (tab 1)
Markers of fetal lung epithelial clusters for fetal and day 21 in vitro sample analyses (tab 2)
Differential expression values comparing day 3 basal-like cluster to in vitro to fetal basal cells and day 21 basal-like cluster to fetal basal cells (tab 3)
Regulons generated from day 3 and day 21 in vitro cells (tab 4)
Supplemental Video 1, related to Figure 2
Functional Multiciliated cells in induced airway organoids treated for 3 days with dual SMAD activation followed by 11 days of dual SMAD inhibition.
Supplemental Video 2, related to Figure 5
Functional Multiciliated cells in induced airway organoids grown from a single GFP+/EGFR+/F3+ basal-like cell.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| *Biotin-Mouse anti-MUC5AC | Abcam | ab79082 |
| *Biotin-Goat anti-P63 | R&D systems | BAF1916 |
| Goat anti-CC10 (SCGB1A1) | Santa Cruz Biotechnology |
sc-9770 |
| Goat anti-Chromogranin A (CHGA) | Santa Cruz Biotechnology |
sc-1488 |
| Goat anti-SOX2 | Santa Cruz Biotechnology |
Sc-17320 |
| Mouse anti-ABCA3 | Seven Hills Bioreagents |
WMAB-17G524 |
| Mouse anti-Acetylated Tubulin (ACTTUB) | Sigma-Aldrich | T7451 |
| Mouse anti-E-Cadherin (ECAD) | BD Transduction Laboratories |
610181 |
| Mouse anti-F3 | Sigma-Aldrich Atlas Antibodies |
AMAb91236 |
| Mouse anti-MUC5B | Abcam | AB77995 |
| Mouse anti-PLUNC | R&D systems | MAP1897 |
| Mouse anti-Surfactant Protein B (SFTPB) | Seven Hills Bioreagents |
Wmab-1B9 |
| Rabbit anti-Clara Cell Secretory Protein (CCSP; SCGB1A1) |
Seven Hills Bioreagents |
Wrab-3950 |
| Rabbit anti-Cleaved Caspase 3 | Cell Signaling | 9664 |
| Rabbit anti-EGFR | Sigma-Aldrich Atlas Antibodies |
HPA018530 |
| Rabbit anti-HOPX | Santa Cruz Biotechnology |
Sc-30216 |
| Rabbit anti-IL33 | Sigma-Aldrich Atlas Antibodies |
HPA024426 |
| Rabbit anti-KRT15 | Sigma-Aldrich Atlas Antibodies |
HPA024554 |
| Rabbit anti-KRT5 | Sigma-Aldrich Atlas Antibodies |
HPA059479 |
| Rabbit anti-NKX2.1 | Abcam | ab76013 |
| Rabbit anti-PDPN | Santa Cruz Biotechnology |
Sc-134482 |
| Rabbit anti-phospho-SMAD1,5,8 | Millipore | AB3848 |
| Rabbit anti-phospho-SMAD2 | Abcam | AB188334 |
| Rabbit anti-Pro-Surfactant protein C (Pro-SFTPC) | Seven Hills Bioreagents |
Wrab-9337 |
| Rabbit anti-SOX9 | Millipore | AB5535 |
| Rabbit anti-Synaptophysin | Abcam | AB32127 |
| Rat anti-KI67 | Biolegend | 652402 |
| Human anti-EGFR-APC (FACS) | Milteny | 130-110-587 |
| Human anti-EGFR-PE (FACS) | Milteny | 130-110-528 |
| Human anti-CD142 (F3)-PE | Milteny | 130-098-743 |
| Human anti-CD142 (F3)-APC | Milteny | 130-115-685 |
| REA control IgG1-APC | Milteny | 130-113-434 |
| mouse IgG1-PE | Milteny | 130-113-762 |
| Bacterial and Virus Strains | ||
| Plasmid for Lentivirus PGK-EGFP-Puro | Addgene | 19070 |
| Plasmid for Lentivirus PGK-mCherry-Puro | Addgene | 21217 |
| Biological Samples | ||
| Human fetal lung tissue, gestational ages 8–24 weeks | University of Washington Laboratory of Developmental Biology | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| N2 supplement | ThermoFisher | 17502048 |
| B27 supplement | ThermoFisher | 17504044 |
| Monothio-glycerol | Sigma | M6145 |
| L-Ascorbic Acid | Sigma | A4544 |
| Recombinant Human Fibroblast Growth Factor 7 | R&D Systems | 251-KG/CF |
| CHIR99021 | Stem Cell Technologies | 72054 |
| All Trans Retinoic Acid | Stemgent | 04-0021 |
| FGF10 | Made in house | |
| A8301 | Stem Cell Technologies | 72024 |
| NOGGIN | R&D Systems | 6057 |
| Y27632 | APExBIO | A30008 |
| LDN212854 | R&D Systems | 6151/10 |
| SB431542 | Stemgent | 04-0010 |
| TGFβ1 | R&D systems | 240-B-002 |
| BMP4 | R&D systems | 314-BP-050 |
| Dexamethasone | Stem Cell Technologies | 72092 |
| IL6 | R&D Systems | 206-IL-010 |
| IL2 | R&D Systems | 202-IL-010 |
| Smoothened Agonist (SAG) | R&D Systems | 4366/1 |
| EGF | R&D Systems | 236-EG-200 |
| IFNγ | R&D Systems | CAA31639 |
| DAPT | R&D Systems | 2634/10 |
| Hydrocortisone | Stem Cell Technologies | 74142 |
| Critical Commercial Assays | ||
| ACDbio RNAscope multiplex fluorescent manual protocol, mRNA in situ hybridization | ACDbio | |
| Neural Tissue Dissociation Kit (for dissociating tissue for scRNA-seq) | Miltenyi | 130-092-628 |
| VILO cDNA kit | Invitrogen | |
| Deposited Data | ||
| Raw scRNA-seq data | EMBL-EBI ArrayExpress database | Accession Number: E-MTAB-8221 |
| Experimental Models: Cell Lines | ||
| All expanding organoid “lines” generated from human fetal tissue sourced from the University of Washington Laboratory for Developmental Biology | ||
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| FOXJ1 | CAACTTCTGCTACTTCCGCC | |
| GAPDH | AATGAAGGGGTCATTGATGG | |
| HOPX | GCCTTTCCGAGGAGGAGAC | |
| KRT5 | CTGGTCCAACTCCTTCTCCA | |
| KRT14 | TCTGCAGAAGGACATTGGC | |
| MUC5AC* | GCACCAACGACAGGAAGGATGAG | |
| TP63 | CCACAGTACACGAACCTGGG | |
| SCGB1A1 | ATGAAACTCGCTGTCACCCT | |
| SOX2 | GCTTAGCCTCGTCGATGAAC | |
| SOX9 | GTACCCGCACTTGCACAAC | |
| SFTPC | AGCAAAGAGGTCCTGATGGA | |
| Recombinant DNA | ||
| Software and Algorithms | ||
| Detailed methods and code for scRNA-seq analysis | https://github.com/qianhuiyu/miller_lung | |
| Other | ||
Highlights.
SMAD activation induces human lung bud tip organoids into an airway fate
Human airway organoids possess basal, secretory, multiciliated and neuroendocrine cells
In vitro organoids were compared to the developing human lung using scRNAseq
In vitro derived basal cells were functional and highly similar to human basal cells
Acknowledgements
This work was supported by a Chan Zuckerberg Initiative Human Cell Atlas Seed Network grant to BT, JGC and JRS, the NIH-NHLBI (R01HL119215) and the Cystic Fibrosis Foundation Therapeutics Epithelial Stem Cell Consortium funding to JRS; AJM was supported by the Ruth L. Kirschstein Predoctoral Individual National Research Service Award NIH-NHLBI F31HL142197; EMH was supported by the Training Program in Basic and Translational Digestive Sciences NIH-NIDDK T32DK094775 and Cellular and Biotechnology Training Program NIH-NIGMS T32GM008353; MC was supported by the Training Program in Organogenesis Fellowship NIH-NICHD T32HD007505. The Laboratory of Developmental Biology, University of Washington, Seattle, WA, United States is supported by NIH-NICHD R24HD000836 to Ian Glass. We thank Judy Opp and the University of Michigan DNA Sequencing Core.
Footnotes
Competing interests
JRS and AJM are co-inventors on patents filed by the Regents Of The University of Michigan relating to the isolation and maintenance of lung bud tip progenitor cells, and the differentiation of lung epithelial progenitor cells into basal cells.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arason AJ, Jonsdottir HR, Halldorsson S, Benediktsdottir BE, Bergthorsson JT, Ingthorsson S, Baldursson O, Sinha S, Gudjonsson T, and Magnusson MK (2014). deltaNp63 Has a Role in Maintaining Epithelial Integrity in Airway Epithelium. PLoS One 9, e88683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniely Y, Liao G, Dixon D, Linnoila RI, Lori A, Randell SH, Oren M, and Jetten AM (2004). Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am. J. Physiol. Physiol. 287, C171–C181. [DOI] [PubMed] [Google Scholar]
- Danopoulos S, Alonso I, Thornton ME, Grubbs BH, Bellusci S, Warburton D, and Al Alam D. (2018). Human lung branching morphogenesis is orchestrated by the spatiotemporal distribution of ACTA2, SOX2, and SOX9. Am. J. Physiol. Cell. Mol. Physiol. 314, L144–L149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BR, Hill DR, Ferguson MA, Tsai Y-H, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, et al. (2015). In vitro generation of human pluripotent stem cell derived lung organoids. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Prasol MS, Estrada J, Baudhuin A, Vranizan K, Choi YG, and Ngai J. (2011). p63 Regulates Olfactory Stem Cell Self-Renewal and Differentiation. Neuron 72, 748–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha A, Vasconcelos M, Cai Y, Yoneda M, Hinds A, Qian J, Li G, Dickel L, Johnson JE, Kimura S, et al. (2012). Neuroepithelial body microenvironment is a niche for a distinct subset of Clara-like precursors in the developing airways. Proc. Natl. Acad. Sci. 109, 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine UI, Munoz EF, Flanders KC, Roberts AB, and Sporn MB (1990). Colocalization of TGF-beta 1 and collagen I and III, fibronectin and glycosaminoglycans during lung branching morphogenesis. Development 109, 29–36. [DOI] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Watkins S, Fuchs E, and Stripp BR (2004). Basal Cells Are a Multipotent Progenitor Capable of Renewing the Bronchial Epithelium. Am. J. Pathol. 164, 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Van de Laar E, Clifford M, Hasenoeder S, Kim BR, Wang D, Lee S, Paterson J, Vu NM, Waddell TK, Keshavjee S, et al. (2014). Cell surface marker profiling of human tracheal basal cells reveals distinct subpopulations, identifies MST1/MSP as a mitogenic signal, and identifies new biomarkers for lung squamous cell carcinomas. Respir. Res. 15, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, and Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney JE, Mori M, Szymaniak AD, Varelas X, and Cardoso WV (2014). The Hippo Pathway Effector Yap Controls Patterning and Differentiation of Airway Epithelial Progenitors. Dev. Cell 30, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Hill DR, Nagy MS, Aoki Y, Dye BR, Chin AM, Huang S, Zhu F, White ES, Lama V, et al. (2018). In Vitro Induction and In Vivo Engraftment of Lung Bud Tip Progenitor Cells Derived from Human Pluripotent Stem Cells. Stem Cell Reports 10, 101–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Dye BR, Ferrer-Torres D, Hill DR, Overeem AW, Shea LD, and Spence JR (2019). Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 14, 518–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. (2018). A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, Zhang B, Solomon GM, Turner B, Bihler H, et al. (2016). Dual SMAD Signaling Inhibition Enables Long-Term Expansion of Diverse Epithelial Basal Cells. Cell Stem Cell 19, 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Ota S, Niki T, Goto A, Li C-P, Ruriko UMR, Ishikawa S, Aburatani H, Kuriyama T, and Fukayama M. (2007). p63 – Key molecule in the early phase of epithelial abnormality in idiopathic pulmonary fibrosis. Exp. Mol. Pathol. 83, 367–376. [DOI] [PubMed] [Google Scholar]
- Nikolić MZ, Caritg O, Jeng Q, Johnson J-A, Sun D, Howell KJ, Brady JL, Laresgoiti U, Allen G, Butler R, et al. (2017). Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Saganta A, Law BM, Tata PR, Villoria J, Saez B, Mou H, Zhao R, and Rajagopal J. (2015). Injury Induces Direct Lineage Segregation of Functionally Distinct Airway Basal Stem/Progenitor Cell Subpopulations. Cell Stem Cell 16, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, and Jaffe AB (2018). A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Ostrowski LE, Randell SH, and Hogan BLM (2007). Lung development and repair: Contribution of the ciliated lineage. Proc. Natl. Acad. Sci. 104, 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Clark CP, Xue Y, and Hogan BLM (2009). The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136, 3741–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, and Hogan BLM (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. 106, 12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockich BE, Hrycaj SM, Shih HP, Nagy MS, Ferguson MAH, Kopp JL, Sander M, Wellik DM, and Spence JR (2013). Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. 110, E4456–E4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N, Papaspyropoulos A, Zomer‐van Ommen DD, Heo I, Böttinger L, Klay D, Weeber F, Huelsz‐Prince G, Iakobachvili N, Amatngalim GD, et al. (2019). Long‐term expanding human airway organoids for disease modeling. EMBO J. 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittke N, Herrick DB, Lin B, Peterson J, Coleman JH, Packard AI, Jang W, and Schwob JE (2015). Transcription factor p63 controls the reserve status but not the stemness of horizontal basal cells in the olfactory epithelium. Proc. Natl. Acad. Sci. 112, E5068–E5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seery JP (2002). Stem cells of the oesophageal epithelium. J. Cell Sci. 115, 1783–1789. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat M-L, Wu L, Lindeman GJ, and Visvader JE (2006). Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88. [DOI] [PubMed] [Google Scholar]
- Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu MM, Piccolo S, Birchmeier W, Whitsett JA, Millar SE, et al. (2005). Wnt/β-catenin signaling acts upstream of Nmyc, BMP4, and FGF signaling to regulate proximal–distal patterning in the lung. Dev. Biol. 283, 226–239. [DOI] [PubMed] [Google Scholar]
- Spence JR, Lange AW, Lin S-CJ, Kaestner KH, Lowy AM, Kim I, Whitsett JA, and Wells JM (2009). Sox17 Regulates Organ Lineage Segregation of Ventral Foregut Progenitor Cells. Dev. Cell 17, 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Gao X, Hong CC, Hotten D, and Hogan BLM (2016). BMP signaling and cellular dynamics during regeneration of airway epithelium from basal progenitors. Development 143, 764–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T. (2004). Defining the Epithelial Stem Cell Niche in Skin. Science (80-. ). 303, 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, et al. (2015). Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M, Yingling JM, Dunn NR, Bellusci S, and Hogan BLM (1999). Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development 126, 4005–4015. [DOI] [PubMed] [Google Scholar]
- Yang Y, Riccio P, Schotsaert M, Mori M, Lu J, Lee D-K, García-Sastre A, Xu J, and Cardoso WV (2018). Spatial-Temporal Lineage Restrictions of Embryonic p63+ Progenitors Establish Distinct Stem Cell Pools in Adult Airways. Dev. Cell 44, 752761.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZ, Guan SP, Liew A-A, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al. (2015). p63+Krt5+ distal airway stem cells are essential for lung regeneration. Nature 517, 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1, related to Figures 1, 3 and 4
Number of cells in fetal and in vitro sc-RNAseq samples (tab 1)
Markers of fetal lung epithelial clusters for fetal and day 21 in vitro sample analyses (tab 2)
Differential expression values comparing day 3 basal-like cluster to in vitro to fetal basal cells and day 21 basal-like cluster to fetal basal cells (tab 3)
Regulons generated from day 3 and day 21 in vitro cells (tab 4)
Supplemental Video 1, related to Figure 2
Functional Multiciliated cells in induced airway organoids treated for 3 days with dual SMAD activation followed by 11 days of dual SMAD inhibition.
Supplemental Video 2, related to Figure 5
Functional Multiciliated cells in induced airway organoids grown from a single GFP+/EGFR+/F3+ basal-like cell.
Data Availability Statement
Data preprocessing and cluster identification
All single-cell RNA-sequencing was performed with an Illumina Hiseq 4000 by the University of Michigan DNA Sequencing core. Reads alignment to human reference genome and generation of gene expression matrix were done using 10x Genomics Cell Ranger v2.1.1–2.2.1 software with provided hg19 reference genome and transcriptome annotation. In each sample, to ensure high data quality for further analysis, cells with more than 20,000 or less than 1,500 genes or mitochondrial transcript fraction over 10% and genes expressed in less than 3 cells were excluded for further analysis. All fetal tissue samples were then combined. Gene expression levels were log-normalized by the total number of unique molecular identifier (UMI) per cell. Cell cycle phase was scored by expression levels of cell cycle related genes. Cellular variance of total number of UMI, mitochondrial transcript fraction and cell cycle phase was regressed out using linear regression. Non-cell cycle related genes showing highly variable expression levels (highly variable genes) were identified in each fetal tissue sample separately. For the combined fetal data, principle component analysis (PCA) was based on z-transformed expression levels of genes that were identified as highly variable genes in at least two samples. Graph-based clustering approach and tSNE dimension reduction were performed based on Euclidean distance between the top 20 PCs. Clusters were classified into epithelial, mesenchymal, endothelial, neuronal, immune cell lineages and red blood cell cluster based on expression patterns of canonical marker genes. To better resolve cellular heterogeneity of epithelial cells, cells with low expression levels of EPCAM in the epithelial lineage were further removed, sub-clustering on the remaining epithelial cells was performed. Identification of positive markers for each cluster was performed with Wilcoxon Rank Sum test. Hierarchical clustering of clusters based on average expression levels of cluster markers was performed. Combined with canonical cell type marker expression patterns, clusters with shared cell type identity were merged. Analyses mentioned in this section were mainly performed with Seurat(Butler et al., 2018).
Characterization of cellular transcriptome heterogeneity in in vitro samples
Each in vitro sample was analyzed separately. Data preprocessing of in vitro data followed the same procedure as fetal tissue data except that minimum number of detected genes in cells of day 0 is 1,000 and cellular variance of cell cycle phase was not regressed out. To quantify the transcriptome similarity between in vitro cells and fetal epithelial sub-clusters, Pearson’s correlation coefficients of log-normalized gene expression levels between in vitro cells and fetal epithelial cell sub-clusters were calculated using the genes for fetal tissue kNN network construction. Clustering and positive marker gene expression identification followed the same procedure as for fetal data. Gene ontology enrichment in cluster markers was performed using DAVID (v6.8)(Huang et al., 2009a, 2009b). Significant enrichment was defined as Benjamini-corrected P < 0.05.


