Abstract
In both plants and animals, nucleotide‐binding leucine‐rich repeat (NLR) immune receptors perceive pathogen‐derived molecules to trigger immunity. Global NLR homeostasis must be tightly controlled to ensure sufficient and timely immune output while avoiding aberrant activation, the mechanisms of which are largely unclear. In a previous reverse genetic screen, we identified two novel E3 ligases, SNIPER1 and its homolog SNIPER2, both of which broadly control the levels of NLR immune receptors in Arabidopsis. Protein levels of sensor NLRs (sNLRs) are inversely correlated with SNIPER1 amount and the interactions between SNIPER1 and sNLRs seem to be through the common nucleotide‐binding (NB) domains of sNLRs. In support, SNIPER1 can ubiquitinate the NB domains of multiple sNLRs in vitro. Our study thus reveals a novel process of global turnover of sNLRs by two master E3 ligases for immediate attenuation of immune output to effectively avoid autoimmunity. Such unique mechanism can be utilized in the future for engineering broad‐spectrum resistance in crops to fend off pathogens that damage our food supply.
Keywords: autoimmunity, E3 ligase, global turnover, NLR immune receptors, plant immunity
Subject Categories: Immunology; Plant Biology; Post-translational Modifications, Proteolysis & Proteomics
Two Arabidopsis E3 ubiquitin ligases act redundantly to suppress immune response and prevent autoimmunity by targeting multiple pathogen effector‐sensing NOD‐like receptors for degradation.

Introduction
The plant innate immune system relies on immune receptors to detect and respond to pathogens. Plasma membrane‐localized pattern‐recognition receptors (PRRs) with extracellular domains perceive conserved microbial patterns to trigger pathogen‐associated molecular patterns (PAMPs)‐triggered immunity (PTI) (Couto & Zipfel, 2016). However, virulent pathogens can deliver effectors into host cells to inhibit PTI. In turn, intracellular sensor nucleotide‐binding leucine‐rich repeat immune receptors (sNLRs) have been evolved to recognize the actions of corresponding effectors directly or indirectly, resulting in activation of effector‐triggered immunity (ETI) to ward off microbial infections (Jones & Dangl, 2006).
Typical sNLRs are further classified into Toll/interleukin 1 receptor (TIR) type (TNLs) or coiled‐coil (CC) type (CNLs) based on their different N‐termini (Jones et al, 2016). Plant sNLRs are highly polymorphic and recognize specific effectors. For example, distinct Arabidopsis RPP1 (Recognition of Peronospora Parasitica 1) alleles can specifically recognize differential ATR1 (Arabidopsis thaliana Recognized 1) effectors from oomycete pathogen Hyaloperonospora arabidopsidis (H.a.) (Steinbrenner et al, 2012). In contrast, helper NLRs (hNLRs), a small subclass of CNLs with an RPW8 (Resistance to Powdery Mildew 8)‐like CC domain, function downstream of diverse sNLRs and are evolutionarily more conserved (Peart et al, 2005; Jubic et al, 2019). There are three hNLR gene families, encoding ADR1s (Activated Disease Resistance 1), NRG1s (N Required Gene 1), and NRCs (NLR protein required for HR‐associated cell death) (Peart et al, 2005; Bonardi et al, 2011; Collier et al, 2011; Gao et al, 2018; Jubic et al, 2019). In addition, hNLRs seem to operate in distinct pathways downstream of TNLs (Castel et al, 2019; Jubic et al, 2019; Wu et al, 2019). Furthermore, TNL signaling generally relies on EDS1 (Enhanced Disease Susceptibility 1), whereas NDR1 (Non‐Race Specific Disease Resistance 1) is required for many CNL‐mediated defense (Aarts et al, 1998). However, the exact molecular functions of hNLRs, EDS1, and NDR1 in NLR signaling remain elusive.
Proper control of NLR homeostasis is critical, as over‐accumulation or aberrant activation of NLR can lead to autoimmunity and growth defects, whereas insufficient NLR accumulation may result in susceptibility to pathogens. For example, snc1, a gain‐of‐function allele of TNL SNC1 (Suppressor of npr1‐1, Constitutive 1), constitutively activates defense responses and exhibits dwarfism due to increased SNC1 accumulation (Zhang et al, 2003; Cheng et al, 2011; Li et al, 2015). Although disturbance of NLR biogenesis or degradation can alter NLR homeostasis (Kapos et al, 2019), the exact mechanisms of how the homeostasis of NLRs is broadly controlled under different conditions are largely unclear.
The ubiquitin–proteasome system (UPS) plays a pivotal role in maintaining protein homeostasis. In the UPS pathway, upon ubiquitin (Ub) activation by E1, Ub ligase (E3) recruits the ubiquitin recipient and facilitates the transfer of ubiquitin from Ub conjugating enzyme (E2) to the targeted substrate (Vierstra, 2009). E3 ligases mostly determine the substrate specificity during ubiquitination. Upon poly‐ubiquitination, proteins are most often degraded via the 26S proteasome (Vierstra, 2009).
Compared with animals, higher plants harbor highly expanded E3 gene families, suggesting key roles they play in diverse biological processes. There are about 1,500 E3 ligases encoded in Arabidopsis genome (Mazzucotelli et al, 2006). The UPS plays important roles in regulating levels of both PRR and NLR immune receptors. For example, Arabidopsis PUB12/13 (Plant U‐Box 12/13) control the turnover of flagellin receptor FLS2 (FLAGELLIN‐SENSING 2) (Lu et al, 2011) and chitin receptor LYK5 (LYSIN MOTIF RECEPTOR KINASE 5) (Liao et al, 2017). The F‐box protein CPR1 is part of the SCFCPR1 complex E3 that targets two sNLRs, SNC1 and RPS2, for ubiquitination and degradation (Cheng et al, 2011; Gou et al, 2012). In parallel, two functionally redundant RING‐type E3 ligases, MUSE1 and MUSE2, are required for the turnover of SNC1's partners, SIKIC1/2/3 (Dong et al, 2018). These E3s show very narrow specificity in regard to substrate recognition.
Coincidentally, higher plant genomes often encode hundreds of sNLRs, which are also massively expanded as compared with that of mammals. During an immune response, broad up‐regulation of NLRs contributes to defense amplification (Ribot et al, 2008; Mohr et al, 2010; Yu et al, 2013; Brechenmacher et al, 2015; Chen et al, 2015). However, how plants universally regulate these NLR homeostasis and whether the UPS plays a role are unknown. Here, we describe the identification of two novel E3 ligases, SNIPER1 and its homolog SNIPER2, which globally control the protein levels of sNLRs. Upon SNIPER1 overexpression, levels of all tested sNLRs are reduced, resulting in enhanced disease susceptibility to pathogens. In contrast, more sNLRs accumulation and enhanced disease resistance were observed when the function of the SNIPERs is attenuated. Such broad regulation seems to be through direct recognition and ubiquitination of the conserved NB domains of sNLRs by the E3 ligase.
Results
SNIPER1 is a complete suppressor of snc1
To search for novel E3s that regulate plant NLR‐mediated immunity, an E3 overexpression snc1‐influencing plant E3 ligase reverse (SNIPER) genetic screen was performed in the autoimmune NLR mutant snc1 background (Tong et al, 2017; Huang et al, 2018). SNIPER1 (AT1G14200) was initially selected as an immunity‐related candidate as DNA of SNIPER1 was 4.2‐fold enriched in the master immune transcription factor SARD1 chromatin immunoprecipitation (ChIP) Sequence (ChIP‐Seq) dataset (Sun et al, 2015). Quantitative real‐time PCR (RT‐PCR) indeed revealed its transcriptional induction during pathogen infection (Appendix Fig S1), suggestive of a role in immune regulation.
Among all SNIPER E3s that could suppress snc1 upon overexpression, SNIPER1 was identified as the only complete snc1 suppressor without any morphological defects (Fig 1A and E). Overexpression of SNIPER1 (SNIPER1 OE) (Fig 1B) fully suppresses snc1‐mediated dwarfism (Fig 1A). In T2 segregants, this dwarfism‐suppression phenotype co‐segregated perfectly with the presence of the transgene, indicating that the morphological suppression was due to the transgene overexpression. Furthermore, enhanced disease resistance to the oomycete pathogen H.a. Noco2 (Fig 1C) and bacterial pathogen Pseudomonas syringae pv. maculicola (P.s.m.) ES4326 (Fig 1D) in snc1 were consistently suppressed upon SNIPER1 overexpression. Interestingly, SNIPER1 OE lines in both snc1 and wild‐type (WT) backgrounds supported higher pathogen growth even when compared to WT (Fig 1B–D and F–H), suggesting that SNIPER1 plays a general negative regulatory role on basal defense. In addition, no observable growth or developmental defects were observed in SNIPER1 OE lines (Fig 1A and E), suggesting that its effects are strictly immunity‐related.
Figure 1. SNIPER1 negatively regulates snc1‐mediated autoimmunity and basal defense.
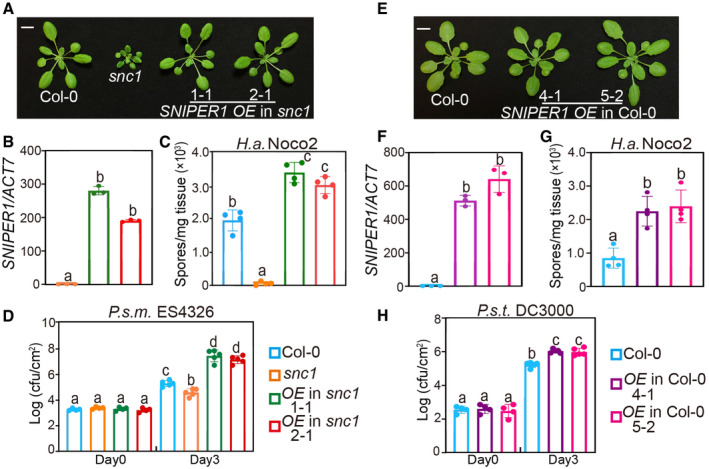
- Morphology of 4‐week‐old soil‐grown plants of Col‐0, snc1, and two independent transgenic lines of SNIPER1 OE into snc1 background. OE stands for overexpression of SNIPER1. Scale bar = 1 cm.
- SNIPER1 gene expression in the indicated plants as determined by RT‐PCR. Error bars represent mean ± SD (n = 3). Two independent experiments were carried out with similar results.
- Quantification of H.a. Noco2 sporulation in the indicated genotypes 7 days post‐inoculation (dpi) with 105 spores per ml water. Error bars represent mean ± SD (n = 4). Three independent experiments were carried out with similar results.
- Growth of P.s.m. ES4326 on 4‐week‐old leaves of the indicated genotypes at 0 and 3 dpi with bacterial inoculum of OD600 = 0.001. Error bars represent mean ± SD (n = 4 for day 0; n = 5 for day 3). Three independent experiments were carried out with similar results.
- Morphology of 4‐week‐old soil‐grown plants of two independent transgenic lines of SNIPER1 OE in Col‐0 background. Scale bar = 1 cm. No difference in growth and development was observed.
- SNIPER1 gene expression in the indicated plants as determined by RT‐PCR. Error bars represent mean ± SD (n = 3). Two independent experiments were carried out with similar results.
- Quantification of H.a. Noco2 sporulation in the indicated genotypes at 7 dpi with 105 spores per ml water. Error bars represent mean ± SD (n = 4). Three independent experiments were carried out with similar results.
- Growth of P.s.t. DC3000 on 4‐week‐old leaves of the indicated genotypes at 0 and 3 dpi with bacterial inoculum of OD600 = 0.001. Error bars represent mean ± SD (n = 4 for day 0; n = 5 for day 3). Three independent experiments were carried out with similar results.
Overexpression of SNIPER1 suppresses NLR‐type autoimmune mutants, but not an RLP autoimmune mutant
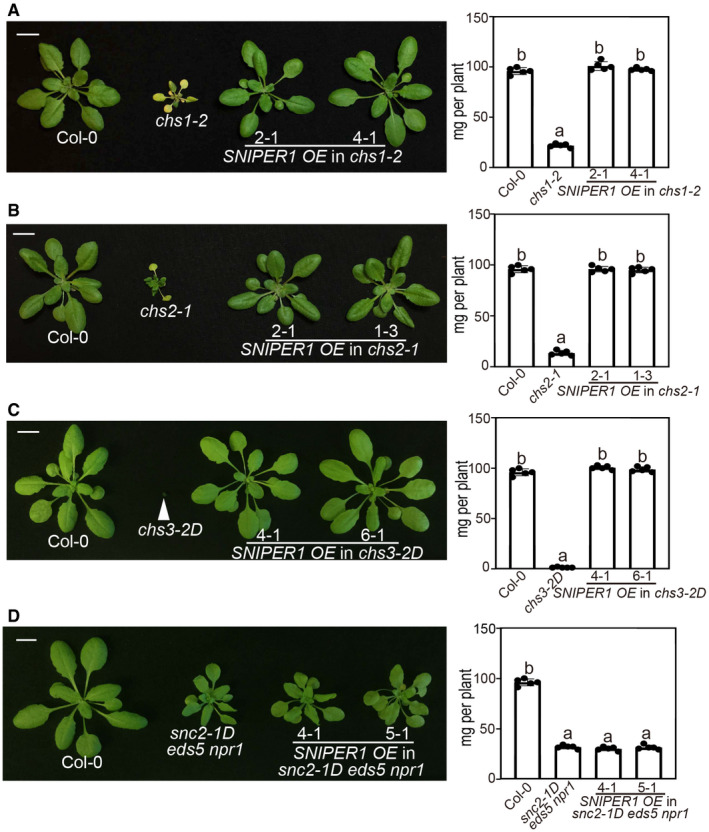
In order to test its specificity, SNIPER1 was overexpressed in three other gain‐of‐function TNL mutants exhibiting the chilling sensitive (CHS) autoimmune phenotypes (Huang et al, 2010; Bi et al, 2011; Wang et al, 2013). Interestingly, the autoimmunity of chs1‐2, chs2‐1 and chs3‐2D was also fully suppressed (Fig 2A–C), indicating that SNIPER1 has a general role in TNL‐mediated immunity. Similar with the SNIPER1 overexpression experiment in snc1 background, these suppression phenotypes were confirmed to be due to the transgene in T2 through co‐segregation analysis. However, overexpression of SNIPER1 (Appendix Fig S2) did not alter the autoimmunity of snc2‐1D eds5 npr1 (Fig 2D), which contains a gain‐of‐function mutation in the receptor‐like protein (RLP) SNC2 and SA‐dependent pathway is blocked by mutations in EDS5 and NPR1 (Zhang et al, 2010; Rekhter et al, 2019), suggesting that SNIPER1 does not contribute to plasma membrane PRR‐mediated immunity.
Figure 2. Overexpression of SNIPER1 suppresses NLR‐type autoimmune mutants, but not an RLP autoimmune mutant.
-
A, BMorphology of 4‐week‐old soil‐grown plants of Col‐0, chs1‐2 (A), chs2‐1 (B), and two independent transgenic lines of SNIPER1 OE into chs1‐2 (A) and chs2‐1 (B) backgrounds. Plants were grown at 16°C under long day conditions. Scale bar = 1 cm.
-
C, DMorphology of 4‐week‐old soil‐grown plants of Col‐0, chs3‐2D (C), snc2‐1D eds5 npr1 (D), and two independent transgenic lines of SNIPER1 OE into chs3‐2D (C) and snc2‐1D eds5 npr1 (D) backgrounds. Scale bar = 1 cm.
We further examined the effect of SNIPER1 on sensor CNLs. To our surprise, the mekk1‐5 ndr1‐mediated autoimmunity, in which the CNL SUMM2 is activated (Zhang et al, 2012), was also fully restored to WT level in morphology upon SNIPER1 overexpression (Fig 3A and B). In T2 populations from independent T1s, all WT‐like individuals were resistant to BASTA which selects the transgene while all dwarfed progenies died, suggesting a causal relationship between the presence of the transgene and the suppression phenotype. Similarly, mekk1‐5 ndr1 plants overexpressing SNIPER1 were more susceptible to H.a. Noco2 (Fig 3C) and P.s.m. ES4326 (Fig 3D) when compared to the parent mekk1‐5 ndr1, indicating a broad effect of SNIPER1 on both TNL‐type and CNL‐type sNLRs.
Figure 3. SNIPER1 is a functional E3 ligase and overexpression of SNIPER1 fully suppresses mekk1‐5 ndr1‐mediated autoimmunity.
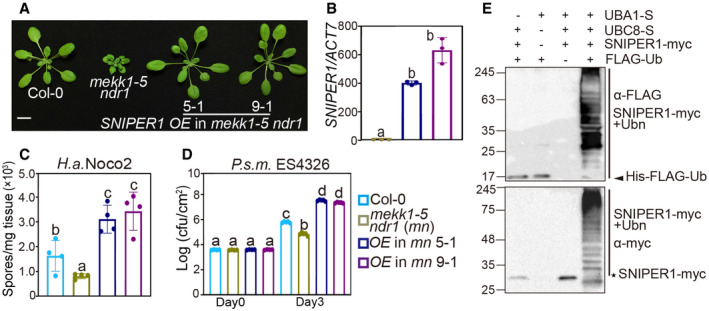
- Morphology of 4‐week‐old soil‐grown plants of Col‐0, mekk1‐5 ndr1, and two independent transgenic lines of SNIPER1 OE into mekk1‐5 ndr1 background. Scale bar = 1 cm. The reason for mekk1 ndr1 double mutant usage instead of the mekk1‐5 single mutant is to overcome the sterility.
- SNIPER1 gene expression in the indicated plants as determined by RT‐PCR. Error bars represent mean ± SD (n = 3). Two independent experiments were carried out with similar results.
- Quantification of H.a. Noco2 sporulation in the indicated genotypes at 7 dpi with 105 spores per ml water. Error bars represent mean ± SD (n = 4). Three independent experiments were carried out with similar results.
- Growth of P.s.m. ES4326 on 4‐week‐old leaves of the indicated genotypes at 0 and 3 dpi with bacterial inoculum of OD600 = 0.001. mn stands for mekk1‐5 ndr1 double mutant. Error bars represent mean ± SD (n = 4). Three independent experiments were carried out with similar results.
- In vitro ubiquitination assay using Escherichia coli (E. coli)‐expressed SNIPER1‐myc (E3), AtUBC8 (E2), AtUBA1 (E1), and/or HIS‐FLAG‐Ubiquitin. The molecular mass markers are indicated on the left (kDa). Asterisk indicates SNIPER1‐myc. Arrow points to His‐FLAG‐Ub and straight lines on the right indicate self‐ubiquitinated SNIPER1.
SNIPER1 encodes an E3 ligase with a RING domain
SNIPER1 encodes a 20.03 kDa small protein with a predicted RING (Really Interesting New Gene) domain (Fig EV1A), which is a signature of a major class of simple E3 ligases that is involved in the interaction with an E2 ubiquitin‐conjugating enzyme. Phylogenetic analysis revealed that SNIPER1‐like proteins are commonly found in dicots, but not monocots (Fig EV1B). To test whether SNIPER1 exhibits E3 activity, an in vitro ubiquitination assay was performed (Han et al, 2017). Higher molecular‐weight polypeptide bands were detected only when SNIPER1 was co‐expressed with Arabidopsis E1 AtUBA1, E2 AtUBC8, and AtUBQ10, suggesting that SNIPER1 has in vitro self‐ubiquitination activity and it is indeed a functional E3 ligase (Fig 3E). The E3 activity of SNIPER1 and its overexpression phenotypes led us to hypothesize that it is likely involved in the degradation of a positive regulator of immunity acting downstream of sNLRs.
Figure EV1. Phylogenetic tree of SNIPER1 in dicots.
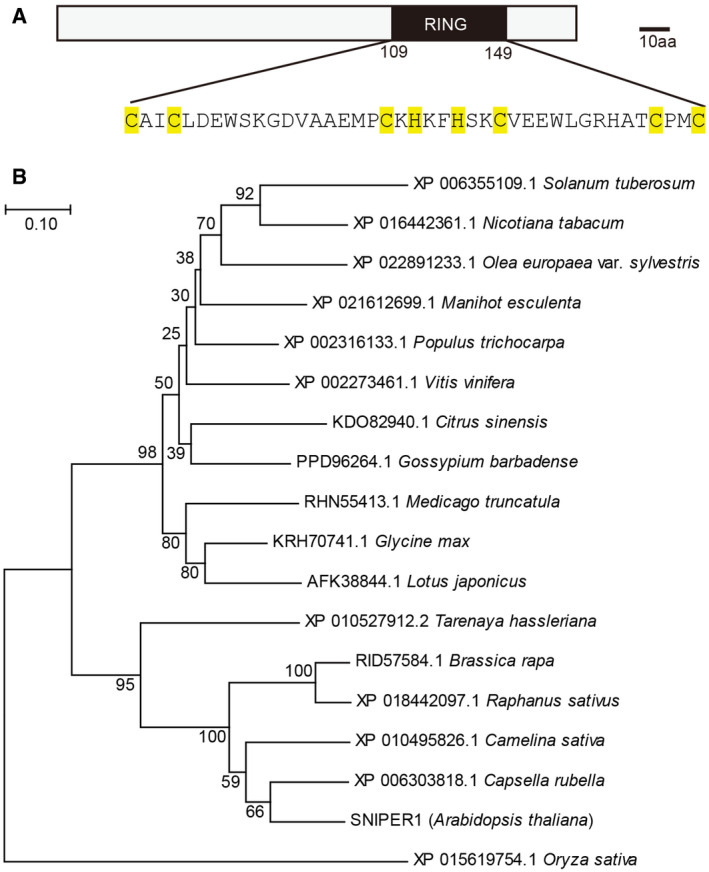
- Schematic structure of SNIPER1. The conserved residues of the RING domain are highlighted in yellow.
- Phylogenetic tree of putative SNIPER1 orthologs in dicots. Putative SNIPER1 othologs were obtained from NCBI using SNIPER1 protein sequence as input. Closest orthology in Oryza sativa was used as an outgroup. Muscle was used for sequences alignment and a neighbor‐joining tree was generated with JTT model and 5,000 bootstrap value in MEGA 7.0.
SNIPER1 does not influence EDS1, PAD4, or ADR1s
Similar to SNIPER1 OE lines, mutations in TNL downstream components EDS1, PAD4, or the hNLRs ADR1s can fully suppress snc1‐mediated autoimmunity and exhibit enhanced disease susceptibility to both avirulent and virulent pathogens (Falk et al, 1999; Feys et al, 2001; Li et al, 2001; Bonardi et al, 2011; Cheng et al, 2011; Dong et al, 2016). Therefore, we hypothesized that SNIPER1 may regulate the turnover of one of these components. However, no obvious change in EDS1 or PAD4 protein levels was observed upon SNIPER1 overexpression (Fig EV2A and B).
Figure EV2. SNIPER1 functions upstream of EDS1, PAD4, and ADR1 family helper NLRs.
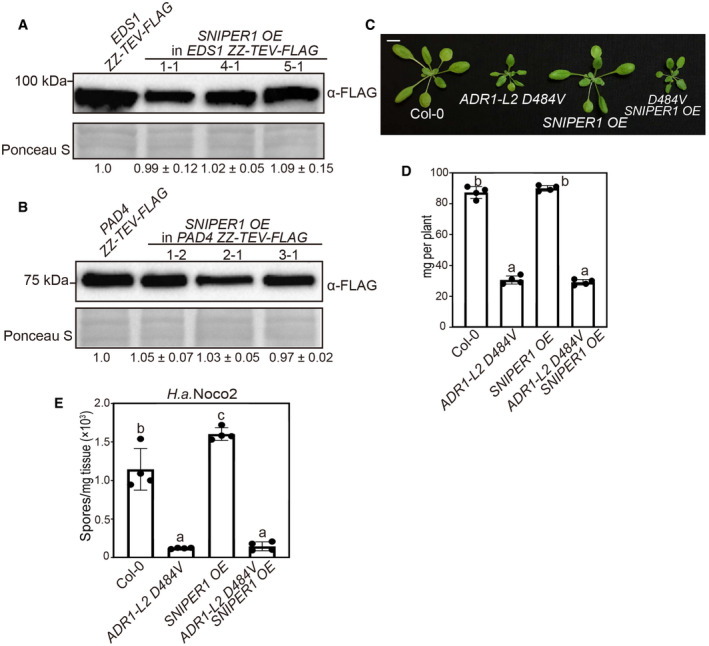
-
A, BEDS1 (A) and PAD4 (B) protein levels in 4‐week‐old soil‐grown plants. Equal loading is shown by Ponceau S staining of a non‐specific band. The numbers below represent the normalized ratio between the intensity of the protein band and the Ponceau S band ± SD (n = 3). Molecular mass markers in kilo Daltons are indicated on the left. Three independent experiments were carried out with similar results.
-
CMorphology of 4‐week‐old soil‐grown plants of Col‐0, ADR1‐L2 D484V, SNIPER1 OE, and ADR1‐L2 D484V SNIPER1 OE. Scale bar = 1 cm.
-
DFresh weights of plants in (C).
-
EQuantification of H.a. Noco2 sporulation in the indicated plants at 7 dpi with 105 spores per ml water.
We further examined the relationship between SNIPER1 and ADR1s by crossing the SNIPER1 OE line with autoimmune transgenic plants expressing ADR1‐L2 D484V, an auto‐active missense variant of ADR1‐L2 that constitutively activates immunity (Roberts et al, 2013). However, overexpression of SNIPER1 has no effect on the autoimmunity mediated by ADR1‐L2 D484V (Fig EV2C–E), suggesting that SNIPER1 functions upstream of the ADR1 family of hNLRs. Therefore, the effects of SNIPER1 seem to be at the sNLR level.
SNIPER1 affects the protein levels of all tested sNLRs
Because of its specific effects on immunity mediated by sNLRs and no effect on hNLRs and downstream components, we examined whether overexpression of SNIPER1 affects the protein or transcript levels of sNLRs. Interestingly, decreased SNC1 protein level, but not transcript level, was observed when SNIPER1 was overexpressed in the snc1 pad4 mutant (Fig 4A and Appendix Fig S3A), where the feedback transcriptional up‐regulation of SNC1 expression due to heightened SA level is blocked by pad4 (Cheng et al, 2011). Overexpression of SNIPER1 in WT and SUMM2‐3HA backgrounds also resulted in reduced accumulation of SNC1 and SUMM2 proteins, respectively, despite that the transcript levels of these sNLRs were not affected (Fig 4B and C, and Appendix Fig S3B and C), suggesting that the regulation by SNIPER1 is post‐translational.
Figure 4. SNIPER1 reduces sNLRs protein levels upon overexpression.
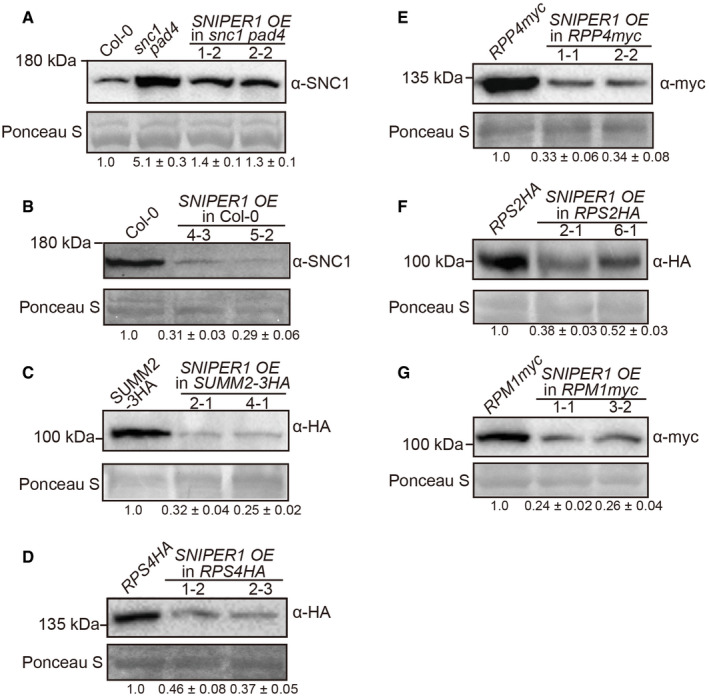
-
A–GImmunoblot analysis of SNC1 (A, B), SUMM2‐3HA (C), RPS4HA (D), RPP4myc (E), RPS2HA (F), and RPM1myc (G) protein levels in the indicated genotypes. For all panels, equal loading is shown by Ponceau S staining of a non‐specific band. The numbers below represent the normalized ratio between the intensity of the protein band and the Ponceau S band ± SD (n = 3). Molecular mass marker in kilo Daltons is indicated on the left. Three independent experiments were carried out with similar results.
Source data are available online for this figure.
To test whether SNIPER1 affects the levels of other sNLRs, lines with epitope tagged TNLs RPS4 and RPP4 were crossed with the SNIPER1 OE line and the protein levels of these sNLRs were investigated (Wirthmueller et al, 2007; Bao et al, 2014). To our amazement, the protein levels of both RPS4 and RPP4 decreased in SNIPER1 OE lines (Fig 4D and E). In addition, CNLs RPS2 and RPM1 protein levels reduced upon overexpression of SNIPER1 (Fig 4F and G) (Grant et al, 1995; Axtell & Staskawicz, 2003). However, the transcript levels of these sNLR genes were not affected in SNIPER1 OE lines (Appendix Fig S3D–G), further supporting that the reduced sNLRs protein levels are regulated by SNIPER1 at post‐translational level.
As SNIPER1 overexpression leads to reduced sNLRs levels, we speculated that more avirulent pathogens might grow in SNIPER1 OE lines. Indeed, when SNIPER1 OE lines were challenged with P.s.t. DC3000 carrying avrRps4, avrRpt2, or avrRpm1, and oomycete H.a. Emwa1, which carries the corresponding cognate effector genes of RPS4, RPS2, RPM1, or RPP4, respectively (Grant et al, 1995; Gassmann et al, 1999; van der Biezen et al, 2002; Axtell & Staskawicz, 2003), varied levels of increased pathogen growth were observed (Fig 5A–D and Appendix Fig S4). These data suggest that SNIPER1 plays a global role in regulating the protein levels of diverse sNLRs, and such reduction leads to general susceptibility to pathogens.
Figure 5. SNIPER1 OE lines show enhanced disease susceptibility to avirulent pathogens.
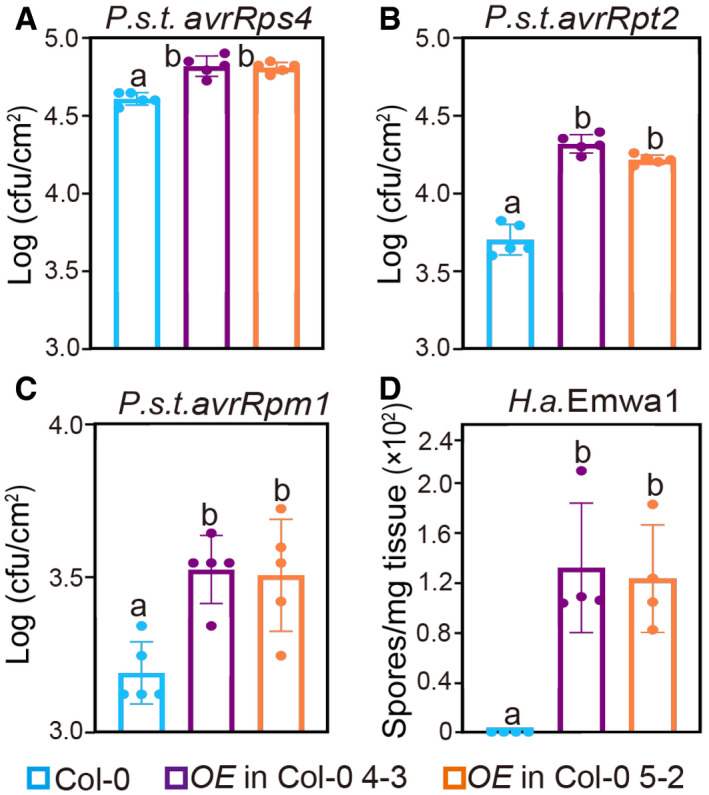
-
A–CGrowth of P.s.t. avrRps4 (A), avrRpt2 (B), and avrRpm1 (C) in 4‐week‐old leaves of the indicated genotypes at 3 dpi with bacterial inoculum of OD600 = 0.0001. Error bars represent mean ± SD (n = 5). Three independent experiments were carried out with similar results. Appendix Fig S4 shows the growth of P.s.t. avrRps4, avrRpt2, and avrRpm1 at 0 dpi.
-
DQuantification of H.a. Emwa1 sporulation in the indicated genotypes at 7 dpi with 105 spores per ml water. Error bars represent mean ± SD (n = 4). Three independent experiments were carried out with similar results.
SNIPER2 has overlapping function with SNIPER1 in ETI, but it is also involved in development
Since overexpression of SNIPER1 E3 ligase leads to decreased accumulation of sNLRs, it is possible that SNIPER1 might directly ubiquitinate the sNLRs. If this is the case, loss of SNIPER1 function may lead to increased accumulation of sNLRs and potentially autoimmunity‐related dwarfism and enhanced resistance against pathogens. We thus tested this prediction by examining the knockout mutant phenotypes of sniper1. As shown in Fig EV3A and B, the exonic T‐DNA insertional mutant of sniper1‐1 (SALK_054376) appeared WT‐like in morphology, with WT level of resistance against P.s.t DC3000 (Fig EV3C) and a similar sNLRs levels as their parents (Fig EV3D). In addition, loss of SNIPER1 does not enhance the morphology of snc1 (Fig EV3A and B).
Figure EV3. Characterization of sniper1‐1 (SALKseq_054376) T‐DNA insertion mutant.
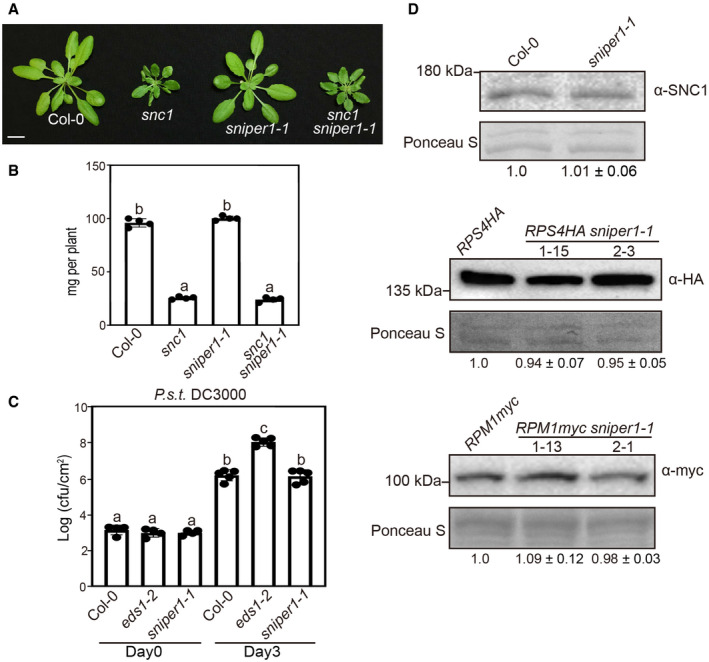
- Morphology of 4‐week‐old Col‐0, snc1, sniper1‐1, and snc1 sniper1‐1. Scale bar = 1 cm.
- Fresh weights of plants in (A). Error bars represent mean ± SD (n = 4).
- Bacterial growth of P.s.t. DC3000 on 4‐week‐old leaves of the indicated genotypes at 0 and 3 dpi with bacterial inoculum of OD600 = 0.001. Error bars represent mean ± SD (n = 4 for day 0; n = 5 for day 3).
- SNC1, RPS4‐HA, and RPM1myc proteins levels in sniper1‐1 plants. Equal loading is shown by Ponceau S staining of a non‐specific band. The numbers below represent the normalized ratio between the intensity of the protein band and the Ponceau S band ± SD (n = 3). Molecular mass markers in kilo Daltons are indicated on the left. Three independent experiments were carried out with similar results.
As there are about 1,500 E3s encoded in the Arabidopsis genome, high level of genetic redundancy is frequently associated with E3‐encoding genes. It is therefore not surprising to be unable to observe any defects in E3 single mutants. A BLAST search with SNIPER1 in Arabidopsis identified its closest paralog, AT1G26800, which shares only 35% amino acid identity and 53% similarity with SNIPER1 (Fig EV4A). Coincidently, this gene was also identified in our SNIPER screen as SNIPER2. It was originally selected as an immune‐related candidate based on its pathogen‐induced expression upon pathogen infection. Like SNIPER1, overexpression of SNIPER2 (AT1G26800) can largely suppress the dwarfism of snc1 (Fig EV4B and C), alluding to a potential overlapping function with SNIPER1. Upon challenge of H.a. Noco2 and P.s.m. ES4326, the enhanced resistance of snc1 was significantly compromised in these transgenic lines (Fig EV4D and E).
Figure EV4. Overexpression of SNIPER2 suppresses snc1‐mediated autoimmunity.
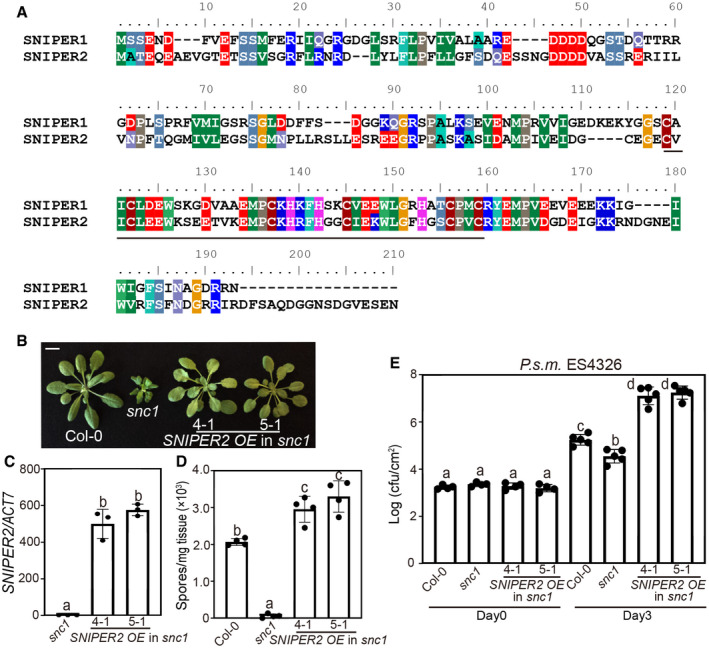
- Full‐length alignment of SNIPER1 and SNIPER2 by ClustalW. The RING domain is indicated by a black line.
- Morphology of 4‐week‐old soil‐grown plants of Col‐0, snc1, and two independent transgenic lines of SNIPER2 OE into snc1 background. Scale bar = 1 cm.
- SNIPER2 gene expression in the indicated plants as determined by RT‐PCR. Error bars represent mean ± SD (n = 3). Two independent experiments were carried out with similar results.
- Quantification of H.a. Noco2 sporulation in the indicated genotypes at 7 dpi with 105 spores per ml water. Error bars represent mean ± SD (n = 4). Three independent experiments were carried out with similar results.
- Growth of P.s.m. ES4326 on 4‐week‐old leaves of the indicated genotypes at 0 and 3 dpi with bacterial inoculum of OD600 = 0.001. Error bars represent mean ± SD (n = 4 for day 0; n = 5 for day 3). Three independent experiments were carried out with similar results.
In addition, the autoimmunity mediated by TNL CHS1, CHS3 and CNL SUMM2 was suppressed by overexpression of SNIPER2 (Appendix Fig S5A–C and E), indicating its similar broad effects on both TNLs and CNLs. Likewise, no effect was observed in RLP SNC2‐mediated autoimmunity although similar amounts of SNIPER2 were overexpressed (Appendix Fig S5D and F). In vitro ubiquitination assay (Zhao et al, 2013) showed the presence of increased molecular‐weight bands above SNIPER2 in the presence of E. coli‐expressed SNIPER2 (E3), AtUBC8 (E2), AtUBA2 (E1) and ubiquitin (Appendix Fig S5G), indicating that SNIPER2 also exhibits E3 activity. Taken together, SNIPER2 seems to play similar roles in immune regulation as SNIPER1.
Besides its roles in plant immunity, SNIPER2 overexpression lines exhibit a distinct morphological phenotype with rounder leaves, longer petioles (Appendix Figs S5A–C, and S6A and B), and late flowering (Appendix Fig S6C and D), indicating that unlike SNIPER1 which does not contribute to growth and development, SNIPER2 regulates both development and immune responses.
SNIPER1 levels are inversely correlated with sNLR levels
To further test the genetic relationship between SNIPER1 and SNIPER2, a cross between sniper1‐1 and sniper2‐1 (SALKseq_63073), an exonic allele of sniper2, was made to generate sniper1‐1 sniper2‐1 double mutant. However, after genotyping 200 F2 plants, none was identified as a double mutant, suggesting a likely lethality phenotype when both genes are knocked out.
To overcome the lethality and redundancy problems, we generated a dominant‐negative (DN) form of SNIPER1, which is expected to affect both the endogenous SNIPER1 as well as its redundant players (Seo et al, 2004). Such strategy is routinely used to study redundant E3s. Interestingly, when a construct expressing DN SNIPER (SNIPER1 H129Y), which contains a Tyr substitution of a conserved His in the RING domain, was transformed into WT, 22 out of 50 independent transformants exhibited dwarfed size and curly leaves (Fig 6A). Enhanced resistance to H.a. Noco2 and increased expression of PR (Pathogenesis Related) defense marker genes were also observed in SNIPER1 H129Y plants (Fig 6B and D), suggesting that overexpression of the DN form of SNIPER1 causes autoimmunity. Double mutant sniper1 sniper2 is lethal; therefore, it is not surprising that the DN‐SNIPER1 phenotype is in between sniper1 knockout and the sniper1 sniper2 double mutant under certain DN‐SNIPER1 expression level. Analysis of RPS4HA, SNC1, RPS2HA, and RPM1myc protein levels in SNIPER1 H129Y background further revealed increased accumulations of these sNLR proteins (Fig 6E–H). Thus, attenuation of functions of SNIPER1 and its redundant player(s) results in global increase of sNLRs levels, leading to constitutive activation of immunity.
Figure 6. Loss‐of‐function of both SNIPER1 and SNIPER2 leads to an EDS1‐dependent autoimmunity and lethality.
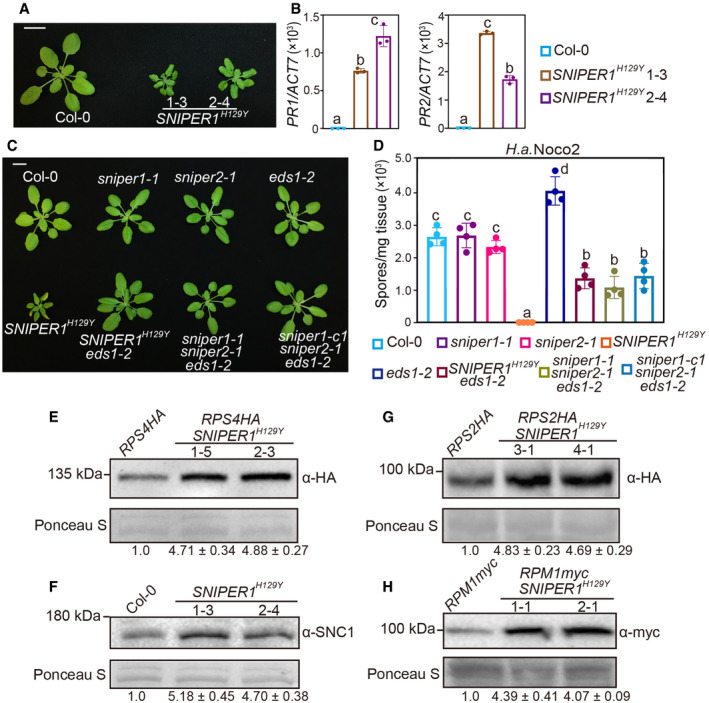
-
AMorphology of 4‐week‐old soil‐grown plants of Col‐0 and two independent transgenic lines of 35S‐SNIPER1 H129Y. Scale bar = 1 cm.
-
BPR1 and PR2 genes expression in the indicated plants as determined by RT‐PCR. Error bars represent mean ± SD (n = 3). Two independent experiments were carried out with similar results.
-
CMorphology of 4‐week‐old soil‐grown plants of Col‐0, sniper1‐1, sniper2‐1, eds1‐2, SNIPER1 H129Y, SNIPER1 H129Y eds1‐2, sniper1‐1 sniper2‐1 eds1‐2, and sniper1‐c1 sniper2‐1 eds1‐2. sniper1‐1 is an exonic T‐DNA insertional mutant. sniper1‐c1 is a SNIPER1 knockout mutant containing 807 bp deletion that was generated by CRISPR‐Cas9 system. Scale bar = 1 cm.
-
DQuantification of H.a. Noco2 sporulation in the indicated plants at 7 dpi with 105 spores per ml water. Error bars represent mean ± SD (n = 4). Two independent experiments were carried out with similar results.
-
E–HImmunoblot of RPS4HA (E), SNC1 (F), RPS2HA (G), and RPM1myc (H) in SNIPER1 H129Y background. Equal loading is shown by Ponceau S staining of a non‐specific band. The numbers below represent the normalized ratio between the intensity of the protein band and the Ponceau S band ± SD (n = 3). Molecular mass marker in kilo Daltons is indicated on the left. Three independent experiments were carried out with similar results.
Loss of both SNIPER1 and SNIPER2 causes EDS1‐dependent lethality
Because SNIPER1 affects the turnover of sNLRs and EDS1 is an essential regulator in TNL‐mediated defense and plays roles in CNL‐mediated pathway (Aarts et al, 1998; Brodersen et al, 2006; Cui et al, 2017), loss‐of‐function mutant of EDS1 (Wagner et al, 2013) was crossed with a SNIPER1 H129Y transgenic line to identify the defense pathways activated by SNIPER1H129Y. Consistent with the effects of SNIPER1 at the sNLRs levels, the dwarfism and enhanced resistance of SNIPER1 H129Y were largely suppressed by eds1‐2 (Fig 6C and D). This further confirms the effects of SNIPER1 upstream of hNLRs and EDS1.
The suppression of SNIPER1 H129Y autoimmunity by eds1‐2 led us to test whether eds1‐2 can also suppress the lethality of sniper1‐1 sniper2‐1 double mutant. We took two approaches. First, a cross was made between sniper1‐1 eds1‐2 and sniper2‐1 eds1‐2 double mutants to generate the triple mutant. In addition, the CRISPR‐Cas9 system was used to delete SNIPER1 in sniper2‐1 eds1‐2 double mutant (Wang et al, 2015) to generate sniper1‐c1 sniper2‐1 eds1‐2 triple mutant (Appendix Fig S7A). Triple mutants were successfully generated in both ways (Appendix Fig S7B), suggesting that sniper1 sniper2 double mutant is indeed lethal and this lethality can be suppressed by eds1‐2. Therefore, SNIPER1 and SNIPER2 seem to be redundant in immune regulation. Furthermore, the sniper1 sniper2 eds1‐2 triple mutants resembled the SNIPER1 H129Y eds1‐2 plant in morphology and resistance to H.a. Noco2 (Fig 6C and D).
Taken together, the inverse correlation between SNIPER1/2 and sNLRs levels suggests that the lethality of sniper1 sniper2 double mutant could be caused by massively accumulated sNLRs due to the loss of these two E3s. As SNIPER2 is functionally redundant with SNIPER1 in regard to immunity, and SNIPER1 does not contribute to development, we chose to focus solely on SNIPER1 for the rest of the biochemical study.
SNIPER1 associates with sNLRs through the NB domain
As SNIPER1 encodes an E3 ligase whose expression inversely correlates with sNLRs protein levels, we further tested the hypothesis that SNIPER1 directly ubiquitinates sNLRs by examining whether SNIPER1 directly interacts with sNLRs. Co‐immunoprecipitation (co‐IP) assays were first performed in Nicotiana (N.) benthamiana using transiently expressed FLAG‐SNIPER1H129Y and different sNLRs with epitope tags. The DN form of the protein is used here since it can stabilize the interaction with the substrate rather than degrading it as in the case of using native E3, defeating the purpose of examination of the protein–protein interactions between E3 and its substrate. Interestingly, the DN form of SNIPER1 could pull down three different selected representative sNLRs including the CNL SUMM2, TNL RPP4, and TN protein CHS1 (Fig EV5A–C), indicating that SNIPER1 can associate with sNLRs in the same protein complex. To further examine whether such association is through direct protein‐protein interaction, split‐luciferase assays were carried out in N. benthamiana by fusing SNIPER1H129Y or sNLRs with the carboxyl‐terminal (CLuc) and amino‐terminal (NLuc) halves of the firefly luciferase, respectively. Apparent luminescence was observed when SNIPER1H129Y‐CLuc was co‐expressed with sNLRs‐NLuc (SUMM2‐NLuc, RPP4‐NLuc, CHS1‐NLuc, or SOC3‐NLuc) (Fig EV5D–G), suggesting that SNIPER1 can directly associate with these sNLRs.
Figure EV5. DN SNIPER1 interacts with diverse sNLRs in planta.
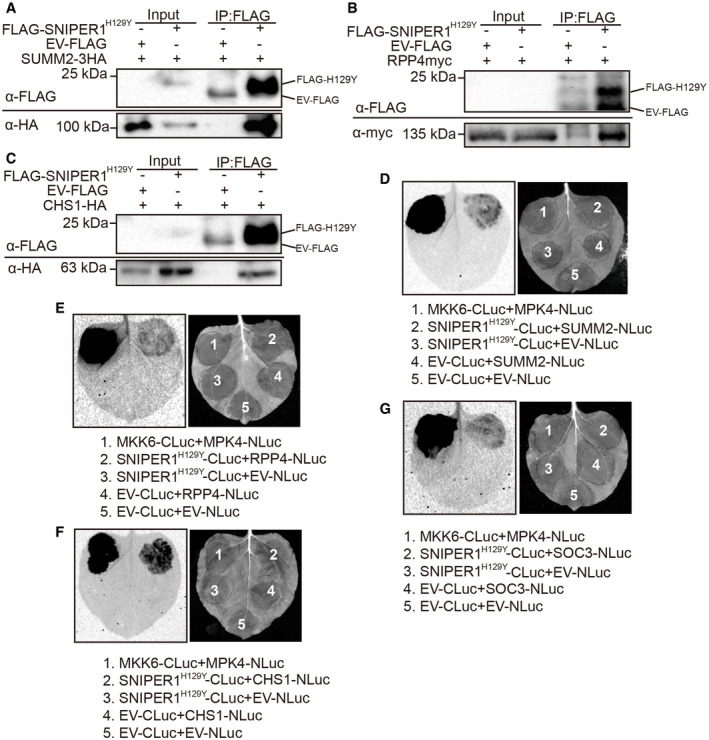
-
A–CImmunoprecipitation of SUMM2‐3HA (A), RPP4myc (B), and CHS1‐HA (C) by FLAG‐SNIPER1H129Y in N. benthamiana. Two biological repeats were carried out with similar results.
-
D–GInteraction of SNIPER1H129Y‐CLuc with SUMM2‐NLuc (D), RPP4‐NLuc (E), CHS1‐NLuc (F), or SOC3‐Nluc (G) as tested by split‐luciferase complementation assay in N. benthamiana. Three biological repeats were carried out with similar results.
Source data are available online for this figure.
Next, we sought to determine which domain of sNLRs specifically interacts with SNIPER1. Since the only domain shared by truncated TN‐type NLR protein CHS1 (which lacks the LRR domain), sensor TNLs (SNC1, RPP4), and sensor CNLs (SUMM2) is the NB domain, we analyzed the interactions between SNIPER1 and the NB domains of these sNLRs using two independent approaches. First, TurboID‐based proximity labeling (Zhang et al, 2019) approach was performed to biotinylate proximal and interacting proteins by TurboID fused protein in the presence of biotin. Such recently developed unbiased method is known to be effective in detecting weak or transient protein‐protein interactions. Indeed, the DN form of SNIPER1 can be pulled down by NB domains of SUMM2, RPP4, or CHS1 and these NB domains can be biotinylated in turn by SNIPER1‐HATurboID (Fig 7A–C), suggestive of a direct interaction between SNIPER1 and the NB domains. Furthermore, strong luciferase activity was observed when SNIPER1H129Y‐CLuc was co‐infiltrated with NB‐NLuc from different sNLRs (Fig 7D–F). Together these data support that SNIPER1 recognizes different sNLR substrates through their common NB domain.
Figure 7. SNIPER1 interacts with diverse sNLRs through the NB domain.
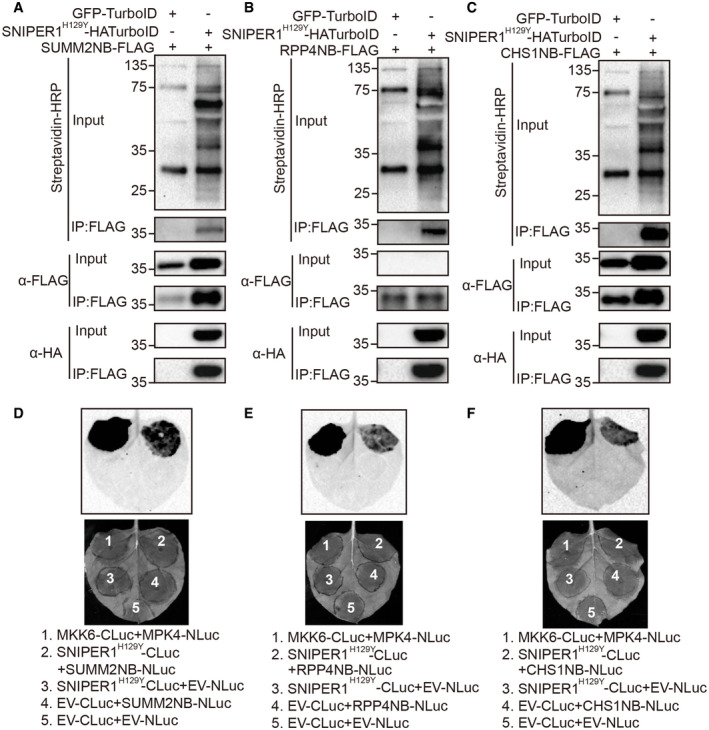
-
A–CImmunoprecipitation and biotinylation of SUMM2NB‐FLAG (A), RPP4NB‐FLAG (B), or CHS1NB‐FLAG (C) by SNIPER1H129Y‐HATurboID in N. benthamiana. Two biological repeats were carried out with similar results.
-
D–FInteraction of SNIPER1H129Y‐CLuc and SUMM2NB‐NLuc (D), RPP4NB‐NLuc (E), or CHS1NB‐NLuc (F) as tested by split‐luciferase complementary assay in N. benthamiana. Three biological repeats with four technical repeats each were carried out with similar results.
Source data are available online for this figure.
In vitro ubiquitination of NB domains of sNLRs by SNIPER1
To further test whether the E3 SNIPER1 could ubiquitinate sNLRs directly, MBP‐NB‐HA proteins were generated, and in vitro ubiquitination assays were performed in E. coli system. As shown in Fig 8, ubiquitinations of NB domains from CNL SUMM2 (Fig 8A), TNL RPP4 (Fig 8B), or TN CHS1 (Fig 8C) were only observed when E1, E2, Ub, and E3 SNIPER1 were present. The absence of any component leads to failed ubiquitination. These results indicate that SNIPER1 can ubiquitinate the NB domains of sNLRs.
Figure 8. In vitro ubiquitination of the NB domains by SNIPER1.
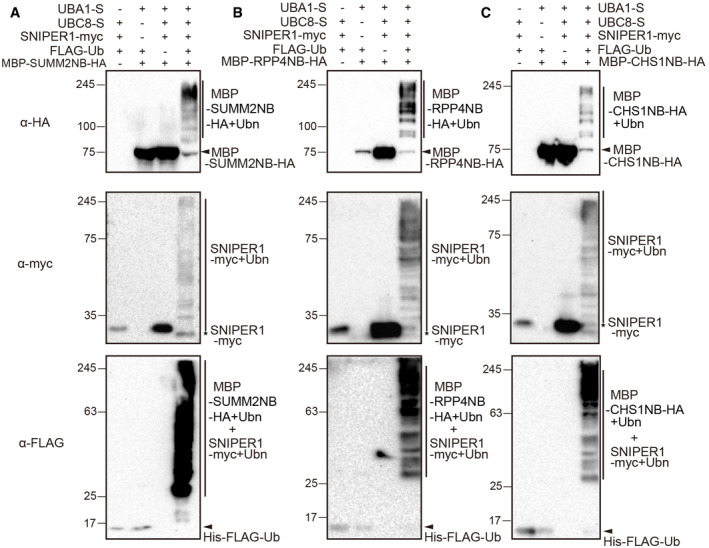
-
A–CUbiquitination of the NB domains from SUMM2 (A), RPP4 (B), and CHS1 (C). Immunoblot of bacterial lysates form E. coli strains expressing AtUBA1‐S, MBP‐NB‐HA, AtUBC8‐S, HIS‐FLAG‐UBQ10, and SNIPER1‐myc, or strains without one of these components. Two biological repeats were carried out with similar results.
Source data are available online for this figure.
Discussion
In all eukaryotes, UPS regulates protein homeostasis and plays pivotal roles in diverse biological processes. The Arabidopsis genome encodes only two E1s and 37 E2s, but around 1,500 E3s, most of which have not been mechanistically studied. Such low research attention is mostly due to their genetic redundancy. Single E3 gene knockout often does not exhibit any phenotypes, posing a technical difficulty for plant biologists.
The E3 overexpression approach is effective to overcome redundancy of both the E3s and their targets, as exemplified with our SNIPER screen. An E3 overexpression can lead to major reduction of the levels of all targeted substrates regardless of their redundancy, often revealing a phenotype which does not appear with single gene knockout of a substrate. On the other hand, E3 overexpression does not eliminate the substrate as in the case of a null knockout, thus could be revealing the phenotype of a partial loss‐of‐function mutant of the target essential gene. Such strategy can help overcome lethality, another big problem in genetic analysis.
Opposite to E3 overexpression, approaches using DN‐E3s were also shown to be effective in solving E3/substrate genetic redundancy. For example, deleting the F‐box domains of redundant F‐box proteins ZEITLUPE (ZTL), FLAVIN‐BINDING, KELCH REPEAT, F‐BOX1 (FKF1), or LOV KELCH PROTEIN2 (LKP2) leads to decoy versions of E3s, enabling genetic analysis of these redundant E3s and their substrates (Lee et al, 2018). Decoy versions of RING‐type E3s can be similarly generated by deleting the RING domain as in the case of redundant MAC3A (PUB59) and MAC3B (PUB60) (Feke et al, 2019).
E3 ligase‐mediated protein ubiquitination and subsequent turnover are often highly specific, with each E3 involved in regulating individual target protein and its close homologs. However, some E3 ligases may target multiple functionally related substrates. For example, AvrPtoB, an E3 ligase containing U‐box motif in P. syringae pv. tomato, can be delivered into the plant cell to degrade distinct PRRs (including FLS2, BAK1 (BRI1‐associated receptor kinase 1), CERK1 (Chitin elicitor receptor kinase 1)) and consequently dampen PTI pathway (Kim et al, 2002). Moreover, some E3 ligases are involved in multiple biological processes by targeting different proteins. PUB13 was reported to play roles in ABA (abscisic acid) signaling, BR (brassinosteroid) signaling, and PTI‐mediated pathway by ubiquitination of ABA co‐receptor ABI1, BR receptor BRI1, flagellin receptor FLS2, and chitin receptor LYK5 (Lu et al, 2011; Kong et al, 2015; Liao et al, 2017; Zhou et al, 2018). PUB13 could target more unrelated substrates in salicylic acid‐mediated pathway, cell death, and flowering time (Li et al, 2012; Liu et al, 2012; Antignani et al, 2015).
In comparison, our current study shows that SNIPER1 serves as a master E3 ligase that can control the turnovers of all tested sNLRs. However, SNIPER1 has no effect on the autoimmunity mediated by gain‐of‐function ADR1‐L2 D484V. Another group of helper NLRs, NRG1s, function in parallel to ADR1s and downstream of many TNLs (Qi et al, 2018; Castel et al, 2019; Wu et al, 2019). No morphological suppression of gain‐of‐function NRG1A D485V was observed by overexpression of SNIPER1. Therefore, SNIPER1 and SNIPER2 seem to broadly control the protein levels of sNLRs, but not hNLRs. The obvious question here is how SNIPER1 achieves such general substrate specificity to regulate the levels of many unrelated sNLRs. Our data suggest a direct E3‐substrate relationship between SNIPER1 and the sNLRs by recognition of the well‐conserved NB domain which exists in all typical plant sNLRs. Another feature that enables the diverse substrate recognition by SNIPER1/2 is their small size, which allows them to move freely in the cell to access sNLRs found in diverse subcellular locations, from plasma membrane to cytosol and nucleus (Boyes et al, 1998; Axtell & Staskawicz, 2003; Deslandes et al, 2003; Burch‐Smith et al, 2007; Wirthmueller et al, 2007; Slootweg et al, 2010; Bi et al, 2011; Xu et al, 2014). Indeed, confocal analysis revealed that both SNIPER1‐GFP and SNIPER2‐GFP proteins were found in both cytosol and nucleus when transiently expressed in N. benthamiana (Appendix Fig S8).
Another question is why dicots require SNIPER1/2 to universally regulate sNLRs. Upon infection, it has been widely observed in different plants that a large number of NLR genes are upregulated as a way of defense amplification (Ribot et al, 2008; Mohr et al, 2010; Yu et al, 2013; Brechenmacher et al, 2015; Chen et al, 2015). However, over‐accumulated NLRs could be detrimental to plant growth and development. Although some NLRs, such as SNC1, SIKIC1/2/3, RPS2, N, and MLAs, were reported to have their own regulatory E3 ligases (Cheng et al, 2011; Gou et al, 2012; Wang et al, 2016; Dong et al, 2018; Zhang et al, 2019), these specific E3s are less efficient in dampening such large‐scale immune up‐regulation. Usage of pathogen‐induced E3s like SNIPER1/2 would be more efficient to reduce global sNLRs levels to prevent autoimmunity. It will be interesting to test whether such mechanism also exists in monocots, and even animals.
The redundancy of SNIPER1 and SNIPER2 is intriguing. On the surface, their low sequence similarity argues against them being redundant. However, our genetic evidence where overexpression of both E3s displays similar immune‐related phenotypes, and EDS1‐dependent lethality of the double knockout mutant with single mutants being WT‐like, supports that these two E3s have overlapping functions in immunity. However, they are apparently not equally redundant, as overexpression of SNIPER2 exhibits distinct developmental defects while SNIPER1 overexpression is fully WT‐like in morphology. SNIPER2 (also named as MPSR1) was previously reported as a cytoplasmic chaperone‐independent protein quality control (PQC) E3 ligase for sensing and ubiquitinating misfolded proteins for their degradation (Kim et al, 2017). In addition, inverse correlation between MPSR1 and chaperone protein was observed for cytoplasmic protein quality control in response to proteotoxic stress (Kim et al, 2019). Since AtHSP90.2 and AtHSP90.3 are known to be involved in snc1 and chs2‐mediated autoimmunity (Bao et al, 2014; Huang et al, 2014), AtHSP90.3 protein level was examined. However, no change in AtHSP90.3 protein level was observed upon SNIPER1 overexpression in snc1 background (Appendix Fig S9). Therefore, the inverse correlation between HSP90 and SNIPER2 seems to be specific during proteotoxic stress, and only for SNIPER2. These independent non‐immune–related PQC activities could possibly contribute to SNIPER2's roles distinct from SNIPER1. The evolutionary distance between SNIPER1 and SNIPER2 also indicates that they may be going through functional diversification, the details of which will be interesting to investigate in the future.
In conclusion, our findings reveal a novel mechanism for global regulation of sNLRs turnover. In addition to strategies such as mutating recessive R genes like MLOs, and engineering NLR diversity and decoys (Innes, 2015; Kim et al, 2016; Kusch & Panstruga, 2017), SNIPER1 could be used in the future as a novel tool for engineering broad‐spectrum resistance in crops due to its global effects on sNLRs.
Materials and Methods
Plant growth conditions
Arabidopsis and N. benthamiana plants were grown at 22°C under 16‐h light/8‐h dark regime, unless otherwise specified.
Construction of plasmids
Overexpression constructs for SNIPER1 and SNIPER1 H129Y were cloned into pHAN vector. SNIPER2 and SNIPER1 H129Y were cloned into pGST1 vector with N terminal HA or FLAG tag. For the split‐luciferase complementation assay, SNIPER1 H129Y was cloned into pCambia1300‐35S‐HA‐CLuc and SUMM2, RPP4, SUMM2NB, RPP4NB, and CHS1NB were cloned into pCambia1300‐35S‐HA‐NLuc (Wu et al, 2017). For the TurboID‐based proximity labeling assay, SNIPER1 H129Y was cloned into pBASTA‐HA‐TurboID. pCambia1300 SUMM2‐3HA, pSuper1300 RPP4‐myc, pCambia1300 CHS1‐HA‐NLuc, pCambia1300 SOC3‐HA‐NLuc, and HA‐FLAG‐SOC3 were described in previous studies (Huang et al, 2010; Liang et al, 2019). CRISPR constructs targeting SNIPER1 were made as previously described (Wang et al, 2015). The primers used are listed in Appendix Table S1.
Arabidopsis stable transformation and transient Nicotiana benthamiana expression
The binary constructs were introduced into Agrobacterium tumefaciens GV3101 by electroporation and subsequently either transformed into Arabidopsis by the floral dipping method (Clough & Bent, 1998) or infiltrated into N. benthamiana leaves (Wu et al, 2019). For snc1, chs1‐2, chs2‐1, and snc2‐1D eds5 npr1, plants were grown at room temperature. For chs3‐2D and mekk1‐5 ndr1 transformation, plants were grown at 28°C under 16‐h light/8‐h dark regime to reduce autoimmunity. Transformants were selected on soil by spraying BASTA (Glufosinate ammonium). Ten transformants were selected each and co‐segregation analysis in T2 generations was carried out to make sure the suppression phenotypes were due to the transgene overexpression. SNIPER1 overexpression lines in SUMM2HA, RPS2HA, and RPM1myc backgrounds were generated by floral transformation with Agrobacterium containing pHAN‐SNIPER1. SNIPER1 overexpression lines in RPS4HA, RPP4myc, and ADR1‐L2 D484V backgrounds were generated by crossing the known SNIPER1‐OE lines 4‐1 with the respective lines due to their incompatible selections.
Pathogen infections
Oomycete and bacterial infection assays were carried out as described previously (Li et al, 2001). In brief, 2‐week‐old soil‐grown seedlings were sprayed with H.a. Noco2 conidia spores at a concentration of 105 spores per ml water. Sporulation was quantified using a hemocytometer after plants were grown at 18°C for 7 days. For bacterial infections, 4‐week‐old plants were infiltrated with bacterial solution at designated concentrations. Leaf disks were collected and ground on the day of infection (Day 0) and 3 days later (Day 3). Colony‐forming units (cfu) were calculated after incubation on LB plates with appropriate antibiotic selection.
Expression analysis
About 0.05 g plate‐grown plant tissue was collected, and RNA was extracted using an RNA isolation kit (Bio Basic; Cat#BS82314). ProtoScript II reverse transcriptase (NEB; Cat#B0368) was used to generate cDNA. Real‐time PCR was performed using a SYBR premix kit (TaKaRa, Cat#RR82LR). The primers used are listed in Appendix Table S1.
TurboID‐based proximity labeling in Nicotiana benthamiana
TurboID‐based proximity labeling assay was performed as described previously (Zhang et al, 2019). In brief, N. benthamiana plants were infiltrated with agrobacterium containing SNIPER1 H129Y‐HATurboID and RPP4NB‐3FLAG, SUMM2NB‐3FLAG, or CHS1NB‐3FLAG constructs. At 48 hpi, biotin was infiltrated, and the plants were incubated at room temperature for 3 h to allow labeling. Leaves were harvested and then followed by immunoprecipitation and Western blot analysis.
Protein extraction, immunoprecipitation, and western blot analysis
100 mg soil‐grown Arabidopsis plant leaves were collected and extracted by buffer (100 mM Tris–HCl pH 8.0, 0.2% SDS, and 2% β‐mercaptoethanol). Loading buffer was added to each protein sample and boiled for 5 min. Protein abundance was quantified using ImageJ (https://imagej.nih.gov/ij/).
Co‐immunoprecipitation assay was performed as previously described (Wu et al, 2017). In brief, N. benthamiana plants were infiltrated with agrobacterium containing SNIPER1 H129Y‐FLAG and HA tagged full‐length RPP4, SUMM2, CHS1 constructs, or NB domain truncated proteins. About 2.5 g N. benthamiana leaves expressing the indicated proteins were harvested at 48 hpi and ground into powder with liquid nitrogen. Extraction buffer containing 25 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.15% Nonidet P‐40, 10% Glycerol, 1 mM PMSF, 1 × Protease Inhibitor Cocktail (Roche; Cat. #11873580001), and 10 mM DTT. The FLAG‐tagged SNIPER1H129Y protein was immunoprecipitated using 20 μl M2 beads (Sigma; Cat. #A2220) for co‐immunoprecipitation. For TurboID‐based proximity labeling assay, the FLAG‐tagged proteins were enriched by M2 beads and biotinylation was detected by Streptavidin‐HRP (Abcam Cat. # ab7403).
The anti‐SNC1 antibody was generated against a SNC1‐specific peptide in rabbit (Li et al, 2010). The anti‐HA antibody was from Roche (Cat. #11867423001). The anti‐FLAG antibody was from Sigma (Cat. #F1804), and the anti‐myc antibody was from Santa cruz Biotechnology (Lot#D1411).
The in vitro ubiquitination assay
The in vitro ubiquitination assay for SNIPER1 was carried as previously described (Li et al, 2010; Han et al, 2017). In brief, E. coli strain BL21 containing the expression vectors were cultured in 2 ml LB liquid medium at 37°C. The expression of recombinant proteins was induced by IPTG. After induction, the bacteria were further grown at 28°C for 11 h before collection of bacteria cells. The bacterial pellet was resuspended with 100 μl 1 × loading buffer, boiled at 95°C for 5 min.
The in vitro ubiquitination of SNIPER2 was carried in a different system (Zhao et al, 2013). Recombinant AtUBA2‐His, glutathione S‐transferase (GST)‐AtUBC8 and GST‐SNIPER2 were expressed in E. coli and purified by Ni‐NTA (Qiagen) or GST affinity chromatography (GSTrap FF, GE Healthcare), respectively. Ubiquitination reactions were performed at 30°C for 2 h and ended by adding 1 × loading buffer, boiled at 95°C for 5 min.
Split‐luciferase complementation assay
Bacteria carrying the respective NLuc or CLuc constructs were co‐infiltrated into 4‐week‐old N. benthamiana leaves, and the infiltrated leaves were incubated with 1 mM luciferin 2 days later. Luminescence was recorded afterward. Three independent biological repeats with four technical repeats were carried out.
Transient expression and confocal microscopy
Transient expression in N. benthamiana was performed as previously described (Wu et al, 2017). Previously infiltrated leaves were cut and mounted in water on microscope slides. Selected confocal images of leaf abaxial epidermis were acquired using oil immersion differential interference contrast objectives on a Perkin‐Elmer Ultraview VoX spinning disk confocal microscope. Confocal images were captured with 488‐nm lasers for GFP excitation.
Statistical analysis
One‐way ANOVA followed by Tukey's post hoc test were performed. The Scheffé multiple comparison was applied for testing correction. Normality test for all data was done in SPSS. Statistical significance was indicated with different letters. P values and sample numbers (n) were detailed in figure legends.
Author contributions
ZW, Data curation, Validation, Investigation, Methodology, Writing—original draft, Project administration; MT, Data curation, Investigation, Methodology; CZ, Initial screen; LT and XLiu, Validation, Methodology; YZ, Conceptualization, Data curation, Formal analysis, Funding acquisition; XLi, Conceptualization, Data curation, Formal analysis, Supervision, Funding acquisition, Writing—original draft, Project administration, Writing—revisions. All authors contributed to revision of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
Drs. Jeff Dangl, Jane Parker, Brian Staskawicz, Shuhua Yang, and Jian‐Min Zhou are cordially thanked for sharing tagged NLR transgenic lines, mutants, and Pseudomonas Avr strains. We thank Dr. Tongjun Sun for discussions and Dr. Dongping Lu for sharing the in vitro ubiquitination assay system. Ms. Wanwan Liang and Mr. Aakar Chatha are thanked for genotyping sniper1‐1 sniper2‐1 double and sniper1‐c1 sniper2‐1 eds1‐2 mutants. Mr. Kevin Ao is sincerely thanked for careful reading of the manuscript. This work was financially supported by CFI‐JELF, the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Program, NSERC‐CREATE PRoTECT program, and the WD Cooper Memorial Fund from UBC.
The EMBO Journal (2020) 39: e104915
This study includes no data deposited in external repositories.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis . Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignani V, Klocko AL, Bak G, Chandrasekaran SD, Dunivin T, Nielsen E (2015) Recruitment of PLANT U‐BOX13 and the PI4Kβ1/β2 phosphatidylinositol‐4 kinases by the small GTPase RabA4B plays important roles during salicylic acid‐mediated plant defense signaling in Arabidopsis . Plant Cell 27: 243–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2‐specified disease resistance in Arabidopsis is coupled to the AvrRpt2‐directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Bao F, Huang X, Zhu C, Zhang X, Li X, Yang S (2014) Arabidopsis HSP90 protein modulates RPP4‐mediated temperature‐dependent cell death and defense responses. New Phytol 202: 1320–1334 [DOI] [PubMed] [Google Scholar]
- Bi D, Johnson KC, Zhu Z, Huang Y, Chen F, Zhang Y, Li X (2011) Mutations in an atypical TIR‐NB‐LRR‐LIM resistance protein confer autoimmunity. Front Plant Sci 2: 71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JD (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR‐NB‐LRR genes and confers downy mildew resistance through multiple signalling components. Plant J 29: 439–451 [DOI] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci USA 108: 16463–16468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Nam J, Dangl JL (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95: 15849–15854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechenmacher L, Nguyen TH, Zhang N, Jun TH, Xu D, Mian MA, Stacey G (2015) Identification of soybean proteins and genes differentially regulated in near isogenic lines differing in resistance to aphid infestation. J Proteome Res 14: 4137–4146 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Bjørn Nielsen H, Zhu S, Newman MA, Shokat KM, Rietz S, Parker J, Mundy J (2006) Arabidopsis MAP kinase 4 regulates salicylic acid‐ and jasmonic acid/ethylene‐dependent responses via EDS1 and PAD4. Plant J 47: 532–546 [DOI] [PubMed] [Google Scholar]
- Burch‐Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh‐Kumar SP (2007) A novel role for the TIR domain in association with pathogen‐derived elicitors. PLoS Biol 5: e68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel B, Ngou PM, Cevik V, Redkar A, Kim DS, Yang Y, Ding P, Jones JDG (2019) Diverse NLR immune receptors activate defence via the RPW8‐NLR NRG1. New Phytol 222: 966–980 [DOI] [PubMed] [Google Scholar]
- Chen J, Pang W, Chen B, Zhang C, Piao Z (2015) Transcriptome analysis of brassica rapa near‐isogenic lines carrying clubroot‐resistant and ‐susceptible alleles in response to Plasmodiophora brassicae during early infection. Front Plant Sci 6: 1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, Li X (2011) Stability of plant immune‐receptor resistance proteins is controlled by SKP1‐CULLIN1‐F‐box (SCF)‐mediated protein degradation. Proc Natl Acad Sci USA 108: 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collier SM, Hamel LP, Moffett P (2011) Cell death mediated by the N‐terminal domains of a unique and highly conserved class of NB‐LRR protein. Mol Plant Microbe Interact 24: 918–931 [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Cui H, Gobbato E, Kracher B, Qiu J, Bautor J, Parker JE (2017) A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol 213: 1802–1817 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong OX, Tong M, Bonardi V, El Kasmi F, Woloshen V, Wünsch LK, Dangl JL, Li X (2016) TNL‐mediated immunity in Arabidopsis requires complex regulation of the redundant ADR1 gene family. New Phytol 210: 960–973 [DOI] [PubMed] [Google Scholar]
- Dong OX, Ao K, Xu F, Johnson KCM, Wu Y, Li L, Xia S, Liu Y, Huang Y, Rodriguez E et al (2018) Individual components of paired typical NLR immune receptors are regulated by distinct E3 ligases. Nat Plants 4: 699–710 [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE (1999) EDS1, an essential component of R gene‐mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feke A, Liu W, Hong J, Li MW, Lee CM, Zhou EK, Gendron JM (2019) Decoys provide a scalable platform for the identification of plant E3 ubiquitin ligases that regulate circadian function. Elife 8: e44558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang W, Zhang T, Gong Z, Zhao H, Han GZ (2018) Out of water: the origin and early diversification of plant. Plant Physiol 177: 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ (1999) The Arabidopsis RPS4 bacterial‐resistance gene is a member of the TIR‐NBS‐LRR family of disease‐resistance genes. Plant J 20: 265–277 [DOI] [PubMed] [Google Scholar]
- Gou M, Shi Z, Zhu Y, Bao Z, Wang G, Hua J (2012) The F‐box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J 69: 411–420 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Han Y, Sun J, Yang J, Tan Z, Luo J, Lu D (2017) Reconstitution of the plant ubiquitination cascade in bacteria using a synthetic biology approach. Plant J 91: 766–776 [DOI] [PubMed] [Google Scholar]
- Huang X, Li J, Bao F, Zhang X, Yang S (2010) A gain‐of‐function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol 154: 796–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Monaghan J, Zhong X, Lin L, Sun T, Dong OX, Li X (2014) HSP90s are required for NLR immune receptor accumulation in Arabidopsis . Plant J 79: 427–439 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhu C, Li X (2018) SCFSNIPER4 controls the turnover of two redundant TRAF proteins in plant immunity. Plant J 95: 504–515 [DOI] [PubMed] [Google Scholar]
- Innes RW (2015) Exploiting combinatorial interactions to expand NLR specificity. Cell Host Microbe 18: 265–267 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones JD, Vance RE, Dangl JL (2016) Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395 [DOI] [PubMed] [Google Scholar]
- Jubic LM, Saile S, Furzer OJ, El Kasmi F, Dangl JL (2019) Help wanted: helper NLRs and plant immune responses. Curr Opin Plant Biol 50: 82–94 [DOI] [PubMed] [Google Scholar]
- Kapos P, Devendrakumar KT, Li X (2019) Plant NLRs: from discovery to application. Plant Sci 279: 3–18 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB (2002) Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589–598 [DOI] [PubMed] [Google Scholar]
- Kim SH, Qi D, Ashfield T, Helm M, Innes RW (2016) Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351: 684–687 [DOI] [PubMed] [Google Scholar]
- Kim JH, Cho SK, Oh TR, Ryu MY, Yang SW, Kim WT (2017) MPSR1 is a cytoplasmic PQC E3 ligase for eliminating emergent misfolded proteins in Arabidopsis thaliana . Proc Natl Acad Sci USA 114: E10009–E10017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Oh TR, Cho SK, Yang SW, Kim WT (2019) Inverse correlation between MPSR1 E3 ubiquitin ligase and HSP90.1 balances cytoplasmic protein quality control. Plant Physiol 180: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Cheng J, Zhu Y, Ding Y, Meng J, Chen Z, Xie Q, Guo Y, Li J, Yang S et al (2015) Degradation of the ABA co‐receptor ABI1 by PUB12/13 U‐box E3 ligases. Nat Commun 6: 8630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch S, Panstruga R (2017) mlo‐Based resistance: an apparently universal “weapon” to defeat powdery mildew disease. Mol Plant Microbe Interact 30: 179–189 [DOI] [PubMed] [Google Scholar]
- Lee CM, Feke A, Li MW, Adamchek C, Webb K, Pruneda‐Paz J, Bennett EJ, Kay SA, Gendron JM (2018) Decoys untangle complicated redundancy and reveal targets of circadian clock F‐box proteins. Plant Physiol 177: 1170–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Clarke JD, Zhang Y, Dong X (2001) Activation of an EDS1‐mediated R‐gene pathway in the snc1 mutant leads to constitutive, NPR1‐independent pathogen resistance. Mol Plant Microbe Interact 14: 1131–1139 [DOI] [PubMed] [Google Scholar]
- Li Y, Li S, Bi D, Cheng YT, Li X, Zhang Y (2010) SRFR1 negatively regulates plant NB‐LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog 6: e1001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Dai L, Wang GL (2012) PUB13, a U‐box/ARM E3 ligase, regulates plant defense, cell death, and flowering time. Plant Signal Behav 7: 898–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kapos P, Zhang Y (2015) NLRs in plants. Curr Opin Immunol 32: 114–121 [DOI] [PubMed] [Google Scholar]
- Liang W, van Wersch S, Tong M, Li X (2019) TIR‐NB‐LRR immune receptor SOC3 pairs with truncated TIR‐NB protein CHS1 or TN2 to monitor the homeostasis of E3 ligase SAUL1. New Phytol 221: 2054–2066 [DOI] [PubMed] [Google Scholar]
- Liao D, Cao Y, Sun X, Espinoza C, Nguyen CT, Liang Y, Stacey G (2017) Arabidopsis E3 ubiquitin ligase PLANT U‐BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol 214: 1646–1656 [DOI] [PubMed] [Google Scholar]
- Liu J, Li W, Ning Y, Shirsekar G, Cai Y, Wang X, Dai L, Wang Z, Liu W, Wang GL (2012) The U‐Box E3 ligase SPL11/PUB13 is a convergence point of defense and flowering signaling in plants. Plant Physiol 160: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucotelli E, Belloni S, Marone D, De Leonardis A, Guerra D, Di Fonzo N, Cattivelli L, Mastrangelo A (2006) The e3 ubiquitin ligase gene family in plants: regulation by degradation. Curr Genomics 7: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr TJ, Mammarella ND, Hoff T, Woffenden BJ, Jelesko JG, McDowell JM (2010) The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Mol Plant Microbe Interact 23: 1303–1315 [DOI] [PubMed] [Google Scholar]
- Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC (2005) NRG1, a CC‐NB‐LRR protein, together with N, a TIR‐NB‐LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol 15: 968–973 [DOI] [PubMed] [Google Scholar]
- Qi T, Seong K, Thomazella DPT, Kim JR, Pham J, Seo E, Cho MJ, Schultink A, Staskawicz BJ (2018) NRG1 functions downstream of EDS1 to regulate TIR‐NLR‐mediated plant immunity in Nicotiana benthamiana . Proc Natl Acad Sci USA 115: E10979–E10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhter D, Lüdke D, Ding Y, Feussner K, Zienkiewicz K, Lipka V, Wiermer M, Zhang Y, Feussner I (2019) Isochorismate‐derived biosynthesis of the plant stress hormone salicylic acid. Science 365: 498–502 [DOI] [PubMed] [Google Scholar]
- Ribot C, Hirsch J, Balzergue S, Tharreau D, Nottéghem JL, Lebrun MH, Morel JB (2008) Susceptibility of rice to the blast fungus, Magnaporthe grisea . J Plant Physiol 165: 114–124 [DOI] [PubMed] [Google Scholar]
- Roberts M, Tang S, Stallmann A, Dangl JL, Bonardi V (2013) Genetic requirements for signaling from an autoactive plant NB‐LRR intracellular innate immune receptor. PLoS Genet 9: e1003465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW et al (2010) Nucleocytoplasmic distribution is required for activation of resistance by the potato NB‐LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrenner AD, Goritschnig S, Krasileva KV, Schreiber KJ, Staskawicz BJ (2012) Effector recognition and activation of the Arabidopsis thaliana NLR innate immune receptors. Cold Spring Harb Symp Quant Biol 77: 249–257 [DOI] [PubMed] [Google Scholar]
- Sun T, Zhang Y, Li Y, Zhang Q, Ding Y (2015) ChIP‐seq reveals broad roles of SARD1 and CBP60 g in regulating plant immunity. Nat Commun 6: 10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong M, Kotur T, Liang W, Vogelmann K, Kleine T, Leister D, Brieske C, Yang S, Lüdke D, Wiermer M et al (2017) E3 ligase SAUL1 serves as a positive regulator of PAMP‐triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol 215: 1516–1532 [DOI] [PubMed] [Google Scholar]
- Vierstra RD (2009) The ubiquitin‐26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker JE (2013) Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14: 619–630 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Wang Z, Zhang X, Yang S (2013) A missense mutation in CHS1, a TIR‐NB protein, induces chilling sensitivity in Arabidopsis . Plant J 75: 553–565 [DOI] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015) Egg cell‐specific promoter‐controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16: 144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Chang C, Gu C, Tang S, Xie Q, Shen QH (2016) An E3 ligase affects the NLR receptor stability and immunity to powdery mildew. Plant Physiol 172: 2504–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller L, Zhang Y, Jones JD, Parker JE (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1‐dependent defense. Curr Biol 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Wu Z, Huang S, Zhang X, Wu D, Xia S, Li X (2017) Regulation of plant immune receptor accumulation through translational repression by a glycine‐tyrosine‐phenylalanine (GYF) domain protein. Elife 6: e23684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Li M, Dong OX, Xia S, Liang W, Bao Y, Wasteneys G, Li X (2019) Differential regulation of TNL‐mediated immune signaling by redundant helper CNLs. New Phytol 222: 938–953 [DOI] [PubMed] [Google Scholar]
- Xu F, Cheng YT, Kapos P, Huang Y, Li X (2014) P‐loop‐dependent NLR SNC1 can oligomerize and activate immunity in the nucleus. Mol Plant 7: 1801–1804 [DOI] [PubMed] [Google Scholar]
- Yu A, Lepère G, Jay F, Wang J, Bapaume L, Wang Y, Abraham AL, Penterman J, Fischer RL, Voinnet O et al (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA 110: 2389–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X (2003) A gain‐of‐function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1‐1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang Y, Fang B, Gannon P, Ding P, Li X (2010) Arabidopsis snc2‐1D activates receptor‐like protein‐mediated immunity transduced through WRKY70. Plant Cell 22: 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y (2012) Disruption of PAMP‐induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB‐LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Song G, Lal NK, Nagalakshmi U, Li Y, Zheng W, Huang PJ, Branon TC, Ting AY, Walley JW et al (2019) TurboID‐based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor‐mediated immunity. Nat Commun 10: 3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Tian M, Li Q, Cui F, Liu L, Yin B, Xie Q (2013) A plant‐specific in vitro ubiquitination analysis system. Plant J 74: 524–533 [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu D, Wang P, Ma X, Lin W, Chen S, Mishev K, Lu D, Kumar R, Vanhoutte I et al (2018) Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U‐box PUB12/PUB13‐mediated ubiquitination. Proc Natl Acad Sci USA 115: E1906–E1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8


