Abstract
Objective
The COVID-19 pandemic poses an unprecedented threat to the health and economic prosperity of the world's population. Yet, because not all regions are affected equally, this research aims to understand whether the relative growth rate of the initial outbreak in early 2020 varied significantly between the US states and counties.
Study design
Based on publicly available case data from across the USA, the initial outbreak is statistically modeled as an exponential curve.
Methods
Regional differences are visually compared using geo maps and spaghetti lines. In addition, they are statistically analyzed as an unconditional model (one-way random effects analysis of variance estimated with HLM 7.03); the bias between state- and county-level models is evidenced with distribution tests and Bland–Altman plots (using SPSS 26).
Results
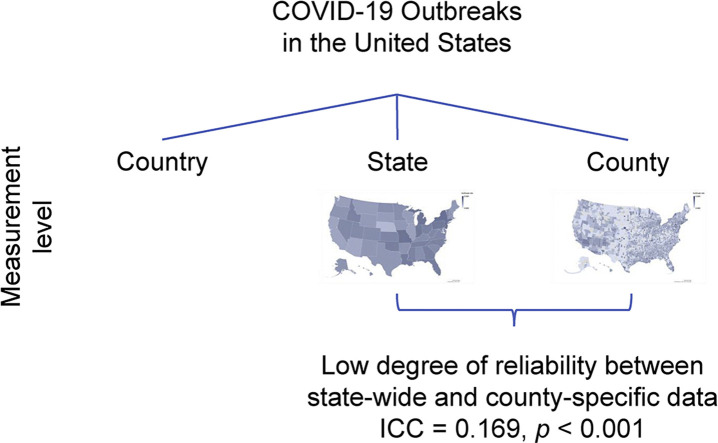
At the state level, the outbreak rate follows a normal distribution with an average relative growth rate of 0.197 (doubling time 3.518 days). But there is a low degree of reliability between state-wide and county-specific data reported (Intraclass correlation coefficient ICC = 0.169, P < 0.001), with a bias of 0.070 (standard deviation 0.062) as shown with a Bland–Altman plot. Hence, there is a significant variation in the outbreak between the US states and counties.
Conclusions
The results emphasize the need for policy makers to look at the pandemic from the smallest population subdivision possible, so that countermeasures can be implemented, and critical resources provided effectively. Further research is needed to understand the reasons for these regional differences.
Keywords: COVID-19, Novel coronavirus, Outbreak, Pandemic, Regional differences
Graphical abstract
Highlights
-
•
The COVID-19 pandemic appears to affect countries or regions in different ways.
-
•
The research uses a statistical model to analyze variations within the US.
-
•
A low degree of reliability between state-wide and county-specific data is found.
-
•
Measures should be implemented at the smallest population subdivision possible.
Introduction
On January 20, 2020, the first case of the novel coronavirus disease 2019 (COVID-19) was reported on the US soil, with cases in the USA growing to over 579,197 as of April 13, 2020.1 In the struggle to contain the pandemic's growth rate, the US government took unprecedented action. At the federal level, international and domestic travel restrictions were imposed, and at a state level, closing down of businesses, stay at home orders, and social distancing mandates were enacted. A community's susceptibility to any virus is determined by a variety of factors, including but not limited to biological determinants, demographic profiles, type of habitat, and socio-economic characteristics.2 As these factors vary significantly across the USA, there is likely to be considerable intra-country variation in the outbreak as well.
In the present study, we examine the relative growth rate of the COVID-19 outbreak and its variation on a state and county levels across the USA. We show, both through visualization and statistical analysis, that the outbreak varies significantly across counties and that an aggregate view at the state level, as it is most often reported in media, hides differences at a lower level. In this article, we show the necessity of analyses on a lower level.
Model and methods
We obtain COVID-19 outbreak data from the China Data Lab published at Harvard Dataverse (as of April 13, 2020) and USA Facts (as of April 14, 2020);1 , 3 we check for consistency between the two databases. Since January 22, 2020, the latter database has aggregated data from the Centers for Disease Control and Prevention and state- and local-level public health agencies, confirming them by referencing state and local agencies directly. For our county-level analysis, we discard cases which USA Facts can only allocate at the state, but not at the county level because of a lack of information. On average, the number of unallocated cases is small, but a few states contribute as many as 4866 (New Jersey), 1300 (both Rhode Island and Georgia), or 1216 (Washington State) unallocated cases, resulting in an average of 308 unallocated cases per state, again as of April 14, 2020.
Following approaches by the Institute for Health Metrics and Evaluation at the University of Washington4 and the COVID-19 Modeling Consortium at the University of Texas at Austin,5 we statistically model the outbreak in the USA at state and county levels using the exponential growth equation , where b is a positive constant called the relative growth rate; it has units of inverse time. Going forward, we simply refer to b as the outbreak rate. Solutions to this differential equation have the form , where a is the initial value of cases y. The doubling time T d can be calculated from the outbreak rate as . This is a statistical, but not an epidemiological model, that is, we are neither trying to model infection transmission nor estimate epidemiological parameters, such as the pathogen's reproductive or attack rate. Instead, we are fitting a curve to observed case data at the state and county levels, so that the estimated outbreak rate is independent of the population in the respective unit. However, it does not control for confounders specific to the habitat. A change-point analysis using the Fisher discriminant ratio as a kernel function does not show any significant change points in the outbreak, and therefore justifies modeling the COVID-19 outbreak as a phenomenon of unrestricted population growth.6 As outbreak rates change over time and their estimation is somewhat sensitive to the starting figure, we alternatively calculate the outbreak rate after it reached 10 and 25 cases in the respective unit, finding a high correlation among the rates. We are aware that testing differences between states may also be important confounds. As the number of tests administered and the number of confirmed cases correlates to varying extents,7 this is, however, difficult to control. A disadvantage of this statistical approach is that we cannot forecast outbreak dynamics, although we do not require extrapolated data in our work.
Statistical results
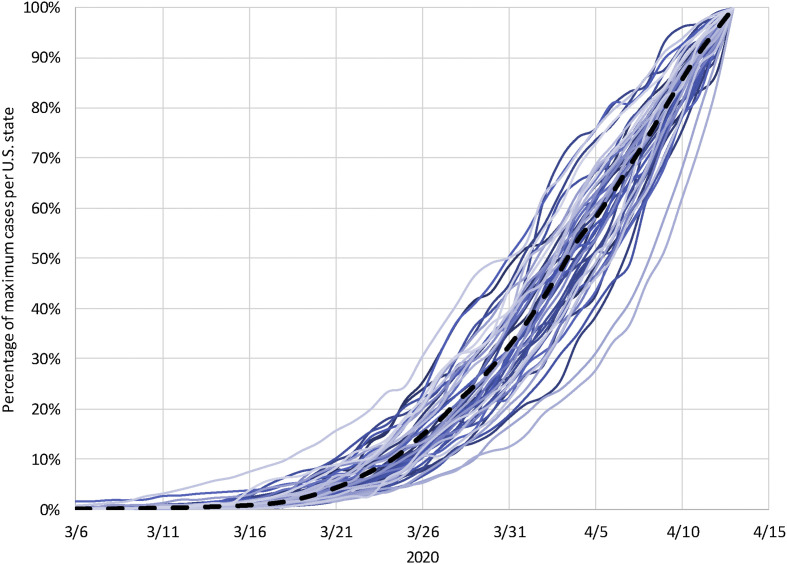
For the entire USA, the outbreak rate is 0.172, which translates to a doubling time of T d = 4.025 days (as of April 13, 2020). At the state level, the average outbreak rate is 0.197 (T d = 3.518) and the median is 0.194. The outbreak rate ranges from 0.085 to 0.282, with a standard deviation of 0.039. The spaghetti lines in Fig. 1 trace the cases as a percentage of the maximum number of cases reported on April 13 at both levels. Across states, the outbreak rates follow a normal distribution, as evidenced by a Shapiro–Wilk test with W (51) = 0.991, P = 0.970. We only identify Nebraska (outbreak rate = 0.085) as a potential outlier. To appropriately report the outbreak at the county level, we first remove all 869 counties where the outbreak has not yet commenced, that is where the growth rate is close to zero or where the number of reported cases is below five. For the remaining counties, the average outbreak rate is 0.134, which translates to T d = 5.172 days. The median outbreak rate is 0.135, standard deviation 0.057, and the maximum outbreak rate 0.426 (Colonial Heights City, Virginia). The outbreak significantly deviates from a normal distribution, W (3145) = 0.982, P < 0.001.
Fig. 1.
Epidemic days at the US country and state levels. The spaghetti lines trace the COVID-19 outbreak in the USA (black dashed line) and the states (straight lines) as a percentage of the cases reported on April 13, 2020, in the entire country and each state, respectively.
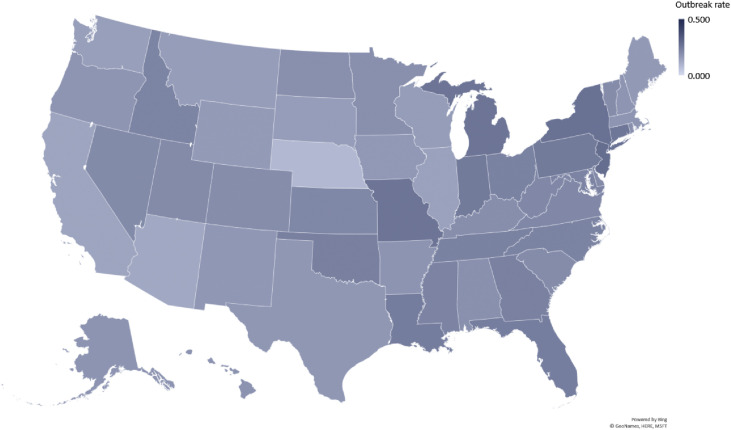
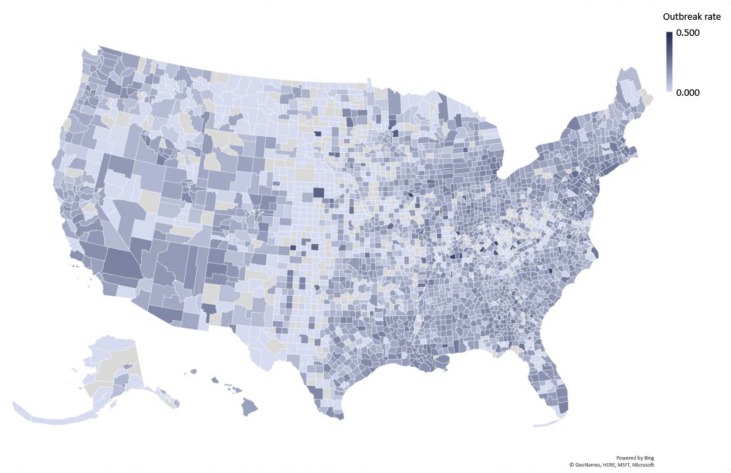
The two geo maps in Figs. 2 and 3 show how the outbreak varies between states and between counties within a state. Statistically, we also find a very low degree of reliability between the state-wide and county-specific breakout rates. Even after removing counties without occurrence of the outbreak, the average intraclass correlation coefficient (ICC) is very low at 0.169, with a 95% confidence interval from −0.080 to 0.360, F(2281,2281) = 1.462, P < 0.001 (unconditional model estimated with HLM 7.03 as a one-way random effects analysis of variance). Similarly, Spearman's ρ = 0.234 and Kendall's τ = 0.160, both P < 0.001. The Bland–Altman plot shows a bias of 0.070 (standard error 0.001 and standard deviation 0.062, calculated with SPSS 26).
Fig. 2.
Variation in outbreak rates at the US state level. This geo map displays the variation in outbreak rates at the US state level. Lighter colors signify that the pandemic has a slower relative growth rate, and darker colors point to a faster growth. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Variation in outbreak rates at the US county level. This geo map reveals the larger variation in outbreak rates at the US county level. The color band is the same as in Fig. 2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Discussion
In the USA, the initial outbreak of the COVID-19 pandemic varied considerably not only between states, but also within the counties of a state. The outbreak rate followed a normal distribution across 50 states plus Washington, D.C. When we extrapolate this to the county data, we find that the outbreak rate significantly deviated from a normal distribution, even when omitting the counties with little to no outbreak. When graphed, this variation in case counts from county to county is easily visible (Fig. 3). In comparison with the state-level depiction (Fig. 2), there is a great variation between the state ranking and the situation in its individual counties. In the USA, most response measures to the pandemic are devised and effected at the state level. Although this is certainly better targeted than an overall response at the federal level, which might spread resources too thinly in some regions, it still may not cater sufficiently for local outbreak differences and resource utilization. For example, although many counties in South Carolina still conveyed a utilized hospital bed capacity of less than 50% (as of April 21, 2020), Lexington County reported 90.6%, followed by Orangeburg and Colleton Counties at 82.2 and 78.0%, respectively.8
Politics and political partisanship play a large role in the resolution of national health emergencies, and have been found to be the strongest predictor of the early adoption of social distancing policies.9 But such policies tend to generalize strategy and target larger populations. Various institutional, societal, and cultural factors influence the development and adoption of these policies, and are important in the analysis of variations in the pandemic's growth rate across states and counties. Between countries at the international level, previous research indicates the association of such contextual factors with the outbreak rate.10 For the USA, we expect comparable findings, and aim to understand potential reasons for the differences in further research.
More generally, our study indicates that governments must track a pandemic's outbreak and tailor appropriate response strategies to the most granular level possible. This would not only increase effectiveness of political policy and response strategy, but also allow for a redistribution of excess resources to areas most vulnerable to the pandemic. This will become increasingly important as the world begins returning to normalcy, and attempts to prevent further waves of the COVID-19 pandemic.
Author statements
Ethical approval
None sought.
Funding
Partly funded through the Center for International Business Education and Research (CIBER) at the University of South Carolina.
Competing interests
None declared.
Acknowledgements
The authors thank the anonymous peer reviewer for their insightful feedbacks that helped to improve the manuscript, and gratefully acknowledge the support of the Darla Moore School of Business and the Center for International Business Education and Research (CIBER) at the University of South Carolina.
References
- 1.China Data Lab . Harvard Dataverse; 2020. US COVID-19 daily cases with basemap. [Internet] Harvard Dataverse, [cited 2020 Apr 14] [DOI] [Google Scholar]
- 2.Chen J.T., Kahn R., Li R., Chen J.T., Krieger N., Buckee C.O. U.S. county-level characteristics to inform equitable COVID-19 response. medRxiv. 2020:1–38. [Google Scholar]
- 3.USA Facts . 2020. Coronavirus locations: COVID-19 map by county and state.https://usafacts.org/visualizations/coronavirus-covid-19-spread-map/ [Internet] [cited 2020 Apr 14]. Available from: [Google Scholar]
- 4.Murray C.J. Forecasting COVID-19 impact on hospital bed-days, ICU-days, ventilator-days and deaths by US state in the next 4 months. medRxiv. 2020:1–26. [Google Scholar]
- 5.Woody S., Tec M., Dahan M., Gaither K., Fox S.J., Meyers L.A. The university of Texas at Austin COVID-19 modeling Consortium. 2020. Projections for first-wave COVID-19 deaths across the U.S. using social-distancing measures derived from mobile phones measures; pp. 1–11.https://www.tacc.utexas.edu/ut_covid-19_mortality_forecasting_model_report [Internet] [cited 2020 Apr 19] [Google Scholar]
- 6.Texier G., Farouh M., Pellegrin L., Jackson M.L., Meynard J.B., Deparis X. Outbreak definition by change point analysis: a tool for public health decision? BMC Med Inf Decis Making. 2016;16(1):1–12. doi: 10.1186/s12911-016-0271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaashoek J., Santillana M. COVID-19 positive cases, evidence on the time evolution of the epidemic or an indicator of local testing capabilities? A case study in the United States. SSRN. 2020:1–14. [Google Scholar]
- 8.SCDHEC . 2020. Hospital bed capacity (COVID-19)https://www.scdhec.gov/infectious-diseases/viruses/coronavirus-disease-2019-covid-19/hospital-bed-capacity-covid-19 [Internet] [cited 2020 Apr 22]. Available from: [Google Scholar]
- 9.Adolph C., Amano K., Bang-Jensen B., Fullman N., Wilkerson J. Pandemic politics: timing state-level social distancing responses to COVID-19. medRxiv. 2020:1–19. doi: 10.1215/03616878-8802162. [DOI] [PubMed] [Google Scholar]
- 10.Messner W. The institutional and cultural context of cross-national variation in COVID-19 outbreaks (forthcoming) Int Public Heal J. 2021;13(2) [Google Scholar]