Abstract
Mitochondria are highly dynamic organelles that constantly undergo fission and fusion events to adapt to changes in the cellular environment. Aberrant mitochondrial fission has been associated with several types of cardiovascular dysfunction; inhibition of pathologically aberrant mitochondrial fission has been shown to be cardioprotective. Pathological fission is mediated by the excessive activation of GTPase dynamin-related protein 1 (Drp1), making it an attractive therapeutic target in numerous cardiovascular diseases. Mitochondrial division inhibitor (mdivi-1) is widely used small molecule reported to inhibit Drp1-dependent fission, elongate mitochondria, and mitigate injury. The purpose of our study was to understand the pleiotropic effects of mdivi-1 on mitochondrial dynamics, mitochondrial respiration, electron transport activities, and macro-autophagy. In this study, we found that mdivi-1 treatment decreased Drp1 expression, proteolytically cleaved L-OPA1, and altered the expression of OXPHOS complex proteins, resulting in increased superoxide production. The altered expression of OXPHOS complex proteins may be directly associated with decreased Drp1 expression, as Drp1 siRNA knockdown in cardiomyocytes showed similar effects. Results from an autophagy flux assay showed that mdivi-1 induced impaired autophagy flux that could be restored by Atg7 overexpression, suggesting that mdivi-1 mediated inhibition of macro-autophagy in cardiomyocytes. Treatment with mdivi-1 resulted in increased expression of p62, which is required for Atg7 overexpression-induced rescue of mdivi-1-mediated impaired autophagy flux. In addition, mdivi-1-dependent proteolytic processing of L-OPA1 was associated with increased mitochondrial superoxide production and altered expression of mitochondrial serine/proteases. Overall, the novel pleiotropic effect of mdivi-1 in cardiomyocytes included proteolytically cleaved L-OPA1, altered expression of OXPHOS complex proteins, and increased superoxide production, which together resulted in defects in mitochondrial respiration and inhibition of macro-autophagy.
Keywords: mdivi-1, Mitochondria fission, Macro-autophagy, Mitochondrial dysfunction, Mitochondrial proteases
Graphical abstract
1. Introduction
The heart muscle contains a large number of mitochondria to meet the high-energy demands of the contracting cardiomyocytes. Mitochondria in cardiomyocytes are highly organized and arranged compactly between contractile filaments (interfibrillar), adjacent to the sarcolemma (subsarcolemmal), and in the perinuclei. Mitochondria are highly dynamic organelles that constantly undergo fission and fusion to maintain their shape, distribution, and size to adapt to changes in the cellular environment. Mitochondrial fission is regulated by cytoplasmic GTPase dynamin-related protein 1 (Drp1). This protein translocates to mitochondrial fission sites on the outer mitochondrial membrane where it interacts with multiple adaptor proteins, including mitochondrial fission 1 (Fis1), mitochondrial dynamics proteins of 49 and 51 kDa (MiD49/51), and mitochondrial fission factor (Mff) [1,2]. Mitochondrial fusion is regulated by optic atrophy type 1 (OPA1) located on the inner mitochondrial membrane and mitofusins (Mfn1 and Mfn2) located on the outer mitochondrial membrane. OPA1 is a dynamin-like GTPase with a mitochondrial import sequence (IMS) that directs the protein to the intermembrane space and cristae. Mitochondria contain various OPA1 isoforms generated by alternative splicing as well as proteolytic processing [3,4]. The cleavage of the mitochondrial targeting sequence produces the long isoforms (L-OPA1) anchored to the inner mitochondrial membrane. Approximately half of these L-OPA1 are constitutively processed by mitochondrial proteases to generate the short isoform (S-OPA1) [[3], [4], [5]]. Overall, there are five isoforms of OPA1, including the long uncleaved OPA1 isoforms (L-OPA1, isoform a and b) and the short, cleaved OPA1 isoforms (S-OPA1: isoform c, d, and e).
Mitochondria contain compartmentally based proteases: mitochondrial matrix localized lon peptidase I (LonP1), caseinolytic mitochondrial matrix peptidase proteolytic subunit (ClpP), and mitochondrial inner membrane AAA family proteases (ATPases associated with diverse cellular activities) including YME1L1 [6] and OMA1 [7]. Both YME1L1 and OMA1 act as critical regulators of key mitochondrial functions, including inner membrane proteostasis and mitochondrial dynamics, by maintaining a balance among the five OPA1 isoforms through proteolytic processing of OPA1. L-OPA1 (a and b) is constitutively cleaved by YME1L1 to create the d isoform, which promotes the maintenance of tubular mitochondrial morphology. Stress-activated OMA1 processes all L-OPA1 isoforms (a and b) to the short isoforms (c and e) that promote mitochondrial fragmentation [8]. Moreover, mitochondrial proteases play a critical role in the regulation of mitochondrial dynamics and are also essential for the surveillance of the OXPHOS machinery, which is constantly susceptible to oxidative damage due to its continuous production of reactive oxygen species (ROS). Therefore, a balance between fission and fusion events is required for mitochondrial homeostasis; an imbalance leads to cardiac pathologies such as ischemia-reperfusion injury, heart failure, and diabetic cardiomyopathy [9]. For instance, excessive Drp1‐mediated fission leads to mitochondrial fragmentation, mitochondrial membrane depolarization, increased ROS generation, and oxidative stress, which lead in turn to the development of mitochondrial dysfunction [10,11]. Therefore, the reduction of mitochondrial fission by inhibiting Drp1 under these conditions is believed to be protective.
Among the mitochondrial fission inhibitors, in a chemical library screen for 23,100 compounds that influence mitochondrial morphology in yeast, mdivi-1 was identified as a cell-permeable quinazolinone derivative inhibitor of Drp1 [12]. However, in this study, mdivi-1 failed to inhibit the GTPase activity of recombinant human Drp1 and its activity with respect to mammalian Drp1 was not determined due to technical issues related to the mammalian Drp1 preparations. Mdivi-1 may impair Drp1 function by acting allosterically and preventing Drp1 oligomerization on the mitochondrial membrane, suggesting that mdivi-1 may prevent Drp1 self-assembly into rings and its association with mitochondria [12]. Since its discovery, mdivi-1 has widely been considered to be a small-molecule inhibitor of Drp1, used extensively in multiple cell types and organs under different disease conditions [13,14]. We also recently reported that inhibition of aberrant mitochondrial fission by mdivi-1 in cardiomyocytes expressing proteotoxic mutant-desmin results in preserved mitochondrial function and decreased apoptotic cell death [15]. Similarly, mdivi‐1 has also been shown to confer cytoprotection in ischemia‐reperfusion injury [9,16] and doxorubicin‐induced cardiomyopathy [17] by reducing the production of ROS, attenuating cytosolic calcium overload, restoring mitochondrial membrane potential, and delaying hypercontracture of cardiomyocytes. Treatment with mdivi-1 resulted in significantly increased mitochondrial content, complex I enzymatic activity, and ATP levels in the db/db mouse hippocampus [18]. However, mdivi-1 treatment in cancer cells decreased proliferation and increased cell death and apoptosis through DRP1 inhibition or a mitochondrial fusion induction-independent mechanism. Similarly, mdivi-1 has been reported to function as a reversible inhibitor of mitochondrial complex I, affecting mitochondrial respiration in COS-7 cells and primary neurons without changing mitochondrial morphology [19]. Additionally, mdivi-1 has also been shown to impair DNA replication and repress mitochondrial respiration independent of Drp1 in multidrug-resistant tumor cells [20]. In contrast, mdivi-1 treatment under high-glucose induced energy stress increased complex I activity and mitochondrial density in human neuronal SK cells [18]. Taken together, the data from these studies demonstrate a context- and cell type-dependent molecular function of mdivi-1 mediated through Drp1 dependent/independent pathways.
The aim of our study was to determine the pleiotropic effects of mdivi-1 that could negatively impact cardiomyocyte function, limiting its long-term use in cardiovascular diseases. We observed the effects of mdivi-1 on the expression of proteins involved in mitochondrial dynamics and OXPHOS regulation, mitochondrial respiration, and autophagic activity in cardiomyocytes. We reported the pleiotropic effects of mdivi-1 on cardiomyocytes including decreased Drp1 expression, L-OPA1 proteolytic cleavage, decreased Complex I protein expression, and inhibition of autophagy activity. We also observed altered expression of the mitochondrial proteases responsible for OPA1 proteolytic processing associated with the degradation of L-OPA1.
2. Material and methods
Materials. The materials used are as follows: αMEM (Gibco), DMEM (Gibco), FBS (Gibco), Cell Lytic M (C2978, Sigma-Aldrich), Antibiotic-antimycotic solution (Gibco), Complete Protease Inhibitor Cocktail (Roche), pre-cast 7.5%–15% Criterion Gels (BioRad), mdivi-1 (M0199, Millipore Sigma), carbonyl cyanide m-chlorophenylhydrazone (CCCP) (C2759, Millipore Sigma), MitoSOX™ Red (M36008, Invitrogen), Bafilomycin A1 (B1793, Sigma-Aldrich), Oligomycin (Sigma-Aldrich), FCCP (Sigma-Aldrich), Rotenone (Sigma-Aldrich), Antimycin A (Sigma-Aldrich), Ponceau S (Acros Organic), and Vectashield Hardset (Vector Labs, H1400).
Animals. Timed-pregnant female Sprague Dawley rats were purchased from Charles River Laboratories International, Inc. (Portage, MI) to isolate primary neonatal rat ventricular cardiomyocytes from 1- to 2-day-old rat pups. All procedures for handling animals complied with the Guide for Care and Use of Laboratory Animals and were approved by the ACUC Committee of LSU Health Sciences Center-Shreveport. All animals were cared for according to the National Institutes of Health guidelines for the care and use of laboratory animals.
Neonatal rat cardiomyocyte (NRC) isolation and culture. NRC were isolated from the ventricles of 1- to 2-day-old Sprague-Dawley rat pups as previously described [15,21]. The ventricular tissues collected from the rat pups were digested with collagenase at 37 °C overnight and then further digested in trypsin. Cardiac fibroblasts were removed by preplating and the isolated cardiomyocytes were plated at 1.5 × 106 cells per 10-cm2 plate in αMEM (Gibco) containing 10% FBS (Gibco) and 1% antibiotic-antimycotic (Gibco). Cells underwent different adenoviral infections or siRNA transfections 24 h after plating and were maintained in DMEM (Gibco) containing 2% FBS and 1% antibiotic-antimycotic solution.
Recombinant adenovirus infection. We prepared adenoviral constructs containing wild-type Atg7 by cloning into a pShuttle-CMV vector; replication-deficient recombinant adenoviruses were made using the AdEasy system (Agilent Technologies) [21]. To overexpress Atg7, NRC were infected with Atg7 carrying an adenoviral vector (0–10 multiplicity of infection (MOI)) for 2 h in DMEM media without serum or antibiotics. After 2 h, media were changed to regular culture media. Plates infected with adenovirus expressing β-galactosidase served as controls for all of the experiments.
siRNA knockdown in cardiomyocytes. A pool of siRNAs (Invitrogen) was tested for their capacity to reduce Drp1 and p62 protein levels in NRC using methods described previously [21]. We used three different siRNAs for both Drp1 and p62 to silence endogenous protein expression. Twenty-four hours after plating, cells were transfected with siRNA with Lipofectamine 2000 (Invitrogen) in OptiMem (Gibco) media overnight [21]. After 16 h, media were changed to regular culture media. A non-specific siRNA was used as a negative control in all silencing experiments. To overexpress Atg7 on p62 siRNA knockdown NRC, cells were first transfected with p62 siRNA and then infected with the Atg7 adenovirus overexpression. Cells were harvested using Cell Lytic M (Sigma-Aldrich) with a protease and phosphatase inhibitor cocktail (Roche) for analysis using Western blot.
2.1. Drp1 siRNA (Invitrogen)
siRNA1: CCGUGACAAAUGAAAUGGUACAUAA and UUAUGUACCAUUUCAUUUGUCACGG; siRNA2: GCCAUGCUGUCAAUUUGCUAGAUGU and ACAUCUAGCAAAUUGACAGCAUGGC; siRNA3: CGAGAACAGCGAGAUUGUGAGGUUA and UAACCUCACAAUCUCGCUGUUCUCG;
2.2. p62 siRNA (Invitrogen)
siRNA1: CGGUGAAGGCCUAUCUACUGGGCAA and UUGCCCAGUAGAUAGGCCU UCACCG;
siRNA2: CAACGUGAUUUGUGAUGGUUGCAAU and AUUGCAACCAUCAC AAAUCACGUUG;
siRNA3: CCUGUGGUGGGAACUCGCUAUAAGU and ACUUAUAG CGAGUUCCCACCACAGG;
mdivi-1, CCCP, and antimycin A treatment. NRC were treated with mdivi-1 (100 μmol/L) or a vehicle 72 h after plating for different time points. A stock solution with a concentration of 40 mmol/L mdivi-1 in dimethyl sulfoxide (DMSO) was used to treat cells, and an equal volume of DMSO was used as the vehicle. For the Atg7 overexpression experiment, NRC were treated with mdivi-1 72 h after infection. In the p62 siRNA knockdown experiments, mdivi-1 treatment was performed 96 h after transfection. CCCP treatment was performed in NRC for 3 h at a concentration of 10 μmol/L. The treatment was performed 21 h after mdivi-1 treatment in NRC with no replacement of media. An equal volume of DMSO was used as the vehicle. In the same way, 21 h after mdivi-1 treatment, NRC were treated with antimycin A (AA) for 3 h at a concentration of 10 μmol/L.
Mitochondrial respiration. The mitochondrial oxygen consumption rate in NRC was measured with an XF24 Extracellular Flux Analyzer (Seahorse Biosciences, North Billerica, MA) using methods described previously [15,22,23]. NRC were seeded at a density of 8 × 104 cells/well into gelatin-coated Seahorse Bioscience XF microplates and grown in DMEM (Gibco) containing 2% FBS (Gibco) and 1% antibiotic-antimycotic solution (Gibco) [15]. To measure the effect of mdivi-1 in mitochondrial OCR, NRC (48 h after plating) were treated with mdivi-1 (100 μmol/L) or vehicle for 24 h. NRC were incubated with DMEM (containing no glucose and pyruvate; Gibco) supplemented with 10 mmol/L glucose and 2 mmol/L pyruvate in a CO2-free incubator at 37 °C for 1 h before loading the plate in the XF24 analyzer. The OCR was measured with the sequential addition of oligomycin (1 μmol/L), FCCP (4 μmol/L), and rotenone (0.5 μmol/L) plus antimycin A (0.5 μmol/L) at specified time points. The OCR values were normalized to total protein content in the corresponding wells and expressed as pmol/min/μg protein [15].
Mitochondrial superoxide production assay. The production of superoxide by mitochondria was visualized using MitoSOX™ Red reagent in conjunction with fluorescence microscopy [24]. Isolated NRC were plated on Lab‐Tek II chamber slides (Thermo Scientific, 154461) at a seeding density of 1 × 105 cells per well. Seventy-two hours after the cells were plated, NRC were treated with mdivi-1 (100 μmol/L) or vehicle dissolved in NRC culture media (DMEM containing 2% FBS and 1% antibiotic-antimycotic solution; Gibco). Twenty-four hours after mdivi-1 treatment, NRC were stained with MitoSOX™ Red reagent (5 μM, M36008, Invitrogen) dissolved in NRC culture medium for 15 min followed by immediate fixation with 4% paraformaldehyde in NRC culture media. Cells were subsequently washed and mounted with Vectashield Hardset antifade mounting media for fluorescence (Vector Laboratories). Cardiomyocytes were then observed using a Nikon A1R high-resolution confocal microscope (Nikon Instruments Inc., Melville, NY) and imaged with Nikon NIS elements software (v4.13.04) with a 60x oil objective lens (NA = 1.4) [21,22]. All image acquisition was performed in an investigator‐blinded manner.
Autophagic flux. For the assessment of autophagic flux, LC3B II protein levels were quantified with and without lysosomal inhibition for each treatment or genetic condition, as described previously [22]. To inhibit lysosomal function, media containing 50 nmol/L Bafilomycin A1 (Sigma) was added to the cells for 3 h; an equal volume of DMSO was used as a vehicle control.
Western blot analyses. Total proteins were extracted from NRC washed with phosphate-buffered saline (PBS) and lysed with Cell Lytic M lysis buffer (Sigma-Aldrich) supplemented with Complete Protease Inhibitor Cocktail (Roche) as described previously [15,21]. The lysed cells were homogenized using sonication and centrifuged at 12,000×g for 15 min to sediment any insoluble material. The protein content of the cardiomyocyte lysates was measured using the modified Bradford protocol/reagent relative to a BSA standard curve (BioRad). Protein lysates were separated on SDS-PAGE using pre-cast 7.5%–15% Criterion Gels (BioRad) and transferred to PVDF membranes (BioRad). Membranes were blocked for 1 h in 5% non-fat dried milk and exposed to primary antibodies overnight. The following primary antibodies were used for immunoblotting: Drp1 (1:500, 14647, Cell Signalling), OPA1 (1:500, 80471, Cell Signalling), MFN2 (1:1000, 9482, Cell Signalling), HSP60 (1:200, sc-13115, Santacruz), LC3B (1:500, 2775, Cell Signalling), p62 (1:500, GP62-C, Progen), OXPHOS (1:1000, ab110413, Abcam), PDH (1:1000, ab110416, Abcam), OMA1 (1:200, sc-515788, Santacruz), YME1L1 (1:1000, 11510-1-AP, Protein Tech), Atg7 (1:500, SAB4200304, Sigma), GAPDH (1:10000, MAB374, EMD Millipore), LonP1 (1:1000, SAB1411647, Sigma), ClpP (1:200, sc-271284, Santacruz), Paraplegin (1:200, sc-514393, Santacruz), HTRA2 (1:500, AF1458, R&D Systems), Rab24 (1:200, sc-136049, Santacruz), and VCP (1:500, 2648, Cell Signalling). Membranes were then washed, incubated with alkaline-phosphatase-conjugated secondary antibodies (Santa Cruz Biotechnology), developed with ECF reagent (Amersham), and imaged using a ChemiDoc™ Touch Imaging System (BioRad). Ponceau S protein staining on the transfer membrane was used as a loading control. Densitometry analysis of the protein bands on the scanned membrane images was carried out using ImageJ software (NIH, Bethesda, MD).
Statistics. Data are expressed as mean ± SEM. All cell culture experiments were repeated in two to three independent experiments. All statistical analysis was conducted in GraphPad Prism 8 software (version 8.4.1 (676)). Data were analyzed for normality using Kolmogorov-Smirnov test. Data passing the normality test were analyzed using two-tailed, unpaired Student's t-test (for 2 groups), one-way ANOVA (for 3 groups or more with one variable) and two- or three-way ANOVA (for 3 groups or more with more than one variable) followed by Tukey's post hoc test. Data failing the normality test were analyzed using Mann Whitney U test and Kruskal Wallis test followed by Bonferroni correction. P values less than 0.05 (95% confidence interval) were considered significant.
3. Results
3.1. Treatment of mdivi-1 decreases expression of the mitochondrial dynamics regulatory protein and suppresses respiration in cardiomyocytes
We performed a temporal study to determine the effects of mdivi-1 on mitochondrial fission and fusion events in cardiomyocytes by treating the neonatal rat primary cardiomyocytes (NRC) with mdivi-1 (100 μmol/L) or vehicle for an extended period of time (0, 3, 6, 12, or 24 h). We selected the concentration of mdivi-1 based on data from an earlier study showing that suppression of Drp1 by mdivi-1 (100 μmol/L) significantly increased the number of cardiomyocytes with elongated mitochondria [11] and also partially mimicked the effect of Drp1 downregulation in cardiomyocytes [11]. Interestingly, we observed a time-dependent decreased expression of Drp1 in cardiomyocytes following mdivi-1 treatment (Fig. 1A and B). Similarly, we also observed proteolytic cleavage of OPA1 isoforms (Fig. 1A and C) and decreased expression of MFN2 in cardiomyocytes after mdivi-1 treatment (Fig. 1A and D). A notable finding of our study is that mdivi-1 treatment proteolytically degrades L-OPA1 (a and b isoforms) in cardiomyocytes in a time dependent manner (Fig. 1A and C).
Fig. 1.
Temporal changes in mitochondrial dynamics regulatory protein expression by mdivi-1 treatment in cardiomyocytes. (A) Representative Western blot showing the temporal changes in the expression of mitochondrial dynamics regulatory proteins (Drp1, OPA1, and MFN2) in cardiomyocytes (NRC) treated mdivi-1 (100 μmol/L) or vehicle at different time points (3, 6, 12, or 24 h). Densitometric quantification showing the expression of (B) Drp1, (C) OPA1, and (D) MFN2 in cardiomyocytes. HSP60 was used as a house keeping protein for mitochondria and GAPDH was used as a loading control to confirm approximately equal loading across the samples (n = 4). Bars represent mean ± SEM. P values were determined by one-way ANOVA followed by Tukey's post hoc test. NS = non-significant.
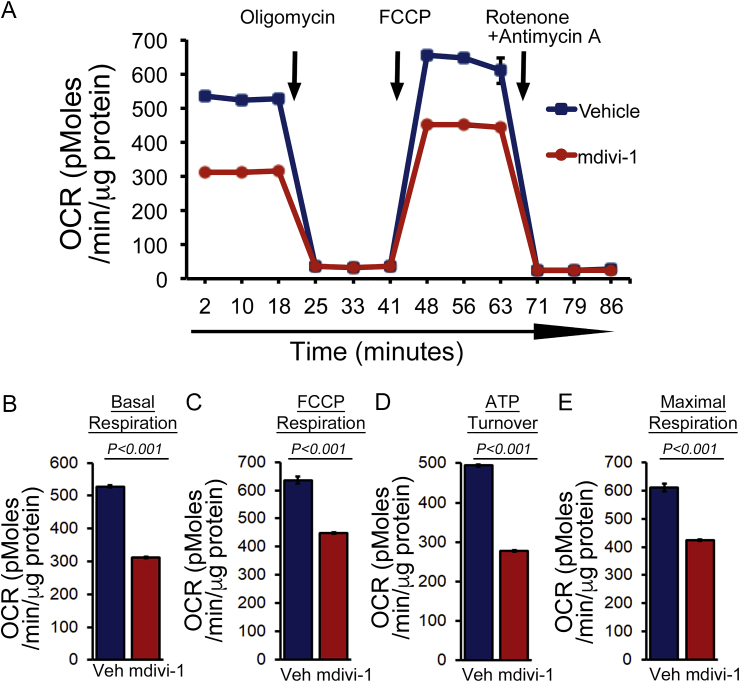
Altered mitochondrial dynamics have been shown to lead to mitochondrial dysfunction. To ascertain this, we treated the cardiomyocytes with mdivi-1 (100 μmol/L) or vehicle for 24 h and monitored mitochondrial respiration. Real‐time monitoring of the mitochondrial oxygen consumption rate (OCR) showed that basal respiration, representing the sum of all physiological mitochondrial oxygen consumption, was decreased in the mdivi-1-treated cardiomyocytes, indicating lower respiratory function compared to those in the vehicle group (Fig. 2A and B). We also observed a decreased FCCP-stimulated OCR. The addition of FCCP uncouples respiration from oxidative phosphorylation, allowing the measurement of maximal OCR, and providing evidence of lower overall mitochondrial activity (Fig. 2C). ATP turnover as measured by subtracting the ATP‐linked respiration from the basal OCR was significantly decreased by mdivi-1 treatment, indicating a minor mitochondrial contribution to the generation of ATP in these cardiomyocytes (Fig. 2D). The maximum respiration calculated by nonmitochondrial respiration subtracted from the FCCP‐stimulated OCR was also significantly lower in mdivi-1 treated cardiomyocytes (Fig. 2E). Overall, we found that treatment with mdivi-1 resulted in suppression of mitochondrial respiration, leading to functionally compromised mitochondria in cardiomyocytes.
Fig. 2.
Suppression of mitochondrial respiration by mdivi-1 in cardiomyocytes. (A) Mitochondrial oxygen consumption rate (OCR) profile measured in cardiomyocytes treated with mdivi-1 (100 μmol/L) or vehicle for 24 h. Arrows indicate sequential addition of oligomycin (1 μmol/L), FCCP ([carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone]; 4 μmol/L), and rotenone (0.5 μmol/L) plus antimycin A (0.5 μmol/L). OCR profiles are expressed as picomoles oxygen per minute per μg of protein. Key parameters of respiration including (B) basal respiration, (C) FCCP-linked respiration, (D) ATP turnover, and (E) maximal respiration were significantly reduced by mdivi-1 treatment (n = 5 wells per group). Bars represent mean ± SEM. P values were determined by two-tailed unpaired Student's t-test.
3.2. Treatment with mdivi-1 as well as Drp1 knockdown in cardiomyocytes alters the expression of both OXPHOS and PDH complex proteins
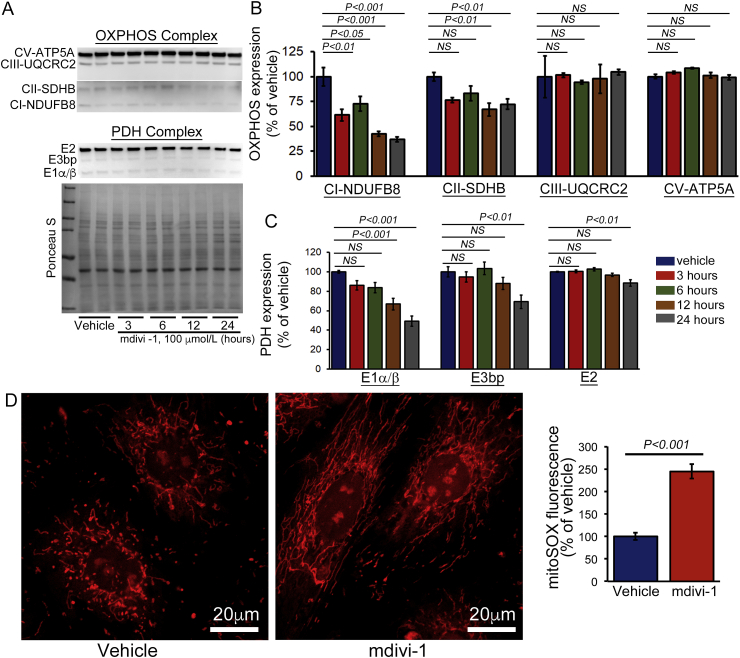
The decreased mitochondrial respiration may be the result of a defect in the electron transport chain (ETC) in the inner mitochondrial membrane. Therefore, we also observed the temporal changes in the expression of the oxidative phosphorylation system (OXPHOS) following mdivi-1 treatment in cardiomyocytes (Fig. 3A). We observed decreased expression of Complex I, and Complex II of the OXPHOS complex after mdivi-1 treatment in cardiomyocytes (Fig. 3A and B). Similar to OXPHOS, the expression of the E1α/β subunit protein of the pyruvate dehydrogenase (PDH) complex was significantly decreased by mdivi-1 treatment. Expression of the E3bp and E2 subunits of PDH complex proteins remain unchanged (Fig. 3A and C). Ponceau S staining of proteins was used to confirm equal loading. We also measured the effect of mdivi-1 on PDH complex expression in isolated rat and mouse adult cardiomyocytes. Similarly to neonatal cardiomyocytes, we observed altered expression of the components of the PDH complex following mdivi-1 treatment (12 h) (Supplementary Fig. S1). Collectively, we found that the defective mitochondrial respiration (Fig. 2) in the mdivi-1 treated cardiomyocytes was associated with altered expression of some components of OXPHOS and PDH complex proteins. We also measured the mdivi-1 induced generation superoxide in cardiomyocytes due to the decreased expression of Complex I. In fact, cardiac-specific complex I knockout mice (Ndufs4) had a decreased Complex I activity resulting in increased production of superoxide [25,26]. Primary fibroblasts isolated from Ndufs4 null mice displayed increased superoxide levels [27]. Measurement of the superoxide in mdivi-1- (for 24 h) or vehicle-treated cardiomyocytes using the MitoSOX Red staining showed a significantly increased level of superoxide generation by mdivi-1 (Fig. 3D).
Fig. 3.
Temporal changes in OXPHOS and PDH complex protein expression by mdivi-1 treatment in cardiomyocytes. (A) Representative Western blot images showing OXPHOS and PDH complex protein expression in vehicle or mdivi-1 treated (100 μmol/L) NRC at different time points (3, 6, 12, or 24 h). Densitometric quantification showing that mdivi-1 induced temporal changes in the expression of (B) OXPHOS complex proteins: Complex I, Complex II, Complex III, and Complex V, and (C) PDH complex proteins: E1α/β, E3bp, and E2. Ponceau S protein staining of the membrane after transfer was used as a loading control to confirm approximately equal loading across the gel (n = 8). (D) Representative fluorescence images of MitoSOX Red-stained cardiomyocytes to visualize mitochondrial superoxide production after mdivi-1 treatment. Data quantification for the fold change in mean fluorescence intensity/μm [2] with respect to the vehicle-treated group (n = 61 cells in vehicle and 85 in mdivi-1). Scale bars = 20 μm. Bars represent mean ± SEM. P values were determined by one-way ANOVA followed by Tukey's post hoc test. NS = non-significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
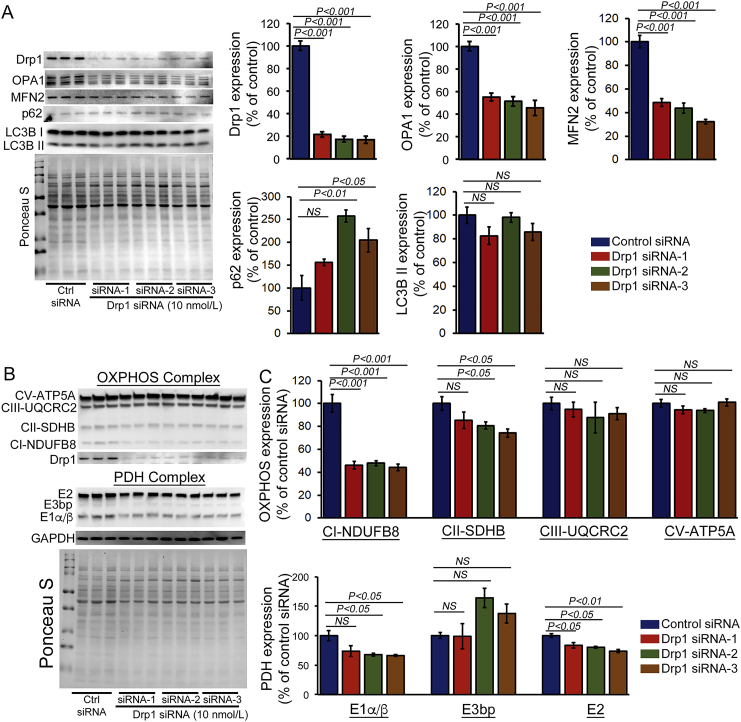
As an earlier study suggested that prolonged suppression of Drp1 by mdivi-1 (100 μmol/L) partially mimics the effect of Drp1 downregulation in cardiomyocytes [11], we also knocked down Drp1 in cardiomyocytes using siRNA transfection. We tested three different types of Drp1 siRNAs. Western blot analysis indicated 80% knockdown of endogenous Drp1 in NRC at 4 days post-transfection with 10 nmol/L Drp1 siRNA (Fig. 4A). Drp1 siRNA knockdown in cardiomyocytes resulted in decreased expression of fusion proteins OPA1 and MFN2. Despite the decreased expression, no proteolytic degradation of L-OPA was observed, unlike what was seen with mdivi-1-treated cardiomyocytes (Fig. 1). We also observed increased expression of autophagy regulatory adapter protein p62. Next, we tested the effects of Drp1 siRNA knockdown on the expression of OXPHOS and PDH complex proteins in cardiomyocytes (Fig. 4B and C). Similar to the effect of mdivi-1 in cardiomyocytes, all of these different Drp1 siRNAs showed a decreased expression of Complex I, and siRNA 2 and 3 showed a decreased level of Complex II (Fig. 4B and C). The expression of the E1α/β subunit protein of the PDH complex was also significantly decreased by Drp1 knockdown, similar to observations following treatment with mdivi-1. Ponceau S staining of proteins was used to confirm equal loading (Fig. 4B and C). Drp1 siRNA knockdown mediated altered expression of OXPHOS and PDH proteins, which suggests that mdivi-1 plays a direct role in Drp1 to alter the expression of these proteins.
Fig. 4.
Drp1 knockdown by siRNA in cardiomyocytes alters OXPHOS and PDH protein expression. (A) Representative Western blot and densitometric quantification showing siRNA knockdown of Drp1 (10 nmol/L) in cardiomyocytes. Western blot showing that siRNA knockdown of Drp1 in cardiomyocytes alters the expression of mitochondrial fusion regulatory proteins (Mfn2 and OPA1) and autophagy proteins (LC3bII and p62). (B) Representative Western blot and densitometric quantification showing the expression of OXPHOS and PDH complex proteins in Drp1 siRNA knockdown cardiomyocytes. Drp1 siRNA knockdown significantly decreased Complex I and Complex II of OXPHOS and E1α/β of PDH complex in cardiomyocytes. Ponceau S protein staining of the membrane after transfer was used as a loading control to confirm approximately equal loading across the gel (n = 6). Bars represent mean ± SEM. P values were determined by one-way ANOVA followed by Tukey's post hoc test. NS = non-significant.
3.3. Treatment with mdivi-1 inhibits macro-autophagy flux in cardiomyocytes
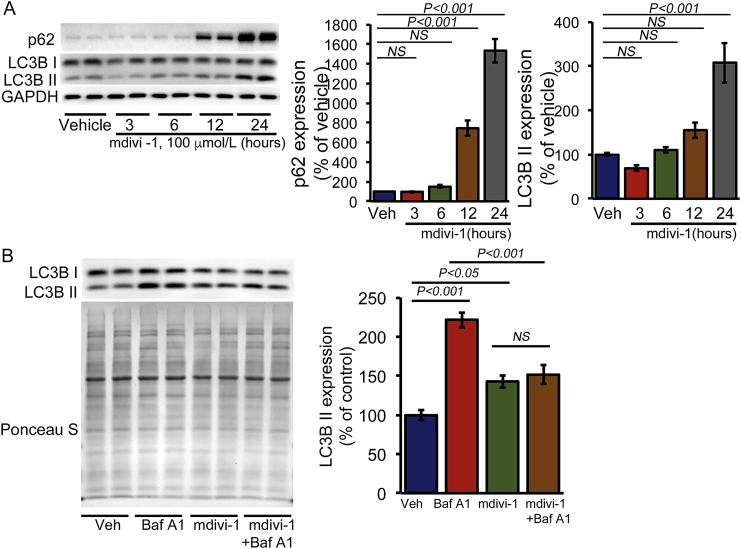
Evidence for Drp1-dependent regulation of macro-autophagy was demonstrated by the observation that Drp1 downregulation in cardiomyocytes significantly reduced Atg7-induced increases in autophagosomes and autolysosomes [11]. Our temporal study of autophagy regulatory protein expression showed gradual accumulation of p62 and LC3B II in cardiomyocytes (Fig. 5A). The mdivi-1-induced time-dependent increase in LC3B II expression indicated the accumulation of autophagosomes and autolysosomes resulting from either activation of autophagosome synthesis or blockade of a downstream autophagic degradation process [22]. To distinguish between these two possibilities, we performed an autophagy flux assay in mdivi-1- or vehicle-treated cardiomyocytes using Bafilomycin A1 (BafA1, 50 nmol/L for 3 h) to inhibit lysosomal degradation followed by determination of LC3B II levels using Western blot analysis (Fig. 5B). Treatment of vehicle-treated cardiomyocytes with BafA1 resulted in a significant increase in LC3B II levels, reflecting autophagic flux in cardiomyocytes under basal conditions. However, mdivi-1-treated cardiomyocytes demonstrated no increase in LC3B II levels following BafA1 treatment, suggesting that accumulation of autophagosomes/autolysosomes occurred due to inhibition of autophagic degradation (Fig. 5B). The autophagy flux assay showed inhibition of autophagic activity in mdivi-1-treated NRC.
Fig. 5.
Mdivi-1 inhibits autophagy flux in cardiomyocytes. (A) Representative Western blot and densitometric quantification of the gradual accumulation of autophagy regulatory proteins (p62 and LC3B) in cardiomyocytes (NRC) treated with mdivi-1 (100 μmol/L) or vehicle at different time points (3, 6, 12, or 24 h). GAPDH was used as a loading control to confirm approximately equal loading across the samples (n = 4). (B) Inhibition of autophagic flux by mdivi-1 (100 μmol/L, 24 h) in NRC examined by immunoblotting of LC3B II. Bafilomycin A1 (50 nmol/L) was added for 3 h to block lysosomal degradation. Ponceau S protein staining of the membrane after transfer was used as a loading control to confirm approximately equal loading across the gel (n = 4). Bars represent mean ± SEM. P values were determined by two-way ANOVA followed Tukey's post hoc test. NS = non-significant.
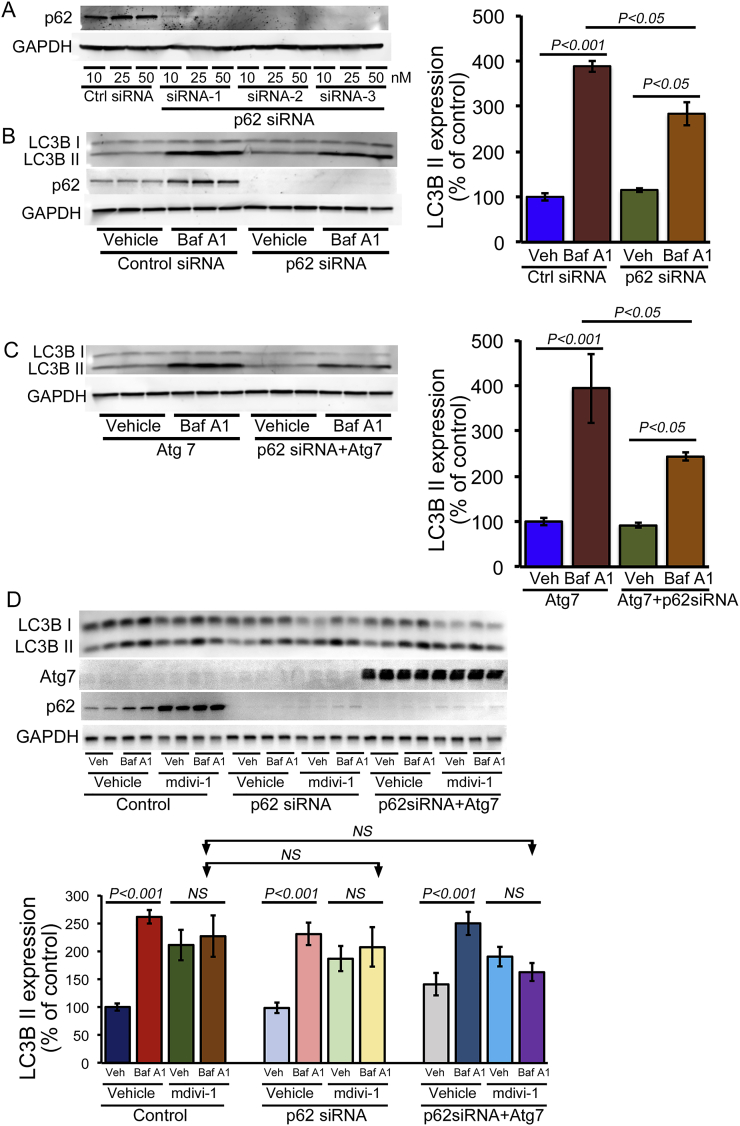
Next, we wanted to confirm whether mdivi-1 directly inhibits macro-autophagy, so we performed a rescue experiment by activating autophagy by overexpressing autophagy-related 7 (Atg7), a critical and rate-limiting autophagy protein [28]. We generated replication-deficient Atg7 adenovirus and confirmed expression levels in NRC by increasing concentrations of adenoviral infections (0–10 multiplicity of infection (MOI)) (Fig. 6A). The autophagy flux assay carried out in Atg7 overexpressed (10 MOI) NRC confirmed that Atg7 overexpression could induce autophagy activity in cardiomyocytes. Interestingly, Atg7 overexpression resulted in an Atg7 expression-dependent increased level of p62 expression in cardiomyocytes (Fig. 6B). To confirm the direct role of mdivi-1 in macro-autophagy, we treated Atg7 overexpressed cardiomyocytes with mdivi-1 (100 μmol/L) or vehicle and then performed the autophagy flux assay using BafA1. The autophagy flux assay showed that adenoviral overexpression of Atg7 in cardiomyocytes could partially rescue the mdivi-1-induced impaired autophagy flux (Fig. 6C). Interestingly, mdivi-1 treatment in Atg7 overexpressed cardiomyocytes showed further increased expression of p62 (Fig. 6C). Overall, these data suggest that mdivi-1 inhibits macro-autophagy, which can be partially rescued by autophagy activation.
Fig. 6.
Induction of autophagy flux by Atg7 overexpression in mdivi-1-treated cardiomyocytes. (A) Western blot showing adenovirus-mediated overexpression of Atg7 in cardiomyocytes at different MOI (0.1, 0.5, 1.0, 2.5, 5.0, and 10.0 MOI). Atg7 overexpression induced p62 expression in NRC, and GAPDH was used as a loading control (n = 3). (B) Induction of autophagic flux by Atg7 overexpression in NRC examined by immunoblotting of LC3B II. Bafilomycin A1 (50 nmol/L) was added for 3 h to block lysosomal degradation. GAPDH was used as a loading control to conform to approximately equal loading across the gel (n = 3). (C) Autophagic flux assay in Atg7 overexpressing cardiomyocytes with or without mdivi-1 treatment (100 μmol/L, 24 h) using BafA1. Representative Western blot and densitometric quantification showing that Atg7 overexpression in cardiomyocytes partially rescued mdivi-1-induced altered autophagy and also further induced p62 expression in NRC. Ponceau S protein staining of the membrane after transfer was used as a loading control to confirm approximately equal loading across the gel (n = 4). Bars represent mean ± SEM. P values were determined by three-way ANOVA followed Tukey's post hoc test. NS = non-significant.
As mdivi-1 treatment, as well as autophagy induction, led to increased p62 expression, we wanted to determine the role of p62 in mdivi-1-mediated impaired macro-autophagy. Therefore, we knocked down p62 in cardiomyocytes with siRNA transfection using three different types of p62 siRNAs. Western blot analysis indicated almost complete knockdown of endogenous p62 in NRC at 5 days post-transfection with p62 siRNA (10, 25, or 50 nmol/L) (Fig. 7A). Autophagy flux assays in p62 siRNA (10 nmol/L) knockdown NRC confirmed that p62 is required to induce autophagic activity in cardiomyocytes (Fig. 7B). Atg7 overexpression resulted in an increased level of p62 along with the activation of autophagy in cardiomyocytes. Therefore, we knocked down p62 using siRNA and then overexpressed Atg7 in cardiomyocytes to monitor the necessity for p62 in autophagy induction by Atg7. Interestingly, the autophagy flux assay showed decreased flux by the p62 siRNA knockdown in Atg7 overexpressing cardiomyocytes (Fig. 7C). Next, we determined the role of p62 in the effect of mdivi-1 on macro-autophagy, by treating cells with mdivi-1 (100 μmol/L) and performing autophagy flux in NRC with the p62 siRNA knockdown as well as in Atg7 overexpressing cardiomyocytes with the p62 siRNA knockdown. In fact, mdivi-1 treatment in p62 siRNA knockdown and p62 siRNA knockdown in Atg7 overexpressing cardiomyocytes prevented induction of autophagy flux (Fig. 7D). Therefore, p62 plays an important role in macro-autophagy activation as p62 siRNA knockdown abrogated the Atg7-induced autophagy flux in cardiomyocytes with or without mdivi-1.
Fig. 7.
p62 required for Atg7-mediated rescue of autophagy in mdivi-1-treated cardiomyocytes. (A) Representative Western blot showing p62 knockdown in cardiomyocytes by siRNA transfection at three different doses (10, 25, or 50 nmol/L). GAPDH was used as a loading control. (B) Decreased autophagic flux by p62 siRNA knockdown in NRC examined by immunoblotting of LC3B II. Bafilomycin A1 (50 nmol/L) was added for 3 h to block lysosomal degradation. GAPDH was used as a loading control to confirm approximately equal loading across the gel (n = 3). (C) Autophagy flux assay in p62 siRNA knockdown (10 nmol/L, siRNA 2) in Atg7 adenoviral (10 MOI) overexpressing cardiomyocytes showed decreased autophagy flux, indicating that p62 is partly required for Atg7-dependent activation of autophagic flux in cardiomyocytes. (D) Autophagy flux assay in p62 siRNA knockdown and p62 knockdown on Atg7 overexpressing cardiomyocytes with or without mdivi-1 treatment (100 μmol/L for 24 h). Representative Western blot and densitometric quantification showing that p62 knockdown was unable to induce autophagy in Atg7 overexpressing cardiomyocytes. Ponceau S protein staining of the membrane after transfer was used as a loading control to confirm approximately equal loading across the gel (n = 4). Bars represent mean ± SEM. P values were determined by three-way ANOVA followed Tukey's post hoc test. NS = non-significant.
3.4. Involvement of altered expression of mitochondrial serine proteases by mdivi-1 in L-OPA1 proteolytic processing
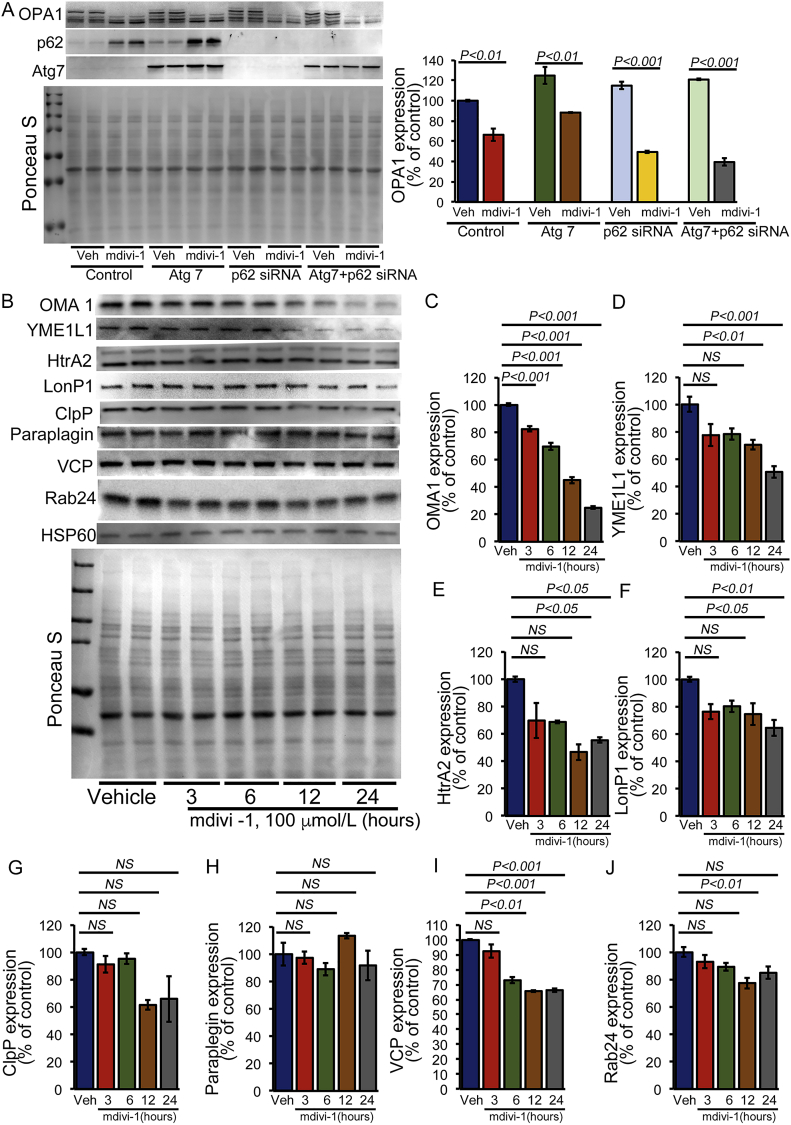
We found that induction of autophagy by Atg7 overexpression as well as p62 siRNA knockdown did not affect the mdivi-1-dependent proteolytic processing of L-OPA1 in cardiomyocytes (Fig. 8A). Treatment with mdivi-1 has been reported to significantly decrease mitochondrial membrane potential (ΔΨ) [29], and we found that it increased mitochondrial superoxide production. Both decreased membrane potential and increased superoxide activation in mitochondria have been reported to activate the serine proteases involved in OPA1 processing [30]. Therefore, we observed the temporal changes in the expression of mitochondrial serine proteases in different mitochondrial compartments, including the mitochondrial inner membrane (OMA1 and YME1L1), the intermembrane space (HtrA2), and the matrix (LonP1, ClpP, and Paraplegin) (Fig. 8B–J). The temporal study demonstrated the time-dependent decrease in the expression of all of these mitochondrial proteases except paraplegin (Fig. 8B–H). We also observed a significant decrease in the expression of AAA-ATPase VCP (valosin-containing protein), which is essential for outer mitochondrial membrane protein turnover [31], and small Rab GTPase Rab24 (Ras-related protein Rab-24), which is involved in mitochondrial fission and activation [32]. Temporal changes in expression of these mitochondrial serine proteases showed a significant decrease in the expression of OMA1 as early as 3 h after mdivi-1 treatment.
Fig. 8.
OPA1 proteolytic cleavage by mdivi-1 treatment associated with altered expression of mitochondrial proteases. (A) Representative Western blot and densitometric quantification showing that mdivi-1 (100 μmol/L, 24 h) induced proteolytic cleavage of L-OPA1 under the autophagy-modifying conditions induced by Atg7 overexpression, p62 siRNA knockdown, and p62 siRNA knockdown on Atg7 overexpression in cardiomyocytes. Ponceau S protein staining of the membrane after transfer was used as a loading control to confirm approximately equal loading across the gel (n = 4). (B) Representative Western blot showing that mdivi-1 (100 μmol/L) induced temporal changes in the expression of different mitochondrial proteases, ATPases, and GTPases in cardiomyocytes at different time points (3, 6, 12, or 24 h). Densitometric quantification showing that mdivi-1 treatment induced temporal changes in (C) OMA1, (D) YME1L1, (E) HtrA2, (F) LonP1, (G) ClpP1, (H) paraplegin, (I) VCP, and (J) Rab24. HSP60 was used as a house keeping protein for mitochondria and Ponceau S protein staining of the membrane after transfer was used as a loading control to confirm approximately equal loading across the gel (n = 4). Bars represent mean ± SEM. P values were determined by one-way ANOVA followed by Tukey's post hoc test.NS = non-significant.
Among these mitochondrial serine proteases, the proteolytic processing of L-OPA1 is mediated by the activity of two mitochondrial inner membrane proteases: OMA1 [7] and YME1L1 [6], both of which play a critical role in the regulation of mitochondrial dynamics. Our study showed a correlation between L-OPA proteolytic degradation and OMA1 processing, both of which decreased significantly as early as 3 h after mdivi-1 treatment. Studies have demonstrated that OMA1 and YME1L1 are activated by the loss of the mitochondrial membrane potential, alterations in the production of ROS, and alterations in cellular levels of ATP. Depolarization of the mitochondrial membrane potential as observed by treatment with the ionophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) results in the loss of L-OPA1 in an OMA1-dependent manner [7]. Similar to the effect of mdivi-1 in cardiomyocytes, CCCP (10 μmol/L) treatment of cardiomyocytes decreases the L-OPA breakdown in a time-dependent manner (Fig. 9A). In contrast, the treatment of cardiomyocytes with antimycin A (AA) (10 μmol/L), which also decreased the mitochondrial membrane potential, did not result in L-OPA processing (Fig. 9B). Earlier studies also showed that the addition of antimycin A (AA) (10 μmol/L) did not efficiently stimulate L-OPA1 processing by OMA1 [33]. Next, we exposed the mdivi-1-treated cardiomyocytes (100 μmol/L, 24 h) to CCCP (10 μmol/L, 3 h) or AA (10 μmol/L, 3 h). Interestingly, both the group treated with CCCP plus mdivi-1 and the group treated with AA-plus mdivi-1 showed proteolytic cleavage of L-OPA1 at a level similar to that observed in the groups treated with either CCCP or mdivi-1 alone. Therefore, addition of either CCCP or AA did not further change the expression of OPA1 compared to results seen in the group treated with mdivi-1 alone. Interestingly, the decreased expression of OMA1 and YME1L1 in these groups was similar to that of the mdivi-1-treated group (Fig. 9D). We used the OMA1 and YME1L1 temporal expression data with mdivi-1 treatment (Fig. 8C and D) as a control group for the effect of CCCP and AA in mdivi-1-treated cardiomyocytes in Fig. 9D. Overall, our data suggest that mdivi-1 may activate OMA1 by autocatalytic proteolysis of OMA1, resulting in OMA1-mediated L-OPA degradation by a mechanism similar to that observed with CCCP [30].
Fig. 9.
Depolarization of the mitochondrial membrane by midivi-1 associated with L-OPA1 proteolytic degradation. (A, B) Representative Western blot showing changes in expression of OPA1, MFN2, and Drp1 in CCCP (10 μmol/L for 1 or 3 h) or vehicle-treated cardiomyocytes. GAPDH was used as a loading control (n = 4). (B) Representative Western blot showing changes in expression of OPA1, MFN2, and Drp1 in Antimycin A (AA) (10 μmol/L for 1 or 3 h) or vehicle-treated cardiomyocytes. GAPDH used as a loading control (n = 4). (C) Representative Western blot and densitometric quantification showing changes in expression levels of OPA1, LC3BII, and p62 in cardiomyocytes treated with mdivi-1 (100 μmol/L for 24 h) followed by CCCP (10 μmol/L for 3 h) or AA (10 μmol/L for 3 h). GAPDH was used as a loading control. (D) Representative Western blot and densitometric quantification showing temporal changes in expression levels of OMA1 and YME1L1 (data from Fig. 8 used as a control) as well as cardiomyocytes treated with mdivi-1 (100 μmol/L, 24 h) along with CCCP (10 μmol/L, 3 h) or AA (10 μmol/L, 3 h). Ponceau S protein staining of the membrane after transfer was used as a loading control to confirm approximately equal loading across the gel (n = 4). Bars represent mean ± SEM. P values were determined by one-way ANOVA followed by Tukey's post hoc test. NS = non-significant.
4. Discussion
In our study, we made the following novel findings: (1) Mdivi-1 treatment in primary neonatal cardiomyocytes decreased Drp1 expression in a time-dependent manner, proteolytically cleaved L-OPA1, and decreased MFN2 expression. Along with the altered mitochondrial dynamics in regulatory protein expression, mdivi-1 treatment results in decreased mitochondrial respiration in cardiomyocytes associated with the decreased expression of Complex I and Complex II of OXPHOS and the E1α/β subunit of the PDH complex. The altered expression of OXPHOS and the PDH complex proteins may be directly associated with decreased Drp1 expression as Drp1 siRNA knockdown in cardiomyocytes showed similar effects. (2) Mdivi-1 increased the accumulation of autophagy protein p62 and LC3B II in a time- and dose-dependent manner. Autophagy flux assay showed autophagy flux impaired by mdivi-1, which can be restored by Atg7 overexpression in cardiomyocytes. However, p62 is required for Atg7 overexpression-induced rescue of mdivi-1-mediated impaired autophagy flux. (3) Mdivi-1 dependent proteolytic processing of L-OPA is associated with increased mitochondrial oxidative stress and altered expression of mitochondrial serine/proteases. Overall, our data show the pleiotropic effects of mdivi-1 in the impairment of mitochondrial respiration and OXPHOS, proteolytic processing of L-OPA, impairment of autophagic activity, and alteration of mitochondrial serine/proteases in cardiomyocytes.
Along with the decreased Drp1 expression in cardiomyocytes, we observed time-dependent proteolytic cleavage in L-OPA and also decreased expression of MFN2. We found that mdivi-1 treatment decreased the expression of Drp1 in cardiomyocytes in a time-dependent manner. In fact, suppression of Drp1 by mdivi-1 (50 or 100 μmol/L) has been shown to significantly increase the number of cardiomyocytes with elongated mitochondria [11]. A higher dose (100 μmol/L) combined with prolonged treatment with mdivi-1 (50 μmol/L every 24 h for 1 week) partially mimics the effect of Drp1 downregulation in cardiomyocytes [11]. Similarly, a recent study showed that mdivi-1 treatment (12.5–100 μmol/L) reduced the expression in a dose-dependent manner of both full-length Drp1 and phosphorylated Drp1, reduced mitochondrial fission, and enhance mitochondrial fusion activity in N2a cells [34]. Moreover, mdivi-1-treated N2a cells showed significantly reduced mitochondrial number and increased length of mitochondria, probably because of reduced GTPase Drp1 enzymatic activity [34]. Despite data supporting the inhibition of mitochondrial fission, studies have also reported that mdivi-1 treatment of COS-7 cells and primary neurons exhibited no effect on the steady-state mitochondrial morphology [19]. Mdivi-1 showed no effect on the mitochondrial size of cortical neurons, nor did it show any effect on mitochondrial morphology following prolonged incubation (1, 2, 6, or 24 h) on COS-7 cells. Additionally, it was unable to prevent mitochondrial fragmentation induced by staurosporine [19]. Moreover, several studies reported that the effects of mdivi-1 were independent of DRP1 inhibition or mitochondrial fusion induction, including alteration of apoptosis, cell division, and viability [29]. Treatment with mdivi-1 has been reported to significantly decreased mitochondrial membrane potential (ΔΨ) in cholangiocarcinoma cell lines [29]. In addition, mdivi-1 retards apoptosis by inhibiting mitochondrial outer membrane permeabilization by blocking Bid-activated Bax/Bak-dependent cytochrome c release from mitochondria [12]. Studies have suggested that mdivi-1 directly regulates mitochondrial outer membrane permeabilization independent of Drp1-mediated division [12]. In several cancer cell lines, mdivi-1 treatment decreased cancer cell proliferation, increasing cell death and apoptosis through the repression of oxidative metabolism, which was independent of DRP1 inhibition or mitochondrial fusion induction [35]. On the other hand, treatment with mdivi-1 also prevented the apoptosis induced by ischemia-reperfusion injury in primary hippocampal cells via inhibition of a reactive oxygen species-activated mitochondrial pathway [36]. All of these studies demonstrate the varied as well as opposing effects of mdivi-1 under different pathological and experimental settings. Therefore, extensive studies focusing on the effects of mdivi-1 in healthy cells are required to understand and minimize the off target and long term effects associated with its use clinically in treatment.
We found that mdivi-1 significantly reduces mitochondrial respiration in cardiomyocytes. Mitochondrial inner membrane ETC participates in oxidative phosphorylation and produces ATP. The reduced respiration is associated with the altered OXPHOS (Complex I and II) and PDH (E1α/β) complex protein expression as the result of treatment with mdivi-1 in cardiomyocytes. Similar to the mdivi-1-mediated altered expression of OXPHOS and the PDH complex, the siRNA knockdown of Drp1 in cardiomyocytes showed a similar trend suggesting a Drp1-dependent effect. A recent study showed mdivi-1-mediated impairment of mitochondrial respiration, suggesting that mdivi-1 reversibly inhibits mitochondrial Complex I-dependent oxygen consumption [19]. The superoxide generation by mdivi-1 may involve the decreased expression of Complex I as supported by a study showing that cardiac-specific Complex I knockout mice (Ndufs4) had a decrease of ~50% in Complex I activity resulting in increased production of superoxide [25,26]. Similar to our findings, MitoSOX staining quantification in MCF7 cells revealed that exposure to different concentrations of mdivi-1 significantly increased mitochondrial superoxide production compared to that seen in vehicle-treated cells [37]. mdivi-1 has also been shown to impair DNA replication and repress mitochondrial respiration independent of Drp1 in multidrug-resistant tumor cells [20]. In contrast, mdivi-1 (10 μmol/L) treatment, as well as exposure to Drp1 siRNA and dominant-negative Drp1 in human neuronal SK cells subjected to high-glucose conditions (50 mmol/L), increased Complex I activity and mitochondrial density [18]. Moreover, treatment with mdivi-1 (25 mg/kg or 10 mg/kg once daily for 2 weeks) resulted in significantly increased mitochondrial content, Complex I enzymatic activity, and ATP levels in the db/db mouse hippocampus [18]. All of these studies demonstrate context- and cell type-dependent molecular functions of mdivi-1 mediated through Drp1 dependent/independent pathways.
Drp1 plays an essential role in mediating Parkin-induced mitophagy in mouse embryonic fibroblast (MEF) cells [38] and Bnip3-induced mitophagy in adult cardiomyocytes [39]. Apart from the mitophagy, Drp1's dependent regulation of macro-autophagy is evident by the finding that Drp1 downregulation in cardiomyocytes significantly reduced Atg7-induced increases in autophagosomes and autolysosomes [11]. In fact, we found that mdivi-1 treatment reduced the Atg7-induced autophagy flux and also induced accumulation of p62 in cardiomyocytes. We found that p62 plays an important role in autophagy activation as p62 siRNA knockdown abrogated the Atg7 induced autophagy flux in cardiomyocytes with or without mdivi-1. Studies support the idea that Drp1 may regulate macro-autophagy through regulation of the Bcl-xL: Beclin 1 interaction [11]. Drp1 has been shown to physically interact with Bcl-xL in neurons [40] and Bcl-xL is known to inhibit autophagy through its binding to Beclin1 [41]. A recent study using coimmunoprecipitation assays also confirmed that Drp1physically interacts with Bcl-2 and Bcl-xL in cardiomyocytes. Using Drp1 overexpression and knockdown in cardiomyocytes, they found that the downregulation of Drp1 augmented the physical interaction between Beclin1 and Bcl-2 or Bcl-xL, whereas increased expression of Drp1 inhibited the interaction [11]. These studies suggest that mdivi-1-mediated inhibition/downregulation of Drp1 may inhibit autophagy through a Bcl-xL-dependent mechanism, most likely by enhancing interaction between Beclin1 and Bcl-xL. However, further studies are required to dissect the molecular mechanism and role of mdivi-1 to differentially regulate macro-autophagy/mitophagy.
Another novel finding of our study is that mdivi-1 treatment proteolytically degraded L-OPA in a time-dependent manner. We found no effect of autophagy activation or inhibition on mdivi-1-induced L-OPA breakdown. Interestingly, we found a gradual decrease in the expression of key serine/proteases residing in the mitochondrial intermembrane space (HtrA2), inner membrane (YME1L1, and OMA1), and matrix (LonP1 and ClpP). Notably, among these mitochondrial serine proteases, the proteolytic processing of OPA1 is mediated by the activity of two mitochondrial inner membrane proteases, including YME1L1 [6] and OMA1 [7]. Studies have demonstrated that cells treated with the protonophore CCCP depolarize mitochondria and reduce cellular ATP, which activates the OMA1 protease involving autocatalytic processing of the OMA1 protease [42], allowing rapid processing of OMA1 substrates such as OPA1 [7,43]. YME1L1 degradation is induced by cellular insults that depolarize mitochondria and deplete ATP through a mechanism involving the mitochondrial protease OMA1 [44]. The decreased YME1L1 may increase cellular sensitivity to subsequent oxidative stress, leading to the development of mitochondrial dysfunction through alterations in mitochondrial proteostasis and quality control [44]. In fact, we observed mdivi-1-induced proteolytic degradation of L-OPA similar to that observed following CCCP treatment in cardiomyocytes. However, mdivi-1 treatment, along with treatment with CCCP or AA, did not further aggravate the OPA1 processing or the expression of OMA1 and YME1L1 in cardiomyocytes. Therefore, the mdivi-1-induced proteolytic processing of L-OPA1 isoforms through differential stress sensitivities of YME1L1 and OMA1 may be an adaptive process developed to maintain mitochondrial morphology.
A potential limitation of the pleiotropic effect of mdivi-1 observed in this study is that it is confined to primary neonatal cardiomyocytes whose physiology differs from that of adult cardiomyocytes and other cell types. Moreover, the molecular mechanism responsible for OPA1/MFN2 altered expression in Drp1 siRNA knockdown cardiomyocytes remains unknown. Future studies are needed to define the molecular signature associated with mdivi-1's mitochondrial division inhibition-independent effects, using comprehensive and unbiased proteomics, RNAseq, and metabolomics studies. Though the disappearance of L-OPA1 caused by midivi-1 suggests proteolytic cleavage by proteases, it is not known whether mdivi-1 also affects L-OPA synthesis or the molecular mechanisms associated with it. However, mdivi-1 provides a very useful tool to study mitochondrial dynamics and our study provides evidence for some new mitochondrial fission-independent effects of mdivi-1 that need to be extensively characterized. Despite these limitations, our findings of the pleiotropic effects of mdivi-1 in altering mitochondrial dynamics, respiration, and autophagy in cardiomyocytes provides increased evidence for caution in their therapeutic use in clinical settings.
In summary, we found some novel pleiotropic effects of mdivi-1 in cardiomyocytes including: 1) decreased expression of OXPHOS complex protein and superoxide production, leading to impaired mitochondrial respiration; 2) impairment of macro-autophagy flux, which can be rescued by autophagy activation, which requires p62 for induction; and 3) an association between L-OPA proteolytic degradation and mitochondrial proteases, including OMA1 and YME1L1, which are known to involved in this process. Although mdivi-1-mediated decreased mitochondrial membrane potential (ΔΨ) [29] and increased superoxide production may be responsible for activating these proteases, further studies are required to dissect the molecular mechanism responsible for the mdivi-1-induced alterations and/or activations of mitochondrial proteases.
Author contributions
R.A. and M.S.B. conceptualized, designed, and wrote the manuscript. R.A., S.A., C.S.A., M.M., S.N., and S.M. performed all experiments and participated in analyses. M.P. and C.G.K. contributed to the analytic tools and reagents used. All co-authors edited and proofread the manuscript and approved the final version.
Sources of funding
This work was supported by the National Institutes of Health grants: HL122354 and HL145753 to M.S.B.; LSUHSC-S CCDS Finish Line Award and Feist Weiller Cancer Center IDEA Grant to M.S.B.; P20GM121307 to C.G.K.; HL141998 to S.M.; AA025744 and AA026708 to M.P.; AHA Postdoctoral Fellowship to S.A.; LSUHSC-S Malcolm Feist and AHA Postdoctoral Fellowship to C.S.A.; and LSUHSC-S Malcolm Feist Pre-doctoral Fellowship to R.A.
Declaration of competing interest
All authors declare no conflicts of interest.
Acknowledgments
We would like to thank the COBRE research core for technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101660.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gao J., Wang L., Liu J., Xie F., Su B., Wang X. Abnormalities of mitochondrial dynamics in neurodegenerative diseases. Antioxidants (Basel) 2017;6(2) doi: 10.3390/antiox6020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loson O.C., Song Z., Chen H., Chan D.C., Fis1 Mff. MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 2013;24(5):659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Dotto V., Mishra P., Vidoni S., Fogazza M., Maresca A., Caporali L., McCaffery J.M., Cappelletti M., Baruffini E., Lenaers G., Chan D., Rugolo M., Carelli V., Zanna C. OPA1 isoforms in the hierarchical organization of mitochondrial functions. Cell Rep. 2017;19(12):2557–2571. doi: 10.1016/j.celrep.2017.05.073. [DOI] [PubMed] [Google Scholar]
- 4.Akepati V.R., Muller E.C., Otto A., Strauss H.M., Portwich M., Alexander C. Characterization of OPA1 isoforms isolated from mouse tissues. J. Neurochem. 2008;106(1):372–383. doi: 10.1111/j.1471-4159.2008.05401.x. [DOI] [PubMed] [Google Scholar]
- 5.MacVicar T., Langer T. OPA1 processing in cell death and disease - the long and short of it. J. Cell Sci. 2016;129(12):2297–2306. doi: 10.1242/jcs.159186. [DOI] [PubMed] [Google Scholar]
- 6.Song Z., Chen H., Fiket M., Alexander C., Chan D.C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007;178(5):749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J.C., Westermann B., Rugarli E.I., Langer T. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 2009;187(7):1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quiros P.M., Ramsay A.J., Sala D., Fernandez-Vizarra E., Rodriguez F., Peinado J.R., Fernandez-Garcia M.S., Vega J.A., Enriquez J.A., Zorzano A., Lopez-Otin C. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 2012;31(9):2117–2133. doi: 10.1038/emboj.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong S.B., Subrayan S., Lim S.Y., Yellon D.M., Davidson S.M., Hausenloy D.J. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121(18):2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- 10.Wu S., Zhou F., Zhang Z., Xing D. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J. 2011;278(6):941–954. doi: 10.1111/j.1742-4658.2011.08010.x. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda Y., Shirakabe A., Maejima Y., Zhai P., Sciarretta S., Toli J., Nomura M., Mihara K., Egashira K., Ohishi M., Abdellatif M., Sadoshima J. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ. Res. 2015;116(2):264–278. doi: 10.1161/CIRCRESAHA.116.303356. [DOI] [PubMed] [Google Scholar]
- 12.Cassidy-Stone A., Chipuk J.E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M.J., Shaw J.T., Hinshaw J.E., Green D.R., Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 2008;14(2):193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosdah A.A., J K.H., Delbridge L.M., Dusting G.J., Lim S.Y. Mitochondrial fission - a drug target for cytoprotection or cytodestruction? Pharmacol. Res. Perspect. 2016;4(3) doi: 10.1002/prp2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith G., Gallo G. To mdivi-1 or not to mdivi-1: is that the question? Dev. Neurobiol. 2017;77(11):1260–1268. doi: 10.1002/dneu.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam S., Abdullah C.S., Aishwarya R., Miriyala S., Panchatcharam M., Peretik J.M., Orr A.W., James J., Robbins J., Bhuiyan M.S. Aberrant mitochondrial fission is maladaptive in desmin mutation-induced cardiac proteotoxicity. J. Am. Heart Assoc. 2018;7(14) doi: 10.1161/JAHA.118.009289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharp W.W., Fang Y.H., Han M., Zhang H.J., Hong Z., Banathy A., Morrow E., Ryan J.J., Archer S.L. Dynamin-related protein 1 (Drp1)-mediated diastolic dysfunction in myocardial ischemia-reperfusion injury: therapeutic benefits of Drp1 inhibition to reduce mitochondrial fission. Faseb. J. 2014;28(1):316–326. doi: 10.1096/fj.12-226225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gharanei M., Hussain A., Janneh O., Maddock H. Attenuation of doxorubicin-induced cardiotoxicity by mdivi-1: a mitochondrial division/mitophagy inhibitor. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0077713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S., Wang Y., Gan X., Fang D., Zhong C., Wu L., Hu G., Sosunov A.A., McKhann G.M., Yu H., Yan S.S. Drp1-mediated mitochondrial abnormalities link to synaptic injury in diabetes model. Diabetes. 2015;64(5):1728–1742. doi: 10.2337/db14-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bordt E.A., Clerc P., Roelofs B.A., Saladino A.J., Tretter L., Adam-Vizi V., Cherok E., Khalil A., Yadava N., Ge S.X., Francis T.C., Kennedy N.W., Picton L.K., Kumar T., Uppuluri S., Miller A.M., Itoh K., Karbowski M., Sesaki H., Hill R.B., Polster B.M. The putative Drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev. Cell. 2017;40(6):583–594 e6. doi: 10.1016/j.devcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian W., Wang J., Roginskaya V., McDermott L.A., Edwards R.P., Stolz D.B., Llambi F., Green D.R., Van Houten B. Novel combination of mitochondrial division inhibitor 1 (mdivi-1) and platinum agents produces synergistic pro-apoptotic effect in drug resistant tumor cells. Oncotarget. 2014;5(12):4180–4194. doi: 10.18632/oncotarget.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alam S., Abdullah C.S., Aishwarya R., Orr A.W., Traylor J., Miriyala S., Panchatcharam M., Pattillo C.B., Bhuiyan M.S. Sigmar1 regulates endoplasmic reticulum stress-induced C/EBP-homologous protein expression in cardiomyocytes. Biosci. Rep. 2017;37(4) doi: 10.1042/BSR20170898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdullah C.S., Alam S., Aishwarya R., Miriyala S., Bhuiyan M.A.N., Panchatcharam M., Pattillo C.B., Orr A.W., Sadoshima J., Hill J.A., Bhuiyan M.S. Doxorubicin-induced cardiomyopathy associated with inhibition of autophagic degradation process and defects in mitochondrial respiration. Sci. Rep. 2019;9(1):2002. doi: 10.1038/s41598-018-37862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdullah C.S., Alam S., Aishwarya R., Miriyala S., Panchatcharam M., Bhuiyan M.A.N., Peretik J.M., Orr A.W., James J., Osinska H., Robbins J., Lorenz J.N., Bhuiyan M.S. Cardiac dysfunction in the Sigma 1 receptor knockout mouse associated with impaired mitochondrial dynamics and bioenergetics. J. Am. Heart Assoc. 2018;7(20) doi: 10.1161/JAHA.118.009775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wojtala A., Bonora M., Malinska D., Pinton P., Duszynski J., Wieckowski M.R. Methods to monitor ROS production by fluorescence microscopy and fluorometry. Methods Enzymol. 2014;542:243–262. doi: 10.1016/B978-0-12-416618-9.00013-3. [DOI] [PubMed] [Google Scholar]
- 25.Pitkanen S., Robinson B.H. Mitochondrial complex I deficiency leads to increased production of superoxide radicals and induction of superoxide dismutase. J. Clin. Invest. 1996;98(2):345–351. doi: 10.1172/JCI118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chouchani E.T., Methner C., Buonincontri G., Hu C.H., Logan A., Sawiak S.J., Murphy M.P., Krieg T. Complex I deficiency due to selective loss of Ndufs4 in the mouse heart results in severe hypertrophic cardiomyopathy. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0094157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valsecchi F., Grefte S., Roestenberg P., Joosten-Wagenaars J., Smeitink J.A., Willems P.H., Koopman W.J. Primary fibroblasts of NDUFS4(-/-) mice display increased ROS levels and aberrant mitochondrial morphology. Mitochondrion. 2013;13(5):436–443. doi: 10.1016/j.mito.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Bhuiyan M.S., Pattison J.S., Osinska H., James J., Gulick J., McLendon P.M., Hill J.A., Sadoshima J., Robbins J. Enhanced autophagy ameliorates cardiac proteinopathy. J. Clin. Invest. 2013;123(12):5284–5297. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tusskorn O., Khunluck T., Prawan A., Senggunprai L., Kukongviriyapan V. Mitochondrial division inhibitor-1 potentiates cisplatin-induced apoptosis via the mitochondrial death pathway in cholangiocarcinoma cells. Biomed. Pharmacother. 2019;111:109–118. doi: 10.1016/j.biopha.2018.12.051. [DOI] [PubMed] [Google Scholar]
- 30.Baker M.J., Lampe P.A., Stojanovski D., Korwitz A., Anand R., Tatsuta T., Langer T. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 2014;33(6):578–593. doi: 10.1002/embj.201386474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu S., Peng G., Wang Y., Fang S., Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol. Biol. Cell. 2011;22(3):291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seitz S., Kwon Y., Hartleben G., Jülg J., Sekar R., Krahmer N., Najafi B., Loft A., Gancheva S., Stemmer K., Feuchtinger A., Hrabe de Angelis M., Müller T.D., Mann M., Blüher M., Roden M., Berriel Diaz M., Behrends C., Gilleron J., Herzig S., Zeigerer A. Hepatic Rab24 controls blood glucose homeostasis via improving mitochondrial plasticity. Nat. Metabol. 2019;1(10):1009–1026. doi: 10.1038/s42255-019-0124-x. [DOI] [PubMed] [Google Scholar]
- 33.Ishihara N., Jofuku A., Eura Y., Mihara K. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem. Biophys. Res. Commun. 2003;301(4):891–898. doi: 10.1016/s0006-291x(03)00050-0. [DOI] [PubMed] [Google Scholar]
- 34.Manczak M., Kandimalla R., Yin X., Reddy P.H. Mitochondrial division inhibitor 1 reduces dynamin-related protein 1 and mitochondrial fission activity. Hum. Mol. Genet. 2019;28(2):177–199. doi: 10.1093/hmg/ddy335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai W., Wang G., Chwa J., Oh M.E., Abeywardana T., Yang Y., Wang Q.A., Jiang L. Mitochondrial division inhibitor (mdivi-1) decreases oxidative metabolism in cancer. Br. J. Canc. 2020;122(9):1288–1297. doi: 10.1038/s41416-020-0778-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J., Wang P., Li S., Wang S., Li Y., Liang N., Wang M. Mdivi-1 prevents apoptosis induced by ischemia-reperfusion injury in primary hippocampal cells via inhibition of reactive oxygen species-activated mitochondrial pathway. J. Stroke Cerebrovasc. Dis. 2014;23(6):1491–1499. doi: 10.1016/j.jstrokecerebrovasdis.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 37.Peiris-Pages M., Bonuccelli G., Sotgia F., Lisanti M.P. Mitochondrial fission as a driver of stemness in tumor cells: mDIVI1 inhibits mitochondrial function, cell migration and cancer stem cell (CSC) signalling. Oncotarget. 2018;9(17):13254–13275. doi: 10.18632/oncotarget.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., Youle R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191(7):1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y., Lee H.Y., Hanna R.A., Gustafsson A.B. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 2011;301(5):H1924–H1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., Alavian K.N., Lazrove E., Mehta N., Jones A., Zhang P., Licznerski P., Graham M., Uo T., Guo J., Rahner C., Duman R.S., Morrison R.S., Jonas E.A. A Bcl-xL-Drp1 complex regulates synaptic vesicle membrane dynamics during endocytosis. Nat. Cell Biol. 2013;15(7):773–785. doi: 10.1038/ncb2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marquez R.T., Xu L. Bcl-2:Beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am. J. Canc. Res. 2012;2(2):214–221. [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K., Li H., Song Z. Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage. EMBO Rep. 2014;15(5):576–585. doi: 10.1002/embr.201338240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rainbolt T.K., Lebeau J., Puchades C., Wiseman R.L. Reciprocal degradation of YME1L and OMA1 adapts mitochondrial proteolytic activity during stress. Cell Rep. 2016;14(9):2041–2049. doi: 10.1016/j.celrep.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rainbolt T.K., Saunders J.M., Wiseman R.L. YME1L degradation reduces mitochondrial proteolytic capacity during oxidative stress. EMBO Rep. 2015;16(1):97–106. doi: 10.15252/embr.201438976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.