Abstract
Background
The number of coronavirus disease 2019 (COVID-19) cases has rapidly increased all over the world. Specific information about immunity in non-survivors with COVID-19 is scarce. This study aimed to analyse the clinical characteristics and abnormal immunity of the confirmed COVID-19 non-survivors.
Methods
In this single-centered, retrospective, observational study, we enrolled 125 patients with COVID-19 who were died between January 13 and March 4, 2020 in Renmin Hospital of Wuhan University. A total of 414 randomly recruited patients with confirmed COVID-19 who were discharged from the same hospital during the same period served as control. The demographic, clinical characteristics and laboratory findings at admission, and treatment used in these patients were collected. The immunity-related risk factors associated with in-hospital death were tested by logistic regression models and Receiver Operating Characteristic (ROC) curve.
Results
Non-survivors (70 years, IQR: 61.5–80) were significantly older than survivors (54 years, IQR: 37–65) (P < 0.001). 56.8% of non-survivors was male. Nearly half of the patients (44.9%) had chronic medical illness. In non-survivors, hypertension (49.6%) was the most common comorbidity, followed by diabetes (20.0%) and coronary heart disease (16.0%). The common signs and symptoms at admission of non-survivors were fever (88%), followed by cough (64.8%), dyspnea (62.4%), fatigue (62.4%) and chest tightness (58.4%). Compared with survivors, non-survivors had higher white blood cell (WBC) count (7.85 vs 5.07 × 109/L), more elevated neutrophil count (6.41 vs 3.08 × 109/L), smaller lymphocyte count (0.69 vs 1.20 × 109/L) and lower platelet count (172 vs 211 × 109/L), raised concentrations of procalcitonin (0.21 vs 0.06 ng/mL) and CRP (70.5 vs 7.2 mg/L) (P < 0.001). This was accompanied with significantly decreased levels of CD3+ T cells (277 vs 814 cells/μl), CD4+ T cells (172 vs 473 cells/μl), CD8+ T cells (84 vs 262.5 cells/μl, P < 0.001), CD19+ T cells (88 vs 141 cells/μl) and CD16+ 56+ T cells (79 vs 128.5 cells/μl) (P < 0.001). The concentrations of immunoglobulins (Ig) G (13.30 vs 11.95 g/L), IgA (2.54 vs 2.21 g/L), and IgE (71.30 vs 42.25 IU/ml) were increased, whereas the levels of complement proteins (C)3 (0.89 vs 0.99 g/L) and C4 (0.22 vs 0.24 g/L) were decreased in non-survivors when compared with survivors (all P < 0.05). The non-survivors presented lower levels of oximetry saturation (90 vs 97%) at rest and lactate (2.40 vs 1.90 mmol/L) (P < 0.001). Old age, comorbidity of malignant tumor, neutrophilia, lymphocytopenia, low CD4+ T cells, decreased C3, and low oximetry saturation were the risk factors of death in patients with confirmed COVID-19. The frequency of CD4+ T cells positively correlated with the numbers of lymphocytes (r = 0.787) and the level of oximetry saturation (r = 0.295), Whereas CD4+ T cells were negatively correlated with age (r =-0.323) and the numbers of neutrophils (r = − 0.244) (all P < 0.001).
Conclusions
Abnormal cellular immunity and humoral immunity were key features of non-survivors with COVID-19. Neutrophilia, lymphocytopenia, low CD4+ T cells, and decreased C3 were immunity-related risk factors predicting mortality of patients with COVID-19.
Keywords: COVID-19, Cellular immunity, Humoral immunity, Mortality
Introduction
Coronavirus disease 2019 (COVID-19) has declared as a pandemic on 12 March, 2020. The pathogen has been identified as a novel enveloped RNA beta-coronavirus that is currently known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, also called 2019-nCoV) [1]. Like severe acute respiratory syndrome (SARS) coronavirus and Middle East respiratory syndrome (MERS) coronavirus, SARS-CoV-2 is a highly pathogenic coronavirus to human. The virus has high transmission capability as well as high morbidity and mortality [2–4]. By April 22, 2020, the acute respiratory disease has affected more than 200 countries around the world. Globally, 2 553 853 cases have been confirmed, with 176 323 deaths have died from this viral infection [5]. However, little is still known about the factors that lead to death by this coronavirus.
Recent reports show that the severely ill patients with confirmed COVID-19 may develop dyspnea and hypoxemia within 1 week after the onset of disease, which may quickly progress to acute respiratory distress syndrome (ARDS) or end-organ failure [6, 7]. The general epidemiological features and clinical characteristics of patients with COVID-19 have been previously reported [2, 4, 8]. However, these studies were based on relatively small sample sizes, specific characterizations of the abnormal immunity of non-survivors with COVID-19 have not yet been well described. In this study, we aimed to explore the clinical characteristics and immunity features in patients with COVID-19 from a hospital in Wuhan, China and evaluate the immunity-related risk factors associated with patient death.
Method
Patients’ involvement and data collection
In this retrospective, single center study, a total of 125 patients who died from confirmed COVID-19 between January 13 and March 4, 2020 in Renmin Hospital of Wuhan University, the designated hospital for treating severe patients with COVID-19 in Wuhan, China. A total of 414 randomly recruited patients with confirmed COVID-19 who were discharged from the same hospital during the same period served as control. All patients were confirmed of COVID-19 according to the World Health Organization interim guidance [9] and Diagnosis and Treatment Guideline for Novel Coronavirus Pneumonia (Trial Version 7.0) [10], and were positive for at least two nucleic acid tests for SARS-CoV-2. The confirmed COVID-19 patients were classified as severe or non-severe cases based on the American Thoracic Society guidelines for community-acquired pneumonia [11]. The patients who received systemic corticosteroid treatment before admission were excluded. The criteria for patient discharge included absence of fever for at least 3 days, substantial improvement in both lungs in chest computed tomography (CT), clinical remission of respiratory symptoms, and negative results for two throat-swab samples obtained at least 24 h apart for SARS-CoV-2 RNA testing.
This study was approved by the Research Ethics Commission of Renmin Hospital of Wuhan University (No.WDRY2020-K143), and written informed consent was waived by the Research Ethics Commission.
Demographic and clinical characteristics (including comorbidities, signs and symptoms), laboratory findings, chest CT scan results and treatment were extracted from electronic medical records system of Renmin Hospital of Wuhan University and analysed by three independent researchers. The access was granted by the director of the hospital. The date of disease onset and admission date, as well as the severity of COVID-19, were also recorded. None of these cases have been previously reported.
Laboratory testing
Patient pharyngeal swab specimens were collected for SARS-CoV-2 nucleic acid detection using real-time reverse transcriptase-polymerase chain reaction (RT-PCR) assay with a standard procedure recommended by Chinese Center for Disease Control and Prevention (China CDC). The viral nucleic acid testing for all patients was performed by the clinical laboratory from Renmin Hospital of Wuhan University, a certified laboratory for SARS-CoV-2 testing. Other laboratory tests at admission, included white blood cell (WBC), neutrophil, lymphocyte, platelet, procalcitonin (PCT), C-reactive protein (CRP), CD3+ T cells, CD4+ T cells, CD8+ T cells, CD19+ T cells, CD16+ 56+ T cells, immunoglobulins (Ig) G, IgM, IgA, IgE, complement proteins (C)3, C4, oximetry saturation, and lactate, were performed in the same hospital and results were retrieved from electronic medical records. Some cytokines such as interleukin (IL)-6, interferon-γ, IL-10 and other cytokines, were not performed in all patients.
Statistical analysis
All statistical analyses were performed using SPSS 24.0 software (SPSS Inc., Chicago, USA). Categorical variables were expressed with frequencies and percentages. Continuous variables were presented as median (interquartile range [IQR]). Means for continuous variables were compared with independent-samples t tests when the data were normally distributed; if not, the Mann-Whitney U test was used. Proportions for categorical variables were compared with the chi-square and Fisher’s exact test. Univariable logistic regression between basic disease or different parameter and demise was performed. Moreover, the main risks related with demise were examined using multivariable logistic regression models adjusted for potential confounders. Receiver Operating Characteristic (ROC) curve model was also used to predict death efficacy. Spearman’s correlation test was used for calculation of correlation between different factors. Statistical significance was determined at P < 0.05.
Results
Demographic and clinical characteristics
A total of 539 patients with confirmed COVID-19 (125 non-survivors and 414 survivors) with comprehensive medical records were included in this study. All patients had pneumonia with abnormal findings on their chest CT scan. The median age of all patients was 58.0 years (IQR: 43.0–69.0), ranging from 16 years to 97 years.
Non-survivors (70 years, IQR: 61.5–80) were significantly older than survivors (54 years, IQR: 37–65) (P < 0.001). Of all COVID-19 patients, the majority was female (52.7%). More than half of non-survivors were male (56.8%). Nearly half of the COVID-19 patients (44.9%) had chronic medical illness, with hypertension being the most common comorbidity, followed by diabetes, coronary heart disease and malignant tumour. In non-survivors, hypertension (49.6%) was the most common comorbidity, followed by diabetes (20.0%) and coronary heart disease (16.0%). The median interval from illness onset to death was 7.0 days (IQR: 4.0–11.0), which is markedly lower than the median time from illness onset to discharge 15 days (IQR: 9.0–21.0). The common signs and symptoms at admission of non-survivors were fever (88%), followed by cough (64.8%), dyspnea (62.4%), fatigue (62.4%) and chest tightness (58.4%).
As for clinical classification, the patients were divided into severe group and non-severe groups. Severe patients (98.4%) were more likely to die compared with non-severe patients and nearly all of non-survivors were classified as severe cases at admission. Almost all the non-survivors received antibiotic treatment (94.4%). Both groups of patients were given antiviral therapy or Chinese Medicine, such as Lianhua Qingwen, oseltamivir, arbidol, and lopinavir/ritonavir. Intravenous human immunoglobulin and corticosteroid were used more frequently in non-survivors than survivors (Table 1).
Table 1.
Demographics, clinical characteristics and treatment of patients with COVID-19
| Total (539) | Non-survivor (125) | Survivor (414) | P value | |
|---|---|---|---|---|
| Age (years) | 58 (43–69) | 70 (61.5–80) | 54 (37–65) | < 0.0001 |
| Gender | 0.015 | |||
| Male | 255 (47.3%) | 71 (56.8%) | 184 (44.4%) | |
| Female | 284 (52.7%) | 54 (43.2%) | 230 (55.6%) | |
| Chronic medical illness | 242 (44.9%) | 88 (70.4%) | 154 (37.2%) | < 0.0001 |
| Chronic obstructive lung disease | 22 (4.1%) | 12 (9.6%) | 10 (2.4%) | < 0.0001 |
| Diabetes | 58 (10.9%) | 25 (20.0%) | 34 (8.2%) | < 0.0001 |
| Hypertention | 140 (26.0%) | 62 (49.6%) | 78 (18.8%) | < 0.0001 |
| Coronary heart disease | 37 (6.9%) | 20 (16.0%) | 17 (4.1%) | < 0.0001 |
| Cerebrovascular diseases | 19 (3.5%) | 15 (12.0%) | 4 (1.0%) | < 0.0001 |
| Hepatitis | 11 (2.0%) | 3 (2.4%) | 8 (1.9%) | 0.746 |
| Malignant tumour | 23 (4.3%) | 11 (8.8%) | 12 (2.9%) | 0.0042 |
| Chronic kidney disease | 8 (1.5%) | 6 (4.8%) | 2 (0.5%) | 0.0005 |
| Immunodeficiency disease | 2 (0.4%) | 0 (0.0%) | 2 (0.5%) | 0.436 |
| Time from illness onset to death or discharge, days | 13 (7–20) | 7 (4–11) | 15 (9–21) | < 0.0001 |
| Signs and symptoms at admission | ||||
| Fever | 420 (77.9%) | 110 (88%) | 310 (74.9) | 0.001 |
| Rhinorrhoea | 6 (1.1%) | 2 (1.6%) | 4 (1.0%) | 0.945 |
| Cough | 332 (61.6%) | 81 (64.8%) | 251 (60.6%) | 0.402 |
| Expectoration | 161 (29.9%) | 48 (38.4%) | 113 (27.3%) | 0.017 |
| Chest tightness | 187 (34.7%) | 73 (58.4%) | 114 (27.5%) | < 0.0001 |
| Dyspnea | 187 (34.7%) | 78 (62.4%) | 98 (23.7%) | < 0.0001 |
| Fatigue | 261 (48.4%) | 78 (62.4%) | 183 (44.2%) | < 0.0001 |
| Nausea | 9 (1.7%) | 5 (4.0%) | 4 (1.0%) | 0.02 |
| Diarrhea | 58 (10.8%) | 8 (6.4%) | 50 (12.1%) | 0.073 |
| Clinical Classification | < 0.0001 | |||
| Non-severe | 133 (24.7%) | 2 (1.6%) | 131 (31.6%) | |
| Severe | 406 (75.3%) | 123 (98.4%) | 283 (68.4%) | |
| Treatment | ||||
| Antibiotic drug | 374 (69.4%) | 118 (94.4%) | 256 (61.8%) | < 0.0001 |
| Antiviral therapy or Chinese Medicine | 225 (41.7%) | 58 (46.4%) | 167 (40.3%) | 0.228 |
| Lianhua Qingwen | 301 (55.8%) | 40 (32.0%) | 261 (63.0%) | < 0.0001 |
| Oseltamivir | 157 (29.1%) | 48 (38.4%) | 109 (26.3%) | 0.0092 |
| Arbidol | 370 (68.6%) | 65 (52.0%) | 305 (73.7%) | < 0.0001 |
| Lopinavir/ritonavir | 32 (5.9%) | 18 (14.4%) | 14 (3.4%) | < 0.0001 |
| Human immunoglobulin | 250 (38.4%) | 91 (72.8%) | 159 (38.4%) | < 0.0001 |
| Alpha-interferon | 170 (31.5%) | 34 (27.2%) | 136 (32.9%) | 0.233 |
| Thymosin | 59 (10.9%) | 20 (16.0%) | 39 (9.4%) | 0.039 |
| Glucocorticoids | 249 (46.2%) | 89 (71.2%) | 160 (38.6%) | < 0.0001 |
Data are median (IQR) and n (%). P values were calculated by Mann-Whitney U test, χ2 test, or Fisher’s exact test, as appropriate. Statistical significance was determined at P<0.05
COVID-19 Coronavirus disease 2019
Blood routine examination, cellular immunity, humoral immunity, and other laboratory findings in non-survivors with COVID-19
Although WBC and platelet count of both non-survivors and survivors were in normal range, the non-survivors had higher WBC count (7.85 × 109/L vs 5.07 × 109/L), more elevated neutrophil count (6.41 × 109/L vs 3.08 × 109/L), smaller lymphocyte count (0.69 × 109/L vs 1.20 × 109/L) and lower platelets count (172 × 109/L vs 211 × 109/L) than survivors (P < 0.001).
Increased levels of infection-related biomarkers, including PCT (0.21 vs 0.06 ng/ml) and CRP (70.5 vs 7.2 mg/L) were found in non-survivors when compared with those of survivors (P < 0.001).
The levels of CD3+ T cells (277 vs 814 cells/μl), CD4+ T cells (172 vs 473 cells/μl), CD8+ T cells (84 vs 262.5 cells/μl, P < 0.001), CD19+ T cells (88 vs 141 cells/μl) and CD16+ 56+ T cells (79 vs 128.5 cells/μl) were significantly decreased in non-survivors when compared with survivors (P < 0.001).
The serum concentrations of IgG (13.30 vs 11.95 g/L, P < 0.001), IgA (2.54 vs 2.21 g/L, P = 0.012), and IgE (71.30 vs 42.25 IU/ml, P < 0.001) were increased, whereas the serum levels of C3 (0.89 vs 0.99 g/L, P < 0.001) and C4 (0.22 vs 0.24 g/L, P = 0.001) were decreased in non-survivors when compared with survivors. The non-survivors presented lower levels of resting oximetry saturation (90 vs 97%, P < 0.001) and lactate (2.40 vs 1.90 mmol/L, P < 0.001) (Table 2). IL-6 concentrations test was just performed in only 135 patients, and the available data illustrated that the levels of IL-6 in non-survivors were significantly higher than survivors (P < 0.001) (Table 3).
Table 2.
Laboratory findings of patients with COVID-19
| Normal Range | Total (539) | No-survivor (125) | Survivor (414) | P value | |
|---|---|---|---|---|---|
| WBC (× 109/L) | 3.5–9.5 | 5.41 (4.1–7.59) | 7.85 (4.74–12.01) | 5.07 (3.96–6.76) | < 0.001 |
| Neutrophils (× 109/L) | 1.8–6.3 | 3.54 (2.42–5.72) | 6.41 (3.77–10.97) | 3.08 (2.29–4.63) | < 0.001 |
| Lymphocytes (× 109/L) | 1.1–3.2 | 1.04 (0.76–1.5) | 0.69 (0.42–0.93) | 1.20 (0.88–1.63) | < 0.001 |
| Haemoglobin (g/L) | 115–150 | 126 (115–138) | 127 (113.5–139.5) | 126 (116–137) | 0.761 |
| Platelets (× 109/L) | 125–350 | 205 (151–261) | 172 (118–235) | 211 (163–271) | < 0.001 |
| CRP (mg/L) | 0–10 | 16.9 (5–61.3) | 70.5 (39.95–133.5) | 7.2 (5–37.95) | < 0.001 |
| PCT (ng/mL) | < 0.1 | 0.07 (0.04–0.25) | 0.21 (0.11–0.95) | 0.06 (0.03–0.12) | < 0.001 |
| CD3+ T cells (cells/μl) | 723–2737 | 667 (356–1010) | 277 (163.5–430) | 814 (516–1088) | < 0.001 |
| CD4+ T cells (cells/μl) | 404–1612 | 392 (200–586) | 172 (99.5–267.5) | 473 (291–657.75) | < 0.001 |
| CD8+ T cells (cells/μl) | 220–1129 | 221 (104–366) | 84 (39.5–155.5) | 262.5 (163–405.25) | < 0.001 |
| CD19+T cells (cells/μl) | 80–616 | 132 (82–202) | 88 (52–151) | 141 (96–219) | < 0.001 |
| CD16+ 56+ T cells (cells/μl) | 84–724 | 114 (70–190) | 79 (39–144) | 128.5 (79–210) | < 0.001 |
| IgG (g/L) | 7.0–16.0 | 12.2 (10.3–14.6) | 13.30 (10.75–16.95) | 11.95 (10.18–13.93) | < 0.001 |
| IgM (g/L) | 0.4–2.3 | 0.95 (0.70–1.3) | 0.94 (0.70–1.26) | 0.95 (0.69–1.31) | 0.658 |
| IgA (g/L) | 0.7–4.0 | 2.26 (1.74–3) | 2.54 (1.81–3.46) | 2.21 (1.73–2.89) | 0.012 |
| IgE (IU/ml) | < 100 | 52.3 (18.3–133) | 71.30 (30.2–214.5) | 42.25 (18.3–120) | < 0.001 |
| C3 (g/L) | 0.9–1.8 | 0.97 (0.82–1.1) | 0.89 (0.74–1.04) | 0.99 (0.84–1.13) | < 0.001 |
| C4 (g/L) | 0.1–0.4 | 0.241 (0.18–0.32) | 0.22 (0.159–0.30) | 0.24 (0.19–0.32) | 0.001 |
| Oximetry saturation (%) | 95–100 | 97 (92–98) | 90 (83–94) | 97 (95–98) | < 0.001 |
| Lactate (mmol/L) | 0.5–1.5 | 2 (1.5–2.53) | 2.40 (1.9–3.65) | 1.90 (1.43–2.38) | < 0.001 |
Data are median (IQR). WBC White blood cell, CRP C-reactive protein, PCT Procalcitonin, Ig immunoglobulins, C Complement proteins. P values were calculated by Mann-Whitney U test, χ2 test, or Fisher’s exact test, as appropriate. Statistical significance was determined at P<0.05
Table 3.
IL-6 concentrations of patients with COVID-19
| Normal Range | Total (135) | No-survivor (33) | Survivor (102) | P value | |
|---|---|---|---|---|---|
| IL-6 (mg/L) | ≤ 20.0 | 7.22 (5.06–22.5) | 60.72 (16.89–146.53) | 5.98 (4.63–12.12) | < 0.001 |
Data are median (IQR)
P values were calculated by Mann-Whitney U test. Statistical significance was determined at P<0.05
Associations between immunity-related indexes and death risk of COVID-19 patients
Univariable logistic regression analysis indicated that old age, comorbidity of malignant tumor, neutrophilia, lymphocytopenia, increased CRP levels, lower CD4+ T cell count, and decreased C3 levels were associated with death (Table 4).
Table 4.
Univariable logistic Regression of death risk of patients with COVID-19
| Wald | P | OR | 95% CI | |
|---|---|---|---|---|
| Age | 27.347 | <0.001 | 0.912 | 0.881–0.944 |
| Malignant tumour | 4.563 | 0.023 | 0.176 | 0.036–0.866 |
| Neutrophils | 5.239 | 0.022 | 0.881 | 0.790–0.982 |
| Lymphocytes | 7.674 | 0.003 | 0.109 | 0.032–0.186 |
| CRP | 4.052 | 0.044 | 0.992 | 0.984–1.000 |
| CD4+ T cells | 9.147 | 0.002 | 1.005 | 1.002–1.009 |
| C3 | 8.033 | 0.005 | 40.209 | 3.326–528.035 |
OR Odds ratio, CI Confidence interval, CRP C-reactive protein, C Complement proteins, COVID-19 Coronavirus disease 2019
Statistical significance was determined at P<0.05
Generalized linear model showed that old age, comorbidity of malignant tumor, neutrophilia, lymphocytopenia, lower CD4+ T cells, decreased C3 and lower oximetry saturation was positively with the risk of death (Table 5).
Table 5.
Generalised linear model of patients with COVID-19
| Wald Chi-Sqare | P | Standard error | |
|---|---|---|---|
| Age | 25.182 | <0.001 | 0.0009 |
| Malignant tumour | 24.070 | <0.001 | 0.0638 |
| Neutrophils | 9.813 | 0.002 | 0.0062 |
| Lymphocytes | 5.831 | 0.016 | 0.0357 |
| CD4+ T cells | 3.602 | 0.048 | 0.00268 |
| C3 | 6.928 | 0.008 | 0.0291 |
| Oximetry saturation | 9.555 | 0.002 | 0.0017 |
C Complement proteins, COVID-19 Coronavirus disease 2019
Statistical significance was determined at P<0.05
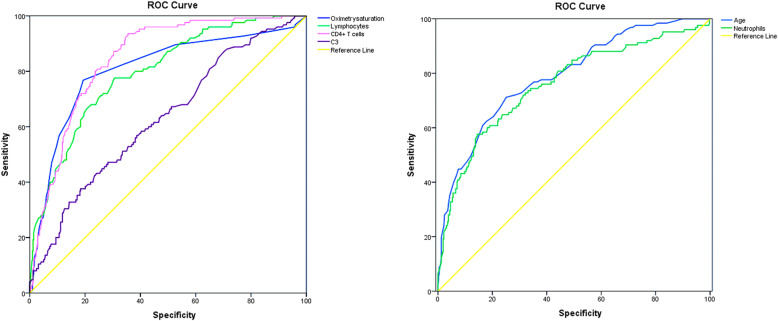
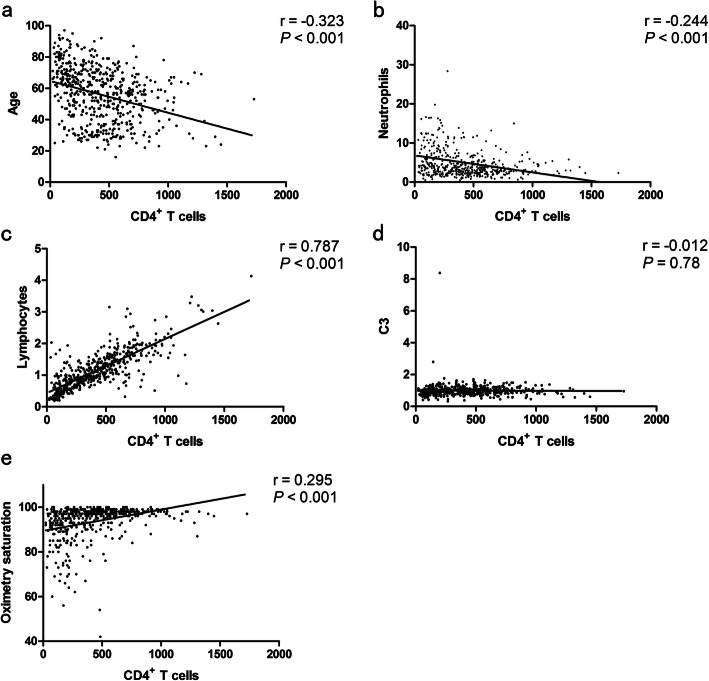
Further, we conducted ROC curve analysis to evaluate the role of these factors in predicting death efficacy. The area under the ROC curve (AUC) was 0.792 (confidence interval [CI]: 0.764–0.837) for age, 0.761(95% CI: 0.709–0.812) for neutrophils, 0.797 (95% CI: 0.753–0.840) for lymphocytes, 0.821 (95% CI: 0.782–0.860) for CRP, 0.848 (95% CI: 0.814–0.883) for CD4+ T cells, 0.630 (95% CI: 0.574–0.685) for C3, and 0.808 (95% CI: 0.759–0.856) for oximetry saturation. Additionally, the cutoff value was 64.5 for age, 5.835 for neutrophils, 0.945 for lymphocytes, 31.4 for CRP, 380.5 for CD4+ T cells, 0.809 for C3, and 94.5 oximetry saturation (Table 6, Fig. 1). The abundance of CD4+ T cells was positively correlated with the numbers of lymphocytes (r = 0.787, P < 0.001) and the level of oximetry saturation (r = 0.295, P < 0.001), respectively. Whereas it was negatively correlated with age (r = -0.323, P < 0.001) or the numbers of neutrophils (r = − 0.244, P < 0.001). However, CD4+ T cell abundance showed no significant correlation with C3 levels (P = 0.78) (Fig. 2).
Table 6.
ROC curve model of patients with COVID-19
| Area | Std.error | P | 95% CI lower | 95% CI upper | Cut off | |
|---|---|---|---|---|---|---|
| Age | 0.792 | 0.023 | 0.000 | 0.764 | 0.837 | 64.5 |
| Neutrophils | 0.761 | 0.026 | 0.000 | 0.709 | 0.812 | 5.835 |
| Lymphocytes | 0.797 | 0.22 | 0.000 | 0.753 | 0.840 | 0.945 |
| CRP | 0.821 | 0.020 | 0.000 | 0.782 | 0.860 | 31.4 |
| CD4+ T cells | 0.848 | 0.018 | 0.000 | 0.814 | 0.883 | 380.5 |
| C3 | 0.630 | 0.028 | 0.000 | 0.574 | 0.685 | 0.809 |
| Oximetry saturation | 0.808 | 0.25 | 0.000 | 0.759 | 0.856 | 94.5 |
ROC Receiver Operating Characteristic, CI Confidence interval, CRP C-reactive protein, C Complement proteins, COVID-19 Coronavirus disease 2019
Statistical significance was determined at P<0.05
Fig. 1.
ROC curve model of age, neutrophils, lymphocytes, CD4+ T cells, C3 and Oximetry saturation in patients with COVID-19.
ROC: Receiver operating characteristic; C: Complement proteins; COVID-19: Coronavirus disease 2019
Fig. 2.
Correlation between CD4+ T cells and age.
(a); CD4+ T cells and neutrophils (b); CD4+ T cells and lymphocytes (c); CD4+ T cells and C3 (d); CD4+ T cells and Oximetry saturation (e) of COVID-19 patients. Spearman’s test was used to evaluate the correlation. Statistical significance was determined at P<0.05. ROC: Receiver operating characteristic; C: Complement proteins; COVID-19: Coronavirus disease 2019
Discussion
In this study, we showed that the non-survivors with confirmed COVID-19 had higher WBC count, neutrophilia, lymphocytopenia, and lower platelet level when compared with survivors at admission. Abnormal immune responses, represented by dysregulation of numerous cellular and humoral immunity markers were found in non-survivors. Old age, comorbidity of malignant tumor, neutrophilia, lymphocytopenia, low CD4+ T cells, decreased C3, and low oximetry saturation were positively correlated with the risk of death in patients with COVID-19.
In this study, we found that non-survivors with confirmed COVID-19 were older, especially above 64.5 years, and more than half were male. Additionally, nearly half of the patients had chronic medical illness with comorbidity of malignant tumor being the risk of death. Previous reports have demonstrated age-dependent defects in T-cell and B-cell function, and excessive production of type 2 cytokines could cause deficiency in control of viral replication and prolonged proinflammatory responses, potentially leading to poor outcome [12]. Furthermore, chronic comorbidities often compromise immune system [13]. In agreement with these previous findings, our data suggested that COVID-19 is more likely to cause death in older men with chronic comorbidities than other patients [14].
In this study, we showed that severely ill patients were more likely to die compared with the non-severe patients even if they were given integrated therapy and that almost all non-survivors were severely ill at admission. Meanwhile, the non-survivors had higher WBC count, neutrophilia, lymphocytopenia and lower platelet levels. In particular, neutrophilia and lymphocytopenia act as markers of high death risks of patients with COVID-19. This is consistent with previous findings showing that severe patients with COVID-19 display higher neutrophil count and lower lymphocyte count during the period of diseases [13, 15] and that neutrophilia and lymphocytopenia are significantly associated with higher risks of the development of ARDS [6]. It has been showed that phagocytosis, release of granular contents, and secretion of cytokines are important effector functions of stimulated neutrophils, suggesting a protective immunity against the virus [16]. However, excessive elevated neutrophils can lead to severe pneumonia and death [16], which have been found in patients with SARS [17, 18] and MERS [19]. As for lymphocytes, it has been reported that the functional exhaustion of cytotoxic lymphocytes is associated with SRAS-CoV-2 infection, suggesting that SARS-CoV-2 infection may break down the antiviral immunity at an early stage [20]. Lymphocytopenia was also found in the MERS cases. Specifically, MERS-CoV can directly infect human primary T lymphocytes and induce T-cell apoptosis through extrinsic and intrinsic apoptosis pathways [21]. Therefore, it is possible that neutrophilia and lymphocytopenia are related, at least in part, to the development of critical illness, with a high mortality rate in the confirmed COVID-19 patients. Future studies to identify the mechanisms underlying these abnormal immune features will enhance our understandings of this disease.
It is well known that subsets of lymphocytes play important roles in the maintenance of immune system function. Here, our results indicated that the numbers of CD3+ T cells, CD4+ T cells, CD8+ T cells, B cells (CD19+), and natural killer (NK) cells (CD16+CD56) were significantly decreased in non-survivors at admission, compared with survivors. Meanwhile, increased serum concentrations of IgG, IgA, and IgE and decreased serum levels of C3 and C4 were found in non-survivors at admission. Taken together, these findings suggested that SARS-CoV-2 infection may impair lymphocytes, especially T lymphocytes, and the humoral immune system on the onset of the disease. The reduced abundance of CD4+ T cells, B cells, and NK cell count is likely a result of the rapidly propagating virus [22]. To mount an antiviral response, the innate immune system recognizes molecular structures that are produced by the invasion of the virus. Helper T (Th) cells orchestrate the overall adaptive response, while cytotoxic T cells are essential for killing of viral infected cells. Humoral immune response plays a protective role by limiting infection at later phase and prevents reinfection in the future. Therefore, SARS-CoV-2 infection can activate both cellular immune response and humoral immune responses in humans [23]. The uncontrolled inflammatory innate responses and impaired adaptive immune responses may lead to harmful tissue damage, both locally and systemically [24]. Patients who survived SARS-CoV and MERS-CoV infections usually had better immune responses than non-survivors [25].
It has been reported that CD4+ T cells and CD8+ T cells play a key antiviral roles by balancing the combat against pathogens and the risk of developing autoimmunity or overwhelming inflammation [26]. Depletion of CD4+ T cells, but not CD8+ T cells, leads to an enhanced immune-mediated interstitial pneumonitis and delayed clearance of SARS-CoV from the lungs in a SARS-CoV-infected mouse model, demonstrating the vital role of CD4+ T cells in preventing SARS-CoV infection [27]. A previous report has shown that higher number of CD4+ T cell may protect patients from developing ARDS in SARS-CoV infection [28]. In our study, we showed that the number of CD4+ T cells positively correlated with the numbers of lymphocytes and the level of oximetry saturation and negatively correlated with age and the numbers of neutrophils in non-survivors. Furthermore, low CD4+ T cell counts was a risk factor of death in patients with confirmed COVID-19. Together, our results provided an evidence showing that decreased number of CD4+ T cells is an immunity-related risk factor predicting death rate of patients with COVID-19. Similar to these findings, autopsy of patients with COVID-19 showed that the counts of peripheral CD4 and CD8 T cells were substantially reduced, which accounts for, in part, the severe immune injury in this patient [29]. High levels of pro-inflammatory cytokines released form CD4+ T cells have been observed in SARS-CoV and MERS-CoV infections as well, suggesting that the cytokine storm may contribute to disease severity [19]. Additionally, recruitment of immune cell populations in the patient’s blood was observed before the resolution of symptoms [30].
Although measurements of IL-6 levels have been performed in only some patients, our results indicated that non-survivors also had higher IL-6 levels as compared to survivors, raising the possibility that abnormal IL-6 levels may also contribute to in-hospital death [14]. IL-6 is a pro-inflammatory regulator of T cells. It can promote differentiation of naïve T cells towards Th17 cell lineage, inhibit the induction of regulatory T-cells (Tregs) [31] Additionally, Th17 cells can induce an inflammatory response through the production of IL-17A and IL-17F [32], which act as key cytokines for neutrophils migration, recruitment and activation [33]. It has been reported that activated neutrophils can phagocytosis, release granular contents, and produce cytokines, therefore contributing to a protective immune response against the virus [16] . However, excessive elevated neutrophils can lead to cytokine release syndrome (CRS) and tissue damage, resulting in severe pneumonia and death [16], which have been found in patients with SARS [17, 18] and MERS [19]. CRS is a systemic inflammatory response that can be triggered by infection and other factors and is characterised by a sharp increase in the level of a large number of pro-inflammatory cytokines [34]. SARS-CoV-2 binds to alveolar epithelial cells, after which the virus activates the innate and adaptive immune systems, resulting in the release of a large number of cytokines, including IL-6 [35]. It has been observed that there is a strong involvement of IL-6 in inducing CRS which corresponds to a higher rate of morbidity associated with SARS-CoV and MERS-CoV infections [36]. The signaling of IL-6 plays an important role in inducing this cytokine storm (both by its classical and trans signaling pathway) in severe COVID patients. Consequently, to control the effect of IL-6 and the resulting abnormal immunity, immunotherapies such as tocilizumab [37], vaccine against SARS-COV-2 [38, 39] and targeting toll-like receptor 5 (TLR5) [40] were developed.
The importance of complement in SARS-CoV pathogenesis is controversial. The complement system is an essential component of the innate immune system, and complement plays an important role in the host antiviral response [41]. C3 is required for protection from the pandemic 2009 H1N1 and the highly pathogenic avian influenza (HPAI) H5N1 influenza virus infections by aiding in viral clearance and regulating lung inflammation [42]. Activation of the complement system also results in an immune reaction capable of destroying pathogens and their products. C3 was an important host mediator of SARS-CoV-induced disease and it regulates a systemic proinflammatory response to SARS-CoV infection [43]. Here, our data showed that non-survivors displayed markedly decreased serum levels of C3 and C4 at admission when compared with survivors. Additionally, our results suggested that decreased serum level of C3 is the immunity-related risk factor predicting mortality of patients with COVID-19.
Our study has some limitations. First, we mainly evaluated the number change of cellular immunity- and humoral immunity-related cell subsets, whereas the function of these cells were no elucidated. Second, due to the retrospective study design, not all laboratory tests, such as IL-6, Interferon-γ, IL-10 and other cytokines, were performed in all patients, and the changes in data after treatment were incomplete.
Conclusions
Abnormal cellular immunity and humoral immunity were key features in non-survivors with COVID-19. Additionally, we determined that neutrophilia, lymphocytopenia, low number of CD4+ T cells, and decreased level of C3 were the immunity-related risk factors that can predict mortality of patients with COVID-19. These new features and markers will shed light on developing new strategies for evaluating the prognosis of patients with COVID-19.
Acknowledgements
We acknowledge all the patients involved in this study, and appreciate all the frontline medical and nursing staff involved in the diagnosis and treatment of patients in Wuhan.
Abbreviations
- COVID-19
Coronavirus disease 2019
- WBC
White blood cell
- CRP
C-reactive protein
- PCT
Procalcitonin
- Ig
Immunoglobulins
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SARS
Severe acute respiratory syndrome
- MERS
Middle East respiratory syndrome
- ARDS
Acute respiratory distress syndrome
- CT
Computed tomograph
- RT-PCR
Reverse transcriptase-polymerase chain reaction
- CDC
Center for disease control and prevention
- C
Complement proteins
- IQR
Interquartile range
- ROC
Receiver operating characteristic
- CI
Confidence interval
- NK
Natural killer
- Th
Helper T
- HPAI
Highly pathogenic avian influenza
- IL
Interleukin
Authors’ contributions
ZY had the idea for and designed the study and take responsibility for the integrity of the data and the accuracy of the data analysis. ZYT, WMM, WT, ZZS, LXC, ZSL and WXJ collected the data. ZY and NHX did the analysis. ZY, NHX and HK drafted the paper, and all authors critically revised the manuscript for important intellectual content and gave final approval for the version to be published. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This study was funded by Emergent tackle key problems Foundation of Science and Techology about Novel Coronavirus Pneumonia of Hubei Province (No. 2020FCA002) and National Natural Science Foundation of China (grant number: 81500022).
Availability of data and materials
The datasets generated and/or analysed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was approved by the Research Ethics Commission of Renmin Hospital of Wuhan University (No.WDRY2020-K143), and written informed consent was waived by the Research Ethics Commission.
Consent for publication
Not applicable.
Competing interests
No conflicts of interest are declared by the authors.
Contributor Information
Yang Zhao, Email: zhaoyangrm@whu.edu.cn.
Han-Xiang Nie, Email: nhxbj@sohu.com.
Ke Hu, Email: huke-rmhospital@163.com.
Xiao-Jun Wu, Email: hxnk2020@163.com.
Yun-Ting Zhang, Email: yuntingzhang@whu.edu.cn.
Meng-Mei Wang, Email: ycsylzx@163.com.
Tao Wang, Email: wangtao20061020@126.com.
Zhi-Shui Zheng, Email: rm000251@whu.edu.cn.
Xiao-Chen Li, Email: lxcwh2002@163.com.
Shao-Lin Zeng, Email: 974397494@qq.com.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of V The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization (WHO). Coronavirus disease (COVID-2019) situation reports. 2020.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed April 25, 2020.
- 6.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:1–11. doi: 10.1001/jamainternmed.2019.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y, Zhang Z, Tian J, Xiong S. Risk factors associated with disease progression in a cohort of patients infected with the 2019 novel coronavirus. Anna Palliat Med. 2020;9:428–436. doi: 10.21037/apm.2020.03.26. [DOI] [PubMed] [Google Scholar]
- 8.Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, Ma K, Xu D, Yu H, Wang H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Clinical management of severe acute respiratory infection ( SARI) when COVID-19 disease is suspected: interim guidance, 13 March 2020. World Health Organization. 2020. https://apps.who.int/iris/handle/10665/331446.
- 10.National Health Commission & State Administration of Traditional Chinese Medicine. Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7). 2020. Accessed March 3, 2020.
- 11.Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, Cooley LA, Dean NC, Fine MJ, Flanders SA, et al. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200:e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Opal SM, Girard TD, Ely EW. The immunopathogenesis of sepsis in elderly patients. Clin Infect Dis. 2005;41(Suppl 7):S504–S512. doi: 10.1086/432007. [DOI] [PubMed] [Google Scholar]
- 13.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020. 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed]
- 14.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y et al. Clinical characteristics of 138 hospitalized patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China.JAMA.2020;7;323(11):1061–1069. [DOI] [PMC free article] [PubMed]
- 16.Lamichhane PP, Samarasinghe AE. The Role of Innate Leukocytes during Influenza Virus Infection. J Immunol Res. 2019;2019:8028725. 10.1155/2019/8028725. [DOI] [PMC free article] [PubMed]
- 17.Wong CK, Lam CW, Wu AK, Ip WK, Lee NL, Chan IH, Lit LC, Hui DS, Chan MH, Chung SS, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong RS, Wu A, To KF. Lee N, Lam CW, Wong CK, Chan PK, Ng MH, Yu LM, Hui DS, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326:1358–1362. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, Xu Y, Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu H, Zhou J, Wong BH, Li C, Chan JF, Cheng ZS, Yang D, Wang D, Lee AC, Li C, et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis. 2016;213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, Damoraki G, Gkavogianni T, Adami ME, Katsaounou P, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baruah V, Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J Med Virol. 2020;92(5):495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20(5):269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arabi YM, Arifi AA, Balkhy HH, Najm H, Aldawood AS, Ghabashi A, Hawa H, Alothman A, Khaldi A, Al RB. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann Intern Med. 2014;160:389–397. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- 26.Cecere TE, Todd SM, Leroith T. Regulatory T cells in arterivirus and coronavirus infections: do they protect against disease or enhance it? Viruses. 2012;4:833–846. doi: 10.3390/v4050833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki SR, Subbarao K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol. 2010;84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, Tang X, Temperton NJ, Weiss RA, Brenchley JM, et al. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE, Jia X, Nicholson S, Catton M, Cowie B, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones BE, Maerz MD, Buckner JH. IL-6: a cytokine at the crossroads of autoimmunity. Curr Opin Immunol. 2018;55:9–14. doi: 10.1016/j.coi.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. 2012;11:763–776. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 33.Deng J, Yu XQ, Wang PH. Inflammasome activation and Th17 responses. Mol Immunol. 2019;107:142–164. doi: 10.1016/j.molimm.2018.12.024. [DOI] [PubMed] [Google Scholar]
- 34.Shimabukuro-Vornhagen A, Godel P, Subklewe M, Stemmler HJ, Schlosser HA, Schlaak M, Kochanek M, Boll B, von Bergwelt-Baildon MS. Cytokine release syndrome. J Immunother Cancer. 2018;6(1):56. doi: 10.1186/s40425-018-0343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, Wu Z, Li JW, Zhao H, Wang GQ. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int J Antimicrob Agents. 2020;55:105954. doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braun GS, Nagayama Y, Maruta Y, Heymann F, van Roeyen CR, Klinkhammer BM, Boor P, Villa L, Salant DJ, Raffetseder U, et al. IL-6 trans-signaling drives murine Crescentic GN. J Am Soc Nephrol. 2016;27:132–142. doi: 10.1681/ASN.2014111147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saha A, Sharma AR, Bhattacharya M, Sharma G, Lee SS, Chakraborty C. Tocilizumab: a therapeutic option for the treatment of cytokine storm syndrome in COVID-19. Arch Med Res. 2020;S0188-4409:30782–30787. doi: 10.1016/j.arcmed.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakraborty C, Sharma AR, Sharma G, Bhattacharya M, Lee SS. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur Rev Med Pharmacol Sci. 2020;24:4016–4026. doi: 10.26355/eurrev_202004_20871. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, Lee SS, Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J Med Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakraborty C, Sharma AR, Bhattacharya M, Sharma G, Lee SS, Agoramoorthy G. Consider TLR5 for new therapeutic development against COVID-19. J Med Virol. 2020. 10.1002/jmv.25997. [DOI] [PMC free article] [PubMed]
- 41.Stoermer KA, Morrison TE. Complement and viral pathogenesis. Virology. 2011;411:362–373. doi: 10.1016/j.virol.2010.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Brien KB, Morrison TE, Dundore DY, Heise MT, Schultz-Cherry S. A protective role for complement C3 protein during pandemic 2009 H1N1 and H5N1 influenza A virus infection. Plos One. 2011;6(3):e17377. doi: 10.1371/journal.pone.0017377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gralinski LE, Sheahan TP, Morrison TE, Menachery VD, Jensen K, Leist SR, Whitmore A, Heise MT, Baric RS. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 2018;9:e01753–e01718. doi: 10.1128/mBio.01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are not publicly available but are available from the corresponding author on reasonable request.